Abstract
Background
Treponema pallidum prevalence and burden at oral and lesion sites in adults with early syphilis were assessed by quantitative polymerase chain reaction (qPCR). Factors associated with oral shedding were also examined.
Methods
Pretreatment oral and lesion swabs were collected from adults with early syphilis in a US multicenter syphilis treatment trial. Oral swabs were collected in the presence and absence of oral lesions. Following DNA extraction, qPCR and whole-genome sequencing (WGS) were performed to assess burden and strain variability.
Results
All 32 participants were male, mean age was 35 years, and 90.6% with human immunodeficiency virus (HIV). T. pallidum oral PCR positivity varied by stage: 16.7% primary, 44.4% secondary, and 62.5% in early latent syphilis. Median oral T. pallidum burden was highest in secondary syphilis at 63.2 copies/µL. Lesion PCR positivity was similar in primary (40.0%) and secondary syphilis (38.5%). Age 18–29 years was significantly associated with oral shedding (vs age 40+ years) in adjusted models. WGS identified 2 distinct strains.
Conclusions
T. pallidum DNA was directly detected at oral and lesion sites in a significant proportion of men with early syphilis. Younger age was associated with oral shedding. Ease of oral specimen collection and increased PCR availability suggest opportunities to improve syphilis diagnostic testing.
Clinical Trials Registration. NCT03637660.
Keywords: early syphilis, lesion swab, oral swab, quantitative PCR (qPCR), shedding, Treponema pallidum, whole-genome sequencing (WGS)
Oral and lesion swabs were collected from adults diagnosed with early syphilis for quantitative T. pallidum PCR testing prior to antibiotic treatment. Oral PCR positivity rates ranged from 17% in primary syphilis to 63% in early latent syphilis.
Syphilis rates in the United States have increased steadily over the past decade, rising to 177 000 reported cases in 2021 [1]. Most early syphilis (defined as primary, secondary, and early latent stages) occurs in men, and many are asymptomatic at the time of diagnosis. Among primary and secondary syphilis cases reported in men who have sex with men (MSM), 45% were people with human immunodeficiency virus (PWH) [1]. Routine syphilis screening is recommended in several groups, including MSM and PWH [2]. Current syphilis diagnostics in symptomatic and asymptomatic patients are based on serology because direct organism detection in patients with visible lesions via darkfield microscopy is rarely available [2, 3].
Diagnostic serologic algorithm performance is limited by long-term persistence of treponemal antibodies after infection among persons with prior syphilis and false-positive nontreponemal antibody testing [4]. As highly sensitive nucleic acid amplification testing (NAAT) for Chlamydia trachomatis and Neisseria gonorrhoeae has become standard of care, there has been growing interest in Treponema pallidum polymerase chain reaction (PCR) testing for syphilis diagnosis [5]. The sensitivity of T. pallidum PCR from primary or secondary syphilis lesions ranges from 69% to 95% compared to microscopy and serology [6–8]. At this time, there are no commercially available Food and Drug Administration-approved molecular diagnostic tests for syphilis.
Early and widespread dissemination of T. pallidum after acquisition is well described, including invasion of the central nervous system in up to 40% of adults with early syphilis [7–9]. T. pallidum shedding detected by NAAT is also described at pharyngeal and rectal sites during early syphilis infection [10]. Interestingly, oral T. pallidum shedding has been detected by PCR among MSM with early syphilis and no visible oropharyngeal lesion [11, 12]. PCR test performance can vary according to sample type, T. pallidum DNA versus RNA target, and genes targeted by the amplification protocol [5, 6]. Quantitative PCR (qPCR) targeting a portion of tp0574, a highly conserved T. pallidum gene encoding for the 47-kDa lipoprotein, is a commonly used approach [13]. Our primary study objective was to quantify and compare the prevalence of T. pallidum detection by qPCR at oral and lesion sites during early syphilis in adults prior to antimicrobial treatment. When possible, we also performed T. pallidum whole-genome sequencing (WGS) to investigate the phylogenetic relationship of strains infecting our patients with previously sequenced strains and isolates. Lastly, we sought to determine clinical and host factors associated with oral T. pallidum shedding.
METHODS
Study Population
This investigation represents a substudy of a multicenter, US syphilis treatment trial that completed enrollment in March 2022 (NCT03637660). In the parent trial, we randomized 249 adults with primary, secondary, or early latent syphilis infection to 1 versus 3 weekly doses of benzathine penicillin G to compare serologic treatment response at 6 and 12 months. Participants were ineligible if they had received antimicrobials with activity against T. pallidum in the past 30 days. Demographic information was collected, and syphilis was staged by experienced providers (J.D. and K.W.) according to physical examination and standard serologic testing with positive treponemal and nontreponemal antibody detection and titer. In this oral swab substudy, consenting participants at the University of Alabama at Birmingham (UAB) and Emory University in Atlanta, Georgia were enrolled between 31 August 2020, and 3 March 2022. All substudy participants had oral and lesion (if present) swab samples collected for PCR testing at baseline prior to treatment with benzathine penicillin G.
Sampling and Testing Procedures
Participants, who had not consumed food or drink in the past 30 minutes, had oral samples collected by study staff with sterile polyester swabs (Puritan Medical Products) obtained in a standardized fashion from the buccal mucosa, inner and outer gingival tissue, posterior pharynx, and bilateral tonsils. Lesion swabs were also collected from any suspicious lesion that was visible on the skin or mucous membranes. Among participants with multiple lesions, the lesion with the most exudate was selected for sampling. Lesions were cleaned with saline and then swabbed vigorously. Swabs were immediately placed in cryotubes containing 1 mL of DNA lysis buffer (10 mM Tris [pH 8.0], 0.1 M EDTA, 0.5% SDS) and stored at −20°C until DNA extraction, performed using the Qiagen DNA Mini Kit per provided protocol. Extracted DNA was stored at −20°C until use.
Amplification and Whole-Genome Sequencing
Extracted DNA was first amplified using primers specific for a fragment of the human β-globin gene to demonstrate the lack of PCR inhibitors, and subsequently with primers specific for the tp0574 gene to quantify T. pallidum DNA. For the β-globin gene, DNA extracted from a healthy donor was used as control, while DNA extracted from the T. pallidum SS14 strain cultivated in the laboratory was used as positive control for the T. pallidum-specific target. The β-globin gene was amplified using sense and antisense primers 5′-CAACTTCATCCACGTTCACC-3′ and 5′-GAAGAGCCAAGGACAGGTA-3′, respectively (amplicon size, 268 bp) as previously described [14]. Quantitative amplification to assess T. pallidum burden targeted the tp0574 gene, which correlates 1:1 with organism load [13]. Results were expressed as log-transformed tp0574 copies/µL considering the volume of sample extracted and DNA elution volume.
Prior to sequencing, precapture libraries were prepared from up to 100 ng input genomic DNA using the Kapa Hyperplus kit (Roche), using a fragmentation time of 8 minutes and standard-chemistry end repair/A-tailing, then ligated to TruSeq adapters (Illumina). Adapter-ligated samples were cleaned with 0.8 × Ampure beads (Beckman Coulter) and amplified with barcoded primers for 14 cycles, followed by another 0.8 × Ampure purification. The capture of T. pallidum genomes was performed according to Integrated DNA Technology's xGen Hybridization Capture protocol as described earlier [13]. Libraries were sequenced with paired-end 151 bp reads on a HiSeqX or Nextseq2000 (Illumina). Reads were quality trimmed and filtered to remove host reads, mapped to the T. pallidum SS14 reference genome (NC_021508.1) using Bowtie2 version 2.4.1 with default parameters, followed by deduplication by MarkDuplicates in Picard version 2.23.3 (http://broadinstitute.github.io/picard). Consensus sequences were called using a custom script available at https://github.com/greninger-lab/Tpallidum_WGS. Genomics data are deposited in GenBank under BioProject PRJNA974070.
Statistical Analyses
Initial exploration of the data began with descriptive statistics of various sociodemographic and clinical characteristics, overall and comparing the outcome of oral T. pallidum shedding. Continuous variables are reported as mean with standard deviation or median with quartiles depending upon the distribution. Categorical variables are reported as frequency and percentages.
Association of oral shedding with various characteristics was examined using unconditional univariable and multivariable logistic regression models. The measure of association was odds ratio (OR, unadjusted and adjusted) with corresponding 95% confidence interval (CI). Based on the clinical importance, the following characteristics were selected a priori for the initial univariable analyses: age, syphilis stage, rapid plasma reagin (RPR) titer, PWH, and CD4 cell count. Due to limited sample size, we included 3 characteristics (age, 18–29, 30–39, and ≥40 years; syphilis stage, primary, secondary, and early latent; and CD4 cell count, <200, 200–349, and ≥350 cells/mm3) in the multivariable analyses. Selection was based on model performance measures such as Akaike information criterion (lower the better), Nagelkerke R2 (higher the better), and C-statistics (higher the better). In multivariable analyses, we built 4 models: model 1: age, stage; model 2: age, CD4; model 3: stage, CD4; model 4: age, stage, and CD4. Model performance was also assessed by Hosmer-Lemeshow goodness of fit (P < .05 indicates poor fit); all the models had P value >.30 indicating good fit. Firth bias correction was needed for models 3 and 4 due to quasi-complete separation. Statistical significance was set at .05 (2-tailed). Analyses were conducted using SAS version 9.4.
Ethics Statement
All participants completed informed consent and the study was approved by the Institutional Review Board (IRB) at the University of Alabama at Birmingham (single IRB 300001726). Swabs sent to the University of Washington were deidentified samples labeled as “oral” or “lesion” and laboratory personnel were blinded to all clinical information, including syphilis stage.
RESULTS
All 32 study participants were male and 90.6% (29/32) were PWH. Mean age was 35 years and 43.8% (14 of 32) had oral T. pallidum shedding detected by qPCR. Among PWH, the median CD4 count was lower in men with oral qPCR positivity (398 cells/mm3; interquartile range [IQR], 140–606 cells/mm3 vs 474 cells/mm3; IQR, 289–695 cells/mm3). Most participants (21 of 32; 66.6%) had at least 1 visible lesion or ulcer and staging categories were 18.8% primary, 56.2% secondary, and 25.0% early latent syphilis. RPR titers were elevated in all participants (ranging from 1:2 to 1:512) and 46.9% (15 of 32) had titers ≥1:128. Although 87.5% of participants reported prior syphilis infections, those with oral T. pallidum qPCR positivity were less likely to report a history of syphilis in the past 12 months (14.3% vs 27.8%) or ever (78.6% vs 94.5%) (Table 1).
Table 1.
Participants With Untreated Early Syphilis by Oral Treponema pallidum qPCR Positivity (n = 32)
| Characteristic | Total (n = 32) |
Oral T. pallidum PCR Positive (n = 14) |
Oral T. pallidum PCR Negative (n = 18) |
|---|---|---|---|
| Age at enrollment, y, mean (SD) | 35 (10) | 31 (8) | 39 (9) |
| Age, y, n (%) | |||
| 18–29 | 11 (34.4) | 9 (64.3) | 2 (11.1) |
| 30–39 | 15 (46.9) | 4 (28.6) | 11 (61.1) |
| 40–49 | 1 (3.1) | 0 (0) | 1 (5.6) |
| ≥50 | 5 (15.6) | 1 (7.1) | 4 (22.2) |
| Male sex, n (%) | 32 (100) | 14 (100) | 18 (100) |
| PWH, n (%) | 29 (90.6) | 11 (78.6) | 18 (100) |
| CD4 in PWH, cells/mm3, median (Q1, Q3) | 453 (285, 651) | 398 (140, 606) | 474 (289, 695) |
| CD4 in PWH, cells/mm3, n (%) | |||
| <200 | 5 (17.9) | 3 (30) | 2 (11.1) |
| 200–350 | 6 (21.4) | 1 (10) | 5 (27.8) |
| ≥350 | 17 (60.7) | 6 (60) | 11 (61.1) |
| Location (study site), n (%) | |||
| Atlanta, Georgia (Emory) | 23 (71.9) | 8 (57.1) | 15 (83.3) |
| Birmingham, Alabama (UAB) | 9 (28.1) | 6 (42.9) | 3 (16.7) |
| Syphilis stage, n (%) | |||
| Primary | 6 (18.8) | 1 (7.1) | 5 (27.8) |
| Secondary | 18 (56.2) | 8 (57.1) | 10 (55.5) |
| Early latent | 8 (25.0) | 5 (35.8) | 3 (16.7) |
| RPR titer, n (%) | |||
| 1:2 | 1 (3.1) | 0 (0) | 1 (5.6) |
| 1:4 | 1 (3.1) | 0 (0) | 1 (5.6) |
| 1:8 | 3 (9.4) | 1 (7.1) | 2 (11.1) |
| 1:16 | 1 (3.1) | 1 (7.1) | 0 (0) |
| 1:32 | 6 (18.8) | 3 (21.4) | 3 (16.7) |
| 1:64 | 5 (15.6) | 2 (14.3) | 3 (16.7) |
| 1:128 | 10 (31.3) | 3 (21.4) | 7 (38.9) |
| 1:256 | 3 (9.4) | 3 (21.4) | 0 (0) |
| 1:512 | 2 (6.3) | 1 (7.1) | 1 (5.6) |
| History of syphilis, n (%) | |||
| Recent (≤12 mo) | 7 (21.9) | 2 (14.3) | 5 (27.8) |
| Past (>12 mo) | 21 (65.6) | 9 (64.3) | 12 (66.7) |
| Never | 4 (12.5) | 3 (32.4) | 1 (5.5) |
| Location of lesion,a n (%) | |||
| Single | 18 (56.2) | 7 (50.0) | 11 (61.1) |
| Penis/scrotum/groin | 6 | 3 | 3 |
| Rectal/perianal | 2 | 0 | 2 |
| Tongue/oropharyngeal | 6 | 2 | 4 |
| Trunk/extremities | 4 | 2 | 2 |
| Multiple | 3 (9.4) | 2 (14.3) | 1 (5.6) |
| Penis/scrotum/groin | 1 | 1 | 0 |
| Tongue/oropharyngeal | 1 | 0 | 1 |
| Trunk/extremities | 1 | 1 | 0 |
Missing data: CD4 was missing in one PWH.
Abbreviations: PWH, people with human immunodeficiency virus; Q1, first quartile; Q3, third quartile; RPR, rapid plasma reagin; SD, standard deviation; UAB, University of Alabama at Birmingham.
aThree participants with secondary syphilis did not have a lesion swab collected.
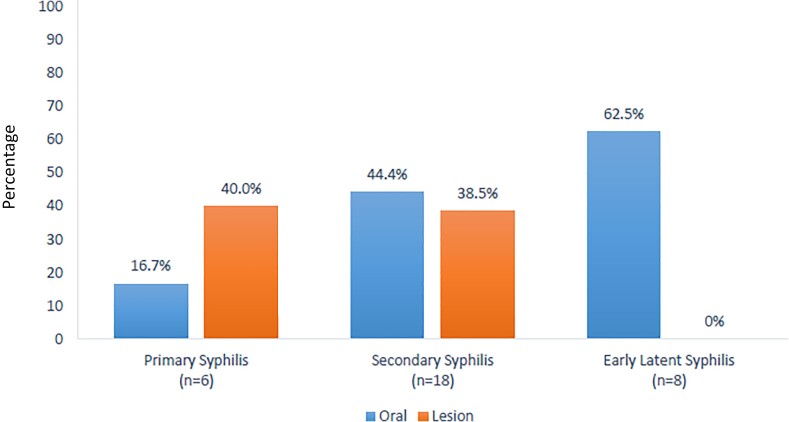
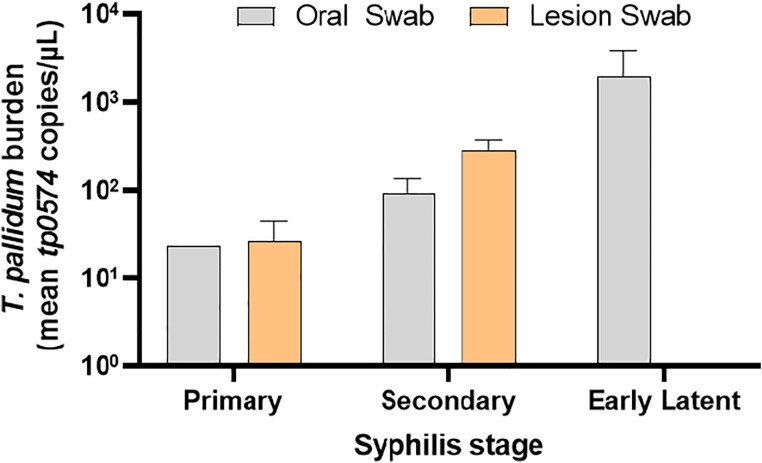
The proportion of swabs positive for T. pallidum DNA by stage and lesion location (oral vs lesion) are shown in Figure 1. T. pallidum oral shedding was detected by qPCR in 43.8% (14 of 32) of participants. Oral detection was more common in persons with secondary and early latent syphilis: 16.7% (1 of 6) in primary, 44.4% (8 of 18) in secondary, and 62.5% (5 of 8) in early latent disease. Among oral swabs performed in the absence of oral lesions, qPCR detection was 37.9% (11 of 29). None of the primary or early latent syphilis cases and 3 of 18 secondary syphilis cases had visible pharyngeal, tonsillar, or oral lesions. Lesion swab T. pallidum qPCR positivity was similar in primary (40%; 2 of 5) and secondary syphilis stages (38.5%; 5 of 13). Median organism burden at the oral site by stage was highest during secondary syphilis with 23.3 copies/µL in primary, 63.2 copies/µL in secondary, and 47.7 copies/µL in early latent syphilis. In comparison, the median T. pallidum burden for lesion swabs was 26.8 copies/µL in primary and 449.5 copies/µL in secondary syphilis (Figure 2).
Figure 1.
Proportion of swabs with Treponema pallidum qPCR positivity by stage and swab location (n = 32). Among all participants, 3 of 18 with secondary syphilis had visible oral lesions.
Figure 2.
Treponema pallidum burden by syphilis stage and swab location (n = 32). Among all participants, 3 of 18 with secondary syphilis had visible oral lesions.
In logistic regression models created to assess demographic and clinical predictors of oral T. pallidum qPCR positivity, age was significantly associated with oral shedding in the univariable analysis (Table 2). Those aged 18–29 years (OR, 22.5; 95% CI, 1.6–314.5; P = .02) had higher odds of shedding compared to age ≥40 years. Although high titer RPR (≥1:32) and secondary and early latent syphilis (vs primary) had higher odds of oral shedding, differences were not statistically significant. Error bars represent standard error of the mean.
Table 2.
Characteristics Associated With Oral Treponema pallidum qPCR Positivity in Men With Early Syphilis: Univariable Logistic Regression Analysis
| Characteristic | Unadjusted OR (95% CI) |
P Value | AIC |
R
2,a % |
C-Statistic, % |
|---|---|---|---|---|---|
| Age, y | .02 | 39 | 38 | 78 | |
| 18–29 | 22.50 (1.61–314.48) | .02 | |||
| 30–39 | 1.82 (.16–20.71) | .63 | |||
| 40+ | Ref | ||||
| Syphilis stage | .27 | 47 | 13 | 66 | |
| Primary | Ref | ||||
| Secondary | 4.00 (.39–41.5) | .25 | |||
| Early latent | 8.33 (.63–110.0) | .11 | |||
| RPR titer | .85 | 50 | 1 | 54 | |
| <1:32 | Ref | ||||
| 1:32–1:64 | 1.67 (.21–13.22) | .63 | |||
| >1:64 | 1.75 (.24–12.64) | .58 | |||
| PWHb | 42 | 15 | 15 | ||
| Yes | 0.09 (.01–2.95) | .18 | |||
| No | Ref | ||||
| CD4 count, cells/mm3 | .36 | 40 | 11 | 65 | |
| <200 | Ref | ||||
| 200–349 | 0.13 (.01–2.18) | .16 | |||
| ≥350 | 0.36 (.05–2.82) | .33 |
P value in bold indicates overall association of a variable using type 3 analysis of effects (Wald χ2) for variables with >2 categories.
Abbreviations: AIC, Akaike information criterion; CI, confidence interval; OR, odds ratio; PCR, polymerase chain reaction; Ref, reference category.
aNagelkerke's max-rescaled R2 reported in PROC LOGISTIC procedure of SAS.
bFirth bias correction used due to zero cell leading to quasi-complete separation.
Multivariable logistic regression analyses and performance measures are presented in Table 3. Model 1 with age and syphilis stage had the best performance with Akaike information criterion 39 and C-statistic 86%. In this adjusted model, age was associated with T. pallidum oral shedding (type 3 P value = .02). Participants aged 18–29 years had the highest adjusted odds of shedding compared to those aged ≥40 years (aOR, 31.6; 95% CI, 1.9–522.4; P = .02). Although the strength of the association of participant characteristics varied across the models, their direction remained the same.
Table 3.
Characteristics Associated With Oral Treponema pallidum qPCR Positivity in Men With Early Syphilis: Multivariable Logistic Regression Analysis
| Characteristic | Model 1 | Model 2 | Model 3 | Model 4 | ||||
|---|---|---|---|---|---|---|---|---|
| Adjusted OR (95% CI) |
P Value | Adjusted OR (95% CI) |
P Value | Adjusted OR (95% CI) |
P Value | Adjusted OR (95% CI) |
P Value | |
| Age, y | .02 | .16 | … | … | .33 | |||
| 18–29 | 31.56 (1.91–522.40) | .02 | 12.16 (.78–189.04) | .07 | … | … | 6.60 (.53–82.92) | .14 |
| 30–39 | 1.69 (.12–24.81) | .70 | 2.42 (.18–32.47) | .50 | … | … | 2.05 (.14–29.78) | .60 |
| 40+ | Ref | Ref | … | … | Ref | |||
| Syphilis stage | .19 | … | … | .33 | .95 | |||
| Primary | Ref | … | … | Ref | Ref | |||
| Secondary | 2.71 (.15–49.24) | .50 | … | … | 4.79 (.18–125.90) | .16 | 3.99 (.12–129.24) | .44 |
| Early latent | 13.73 (.64–296.55) | .10 | … | … | 11.99 (.38–383.26) | .35 | 10.59 (.33–341.07) | .18 |
| CD4 count, cells/mm³ | … | … | .55 | .83 | .36 | |||
| <200 | … | … | Ref | Ref | Ref | |||
| 200–349 | … | … | 0.19 (.01–4.15) | .29 | 0.42 (.03–6.81) | .54 | 0.59 (.03–13.96) | .74 |
| ≥350 | … | … | 0.38 (.04–3.68) | .40 | 0.71 (.08–6.21) | .75 | 0.84 (.08–8.62) | .88 |
| AIC | 39 | 40 | 40 | 41 | ||||
| R 2,a % | 49 | 29 | 23 | 35 | ||||
| C-statistics, % | 86 | 76 | 74 | 83 | ||||
| H-L fit, P value | .89 | .47 | .32 | .61 | ||||
| Firth bias correctionb | No | No | Yes | Yes | ||||
P value in bold indicates overall association of a variable using type 3 analysis of effects (Wald χ2).
Abbreviations: AIC, Akaike information criterion; CI, confidence interval; OR, odds ratio; PCR, polymerase chain reaction; Ref, reference category.
aNagelkerke's max-rescaled R2 reported in PROC LOGISTIC procedure of SAS.
bFirth bias correction used due to zero cell leading to quasi-complete separation.
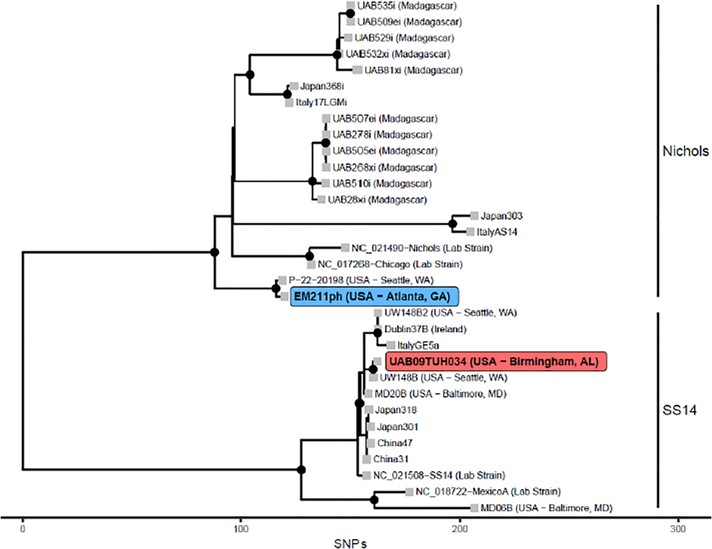
T. pallidum genomics data could be collected for 6 strains, with a full genome obtained from 2 samples: UAB09TUH034 collected in Birmingham, Alabama and EM211ph collected in Atlanta, Georgia. The remaining 4 samples yielded partial genomic data that, however, were sufficient to assign each strain to a T. pallidum clade. The UAB09TUH034 strain was found to belong to the SS14 omega clade (Figure 3; highlighted in red). This clade encompasses most T. pallidum strains circulating worldwide [15]. Accordingly, UAB09TUH034 was found to be resistant to macrolides via the A2058G mutation on the 23S rRNA gene, common to most of the SS14 clade strains. In contrast, all the samples collected in Atlanta (including EM211ph) belonged to the Nichols strain based on 1 complete genome sequence (Figure 3; highlighted in blue) or the sequences of the polymorphic tp0136 and tp01031 loci, used for T. pallidum molecular typing [16]. Interestingly, the EM211ph genome was found to be nearly identical to a strain recently sequenced in Seattle (P-22-20198, 99.9994% identical; Figure 3) [17]. Both strains appear to belong to a new Nichols subclade (Nichols F) not previously described. Except for 1 strain, for which genomics data did not allow us to resolve the sequence of the rRNA genes, all the strains from Atlanta were also resistant to macrolides via the same A2058G mutation described in the UAB09TUH034 strain. This differs from most previously published strains belonging to the Nichols clade, which were reported as macrolide sensitive [13]. All genomics data are deposited in GenBank under Bioproject PRJNA974070.
Figure 3.
Phylogenetic tree of Treponema pallidum genomes showing the evolutionary relationships between the EM211ph (collected in Georgia, US) and UAB09TUH034 (collected in Birmingham, US) strains in participants and selected publicly available genomes from historical (eg, Nichols, Chicago, SS14, MexicoA) laboratory strains and modern global strains specified either by the strain name (eg, Italy, Japan, etc.) or indicated next to the strain name. The x axis units are the number of single nucleotide polymorphisms (SNPs) or mismatches that distinguish strains of T. pallidum.
DISCUSSION
Successful management of syphilis in clinical settings depends on the availability and performance of quality diagnostic testing. According to 2021 Centers for Disease Control and Prevention sexually transmitted diseases treatment guidelines, “Darkfield examinations and molecular tests for detecting T. pallidum directly from lesion exudate or tissue are the definitive methods for diagnosing early syphilis…” [3]. Although darkfield microscopy is rarely available, NAAT testing for T. pallidum is increasingly available. This study conducted in a small number of men with early syphilis, most PWH, demonstrates a substantial proportion with T. pallidum qPCR positivity at the oral site, even in the absence of lesions. These findings suggest that qPCR testing of swab specimens from the oral cavity could enhance syphilis diagnosis through direct detection of the organism. The highest proportion with oral shedding occurred in younger men with secondary and early latent syphilis. This association between oral shedding and stage is consistent with prior studies from Australia and China, while the association with age has not been previously reported [11, 12]. In the setting of increasing US syphilis rates and the limitations of serologic testing, the potential utility of T. pallidum direct detection as a diagnostic tool for syphilis is appealing.
Limitations of currently recommended syphilis serologic testing include false-positive treponemal and nontreponemal screening tests as well as persistent reactivity related to prior syphilis in high-prevalence populations. Treponemal antibodies usually remain persistently positive and do not distinguish between treated and untreated infection. In the traditional diagnostic algorithm that includes a nontreponemal RPR screening test, false-positive testing occurs in 5%–15% of PWH and others with chronic viral hepatitis, pregnancy, and autoimmune disease [18]. Furthermore, RPR can remain elevated despite appropriate treatment and the serofast state frequently complicates the clinical interpretation of serologic tests [19, 20]. Direct detection of T. pallidum can be useful to establish the diagnosis and potential transmission risk to partners. Primary syphilis chancres are typically painless, may be missed on examination, especially with lesions in the mouth or internal anogenital location and have clinical manifestations that resemble other conditions. The ability to detect oral shedding may be particularly useful in evaluation of patients with latent syphilis who have no visible lesions but demonstrate qPCR positivity on oral swabs, as documented in this study.
Published studies documenting the performance of T. pallidum PCR testing in patients with confirmed or probable syphilis include qualitative PCR with pooled sensitivity of 78% (95% CI, 60%–89%) and qPCR with pooled sensitivity of 68% (95% CI, 55%–79%) in the setting of primary and secondary syphilis [6]. We suspect that our lesion swab PCR positivity rate was somewhat lower because investigators were encouraged to swab any lesion, even if it lacked the appearance of a typical primary ulcer or secondary syphilitic rash [21]. Studies vary regarding target genes and sample type and T. pallidum detection test performance in whole blood is limited because the duration of spirochetemia is relatively brief [22]. The yield of PCR testing on syphilitic cutaneous lesions compared to darkfield microscopy averages 70%–80% and the recognition of T. pallidum shedding at the oral site during early syphilis has only relatively recently been described [10–12, 23]. Among 200 MSM in Australia with early syphilis, 24% had T. pallidum detected by qPCR performed on oral swab or oral rinse [11]. Oral T. pallidum shedding in Australia was more frequent in men with secondary syphilis (44% vs 7% with other stages; P < .01) and with RPR titer ≥1:64 (32% vs 13% for lower RPR titer; P < .01).
Current tests including the tp0574 target gene used in this study readily distinguish T. pallidum subspecies from oral treponemes that are considered nonpathogenic but inhabit the oral mucosa [24]. Although tp0574 amplification does not discriminate between T. pallidum subspecies, given the demographics of our cohort and the epidemiology and mode of transmission of endemic treponematoses, the potential exposure to yaws or bejel instead of syphilis are negligible. Findings from this study also provide insights about T. pallidum dissemination. Wide dissemination in syphilis is typically considered via hematogenous and lymphatic spread with early invasion of the central nervous system, placenta (when present), and organ tissues. Asymptomatic oral and anal shedding in adults with early syphilis has only recently been recognized and has yet to be characterized in terms of timing, organism burden, immune responses, and transmission dynamics. In this study, we are not able to determine the precise location of T. pallidum oral shedding (whether from gingival tissue, tonsils, or another site) because participants had multiple oral areas sampled with the same swab.
In terms of predictors of oral shedding, we hypothesized that secondary syphilis would have the highest rates and the median T. pallidum burden at the oral site would be highest with secondary syphilis. Although mean burden was higher in early latent syphilis, median values are preferred for comparison because there was an outlier with high burden in the early latent stage. Although it is possible that oral shedding may continue throughout a prolonged period after acquisition, our understanding of shedding duration cannot be informed by this cross-sectional study. In unadjusted and adjusted models (for stage), younger age was associated with T. pallidum oral shedding. This may suggest acquired T. pallidum immunity with age that reduces the rate of shedding at the oral site. Additional evidence in support of potential immunity is also suggested by the fact that participants with oral shedding were less likely to report a lifetime history of syphilis or recent infection in the past 12 months. Because most participants in the study were PWH, we are not able to draw conclusions about a relationship between HIV and T. pallidum shedding. CD4 count did not predict shedding in our models.
Although very few samples were sufficiently concentrated in T. pallidum DNA to yield genome sequences or genomic information, it is interesting that the strain isolated from a participant in Atlanta, GA belongs to the Nichols clade, while most T. pallidum strains circulating in the United States are in the SS14 clade. In particular, the EM211ph strain from Atlanta isolated in this study along with a strain collected in 2022 from an individual in Seattle, WA appear to define a new Nichols subclade, F (Figure 3). This strain is distantly related (>50 SNPs) to other Nichols clinical and reference strains, which supports the continuous intraclade microevolution of the syphilis spirochete, as recently reported [25].
Study limitations include the small sample size and lack of representation of women. Because all participants had early syphilis, and most had high titer RPR and were PWH, these findings may not extend to adults with low titer RPR infection, late latent syphilis, or those without HIV. Also, classification bias is possible due to imperfect syphilis staging because clinical manifestations of primary and secondary syphilis may be missed or can overlap. We tried to reduce this risk by relying on experienced infectious diseases clinicians for staging. Study strengths include standardized collection of oral swabs as part of a multicenter randomized syphilis treatment trial and laboratory testing performed by blinded staff with a highly sensitive and specific qPCR test for detection of T. pallidum.
Future studies are needed to replicate these findings in a larger sample size that includes women, adolescents, and all stages of syphilis (including neurosyphilis, latent syphilis, and infection in pregnancy and infants). Information about sites of exposure, sex partners, and symptom status is also important. Potential immune protection should be investigated among people with varying age, levels of prior syphilis exposure, people with and without HIV, and a range of CD4 counts. The precise oral location of T. pallidum replication and shedding during each stage is yet to be defined. Finally, oral shedding of T. pallidum warrants investigation to characterize the potential transmission risk to partners.
CONCLUSION
Study findings suggest a new role for oral and lesion swabs for the molecular detection of T. pallidum even in the absence of visible lesions in persons with early syphilis. Increasing availability of qPCR, the relative ease of specimen collection, the significant proportion of adults with T. pallidum positivity demonstrated at oral and/or lesion sites, and the opportunity to directly demonstrate the presence of T. pallidum during all stages of early syphilis suggest the opportunity for qPCR testing to enhance and refine current syphilis diagnostic testing.
Contributor Information
Jodie A Dionne, Department of Medicine, Division of Infectious Diseases, University of Alabama at Birmingham, Birmingham, Alabama, USA.
Lorenzo Giacani, Department of Medicine, Division of Allergy and Infectious Diseases, University of Washington, Seattle, Washington, USA; Department of Global Health, University of Washington, Seattle, Washington, USA.
Ashutosh Tamhane, Department of Medicine, Division of Infectious Diseases, University of Alabama at Birmingham, Birmingham, Alabama, USA.
Kimberly Workowski, Department of Medicine, Division of Infectious Diseases, Emory University, Atlanta, Georgia, USA.
Nicole A P Lieberman, Department of Laboratory Medicine and Pathology, University of Washington School of Medicine, Seattle, Washington, USA.
Alexander L Greninger, Department of Laboratory Medicine and Pathology, University of Washington School of Medicine, Seattle, Washington, USA; Vaccine and Infectious Disease Division, Fred Hutchinson Cancer Research Center, Seattle, Washington, USA.
Charlotte Perlowski, FHI 360, Durham, North Carolina, USA.
Lori Newman, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Rockville, Maryland, USA.
Edward W Hook, III, Department of Medicine, Division of Infectious Diseases, University of Alabama at Birmingham, Birmingham, Alabama, USA.
Notes
Disclaimer . The views and opinions presented do not reflect those of the funding agency.
Financial support. This work was supported by the National Institute of Allergy and Infectious Diseases (NIAID) Sexually Transmitted Infections Clinical Trials Group (grant number HHSN272201300012I). Genome sequencing was supported by the National Institutes of Health, NIAID (grant number U19AI144133 to L. G. and A. L. G.).
References
- 1. Centers for Disease Control and Prevention . Sexually transmitted infection surveillance 2021. https://www.cdc.gov/std/statistics/2021/default.htm. Accessed 7 January 2023.
- 2. Mangione CM, Barry MJ, Nicholson WK, et al. Screening for syphilis infection in nonpregnant adolescents and adults: US preventive services task force reaffirmation recommendation statement. JAMA 2022; 328:1243–9. [DOI] [PubMed] [Google Scholar]
- 3. Workowski KA, Bachmann LH, Chan PA, et al. Sexually transmitted infections treatment guidelines, 2021. MMWR Recomm Rep 2021; 70:1–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dionne-Odom J, Van Der Pol B, Boutwell A, Biligowda N, Schmid DG, Hook EW III. Limited utility of reverse algorithm syphilis testing in HIV clinic among men who have sex with men. Sex Transm Dis 2021; 48:675–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pillay A. Centers for disease control and prevention syphilis summit-diagnostics and laboratory issues. Sex Transm Dis 2018; 45(9S Suppl 1):S13–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Simpore A, Bazie BV, Zoure AA, et al. Performance of molecular tests in the diagnosis of syphilis from 2009 to 2019: a systematic review and meta-analysis. Sex Transm Dis 2022; 49:469–76. [DOI] [PubMed] [Google Scholar]
- 7. Peeling RW, Mabey D, Kamb ML, Chen XS, Radolf JD, Benzaken AS. Syphilis. Nat Rev Dis Primers 2017; 3:17073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Peeling RW, Hook EW III. The pathogenesis of syphilis: the great mimicker, revisited. J Pathol 2006; 208:224–32. [DOI] [PubMed] [Google Scholar]
- 9. Lukehart SA, Hook EW III, Baker-Zander SA, Collier AC, Critchlow CW, Handsfield HH. Invasion of the central nervous system by Treponema pallidum: implications for diagnosis and treatment. Ann Intern Med 1988; 109:855–62. [DOI] [PubMed] [Google Scholar]
- 10. Golden M, O’Donnell M, Lukehart S, et al. Treponema pallidum nucleic acid amplification testing to augment syphilis screening among men who have sex with men. J Clin Microbiol 2019; 57:e00572-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Towns JM, Leslie DE, Denham I, et al. Treponema pallidum detection in lesion and non-lesion sites in men who have sex with men with early syphilis: a prospective, cross-sectional study. Lancet Infect Dis 2021; 21:1324–31. [DOI] [PubMed] [Google Scholar]
- 12. Yang CJ, Chang SY, Wu BR, et al. Unexpectedly high prevalence of Treponema pallidum infection in the oral cavity of human immunodeficiency virus-infected patients with early syphilis who had engaged in unprotected sex practices. Clin Microbiol Infect 2015; 21:787.e1–e7. [DOI] [PubMed] [Google Scholar]
- 13. Lieberman NAP, Lin MJ, Xie H, et al. Treponema pallidum genome sequencing from six continents reveals variability in vaccine candidate genes and dominance of Nichols clade strains in Madagascar. PLoS Negl Trop Dis 2021; 15:e0010063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Aldrete S, Kroft SH, Romeis E, et al. Whole genome sequence of a Treponema pallidum strain from a formalin-fixed paraffin-embedded fine needle aspirate of a cervical lymph node. Sex Transm Dis 2023; 50:550–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Arora N, Schuenemann VJ, Jäger G, et al. Origin of modern syphilis and emergence of a pandemic Treponema pallidum cluster. Nat Microbiol 2016; 2:16245. [DOI] [PubMed] [Google Scholar]
- 16. Vrbová E, Grillová L, Mikalová L, et al. MLST typing of Treponema pallidum subsp. pallidum in the Czech republic during 2004–2017: clinical isolates belonged to 25 allelic profiles and harbored 8 novel allelic variants. PLoS One 2019; 14:e0217611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lieberman NAP, Avendaño CC, Bakhash SAKM, et al. Genomic epidemiology of Treponema pallidum and circulation of strains with diminished tprK antigen variation capability in Seattle, 2021–2022. J Infect Dis 2024; 229:866–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sena AC, Zhang XH, Li T, et al. A systematic review of syphilis serological treatment outcomes in HIV-infected and HIV-uninfected persons: rethinking the significance of serological non-responsiveness and the serofast state after therapy. BMC Infect Dis 2015; 15:479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sena AC, Wolff M, Behets F, et al. Response to therapy following retreatment of serofast early syphilis patients with benzathine penicillin. Clin Infect Dis 2013; 56:420–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Seña AC, Wolff M, Martin DH, et al. Predictors of serological cure and serofast state after treatment in HIV-negative persons with early syphilis. Clin Infect Dis 2011; 53:1092–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gayet-Ageron A, Ninet B, Toutous-Trellu L, et al. Assessment of a real-time PCR test to diagnose syphilis from diverse biological samples. Sex Transm Infect 2009; 85:264–9. [DOI] [PubMed] [Google Scholar]
- 22. Ho EL, Lukehart SA. Syphilis: using modern approaches to understand an old disease. J Clin Invest 2011; 121:4584–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gayet-Ageron A, Combescure C, Lautenschlager S, Ninet B, Perneger TV. Comparison of diagnostic accuracy of PCR targeting the 47-kilodalton protein membrane gene of Treponema pallidum and PCR targeting the DNA polymerase I gene: systematic review and meta-analysis. J Clin Microbiol 2015; 53:3522–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Giacani L, Lukehart SA. The endemic treponematoses. Clin Microbiol Rev 2014; 27:89–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Edmondson DG, De Lay BD, Hanson BM, Kowis LE, Norris SJ. Clonal isolates of Treponema pallidum subsp. pallidum Nichols provide evidence for the occurrence of microevolution during experimental rabbit infection and in vitro culture. PLoS One 2023; 18:e0281187. [DOI] [PMC free article] [PubMed] [Google Scholar]