Abstract
We have demonstrated previously that the adenovirus L1 52/55-kDa protein binds to the viral IVa2 protein in infected cells. The significance of this interaction was unclear, however, based on the known functions of these two proteins: the 52/55-kDa protein is required for viral DNA packaging, while the IVa2 protein is a transactivator of the major late promoter (MLP). In this report, we have attempted to elucidate a role for each of the two proteins in the other's known function. There is no apparent effect of the 52/55-kDa protein on the interaction of the IVa2 protein with the MLP. Surprisingly, however, we found that the IVa2 protein can interact with the adenoviral packaging signal and that this interaction involves DNA sequences that have previously been demonstrated to be required for packaging.
Adenovirus has been used for many years as a model system to study DNA replication, gene expression, and virus-host interactions. In recent years, interest has also been focused on adenovirus because of its potential as a vector for gene therapy. However, the detailed mechanisms of how virus particles are assembled and viral DNA is specifically packaged are still not fully understood. The assembly of adenovirus virions starts with polymerization of the hexon to form the capsomers that are the basic structural unit of the capsids (24). Two populations of virus particles, called light and heavy particles, can be distinguished by CsCl equilibrium centrifugation (26). The light particles, with a density of 1.315 g/cm2, are premature assembly intermediates containing no DNA, or, in some cases, part of the viral genome, and protein components, including the hexon, penton, and fiber proteins and precursors of proteins VI and VIII. The partial genome in these intermediates may be the result of shearing of viral DNA outside of the particle during virus preparation (4) and is predominantly derived from the left end of the viral genome, indicating that packaging of viral DNA starts from the left end of the viral genome (30). Core proteins V and VII are not present in the empty capsids (5). Pulse-chase experiments indicate that DNA and these core proteins are inserted into the empty capsid to generate the heavier particles, consisting of young and mature virions (5). The final maturation process involves the proteolytic cleavage of precursor proteins in the young virion, including pIIIa, pTP, pVI, pVIII, and pVII.
The mechanism of adenovirus DNA encapsidation is not known. Specific packaging of viral DNA has been shown to be mediated by the packaging sequence, which is located at the left end of the viral genome (nucleotides 194 to 382 in Ad5) (17, 21). This region contains at least five functionally redundant domains, the A repeats, with AI, AII, AV, and AVI being the most important elements (7, 8). Each of the A repeats fits a consensus motif and can function independently (28). The proteins that are involved in DNA packaging are not known, although cellular components have been shown to bind to sequences in the packaging domains that are required for packaging (29). Among the viral proteins, only the 52/55-kDa protein has been shown to date to be required for viral DNA packaging (15), though pIX has been shown to affect the ability of the virion to package full-length genomes (1, 2, 6). Early studies demonstrated that the 52/55-kDa protein is present in assembly intermediates, but not in mature virions, and is required for virus assembly, suggesting that the 52/55-kDa protein is a scaffolding protein (19). Recently, we demonstrated that the 52/55-kDa protein is required for viral DNA encapsidation and does not function as a scaffolding protein, because a mutant virus (pm8001) that does not express this protein accumulates empty capsids with no viral DNA (15). However, the mechanism by which the 52/55-kDa protein directs viral DNA encapsidation is not clear. Presumably, the 52/55-kDa protein must bind directly to the packaging sequences or interact with other viral or cellular proteins that bind to the packaging sequences to mediate specific DNA packaging.
Using the yeast two-hybrid system, we demonstrated previously that the 52/55-kDa protein interacts with the adenovirus IVa2 protein (16). The IVa2 protein is a transcriptional activator of the major late promoter (MLP), acting as a component of two factors that bind to the downstream element (DE) of the MLP (25, 27, 31), DEF-A and DEF-B. DEF-B is a homodimer of IVa2 and binds to a site referred to as DE2b (23). DEF-A is a heterodimer, of which the IVa2 protein is one component, and binds to the DE1 and DE2a sites. Binding of DEF-A to the DE2a site requires the cooperation of DEF-B. The interaction of the 52/55-kDa and IVa2 proteins suggests that the 52/55-kDa protein may be involved in regulating the MLP, possibly as a component of DEF-A, since delayed expression of viral late proteins was detected in pm8001-infected cells (15), although the 52/55-kDa protein itself has no known DNA binding ability. On the other hand, the DNA binding property of the IVa2 protein, along with its ability to bind the 52/55-kDa protein, suggests the possibility that the IVa2 protein is involved in DNA encapsidation. Moreover, the IVa2 protein has been shown to be a component of both assembly intermediates and mature virions, further suggesting a possible role of IVa2 protein in virus assembly (32).
In this report, we describe experiments to test these possibilities. We have found that the 52/55-kDa protein does not appear to be involved in binding of the IVa2 protein to the DE. While examining the DE and packaging sequence A repeats, however, we found significant sequence homology among these elements. We have demonstrated, then, that the IVa2 protein can bind to the A repeats. Moreover, mutations in conserved functional motifs of the A repeats eliminate the binding of the IVa2 protein. These results indicate that the IVa2 protein interaction with the 52/55-kDa protein may play a role in the encapsidation of viral DNA.
MATERIALS AND METHODS
Cells and viruses.
293 cells are human embryonic kidney cells expressing adenovirus E1A and E1B proteins (10). 293-L1 cells are 293 cells that stably express the 52/55-kDa protein, which are used as a helper cell line for growing the 52/55-kDa mutant virus, pm8001 (15). All of these cells were maintained in Dulbecco's modified Eagle's medium with 10% fetal bovine serum. For 293-L1 cells, 0.5 mg of G418 per ml was added to the medium. Wild-type Ad5 virus was propagated on 293 cells as described previously (9).
Nuclear extracts.
293 cells were infected with Ad5 or pm8001 at a multiplicity of infection of 5 PFU/cell. Twenty-four hours after infection, nuclear proteins were extracted as described previously (3). Briefly, cells were washed twice with phosphate-buffered saline, resuspended in 4 pellet volumes of buffer A (10 mM HEPES [pH 7.9]; 10 mM KCl; 1.5 mM MgCl2; 5 mM dithiothreitol [DTT]; 0.5 mM phenylmethylsulfanyl fluoride; 5 μg [each] of aprotinin, leupeptin, and pepstatin per ml) and incubated on ice for 1 h. The cells were then transferred to a glass Dounce homogenizer and lysed with 20 strokes of a tight-fitting pestle. The nuclei were centrifuged at 2,000 × g for 5 min at 4°C. After being washed once with 1 ml of buffer A, the nuclei were resuspended in 3 pellet volumes of buffer B (20 mM HEPES [pH 7.9]; 20% glycerol; 420 mM NaCl; 1.5 mM MgCl2; 0.2 mM EDTA; 5 mM DTT; 0.5 mM phenylmethylsulfonyl fluoride; 5 μg [each] of aprotinin, leupeptin, and pepstatin per ml) and incubated on ice for 30 min. The supernatant was collected after centrifugation at 12,000 × g for 30 min, snap frozen on dry ice, and stored at −80°C.
Electrophoretic mobility shift assay.
DNA binding assays were performed as described previously (18), with the following modifications. Four micrograms of nuclear extract was added to 13 μl of reaction mixture containing 10 mM HEPES [pH 7.9], 20 mM KCl, 3 mM MgCl2, 10 mM EDTA, 12% glycerol, 300 μg of bovine serum albumin per ml, 1 μg of poly(dI-dC), 1 mM DTT, and 100,000 cpm of 32P-labeled probe. The probes were oligonucleotides which were annealed from synthetic complementary single-stranded DNA (see Table 1 for sequences). One hundred nanograms of double-stranded DNA was end labeled with 1 U of T4 DNA kinase and 50 μCi of [γ-32P]ATP in 30 μl of buffer at 37°C for 1 h and then purified on a Sephadex G-25 spin column. The binding reaction mixtures were incubated at room temperature for 15 min and then were resolved in a 4.5% polyacrylamide (40:1 ratio of acrylamide to bisacrylamide) gel in 0.5× Tris-borate-EDTA buffer for 4 h at 150 V and 10°C. Unlabeled oligonucleotides or a DNA fragment containing the complete packaging sequence (prepared by PCR) was added to the mixtures as a competitor in some assays. For supershift assays, the nuclear extracts were incubated with purified monoclonal anti-IVa2 or polyclonal anti-52/55-kDa antibodies on ice for 15 min and then added to the reaction mixture for further incubation. Purified monoclonal anti-simian virus 40 (SV40) T antigen (TAg) and polyclonal anti-Rb antibodies were used as controls in the supershift assays.
TABLE 1.
DNA sequences used in mobility shift assays
| A repeat | Sequencea | Ad5 nucleotide position |
|---|---|---|
| DE | GATTGTCAGTTTCCAAAAACGAGGAGGATTTGATATTCACCTGG | 6132–6175 |
| PS | 195–382 | |
| AI-II | TAGTAAATTTGGGCGTAACCGAGTAAGATTTGGCCATTTTCG | 238–283 |
| AI-II-m1 | TAGTAAATTacGGCGTAACCGAGTAAGATTacGCCATTTTCG | 238–283 |
| AI-II-m2 | TAGTAAATTTGGGCGTAACtaAtTAAGATTTGGCCATTTTCG | 238–283 |
| AII-a | AACCGAGTAAGATTTGGC | 254–271 |
| AII-b | TAACCGAGTAAGATTTGGCCATTTTCGCGGG | 253–287 |
| AIV-V | ATTTTGTGTTACTCATAGCGCGTAATATTTGTCTAGGGCCGCG | 316–358 |
| AVI | CGCGGGGACTTTGACCGTTTACGTGG | 355–380 |
The conserved TTTG and CGXG motifs are underlined. The mutated nucleotides in AI-II-m1 and AI-II-m2 are in lowercase letters.
RESULTS
The 52/55-kDa protein does not bind to the DE.
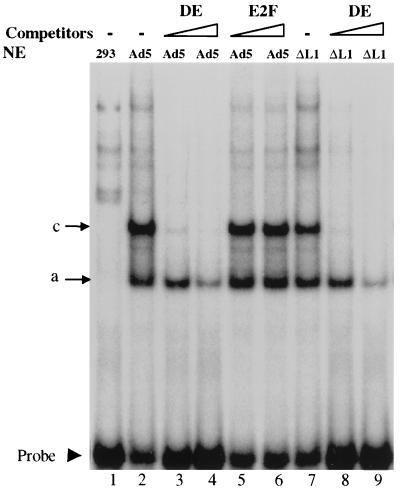
The interaction of the 52/55-kDa protein with the IVa2 protein, one of the activators of MLP, led us to investigate a potential role of the 52/55-kDa protein in regulating the MLP. Although the 52/55-kDa protein is not required for the full activation of the MLP, the slightly delayed activation of the MLP in cells infected with pm8001 (15) suggested that perhaps the 52/55-kDa protein up-regulates the MLP during early times of infection. This effect could be mediated through its interaction with the IVa2 protein, which binds to the DE. For example, it is possible that the 52/55-kDa protein or its previously reported 40-kDa degradation product (19) is a component of the DEF-A complex. To test this possibility, we performed electrophoretic mobility shift assays using nuclear extracts from 293 cells infected with wild-type Ad5 or pm8001, which does not express the 52/55-kDa protein. The probe was a 32P-labeled DE DNA fragment (Table 1). We detected two dominant virus-specific complexes (compare lanes 1 and 2), which correspond to the “a” and “c” complexes reported previously (23) (Fig. 1). The binding is specific, because cold homologous probe could compete for complex formation, whereas an E2F binding sequence could not. The band pattern in the extract from the pm8001-infected cells (lane 7) was the same as that of the Ad5-infected cells (lane 2), indicating that the 52/55-kDa protein was not a component in these complexes. Supershift analysis with antibodies specific for the IVa2 or 52/55-kDa proteins showed that the IVa2 protein, as previously reported, was present in these complexes, while the 52/55-kDa protein was not (data not shown). Furthermore, the formation of the two complexes appeared at the same time, 12 h postinfection of both viruses, and increased thereafter (data not shown), indicating that the formation of the complexes paralleled the activation of the MLP. Thus, it appears that the 52/55-kDa protein does not influence binding of the IVa2 protein to the MLP.
FIG. 1.
The L1 52/55-kDa protein is not a component of the complexes binding to the DE. Electrophoretic mobility shift assays were performed with nuclear extracts (NE) prepared from uninfected 293 cells (293), Ad5-infected 293 cells (Ad5), or pm8001-infected cells (ΔL1) and a labeled DE probe, with or without 20× or 200× molar excess cold DE or the E2F binding sequence in the adenovirus E2 promoter (E2F) as competitors. The arrows indicate the two dominant virus-specific complexes and free DE probe.
The packaging sequence competes with DE for binding to DEF.
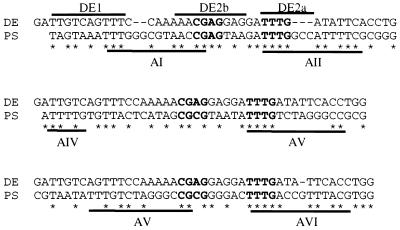
We next investigated whether the interaction between the 52/55-kDa and IVa2 proteins is involved in viral DNA packaging. We examined the sequences in the packaging domain to determine if there were potential binding sites for the IVa2 protein. There is 60.9% sequence identity between the AI-II repeats and DE, 51.2% identity between the AIV-V repeats and DE, and 53.5% identity between the AV-VI repeats and DE. A consensus sequence, CGXGN5TTTG, which is in the four dominant A repeats (I, II, V, and VI), is also present in the DE (Fig. 2). This consensus sequence covers two regions of the DE, DE2b, and DE2a, which have been shown to be the binding sites of DEF-B and DEF-A, respectively.
FIG. 2.
Sequence similarity between the DE of the major late promoter and the PS. The binding sites for DEF-A and -B are indicated by the lines above the DE sequence, and the packaging A repeats are indicated by the lines below the PS. Identical nucleotides are indicated by asterisks. The conserved CGXG and TTTG motifs are indicated in boldface type.
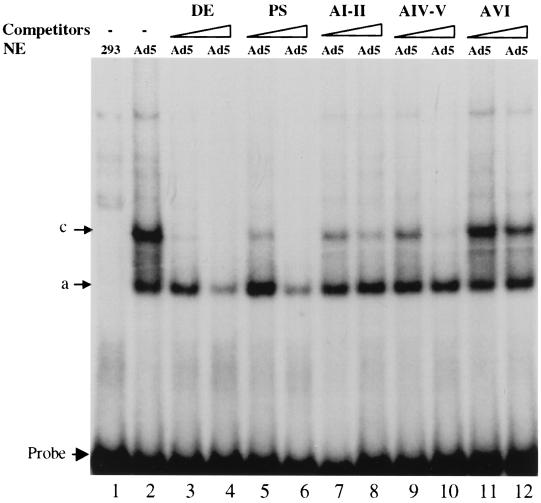
This sequence homology between the DE and the A repeats, especially the presence of the DEF binding sites, suggested that DEF-A and/or B could bind to the packaging sequence. We therefore performed mobility shift assays to test if the packaging sequence could compete with the DE probe for binding to the DEFs. The sequences of the A repeats used in the assay are listed in Table 1. The entire 192-bp packaging sequence (PS) competed for binding to the DE probe (Fig. 3). The AI-II and AIV-V domains also could block the formation of complex c, but not complex a, whereas AVI had little effect on the formation of both complexes. These results suggested that two A repeats together could bind to at least one of the components of the DEFs.
FIG. 3.
Packaging sequences compete with DE for binding to DEF complexes. Electrophoretic mobility shift assays were performed as described in the legend to Fig. 1, using the indicated competitors.
Binding of viral proteins to the AI-II repeats.
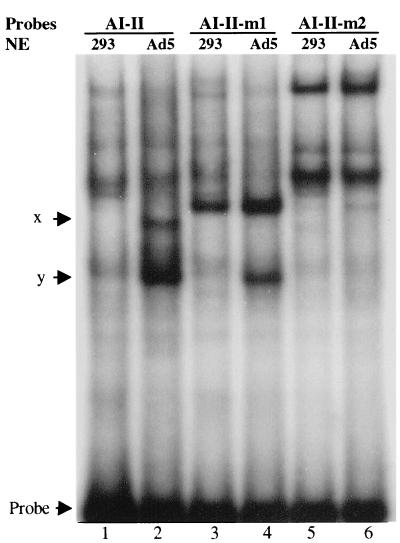
To prove further that the packaging sequence could be bound by virus-specific proteins, assays were performed with a 32P-labeled AI-II probe (Fig. 4). In a comparison of nuclear extracts from uninfected 293 cells (lane 1) and Ad5-infected 293 cells (lane 2), two virus-specific complexes were formed with the AI-II probe, which we have designated as complexes x and y. Cold DE blocked the formation of both complexes, further indicating that a common component, most likely the IVa2 protein, was involved in the DE and AI-II complexes. Complex x was inhibited by all of the packaging A repeats tested, with AVI alone being the weakest competitor. Complex y was only partially competed by the packaging A repeats at the indicated molar excess. An E2F binding sequence did not compete for either complex, however, indicating that the competition by the A repeats was specific (data not shown). We also tested two smaller oligonucleotides that span the AII repeat, AII-a and AII-b, as competitors. AII-a did not compete, and AII-b competed less efficiently than cold AI-II (data not shown). This indicated that the required binding region was larger than AII-a and that perhaps cooperative binding might occur. In addition, a 32P-labeled AIV-V probe was tested for binding, and the same two complexes were detected (data not shown).
FIG. 4.
Binding of viral proteins to the AI-II repeats. Electrophoretic mobility shift assays were performed as described in the legend to Fig. 1, except that a labeled AI-II probe was used, along with the indicated competitors.
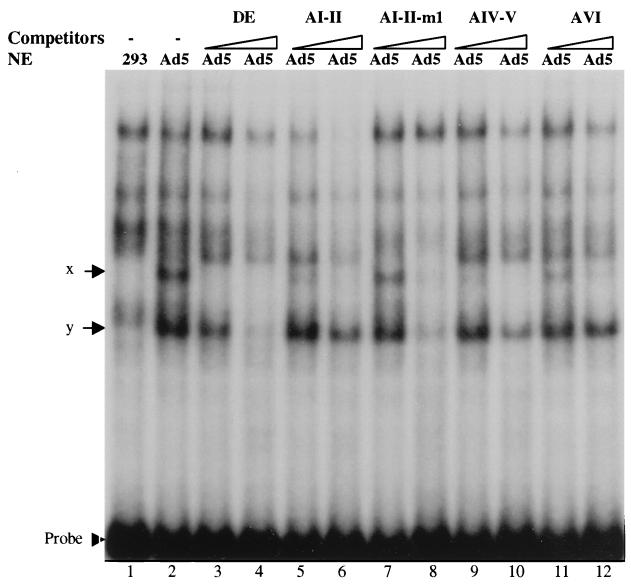
To determine if the conserved CGXGN5TTTG motif in the A repeats was the binding site for complexes x and y, we introduced mutations into this sequence. First, we mutated the TTTG to TTAC in both the AI and AII repeats of the AI-II probe, since TTTG has been defined as a critical component of the DE binding site for DEF-A to form complex c and is also an important motif in the A repeats for viral DNA packaging. This mutant oligonucleotide, AI-II-m1, was used in the assay as a competitor. Compared with wild-type AI-II competitor (lanes 5 and 7 in Fig. 4), AI-II-m1 did not block the formation of complex x efficiently at 20-fold molar excess, indicating that the mutation might affect the formation of complex x. However, AI-II-m1 did block the formation of both complexes x and y efficiently at 200-fold molar excess.
To determine the binding sites for complexes x and y further, different mutations were made, changing the CGAG to TAAT in the AI-II probe to generate AI-II-m2. This region was also shown previously to be important for DNA packaging. Direct binding of 32P-labeled wild-type AI-II, AI-II-m1, and AI-II-m2 probes to viral proteins was compared. Complex x, but not y, disappeared (compare lanes 2 and 4) when the AI-II-m1 probe was used (Fig. 5), indicating that TTTG is required to form complex x, whereas formation of complex y does not require TTTG. This is in agreement with the results of the previous experiment in which AI-II-m1 did not compete efficiently for complex x. However, neither virus-specific complex was detected with the AI-II-m2 probe (compare lanes 2, 4, and 6), indicating that CGAG is required for the formation of both complexes x and y.
FIG. 5.
Mutational analysis of the binding sites for complexes x and y. Electrophoretic mobility shift assays were performed with wild-type AI-II, AI-II-m1, and AI-II-m2 probes.
The IVa2 protein is a component of complexes x and y.
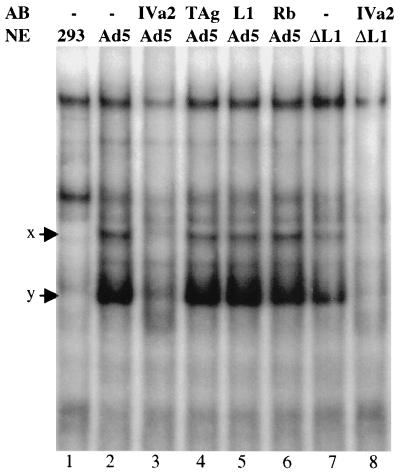
The next question we wished to address is whether the IVa2 protein or 52/55-kDa protein is present in complexes x and y. We performed supershift assays using antibodies to the IVa2 and 52/55-kDa proteins and a 32P-labeled wild-type AI-II probe. When nuclear extracts from Ad5-infected 293 cells were mixed with a monoclonal anti-IVa2 antibody, both complexes x and y disappeared (Fig. 6), indicating that both complexes contained the IVa2 protein. Nuclear extracts mixed with anti-L1 antibody or two control antibodies, anti-SV40 large TAg and anti-Rb (lanes 4 to 6), gave the same binding patterns as the untreated extract (lane 2). This indicated that the 52/55-kDa protein might not be present in these two complexes. This conclusion was further supported by the fact that both complexes x and y were still present in nuclear extracts from pm8001-infected cells, which do not contain any 52/55-kDa protein (lane 7).
FIG. 6.
The IVa2 protein is a component of complexes x and y. Supershift assays were performed with nuclear extracts (NE) prepared from uninfected 293 cells (293), Ad5-infected 293 cells (Ad5), or pm8001-infected 293 cells (ΔL1) with anti-IVa2, L1, SV40 TAg, or Rb protein antibodies (AB) and a labeled AI-II probe.
DISCUSSION
In this report, we investigated the possible significance of the interaction between the adenovirus 52/55-kDa and IVa2 proteins. Although we were unable to establish a definitive role for this interaction, we obtained the unexpected result that the IVa2 protein is not only an activator of MLP, but may also play a significant role in viral DNA packaging.
The first possibility we tested was whether the 52/55-kDa protein is involved in the activation of the MLP. The previous observation that there is a slightly delayed expression of late viral proteins in pm8001-infected cells indicated that the 52/55-kDa protein might have an effect on MLP at early times. We were unable to find the 52/55-kDa protein in complexes formed with the DE, however, as judged by either antibody supershift analysis or differences in complex formation in extracts from cells infected with pm8001. While this does not definitively prove that the 52/55-kDa protein does not affect transcription from the MLP, it argues strongly against it. It remains possible that the 52/55-kDa protein could have an effect on the binding of the DEFs to DE when IVa2 is expressed at low levels during early times of the infection. Another possibility is that the 52/55-kDa protein interacts with complexes a and c to form a larger complex in vivo, which may be unstable in the in vitro assays.
Sequence homology between the DE and the packaging sequence led us to begin to investigate the possibility that the IVa2 protein is involved in viral DNA packaging. Our discovery that the IVa2 protein is a component of two complexes binding to the packaging A repeats provides the first evidence that a viral protein-DNA interaction may be involved in adenovirus DNA packaging. There are two predominant virus-specific complexes formed with the DE and the AI-II probes, and they share some common properties. First, they share a common component, the IVa2 protein. Second, they both require common motifs for binding: the TTTG motif is the binding site for DEF-A to form complex c and is also required for the formation of complex x, and the CGXG motif is in the binding site of DEF-B and is also required for the formation of complex y (27). Third, formation of complex x requires both the TTTG and CGXG motifs, similar to their requirement for the formation of complex c on the DE (27). Based on the similarities between the complexes formed with the DE and the A repeats, we speculate that the factor that makes up complex y on the packaging AI-II probe may be DEF-B, and complex x may be DEF-B together with DEF-A.
The TTTG and CGXG motifs in the A repeats that are required for the formation of complexes x and y have also been shown to be critical for viral DNA packaging (28), suggesting that complexes x and y play roles in viral DNA packaging. In addition to these virus-specific complexes, we also detected cellular components present in uninfected extracts that bind the A repeats. These may be the same as the P-complex, which has been shown previously to bind to the A repeats (29). It is interesting to note, however, that binding of the P-complex only requires the TTTG motif and not the CGXG motif. Since mutations and insertions in the CGXG sequence have been shown to dramatically reduce the efficiency of viral DNA packaging (28), it appears that CGXG and its flanking sequence are required for the binding of a factor or factors that are critical for packaging. Our data demonstrate that this sequence is essential for the formation of both of the virus-specific, IVa2-containing complexes, indicating that the IVa2 protein is likely involved in mediating viral DNA packaging. Preliminary analysis of an IVa2 mutant virus suggests that the IVa2 protein might be involved in assembly (H. Young, personal communication).
To our surprise, however, the 52/55-kDa protein does not appear to be a component of these complexes. The requirement of the 52/55-kDa protein for viral DNA encapsidation indicates that this protein is involved in one of the steps of this process. Virus particles from a temperature-sensitive mutant, ts369, which has a single-amino-acid change in the 52/55-kDa protein, have been shown to contain only the left end of the viral genome at the nonpermissive temperature (20). In addition, an L1-null mutant does not encapsidate any DNA (15). These results indicate that the 52/55-kDa protein may play roles in both the initial association of the viral DNA with empty capsids and the subsequent translocation of the viral DNA. One explanation for why the 52/55-kDa protein is undetectable in the in vitro binding assays is that this protein may need other structural proteins in the intact empty capsid to interact stably with the IVa2-DNA complexes and mediate the encapsidation of the viral DNA. Another possibility is that the interaction of the 52/55-kDa protein with the IVa2-DNA complexes is not detectable under the in vitro assay conditions.
Bacteriophages λ and φ29 have been excellent model systems for studying viral DNA packaging. Adenoviruses share some similarities with the phage systems. First, the genomic, double-stranded DNAs of these viruses are inserted into an empty viral capsid. Second, the DNAs have a single packaging signal located at one end of the genome (22). Third, both adenovirus and phage φ29 have a protein covalently bound to the 5′ end of the DNA, the preterminal protein, and gp3, respectively. The protein-protein and protein-DNA interactions that are necessary for the translocation of viral DNA into the capsid have been well defined in the phage systems, but not in adenovirus. The virus-encoded terminase of phage λ and gp16 of phage φ29 are the proteins that interact with the phage DNA and mediate the translocation of the viral DNA (11, 12, 33, 34). Both the terminase and gp16 function as ATPases, and DNA translocation is driven by ATP hydrolysis (13). Furthermore, a packaging RNA is required for the translocation of viral DNA in the phage φ29 system (14). It will be of interest to determine if the adenovirus 52/55-kDa and IVa2 proteins play similar roles in the encapsidation of its DNA.
ACKNOWLEDGMENTS
We thank the members of the Imperiale laboratory for help with this work, Erle Robertson for suggestions on the mobility shift assays and the manuscript, Hamish Young for communicating results before publication, Claude Kedinger for the anti-IVa2 antibody, and Xinyun Lu for printing the figures.
This work was supported by PHS grant GM34902 from the NIH.
REFERENCES
- 1.Bett A J, Prevec L, Graham F L. Packaging capacity and stability of human adenovirus type 5 vectors. J Virol. 1993;67:5911–5921. doi: 10.1128/jvi.67.10.5911-5921.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caravokyri C, Leppard K N. Constitutive episomal expression of polypeptide IX (pIX) in a 293-based cell line complements the deficiency of pIX mutant adenovirus type 5. J Virol. 1995;69:6627–6633. doi: 10.1128/jvi.69.11.6627-6633.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Edvardsson B, Everitt E, Jörnvall H, Prage L, Philipson L. Intermediates in adenovirus assembly. J Virol. 1976;19:533–547. doi: 10.1128/jvi.19.2.533-547.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Everitt E, Sundquist B, Pettersson U, Philipson L. Structural proteins of adenoviruses. X. Isolation and topography of low molecular weight antigens from the virion of adenovirus type 2. Virology. 1973;52:130–147. doi: 10.1016/0042-6822(73)90404-2. [DOI] [PubMed] [Google Scholar]
- 6.Ghosh-Choudhury G, Haj-Ahmad Y, Graham F L. Protein IX, a minor component of the human adenovirus capsid, is essential for the packaging of full length genomes. EMBO J. 1987;6:1733–1739. doi: 10.1002/j.1460-2075.1987.tb02425.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grable M, Hearing P. Adenovirus type 5 packaging domain is composed of a repeated element that is functionally redundant. J Virol. 1990;64:2047–2056. doi: 10.1128/jvi.64.5.2047-2056.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gräble M, Hearing P. cis and trans requirements for the selective packaging of adenovirus type 5 DNA. J Virol. 1992;66:723–731. doi: 10.1128/jvi.66.2.723-731.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Graham F L, Prevec L. Manipulation of adenovirus vectors. Methods Mol Biol. 1991;7:109–128. doi: 10.1385/0-89603-178-0:109. [DOI] [PubMed] [Google Scholar]
- 10.Graham F L, Smiley J, Russell W C, Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol. 1977;36:59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 11.Guo P, Grimes S, Anderson D. A defined system for in vitro packaging of DNA-gp3 of the Bacillus subtilis bacteriophage phi 29. Proc Natl Acad Sci USA. 1986;83:3505–3509. doi: 10.1073/pnas.83.10.3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo P, Peterson C, Anderson D. Initiation events in in-vitro packaging of bacteriophage phi 29 DNA-gp3. J Mol Biol. 1987;197:219–228. doi: 10.1016/0022-2836(87)90120-3. [DOI] [PubMed] [Google Scholar]
- 13.Guo P, Peterson C, Anderson D. Prohead and DNA-gp3-dependent ATPase activity of the DNA packaging protein gp16 of bacteriophage phi 29. J Mol Biol. 1987;197:229–236. doi: 10.1016/0022-2836(87)90121-5. [DOI] [PubMed] [Google Scholar]
- 14.Guo P, Zhang C, Chen C, Garver K, Trottier M. Inter-RNA interaction of phage phi 29 pRNA to form a hexameric complex for viral DNA transportation. Mol Cell. 1998;2:149–155. doi: 10.1016/s1097-2765(00)80124-0. [DOI] [PubMed] [Google Scholar]
- 15.Gustin K E, Imperiale M J. Encapsidation of viral DNA requires the adenovirus L1 52/55-kilodalton protein. J Virol. 1998;72:7860–7870. doi: 10.1128/jvi.72.10.7860-7870.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gustin K E, Lutz P, Imperiale M J. Interaction of the adenovirus L1 52/55-kilodalton protein with the IVa2 gene product during infection. J Virol. 1996;70:6463–6467. doi: 10.1128/jvi.70.9.6463-6467.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hammarskjold M L, Winberg G. Encapsidation of adenovirus 16 DNA is directed by a small DNA sequence at the left end of the genome. Cell. 1980;20:787–795. doi: 10.1016/0092-8674(80)90325-6. [DOI] [PubMed] [Google Scholar]
- 18.Harris K F, Christensen J B, Radany E H, Imperiale M J. Novel mechanisms of E2F induction by BK virus large-T antigen: requirement of both the pRb-binding and the J domains. Mol Cell Biol. 1998;18:1746–1756. doi: 10.1128/mcb.18.3.1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hasson T B, Ornelles D A, Shenk T. Adenovirus L1 52- and 55-kilodalton proteins are present within assembling virions and colocalize with nuclear structures distinct from replication centers. J Virol. 1992;66:6133–6142. doi: 10.1128/jvi.66.10.6133-6142.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hasson T B, Soloway P D, Ornelles D A, Doerfler W, Shenk T. Adenovirus L1 52- and 55-kilodalton proteins are required for assembly of virions. J Virol. 1989;63:3612–3621. doi: 10.1128/jvi.63.9.3612-3621.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hearing P, Shenk T. The adenovirus type 5 E1A transcriptional control region contains a duplicated enhancer element. Cell. 1983;33:695–703. doi: 10.1016/0092-8674(83)90012-0. [DOI] [PubMed] [Google Scholar]
- 22.Hendrix R W. Bacteriophage DNA packaging: RNA gears in a DNA transport machine. Cell. 1998;94:147–150. doi: 10.1016/s0092-8674(00)81413-0. [DOI] [PubMed] [Google Scholar]
- 23.Jansen-Durr P, Mondésert G, Kédinger C. Replication-dependent activation of the adenovirus major late promoter is mediated by the increased binding of a transcription factor to sequences in the first intron. J Virol. 1989;63:5124–5132. doi: 10.1128/jvi.63.12.5124-5132.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leibowitz J, Horwitz M S. Synthesis and assembly of adenovirus polypeptides. III. Reversible inhibition of hexon assembly in adenovirus type 5 temperature-sensitive mutants. Virology. 1975;66:10–24. doi: 10.1016/0042-6822(75)90175-0. [DOI] [PubMed] [Google Scholar]
- 25.Lutz P, Kedinger C. Properties of the adenovirus IVa2 gene product, an effector of late-phase-dependent activation of the major late promoter. J Virol. 1996;70:1396–1405. doi: 10.1128/jvi.70.3.1396-1405.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maizel, J. V., Jr., D. O. White, and M. D. Scharff. 1968. The polypeptides of adenovirus. II. Soluble proteins, cores, top components and the structure of the virion. Virology 36:126–136. [DOI] [PubMed]
- 27.Mondesert G, Tribouley C, Kedinger C. Identification of a novel downstream binding protein implicated in late-phase-specific activation of the adenovirus major late promoter. Nucleic Acids Res. 1992;20:3881–3889. doi: 10.1093/nar/20.15.3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmid S I, Hearing P. Bipartite structure and functional independence of adenovirus type 5 packaging elements. J Virol. 1997;71:3375–3384. doi: 10.1128/jvi.71.5.3375-3384.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmid S I, Hearing P. Cellular components interact with adenovirus type 5 minimal DNA packaging domains. J Virol. 1998;72:6339–6347. doi: 10.1128/jvi.72.8.6339-6347.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tibbetts C. Viral DNA sequences from incomplete particles of human adenovirus type 7. Cell. 1977;12:243–249. doi: 10.1016/0092-8674(77)90202-1. [DOI] [PubMed] [Google Scholar]
- 31.Tribouley C, Lutz P, Staub A, Kedinger C. The product of the adenovirus intermediate gene IVa2 is a transcriptional activator of the major late promoter. J Virol. 1994;68:4450–4457. doi: 10.1128/jvi.68.7.4450-4457.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Winter N, D'Halluin J-C. Regulation of the biosynthesis of subgroup C adenovirus protein IVa2. J Virol. 1991;65:5250–5259. doi: 10.1128/jvi.65.10.5250-5259.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang Q, de Beer T, Woods L, Meyer J D, Manning M C, Overduin M, Catalano C E. Cloning, expression, and characterization of a DNA binding domain of gpNu1, a phage lambda DNA packaging protein. Biochemistry. 1999;38:465–477. doi: 10.1021/bi981271d. [DOI] [PubMed] [Google Scholar]
- 34.Yang Q, Hanagan A, Catalano C E. Assembly of a nucleoprotein complex required for DNA packaging by bacteriophage lambda. Biochemistry. 1997;36:2744–2752. doi: 10.1021/bi9622682. [DOI] [PubMed] [Google Scholar]