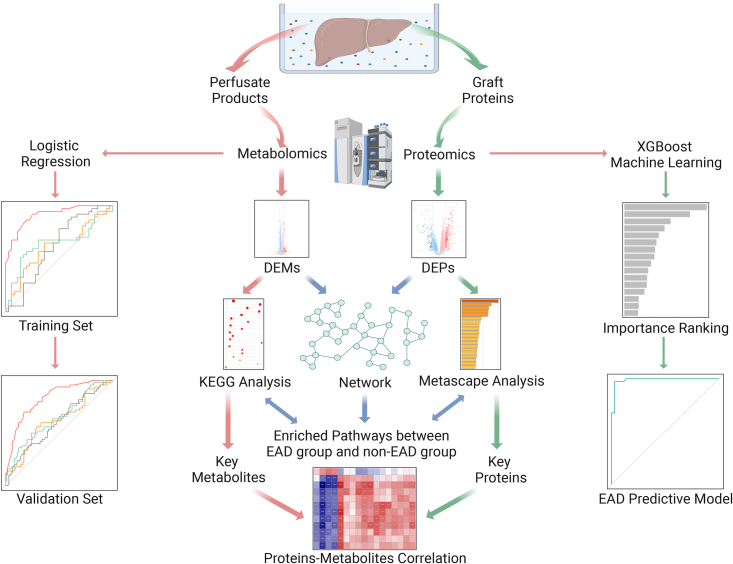
Abstract
Background:
Early allograft dysfunction (EAD) is a common complication after liver transplantation (LT) and is associated with poor prognosis. Graft itself plays a major role in the development of EAD. We aimed to reveal the EAD-specific molecular profiles to assess graft quality and establish EAD predictive models.
Methods:
A total of 223 patients who underwent LT were enrolled and divided into training (n=73) and validation (n=150) sets. In the training set, proteomics was performed on graft biopsies, together with metabolomics on paired perfusates. Differential expression, enrichment analysis, and protein–protein interaction network were used to identify the key molecules and pathways involved. EAD predictive models were constructed using machine learning and verified in the validation set.
Results:
A total of 335 proteins were differentially expressed between the EAD and non-EAD groups. These proteins were significantly enriched in triglyceride and glycerophospholipid metabolism, neutrophil degranulation, and the MET-related signaling pathway. The top 12 graft proteins involved in the aforementioned processes were identified, including GPAT1, LPIN3, TGFB1, CD59, and SOS1. Moreover, downstream metabolic products, such as lactate dehydrogenase, interleukin-8, triglycerides, and the phosphatidylcholine/phosphorylethanolamine ratio in the paired perfusate displayed a close relationship with the graft proteins. To predict the occurrence of EAD, an integrated model using perfusate metabolic products and clinical parameters showed areas under the curve of 0.915 and 0.833 for the training and validation sets, respectively. It displayed superior predictive efficacy than that of currently existing models, including donor risk index and D-MELD scores.
Conclusions:
We identified novel biomarkers in both grafts and perfusates that could be used to assess graft quality and provide new insights into the etiology of EAD. Herein, we also offer a valid tool for the early prediction of EAD.
Keywords: early allograft dysfunction, liver transplantation, metabolomics, predictive model, proteomics
Introduction
Highlights
Early allograft dysfunction (EAD) negatively impacts graft and patient outcomes.
Graft proteomics showed that grafts with lipid disorders and activated inflammatory infiltration were susceptible to EAD.
The downstream metabolic products in perfusate displayed a close connection with graft proteins.
An Integrated-Model based on perfusate metabolic products and clinical parameters could excellently identify liver recipients with a high risk of EAD.
Liver transplantation (LT) is a life-saving treatment for patients with acute liver failure and end-stage liver diseases. However, the disparity between the need for LT and the shortage of donor organs leads to the use of extended criteria donors (ECD), which increases the risk of poor graft function and reduces graft survival1. Early allograft dysfunction (EAD), which has an incidence ranging from 27 to 55%1–4, represents marginal graft function during the first week following LT and is associated with adverse outcomes. First, EAD is associated with inferior survival2,5. For instance, in a large-center retrospective study conducted by the Mayo Clinic that included 1950 cases of LT, EAD significantly decreased the 1-, 3-, and 5-year graft and patient survivals5. Second, EAD causes dysfunction in other organs. Several studies have demonstrated that EAD independently increases the risk of acute kidney injury and chronic kidney failure in LT recipients4,6. EAD has also been reported to correlate with post-transplant ascites, sepsis, and multiple organ failure7,8. Third, EAD prolongs the duration of mechanical ventilation, intensive care unit (ICU) stay, and hospitalization5,6. In addition, EAD increases total hospital costs9. Graft quality is believed to play a dominant role in early graft function, thereby dramatically influencing graft survival and mortality10. Therefore, in the era of ECD, there has been a continued effort to establish effective systems to assess donor liver quality and to rapidly predict the risk of EAD11.
The relationship between clinical risk factors (e.g. advanced donor age, severe steatosis, and prolonged ischemia time) and the occurrence of EAD has been demonstrated12,13. Currently, a series of scoring systems, such as the donor risk index (DRI)14, early allograft failure simplified estimation score15, and liver graft assessment following transplantation score12, have been established to predict early graft outcomes. Furthermore, newly developed biological and bioinformatics technologies revealed the molecular features of grafts, providing insights for EAD prediction. Kurian et al.16 found an alteration in inflammatory and metabolic responses in grafts developing EAD using an RNA microarray and constructed a gene expression classifier with good diagnostic accuracy. Cortes et al.17 and Faitot et al.18 performed different metabolomics studies on donor liver grafts and established graft metabolic scoring systems to predict EAD. Moreover, integrating molecular features into clinical models could achieve better accuracy in predicting EAD. We previously investigated metabolic features of the primary nonfunction of grafts, a severe type of graft dysfunction, and built an integrated model, presenting an area under the diagnostic curve of 0.93019. Therefore, a better understanding of graft molecular profiles could help in providing more accurate predictions and earlier prophylaxis of EAD following LT.
Functionally, proteins participate in most physiological processes, and specific alterations in protein levels could influence and predict disease onset and progression20. Previous studies have found that some graft proteins, such as CXCL116, CEACAM121, VEGF22, and occult collagen23, might be linked to the development of EAD. Facilitated by recent advances in high-throughput technologies, mass spectrometry (MS)-based proteomics and metabolomics have been greatly developed and provide a powerful platform for biomarker identification and disease mechanism exploration20. Proteomics could be used to rapidly identify and quantify proteins in the liver, whereas metabolomics could help determine the levels of metabolites, which are the end products of cellular regulatory processes and are considered the ultimate response of biological systems to pathophysiological changes. Protein levels influence the metabolic profile and metabolite concentrations, which in turn affect protein expression24. Therefore, integrative omics could provide new insights into biological entities and disease management strategies.
In this study, we used a proteomics approach to identify the molecular features of grafts that had developed EAD. Moreover, we detected metabolic products in perfusate, which were released from the grafts as the end products of cellular regulatory processes and roughly reflect the pathophysiological changes in donor grafts during organ preservation25. We aimed to provide a global view of the biological processes, screen for key molecules that contribute to EAD development, and construct integrated models to predict EAD prior to LT.
Methods
Patients and sample procurement
We enrolled adult patients who received LT between April 2020 and August 2021 at our center. Re-transplantations and split, domino, and multi-organ transplantations, as well as cases with insufficient data, were excluded. Ultimately, 223 patients were included in this study. All graft tissues and perfusates were obtained before implantation and rapidly stored at −80°C until analysis. The study design is illustrated in Supplementary Figure S1 (Supplemental Digital Content 1, http://links.lww.com/JS9/C107). Out of the 223 cases, 90 paired liver samples were obtained and sent for proteomics analysis. To avoid the impact of fatty change in proteomics, we excluded grafts with fatty liver (macrovesicular steatosis >30%) (n=2, 1 developed EAD following LT). In addition, 15 samples failed in the quality control of proteomics detection. Finally, 73 available liver samples were applied for proteomics analysis. Then, the paired 73 perfusates were used for metabolomics analysis. Next, we performed integrative omics analysis to reveal the EAD-specific molecular features. As for model construction, 73 cases of LT were enrolled as a training set, while the other 150 cases were considered as a validation set. Patients with hepatitis B virus (HBV) infections received a standard antiviral protocol (nucleoside analogs combined with low-dose immunoglobulin therapy), as previously described26. A tacrolimus-based immunosuppressive protocol was applied to LT recipients, as previously reported27. This study was approved by the Ethics Committee of our hospital according to the Regulations on Human Organ Transplant and national legal requirements. This study complied with the guidelines of the China Ethical Committee and the Declaration of Helsinki. No organs were obtained from executed prisoners. Written informed consent was obtained from the patients for publication and any accompanying images. Moreover, the work has been reported in line with the STROCSS criteria28 (Supplemental Digital Content 2, http://links.lww.com/JS9/C108).
Endpoint definition
EAD was defined as total bilirubin ≥10 mg/dl (171 µmol/l) or an international normalized ratio ≥1.6 on postoperative day 7, or alanine transferase (ALT) or aspartate transferase (AST) levels >2000 U/l within the first 7 postoperative days29.
Measurement and data analysis of graft proteomics using 4D-DIA-MS
Each graft tissue sample was used for four-dimensional data-independent acquisition (4D-DIA)-based proteomics analysis. Details of the measurement process are described in the Supplementary Materials (Supplemental Digital Content 1, http://links.lww.com/JS9/C107). Original data were obtained through database retrieval and proteins with an expression value ≥50% in any of the sample groups were retained. Proteins with a missing value <50% were filled in with the mean value from the same sample group30,31. Through median normalization and log2 conversion, credible proteins were obtained, whereafter principal component analysis (PCA) was performed. The fold change (FC) and P-value were calculated using the t-test results. Differentially expressed proteins (DEPs) were defined as proteins with |log2 FC| > log21.5 and P-value <0.05 between the EAD and non-EAD groups. Heatmaps were generated using the Euclidean Distance Matrix Hierarchical Clustering method. Metascape online analysis (https://metascape.org/) was used for pathway analysis. The molecular complex detection (MCODE) network is provided at https://metascape.org/. A protein–protein interaction (PPI) network was constructed using the STRING database (https://string-db.org/), and the Degree method of the CytoHubba plug-in in Cytoscape was used to screen hub proteins.
Measurement and data analysis of perfusate metabolomics using LC–MS and GC–MS
LC–MS and GC–MS-metabolomics analyses were performed on each perfusate sample in the training set. We performed PCA according to the categories of the EAD and non-EAD groups. Differentially expressed metabolites (DEMs) were selected based on the combination of a variable influence on projection (VIP) value >1 and P-value <0.05. Metabolomics pathway analysis was performed using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. Moreover, an integrative analysis of graft proteomics and perfusate metabolomics was conducted based on the KEGG database and KEGG Markup Language (KGML) network analysis.
Machine learning for feature selection and model establishment
We employed the eXtreme Gradient Boosting (XGBoost) method to analyze the contribution of each DEP, and SHapley Additive exPlanations (SHAP) values were processed to select features32. Finally, these variables were included to construct the XGBoost algorithm model. For increased confidence, this process was repeated 20 times. To control the overfitting, we applied early stopping with 10 rounds33. To assess the performance of the model, the receiver operating characteristic (ROC) curve, the area under the curve (AUC), accuracy, specificity, and sensitivity were calculated.
Statistical analysis
Quantitative variables were presented as mean±SD or median (interquartile range). Student’s t-test or Wilcoxon’s rank-sum test was used to compare quantitative variables between the two groups. Categorical variables are presented as values (percentages) and compared using the Chi-square or Fisher’s exact tests. Statistically significant clinical and perfusate parameters were entered into the risk factor analysis. Moreover, several clinical risk factors [e.g. donor ALT/AST levels, warm/cold ischemia time, and model for end-stage liver disease (MELD) score] reported in previous studies were included in the analysis19,34. The cutoff value was selected according to the diagnostic sensitivity, specificity, and clinical value. A risk factor analysis was performed using a logistic regression model. Clinical and perfusate biochemical variables with P<0.05 in the univariate analysis were entered into a step-by-step multivariate regression analysis. The establishment of predictive models and the calculation of risk scores have been described in our previous study19. Correlations between the parameters were assessed using Spearman or Pearson correlation analyses. The 5-fold cross-validation analysis was applied to evaluate the performance of the predictive models35. The ROC curve and AUC were calculated to evaluate the diagnostic accuracy. The calibration of the model was assessed using a calibration curve and the Brier score. Internal validation was performed using 1000 bootstrap samples, and discrimination of the model was evaluated. A nomogram was constructed using the predictive model as a graphical representation. All statistical analyses were conducted with SPSS v.25.0 (SPSS Inc., Chicago, IL, USA) and R v.4.2.0 software. Statistical significance was set at P<0.05.
Results
Incidence, prognosis, and clinical characteristics of early allograft dysfunction
Of the 223 LT recipients, 39.0% (87/223) developed EAD. Among patients with EAD, 19.5% (17/87) died within one year, whereas only 4.4% (6/136) of those without EAD died (P<0.001). Basic clinical characteristics of the donors, recipients, and surgical procedures are presented in Supplementary Table S1 (Supplemental Digital Content 1, http://links.lww.com/JS9/C107). Compared with those without EAD, patients with EAD had a longer ICU stay (11.0 days vs. 6.0 days; P<0.001), hospital stay (28.0 days vs. 23.0 days; P=0.011), and mechanical ventilation time (2.0 days vs. 1.0 days; P<0.001).
There were 73 graft biopsies available for omics analysis, which served as the training set. Of the 73 patients, 37 developed EAD and 36 did not. As shown in Table 1, age, sex, graft source, graft histopathology, ABO mismatch, recipient HBV status, and surgical characteristics were comparable between the EAD and non-EAD groups. The EAD group showed more grafts with higher donor gamma-glutamyl transpeptidase levels (100.0 U/l vs. 49.5 U/l; P=0.020), heavier graft weights (1.5 kg vs. 1.2 kg; P=0.002), and longer graft warm ischemia times (39.0 min vs. 25.5 min; P=0.040).
Table 1.
The comparison of clinical factors in the training set.
| EAD group (n=37) | Non-EAD group (n=36) | P | |
|---|---|---|---|
| Donor characteristic | |||
| Age (years) | 47.0±13.0 | 47.0±15.0 | 0.961 |
| Male sex (n/%) | 30 (81.1%) | 28 (77.8%) | 0.727 |
| Source of organs (n/%) | 0.565 | ||
| DCD | 25 (67.6%) | 22 (61.1%) | |
| DBD | 12 (32.4%) | 14 (38.9%) | |
| Cause of death (n/%) | 0.713 | ||
| Trauma | 17 (45.9%) | 14 (38.9%) | |
| CVA | 16 (43.8%) | 16 (44.4%) | |
| Other | 4 (13.7%) | 6 (16.7%) | |
| Blood biochemistry | |||
| Cr (μmol/l) | 103.5 (62.0, 188.0) | 79.0 (54.0, 153.7) | 0.168 |
| TB (μmol/l) | 15.5 (9.4, 23.6) | 15.0 (10.4, 29.0) | 0.695 |
| ALT (U/l) | 42.0 (22.0, 121.0) | 35.6 (21.5, 65.8) | 0.150 |
| AST (U/l) | 63.0 (31.5, 109.0) | 45.0 (34.8, 80.0) | 0.363 |
| γGTP (U/l) | 100.0 (41.0, 133.5) | 49.5 (27.0, 81.5) | 0.020 |
| Graft weight (kg) | 1.5±0.3 | 1.2±0.3 | 0.002 |
| Hepatocellular swelling | 0.994 | ||
| None or mild | 20 (54.1%) | 19 (52.8%) | |
| Moderate | 11 (29.7%) | 11 (30.5%) | |
| Severe | 6 (16.2%) | 6 (16.7%) | |
| Hepatocellular necrosis (n/%) | 0.620 | ||
| None | 35 (94.6%) | 33 (91.7%) | |
| Spotty necrosis | 2 (5.4%) | 3 (8.3%) | |
| Zonal necrosis | 0 | 0 | |
| Inflammatory infiltration (n/%) | 0.930 | ||
| None | 13 (35.1%) | 12 (33.3%) | |
| Mild | 19 (51.4%) | 18 (50.0%) | |
| Moderate | 5 (13.5%) | 6 (16.7%) | |
| Graft fibrosis (n/%) | 0.713 | ||
| 0 | 20 (54.1%) | 21 (58.3%) | |
| 1 | 13 (35.1%) | 13 (36.1%) | |
| 2 | 4 (10.8%) | 2 (5.6%) | |
| Graft macrosteatosis >20% (n/%) | 1 (2.7%) | 1 (2.8%) | 0.984 |
| Positive culture of perfusate (n/%)a | 11 (29.7%) | 11 (30.6%) | 0.939 |
| ABO mismatch (n/%) | 6 (16.2%) | 6 (16.7%) | 0.959 |
| Surgical characteristics | |||
| DWIT (min) | 8.0 (0, 12.0) | 6.0 (0, 13.0) | 0.541 |
| DCIT (h) | 8.3 (5.8, 9.5) | 7.7 (6.1, 9.2) | 0.908 |
| GWIT (min) | 39.0 (0, 49.0) | 25.5 (0, 35.0) | 0.040 |
| Anhepatic time (min) | 55.0 (50.5, 62.0) | 52.0 (43.5, 62.5) | 0.189 |
| Operation time (h) | 5.7 (4.8, 6.8) | 5.1 (4.6, 6.1) | 0.108 |
| Blood loss (l) | 1.5 (1.0, 2.1) | 1.2 (0.8, 2.5) | 0.280 |
| RBC transfusion (U) | 6.0 (4.0, 10.3) | 6.3 (2.4, 10.0) | 0.370 |
| Recipient characteristic | |||
| Age (years) | 54.0±9.0 | 53.0±10.0 | 0.575 |
| Male sex (n/%) | 29 (78.4%) | 27 (75.0%) | 0.733 |
| BMI (kg/m2) | 21.1 (18.8, 23.1) | 21.5 (20.0, 23.2) | 0.616 |
| Etiology of liver disease (n/%) | 0.405 | ||
| HBV-related cirrhosis | 21 (56.8%) | 26 (72.2%) | |
| HCC | 14 (37.8%) | 13 (36.1%) | |
| Alcoholic cirrhosis | 2 (5.4%) | 6 (16.7%) | |
| MELD score | 33.0 (27.0, 40.0) | 31.0 (15.0, 40.0) | 0.182 |
| Child–Pugh score | 11.0 (9.0, 11.0) | 10.0 (8.0, 12.0) | 0.987 |
Positive was defined as the growth of any microorganism in perfusate culture.
ALT, alanine transferase; AST, aspartate transferase; BMI, body mass index; Cr, creatinine; CVA, cerebrovascular accident; DBD, donation after brain death; DCD, donation after cardiac death; DCIT, donor cold ischemia time; DWIT, donor warm ischemia time; GWIT, graft warm ischemia time; HBV, hepatitis B virus; HCC, hepatocellular carcinoma; MELD, model for end-stage liver disease; RBC, red blood cell; TB, total bilirubin; γGTP, gamma-glutamyl transpeptidase.
Graft proteomic profiles of early allograft dysfunction
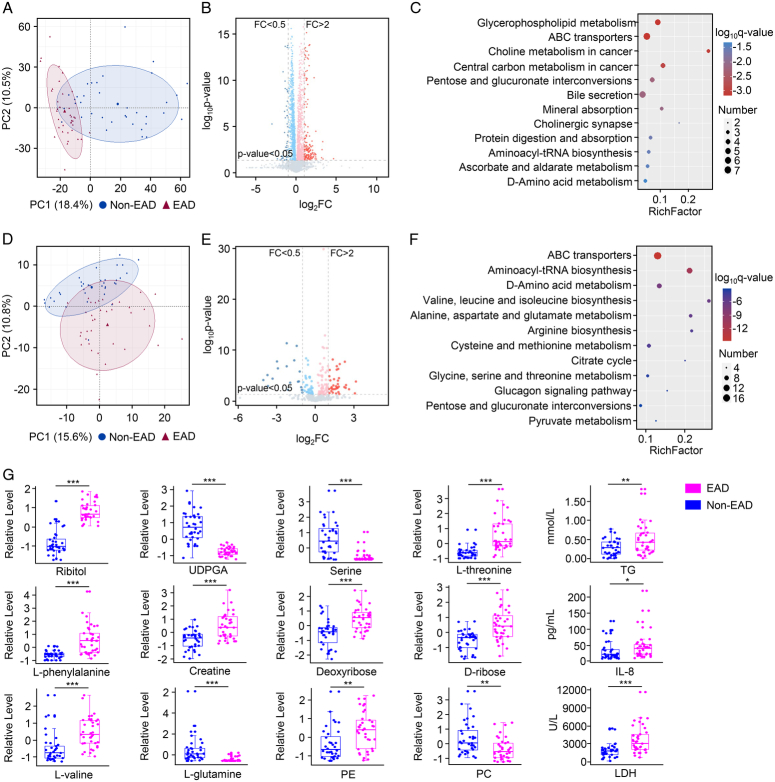
In the training set (n=76), 4D-DIA-based proteomics analysis was performed to identify the molecular signatures of EAD. According to the PCA results, there was a significant segregation between the proteins identified in the EAD and non-EAD groups (Fig. 1A). A total of 5926 proteins were identified in all 73 samples. Compared with those of the non-EAD group, the EAD group showed 335 DEPs with |FC| >1.5 and P<0.05 (Fig. 1B). Among these proteins, 214 were upregulated and 121 downregulated.
Figure 1.
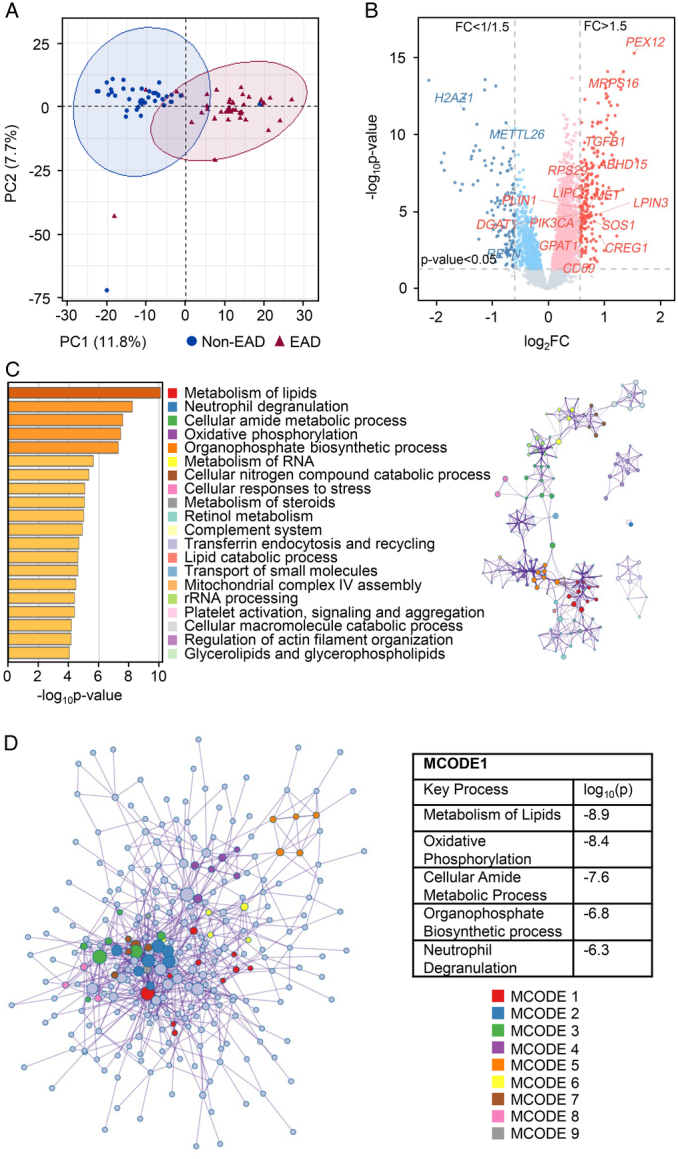
Graft proteomic features of EAD. (A) Principal component analysis plot of the EAD and non-EAD groups. (B) The volcano plot for the DEPs. Red represented FC >1.5 and P<0.05; blue represented FC <1/1.5 and P<0.05. (C) The top 20 enrichment clusters were identified by Metascape online analysis (left) and a subset of representative terms from the full cluster and converted into a network layout (right). Each term was represented by a circle node, where its size was proportional to the number of input genes that fall under that term, and its color represented the cluster to it belonged. Terms with a similarity score >0.3 were linked by an edge (the thickness of the edge represents the similarity score). (D) All DEPs were extracted to form a PPI network. Using the MCODE plug-in method, nine densely connected network components were identified. MCODE1 was the most significant cluster in the network. DEP, differentially expressed protein; EAD, early allograft dysfunction; FC, fold change; MCODE, molecular complex detection.
To identify the proteomic features of EAD, the 335 DEPs were analyzed using Metascape online analysis. The enrichment clusters are shown in Figure 1C. We also selected a subset of representative terms from the full clusters and converted them into a network layout, demonstrating that these terms could be classified into the top 20 enrichment clusters. Moreover, 335 DEPs were extracted to form a PPI network that was divided into nine functional clusters using the MCODE plug-in (Fig. 1D). Notably, the top five enrichment clusters identified in the Metascape analysis (metabolism of lipids, neutrophil degranulation, cellular amide metabolic process, oxidative phosphorylation, and organophosphate biosynthetic process) overlapped with the key processes in MCODE1 (the most significant cluster in the network).
Next, we focused on the above five processes and the 80 DEPs involved, which effectively separated the two groups (Fig. 2A). Among the DEPs involved in lipid metabolism (Supplementary Fig. S2A, Supplemental Digital Content 1, http://links.lww.com/JS9/C107) and organophosphate biosynthetic process (Supplementary Fig. S2B, Supplemental Digital Content 1, http://links.lww.com/JS9/C107), the top five upregulated lipoproteins (DGAT1, LIPC, PLIN1, LPIN3, and GPAT1) were identified as key targets for triglyceride (TG) or glycerophospholipid (GPL) metabolism36–38. Regarding neutrophil degranulation (Supplementary Fig. S3, Supplemental Digital Content 1, http://links.lww.com/JS9/C107), RETN downregulation and CREG1 and CD59 upregulation were identified as the top three targets that could protect against hepatic inflammation during acute liver injury39–41. Furthermore, DEPs in both the cellular amide metabolic (Supplementary Fig. S4A, Supplemental Digital Content 1, http://links.lww.com/JS9/C107) and oxidative phosphorylation processes (Supplementary Fig. S4B, Supplemental Digital Content 1, http://links.lww.com/JS9/C107) were significantly clustered in the MET-related signaling pathway. Four MET-associated proteins (PIK3CA, TGFB1, MET, and SOS1) were dramatically upregulated in the EAD group compared with those in the non-EAD group. MET could interplay with PIK3CA, TGFB1, or SOS1 for reciprocal activation, which would trigger diverse signaling pathways (e.g. PI3K/Akt/mTOR, Ras signaling pathways) and enhance lipid synthesis and hepatic regeneration42,43. The key pathways and proteins associated with EAD are summarized in Table 2. They were not significantly correlated with the clinical parameters (Supplementary Fig. S5, Supplemental Digital Content 1, http://links.lww.com/JS9/C107). The relative expression levels of the 12 DEPs are shown in Figure 2B.
Figure 2.
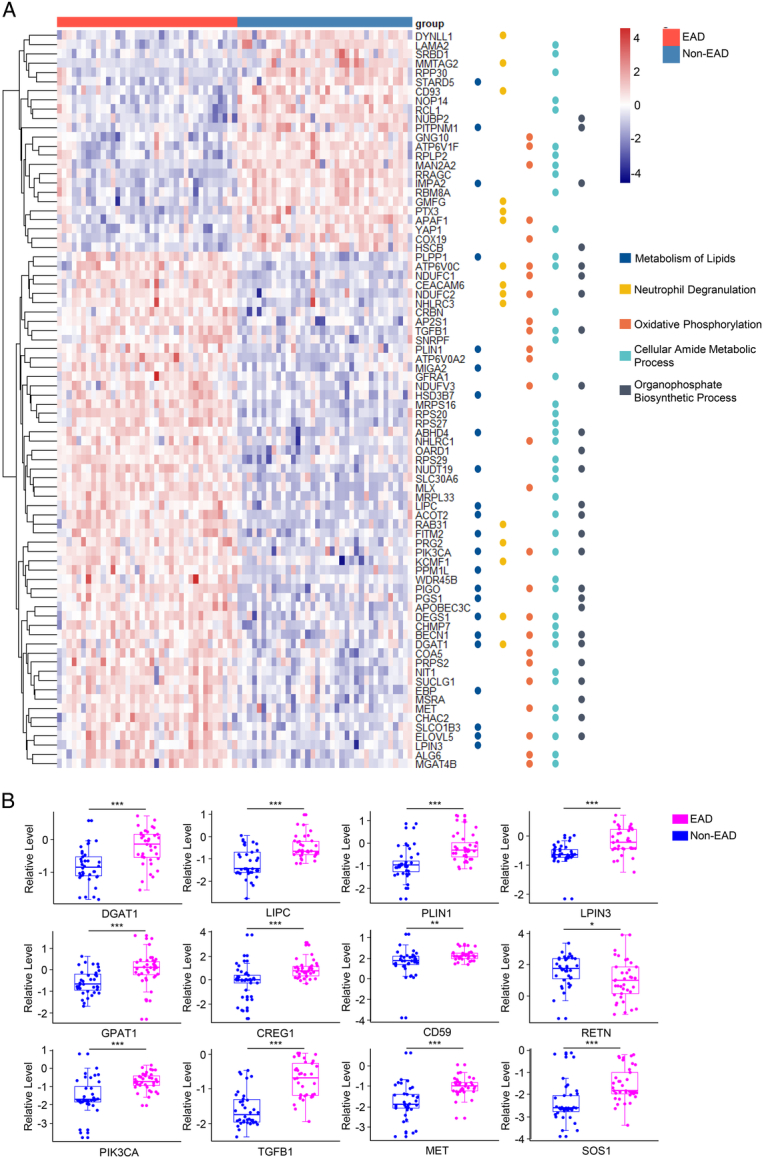
Graft proteins that are involved in the key enrichment pathways. (A) Heatmap of 80 selected DEPs (according to P-value) involved in the five processes identified by Metascape online analysis. (B) The relative expression level (normalized original value) of 12 key proteins between the EAD group and non-EAD group (***P<0.001; **P<0.01; *P<0.05), which participated in the process of triglyceride and glycerophospholipid metabolism (DGAT1, LIPC, PLIN1, LPIN3, and GPAT1), neutrophil degranulation (RETN, CREG1, and CD59), and MET-related signaling pathway (PIK3CA, TGFB1, MET, and SOS1). DEP, differentially expressed protein; EAD, early allograft dysfunction.
Table 2.
Central pathways and proteins contributed to the development of early allograft dysfunction.
| Process | Key pathways | TOP proteins | Functions |
|---|---|---|---|
| Metabolism of lipids | TG metabolism | DGAT1, LPIN3, GPAT1 | TG synthesis36 |
| PLIN1 | TG accumulation38 | ||
| LIPC | TG hydrolysis36 | ||
| GPL metabolism | LPIN3, GPAT1 | GPL synthesis37 | |
| Organophosphate biosynthetic process | GPL metabolism | GPAT1 | GPL synthesis37 |
| Neutrophil degranulation | Neutrophil degranulation | RETN, CREG1, CD59 | Protections against IRI39–41 |
| Cellular amide metabolic process | MET-related signaling pathway | PIK3CA, TGFB1, MET, SOS1 | Lipid regulation42
Hepatic regeneration43 |
| Oxidative phosphorylation |
GPL, glycerophospholipid; IRI, ischemia-reperfusion injury; TG, triglyceride.
Perfusate metabolomic profiles of early allograft dysfunction
Perfusates and liver grafts are closely linked during organ preservation. For instance, grafts require the nutrients present in perfusates, which they release metabolic products into. Therefore, evaluation of graft status may be achieved through the detection of metabolic products in the perfusate. MS-based metabolomics was performed on 73 paired perfusate samples. In the LC–MS-based platform, there was significant segregation in the PCA results between the EAD and non-EAD groups (Fig. 3A). Based on the Human Metabolome Database, Lipidmaps (v2.3), and METLIN Database, we quantified 3354 metabolites in the perfusate (Fig. 3B), of which 193 were DEMs (112 upregulated and 81 downregulated) with VIP >1 and P<0.05. KEGG pathway analysis revealed that these DEMs were enriched in GPL metabolism, ABC transporters, and pentose and glucuronate interconversions (Fig. 3C). In the GC–MS-based platform, the PCA plot showed significant differences between the EAD and non-EAD groups (Fig. 3D). A total of 425 metabolites were shown (Fig. 3E), and 94 DEMs (62 upregulated and 32 downregulated) with VIP >1 and P<0.05 identified. KEGG pathway analysis indicated that these DEMs were clustered in ABC transporters, amino acid-related pathways (e.g. aminoacyl-tRNA biosynthesis, d-amino acid metabolism, and valine/leucine/isoleucine biosynthesis), the citrate cycle, and pentose and glucuronate interconversions (Fig. 3F). Next, we detected biochemical profiles and cytokines using a panel and found significant differences between the EAD and non-EAD groups (Supplementary Table S2, Supplemental Digital Content 1, http://links.lww.com/JS9/C107). The EAD group had higher levels of ALT, AST, lactate dehydrogenase (LDH), TG, and interleukin-8 (IL-8). In addition, the metabolic products in the perfusate did not differ significantly between positive and negative culturing of perfusates (Supplementary Fig. S6, Supplemental Digital Content 1, http://links.lww.com/JS9/C107). The representative differentially expressed metabolic products are shown in Figure 3G.
Figure 3.
Perfusate metabolomic features of EAD. (A) Principal component analysis plot of the EAD and non-EAD groups in LC–MS-metabolomics. (B) The volcano plot for the DEMs in LC–MS-metabolomics. Red represented FC >2 and P<0.05; blue represented FC <0.5 and P<0.05. (C) KEGG pathway analysis between the EAD and non-EAD groups in LC–MS-metabolomics. (D) Principal component analysis plot of the EAD and non-EAD groups in GC–MS-metabolomics. (E) The volcano plot for the DEMs in GC–MS-metabolomics. Red represented FC >2 and P<0.05; blue represented FC <0.5 and P<0.05. (F) KEGG pathway analysis between the EAD and non-EAD groups in GC–MS-metabolomics. (G) The representative differential expressed metabolic products between the EAD and non-EAD groups (***P<0.001; **P<0.01; *P<0.05). DEMs, differentially expressed metabolites; EAD, early allograft dysfunction; FC, fold change; IL-8, interleukin-8; LDH, lactate dehydrogenase; PC, phosphatidylcholine; PE, phosphorylethanolamine; TG, triglyceride; UDPGA, UDP glucuronic acid.
Integrated proteomics and metabolomics approach for assessing graft quality
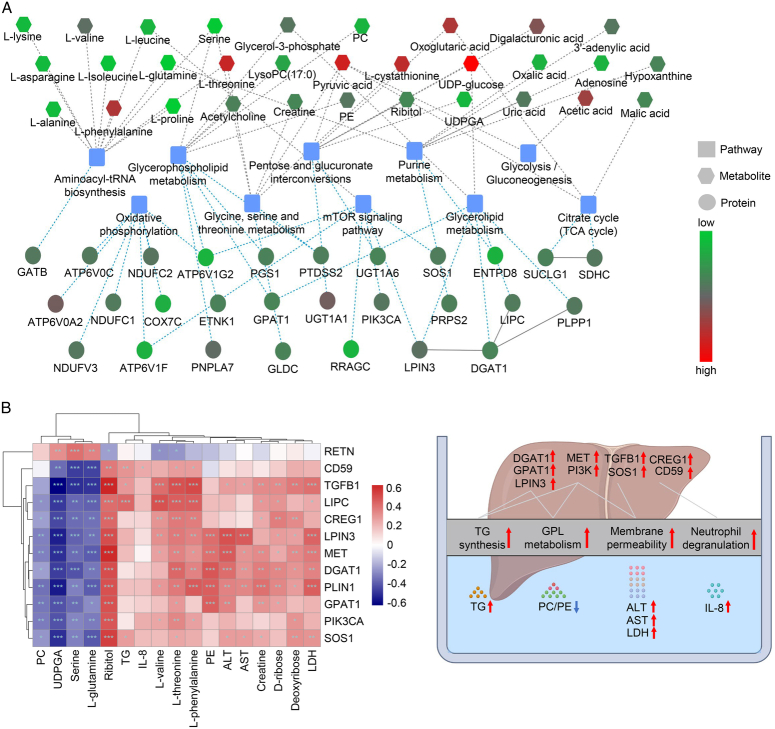
To further reveal the EAD-specific molecular features and assess graft quality, we performed an integrative analysis of graft proteomics and perfusate metabolomics based on the KEGG database and KGML network analysis. Pathway enrichment analysis revealed that the DEPs and DEMs were significantly correlated and enriched in some common processes, such as GPL metabolism, oxidative phosphorylation, the mTOR signaling pathway, glycolysis, pentose and glucuronate interconversions, and glycerolipid metabolism (Fig. 4A).
Figure 4.
Integrative analysis of graft proteomics and perfusate metabolomics. (A) The network showed the interaction of differentially abundant proteins and metabolites and the related metabolic pathways. (B) The correlation plot between the TOP DEMs and DEPs (***P<0.001; **P<0.01; *P<0.05). (C) The key graft proteins and perfusate metabolic products reflected the graft status, including TG and GPL metabolism, cellular membrane permeability, and neutrophil degranulation, which were associated with EAD. ALT, alanine transferase; AST, aspartate transferase; EAD, early allograft dysfunction; FC, fold change; GPL, glycerophospholipid; IL-8, interleukin-8; LDH, lactate dehydrogenase; PC, phosphatidylcholine; PE, phosphorylethanolamine; TG, triglyceride; UDPGA, UDP glucuronic acid.
According to the integrative analysis, we selected the top DEMs, including ribitol, l-threonine, l-phenylalanine, l-valine, phosphorylethanolamine (PE), phosphatidylcholine (PC), creatine, and TG. Correlation analysis between DEPs in the graft and metabolic products in the perfusate showed a good correlation (Fig. 4B). For instance, the PC/PE ratio was negatively correlated with GPL synthesis-related proteins (e.g. GPAT1 and LPIN3); LDH significantly correlated with TG/GPL synthesis proteins (e.g. LPIN3 and PLIN1) and MET-related proteins (e.g. MET and TGFB1); TG positively correlated with LIPC, CD59, TGFB1, and SOS1; and IL-8 positively correlated with CD59.
Taken together, these results showed an EAD-specific signature, including the activation of TG and GPL metabolism, neutrophil degranulation, and the MET-related signaling pathway. The hypothesis of the development of EAD is shown in Figure 4C. For instance, grafts with upregulated GPAT1 and LPIN3 protein contents indicated an active TG and GPL metabolism and toxicity, which was reflected by the reduced PC/PE ratio and increased TG and cytokine levels in the perfusate.
Prediction models of early allograft dysfunction
We first employed a decision tree to select and rank the DEPs that could be used to construct a predictive model (Supplementary Fig. S7A, Supplemental Digital Content 1, http://links.lww.com/JS9/C107). We further trained an integrated XGBoost machine learning model (Proteomics-Model) based on the DEPs, which showed an AUC of 0.988, accuracy of 0.973, specificity of 0.972, and sensitivity of 0.973 (Supplementary Fig. S7B, Supplemental Digital Content 1, http://links.lww.com/JS9/C107). The calibration curves of the Proteomics-Model showed excellent concordance between the predicted and actual probabilities (Supplementary Fig. S7C, Supplemental Digital Content 1, http://links.lww.com/JS9/C107).
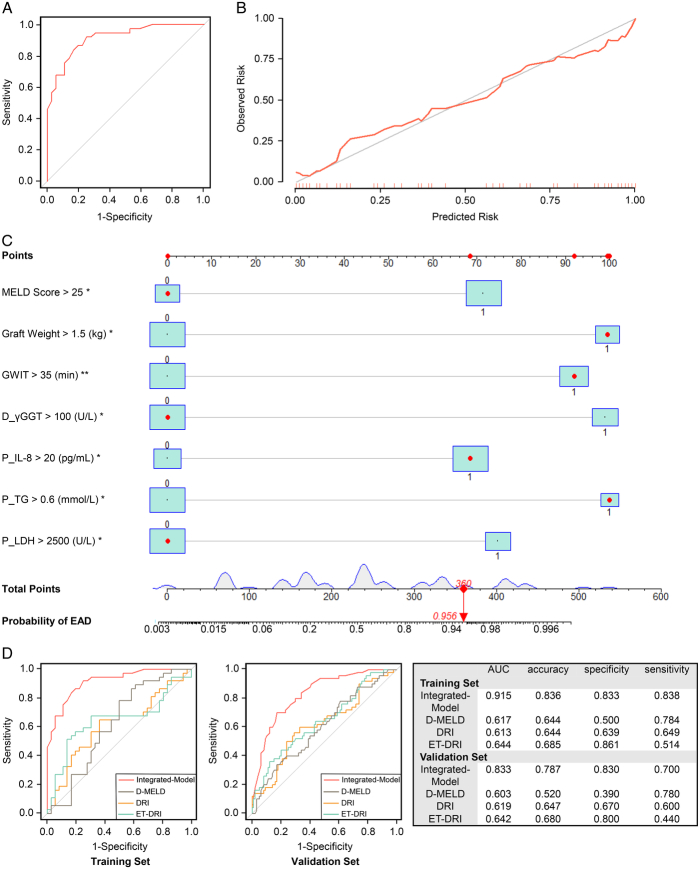
Given the simplification of the detection of perfusate biochemistry and cytokine levels, as well as the close interaction between the graft and perfusate, we further considered perfusate metabolic products as potential biomarkers for predicting EAD. In multivariate analysis, we found that perfusate LDH, TG, and IL-8 levels were independent factors influencing the development of EAD (Table 3). Accordingly, we built a predictive model using clinical parameters and perfusate metabolic products (Integrated-Model), which displayed an AUC of 0.915, accuracy of 0.836, specificity of 0.833, and sensitivity of 0.838 in the training set (Fig. 5A). Calibration curves of the Integrated-Model exhibited good concordance between the predicted and actual probabilities (Fig. 5B). A nomogram was constructed using the coefficients of this model (Fig. 5C), and the probability of developing EAD calculated using the nomogram formula.
Table 3.
Risk factors of early allograft dysfunction.
| Univariate | Multivariate | |||
|---|---|---|---|---|
| OR (95% CI) | P | OR (95% CI) | P | |
| Donor, surgical, and recipient characteristics | ||||
| γGTP >100 (U/l) | 3.925 (1.424, 11.80) | 0.011 | 10.78 (2.032, 89.68) | 0.011 |
| ALT >100 (U/l) | 3.840 (1.178, 15.07) | 0.034 | ||
| AST >60 (U/l) | 2.322 (0.916, 6.073) | 0.079 | ||
| Graft weight >1.5 (kg) | 3.156 (1.137, 9.499) | 0.032 | 10.92 (2.014, 94.18) | 0.013 |
| GWIT >35 (min) | 4.594 (1.708, 13.34) | 0.003 | 9.106 (2.202, 50.30) | 0.005 |
| DWIT >7.5 (min) | 1.647 (0.656, 4.212) | 0.291 | ||
| DCIT >8 (h) | 1.868 (0.737, 4.848) | 0.191 | ||
| MELD score >25 | 2.900 (1.067, 8.390) | 0.041 | 5.573 (1.194, 34.55) | 0.041 |
| Perfusate parameters | ||||
| ALT >400 (U/l) | 5.372 (2.032, 15.18) | <0.001 | ||
| AST >850 (U/l) | 5.417 (2.044, 15.37) | <0.001 | ||
| LDH >2500 (U/l) | 5.278 (1.860, 16.81) | 0.003 | 6.026 (1.206, 41.03) | 0.041 |
| TG >0.6 (mmol/l) | 5.280 (1.490, 25.02) | 0.017 | 11.05 (1.547, 113.2) | 0.026 |
| IL-8 >20 (pg/ml) | 3.111 (1.174, 8.718) | 0.025 | 5.182 (1.249, 26.39) | 0.031 |
The cutoff value was selected according to the diagnostic specificity and sensitivity.
ALT, alanine transferase; AST, aspartate transferase; DCIT, donor cold ischemia time; DWIT, donor warm ischemia time; GWIT, graft warm ischemia time; IL-8, interleukin-8; LDH, lactate dehydrogenase; MELD, model for end-stage liver disease; TG, triglyceride; γGTP, gamma-glutamyl transpeptidase.
Figure 5.
Prediction of EAD. (A) The receiver operating characteristic curve of the Integrated-Model in the training set. (B) The calibration curves of the Integrated-Model in the training set. (C) The nomogram of the Integrated-Model. (D) The receiver operating characteristic curves of Integrated-Model, DRI, ET-DRI, and D-MELD scores in the training set and validation set. AUC, the area under the curve; D-MELD, donor age and model for end-stage liver disease; DRI, donor risk index; EAD, early allograft dysfunction; ET-DRI, eurotransplant-donor risk index; MELD, model for end-stage liver disease.
EAD occurred in 33.3% of the LT recipients in the validation set (n=150). We found that the Integrated-Model could stratify patients with EAD from those without, with an AUC of 0.833, accuracy of 0.787, specificity of 0.830, and sensitivity of 0.700. Furthermore, we compared the diagnostic efficacy of the Integrated-Model and current existing models, including the DRI, eurotransplant-donor risk index (ET-DRI), and D-MELD scores in both the training and validation sets (Fig. 5D). The integrated model showed superior predictive efficacy compared with that of the other models.
Discussion
To the best of our knowledge, this is a pioneering study as it was the first to explore the EAD-specific liver graft proteomic profiles and associated downstream metabolic features in the perfusate. Compared with that of grafts without EAD, those developing EAD displayed significant activation of TG and GPL metabolism, neutrophil degranulation, and the MET-related signaling pathway. To provide more evidence to support this, we identified the metabolic products present in the paired perfusate, which not only presented an EAD-specific distinction but also uncovered the close connection between the liver graft and perfusate. Based on the cross-omics analysis, we suggest that grafts with lipid disorders (TG and GPL upregulation) and inflammatory infiltration may be susceptible to EAD. Key molecules, including GPAT1, LPIN3, TG, PC/PE, LDH, and IL-8, which are involved in the above-mentioned processes, could serve as potential biomarkers for assessing graft quality.
This study emphasized the concern of using liver grafts in patients with lipid metabolism disorders. It is well-known that a hepatic macrosteatosis level above 60% increases the incidence of EAD1. Herein, we showed that liver grafts with overactivation of TG metabolism, both the synthesis (e.g. DGAT1, LPIN3, and GPAT1) and accumulation (e.g. PLIN1), could increase the risk of EAD, even though steatosis has not been pathologically presented. As the first rate-limiting enzyme in TG synthesis, GPAT1 acylates glycerol-3-phosphate into phosphatidic acid in the mitochondria, which is then dephosphorylated by LPIN3 to produce diacylglycerol (DAG)36. DAG is then esterified by DGAT1 to ultimately form TG44. Previous studies have shown a close relationship between increased GPAT1 and DGAT1 levels and higher liver TG concentrations and plasma lipid levels45. Genetic variation in GPAM, which encodes GPAT1, is associated with poor liver function in various chronic liver diseases (e.g. alcohol-related liver disease and cirrhosis)44,46. In addition, PLIN1 promotes TG accumulation by inducing lipid droplet formation and structural modifications and inhibiting lipolysis38. In non-alcoholic fatty liver disease, the overexpression of PLIN1 has relevance with oxidative injury and hepatocyte ballooning47. TG itself is straightforwardly toxic to hepatocytes and indirectly leads to hepatic damage by promoting the overproduction of lipotoxins (e.g. free Fas and lysophosphatidic acid)48. Consistent with the proteomic findings, we observed a dramatically increased TG concentration in the perfusate of grafts that developed EAD. Therefore, we highlighted the potential value of TG metabolism-related proteins as early warning signs of EAD.
Our previous metabolomics-based49 and the present cross-omics studies showed that activated GPL metabolism (e.g. GPAT1 and LPIN3) may be linked to the development of EAD. Two key proteins, GPAT1 and LPIN3, play dominant roles in GPL metabolism and increase the production of PC and PE37. Balancing the PC/PE ratio is essential to maintain cellular membrane integrity50. In this study, EAD was associated with downregulated PC and upregulated PE levels, leading to a reduced PC/PE ratio in the perfusate. The downregulation of PC and the reduced PC/PE ratio could increase membrane permeability, leading to the leakage of hepatocellular content into the extracellular environment51. Injured hepatocyte membrane integrity was confirmed by a sharp increase observed in ALT and AST levels in the perfusates. This process could further activate Kupffer cells and trigger the infiltration of other immune cells (e.g. neutrophils), thereby leading to cytokine-mediated injury in hepatocytes51,52. Therefore, we hypothesized that GPL-related toxicity plays a role in EAD etiology. The inhibition of GPAT1 or LPIN3 could reduce the production of GPLs, which may provide a potential therapeutic strategy for grafts developing EAD.
There is also evidence of a close link between EAD and graft lipid disorders. Liver grafts developing EAD displayed upregulation of MET-associated proteins (e.g. MET and PIK3CA) and activation of the MET/PI3K/Akt/mTOR signaling pathway, according to the combined proteomics and metabolomics analyses. MET is a receptor of the hepatocyte growth factor (HGF), which is secreted by Kupffer, hepatic stellate, and vascular endothelial cells43. Upon binding to HGF, MET is activated and recruits PIK3CA to stimulate the PI3K/Akt/mTOR signaling pathway, thereby functioning as a crucial regulator of metabolism, including nucleic acids, lipids, proteins, and carbohydrates42. By elevating glucose flux into the pentose phosphate pathway, the activated PI3K/Akt/mTOR signaling pathway provides sufficient NADPH for lipid synthesis, particularly of TG and GPL, and promotes lipid droplet formation42. Moreover, activation of the MET/PI3K/Akt/mTOR signaling pathway could inhibit lipolysis and lipid oxidation, leading to lipid accumulation42,53.
In addition to regulating lipid metabolism, the HGF/MET-associated signaling pathway is indispensable for liver regeneration and repair through triggering diverse pathways, such as the PI3K/Akt and Ras pathways43,54. When MET is activated, SOS1 is recruited to stimulate the Ras pathway, promoting cell proliferation and growth54. In contrast, TGFB1 can block activation of the HGF/MET axis via binding to HGF, restoring hepatocytes55. Thus, the coordination and balance between MET, PIK3CA, SOS1, and TGFB1 are essential for repair after liver injury. Previous studies have demonstrated that the HGF/MET axis can be rapidly activated in cases of acute liver failure and partial hepatectomy56,57. In rat LT and ischemia-reperfusion injury (IRI) models, HGF pretreatment of recipients resulted in improved graft function and reduced mortality58. In this study, the grafts that developed EAD showed significant activation of MET-related proteins, indicating active hepatic regeneration and severe organ injury.
Enrichment of the neutrophil degranulation pathway further suggests the involvement of inflammatory regulation in EAD development. It is well-known that the inflammatory phase is inevitable during organ procurement, preservation, and implantation. Several clinical situations such as marginal donor livers, prolonged ischemia time, and massive blood loss, may worsen the inflammatory phase59. Moreover, the comprehensive remodeling of liver immunity via donor-recipient interplay causes inflammation60. As the first responders to inflammation and infection, neutrophils can be recruited to serve as a signal for the severity of injury61. Our proteomics analysis demonstrated that grafts developing EAD displayed dysregulation of proteins involved in neutrophil degranulation, such as RETN, CREG1, and CD59. It has been reported that RETN, CREG1, and CD59 protect against hepatic IRI, a well-known cause of EAD, through the PI3K/Akt pathway39, mitogen-activated protein kinase signaling pathway40, and complement response41, respectively. In the perfusate, IL-8, a potent human neutrophil-attracting chemokine62, was significantly upregulated in the EAD group compared with that in the non-EAD group and showed a positive correlation with graft CD59. Herein, we highlighted the vital role of these neutrophil degranulation-related proteins and perfusate IL-8 in the early warning signs of EAD. In addition, graft proteomics revealed the potential mechanism underlying acute kidney injury after LT. Norén et al.63 performed proteomics analysis in a liver graft and found significantly differential expressed proteins, which were related to immune and inflammatory responses, host defense, and neutrophil degranulation, between the acute kidney injury group and the non-acute kidney injury group. Therefore, a more severe graft inflammation status, possibly as a consequence of donation after circulatory death (DCD), steatosis, or long-distance traveling liver graft, would be associated with poor organ function after LT. Strategies aiming to alleviate sterile inflammation such as pharmacological therapies and machine perfusion have been developed to preserve organ function and improve long-term transplant outcomes, particularly for expanded criteria donors like DCD64.
From bench to bedside, accurate prediction of EAD would be of great help in early intervention and improving treatment outcomes. We believe that the integrative analysis of proteomics and metabolomics could not only reveal the early biological processes leading to EAD but also identify biomarkers for disease prediction. We established a Proteomics-Model that could effectively disaggregate the EAD cases from those of the others with extremely high accuracy. Graft biopsies can be easily collected during graft preservation and are available for histological assessment and accurate injury quantification, thus serving as potential tools for assessing graft quality and predicting treatment outcomes.
To predict EAD in a timely manner, we focused on the biochemistry and cytokine levels in the perfusate, which can be obtained noninvasively and rapidly. We found that metabolic products, such as LDH, TG, and IL-8, were independent risk factors for the development of EAD. These results were consistent with the proteomics findings showing that perfusates were more enriched in the products of oxidative stress, inflammation, and TG and GPL metabolism in the EAD group than in the non-EAD group. We further established an integrated model using perfusate metabolic products and clinical parameters. The model could triage grafts that were susceptible to EAD with an AUC >0.8 in both the training and validation sets. The model also showed better predictive efficiency than that of existing models.
This study had pros and cons. The major advantage was the integrative omics approach employed. Liver proteomics and perfusate metabolomics were well connected. Proteomics analysis could rapidly identify and quantify graft proteins, whereas metabolomics could determine the levels of perfusate metabolites, which are the end products of cellular regulatory processes in the liver. Therefore, the integrative omics analysis comprehensively proposed a coordinated mechanism for the development of EAD in different biological layers and identified the key molecules involved. Integration of key molecules into the clinical model significantly improved its diagnostic efficacy. However, this study had some limitations. First, protein detection, quantitation, and analysis using common technologies, such as immunology-based methods, are time-consuming. Therefore, it still lacks efficacy for timely decision-making regarding the use or discarding of donor allografts. Second, the predictive efficacy of our models needs to be verified in large cohorts from different regions and patient populations. The mechanisms underlying the roles of key proteins and pathways in graft function require further exploration. Third, the use of perfusates has some limitations, as mentioned in the International Liver Transplantation Society Consensus Guidelines65. For instance, the microorganisms and surgeons involved in the backbench and retrieval would influence the levels of perfusate molecules. The perfusate samples, which were obtained in a static storage setting, may not include all the metabolic products. Some metabolites may not have diffused out in the storage bag, and variable degrees may still retained in the liver. In addition, single-cell technologies, including single-cell transcriptomics, proteomics, and metabolomics, could effectively identify the molecular landscape of signal cells in organisms and allow an understanding of the heterogeneity and relevance between cells66. These technologies may better elucidate the differences between grafts that develop EAD and those that do not.
In summary, this study provides a better understanding of the etiology of EAD from the perspective of graft proteomics and its close correlation with perfusate metabolic products. We demonstrated that grafts susceptible to EAD showed overactivated TG and GPL metabolism and inflammatory infiltration. Graft proteins, such as LPIN3, TGFB1, CD59, and SOS1, could be used as biomarkers for assessing graft quality and may also serve as potential therapeutic targets. Perfusate metabolic products, including LDH, IL-8, TG, and PC/PE, could help decipher the pathophysiology of grafts and effectively predict the risk of EAD.
Ethical approval
This study was approved by the Ethics Committee of the First Affiliated Hospital, Zhejiang University School of Medicine (No. 2022-1550).
Consent
Written informed consent was obtained from the patient for publication and any accompanying images. A copy of the written consent is available for review by the Editor-in-Chief of this journal on request.
Sources of funding
This work was supported by grants from the National Natural Science Foundation of China (Nos. 82171757 and 82241215), the Natural Science Foundation of Zhejiang Province (Nos. LZ22H030004 and LQ20H160048), and the Research Project of Jinan Microecological Biomedicine Shandong Laboratory (JNL-2023006C).
Author contribution
Q.L.: conceived the paper; Y.L., J.C., K.Z., R.C., J.J., X.Y., and S.F.: collected the samples and clinical data; Y.L.: performed the bioinformatic analysis and wrote the original draft; Y.L. and H.H.: generated the figures; Q.L., H.H., J.L., and S.Z.: reviewed and edited the paper. All the authors agreed to publish the manuscript.
Conflicts of interest disclosure
The authors of this manuscript have no conflicts of interest to disclose.
Research registration unique identifying number (UIN)
Our UIN is “researchregistry9532”.
Guarantor
Qi Ling.
Data availability statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Provenance and peer review
Our paper was not invited.
Presentation
None.
Supplementary Material
Footnotes
Sponsorships or competing interests that may be relevant to content are disclosed at the end of this article.
Supplemental Digital Content is available for this article. Direct URL citations are provided in the HTML and PDF versions of this article on the journal's website, www.lww.com/international-journal-of-surgery.
Published online 19 March 2024
Contributor Information
Yimou Lin, Email: 22118038@zju.edu.cn.
Haitao Huang, Email: 12018489@zju.edu.cn.
Jiaying Cao, Email: 22218547@zju.edu.cn.
Ke Zhang, Email: 21918367@zju.edu.cn.
Ruihan Chen, Email: rhchen@zju.edu.cn.
Jingyu Jiang, Email: jyjiang162003@zju.edu.cn.
Xuewen Yi, Email: xuewenyi@zju.edu.cn.
Shi Feng, Email: fengshi@zju.edu.cn.
Jimin Liu, Email: nliu@haltonhealthcare.com.
Shusen Zheng, Email: shusenzheng@zju.edu.cn.
Qi Ling, Email: lingqi@zju.edu.cn;lingqilynch@163.com.
References
- 1.Croome KP, Mathur AK, Mao S, et al. Perioperative and long-term outcomes of utilizing donation after circulatory death liver grafts with macrosteatosis: a multicenter analysis. Am J Transplant 2020;20:2449–2456. [DOI] [PubMed] [Google Scholar]
- 2.Lee DD, Singh A, Burns JM, et al. Early allograft dysfunction in liver transplantation with donation after cardiac death donors results in inferior survival. Liver Transpl 2014;20:1447–1453. [DOI] [PubMed] [Google Scholar]
- 3.Nie Y, Huang JB, He SJ, et al. Validation and performance of three scoring systems for predicting primary non-function and early allograft failure after liver transplantation. Hepatobiliary Pancreat Dis Int 2023;Aug 23:S1499-3872(23)00134-0; [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 4.Wadei HM, Lee DD, Croome KP, et al. Early allograft dysfunction after liver transplantation is associated with short- and long-term kidney function impairment. Am J Transplant 2016;16:850–859. [DOI] [PubMed] [Google Scholar]
- 5.Lee DD, Croome KP, Shalev JA, et al. Early allograft dysfunction after liver transplantation: an intermediate outcome measure for targeted improvements. Ann Hepatol 2016;15:53–60. [DOI] [PubMed] [Google Scholar]
- 6.Hudcova J, Scopa C, Rashid J, et al. Effect of early allograft dysfunction on outcomes following liver transplantation. Clin Transplant 2017;31:e12887. [DOI] [PubMed] [Google Scholar]
- 7.Bloom PP, Gilbert T, Santos-Parker K, et al. The incidence and natural history of ascites after liver transplantation. Hepatol Commun 2023;7:e0158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pomposelli JJ, Goodrich NP, Emond JC, et al. Patterns of early allograft dysfunction in adult live donor liver transplantation: the A2ALL experience. Transplantation 2016;100:1490–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moosburner S, Sauer IM, Forster F, et al. Early allograft dysfunction increases hospital associated costs after liver transplantation-a propensity score-matched analysis. Hepatol Commun 2021;5:526–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Agopian VG, Petrowsky H, Kaldas FM, et al. The evolution of liver transplantation during 3 decades analysis of 5347 consecutive liver transplants at a single center. Ann Surg 2013;258:409–421. [DOI] [PubMed] [Google Scholar]
- 11.Caimano M, Bianco G, Coppola A, et al. Indocyanine green clearance tests to assess liver transplantation outcomes: a systematic review. Int J Surg 2024;110:431–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Agopian VG, Markovic D, Klintmalm GB, et al. Multicenter validation of the liver graft assessment following transplantation (L-GrAFT) score for assessment of early allograft dysfunction. J Hepatol 2021;74:881–892. [DOI] [PubMed] [Google Scholar]
- 13.Bastos-Neves D, Salvalaggio PRO, Almeida MD. Risk factors, surgical complications and graft survival in liver transplant recipients with early allograft dysfunction. Hepatobiliary Pancreat Dis Int 2019;18:423–429. [DOI] [PubMed] [Google Scholar]
- 14.Feng S, Goodrich NP, Bragg-Gresham JL, et al. Characteristics associated with liver graft failure: the concept of a donor risk index. Am J Transplant 2006;6:783–790. [DOI] [PubMed] [Google Scholar]
- 15.Avolio AW, Franco A, Schlegel A, et al. Development and validation of a comprehensive model to estimate early allograft failure among patients requiring early liver retransplant. JAMA Surg 2020;155:e204095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kurian SM, Fouraschen SMG, Langfelder P, et al. Genomic profiles and predictors of early allograft dysfunction after human liver transplantation. Am J Transplant 2015;15:1605–1614. [DOI] [PubMed] [Google Scholar]
- 17.Cortes M, Pareja E, Garcia-Canaveras JC, et al. Metabolomics discloses donor liver biomarkers associated with early allograft dysfunction. J Hepatol 2014;61:564–574. [DOI] [PubMed] [Google Scholar]
- 18.Faitot F, Besch C, Battini S, et al. Impact of real-time metabolomics in liver transplantation: graft evaluation and donor-recipient matching. J Hepatol 2018;68:699–706. [DOI] [PubMed] [Google Scholar]
- 19.Zhang XY, Zhang C, Huang HT, et al. Primary nonfunction following liver transplantation: learning of graft metabolites and building a predictive model. Clin Transl Med 2021;11:e483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aebersold R, Mann M. Mass-spectrometric exploration of proteome structure and function. Nature 2016;537:347–355. [DOI] [PubMed] [Google Scholar]
- 21.Nakamura K, Kageyama S, Kaldas FM, et al. Hepatic CEACAM1 expression indicates donor liver quality and prevents early transplantation injury. J Clin Invest 2020;130:2689–2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xie H, Zhang L, Guo D, et al. Protein profiles of pretransplant grafts predict early allograft dysfunction after liver transplantation from donation after circulatory death. Transplantation 2020;104:79–89. [DOI] [PubMed] [Google Scholar]
- 23.Hirao H, Ito T, Kadono K, et al. Donor hepatic occult collagen deposition predisposes to peritransplant stress and impacts human liver transplantation. Hepatology 2021;74:2759–2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu S, Gui Y, Wang MS, et al. Serum integrative omics reveals the landscape of human diabetic kidney disease. Mol Metab 2021;54:101367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Verhoeven CJ, Farid WR, de Jonge J, et al. Biomarkers to assess graft quality during conventional and machine preservation in liver transplantation. J Hepatol 2014;61:672–684. [DOI] [PubMed] [Google Scholar]
- 26.Huang HT, Zhang XY, Zhang C, et al. Predicting dyslipidemia after liver transplantation: a significant role of recipient metabolic inflammation profile. World J Gastroenterol 2020;26:2374–2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ling Q, Huang H, Han Y, et al. The tacrolimus-induced glucose homeostasis imbalance in terms of the liver: from bench to bedside. Am J Transplant 2020;20:701–713. [DOI] [PubMed] [Google Scholar]
- 28.Mathew G, Agha R, Albrecht J, et al. STROCSS 2021: Strengthening the reporting of cohort, cross-sectional and case–control studies in surgery. Int J Surg 2021;96:106165. [DOI] [PubMed] [Google Scholar]
- 29.Olthoff KM, Kulik L, Samstein B, et al. Validation of a current definition of early allograft dysfunction in liver transplant recipients and analysis of risk factors. Liver Transpl 2010;16:943–949. [DOI] [PubMed] [Google Scholar]
- 30.Chen Z, Zhou B, Wang X, et al. Synergistic effects of mechanical stimulation and crimped topography to stimulate natural collagen development for tendon engineering. Acta Biomater 2022;145:297–315. [DOI] [PubMed] [Google Scholar]
- 31.Feng Q, Xia W, Dai G, et al. The aging features of thyrotoxicosis mice: malnutrition, immunosenescence and lipotoxicity. Front Immunol 2022;13:864929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tseng PY, Chen YT, Wang CH, et al. Prediction of the development of acute kidney injury following cardiac surgery by machine learning. Crit Care 2020;24:478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bader JM, Geyer PE, Muller JB, et al. Proteome profiling in cerebrospinal fluid reveals novel biomarkers of Alzheimer’s disease. Mol Syst Biol 2020;16:e9356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang K, Lu D, Liu Y, et al. Severity of early allograft dysfunction following donation after circulatory death liver transplantation: a multicentre study. Hepatobiliary Surg Nutr 2021;10:9–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tran D, Cooke S, Illingworth PJ, et al. Deep learning as a predictive tool for fetal heart pregnancy following time-lapse incubation and blastocyst transfer. Hum Reprod 2019;34:1011–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Coleman RA, Mashek DG. Mammalian triacylglycerol metabolism: synthesis, lipolysis, and signaling. Chem Rev 2011;111:6359–6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ecker J, Liebisch G. Application of stable isotopes to investigate the metabolism of fatty acids, glycerophospholipid and sphingolipid species. Prog Lipid Res 2014;54:14–31. [DOI] [PubMed] [Google Scholar]
- 38.Ju L, Han J, Zhang X, et al. Obesity-associated inflammation triggers an autophagy-lysosomal response in adipocytes and causes degradation of perilipin 1. Cell Death Dis 2019;10:121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jimenez-Castro MB, Casillas-Ramirez A, Mendes-Braz M, et al. Adiponectin and resistin protect steatotic livers undergoing transplantation. J Hepatol 2013;59:1208–1214. [DOI] [PubMed] [Google Scholar]
- 40.Yang L, Wang W, Wang X, et al. Creg in hepatocytes ameliorates liver ischemia/reperfusion injury in a TAK1-dependent manner in mice. Hepatology 2019;69:294–313. [DOI] [PubMed] [Google Scholar]
- 41.Zhang J, Hu W, Xing W, et al. The protective role of CD59 and pathogenic role of complement in hepatic ischemia and reperfusion injury. Am J Pathol 2011;179:2876–2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simcox J, Lamming DW. The central moTOR of metabolism. Dev Cell 2022;57:691–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao Y, Ye W, Wang YD, et al. HGF/c-Met: a key promoter in liver regeneration. Front Pharmacol 2022;13:808855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen G, Harwood JL, Lemieux MJ, et al. Acyl-CoA:diacylglycerol acyltransferase: properties, physiological roles, metabolic engineering and intentional control. Prog Lipid Res 2022;88:101181. [DOI] [PubMed] [Google Scholar]
- 45.Neschen S, Morino K, Hammond LE, et al. Prevention of hepatic steatosis and hepatic insulin resistance in mitochondrial acyl-CoA:glycerol-sn-3-phosphate acyltransferase 1 knockout mice. Cell Metab 2005;2:55–65. [DOI] [PubMed] [Google Scholar]
- 46.Hakim A, Moll M, Brancale J, et al. Genetic variation in the mitochondrial glycerol-3-phosphate acyltransferase is associated with liver injury. Hepatology 2021;74:3394–3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fujii H, Ikura Y, Arimoto J, et al. Expression of perilipin and adipophilin in nonalcoholic fatty liver disease; relevance to oxidative injury and hepatocyte ballooning. J Atheroscler Thromb 2009;16:893–901. [DOI] [PubMed] [Google Scholar]
- 48.Semova I, Biddinger SB. Triglycerides in nonalcoholic fatty liver disease: guilty until proven innocent. Trends Pharmacol Sci 2021;42:183–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu Z, Zhu H, Wang W, et al. Metabonomic profile of macrosteatotic allografts for orthotopic liver transplantation in patients with initial poor function: mechanistic investigation and prognostic prediction. Front Cell Dev Biol 2020;8:826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Phillips R, Ursell T, Wiggins P, et al. Emerging roles for lipids in shaping membrane-protein function. Nature 2009;459:379–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li Z, Agellon LB, Allen TM, et al. The ratio of phosphatidylcholine to phosphatidylethanolamine influences membrane integrity and steatohepatitis. Cell Metab 2006;3:321–331. [DOI] [PubMed] [Google Scholar]
- 52.Ling J, Chaba T, Zhu LF, et al. Hepatic ratio of phosphatidylcholine to phosphatidylethanolamine predicts survival after partial hepatectomy in mice. Hepatology 2012;55:1094–1102. [DOI] [PubMed] [Google Scholar]
- 53.Zhang Y, Ma KL, Ruan XZ, et al. Dysregulation of the low-density lipoprotein receptor pathway is involved in lipid disorder-mediated organ injury. Int J Biol Sci 2016;12:569–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu J, Chen GG, Lai PBS. Targeting hepatocyte growth factor/c-mesenchymal–epithelial transition factor axis in hepatocellular carcinoma: rationale and therapeutic strategies. Med Res Rev 2021;41:507–524. [DOI] [PubMed] [Google Scholar]
- 55.Kogure K, Zhang YQ, Maeshima A, et al. The role of activin and transforming growth factor-beta in the regulation of organ mass in the rat liver. Hepatology 2000;31:916–921. [DOI] [PubMed] [Google Scholar]
- 56.Sasturkar SV, David P, Sharma S, et al. Serial changes of cytokines and growth factors in peripheral circulation after right lobe donor hepatectomy. Liver Transpl 2016;22:344–351. [DOI] [PubMed] [Google Scholar]
- 57.Wang K, Li Y, Zhu T, et al. Overexpression of c-Met in bone marrow mesenchymal stem cells improves their effectiveness in homing and repair of acute liver failure. Stem Cell Res Ther 2017;8:162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Oe S, Hiros T, Fujii H, et al. Continuous intravenous infusion of deleted form of hepatocyte growth factor attenuates hepatic ischemia-reperfusion injury in rats. J Hepatol 2001;34:832–839. [DOI] [PubMed] [Google Scholar]
- 59.Chen G, Hu X, Huang Y, et al. Role of the immune system in liver transplantation and its implications for therapeutic interventions. MedComm 2023;4:e444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Robinson MW, Harmon C, O’Farrelly C. Liver immunology and its role in inflammation and homeostasis. Cell Mol Immunol 2016;13:267–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hirao H, Nakamura K, Kupiec-Weglinski JW. Liver ischaemia-reperfusion injury: a new understanding of the role of innate immunity. Nat Rev Gastroenterol Hepatol 2022;19:239–256. [DOI] [PubMed] [Google Scholar]
- 62.Cambier S, Gouwy M, Proost P. The chemokines CXCL8 and CXCL12: molecular and functional properties, role in disease and efforts towards pharmacological intervention. Cell Mol Immunol 2023;20:217–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Norén A, Oltean M, Friman S, et al. Liver graft proteomics reveals potential incipient mechanisms behind early renal dysfunction after liver transplantation. Int J Mol Sci 2022;23:11929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kahan R, Cray PL, Abraham N, et al. Sterile inflammation in liver transplantation. Front Med (Lausanne) 2023;10:1223224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Martins PN, Rizzari MD, Ghinolfi D, et al. Design, analysis, and pitfalls of clinical trials using ex situ liver machine perfusion: The International Liver Transplantation Society Consensus Guidelines. Transplantation 2021;105:796–815. [DOI] [PubMed] [Google Scholar]
- 66.Huang H, Chen R, Lin Y, et al. Decoding single-cell landscape and intercellular crosstalk in the transplanted liver. Transplantation 2023;107:890–902. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.