Abstract
Background:
Sleeve gastrectomy (SG) is known to alleviate non-alcoholic fatty liver disease (NAFLD) and restore liver function; however, its underlying mechanism remains unclear.
Materials and methods:
We investigated the effect of SG on the metabolic phenotype of diet-induced obese (DIO) mice. Postoperative stained liver images were analyzed to determine the hepatocyte proliferation phenotype. Single-cell RNA sequencing was used to characterize the regeneration signals of the liver after SG in DIO mice, and real-time quantitative reverse transcription PCR was performed to verify the above results. We employed Olink proteomics to capture serum element changes and investigated the role of Yes1 protein in liver regeneration and carcinogenesis through the Hippo–YAP pathway. DIO mice were treated with YAP inhibitor verteporfin after SG mice to clarify whether SG-induced liver regeneration is related to the YAP signaling pathway.
Results:
SG significantly reduced NAFLD-associated dysfunction in hepatocytes and replaced them with fully functional hepatocytes, which have a high regenerative capacity across the entire liver. SG also enhanced the hepatic regenerative capacity, as demonstrated by SG combined with hepatic lobectomy in healthy mice. Yes1 protein was identified as the signaling molecule most closely related to classical regeneration signals. Our study showed that SG-enhanced proliferation and improved metabolism did not depend on YAP signaling.
Conclusion:
SG can enhance hepatic regenerative capacity and improve liver metabolism. This study provides a better understanding of the mechanisms underlying SG-induced metabolic improvements.
Keywords: hepatocyte, liver regeneration, NAFLD, sleeve gastrectomy
Introduction
Highlights
First report of sleeve gastrectomy (SG)-induced liver regeneration.
Independent mechanism of SG-induced liver regeneration from the improvement of fatty liver and metabolism.
New mechanistic insights into the improvement of hepatic function by SG.
The enhanced liver regeneration effect induced by SG is independent of the Yes1 protein.
Over the past few decades, non-alcoholic fatty liver disease (NAFLD) has gradually become the most prevalent chronic liver disease worldwide, affecting nearly 30% of the adult population and imposing an increasingly severe global health burden1,2. NAFLD is defined as the presence of more than 5% hepatocytes with steatosis, associated with metabolic risk factors, and the absence of excessive alcohol consumption (≥30 g/day in men and ≥20 g/day in women) or other chronic liver diseases3,4. Non-alcoholic steatohepatitis (NASH), liver fibrosis, and hepatocellular carcinoma (HCC) may develop sequentially as NAFLD progresses. These conditions often result in significant impairment of liver function. Fortunately, appropriate interventions, particularly bariatric surgery, have been demonstrated to a certain degree. Consequently, NAFLD is no longer perceived solely as a public health burden but also as a critical window for intervention to preserve patients’ lives and improve their health status before irreversible damage occurs5,6.
Bariatric surgery, particularly sleeve gastrectomy (SG) and Roux-en-Y gastric bypass (RYGB) play a significant role in improving liver dysfunction in NAFLD7. Several studies have demonstrated that after bariatric surgery in patients with NAFLD, liver steatosis is reduced, as evidenced by a significant decrease in the number of balloon cells and related biochemical indicators in liver tissue sections6. Moreover, bariatric surgery effectively alleviated the insulin resistance associated with NAFLD, thereby improving impaired glucose tolerance and insulin sensitivity. Among the various bariatric surgery methods, SG has gained popularity owing to its relatively simple procedure and reliable surgical outcomes8. Previous studies have extensively investigated the mechanisms through which SG improves glucose tolerance. The prevailing theory suggests that these metabolic benefits can be attributed to postoperative changes in the gut microbiota and bile acid profiles, which increase the secretion of GLP-1 by intestinal L cells located in the terminal ileum, consequently restoring insulin sensitivity9–11. However, the precise mechanism by which SG improves hepatic steatosis remains unclear.
In our study, we observed that the histological distribution of well-functioning hepatocytes after SG in mice resembled the regions where liver regeneration occurred. While most solid organs possess a certain ability for self-repair and regeneration12, the liver has a distinct advantage over other solid organs. Existing research has demonstrated that the liver maintains a functional mass-to-body weight ratio of 100% to ensure homeostasis through regenerative repair mechanisms12. Whenever liver damage causes the relative mass and function to fall below 100%, liver regeneration is initiated to restore homeostasis. Self-surveillance and regeneration mechanisms appear relentless. It is more accurate to state that the liver cannot remain in a state where its function is below 100% and must strive to return to that level. If the regenerative process cannot be completed, such as when both the classical and alternative pathways are blocked, liver failure occurs with dire consequences13.
Here, we present a novel physiological phenomenon observed after SG that involves a significant enhancement in liver regeneration activity. During the transition from NAFLD to a healthy state, the involvement of newborn hepatocytes in the reversal of liver function may serve as an upstream factor for changes in metabolism-related target genes.
In this study, we aimed to investigate the effect of SG on the remission of hepatic dysfunction by reinforcing liver regeneration and to explore the possible origin of regenerated hepatic cell types. Additionally, we identified the key signaling pathways involved in the regeneration process and discovered a link between liver regeneration and metabolic improvement. Consequently, we determined that SG improves liver dysfunction by enhancing the regeneration program.
Materials and methods
Animals
Four-week male, C57Bl/6N mice were fed a high-fat diet (HFD, 60% kcal fat; RD12492; Research Diets, NJ, USA) in an SPF (specific pathogen-free) room and housed in a climate-controlled environment with 12-h light/dark cycles until they reached 20 weeks of age (16 wk HFD provided). Euthanasia was performed in a fasted state to minimize the confounding effects of varying food intake between mice and allow for accurate fasting blood sample collection. In the YAP inhibition experiment, starting on the 7th day after surgery, mice were intraperitoneally injected with 100 mg/kg verteporfin (HY-B0146, MCE) or an equivalent volume of corn oil for 3 weeks, after which liver samples were collected.
This study involved a total of three experimental groups and control groups of mice. The groups included:
Firstly, diet-induced obese (DIO) mice undergoing either SG or sham surgery. In the SG group, 80% of the stomach was removed, while in the Sham group, mice underwent a similar laparotomy, peri-gastric dissection, and short gastric artery isolation without stomach resection. Both groups consisted of DIO mice, totaling 30, randomly divided into two groups for SG and sham surgery. The SG group served as the experimental group, while the Sham group served as the control group.
Additionally, there were combined surgery experiments involving C57Bl/6N mice. The purpose of this experiment was to determine whether the enhanced liver regeneration effect caused by SG was a result of NAFLD alleviation, thereby eliminating the unrelated variable of ‘NAFLD relief.’ The mice used in this part were 20-week-old C57Bl/6N mice, matching the age of the mice receiving surgical intervention in Experiment A. However, these mice were fed a chow diet (CD). The experiment included 12 mice, randomly divided into two groups: SG+L group undergoing SG and liver lobectomy (experimental group) and Sham+L group undergoing sham surgery and liver lobectomy (control group). The SG+L group underwent the same SG procedure as the SG group in Experiment A, and additionally, a left lateral liver lobectomy. The Sham+L group underwent the same sham procedure as the Sham group in Experiment A, and additionally, a left lateral liver lobectomy.
Lastly, there were combined surgery experiments involving DIO mice. The purpose of this part was to determine the appropriate observation window for assessing the liver regeneration status of mice after combined surgery. The mice in this experiment were the same as the DIO mice in Experiment A, all 20 weeks old and subjected to 16 weeks of high-fat diet feeding. The experiment included 12 mice, randomly divided into two groups: SG+L group and Sham+L group, with the former as the experimental group and the latter as the control group. The procedures were the same as in Experiment B, with the distinction that this group of mice was DIO mice, whereas the mice in Experiment B were on a chow diet.
Surgery
Sham or SG procedure
SG was performed through a 1.5 cm laparotomy under isoflurane anesthesia. The stomach was dissected free from its attachments and the vessels between the spleen and stomach (short gastric vessels) were ligated and cut off. The gap between the gastric antrum and the pancreas was completely dissected. The blood vessels between the pancreas and distal gastric antrum were ligated and excised. The dorsal pancreatic artery was preserved to ensure a sufficient blood supply to the pancreas. Then, the greater curvature of the stomach has been fully exposed to ensure subsequent efficient gastric body resection, and a tubular stomach was created by removing 80% of the stomach. The gastric sleeve is performed by an inner running suture along the incision site using 8-0 Prolene (W2777; Johnson & Johnson Services, Inc., USA). The sham procedure involved a similar dissection of the stomach and pancreas, followed by the manual application of pressure with blunt forceps along the same cutting line as in the SG procedure. For the first day after surgery, the mice were fed a liquid diet, and HFD was restarted on the morning of postoperative day 2. Mice were sacrificed 6 weeks post-surgery after overnight fasting (20:00 to 08:00).
Lobectomy
Lobectomy was performed under isoflurane anesthesia. To perform 1/3 partial hepatectomy, the left lateral lobe was resected via one-step ligature using a 5-0 polyester suture tie. Thereafter, Sham or SG procedures were performed.
Western blot
Nuclear and cytoplasmic proteins were extracted using a Nuclear and Cytoplasmic Protein Extraction Kit (Beyotime, Catalog No. P0028) according to the manufacturer’s instructions. The collected protein samples were quantified using a Pierce BCA Protein Assay Kit (Catalog No. 23225, Thermo Fisher Scientific). Aliquots of liver protein were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and then transferred onto Immobilon-P Transfer Membranes (PVDF membrane, Millipore). After blocking with Intercept (TBS) Blocking Buffer (Part No. 927-60001, LI-COR), the membrane was incubated with primary antibodies overnight at 4°C. Subsequently, the membranes were incubated with horseradish peroxidase-conjugated secondary antibodies for 1 h. Protein bands were visualized using electrogenerated chemiluminescence. Primary antibodies included Stat3 (Cell Signaling Technology Cat# 12640, RRID: AB_2629499), Phospho-Stat3 (Cell Signaling Technology Cat# 9138, RRID: AB_331262), YAP (Cell Signaling Technology Cat# 14074, RRID: AB_2650491), Phospho-YAP (Cell Signaling Technology Cat# 13008, RRID: AB_2650553), Lats1 (Cell Signaling Technology Cat# 3477, RRID: AB_2133513), Phospho-lats1 (Cell Signaling Technology Cat# 9157, RRID: AB_2133515), Mst1 (Cell Signaling Technology Cat# 3682, RRID: AB_2144632), Phospho-mst1 (Cell Signaling Technology Cat# 49332, RRID: AB_2799355), Taz (Cell Signaling Technology Cat# 83669, RRID: AB_2800026) were purchased from Cell Signaling Technology (Danvers, MA, USA). Primary antibodies, including proliferating cell nuclear antigen (PCNA) (Proteintech Cat# 10205-2-AP, RRID: AB_2160330), Laminb1 (Proteintech Cat# 12987-1-AP, RRID: AB_2136290), and β-actin (Proteintech Cat# 66009-1-Ig, RRID: AB_2687938) were purchased from Proteintech (Philadelphia, PA, USA).
Immunofluorescence staining
Tissues were fixed in formalin for 24 h, embedded in paraffin, and sectioned. De-paraffinized tissue sections were serially incubated in xylene; 100%, 95%, 90%, 80%, and 70% ethanol; and distilled deionized water. Heat-mediated antigen retrieval was performed using Tris-ethylenediaminetetraacetic acid buffer (pH 9.0). Sections were blocked with 30% goat serum for 1 h at room temperature and incubated with primary antibodies overnight at 4°C. Primary antibodies against Ki67 (Abcam, Cat# ab15580, RRID: AB_443209), Phospho-stat3 (Abcam, Cat# ab32143, RRID: AB_2286742) were purchased from Abcam (Cambridge, UK). Primary antibodies including Albumin (Proteintech Cat# 16475-1-AP, RRID: AB_2242567), Cd326 (Proteintech Cat# 21050-1-AP, RRID: AB_10693684), α-SMA (Proteintech Cat# 14395-1-AP, RRID: AB_2223009) were purchased from Protein-Tech (Philadelphia, PA, USA). The primary antibody YAP (Cell Signaling Technology Cat# 14074, RRID: AB_2650491) was purchased from Cell Signaling Technology (Danvers, MA, USA). The sections were then washed three times with washing buffer and incubated with the secondary antibody. Images were acquired using an Olympus IX73 Inverted Microscope and analyzed using the ImageJ software.
Immunohistochemical staining
The process of dewaxing and antigen retrieval of paraffin sections was the same as that used for immunofluorescence staining. The slices were placed in 3% hydrogen peroxide solution, incubated at room temperature in the dark for 25 min, placed in PBS (pH 7.4) and shaken, and washed three times on a decolorizing shaker for 5 min each. Sections were blocked with 30% goat serum for 1 h at room temperature and incubated with primary antibodies overnight at 4°C. Primary antibody PCNA (Proteintech Cat# 10205-2-AP, RRID: AB_2160330) was purchased from Protein-tech (Philadelphia, PA, USA). The slides were placed in PBS (pH 7.4) and shaken and washed thrice on a decolorizing shaker for 5 min each. The secondary antibody [horseradish peroxidase (HRP)-labeled goat anti-rabbit] was added dropwise to the circle to cover the tissue and incubated at room temperature for 1 h. The slides were placed in PBS (pH 7.4) and shaken and washed thrice on a decolorizing shaker for 5 min each. Freshly prepared DAB (3,3′-diaminobenzidine) color development solution was added dropwise to the circle, and the color development time was controlled under a microscope. The positive color was brownish yellow, and the sections were rinsed with tap water to stop color development. The sections were counterstained with hematoxylin for 3 min, washed with tap water, differentiated in hematoxylin differentiation solution for a few seconds, rinsed with tap water, stained blue with hematoxylin solution, and then rinsed with running water. The slices were placed in 75% alcohol for 5 min, 85% alcohol for 5 min, absolute ethanol I for 5 min, absolute ethanol II for 5 min, and xylene I for 5 min for dehydration and transparency. Slices were removed from xylene, dried slightly, and sealed with mounting glue. Images were acquired using an Olympus IX73 Inverted Microscope and analyzed using the ImageJ software.
qRT-PCR analysis
Total RNA was extracted from the liver tissues using an RNA extraction kit (Fastagen RNAFast200, Shanghai, China) according to the manufacturer’s instructions. Complementary DNA (cDNA) was synthesized by reverse transcription using a PrimeScript RT Reagent Kit (Catalog No. RR074A). Real-time PCR was performed using a LightCycler 480 II System with FastStart Essential DNA Green Master Mix (SYBR Green, Roche, Catalog No. 06924204001). β-Actin was used to normalize the messenger RNA (mRNA) content, and the relative expression of mRNA was calculated using the 2−ΔΔCt method. The primer sequences were as follows:
| Gene | Sequence |
|---|---|
| Socs3 | |
| Forward | GGACCAAGAACCTACGCATCCA |
| Reverse | CACCAGCTTGAGTACACAGTCG |
| Myc | |
| Forward | TCGCTGCTGTCCTCCGAGTCC |
| Reverse | GGTTTGCCTCTTCTCCACAGAC |
| Sox9 | |
| Forward | CACACGTCAAGCGACCCATGAA |
| Reverse | TCTTCTCGCTCTCGTTCAGCAG |
| Egfr | |
| Forward | GGACTGTGTCTCCTGCCAGAAT |
| Reverse | GGCAGACATTCTGGATGGCACT |
| Fn14 | |
| Forward | GACCTCGACAAGTGCATGGACT |
| Reverse | CGCCAAAACCAGGACCAGACTA |
| Ifnγ | |
| Forward | CAGCAACAGCAAGGCGAAAAAGG |
| Reverse | TTTCCGCTTCCTGAGGCTGGAT |
| Flt1 | |
| Forward | TGGATGAGCAGTGTGAACGGCT |
| Reverse | GCCAAATGCAGAGGCTTGAACG |
| Stat3 | |
| Forward | AGGAGTCTAACAACGGCAGCCT |
| Reverse | GTGGTACACCTCAGTCTCGAAG |
| C/ebpβ | |
| Forward | CAACCTGGAGACGCAGCACAAG |
| Reverse | GCTTGAACAAGTTCCGCAGGGT |
| Atgl | |
| Forward | GGAACCAAAGGACCTGATGACC |
| Reverse | ACATCAGGCAGCCACTCCAACA |
| Nr1h3 | |
| Forward | ATCGCCTTGCTGAAGACCTCTG |
| Reverse | CTGCTTTGGCAAAGTCTTCCCG |
| Fabp1 | |
| Forward | AGGAGTGCGAACTGGAGACCAT |
| Reverse | GTCTCCATTGAGTTCAGTCACGG |
| Fasn | |
| Forward | CACAGTGCTCAAAGGACATGCC |
| Reverse | CACCAGGTGTAGTGCCTTCCTC |
| Scd1 | |
| Forward | GCAAGCTCTACACCTGCCTCTT |
| Reverse | CGTGCCTTGTAAGTTCTGTGGC |
| Ucp2 | |
| Forward | TAAAGGTCCGCTTCCAGGCTCA |
| Reverse | ACGGGCAACATTGGGAGAAGTC |
| Cd36 | |
| Forward | GGACATTGAGATTCTTTTCCTCTG |
| Reverse | GCAAAGGCATTGGCTGGAAGAAC |
| HMGCS2 | |
| Forward | TGCTATGCAGCCTACCGCAAGA |
| Reverse | GCCAGGGATTTCTGGACCATCT |
| Pdk4 | |
| Forward | GTCGAGCATCAAGAAAACCGTCC |
| Reverse | GCGGTCAGTAATCCTCAGAGGA |
| Srebf1 | |
| Forward | CGACTACATCCGCTTCTTGCAG |
| Reverse | CCTCCATAGACACATCTGTGCC |
| Thrsp | |
| Forward | AAACCAGCGAGGCTGAGAACGA |
| Reverse | CAGGTGGGTAAGGATGTGATGG |
| Dgat2 | |
| Forward | CTGTGCTCTACTTCACCTGGCT |
| Reverse | CTGGATGGGAAAGTAGTCTCGG |
| Mt2 | |
| Forward | CGTGGGCTGTGCGAAGTGCTC |
| Reverse | AAAGGCTAGGCTTCTACATGGTC |
| Saa2 | |
| Forward | GGAGTCTGGGCTGCTGAGAAAA |
| Reverse | TGTCTGTTGGCTTCCTGGTCAG |
| Hmgcs1 | |
| Forward | GGAAATGCCAGACCTACAGGTG |
| Reverse | TACTCGGAGAGCATGTCAGGCT |
| Fabp2 | |
| Forward | TCCCTACAGTCTAGCAGACGGA |
| Reverse | CCAGAAACCTCTCGGACAGCAA |
| Atp5g1 | |
| Forward | CATCGACACAGCAGCCAAGTTC |
| Reverse | CCAGAATGGCATAGGAGAAGAGC |
| Mlxipl | |
| Forward | GAGTGCTTGAGCCTGGCTTACA |
| Reverse | GCTCTCCAGATGGCGTTGTTCA |
| Gcgr | |
| Forward | CAATGCCACCACAACCTAAGCC |
| Reverse | GGCAGGAAATGTTGGCAGTGGT |
| Xbp1 | |
| Forward | TGGACTCTGACACTGTTGCCTC |
| Reverse | TAGACCTCTGGGAGTTCCTCCA |
| Man2a1 | |
| Forward | GCAAAGTTCACAAGATTCTCTGCC |
| Reverse | GATGGACACCACTGAATGCCGT |
| Jun | |
| Forward | CAGTCCAGCAATGGGCACATCA |
| Reverse | GGAAGCGTGTTCTGGCTATGCA |
| Sdhb | |
| Forward | TGCGGACCTATGGTGTTGGATG |
| Reverse | CCAGAGTATTGCCTCCGTTGATG |
| Cox6b1 | |
| Forward | GAACTGTTGGCAGAACTACCTGG |
| Reverse | ATGACACGGGACAGAGGGACTT |
| Ndufa13 | |
| Forward | TGCTGATTGAGGACTTGGAGGC |
| Reverse | GTGGTATGGAACACAGACTCGC |
| Ndufs5 | |
| Forward | CGGGCGAAAAAGGAGTGCAAGA |
| Reverse | GCCCTCTTTCATTAGCTTCTCCC |
| Acaa1b | |
| Forward | GGAGAATGTGGCTGAGCGGTTT |
| Reverse | AGGACAGTGGTTGTCACAGGCA |
| Srebp1 | |
| Forward | CGACTACATCCGCTTCTTGCAG |
| Reverse | CCTCCATAGACACATCTGTGCC |
| Orm1 | |
| Forward | CCTTCATGCTTGCCTTTGACCTC |
| Reverse | CGTGTGTGACAGCCTTCTGGAA |
BrdU labeling and staining
Mouse hepatic cells were labeled using a BD Pharmingen BrdU In Situ Detection Kit II (BD Pharmingen, Catalog No. 551321, BrdU, BD Pharmingen, Catalog No. 550891) following the Instruction Manual in vivo Labeling Intraperitoneal Method (1 ml/1 mg BrdU i.p., 48 h before sacrifice) to estimate hepatic cell proliferation. Subsequently, immunostaining for BrdU was performed according to the instruction manual.
H&E staining
Dewaxing was performed as follows: Xylene I for 20 min; Xylene II for 20 min; 100% ethanol I for 5 min; 100% ethanol II for 5 min; and 75% ethanol for 5 min. Rinse with tap water. The sections were stained with hematoxylin solution for 3–5 min and rinsed with tap water. The sections were treated with a Hematoxylin Differentiation solution and rinsed with tap water. The sections were treated with hematoxylin and Scott tap blue and rinsed with tap water – 85% ethanol for 5 min and 95% ethanol for 5 min. Finally, the sections were stained with eosin for 5 min. The samples were dehydrated as follows:100% ethanol I for 5 min, 100% ethanol II for 5 min, 100% ethanol III for 5 min, Xylene I for 5 min, and Xylene II for 5 min. Finally, the seal was sealed with a neutral gum. Images were acquired with an IX73 inverted microscope (Olympus).
Oil Red O staining
The frozen slices were reheated, dried, fixed in a fixative solution for 15 min, washed with tap water, and dried. Sections were stained with Oil Red O solution for 8–10 min in the dark and covered with a lid during dyeing. The slices were removed, stayed for 3 s, and then immersed in two cups of 60% isopropanol for differentiation for 3 s and 5 s, respectively. Slices were immersed in two cups of pure water for 10 s each. The slices were removed and immersed in hematoxylin for 3–5 min after 3 s and then rinsed in three cups of pure water for 5 s, 10 s, and 30 s in turn. The cells were treated with the differentiation solution (60% alcohol as a solvent) for 2–8 s, two cups of distilled water for 10 s each, and Scott Tap Bluing for 1 s. The slices were lightly dipped in two cylinders of tap water for 5 s and 10 s in turn, and the staining effect was examined using a microscope. The slices were then sealed with glycerin-gelatin. Images were acquired with an IX73 inverted microscope (Olympus).
Sirius red staining
Dewaxing was performed as follows: Xylene I for 20 min; Xylene II for 20 min; 100% ethanol I for 5 min; 100% ethanol II for 5 min; and 75% ethanol for 5 min. Rinse with tap water. The sections were stained with Sirius Red solution for 8 min and then quickly dehydrated with two or three cups of anhydrous ethanol. Xylene for 5 min and sealed with neutral gum. Images were acquired with an IX73 inverted microscope (Olympus).
BODIPY staining
Frozen sections were rewarmed at room temperature, water was controlled, and immunohistochemical strokes were circled. BODIPY was diluted in the tissues and incubated at room temperature for 20 min. The slides were placed in PBS (pH 7.4) and shaken and washed thrice on a decolorizing shaker for 5 min each. DAPI (4′,6-diamidino-2-phenylindole) staining solution was added dropwise and incubated at room temperature in the dark for 10 min. The slides were placed in PBS (pH 7.4) and shaken and washed thrice on a decolorizing shaker for 5 min each. The slides were mounted using an anti-fade mounting medium. Images were acquired with an IX73 inverted microscope (Olympus).
Single-cell RNA-seq
Single-cell dissociation
Single-cell RNA-seq experiments were performed by personnel in the laboratory of NovelBio Co. Ltd. The tissues were surgically removed and stored in MACS Tissue Storage Solution (Miltenyi Biotec) until processing. Tissue samples were processed as described below. Briefly, the samples were washed with phosphate-buffered saline (PBS), minced into small pieces (~1 mm3) on ice, and enzymatically digested with collagenase I (Worthington) and DNase I (Worthington) for 45 min at 37°C with agitation. After digestion, the samples were sieved through a 70 μm cell strainer and centrifuged at 300g for 5 min. After the supernatant was removed, the pelleted cells were suspended in red blood cell lysis buffer (Miltenyi Biotec) to lyse their red blood cells. After washing with PBS containing 0.04% BSA, the cell pellets were resuspended in PBS containing 0.04% BSA and re-filtered through a 35 μm cell strainer. Dissociated single cells were stained with AO/PI for viability assessment using a Countstar Fluorescence Cell Analyzer. The single-cell suspension was further enriched with a MACS dead cell removal kit (Miltenyi Biotec).
Single-cell sequencing
The scRNA-Seq libraries were generated using the 10X Genomics Chromium Controller Instrument and Chromium Single Cell 3′ V3 Reagent Kits (10X Genomics, Pleasanton, CA). Briefly, cells were concentrated to 1000 cells/μl, and ~8000 cells were loaded into each channel to generate single-cell Gel Bead-In-Emulsions (GEMs), which resulted in the expected mRNA barcoding of 5000 single cells for each sample. After RT, the GEMs were broken, and barcoded cDNA was purified and amplified. The amplified barcoded cDNA was fragmented, A-tailed, ligated with adaptors, and amplified by PCR. The final libraries were quantified using a Qubit High-Sensitivity DNA assay (Thermo Fisher Scientific), and the size distribution of the libraries was determined using a High-Sensitivity DNA chip on a Bioanalyzer 2200 (Agilent). All libraries were sequenced using an Illumina sequencer (Illumina, San Diego, CA, USA) on a 150 bp paired-end run.
Single-cell RNA statistical analysis
GO analysis: Gene ontology (GO) analysis was performed to elucidate the biological implications of unique genes in the significant or representative profiles of the differentially expressed genes in experiment14. GO annotations were downloaded from NCBI (http://www.ncbi.nlm.nih.gov/), UniProt (http://www.uniprot.org/), and Gene Ontology (http://www.geneontology.org/). Fisher’s exact test was applied to identify significant GO categories, and FDR (false discovery rate) was used to correct P values.
KEGG pathway analysis: Pathway analysis was used to determine the significant pathways of the differential genes according to the KEGG database. Fisher’s exact test was used to select the significant pathway, and the threshold of significance was defined by the P-value and FDR15.
QuSAGE analysis: To characterize the relative activation of a given gene set, the R package QuSAGE was used as described for gene set enrichment analysis to achieve enrichment status and to enrich the significance of each gene set16.
Spatial transcriptomic analysis
Tissue section preparation
The tissues were surgically removed and embedded in an optimal cutting temperature (OCT) compound (SAKURA). The tissues containing the OCT compound were immediately frozen on dry ice and stored at −80°C until cryosectioning. Cryosectioning was performed in a cryostat (Leica, CM1950) to cryosection the OCT-embedded tissue blocks into appropriately sized sections for Visium Spatial slides while keeping the samples frozen. Tissue sections were 10 μm thick. Tissue sections were placed within the frames of Capture Areas on Visium Spatial slides (10X Genomics).
Fixation, staining, and imaging
Tissue section slides were incubated for 1 min at 37°C and then fixed in methanol at −20°C for 30 min. For staining, the slides were incubated in hematoxylin for 7 min and in Bluing Buffer for 2 min. Eosin was then added to the slides and incubated for 1 min. After each staining step, slides were washed with DNase-free and RNase-free water. Stained tissue sections were imaged using a microscope (Pannoramic MIDI, 3DHISTECH).
Tissue pre-permeabilization
Pre-permeabilization was performed to optimize the permeabilization time. Visium Spatial Tissue Optimization Slides & Reagent Kits (10X Genomics) were used for pre-permeabilization. The tissues were permeabilized with Permeabilization Enzyme for varying amounts of time, and then Fluorescent RT Master Mix was added to the tissue sections. For tissue removal, tissue sections were incubated in the Tissue Removal Mix for 60 min at 56°C. The optimal permeabilization time was selected using a fluorescence microscope (Pannoramic MIDI, 3DHISTECH).
Tissue permeabilization and spatial transcriptomic sequencing
Tissue permeabilization and spatial transcriptomic sequencing were performed using Visium Spatial Gene Expression Slides and Reagent Kits. The stained slides were incubated in RT Master Mix for 45 min at 53°C to preserve transcription after permeabilization for an appropriate time. Next, the Second Strand Mix was added to the tissue sections on the slide and incubated for 15 min at 65°C to initiate the second strand synthesis. After transferring the cDNA from the slides, barcoded cDNA was purified and amplified. The amplified barcoded cDNA was fragmented, A-tailed, ligated with adaptors, and amplified by PCR. The final libraries were quantified using a Qubit High-Sensitivity DNA assay (Thermo Fisher Scientific), and the size distribution of the libraries was determined using a High-Sensitivity DNA chip on a Bioanalyzer 2200 (Agilent). All libraries were sequenced using an Illumina sequencer (Illumina, San Diego, CA, USA) on a 150 bp paired-end run.
Spatial transcriptomics statistical analysis
We applied fastp with default parameter filtering of the adaptor sequence and removed low-quality reads to obtain clean data. Feature-barcode matrices were then obtained by aligning reads to the human genome (GRCh38) using SpaceRanger v1.2.2. To minimize the sample batch, we applied down-sample analysis among samples sequenced according to the mapped barcoded reads per spot of each sample, and finally achieved the aggregated matrix. The Seurat package (version3.2, https://satijalab.org/seurat/) was used for spot normalization and regression analyses. Principal component analysis (PCA) was constructed based on scaled data with all highly variable genes, and the top 10 principal components were used for tSNE (t-distributed stochastic neighbor embedding) construction. Utilizing the graph-based cluster method, we acquired the unsupervised cell cluster result based on the top 10 principal components of the PCA and calculated the marker genes using the FindAllMarkers function with the Wilcoxon rank sum test algorithm under the following criteria: (1) log FC >0.25; (2) P value <0.05; (3) min.pct >0.1. Spatial feature expression plots were generated using the SpatialFeaturePlot function in Seurat (version 3.1.3) and the STUtility R package (version 1.0.0).
Go analysis
Gene ontology (GO) analysis was performed to elucidate the biological implications of marker genes and differentially expressed genes. GO annotations were downloaded from NCBI (http://www.ncbi.nlm.nih.gov/), UniProt (http://www.uniprot.org/), and Gene Ontology (http://www.geneontology.org/). Fisher’s exact test was applied to identify significant GO categories, and FDR was used to correct P values.
Pathway analysis
Pathway analysis was used to identify significant pathways of the marker genes and differentially expressed genes according to the KEGG database. Fisher’s exact test was used to select the significant pathways, and the threshold of significance was defined by the P-value and FDR.
Olink proteome
Olink proteome analysis was performed using Gene Denovo (Gene Denovo, China). Mouse serum samples were preserved in liquid nitrogen for link proteomic detection using the Gene Denovo. The grouping of serum samples was single-blinded to the Gene Denovo. After the test, we performed regrouping and personalized analysis on the analysis website provided by Gene Denovo (https://www.omicsmart.com/home.html#/).
Glucose tolerance test and insulin tolerance test
After overnight fasting, intraperitoneal glucose tolerance testing (IPGTT) and insulin tolerance testing (ITT) were performed at postoperative weeks 4 and 5. During the IPGTT, the mice received 1 mg/g of intraperitoneal glucose. Serum glucose levels were measured in the tail vein at 15, 30, 60, and 120 min. ITT was performed by intraperitoneal insulin injection (0.5 IU/kg; Tonghua Dongbao Pharmacy, Gansu, China). Serum glucose levels were measured at 15, 30, 60, and 120 min. Baseline glucose levels were measured before medication administration.
Insulin ELISA measurement
An Ultrasensitive Rodent Insulin ELISA kit (Mercodia, Uppsala, Sweden) was used to measure mouse serum insulin levels according to the manufacturer’s instructions.
Primary hepatocytes isolation
Isolation of primary hepatocytes from postoperative mice liver. In brief, primary liver cells were isolated by perfusion with a 2% balanced salt solution (25 mM HEPES, 121 mM NaCl, 4.7 mM KCl, 1.2 mM MgSO4, 5 mM NaHCO3, 2 mM CaCl2, 10 mM glucose, pH 7.4). Collagenase IV (Gibco, USA) was administered through the portal vein after mouse anesthesia. Cell debris was removed by gradient centrifugation using Dulbecco’s Modified Eagle Medium (DMEM) containing 50% Percoll (Sigma, USA).
Serum biochemical analysis
Biochemical analysis and detection of mouse serum samples were performed by Beijing Lab-test Biotechnology Co., Ltd., using the iChem-340 fully automated biochemical analyzer for serum sample composition analysis.
Liver ultrasound scan
For both SG and Sham groups of mice, within the 3 days preceding euthanasia, liver ultrasound examinations were conducted in a gas-anesthetized state using the Vevo 3100 scanner (FUJIFILM VisualSonics, Inc.). The parameters were set as follows: transmit frequency 40 MHz, acquisition frame rate 65, and acquisition gain 35.0 dB. The probe was positioned with a 30° upward tilt to the abdominal plane beneath the right costal margin of the mouse to acquire ultrasound image data. The images were exported using the Vevo Lab software.
Statistical analysis
Data are presented as the mean±standard error. Body weight curves were analyzed using RM ANOVA with Tukey’s post-hoc test. The ITT and IPGTT results were analyzed using two-tailed Student’s t-test. Other data were analyzed using two-tailed Welch’s t-test. Data were analyzed using Prism software (Graph Pad, San Diego, CA, USA), with the critical value set at P<0.05.
ARRIVE guidelines statement
The work has been reported in accordance with the ARRIVE guidelines17 (Supplemental Digital Content 1, http://links.lww.com/JS9/C347).
Results
SG generates weight loss and insulin resistance remission in DIO mice
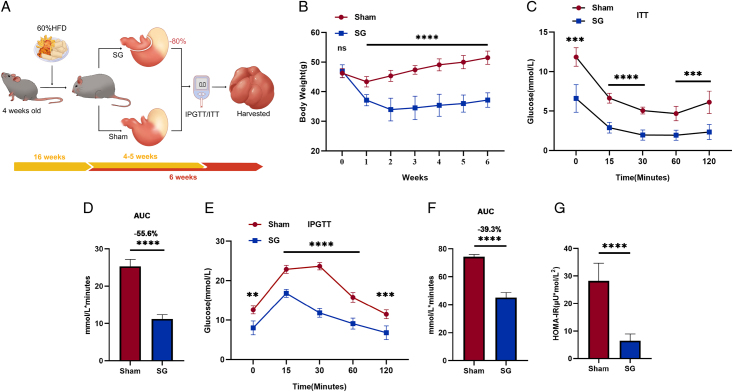
We conducted SG and sham procedures on DIO mice as described (Fig. 1A). Following surgical intervention, mice in the SG group exhibited a rapid and significant decrease in body weight, which was consistently lower than that of mice in the sham operation group throughout the observation period (Fig. 1B). Furthermore, the results of the insulin tolerance test (ITT) and the IPGTT, conducted during the fourth and fifth weeks after surgery, respectively, demonstrated that SG mice exhibited significantly improved glucose tolerance and increased insulin sensitivity (Fig. 1C–F). Additionally, the significant decrease in the homeostatic model assessment of insulin resistance (HOMA-IR) in SG mice further supports the metabolic improvements induced by the SG procedure (Fig. 1G).
Figure 1.
SG generates weight loss and insulin resistance remission in DIO mice. (A) Flowchart about surgical interventions and postoperative assessments. (B) SG mice lose weight and maintain this weight loss relative to the Sham group. ITT test (C) and IPGTT test (E) were performed in the fourth and fifth week after surgery, respectively. (D, F) Corresponding area under the blood glucose curve was reduced in SG compared to sham-operated mice. Mice were sacrificed in a fasted state 6 weeks after surgery. Serum and liver samples were collected. (G) The HOMA-IR value was calculated by detecting the insulin and blood glucose values in the serum. (Data are shown as mean±SE. *=P<0.05, **=P<0.01, ***=P<0.001, and ****=P<0.0001. For panel B, n=12 Sham, 8 SG. For panel C, n=5 Sham and SG. For panels E and G, n=6 Sham and SG). DIO, diet-induced obese; IPGTT, intraperitoneal glucose tolerance testing; SG, Sleeve gastrectomy; ITT, insulin tolerance testing.
SG significantly alleviates liver dysfunction in DIO mice
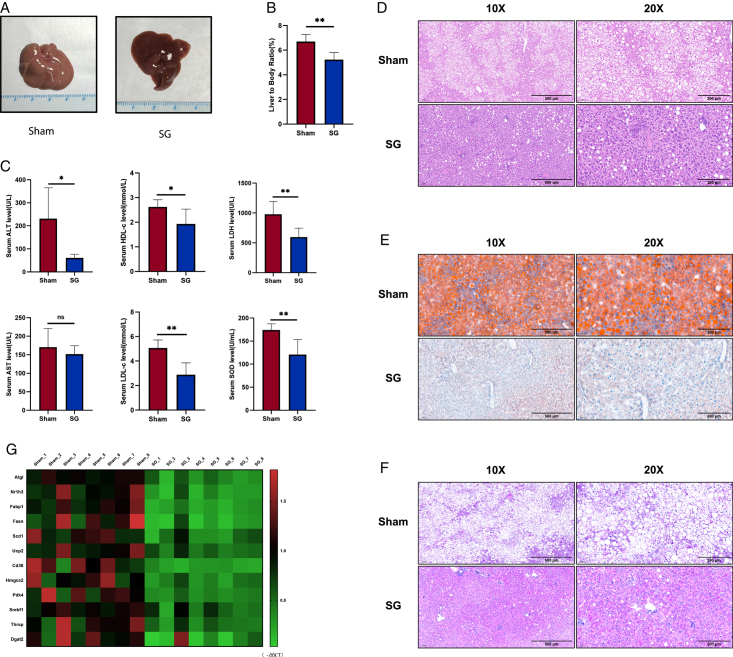
A gross examination of the liver specimens from the SG group revealed obvious reversal and improvement in fatty changes. In contrast, livers from the sham group displayed a distinct pale-yellow texture, whereas the livers of SG mice appeared more compact and smaller (Fig. 2A). Additionally, we observed a lower liver-to-body ratio in SG mice (Fig. 2B), which is an important phenotype that indicates improvement in liver function18. Similarly, serological indicators reflected enhanced liver function after SG. We measured the levels of several biochemical markers used to assess liver function in mouse serum and observed significant reductions in serum levels of alanine aminotransferase (ALT), lactate dehydrogenase (LDH), and superoxide dismutase (SOD) after SG. Moreover, the significant postoperative decrease in HDL-c and LDL-c levels indicated that SG induced systemic improvement in lipid-related metabolism, which was consistent with the amelioration of hepatic steatosis (Fig. 2C).
Figure 2.
SG significantly alleviates liver dysfunction in DIO mice. (A) Liver gross view of SG and Sham mice. (B) Liver-to-body ratio of SG and Sham mice. (C) Serum levels of major metabolic enzymes and apolipoproteins reflecting liver function changed after SG. (D) Histological examination using H&E staining revealed fat degeneration below 33%, no evidence of lobular inflammation, and NAS scores of 0–1. In the Sham group, mice displayed strong liver ultrasound signals and larger liver volume. H&E staining indicated fat degeneration exceeding 66%, with no Mallory bodies observed in either group. The Sham group received NAS scores of 4–5. (E) Liver section oil red O staining. (F) Liver section PAS staining. (G) The changes in the expression level of metabolism-related genes were shown by qRT-PCR results in the form of a heatmap. (Data are shown as mean±SE. ns=P>0.05, *=P<0.05, and **=P<0.01. For panels B and C, n=6 Sham and SG). DIO, diet-induced obese; SG, sleeve gastrectomy.
Furthermore, we evaluated the ability of SG to improve liver steatosis histologically. Based on the H&E stain and balloon cell counting results, a grading system was applied to assess hepatic steatosis in the livers of mice from the SG and Sham groups19. Mice in the SG group were classified between grade 0 and 1, while those in the Sham group were classified as grade 3. If using the NAS (NAFLD Activity Score) system20, the SG group aligns with NAS scores of 0–1, while the Sham group should have NAS scores in the range of 4–5. It is evident that SG surgery significantly alleviates the NAFLD condition in DIO mice. Oil Red O staining of frozen sections demonstrated a marked decrease in lipid accumulation (Fig. 2D, E and Supplementary Fig. 1, Supplemental Digital Content 2, http://links.lww.com/JS9/C348). PAS (periodic acid-Schiff) staining revealed a significant enhancement in the liver glycogen synthesis capacity after SG, consistent with the improvement in glucose tolerance observed in the aforementioned tolerance tests (Fig. 2F). Liver ultrasound images of mice showed that mice in the SG group exhibited a significantly decreased liver ultrasound signal compared to those in the Sham group. This suggests a noticeable alleviation of hepatic steatosis in the SG group. Additionally, the liver volume was significantly reduced in the SG group (Supplementary Fig. 3, Supplemental Digital Content 2, http://links.lww.com/JS9/C348). In addition, we compared the expression of several crucial metabolism-related genes in the mouse liver after SG and found that SG significantly improved the metabolic function of DIO mouse liver (Fig. 2G).
The liver regeneration signal was significantly enhanced after SG
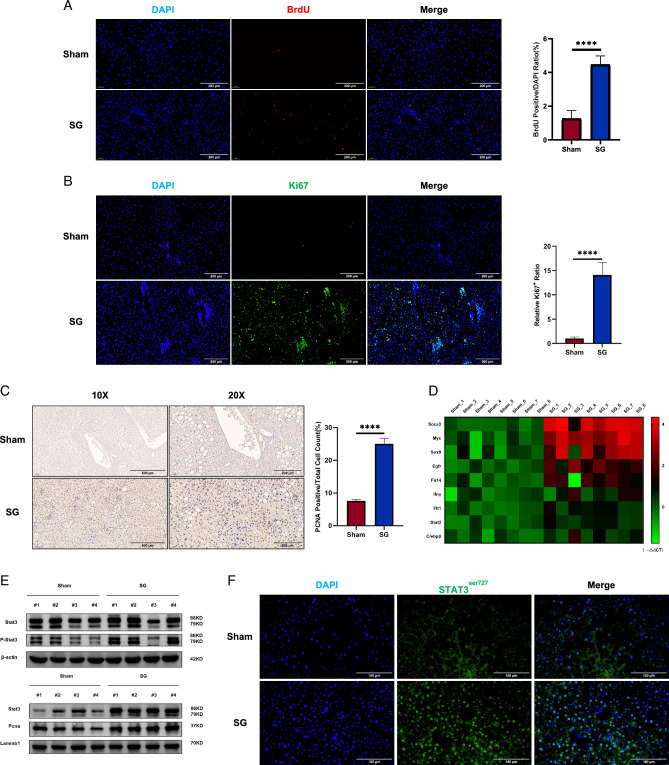
The H&E and PAS staining results provided valuable histological information. Following SG, a characteristic arrangement of healthy and functional hepatocytes was observed radiating from the portal vein to the central vein, reminiscent of the histological pattern of hepatocyte regeneration21. We investigated whether liver regeneration processes were involved in the recovery of liver function after SG. To minimize the potential toxicity of BrdU, we labeled the operated mice with BrdU via intraperitoneal injection 48 h before sacrifice. Interestingly, BrdU staining results demonstrated amplified liver regeneration signals in SG mice, which was further supported by the statistically significant increase in the ratio of BrdU-positive cell counts (Fig. 3A, B). Subsequently, we performed Ki67 immunofluorescence and PCNA IHC staining, which revealed enhanced proliferation capacity of the liver after SG (Fig. 3C). Moreover, liver mRNA level analysis through qRT-PCR assays showed the upregulation of various liver regeneration signals associated with SG, including Stat3, Egfr, Tgfβ1, Myc, Socs3, Sox9, Fn14, Flt1, and C/ebpβ (Fig. 3D). STAT3 and NF-κB have been extensively studied in the field of liver regeneration and are known to be crucial molecules in this process22. We observed that after SG, the phosphorylation level of the STAT3 protein in the liver increased, along with its nuclear localization (Fig. 3E). Immunofluorescence staining of the phosphorylated form of STAT3 supported this subcellular localization feature (Fig. 3F). Collectively, these findings provide evidence that liver regeneration in DIO mice was significantly enhanced after SG.
Figure 3.
The liver regeneration signal was significantly enhanced after SG. (A) BrdU staining, (B) Ki67 immunofluorescence, and (C) PCNA immunohistochemistry showed enhanced liver regeneration signals after SG. (D) The changes in the expression level of regeneration-related genes were shown by qRT-PCR results in the form of a heatmap. (E) Liver tissue Western blot results showed that Stat3, a classical signal of liver regeneration, was significantly up-regulated after SG. (F) Immunofluorescence staining of Stat3 in its phosphorylated form demonstrated whole-liver activation of Stat3-mediated regeneration signaling after SG. (Data are shown as mean±SE. ****=P<0.0001. For panels A–C, seven visual fields were collected to calculate BrdU, Ki67, and PCNA positive rates, respectively). DIO, diet-induced obese; SG, sleeve gastrectomy; PCNA, proliferating cell nuclear antigen.
Cellular and histological features of SG-generated enhanced liver regeneration
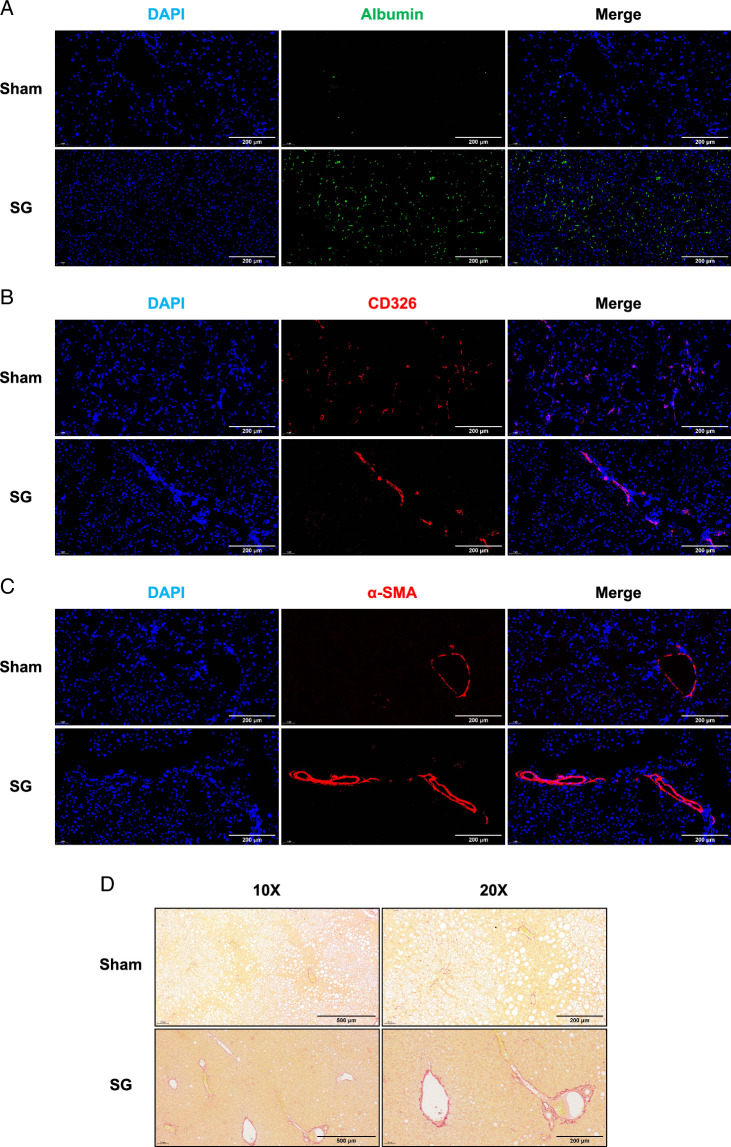
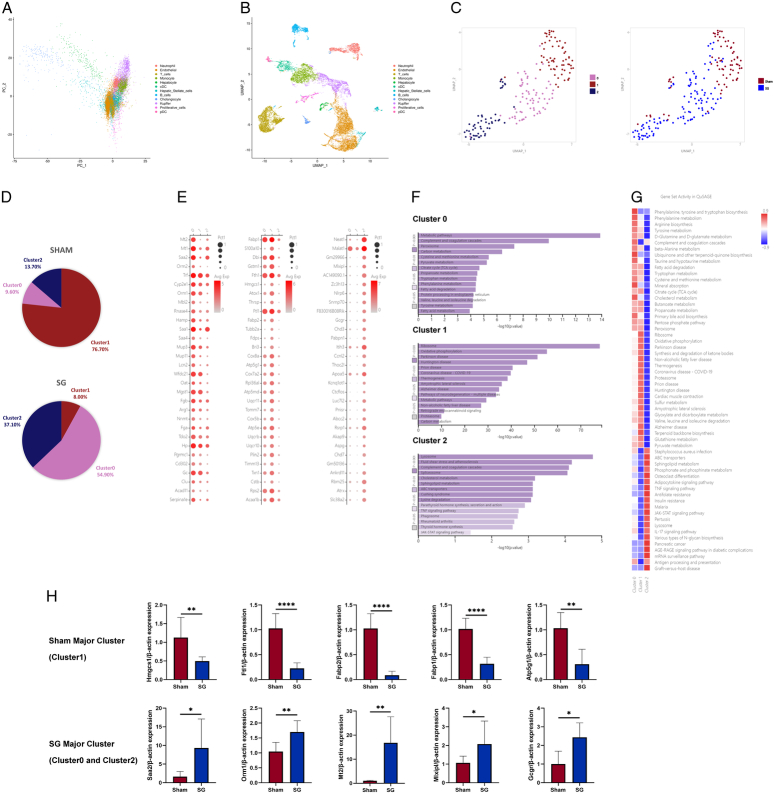
These results indicate an overall enhancement of liver regeneration signals following SG. To further investigate the detailed process and mechanism of this enhanced liver regeneration, we conducted the following experiments: According to the existing theory of liver regeneration, hepatocytes (HCs) and biliary epithelial cells can either proliferate conservatively or undergo transdifferentiation during liver regeneration12,23,24. Additionally, alterations and remodeling of the extracellular matrix (ECM) associated with hepatic stellate cells (HSCs) also play a significant role25. Immunofluorescence staining using characteristic cell markers allowed us to distinguish between these cell types. The results revealed a significant increase in functional hepatocytes and a relative decrease in BECs after SG (Fig. 4A, B). Moreover, changes in ECM were associated with activated HSCs (Fig. 4C, D). These findings suggest that the complete process of liver regeneration may have been enhanced after SG. To provide a more detailed description of this enhancement phenomenon, we performed single-cell RNA sequencing (scRNA-seq) on fresh liver samples from two groups of mice and employed the UMAP (Uniform Manifold Approximation and Projection) algorithm for dimensionality reduction, resulting in the separation of 12 cell clusters (Fig. 5A, B). PCA analysis successfully distinguished hepatocytes, leading us to re-cluster cells within the hepatocyte cluster, resulting in the identification of three new subclusters (Fig. 5C). The re-clustered results exhibited notable differences between the SG and Sham groups, with Cluster 0 and Cluster 2 dominating hepatocytes from the SG group, whereas Cluster 1 was the dominant cell type in the Sham group (Fig. 5D). To provide detailed characterization of the functional characteristics of the three hepatocyte subclusters, we examined the functional characteristics of the marker genes used to distinguish each cluster. Cluster 0, which showed significant differences from other subclusters, was primarily associated with normal metabolic processes in the liver. For instance, Serpinale plays an important role in liver gluconeogenesis26. The marker genes of Cluster 1 were highly correlated with NAFLD, including Fabp1, Fabp2, and Thrsp27,28. Cluster 2 represented the main proliferating cell type, whereas Cluster 0 was defined as a cell type with normal metabolic function (Fig. 5E). Pathway analysis of the marker genes in each subcluster revealed that the TNF and JAK-STAT signaling pathways in Cluster 2 play important roles in liver regeneration processes. Cluster 1 exhibited a high correlation with NAFLD, consistent with its dominant cell type in the sham group, which was aligned with the gross appearance and pathological state. Cluster 0 reflected the functional metabolic characteristics of normal hepatocytes (Fig. 5F). The results of the QuSAGE analysis mirrored the findings described above, with TNF, JAK-STAT, and other regeneration-related gene sets that were highly activated in Cluster 2. NAFLD-related gene sets were significantly more activated in Cluster 1, and various metabolism-related gene sets were predominantly activated in Cluster 0 (Fig. 5G). Based on the sequencing results mentioned above, we isolated primary hepatocytes from the livers of mice after SG and sham surgery. Real-time quantitative PCR assays were conducted on marker genes of different subclusters (Fig. 5H). The results closely aligned with the differences observed in the sequencing data.
Figure 4.
Histological features of SG-generated enhanced liver regeneration. The three main cell populations involved in liver regeneration include HCs, BECs, and HSCs. Select the corresponding cell markers (albumin (A), Cd326 B), and α-SMA (C)) to track major cell populations or their effects. (D) The results of Sirius red staining indirectly showed enhanced activation of HSCs with increased type I collagen after SG. BECs, biliary epithelial cells; SG, sleeve gastrectomy.
Figure 5.
Single-cell transcriptomics reveals changes in liver cell populations after SG. (A) PCA analysis was not suitable for analyzing scRNA-seq data, so (B) the UMAP algorithm was chosen for dimensionality reduction, dividing liver cells into 12 cell clusters and (C) re-clustering liver cell populations to obtain 3 subclusters, with (D) significant differences in the distribution of 3 subclusters between the SG and Sham groups. (E) The functions of each subcluster were defined based on the dominant expressed genes. (F) Pathway analysis of the 3 subclusters showed that the metabolic ability of Cluster 0 was significantly different, Cluster 1 was closely related to NAFLD, and Cluster 2 had obvious regeneration signals. (G) The QuSAGE analysis was consistent with the above conclusion. (H) The qRT-PCR results of isolated postoperative mouse hepatocytes validate the expression changes in different hepatocyte subclusters indicated by scRNA-seq. (Data are shown as mean±SE. *=P<0.05, **=P<0.01, and ****=P<0.0001. For panel H, n=8 Sham, 8 SG). NAFLD, non-alcoholic fatty liver disease; PCA, principal component analysis; SG, sleeve gastrectomy.
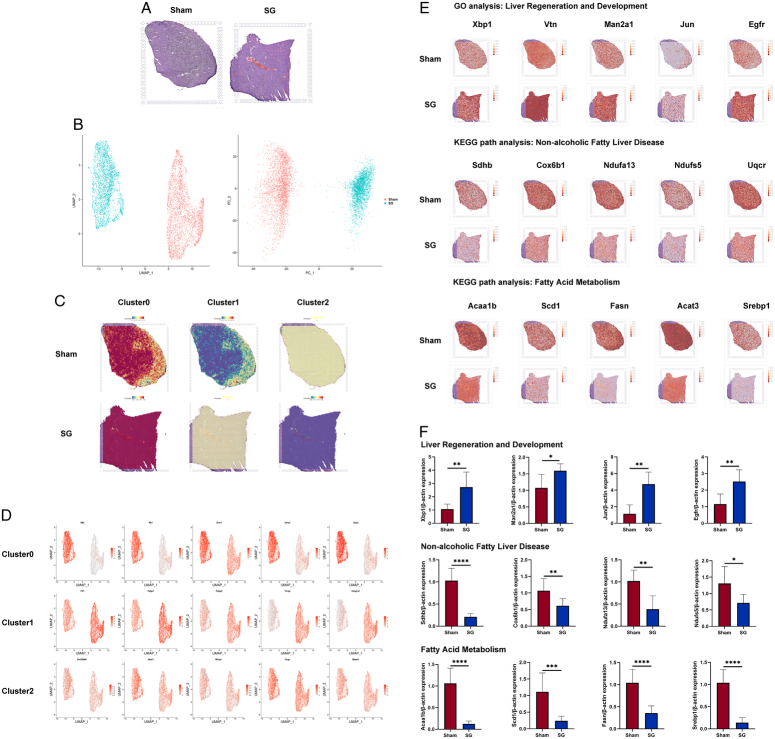
Considering the potential information loss associated with the aforementioned scRNA-seq, which could lead to inaccuracies in the analysis results, we added spatial transcriptomics (ST) analysis to obtain a broader perspective of the data observations. This enhances the credibility of the scRNA-seq results and better characterizes the improvement in proliferation and metabolism. Representative H&E slices were selected, followed by PCA and UMAP dimensionality reduction analyses. The results demonstrated significant differences in liver gene expression and spatial distribution between the SG and Sham groups (Fig. 6A, B). We mapped the marker gene sets of the three hepatocyte subclusters from the scRNA-seq to the ST results, revealing an overall metabolic improvement in the liver after SG accompanied by widespread proliferative signals (Fig. 6C). The spatial distribution of hepatocytes within the liver tissue interspersed with various other cell types may have affected the resolution of the spatial transcriptomic data. We spatially characterized the representative marker genes of the three hepatocyte subclusters, and the results were consistent with those obtained from the single cells. SG resulted in significant improvements in the metabolism, activation of proliferation, and alleviation of NAFLD (Fig. 6D). Additionally, we extracted key genes from the GO and KEGG pathway analyses of the scRNA-seq results and mapped them to the spatial transcriptome data. The findings demonstrated significant improvements in the metabolic and proliferation-related pathways in the mouse liver after SG (Fig. 6E). Similarly, we conducted qRT-PCR assays on isolated primary hepatocytes from postoperative mice, and the results were highly consistent with the findings from the ST analysis (Fig. 6F). Collectively, these data indicate marked alterations in hepatocyte populations after SG and widespread improvements in both proliferation and metabolism throughout the liver.
Figure 6.
Spatial transcriptomics unveils extensive changes in hepatocyte expression induced by SG. (A) The H&E staining results of the liver samples used in ST were used to provide spatial location information. (B) Both PCA analysis and UMAP dimensionality reduction analysis showed significant differences between the two groups of liver spots. (C) Marker genes for re-clusters in scRNA-seq were used to stain the ST spots, and the results were consistent with the distribution of the above re-clusters results. (D) However, since each spot often contains multiple cells, it caused a certain degree of specificity reduction, so the functional representative genes of the 3 subclusters were extracted and stained on the ST results. (E) Meanwhile, genes related to liver regeneration and development, NAFLD, and fatty acid metabolism pathways in scRNA-seq were extracted and stained on ST results, showing that the liver regeneration and fatty acid metabolism abilities were significantly enhanced after SG, and there were significant differences in the spatial distribution of NAFLD-related cells between the SG and Sham groups in the liver. (F) The qRT-PCR results of isolated postoperative mouse hepatocytes validate the pathway analysis results of GO and KEGG provided by ST. (Data are shown as mean±SE. *=P<0.05, **=P<0.01, ***=P<0.001, and ****=P<0.0001. For panel F, n=8 Sham, 8 SG). NAFLD, non-alcoholic fatty liver disease; PCA, principal component analysis; SG, sleeve gastrectomy; ST spatial transcriptomics
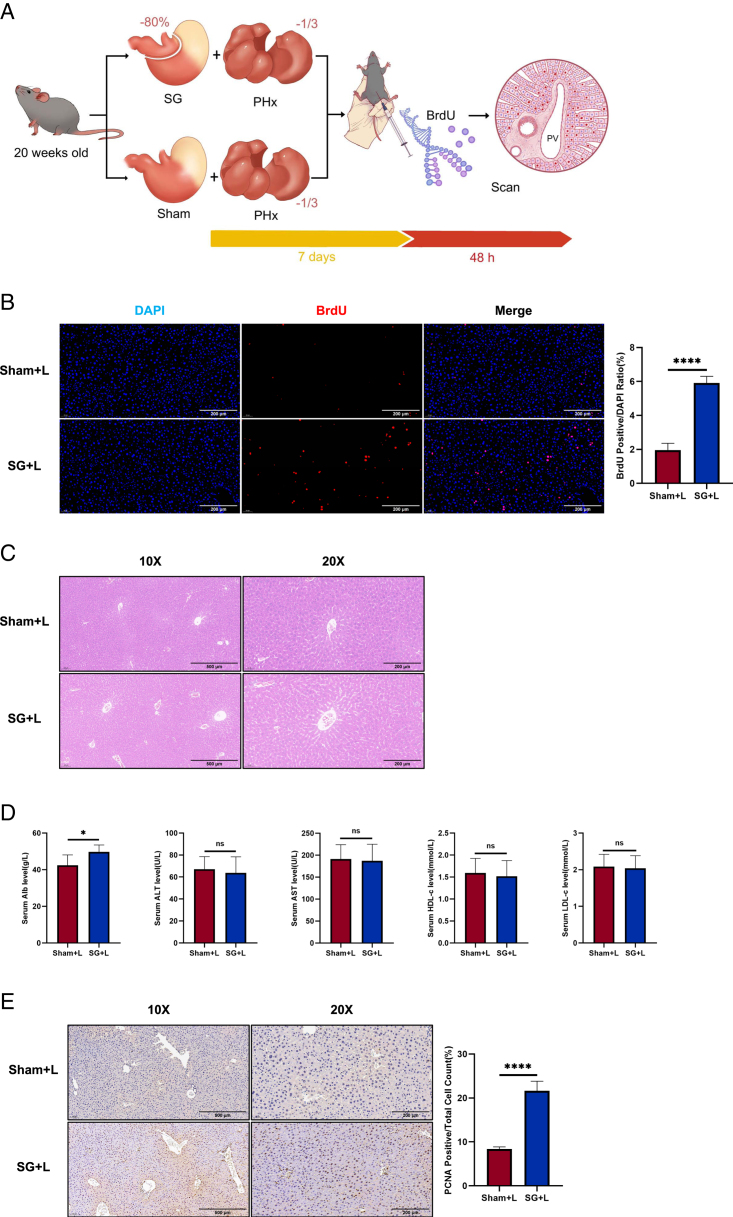
Enhanced liver regeneration induced by SG is not a consequence of NAFLD remission
To investigate whether the enhancement of liver regeneration after SG is solely a consequence of NAFLD remission, we conducted SG combined with hepatic left lateral lobectomy (SG+L) in mice fed a chow diet, while the control group underwent a sham procedure with hepatic left lateral lobectomy (Sham+L) (Fig. 7A). On the ninth day post-surgery, liver regeneration was more active in the SG+L group than in the Sham+L group (Fig. 7B). The results of liver H&E staining, as well as serum biochemical indicators, suggest that the livers of both mouse groups were in nearly identical health states at the observed time point. The higher levels of Alb in the SG+L group may reflect a more intense regenerative process (Fig. 7C, D). PCNA immunohistochemical staining further revealed a more robust state of liver regeneration in the SG+L group (Fig. 7E). These findings suggest that the enhancement of liver regeneration ability after SG is not merely a consequence of NAFLD remission resulting from surgery itself. Based on these observations, we can reasonably assume that SG enhances the regenerative capacity of the liver, leading to the generation of revitalized hepatocytes that contribute to overall hepatic metabolic improvements and restoration of liver function.
Figure 7.
Enhanced liver regeneration induced by SG is not a consequence of NAFLD remission. (A) Experimental flowchart of SG/Sham combined hepatectomy. (B) BrdU staining results showed that SG can directly enhance liver regeneration. A comparison of SG versus Sham results indicates that the enhanced liver regeneration by SG is not caused by the alleviation of fatty liver caused by SG itself. (C) The H&E results showed that the liver status of both groups was basically the same, and there was no histological damage caused by the surgery. (D) Liver function blood biochemistry results indicate that the physiological condition of the livers in both mouse groups is in a similar healthy state. (E) The results of PCNA immunohistochemical staining also confirmed the presence of enhanced liver regeneration. (Data are shown as mean±SE. ns=P>0.05, *=P<0.05, and ****=P<0.0001. For panel D, n=5 Sham, 5 SG. For panels B and E, seven visual fields were collected to calculate BrdU and PCNA positive rates, respectively). NAFLD, non-alcoholic fatty liver disease; SG, sleeve gastrectomy.
Enhanced regeneration induced by SG is independent of the Yap function
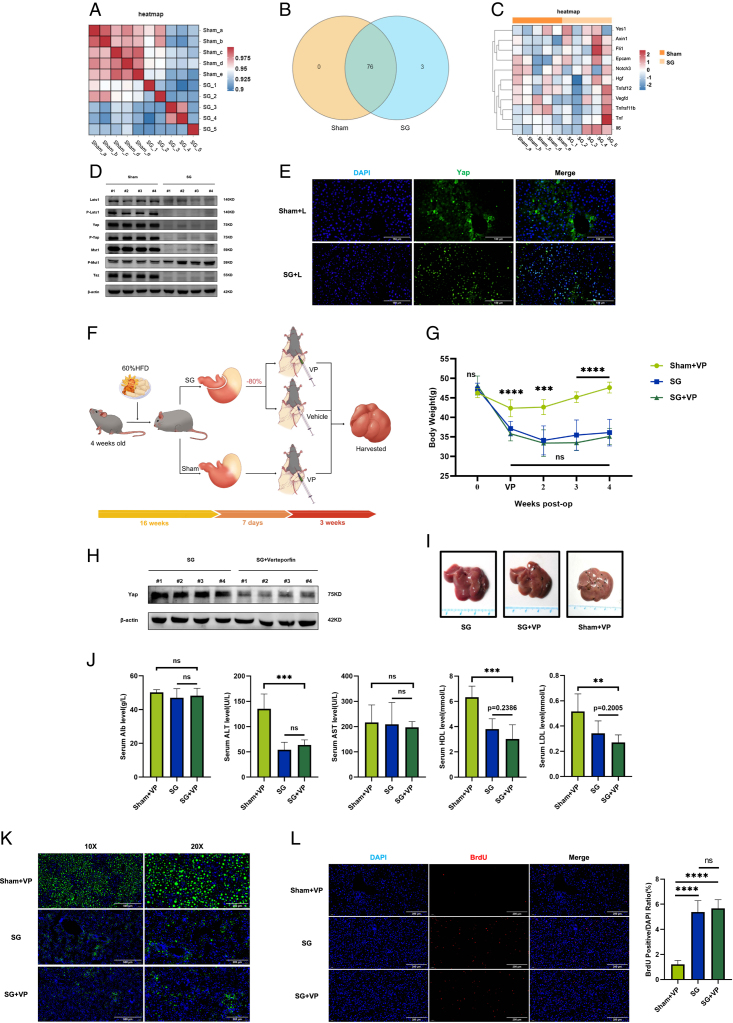
Given the enhanced liver regeneration signal observed after SG, concerns regarding the safety of this procedure arise because of the similarities between liver regeneration and the pathogenesis of hepatic malignancies. To assess the safety risk as much as possible, we collected serum samples from SG and Sham mice 6 weeks after the operation and performed an Olink Proximity Extension Assay to analyze 84 indicators. The results revealed relatively close serological similarity between the two groups of mice (Fig. 8A). Of the 79 detected serum proteins, three differentially expressed proteins were identified in the SG group compared with the sham group: Axin1, Epcam, and Fli1 (Fig. 8B). Correlation analysis between these differential proteins and classical liver regeneration-related proteins demonstrated a stronger association with the Yes1 protein than with the classic TNF and IL6 signals (Fig. 8C).
Figure 8.
Enhanced regeneration induced by SG is independent of the Yap function. (A) The correlation analysis heatmap of serum from SG and Sham groups of mice showed high similarity in serum composition, but differences existed between SG and Sham groups. (B) A total of 79 serum proteins were detected, and the Venn diagram showed that there were three differentially expressed proteins in SG compared to the Sham group. (C) Target protein screening was conducted, and the target protein needed to meet the following criteria: closely related to classical liver regeneration signals IL6 and Tnf, more closely related to the three differentially expressed proteins detected above, and able to mediate tumor occurrence. The result showed that Yes1 was the most suitable candidate based on the screening criteria. (D) Western blot was used to detect the Hippo–Yap signal in SG versus Sham groups, and the results showed that the Yap signal in the mouse liver was strongly inhibited 6 weeks after SG. (E) In order to evaluate the effect of SG on the Yap signal throughout the postoperative period, Yap immunofluorescence staining was performed on mice that underwent combined surgery 7 days after the operation, and the results showed that SG seemed to enhance Yap function in the early postoperative period. (F) In order to verify whether Yap is a necessary factor for SG to produce liver regeneration enhancement and metabolic improvement during the long-term postoperative process, verteporfin was used to inhibit Yap function in mice, and surgical intervention and a control group were set up simultaneously. (G) The weight loss effect induced by SG was not influenced by verteporfin treatment. (H) The Western blot results demonstrated that verteporfin effectively suppressed the expression levels of YAP. (I) Gross observation of the liver showed that compared to the Sham+VP group, the other two groups had significant and almost similar relief of steatosis. (J) Serological test results indicate that verteporfin did not affect the beneficial effects of liver function improvement induced by SG. (K) The results of BODIPY staining showed that Yap inhibition did not affect the ability of SG to relieve fatty liver, and (L) the results of BrdU staining showed that the enhancement of liver regeneration produced by SG was not affected by Yap inhibition. (Data are shown as mean±SE. ns=P>0.05, **=P<0.01, ***=P<0.001, and ****=P<0.0001. For panels B and D, nine visual fields were collected to calculate the BrdU-positive rate. For panel G, n=6 Sham+VP, 8 SG, 7 SG+VP. For panel J, n=5 Sham+VP, 5 SG, 5 SG+VP). SG, sleeve gastrectomy.
Notably, Yes1 is closely linked to the transcriptional coactivator Yap, which plays a role in the transformation of hepatocytes into biliary epithelial cells via EpCAM29. Axin1 is also implicated in the occurrence and proliferation of YAP and CTNNB1-mediated hepatocellular carcinoma (HCC) occurrence and proliferation30. Fli1 is associated with YAP in endothelial cell differentiation31. Considering the relevance of YAP in liver regeneration and its interaction with Myc/Max/Mad encoded by Mlxipl, which is a top marker gene in Cluster 2 of the scRNA-seq results, we examined YAP signaling activation in the liver after SG and found that YAP function appeared to be strongly suppressed (Fig. 8D). However, considering the low expression level of YAP in the liver and its reliance on subcellular localization, we performed YAP immunofluorescent staining in the livers of mice that underwent combined resection (Sham+L vs. SG+L). The results indicated distinct intranuclear localization of YAP signals in the livers of mice in the SG+L group (Fig. 8E).
To investigate the long-term effects of YAP signaling beyond one week postoperatively, we administered a YAP signaling blockade using verteporfin one week after the operation (Fig. 8F). The results indicate that mice in the SG group, after receiving long-term injections of verteporfin, showed no significant difference in weight change compared to those not treated with verteporfin. In contrast, mice undergoing sham surgery and receiving verteporfin exhibited noticeable weight changes. This suggests that verteporfin, in the process of inhibiting Yap, does not impact the weight loss effect associated with SG (Fig. 8G). In addition, in the presence of verteporfin, which exerted a stable inhibitory effect on Yap signaling in the mouse liver, both groups of mice that underwent SG surgery exhibited noticeable and consistent remission of steatosis and volume changes in the liver (Fig. 8H, I). During the inhibition of Yap, the serological results indicate that SG still demonstrates the ability to improve the physiological status of the liver. Furthermore, this ameliorative effect is comparable to mice that underwent SG surgery without receiving verteporfin treatment (Fig. 8J). BODIPY and BrdU staining results indicated that YAP inhibition did not significantly affect the degree of hepatic steatosis or liver regeneration (Fig. 8K, L).
Discussion
SG, an increasingly popular type of bariatric surgery, has generated significant excitement in the field of metabolic research owing to its remarkable metabolic improvements. One particular aspect that has garnered attention is the ability of surgery to effectively improve the NAFLD status often associated with obese patients, leading to the restoration of liver function. However, the mechanism underlying this therapeutic effect remains largely unknown.
In our recent study, we made a novel discovery regarding the regenerative activity of the liver, which was significantly enhanced after SG. This finding provides a new theoretical and investigative direction to explain the alleviation of liver dysfunction observed in patients with NAFLD following surgery. To explore this phenomenon, we employed DIO mice as a model animal, as they closely resemble the NAFLD state observed in humans when subjected to prolonged induction with a high-fat diet. This animal model effectively mimicked the conditions experienced by patients who underwent SG. In our study, we performed SG on DIO mice and compared them to a control group of sham-operated mice. By carefully observing stained tissue sections, we not only replicated the widely reported metabolic improvement phenotype in mouse models but also discovered an intriguing new phenotype, an enhanced state of liver regeneration. The liver possesses unique regenerative and repair capabilities and serves as a vital metabolic organ within the body. The mechanisms governing liver regeneration are highly intricate, with the ability to initiate the process within minutes when liver function is impaired32,33. Extensive research has already shed light on liver regeneration by identifying key molecules such as STAT3 and NF-κB, which play crucial roles in this process34,35. Our study provides substantial evidence for the enhancement of liver regeneration following SG using various approaches. We used techniques such as scRNA-Seq and spatial transcriptome analysis to validate the existence of this phenomenon.
After regeneration, the liver exhibited a higher number of hepatocytes with intact metabolic function. Notably, histological analyses, including H&E staining, Oil Red O staining for lipid droplet clearance, and PAS staining for glycogen, demonstrated a significant reduction in balloon-like cells with metabolic dysfunction. Additionally, increased glycogen storage indicated a metabolic state similar to that of a newborn liver. While these experimental findings are highly promising, several additional questions warrant further investigation. For instance, it remains unclear whether the enhancement of liver regeneration is an additional phenomenon that occurs after SG improves the metabolic state of the liver or if SG acts as an initiating factor for liver regeneration. Numerous studies have indicated that the liver regeneration process cannot be completely halted, which prevented us from investigating whether SG can improve liver metabolism in the absence of liver regeneration12,22,36.
Nonetheless, our findings provide valuable insights into the mechanisms underlying metabolic improvements observed after SG. Further research is necessary to unravel the intricacies of liver regeneration enhancement and its relationship with the metabolic state of the liver following bariatric surgery. For these reasons, we were unable to determine whether SG directly induced liver regeneration. However, we designed experiments to demonstrate that SG enhances liver regeneration independently of the metabolic improvements it causes. To eliminate the effect of SG on liver steatosis, we performed SG in mice fed a chow diet and simultaneously resected the left lateral lobe of the liver. On the seventh day after surgery, we performed BrdU labeling and observed a difference in liver regeneration intensity 48 h after labeling. Liver regeneration after partial liver resection starts within 1 min and continues for ~14 days, during which the process gradually diminishes32. We aimed to observe the effect of SG on liver regeneration during this period. Although early traumatic stress after SG might affect liver regeneration, we can conclude that the recovery ability of DIO mice is weaker than that of mice fed a chow diet. This difference is a key factor in solving the aforementioned problems. By performing combined surgery on DIO mice and monitoring their daily body weight, we observed that around 7 days after surgery, the body weight curve of the SG+L group reached a plateau, similar to that of the Sham+L group. Moderate weight loss was followed by a gradual increase in body weight, indicating a recovery process (Supplementary Fig. 2, Supplemental Digital Content 2, http://links.lww.com/JS9/C348). DIO mice have a slower recovery after surgery because of their physiological liver conditions. Therefore, the recovery time point for chow diet-fed mice after SG+L surgery should be within 7 days, still within the 14-day window of liver regeneration caused by liver lobectomy. This critical time point allowed us to study the enhancement of liver regeneration as early as possible.
To reveal the histological and cellular manifestations of enhanced liver regeneration by SG, we performed scRNA-Seq and spatial transcriptomic analyses. The results showed a change in the grouping of hepatocytes after SG, with three clusters of hepatocytes in different states. Cluster 0 represented hepatocytes with normal metabolic function, Cluster 2 exhibited proliferative activity, and Cluster 1 was highly related to NAFLD. These findings indicate that SG not only enhances regenerative ability but also significantly improves liver metabolic function. The QuSAGE analysis and spatial transcriptomic results further support these findings. According to the results of scRNA-seq, the hepatocytes of mice in the SG group exhibit predominant clusters, namely Clusters 0 and 2, while the predominant cluster for Sham group mice is Cluster 1. To provide more robust evidence, extensive qRT-PCR validation was conducted on marker genes for each predominant cluster. To eliminate potential interference from a large number of stromal and immune cells present in the liver, we isolated mouse hepatocytes and conducted the aforementioned experiments. Not only that but to restore the spatial information lost in scRNA-seq, we supplemented it with spatial transcriptomics (ST). ST preserves histological spatial information, providing an intuitive distribution of transcriptional differences in liver cells. The combination of these two sequencing methods, along with the results from GO and KEGG, aligns with the observed phenomena of enhanced liver regeneration and metabolic improvement. Similarly, ST has the weakness of lower resolution, where transcription signals may simultaneously include both hepatocytes and surrounding non-hepatocytes. Therefore, we conducted qRT-PCR validation on the pathway results characterized by ST, using isolated primary hepatocytes from postoperative mouse livers to eliminate interference from other cell types on ST results.
Considering the overlap between HCC and liver regeneration pathways, we made a preliminary attempt to explore the safety of SG. We performed Olink sequencing of mouse serum samples to detect faint small-molecule signals. This technique identified three differential proteins, Axin1, Fli1, and Epcam, which were all present in the SG group. These proteins are closely related to canonical liver regeneration signals. Among them, Yes1 was found to be associated with three proteins, and its related transcription factor, Yap, has been extensively studied in tumorigenesis and liver regeneration. We examined YAP signaling in DIO mice 6 weeks after surgery and found that YAP was strongly suppressed after SG. To investigate the role of YAP in the early regeneration process, we stimulated YAP activation by reducing liver volume and observed its effect. Additionally, we conducted fluorescent staining of YAP to determine the functional state of nuclear translocation37–39. The results showed that YAP nuclear translocation appeared to be activated in the SG+L group, while YAP in the Sham+L group was predominantly located in the cytoplasm. To further explore the function of YAP, we administered the YAP inhibitor verteporfin 1 week after surgery and collected samples 3 weeks later. The results demonstrated that YAP inhibition did not affect the improvement of liver steatosis or the enhancement of liver regeneration by SG. Therefore, we conclude that Yap is not necessary for SG to enhance liver regeneration and improve metabolic function. However, our study is merely preliminary and exploratory. We have only observed the weight loss and improved physiological status associated with SG, which is seemingly unrelated to Yap. Moreover, Yap appears to be suppressed in the long-term process following SG. Although this prolonged suppression is unfavorable for tumor occurrence, it does not provide a definitive conclusion on the safety of SG. Tumorigenesis and carcinogenesis are highly complex processes, and the information obtained from our study is very limited and, in some respects, inconclusive. Further, more comprehensive research is needed to thoroughly address the safety concerns associated with SG.
Conclusion
Our study suggests that SG enhances liver regeneration capacity in DIO mice, contributing to a better understanding of why SG can yield such robust metabolic benefits. Importantly, we have determined that this enhanced liver regenerative activity is unrelated to the improvement of hepatic steatosis. These findings provide new insights into the mechanisms underlying the metabolic benefits of SG, offering researchers and healthcare professionals a fresh perspective on understanding the impact of SG procedures.
Ethical approval
This research involves no patient.
Sources of funding
This work was supported by the National Natural Science Foundation of China (NSFC) (No.8227033969) and The Major Basic Research Project of Shandong Province (No. ZR2020ZD15).
Author contribution
S.H. and X.M.: designed the project and revised the manuscript; T.Y.: performed the experiments and drafted the original manuscript; Y.C. and Z.W.: participated in the completion of the animal experiment; G.Z.: participated in data analysis; H.D. and J.Y.: processed and analyzed the experimental data; Y.W.: completed the drawing of the experimental flowchart.
Conflicts of interest disclosure
The authors declare that they have no conflicts of interest.
Research registration unique identifying number (UIN)
This is not a human study.
Guarantor
Sanyuan Hu.
Data availability statement
All datasets used and analyzed during the current study are available from the corresponding author upon reasonable request.
Provenance and peer review
Not applicable.
Supplementary Material
Footnotes
Tianming Yu, Xiaomin Ma, and Yang Cheng contributed equally to this article.
Sponsorships or competing interests that may be relevant to content are disclosed at the end of this article.
Supplemental Digital Content is available for this article. Direct URL citations are provided in the HTML and PDF versions of this article on the journal's website, www.lww.com/international-journal-of-surgery.
Published online 4 April 2024
Contributor Information
Tianming Yu, Email: ikemind@163.com.
Xiaomin Ma, Email: 15910098683@163.com.
Yang Cheng, Email: cysdu0415@163.com.
Zeyu Wang, Email: 15650512858@163.com.
Guangyong Zhang, Email: guangyongzhang@hotmail.com.
Huanxin Ding, Email: sdhuanxinding@outlook.com.
Jialuo Yin, Email: yurike2022@163.com.
Yifei Wang, Email: 17865195352@163.com.
Sanyuan Hu, Email: drsanyuanhu@163.com.
References
- 1.Powell EE, Wong VW-S, Rinella M. Non-alcoholic fatty liver disease. Lancet 2021;397:2212–2224. [DOI] [PubMed] [Google Scholar]
- 2.Targher G, Tilg H, Byrne CD. Non-alcoholic fatty liver disease: a multisystem disease requiring a multidisciplinary and holistic approach. Lancet Gastroenterol Hepatol 2021;6:578–588. [DOI] [PubMed] [Google Scholar]
- 3.European Association for the Study of the Liver (EASL), European Association for the Study of Diabetes (EASD), and European Association for the Study of Obesity (EASO) . EASL–EASD–EASO Clinical Practice Guidelines for the management of non-alcoholic fatty liver disease. Obes Facts 2016;9:65–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Geurtsen ML, Santos S, Felix JF, et al. Liver fat and cardiometabolic risk factors among school-age children. Hepatology 2020;72:119–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vilar-Gomez E, Martinez-Perez Y, Calzadilla-Bertot L, et al. Weight loss through lifestyle modification significantly reduces features of nonalcoholic steatohepatitis. Gastroenterology 2015;149:367–78.e5. [DOI] [PubMed] [Google Scholar]
- 6.Lassailly G, Caiazzo R, Ntandja-Wandji L-C, et al. Bariatric surgery provides long-term resolution of nonalcoholic steatohepatitis and regression of fibrosis. Gastroenterology 2020;159:1290–1301.e5. [DOI] [PubMed] [Google Scholar]
- 7.Seeberg KA, Borgeraas H, Hofsø D, et al. Gastric bypass versus sleeve gastrectomy in type 2 diabetes: effects on hepatic steatosis and fibrosis: a randomized controlled trial. Ann Intern Med 2022;175:74–83. [DOI] [PubMed] [Google Scholar]
- 8.Arterburn DE, Telem DA, Kushner RF, et al. Benefits and risks of bariatric surgery in adults: a review. JAMA 2020;324:879–887. [DOI] [PubMed] [Google Scholar]
- 9.Chaudhari SN, Harris DA, Aliakbarian H, et al. Bariatric surgery reveals a gut-restricted TGR5 agonist with anti-diabetic effects. Nat Chem Biol 2021;17:20–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chaudhari SN, Luo JN, Harris DA, et al. A microbial metabolite remodels the gut–liver axis following bariatric surgery. Cell Host Microbe 2021;29:408–424.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ryan KK, Tremaroli V, Clemmensen C, et al. FXR is a molecular target for the effects of vertical sleeve gastrectomy. Nature 2014;509:183–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Michalopoulos GK, Bhushan B. Liver regeneration: biological and pathological mechanisms and implications. Nat Rev Gastroenterol Hepatol 2021;18:40–55. [DOI] [PubMed] [Google Scholar]
- 13.Michalopoulos GK, DeFrances MC. Liver regeneration. Science 1997;276:60–66. [DOI] [PubMed] [Google Scholar]
- 14.Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000;25:25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Draghici S, Khatri P, Tarca AL, et al. A systems biology approach for pathway level analysis. Genome Res 2007;17:1537–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yaari G, Bolen CR, Thakar J, et al. Quantitative set analysis for gene expression: a method to quantify gene set differential expression including gene–gene correlations. Nucleic Acids Res 2013;41:e170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kilkenny C, Browne WJ, Cuthill IC, et al. Improving bioscience research reporting: the ARRIVE guidelines for reporting animal research. PLoS Biol 2010;8:e1000412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hou X, Yin S, Ren R, et al. Myeloid-cell-specific IL-6 signaling promotes microRNA-223-enriched exosome production to attenuate NAFLD-associated fibrosis. Hepatology 2021;74:116–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bedossa P. Pathology of non-alcoholic fatty liver disease. Liver Int 2017;37(Suppl 1):85–89. [DOI] [PubMed] [Google Scholar]
- 20.Kleiner DE, Brunt EM, Van Natta M, et al. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology 2005;41:1313–1321. [DOI] [PubMed] [Google Scholar]
- 21.Rabes HM. Kinetics of hepatocellular proliferation as a function of the microvascular structure and functional state of the liver. Ciba Found Symp 1977:31–53. [DOI] [PubMed] [Google Scholar]
- 22.Fausto N, Campbell JS, Riehle KJ. Liver regeneration. Hepatology 2006;43(2 Suppl 1):S45–S53. [DOI] [PubMed] [Google Scholar]
- 23.Limaye PB, Bowen WC, Orr AV, et al. Mechanisms of hepatocyte growth factor-mediated and epidermal growth factor-mediated signaling in transdifferentiation of rat hepatocytes to biliary epithelium. Hepatology 2008;47:1702–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Michalopoulos GK, Khan Z. Liver stem cells: experimental findings and implications for human liver disease. Gastroenterology 2015;149:876–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rudolph KL, Trautwein C, Kubicka S, et al. Differential regulation of extracellular matrix synthesis during liver regeneration after partial hepatectomy in rats. Hepatology 1999;30:1159–1166. [DOI] [PubMed] [Google Scholar]
- 26.Yuan Y, Zhu C, Wang Y, et al. α-Ketoglutaric acid ameliorates hyperglycemia in diabetes by inhibiting hepatic gluconeogenesis via serpina1e signaling. Sci Adv 2022;8:eabn2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lehner R, Quiroga AD. Chapter 5 - Fatty acid handling in mammalian cells. In: Ridgway ND, McLeod RS, editors. Biochemistry of Lipids, Lipoproteins and Membranes (7th ed). Elsevier; 2021:161–200. [Google Scholar]
- 28.Wu J, Wang C, Li S, et al. Thyroid hormone-responsive SPOT 14 homolog promotes hepatic lipogenesis, and its expression is regulated by liver X receptor α through a sterol regulatory element-binding protein 1c-dependent mechanism in mice. Hepatology 2013;58:617–628. [DOI] [PubMed] [Google Scholar]
- 29.Lee S-H, So J, Shin D. Hepatocyte-to-cholangiocyte conversion occurs through transdifferentiation independently of proliferation in zebrafish. Hepatology 2023;77:1198–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liang B, Wang H, Qiao Y, et al. Differential requirement of Hippo cascade during CTNNB1 or AXIN1 mutation-driven hepatocarcinogenesis. Hepatology 2023;77:1929–1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quan Y, Shan X, Hu M, et al. YAP inhibition promotes endothelial cell differentiation from pluripotent stem cell through EC master transcription factor FLI1. J Mol Cell Cardiol 2022;163:81–96. [DOI] [PubMed] [Google Scholar]
- 32.Mars WM, Liu ML, Kitson RP, et al. Immediate early detection of urokinase receptor after partial hepatectomy and its implications for initiation of liver regeneration. Hepatology 1995;21:1695–1701. [PubMed] [Google Scholar]
- 33.Kim TH, Mars WM, Stolz DB, et al. Expression and activation of pro-MMP-2 and pro-MMP-9 during rat liver regeneration. Hepatology 2000;31:75–82. [DOI] [PubMed] [Google Scholar]
- 34.FitzGerald MJ, Webber EM, Donovan JR, et al. Rapid DNA binding by nuclear factor kappa B in hepatocytes at the start of liver regeneration. Cell Growth Differ 1995;6:417–427. [PubMed] [Google Scholar]
- 35.Cressman DE, Diamond RH, Taub R. Rapid activation of the Stat3 transcription complex in liver regeneration. Hepatology 1995;21:1443–1449. [PubMed] [Google Scholar]
- 36.Higgins GM, Anderson RM, Higgins G, et al. Experimental pathology of the liver. Arch Pathol 1931;12:186–202. [Google Scholar]
- 37.Camargo FD, Gokhale S, Johnnidis JB, et al. YAP1 increases organ size and expands undifferentiated progenitor cells. Curr Biol 2007;17:2054–2060. [DOI] [PubMed] [Google Scholar]
- 38.Song H, Mak KK, Topol L, et al. Mammalian Mst1 and Mst2 kinases play essential roles in organ size control and tumor suppression. Proc Natl Acad Sci U S A 2010;107:1431–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heallen T, Zhang M, Wang J, et al. Hippo pathway inhibits Wnt signaling to restrain cardiomyocyte proliferation and heart size. Science 2011;332:458–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All datasets used and analyzed during the current study are available from the corresponding author upon reasonable request.