Abstract
Gastric cancer typically originates from the abnormal proliferation of normal cells within the gastric mucosa, eventually forming tumors. The roles of sperm-associated antigen 5 (SPAG5) and abnormal spindle-like microcephaly (ASPM) associated genes in gastric cancer are not yet clear. Gastric cancer datasets GSE51575 and GSE36076 profiles were downloaded from the GPL13607 and GPL570-generated gene expression omnibus database. The analysis included filtering for differentially expressed genes, weighted gene co-expression network analysis, functional enrichment analysis, gene set enrichment analysis, immune infiltration analysis, construction and analysis of the protein–protein interaction network, survival analysis, and Comparative Toxicogenomics Database analysis. Heatmaps of gene expression were also created. A total of 1457 differentially expressed genes were identified. According to gene ontology analysis, they are primarily enriched in the metabolic processes of organic acids, condensed chromosome centromere regions, and oxidoreductase activity. Kyoto Encyclopedia of Gene and Genome analysis showed they are mainly involved in metabolic pathways, P53 signaling pathway, and PPAR signaling pathway. The soft threshold power for weighted gene co-expression network analysis was set to 8. Three core genes (CENPE, SPAG5, and ASPM) were identified. Heatmaps of core gene expression revealed that SPAG5 and ASPM are highly expressed in gastric cancer samples and low in normal samples. Comparative Toxicogenomics Database analysis indicated that the core genes (CENPE, SPAG5, and ASPM) are associated with gastric tumors, gastric diseases, gastritis, gastric ulcers, tumors, inflammation, and necrosis. The SPAG5 and ASPM genes are overexpressed in gastric cancer tissues, and higher expression levels are associated with worse prognosis, may serve as potential prognostic markers.
Keywords: ASPM, differentially expressed genes, gastric cancer, SPAG5
1. Introduction
Gastric cancer is a malignant tumor originating from abnormal proliferation within gastric tissues, typically starting in the inner lining of the gastric mucosa. Over time, it can penetrate deeper into the gastric wall and spread to surrounding tissues and organs.[1] The growth patterns of gastric cancer can be polypoid, invasive, or ulcerative, and its pathological characteristics include tissue type, grade, depth of invasion, and lymph node metastasis.[2] Clinical manifestations of gastric cancer may vary due to individual differences, with early stages often presenting no symptoms or showing nonspecific symptoms such as indigestion and loss of appetite. As the disease progresses, symptoms like abdominal pain, nausea, vomiting, weight loss, and melena may occur. Advanced gastric cancer often accompanies severe complications, such as gastric bleeding and perforation.[3] The incidence of gastric cancer shows significant geographical differences globally, with higher rates in Asia compared to North America and Europe. The incidence increases with age, typically more common in individuals over 50, with males being more susceptible than females.[4] Gastric cancer can invade surrounding blood vessels, including veins and arteries, which increases the risk of metastasis and recurrence. Lymph node metastasis is also common and impacts disease staging and prognosis. Gastric cancer cells typically exhibit anaplasia, with irregular shapes and sizes and active nuclear division. Nuclear atypia is a significant pathological feature of gastric cancer.[5] Diagnosis is usually made through endoscopic examination coupled with tissue biopsy. Based on tumor size, depth of invasion, and lymph node involvement, gastric cancer is often staged to guide treatment and prognosis assessment. Treatment generally includes surgery, chemotherapy, radiation, or a combination of these, with the depth of gastric wall invasion being a crucial indicator for assessing tumor aggressiveness and choosing treatment methods.[6] High intake of salty and pickled foods is associated with increased gastric cancer rates. Additionally, smoking, alcohol consumption, and a lack of healthy dietary habits such as consuming fresh fruits and vegetables are also linked to increased risks of gastric cancer. However, the exact causes of gastric cancer remain unclear, making it crucial to study its molecular mechanisms more deeply.
Sperm-associated antigen 5 (SPAG5) is a gene that encodes a protein primarily expressed in germ cells. The protein encoded by this gene plays a crucial role in sperm formation and motility. The SPAG5 protein is involved in essential processes such as cell division and microtubule dynamics, which are vital for maintaining normal cellular function. Aberrant expression or mutations of SPAG5 may be associated with the occurrence and development of certain diseases.[7] Abnormal spindle-like microcephaly associated (ASPM) gene encodes a protein that plays an important role in embryonic development. This gene is closely associated with normal development of intelligence and cortical morphology in the brain.[8] Mutations or abnormal function of ASPM are linked to Microcephaly, a rare neurodevelopmental disorder characterized by abnormally small head size and delayed intellectual development.[9] Research on these 2 genes provides important insights into cell biology, reproductive biology, and neurodevelopment, with potential clinical significance for the diagnosis and treatment of diseases related to these genes.
Bioinformatics technology is an interdisciplinary field that applies computer science and information technology to biology, encompassing a variety of techniques and methods for processing, analyzing, and interpreting biological data.[10] The application of bioinformatics in the life sciences is becoming increasingly widespread, providing powerful tools and methods that accelerate scientific research and advance the biomedical field.[11] With continuous innovation and development, bioinformatics is expected to maintain its significant role in the future, contributing further breakthroughs and advancements in human health and biological resource utilization. The advantages of bioinformatics include efficiency, precision, comprehensiveness, visualization, predictability, systematic nature, standardization, and broad applicability, all of which significantly propel and facilitate research and applications in the life sciences.[12,13]
However, the relationship between SPAG5 and ASPM genes and gastric cancer is not yet clear. Therefore, this paper aims to use bioinformatics techniques to identify core genes between gastric cancer and normal tissues and to perform enrichment and pathway analyses. Public datasets will be used to validate the significant role of SPAG5 and ASPM genes in gastric cancer.
2. Methods
2.1. Gastric cancer dataset
In this study, gastric cancer datasets GSE51575 and GSE36076 profiles were downloaded from the gene expression omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/), generated from GPL13607 and GPL570 platforms. GSE51575 includes samples from 26 gastric cancer cases and 26 normal tissues, GSE36076 includes 3 gastric cancer and 10 normal tissue samples. These datasets were utilized to identify differentially expressed genes (DEGs) in gastric cancer.
2.2. Selection of DEGs
Initially, log2 transformation was applied separately to gastric cancer datasets GSE51575 and GSE36076. Multivariate linear regression was conducted using the lmFit function, with empirical Bayes moderation adjusting the standard errors toward a common value to calculate the adjusted t statistic, adjusted f statistic, and log-odds of differential expression. The R package “limma” was used for probe summarization and background correction of the combined matrices of GSE51575 and GSE36076. The Benjamini–Hochberg method was employed to adjust raw P values. Fold change (FC) was calculated using the false discovery rate (FDR). The cutoff for DEGs was set at P < .05 and FC > 1.5. Volcano plots were then generated to illustrate the significance of gene differences. An intersection of differential genes between the GSE51575 and GSE36076 datasets was taken to obtain the DEGs.
2.3. Weighted gene co-expression network analysis (WGCNA)
Using the batch-corrected combined gene expression matrix from the GSE51575 and GSE36076 datasets, the Median Absolute Deviation (MAD) for each gene was calculated, and the genes in the lowest 50% of MAD values were excluded. Outlier genes and samples were removed using the good samples genes method of the WGCNA R package, and a scale-free co-expression network was constructed using WGCNA. To categorize genes with similar expression patterns into modules, average linkage hierarchical clustering based on the topological overlap matrix (TOM) similarity measure was performed, with a minimum size (gene group) for the gene dendrogram set at 30. Sensitivity was set to 3. For further module analysis, module eigengene dissimilarity was calculated, a cutline was selected for the module dendrogram, and some modules were merged. Additionally, modules with a distance of <0.25 were merged; notably, the gray module is considered a collection of genes that could not be assigned to any module.
2.4. Functional enrichment analysis
Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses are computational methods for assessing gene functions and biological pathways. In this study, the Venn diagram-filtered list of differential genes was input into the KEGG API (https://www.kegg.jp/kegg/rest/keggapi.html) to obtain the latest KEGG pathway gene annotations, which served as a background for mapping genes to the background set. Enrichment analysis was conducted using the R package clusterProfiler (version 3.14.3). GO annotations of genes were obtained from the R package org.Hs.e.g..db (version 3.1.0), with a minimum gene set of 5 and a maximum of 5000. A P value of <.05 and an FDR of <0.25 were considered statistically significant.
Moreover, the metascape database (http://metascape.org/gp/index.html) provided comprehensive gene list annotation and analysis resources and was used to perform functional enrichment analysis and export visualizations for the differential gene list.
2.5. Gene set enrichment analysis (GSEA)
For GSEA, GSEA software (version 3.0) was obtained from the GSEA website (DOI:10.1073/pnas.0506580102, http://software.broadinstitute.org/gsea/index.jsp). Two sample groups, disease and normal tissues, were separately divided into 2 groups. The c2.cp.kegg.v7.4.symbols.gmt subset was downloaded from the Molecular Signatures Database (DOI:10.1093/bioinformatics/btr260, http://www.gsea-msigdb.org/gsea/downloads.jsp) to evaluate relevant pathways and molecular mechanisms. Gene expression profiles and phenotype grouping were utilized, with the minimum gene set to 5, the maximum gene set to 5000, and 1000 permutations. A significance threshold of P value < .05 and FDR < 0.25 was considered statistically significant. GO and KEGG analyses were also conducted on the entire genome as per GSEA.
2.6. Immune infiltration analysis
CIBERSORT (http://CIBERSORT.stanford.edu/) is a widely used method for calculating immune cell infiltration. The LM22 gene file defines 22 immune cell subtypes. Integrated bioinformatics methods were applied using the CIBERSORT package to analyze gene expression matrices of gastric cancer datasets GSE51575 and GSE36076. Linear support vector regression was employed to deconvolute the expression matrix of immune cell subtypes to estimate immune cell abundance. Samples with a confidence threshold of P < .05 were selected.
2.7. Protein–protein interaction (PPI) network construction and analysis
The Search Tool for the Retrieval of Interacting Genes (STRING) database (http://string-db.org/) collects, scores, and integrates all publicly available protein–protein interaction information sources, supplemented by predicted interactions. The list of differentially expressed genes was input into the STRING database to construct a predicted core gene PPI network (confidence > 0.4). Cytoscape software was used for biological network analysis and 2-dimensional (2D) visualization. The PPI network from the STRING database was imported into Cytoscape software for visualization. Modules with the best correlations were identified using MCODE, and 2 algorithms (MCC, DMNC) were used to calculate genes with the best correlations and intersected. The resulting core gene list was visualized and exported.
2.8. Survival analysis
Clinical survival data for gastric cancer were obtained from TCGA. The R package maxstat (version: 0.7-25) was used to calculate the optimal cutoff value for RiskScore of core genes. Groups with a minimum of 25% and a maximum of 75% of samples were set. The patients were divided into high and low groups based on this cutoff, and the survival differences between the 2 groups were further analyzed using the survfit function from the survival R package. The log-rank test was used to evaluate the significance of survival differences between different groups. The forest plot of core genes was created using the R package forest to observe the significant impact of each independent core gene on gastric cancer prognosis.
2.9. Gene expression heatmap
A heatmap of gene expression levels of core genes found in the PPI network was generated using the R package heatmap, visualizing the expression differences between gastric cancer datasets GSE51575 and GSE36076 and normal tissue samples.
2.10. CTD analysis
The Comparative Toxicogenomics Database (CTD) integrates extensive data on interactions between chemicals, genes, phenotypes, and diseases, providing great convenience for studying disease-related environmental exposure factors and potential drug mechanisms. Core genes were input into the CTD website to find the most relevant diseases associated with core genes. Radar plots of gene expression differences were drawn using Excel.
3. Result
3.1. Analysis of DEGs
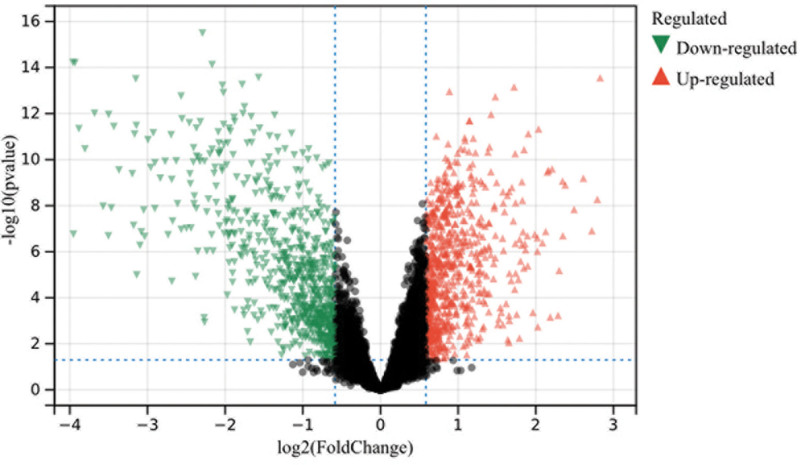
In this study, we identified differential gene expression by batch-corrected merging matrices from GSE51575 and GSE36076, using a preset cutoff value (P < .05). A total of 1457 DEGs were identified through R software, and a volcano plot was generated (Fig. 1).
Figure 1.
Differentially expressed genes (DEGs). A total of 1457 DEGs.
3.2. Functional enrichment analysis
3.2.1. DEGs
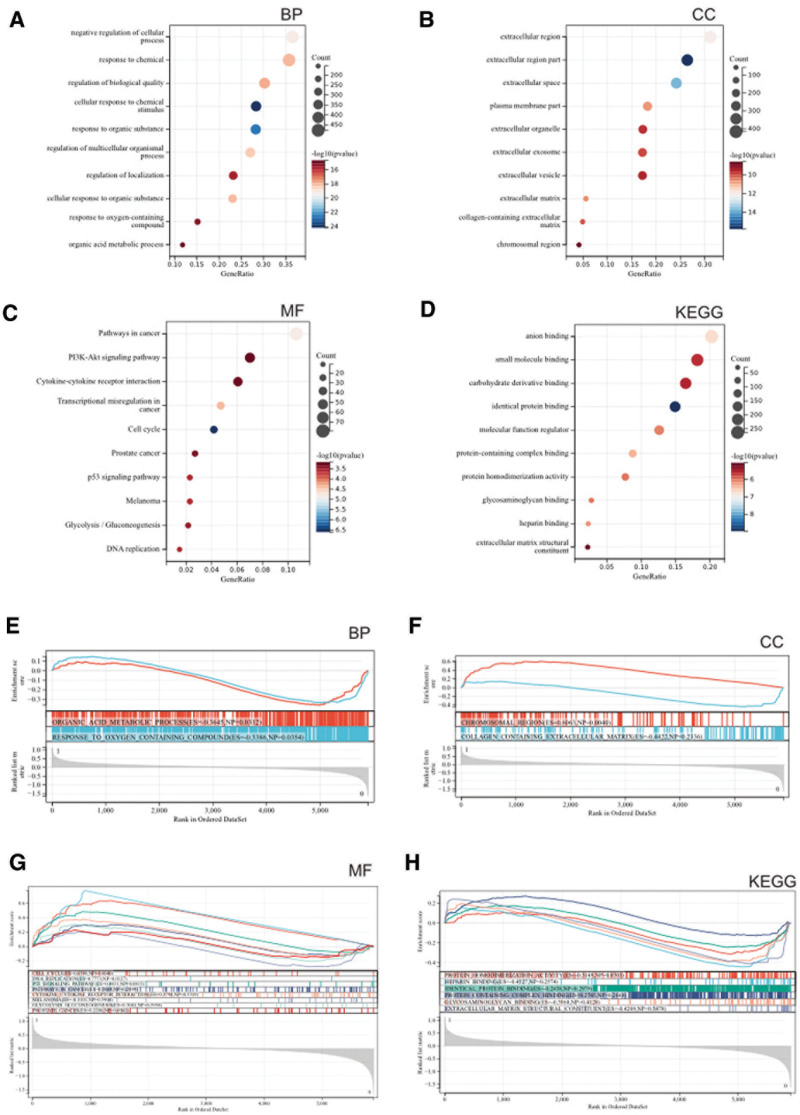
GO and KEGG analyses were performed on these DEGs. According to the GO analysis, they were mainly enriched in organic acid metabolic processes, condensed chromosome kinetochores, and oxidoreductase activity (Fig. 2A–C). In the KEGG analysis, they were primarily concentrated in metabolic pathways, the P53 signaling pathway, and the PPAR signaling pathway (Fig. 2D).
Figure 2.
(A–D) GOKEGG enrichment analysis of DEGs. (A) Biological process analysis. (B) Cellular component analysis. (C) Molecular function analysis. (D) KEGG enrichment analysis. (E–H) GSEA of DEGs. (E) Biological process analysis. (F) Cellular component analysis. (G) Molecular function analysis. (H) KEGG enrichment analysis. DEGs = differentially expressed genes, GSEA = gene set enrichment analysis, KEGG = Kyoto Encyclopedia of Gene and Genome.
3.2.2. GSEA
Additionally, we conducted GSEA on the entire genome to identify potential enrichment items among non-differentially expressed genes and validate the results of differentially expressed genes. The enrichment items were consistent with the GO and KEGG enrichment results of the DEGs, showing enrichment in organic acid metabolic processes, the P53 signaling pathway, and the PPAR signaling pathway (Fig. 2E–H).
3.2.3. Metascape enrichment analysis
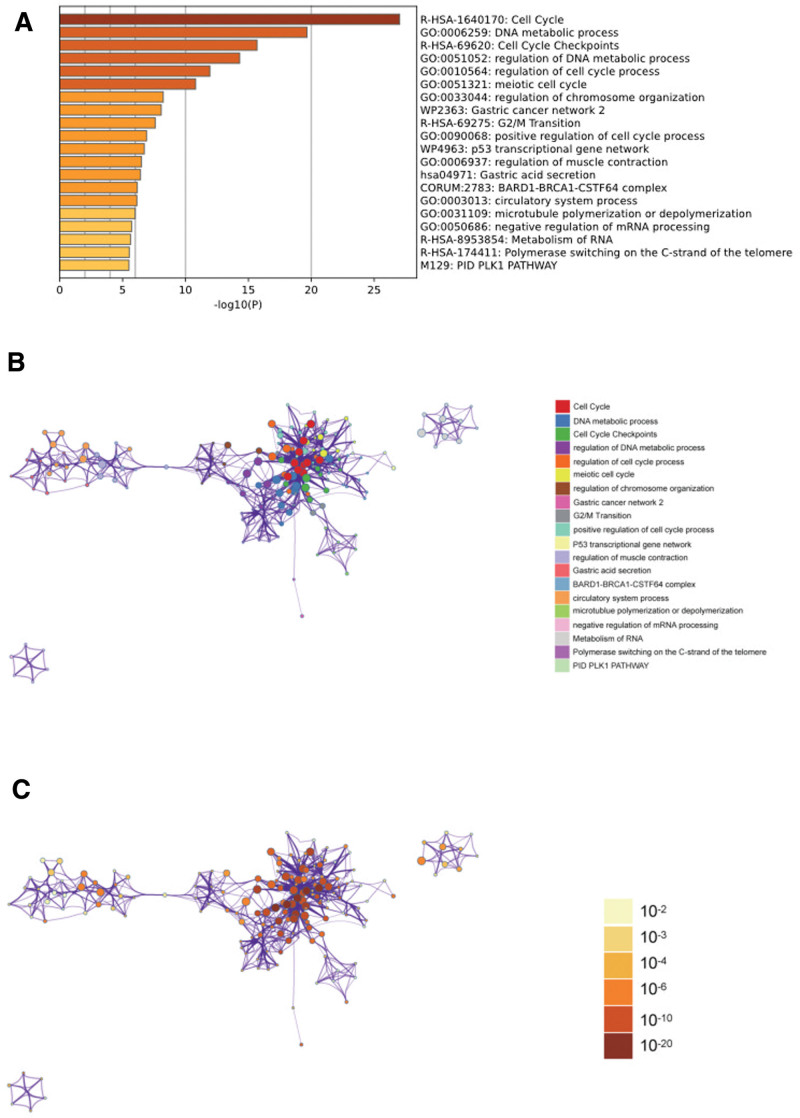
In the metascape enrichment analysis, the GO enrichment item of skeletal system development was observed (Fig. 3A). We also generated enrichment networks colored by enrichment terms and P values, providing visual representations of the associations and confidence levels of various enrichment items (Figs. 3B, C, and 4).
Figure 3.
Metascape enrichment analysis. (A) Bar graph of enriched terms across input gene lists, colored by P values. (B) Network of enriched terms: colored by cluster ID, where nodes that share the same cluster ID are typically close to each other. (C) Colored by P value, where terms containing more genes tend to have a more significant P value.
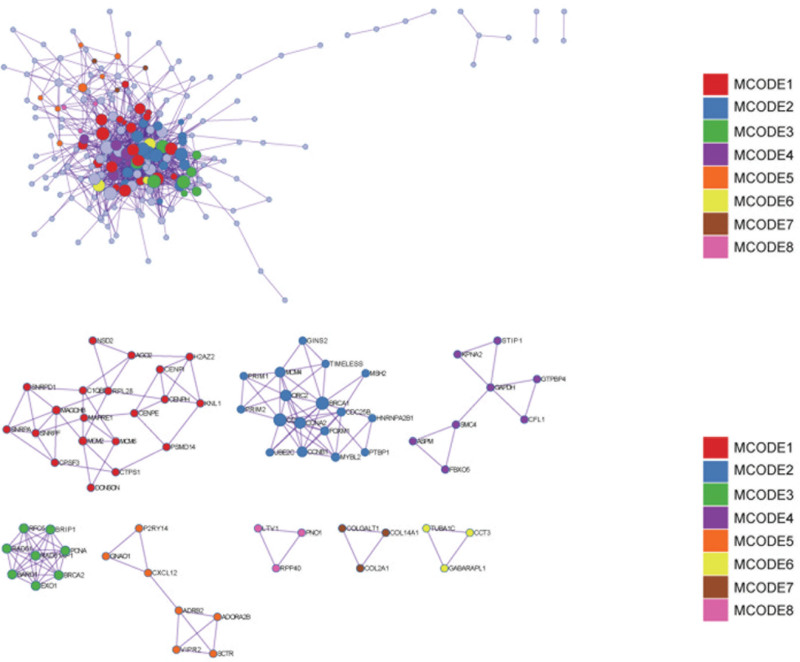
Figure 4.
Protein–protein interaction network. And MCODE components identified in the gene lists.
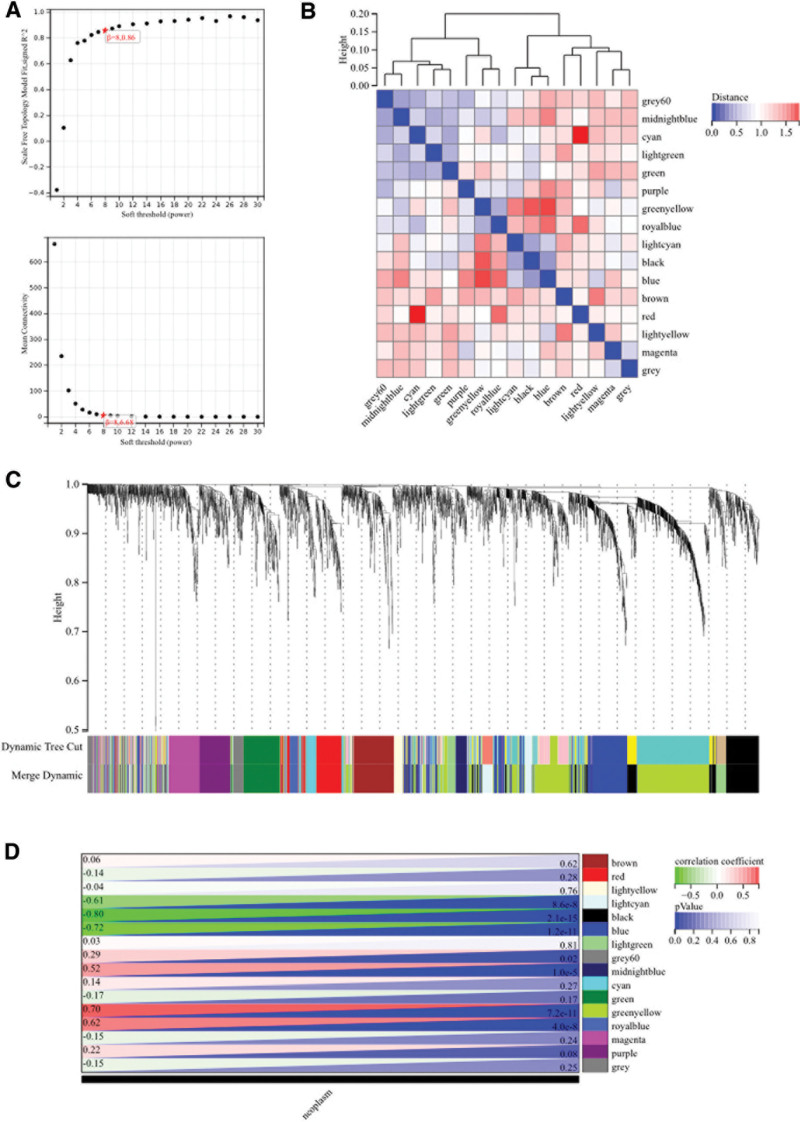
3.3. WGCNA
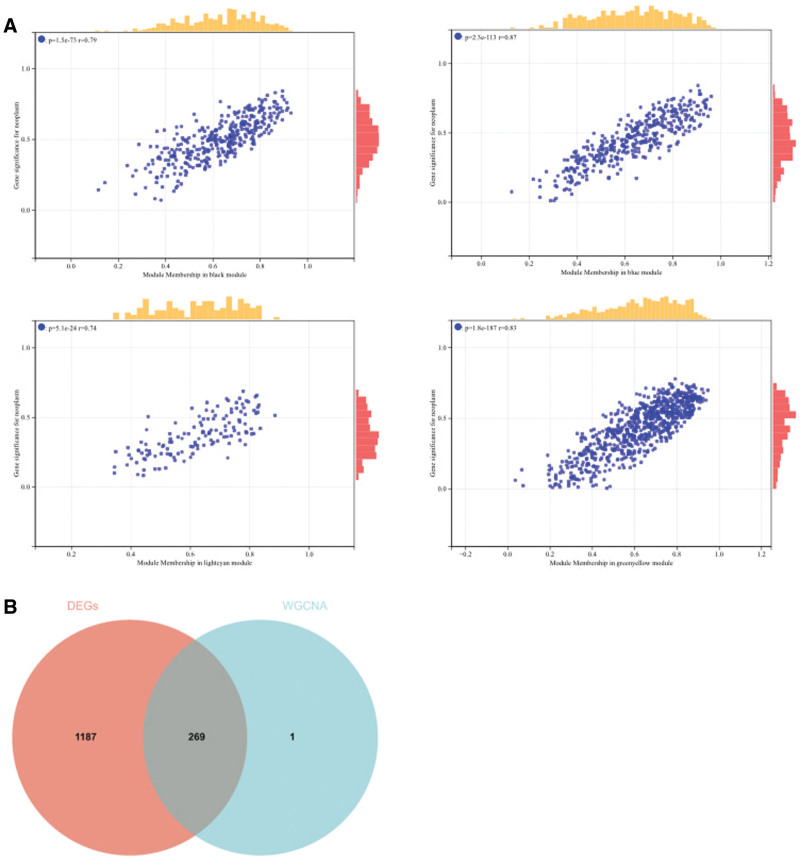
The selection of the soft threshold power is an important step in WGCNA. We performed network topology analysis to determine the soft threshold power, which was set to 8 (Fig. 5A). A total of 16 modules were constructed based on hierarchical clustering of all genes (Fig. 5B), and interactions between important modules were analyzed (Fig. 5C). Furthermore, we generated a heatmap of module-trait correlations (Fig. 5D) and a scatter plot of the correlation between gene significance (GS) and module membership (MM) for hub genes (Fig. 6A). We identified 4 hub genes in clinically significant modules based on the cutoff criterion (|MM| > 0.8). We also generated a Venn diagram by intersecting WGCNA with the DEGs to visualize the overlap (Fig. 6B).
Figure 5.
WGCNA. (A) β = 8,0.86. β = 8, 6.68. (B, C) The hierarchical clustering tree of all genes was constructed, and 38 important modules were generated. (D) The heatmap of correlation between modules and phenotypes.
Figure 6.
(A) The scatter map of correlation between GS and MM of related hub genes. (B) The DEGs screened by WGCNA and DEGs was used to obtain Venn map. Two hundred sixty nine intersection genes were obtained. DEGs = differentially expressed genes, GS = gene significance, MM = module membership, WGCNA = weighted gene co-expression network analysis.
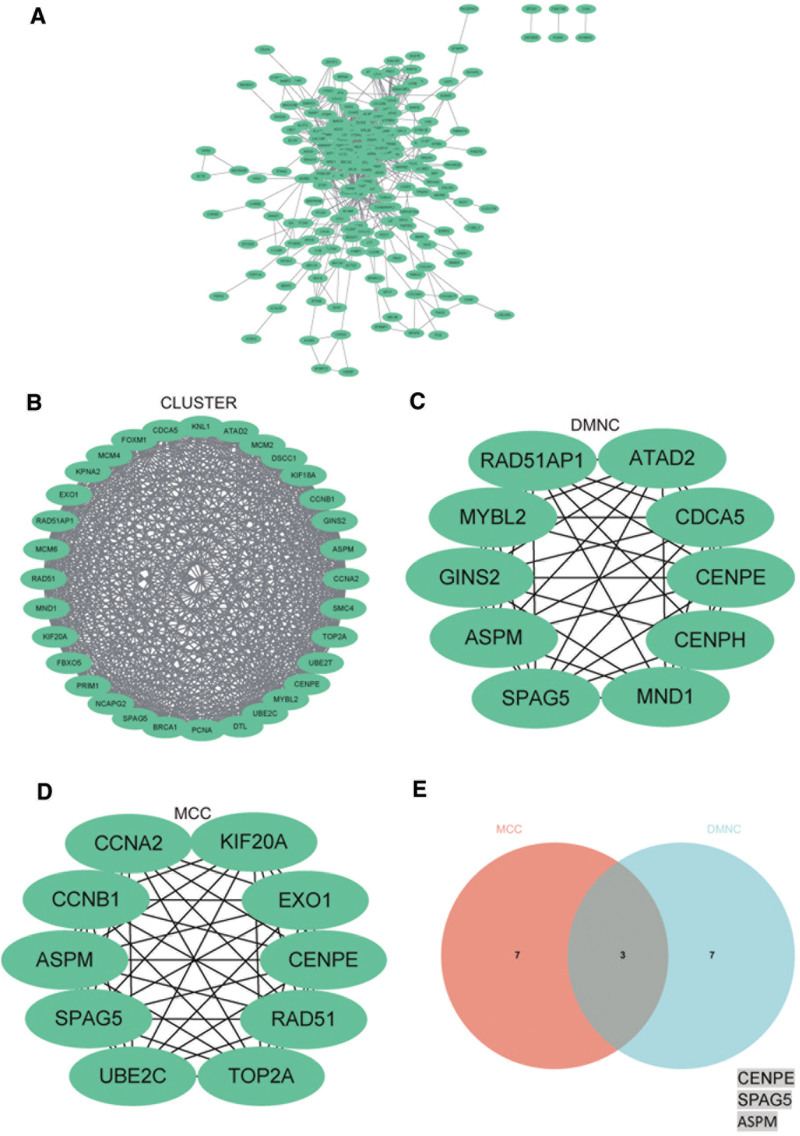
3.4. PPI network construction and analysis
The PPI network of DEGs was constructed using the STRING online database and analyzed using Cytoscape software (Fig. 7A). The core gene cluster was obtained (Fig. 7B). Subsequently, central genes were identified using the MCC and DMNC algorithms (Fig. 7C, D), and a Venn diagram was generated to obtain the union as core genes (Fig. 7E), finally obtaining 3 core genes (CENPE, SPAG5, ASPM).
Figure 7.
Construction and analysis of protein–protein interaction (PPI) networks. (A) PPI network of DEGs. (B) CLUSTER was used to identify the central gene. (C) DMNC was used to identify the central gene. (D) MCC was used to identify the central gene. (E) Three core genes (CENPE, SPAG5, ASPM) were obtained by merging using Venn diagrams. ASPM = abnormal spindle-like microcephaly associated, DEGs = differentially expressed genes, SPAG5 = sperm-associated antigen 5.
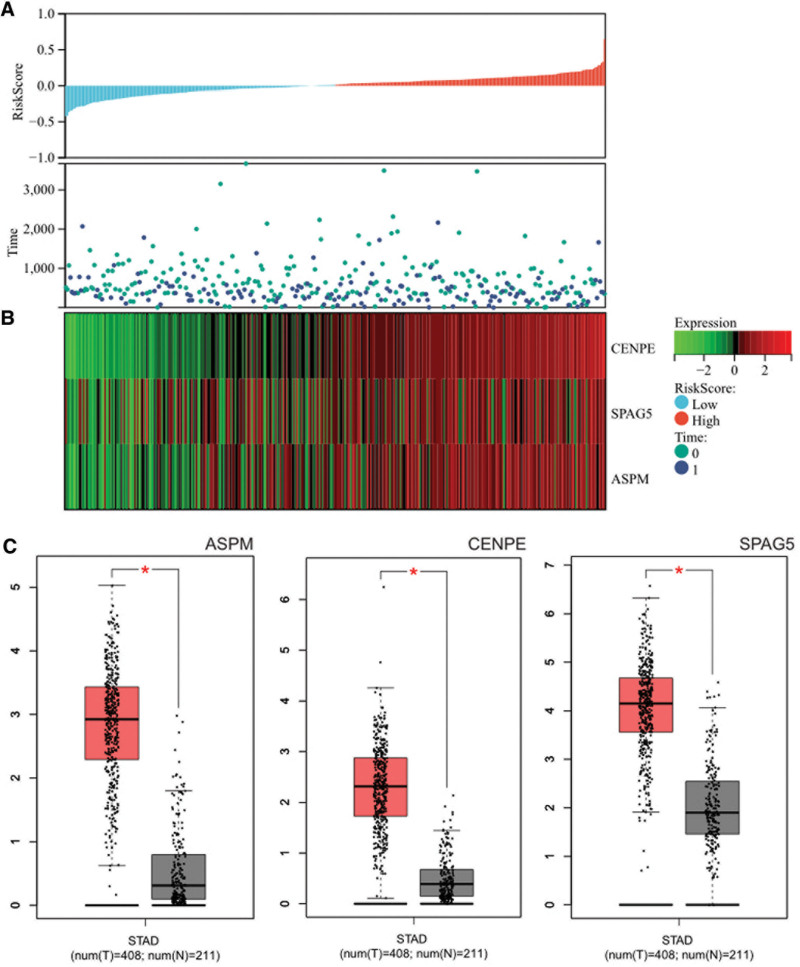
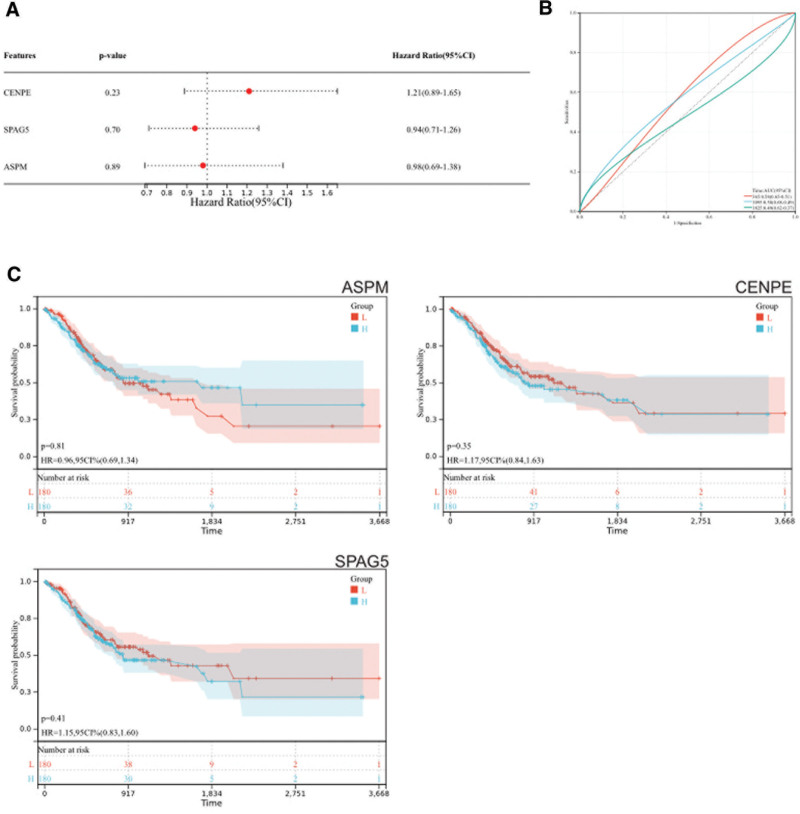
3.5. Survival analysis
We obtained prognosis scoring relationship plots from gastric cancer survival data downloaded from TCGA, showing a significant decrease in patient survival rate with increasing risk scores, with higher survival time and rate in the low-risk group compared to the high-risk group (Fig. 8A). Visualization of the expression level heatmap of core genes in gastric cancer survival data indicated that core genes (CENPE, SPAG5, ASPM) were risk factors, showing an upward trend in expression with increasing risk scores (Fig. 8B). Box plots of core genes in gastric cancer demonstrated significant differences between cancer and normal samples (Fig. 8C). We also obtained forest plots (Fig. 9A), ROC curves (Fig. 9B), and KM survival curves (Fig. 9C) related to core genes (CENPE, SPAG5, ASPM), suggesting a significant association with survival rate and potential prognostic roles in gastric cancer patients.
Figure 8.
Survival analysis. (a) Effect of GSE51575 and GSE36076 on survival time and survival in patients with gastric cell carcinoma. (b) Heatmap of GSE51575 and GSE36076 core gene expression in liver cancer survival data. (c) Box plot of core genes in gastric cancer.
Figure 9.
(A) Overall survival of patients with gastric cancer. (B) ROC risk score curve. (C) Knowledge management survival curve for risk score.
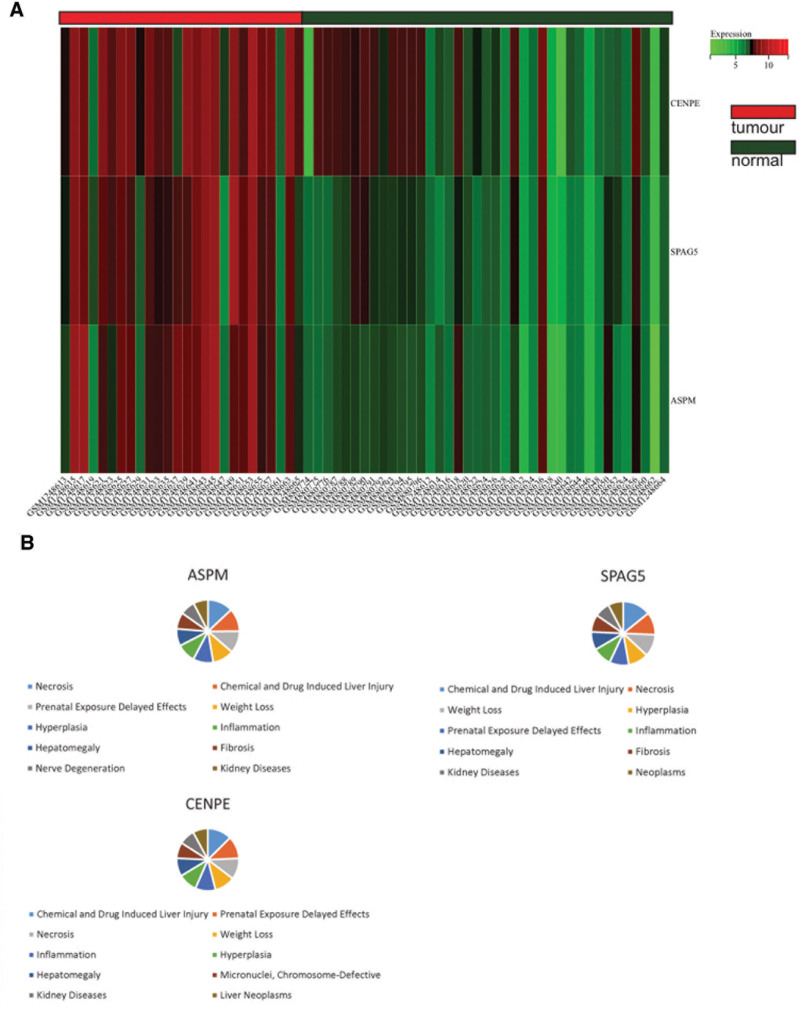
3.6. Heatmap of core gene expression
The expression levels of core genes in the merged matrices of gastric cancer datasets GSE51575 and GSE36076 were visualized and depicted as heatmaps (Fig. 10A). It was observed that core genes (SPAG5, ASPM) were highly expressed in gastric cancer samples compared to normal samples, suggesting their potential regulatory roles in gastric cancer.
Figure 10.
(A) The heatmap depicting the expression levels of differentially expressed genes related to ferroptosis in the merged matrix of GSE51575 and GSE36076 datasets. (B) CTD analysis. Three core genes (CENPE, SPAG5, ASPM) are associated with gastric cancer, gastric cell carcinoma, invasive tumor and end-stage liver disease. ASPM = abnormal spindle-like microcephaly associated, CTD = Comparative Toxicogenomics Database, SPAG5 = sperm-associated antigen 5.
3.7. CTD analysis
In this study, we inputted the hub gene list into the CTD website to identify diseases associated with core genes, enhancing our understanding of the gene-disease associations. Core genes (CENPE, SPAG5, ASPM) were found to be associated with gastric tumors, gastric diseases, gastritis, gastric ulcers, tumors, inflammation, and necrosis (Fig. 10B).
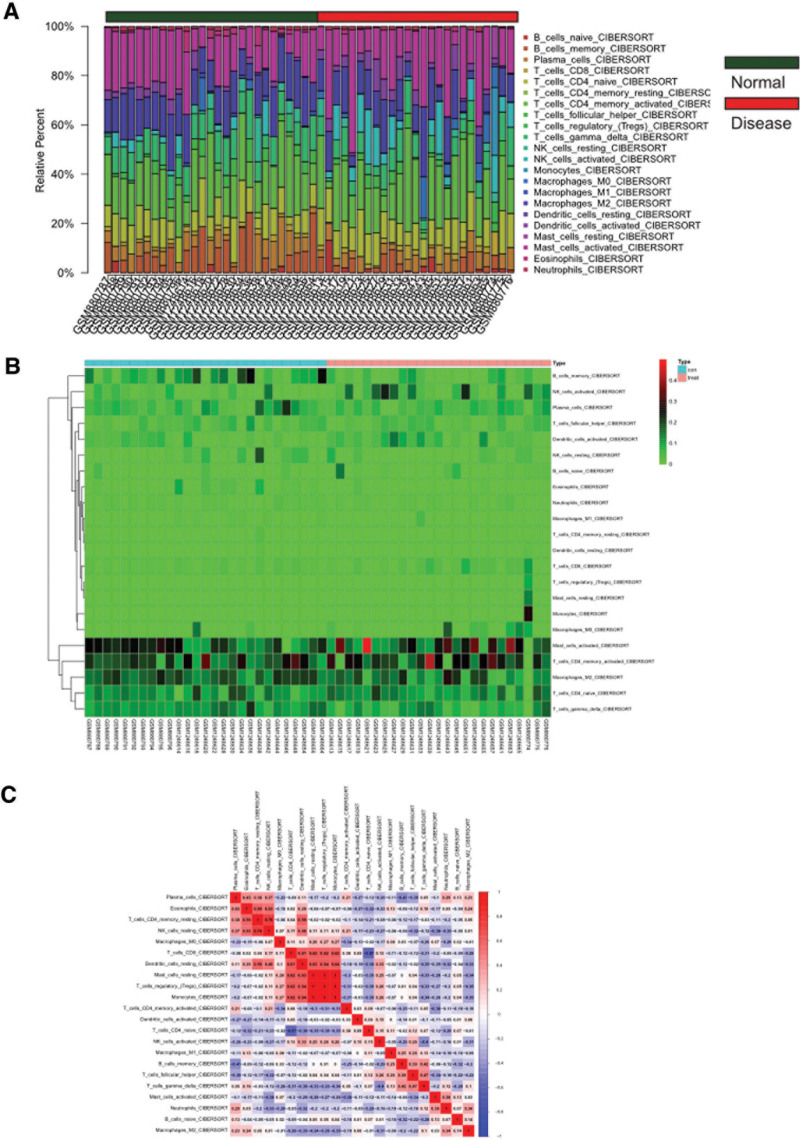
3.8. Immune infiltration analysis
Using the CIBERSORT package, we analyzed the gene expression matrices of gastric cancer datasets GSE51575 and GSE36076 and obtained the proportions of immune cells in the entire gene expression matrix at a 95% confidence level. The results indicated a higher proportion of Mast_cells_activated in the samples (Fig. 11A). Heatmaps of immune cell expression levels in the dataset showed higher expression of Mast_cells_activated in gastric cancer samples (Fig. 11B). Furthermore, we conducted correlation analysis of infiltrating immune cells and obtained a co-expression pattern map of immune cell components, suggesting a highly positive correlation between Monocytes and Mast_cells_resting when Monocytes expression was high, which may affect the progression of gastric cancer (Fig. 11C).
Figure 11.
Immunoinfiltration analysis. (A) Whole gene expression matrix results in proportion of immune cells. (B) Heatmap of immune cell expression in data set. (C) Map of co-expression patterns between immune cell components.
4. Discussion
Gastric cancer is a malignant tumor with serious consequences, posing a threat not only to the health and life of patients but also potentially affecting their psychological well-being, economic status, and social functioning.[14] The main findings of this research indicate that the SPAG5 and ASPM genes are overexpressed in gastric cancer, and higher expression levels of SPAG5 and ASPM correlate with poorer prognosis.
Sperm-associated antigen 5 (SPAG5) is found in various tissues of the human body, including the testes, initially discovered in association with sperm cells. SPAG5 is a spindle apparatus protein involved in cell division, particularly crucial during mitosis, aiding in the organization of the spindle apparatus and chromosome alignment, regulating the separation of sister chromatids to daughter cells.[15,16] Dysfunction or dysregulation of SPAG5 is associated with cancer, with high levels of SPAG5 expression linked to increased proliferation, migration, and invasion of tumor cells, potentially leading to rapid tumor growth and spread.[17] In terms of tumor immunity, high expression of SPAG5 correlates with an immunosuppressive tumor microenvironment and immune therapy efficacy indicators. SPAG5 serves as a diagnostic, prognostic, and immunological marker in various cancers and may offer new targets for tumor immunotherapy.[18]
Related studies indicate that SPAG5 is overexpressed in lung adenocarcinoma tissues, positively correlated with clinical staging, overall survival, recurrence-free survival, and progression-free survival of patients. SPAG5 may be involved in regulating processes such as cell cycle, proliferation, invasion, DNA damage and repair, and tumor immune suppression, playing a role in promoting malignant phenotypes and immune suppression in lung adenocarcinoma.[19] Similarly, SPAG5 is upregulated in osteosarcoma tissues, and its overexpression is significantly associated with malignant phenotypes and poor survival of patients. Knockdown of SPAG5 markedly inhibits the in vitro invasion and migration ability of osteosarcoma cells and the epithelial–mesenchymal transition (EMT) process, while lowering the expression of SPAG5 increases the survival rate of osteosarcoma cells.[20] SPAG5 also promotes proliferation, migration, invasion, and EMT in colon cancer cells through activation of the PI3K/AKT signaling pathway.[21] SPAG5 is highly expressed in lung adenocarcinoma tissues, with its high expression associated with poor prognosis in patients. SPAG5 promotes proliferation, migration, invasion, and autophagy of A549 cells, inhibiting apoptosis.[22] Compared to normal lung epithelial cells, SPAG5 is upregulated in most lung adenocarcinoma cell lines. Knockdown of SPAG5 suppresses the in vitro proliferation, colony formation, and migration of lung adenocarcinoma A549 cells, as well as inhibiting tumor growth in vivo.[23] In breast cancer, SPAG5 silencing inhibits proliferation and invasion of cancer cells, while overexpression of SPAG5 promotes proliferation and invasion. SPAG5 activates the Wnt/β-catenin signaling pathway by upregulating Wnt3 expression, thereby promoting proliferation and invasion of breast cancer cells.[24]
The expression level of SPAG5 is closely associated with the prognosis of gastric cancer patients. Studies have shown that SPAG5 activates the PI3K/AKT pathway, promoting tumor progression and chemoresistance in gastric cancer. High expression of SPAG5 is correlated with poorer survival rates and prognosis. By regulating the PI3K/AKT signaling pathway, SPAG5 promotes the growth of gastric cancer cells and holds promise as a potential target gene for gastric cancer treatment.[25] Furthermore, research indicates that SPAG5 is highly expressed in gastric cancer tissues and associated with poor survival rates in patients. SPAG5 plays a crucial oncogenic role in the development of gastric cancer by activating the Wnt/β-catenin pathway, suggesting its potential as a prognostic and therapeutic target for gastric cancer patients.[26]
Abnormal spindle-like microcephaly associated (ASPM) is a gene in the human genome, encoding a protein related to neurodevelopment.[27] ASPM regulates mitosis, cell proliferation, replication stress response, DNA repair, and tumorigenesis.[8] Mutations or abnormalities in the ASPM gene are associated with microcephaly, a neurodevelopmental disorder characterized by significantly smaller head size than normal.[9] ASPM is a multifunctional gene, with its mutations closely linked to intelligence, neurodevelopment, and tumor occurrence and development.[28]
ASPM interacts with components of DNA repair and replication mechanisms, essential for maintaining chromosome DNA stability.[29] ASPM is associated with the occurrence and development of tumors, with its abnormal expression observed in various cancers. ASPM plays a significant role in regulating cell proliferation and differentiation, potentially promoting cancer development by enhancing tumor cell proliferation and invasion.[30] In multiple tumors, ASPM is crucial for cell replication and tumor progression during the process of mitosis spindle function. ASPM promotes migration and invasion of thyroid undifferentiated carcinoma by stabilizing KIF11.[31] Research indicates that ASPM activates the Hedgehog and Wnt signaling pathways to promote the stemness and progression of small cell lung cancer.[32] ASPM promotes the growth of glioblastoma by regulating the G1 restriction point precursor.[33]
ASPM plays multiple roles in mitosis, cell cycle progression, and DNA double-strand break repair, with tumorigenic ASPM serving as a regulatory hub for development and signaling in cancer.[34] ASPM has synergistic effects with PARP inhibition in killing tumor cells, making it a potential new target for lethal tumor combination therapy.[35] Whole-genome analysis reveals that upregulation of ASPM is associated with gastric cancer, and the E2F1-ASPM axis may represent a new mechanism for gastric cancer occurrence.[36] ASPM may serve as an overexpressed gastric stem/progenitor cell marker in cancer, with expression profiling and immunohistochemistry demonstrating increased expression of ASPM and transcription factor E2F1 in human gastric cancer cell lines, showing correlation between them. The association between ASPM and transcription factor E2F1 in gastric tissue is relevant, indicating their involvement in crucial cell fate regulation mechanisms. ASPM, as a potential new stem/progenitor cell marker, may participate in normal gastric physiology and gastric cancer occurrence.[37] High levels of ASPM expression correlate with enhanced tumor invasiveness and metastatic ability, suggesting its involvement in the development and progression of gastric cancer.
4.1. Future perspectives
Despite conducting rigorous bioinformatics analysis, this study still has some limitations. Animal experiments involving gene overexpression or knockout were not performed to further validate their functions. Given the significant roles of SPAG5 and ASPM in gastric cancer, future research could delve into exploring to deepen understanding and potentially improve clinical management. Further elucidating the molecular mechanisms through which SPAG5 and ASPM promote gastric cancer initiation, progression, and metastasis could uncover new therapeutic targets. Investigating their interactions with other genes, proteins, and signaling pathways involved in gastric cancer onset could provide valuable insights. Evaluating the potential of SPAG5 and ASPM as diagnostic biomarkers for gastric cancer, especially in conjunction with other established markers, could strengthen early detection strategies. Additionally, assessing their utility as prognostic indicators to predict patient outcomes and guide treatment decisions could be valuable. Translating the efficacy and safety of SPAG5 and ASPM-targeted therapies into clinical trials in gastric cancer patients is a crucial step. Collaboration among researchers, clinicians, and industry partners in multidisciplinary teams is essential for successful clinical translation. Exploring the potential of SPAG5 and ASPM as targets for personalized therapeutic interventions based on individual tumor molecular characteristics could enhance the effectiveness of treatment strategies.
5. Conclusion
In summary, the expression levels of SPAG5 and ASPM genes are elevated in gastric cancer. SPAG5 and ASPM genes may play crucial roles in the occurrence and development of gastric cancer, potentially serving as potential prognostic markers. Thus, targeting the expression regulation of SPAG5 and ASPM genes may become an important direction for the treatment and prognosis assessment of gastric cancer.
Author contributions
Conceptualization: Mei Xue, Chao Ma, HaiFeng Shan.
Methodology: Mei Xue, Chao Ma, HaiFeng Shan, Shiyang Hou, Chunbo Kang.
Validation: Mei Xue, Chao Ma, HaiFeng Shan.
Visualization: Mei Xue, Chao Ma, HaiFeng Shan, Shiyang Hou, Chunbo Kang.
Writing – review & editing: Mei Xue, Chao Ma, HaiFeng Shan.
Software: HaiFeng Shan.
Data curation: Shiyang Hou, Chunbo Kang.
Writing – original draft: Shiyang Hou, Chunbo Kang.
Formal analysis: Chunbo Kang.
Abbreviations:
- ASPM
- abnormal spindle-like microcephaly associated
- CTD
- Comparative Toxicogenomics Database
- DEGs
- differentially expressed genes
- FC
- fold change
- FDR
- false discovery rate
- GEO
- gene expression omnibus
- GO
- gene ontology
- GS
- gene significance
- GSEA
- gene set enrichment analysis
- KEGG
- Kyoto Encyclopedia of Gene and Genome
- MAD
- Median Absolute Deviation
- MM
- module membership
- PPI
- protein–protein interaction
- SPAG5
- sperm-associated antigen 5
- STRING
- Search Tool for the Retrieval of Interacting Genes
- TOM
- topological overlap matrix
- WGCNA
- weighted gene co-expression network analysis.
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
The authors have no funding and conflicts of interest to disclose.
The data in this article are from public databases and are exempt from ethical review.
How to cite this article: Xue M, Ma C, Shan H, Hou S, Kang C. SPAG5 and ASPM play important roles in gastric cancer: An observational study. Medicine 2024;103:24(e38499).
MX, CM, and HS contributed equally to this work.
Contributor Information
Mei Xue, Email: xmacq@163.com.
Chao Ma, Email: mysicwal@sina.com.
HaiFeng Shan, Email: 13811945898@163.com.
Shiyang Hou, Email: houshiyang23117@163.com.
References
- [1].Smyth EC, Nilsson M, Grabsch HI, van Grieken NC, Lordick F. Gastric cancer. Lancet. 2020;396:635–48. [DOI] [PubMed] [Google Scholar]
- [2].Röcken C. Predictive biomarkers in gastric cancer. J Cancer Res Clin Oncol. 2023;149:467–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].López MJ, Carbajal J, Alfaro AL, et al. Characteristics of gastric cancer around the world. Crit Rev Oncol Hematol. 2023;181:103841. [DOI] [PubMed] [Google Scholar]
- [4].Machlowska J, Baj J, Sitarz M, Maciejewski R, Sitarz R. Gastric cancer: epidemiology, risk factors, classification, genomic characteristics and treatment strategies. Int J Mol Sci . 2020;21:4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Thrift AP, El-Serag HB. Burden of gastric cancer. Clin Gastroenterol Hepatol. 2020;18:534–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Guan WL, He Y, Xu RH. Gastric cancer treatment: recent progress and future perspectives. J Hematol Oncol. 2023;16:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Gao X, Bu H, Gao X, Wang Y, Wang L, Zhang Z. Pan-cancer analysis: SPAG5 is an immunological and prognostic biomarker for multiple cancers. FASEB J. 2023;37:e23159. [DOI] [PubMed] [Google Scholar]
- [8].Wu X, Li Z, Wang ZQ, Xu X. The neurological and non-neurological roles of the primary microcephaly-associated protein ASPM. Front Neurosci. 2023;17:1242448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Verloes A, Drunat S, Passemard S. ASPM Primary Microcephaly. Seattle, WA: GeneReviews(®); 2020. [PubMed] [Google Scholar]
- [10].Wang Y, Zhao Y, Bollas A, Wang Y, Au KF. Nanopore sequencing technology, bioinformatics and applications. Nat Biotechnol. 2021;39:1348–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Azad RK, Shulaev V. Metabolomics technology and bioinformatics for precision medicine. Brief Bioinform. 2019;20:1957–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Uesaka K, Oka H, Kato R, et al. Bioinformatics in bioscience and bioengineering: recent advances, applications, and perspectives. J Biosci Bioeng. 2022;134:363–73. [DOI] [PubMed] [Google Scholar]
- [13].Gauthier J, Vincent AT, Charette SJ, Derome N. A brief history of bioinformatics. Brief Bioinform. 2019;20:1981–96. [DOI] [PubMed] [Google Scholar]
- [14].Petryszyn P, Chapelle N, Matysiak-Budnik T. Gastric cancer: where are we heading. Dig Dis. 2020;38:280–5. [DOI] [PubMed] [Google Scholar]
- [15].Li H, Qin Y, Huang Y, Wang J, Ren B. SPAG5, the upstream protein of Wnt and the target of curcumin, inhibits hepatocellular carcinoma. Oncol Rep. 2023;50:86–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Zhu C, Menyhart O, Győrffy B, He X. The prognostic association of SPAG5 gene expression in breast cancer patients with systematic therapy. BMC Cancer. 2019;19:1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Wang C, Su H, Cheng R, Ji H. SPAG5 is involved in human gliomagenesis through the regulation of cell proliferation and apoptosis. Front Oncol. 2021;11:673780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Chen W, Chen X, Li S, Ren B. Expression, immune infiltration and clinical significance of SPAG5 in hepatocellular carcinoma: a gene expression-based study. J Gene Med. 2020;22:e3155. [DOI] [PubMed] [Google Scholar]
- [19].Xiao G, Xu X, Chen Z, Zeng J, Xie J. SPAG5 expression predicts poor prognosis and is associated with adverse immune infiltration in lung adenocarcinomas. Clin Med Insights Oncol. 2023;17:11795549231199915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Li Z, Li H, Chen J, et al. SPAG5 promotes osteosarcoma metastasis via activation of FOXM1/MMP2 axis. Int J Biochem Cell Biol. 2020;126:105797. [DOI] [PubMed] [Google Scholar]
- [21].Zhang X, Wu W, Li X, He F, Zhang L. SPAG5 promotes the proliferation, migration, invasion, and epithelial–mesenchymal transformation of colorectal cancer cells by activating the PI3K/AKT signaling pathway. Chin J Physiol. 2023;66:365–71. [DOI] [PubMed] [Google Scholar]
- [22].Huang R, Li A. SPAG5 is associated with unfavorable prognosis in patients with lung adenocarcinoma and promotes proliferation, motility and autophagy in A549 cells. Exp Ther Med. 2020;20:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Wang T, Li K, Song H, et al. p53 suppression is essential for oncogenic SPAG5 upregulation in lung adenocarcinoma. Biochem Biophys Res Commun. 2019;513:319–25. [DOI] [PubMed] [Google Scholar]
- [24].Jiang J, Wang J, He X, et al. High expression of SPAG5 sustains the malignant growth and invasion of breast cancer cells through the activation of Wnt/β-catenin signalling. Clin Exp Pharmacol Physiol. 2019;46:597–606. [DOI] [PubMed] [Google Scholar]
- [25].An J, Yang L, Pan Y, et al. SPAG5 Activates PI3K/AKT Pathway and promotes the tumor progression and chemo-resistance in gastric cancer. DNA Cell Biol. 2022;41:893–902. [DOI] [PubMed] [Google Scholar]
- [26].Liu G, Liu S, Cao G, et al. SPAG5 contributes to the progression of gastric cancer by upregulation of Survivin depend on activating the wnt/β-catenin pathway. Exp Cell Res. 2019;379:83–91. [DOI] [PubMed] [Google Scholar]
- [27].Vange P, Bruland T, Bakke I. Authors’ reply – Re: Wang et al. Controversial role of the possible oxyntic stem cell marker ASPM in gastric cancer. J Pathol. 2017;241:562–3. [DOI] [PubMed] [Google Scholar]
- [28].Wang F, Li J, Liu J, Zhao Q. Controversial role of the possible oxyntic stem cell marker ASPM in gastric cancer. J Pathol. 2017;241:559–61. [DOI] [PubMed] [Google Scholar]
- [29].Razuvaeva AV, Graziadio L, Palumbo V, et al. The multiple mitotic roles of the ASPM orthologous proteins: insight into the etiology of ASPM-Dependent Microcephaly. Cells. 2023;12:922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Hsu CC, Liao WY, Chang KY, et al. A multi-mode Wnt- and stemness-regulatory module dictated by FOXM1 and ASPM isoform I in gastric cancer. Gastric Cancer. 2021;24:624–39. [DOI] [PubMed] [Google Scholar]
- [31].Fang Q, Li Q, Qi Y, Pan Z, Feng T, Xin W. ASPM promotes migration and invasion of anaplastic thyroid carcinoma by stabilizing KIF11. Cell Biol Int. 2023;47:1209–21. [DOI] [PubMed] [Google Scholar]
- [32].Cheng LH, Hsu CC, Tsai HW, et al. ASPM activates hedgehog and wnt signaling to promote small cell lung cancer stemness and progression. Cancer Res. 2023;83:830–44. [DOI] [PubMed] [Google Scholar]
- [33].Chen X, Huang L, Yang Y, et al. ASPM promotes glioblastoma growth by regulating G1 restriction point progression and Wnt-β-catenin signaling. Aging (Albany NY). 2020;12:224–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Tsai KK, Bae BI, Hsu CC, Cheng LH, Shaked Y. Oncogenic ASPM is a regulatory hub of developmental and stemness signaling in cancers. Cancer Res. 2023;83:2993–3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Xu S, Wu X, Wang P, Cao SL, Peng B, Xu X. ASPM promotes homologous recombination-mediated DNA repair by safeguarding BRCA1 stability. iScience. 2021;24:102534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Zhu HH, Zhuang G, Gao WQ. A candidate gastric stem/progenitor cell marker revealed by genome-wide analysis. J Pathol. 2016;238:3–6. [DOI] [PubMed] [Google Scholar]
- [37].Vange P, Bruland T, Beisvag V, et al. Genome-wide analysis of the oxyntic proliferative isthmus zone reveals ASPM as a possible gastric stem/progenitor cell marker over-expressed in cancer. J Pathol. 2015;237:447–59. [DOI] [PMC free article] [PubMed] [Google Scholar]