Fig. 1. Programmable deletions of RNA with RNA-guided type III-A CRISPR complexes.
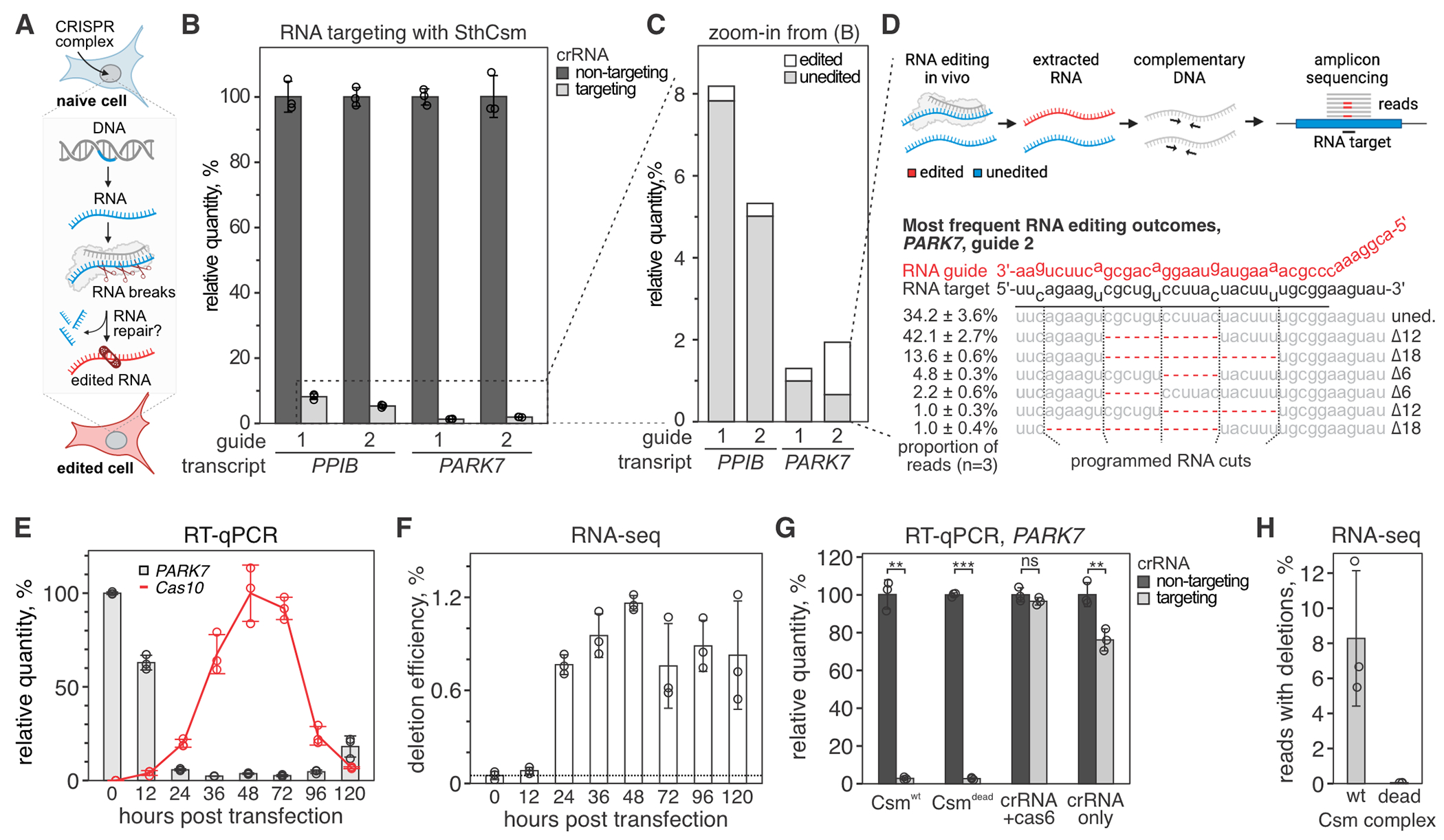
(A) Diagram of RNA editing in eukaryotic cells through sequence-specific RNA cleavage and RNA repair. (B) Human cells (293T) were transfected with plasmids encoding for NLS-tagged type III CRISPR complex of Streptococcus thermophilus (SthCsm) and RNA guides targeting PPIB or PARK7 messenger RNAs. Target transcripts were quantified with RT-qPCR, and the qPCR signal was normalized to ACTB and a non-targeting guide RNA control. Data is shown as mean ± SD of three biological replicates. (C) Zoom-in from panel B. Deep sequencing was used to quantify the proportion of signal that is derived from edited RNA. (D) Top: schematics of deep sequencing approach used to quantify RNA editing. Bottom: top five most frequent RNA editing outcomes in PARK7 transcript (guide 2). Dotted lines indicate the positions of RNA breaks by the SthCsm complex. Data is shown as mean ± SD. (E) Kinetics of PARK7 mRNA knockdown (guide 2) vs. NLS-tagged SthCsm complex expression. PARK7 qPCR signal was normalized to ACTB and 0 h time point. Cas10 expression was normalized to ACTB and maximum expression level at 48 h. Data is shown as mean ± SD of three biological replicates. (F) PARK7 qPCR products in (E) were sequenced, and deletion efficiency was calculated as [relative quantity] × [fraction of reads with deletions]. (G) PARK7 knockdown efficiencies with the “wildtype” NLS-tagged SthCsm complex (Csmwt), catalytically inactive NLS-tagged SthCsm complex (Csmdead), crRNA expressed only with Cas6 gene (no csm genes), and cRNA alone were measured using RT-qPCR. Welch’s t-test was used to compare samples expressing targeting and non-targeting crRNA. ** - p < 0.01, *** - p < 0.001, ns – non-significant. (H) RT-qPCR products in (G) were sequenced, and a fraction of reads with programmable deletions was quantified.
