Abstract
Jembrana disease virus (JDV) is a bovine lentivirus genetically similar to bovine immunodeficiency virus; it causes an acute and sometimes fatal disease in infected animals. This virus carries a very potent Tat that can strongly activate not only its own long terminal repeat (LTR) but also the human immunodeficiency virus (HIV) LTR. In contrast, HIV Tat cannot reciprocally activate the JDV LTR (H. Chen, G. E. Wilcox, G. Kertayadnya, and C. Wood, J. Virol. 73:658–666, 1999). This indicates that in transactivation JDV Tat may utilize a mechanism similar to but not the same as that of the HIV Tat. To further study the similarity of JDV and HIV tat in transactivation, we first tested the responses of a series of HIV LTR mutants to the JDV Tat. Cross-transactivation of HIV LTR by JDV Tat was impaired by mutations that disrupted the HIV type 1 transactivation response element (TAR) RNA stem-loop structure. Our results demonstrated that JDV Tat, like HIV Tat, transactivated the HIV LTR at least partially in a TAR-dependent manner. However, the sequence in the loop region of TAR was not as critical for the function of JDV Tat as it was for HIV Tat. The competitive inhibition of Tat-induced transactivation by the truncated JDV or HIV Tat, which consisted only of the activation domain, suggested that similar cellular factors were involved in both JDV and HIV Tat-induced transactivation. Based on the one-round transfection assay with HIV tat mutant proviruses, the cotransfected JDV tat plasmid can functionally complement the HIV tat defect. To further characterize the effect of JDV Tat on HIV, a stable chimeric HIV carrying the JDV tat gene was generated. This chimeric HIV replicated in a T-cell line, C8166, and in peripheral blood mononuclear cells, which suggested that JDV Tat can functionally substitute for HIV Tat. Further characterization of this chimeric virus will help to elucidate how JDV Tat functions and to explain the differences between HIV and JDV Tat transactivation.
Lentiviruses are a group of retroviruses usually associated with slowly developing diseases with a number of different clinical manifestations of infection in different virus-host systems (13, 14). For example, human immunodeficiency virus (HIV) causes immunodeficiency and ultimately leads to the death of infected patients several years after infection (14). In contrast, a recently characterized bovine lentivirus, Jembrana disease virus (JDV), causes an acute disease in infected animals after a short incubation period (7, 60, 61). It is probable that an understanding of the differences in molecular mechanisms involved in the pathogenesis of different lentiviruses will provide important insights into methods of controlling lentivirus-induced diseases such as AIDS.
The genomes of all lentiviruses contain three major structural genes, gag, pol, and env, as well as several accessory and regulatory genes flanked by the two long terminal repeats (LTRs) (14). In HIV, at least six accessory and regulatory genes, tat, rev, nef, vif, vpr, and vpu, have been identified previously (14). Studies of these accessory genes have suggested that most of them are involved in viral replication and pathogenesis (19, 33, 39, 42, 44, 46, 57). The HIV tat gene has been extensively studied and is an essential determinant of viral replication and the pathogenesis of infection (3, 4, 12, 16, 18, 31, 58, 62). Tat is a potent transactivator for HIV gene expression (3, 4) and has also been shown to modulate the expression of cellular genes, such as those for major histocompatibility complex class I (30), tumor necrosis factor alpha (8, 48), interleukin-2 (IL-2) (59), IL-6 (53), and several extracellular matrix proteins (54). The Tat protein is released from HIV type 1 (HIV-1)-infected cells and can be detected in sera of HIV-1-infected individuals (24). Extracellular Tat may be involved in suppression of the host immune response and cellular disorders associated with AIDS pathology (2, 23). Tat can synergize with cellular basic fibroblast growth factor and may be involved in the induction of Kaposi's sarcoma lesions (6, 22, 23). In addition, Tat may participate in the induction of apoptosis in lymphocytes and contribute to the depletion of the CD4+ T cells in AIDS patients (40, 43, 58). The role of Tat in HIV pathogenesis may not be limited to its transactivation function (31). Tat may also play a role in viral replication. Indeed, Tat has recently been shown to be required for efficient HIV-1 reverse transcription (27, 28, 29). Thus, there is ample evidence to suggest that the HIV Tat is a pleiotropic protein, but how Tat is involved in HIV pathogenesis is still an enigma. Further understanding of the molecular basis for how Tat plays a role in viral replication and viral pathogenesis would be of fundamental importance to our understanding of lentiviruses.
JDV is a recently identified lentivirus (9, 10, 61) related to bovine immunodeficiency virus (BIV), but in contrast to BIV, it causes an acute disease atypical of most lentivirus infections (7, 60, 61). JDV infection of cattle can lead to death of the animal within 1 to 2 weeks (60, 61). The virus replicates to extremely high titers in infected animals, and since Tat plays an important role in viral replication, we hypothesized that JDV encodes a potent transactivator, Tat, responsible for its robust replication activity in vivo. In previous studies, we have shown that JDV encodes a Tat that can strongly activate not only the JDV LTR but also the LTRs of other lentiviruses, including the HIV LTR (11). Other than BIV Tat, JDV Tat is the only other nonprimate viral Tat to effectively transactivate the HIV LTR (41), which implies that similar mechanisms may be shared between HIV and the bovine lentivirus Tat proteins. It also suggests that bovine lentiviruses may have a close evolutionary relationship with primate lentiviruses, at least in terms of transactivation mechanism, and may be used as a model to further dissect the molecular mechanism of Tat transactivation and its role in the pathogenesis of lentivirus infections.
To further study the similarities and differences between the JDV and the HIV Tat, we investigated the transactivation of the HIV LTR by the JDV Tat. As expected, JDV Tat transactivated HIV LTR at least partially in a TAR-dependent manner, and this may involve similar mechanisms and common cellular factors in the two systems. Moreover, we demonstrated that JDV Tat functionally substituted for the HIV Tat when the JDV tat gene was introduced into the HIV genome. Our study provides additional evidence that the JDV Tat is a potent transactivator and indicates that further characterization of the molecular mechanism involved in transactivation by JDV Tat is warranted.
MATERIALS AND METHODS
Plasmids.
Several HIV and JDV LTR promoter constructs were used in this study. A series of HIV LTR mutants with deletions and mutations in the TAR region of the LTR, TARCAT and Mod1CAT to Mod7CAT, were used. The clone TARCAT, used as a positive control, contained the HIV 5′ LTR region (nucleotide [nt] −121 to +82, encompassing the intact TAR sequence) cloned in front of the indicator chloramphenicol acetyltransferase (CAT). Additional mutant HIV LTR promoter constructs used were derivatives of TARCAT; these were used previously to test their responsiveness to Tat transactivation (25). A pHIV-CAT clone that contained the intact HIV U3R region was described previously (35). The constructs pHTLV-CAT and pCMV-Tax were a kind gift from Fatah Kashanchi (37). The mutant HIV Tat construct pΔCys1-4 used was described previously (26). The JDV promoter construct, pJU3R-CAT, was constructed as previously described (14); this clone was generated by insertion of the CAT gene into a site 3′ of the U3R region of the JDV LTR.
Various HIV and JDV tat expression constructs were generated using the expression plasmid pUC-RSV. The pUC-RSV plasmid was generated by inserting the Rous sarcoma virus LTR and the bovine growth hormone polyadenylation signal sequence from pRc/RSV (Invitrogen, Carlsbad, Calif.) into pUC18. The HIV or the JDV tat exon 1 was PCR amplified and inserted into vector pUC-RSV to generate pUC-Htat and pUC-Jtat, both of which encoded the transactivation-competent Tat peptides. The truncated HIV tat (corresponding to amino acids 1 to 50) and the truncated JDV tat sequences (corresponding to amino acids 1 to 67) were cloned into pUC-RSV to generate plasmids pUC-H50 and pUC-J67. These two plasmids encoded truncated peptides which had lost their transactivation abilities. Two tat mutant proviral clones, pMtat(−) and pMtat30, were obtained from the AIDS Research and References Program National Institute of Allergy and Infectious Diseases [NIAID], National Institutes of Health, Bethesda, Md.): pMtat(−) contains a termination codon (TGA) in place of the ATG initiation codon and is unable to synthesize Tat (49); pMtat30, which carries a Cys→Gly substitution within the cysteine-rich region, produces a missense Tat (50).
The HIV infectious proviral clone pNL43 was obtained from the AIDS Repository (NIAID) (1). This proviral clone was used to generate other modified HIV proviral clones. The tat gene mutant pNL-tatm proviral plasmid was generated by introducing a stop codon into the tat gene of pNL43 using a PCR-based mutagenesis method (5). This mutation was verified by EcoRI restriction enzyme digestion, since a new EcoRI site was generated in the mutated tat gene (TGG AAG CAT mutated to TGA ATT CAT). Using this plasmid, an NdeI restriction site was introduced into the ATG site of the nef gene and was then used to construct another modified proviral clone that carried the JDV tat gene. The plasmid pNL-Jtat contained the JDV tat gene, in which the NdeI/XhoI fragment of the nef gene of pNL-tatm was replaced by the JDV tat gene exon 1 sequences. In addition, a random stretch of sequence (32 bp) was introduced into the nef gene of pNL-tatm to generate pNL-nef, which was used as the negative control in the various experiments. As a control, the plasmid Tat(−)1ex, which contains the HIV tat in place of the nef gene, was constructed using a similar strategy (45), and was kindly provided by K. T. Jeang (NIAID).
Transfection, virus stock, and CAT ELISA.
CV-1, HeLa, and 293T cells were maintained in Dulbecco's modified Eagle's medium with 10% fetal calf serum. As described previously (11), 2 × 105 cells were seeded into 60-mm-diameter dishes 24 h prior to transfection. The cells were transfected with 1 to 2 μg of CsCl-purified plasmid DNA using Lipofectamine (GIBCO BRL, Grand Island, N.Y.). At 48 h after transfection, the cells were harvested and lysed, and the amount of protein in the lysate was determined using a bicinchoninic acid protein assay kit (Pierce, Rockford, Ill.). For cells transfected with the promoter CAT construct plasmids, the level of CAT expression was determined by a commercial CAT enzyme-linked immunosorbent assay (ELISA) (Boehringer Mannheim, Indianapolis, Ind.) according to the protocols described by the manufacturer.
Stocks of infectious virus were produced by transfection of cells with proviral DNA. The medium from the transfected cells was collected 48 h posttransfection, the cells and cellular debris were removed by centrifugation, and the supernatant was used as a source of infectious virus. The titer of virus was measured by the p24 assay according to protocols described by the manufacturer (NEN Life Sciences Products, Boston, Mass.).
Viral infection and p24 assays.
Human peripheral blood mononuclear cells (PBMC) were maintained in RPMI 1640 with 10% fetal bovine serum and IL-2 (2 U/ml) (Pharmacia, Piscataway, N.J.) and stimulated with phytohemagglutinin (at a final concentration of 250 ng/ml) for 1 to 3 days prior to infection. For each infection, 2 × 106 PBMC in a 0.5-ml volume were incubated with virus at 37°C for 2 h. The virus-containing medium was then decanted and replaced with fresh RPMI medium containing IL-2 (2 U/ml). The infected cultures were sampled every 6 to 7 days, and the amount of virus produced was measured by p24 assay.
Sequence analysis.
The alignment of Tat amino acid sequences was performed using the Clustal W program in the GCG package (Madison, Wis.). Phylogenetic reconstruction trees were generated using the Evolution program in the GCG package. The Distance program (with Kimura's formula) was used to generate a pairwise matrix of the evolutionary distance of the amino acid sequences. Phylogenetic trees were constructed from the same distance matrices with the Grow-tree program (neighbor-joining algorithm or unweighted pair group method with averages [UPGMA]).
RESULTS
Transactivation of HIV LTR by JDV Tat is TAR dependent.
Our previous study demonstrated that JDV Tat was a potent transactivator which could significantly increase the LTR-directed gene expression of different animal and primate lentiviruses, including HIV (11, 14). We found that the JDV Tat not only stimulated its own LTR significantly but also stimulated HIV LTR-directed CAT expression to high levels equivalent to those obtained with HIV Tat (11). The activation of JDV LTR by JDV Tat was mediated by TAR-like elements located in the LTR, and the loop region in the TAR seemed to be less critical for JDV-mediated transactivation.
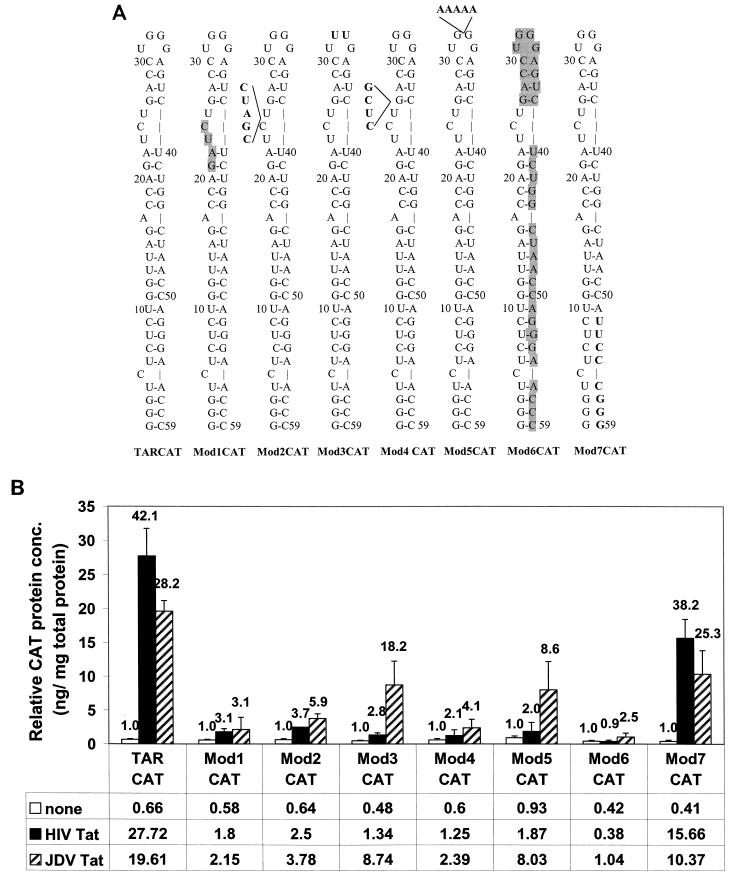
To further investigate the activation of HIV LTR by JDV Tat and whether similar mechanisms of transactivation were involved, a series of HIV TAR and LTR mutants were studied. These constructs were generated previously (25) and consisted of either deletions or mutations in the stem, the bulge, or the loop region of the HIV TAR. These mutants are summarized in Fig. 1A and were designated TARCAT, Mod1CAT, Mod2CAT, Mod3CAT, Mod4CAT, Mod5CAT, Mod6CAT, and Mod7CAT. Each plasmid construct was cotransfected into CV-1 cells with the JDV tat expression plasmid, pUC-Jtat, and tested for its ability to be activated by the JDV Tat (Fig. 1B). TARCAT containing the intact TAR sequence was used as the wild-type LTR. Mod7CAT, which contained the minimum HIV TAR required for HIV Tat transactivation, showed wild-type levels of transactivation in the presence of either JDV or HIV Tat. The responsiveness of the remaining HIV LTR mutants to transactivation by HIV Tat and JDV Tat was severely impaired and varied. The mutants Mod1CAT, Mod2CAT, Mod4CAT, and Mod6CAT, with mutations in the TAR stem and bulge region, were activated by JDV Tat about 60 to 80% less than was the wild-type HIV LTR. However, we consistently observed that these mutant promoters still responded better to the JDV Tat than to the HIV Tat, even though the activities were much lower than that with the wild-type LTR. The Mod3CAT mutant, which had a 2-base substitution in the loop region, retained most of its responsiveness to JDV Tat, but not to HIV Tat. Mod5CAT, which had a 5-base insertion in the loop region, was impaired in its ability to be activated although it has a relatively higher basal level of promoter activity. The data demonstrated that the JDV Tat transactivated the HIV LTR at least partially in a TAR-dependent manner, and sequences in the loop region of HIV LTR seemed to be less important. The data further support our earlier results that showed that activation of the JDV LTR by JDV Tat did not involve the loop sequence of the TAR region (11).
FIG. 1.
Effects of HIV TAR mutations on the transactivation of HIV LTR by both JDV Tat and HIV Tat. (A) Schematic presentation of the HIV LTR and mutant TAR constructs which were described in detail previously (25). TARCAT represents the wild-type LTR. All other modifications are as follows: Mod1CAT, 4-base deletion (GAUC) in the bulge region; Mod2CAT, 5-base insertion (CGAUC) in the bulge region; Mod3CAT, 2-base substitution (GG mutated to UU) in the loop region; Mod4CAT, 4-base insertion (CUCG) between the bulge and the upper stem-loop region; Mod5CAT, 5-base insertion (AAAAA) in the loop region; Mod6CAT, 35-base deletion, including the bulge and the downstream sequences; Mod7CAT, 8-base substitution (UUCCCGGG) in the lower stem-loop region. The shaded letters represent the deleted residues, and the boldface letters represent the substituted residues. (B) A 0.05-μg amount of LTR mutants was cotransfected with 0.5 μg of either pUC-Jtat or pUC-Htat. At 48 h posttransfection, cell lysates were collected and subjected to bicinchoninic acid assay. By using the same amount of total cellular protein, the expressed CAT protein in the transfected cell lysate was measured with the CAT ELISA kit as described in Materials and Methods. The relative CAT concentrations (conc.) shown in the figure represent the results from at least three independent experiments. The number above each bar represents fold activation for each LTR, and the transactivation fold was obtained by dividing the amount of CAT protein in the presence of Tat protein by the amount of CAT in the absence of Tat (11).
JDV and HIV Tat activation of the LTR can be competitively inhibited by coexpression of the HIV or JDV Tat activation domain.
HIV Tat has been shown to transactivate the LTR by recruiting the cyclin T-CDK9 complex to the TAR region which then phosphorylates the C-terminal domain of RNA polymerase II (15, 32, 34, 47, 56). In comparing the JDV Tat sequence with HIV Tat, we found that the JDV Tat contained a very conserved cysteine-rich domain and a core domain, which are both involved in the HIV Tat binding of cellular factors. Therefore, it is likely that JDV Tat transactivates HIV LTR by a similar mechanism and might involve similar cellular factors.
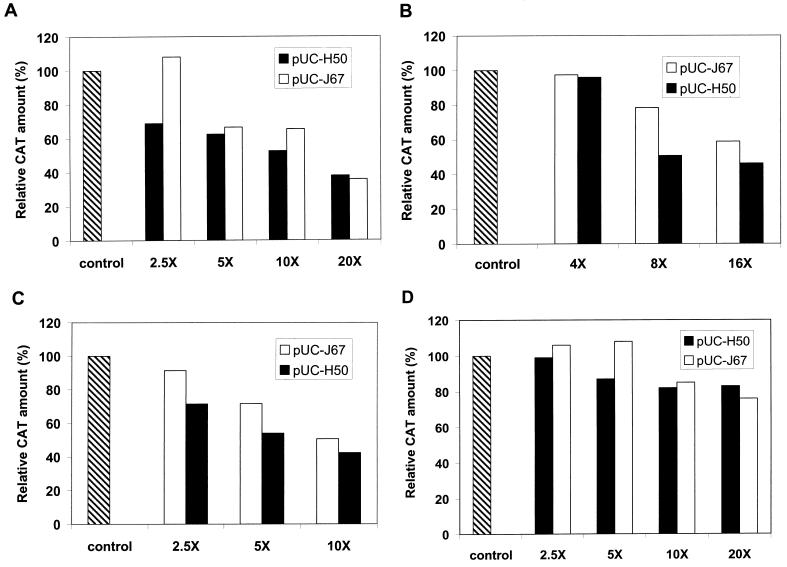
We constructed two truncated tat plasmids, pUC-J67, which encoded a JDV Tat peptide, JTat67 (amino acids 1 to 67), and pUC-H50, which encoded an HIV Tat activation domain, HTat50 (amino acids 1 to 50). We first examined the ability of the expressed truncated Tat peptide JTat67 and HTat50 to inhibit the transactivation function of HIV Tat and the effect of intact JDV Tat on the HIV LTR (Fig. 2A and B). These truncated proteins lost their abilities to transactivate HIV LTR and JDV LTR and did not inhibit the basal level of LTR expression (data not shown). We then cotransfected CV-1 cells with the target plasmid pHIV-CAT, effector plasmid pUC-Htat or pUC-Jtat, and varying concentrations of the competitor plasmid pUC-H50 or pUC-J67 (Fig. 2A and B); the total amount of DNA in each transfection was normalized by cotransfection of the parent vector pUC18. As shown in Fig. 2A and B, JTat67 and HTat50 efficiently inhibited transactivation of the HIV LTR by both HIV Tat and JDV Tat, in a dose-dependent manner. A 20-fold excess of the competitor plasmid pUC-J67 or pUC-H50 inhibited HIV Tat activation of the HIV LTR by about 65% (Fig. 2A), whereas a 10-fold excess of pUC-J67 inhibited HIV Tat transactivation of the HIV LTR by about 40% and a 10-fold excess of pUC-H50 resulted in a 50% decrease in activity (Fig. 2A). However, when only a 2.5-fold excess of competitor was added, the JDV peptide JTat67 was ineffective while the HIV peptide inhibited activation about 30%. The inhibition of HIV LTR activation by JDV Tat was similar (Fig. 2B): a 16-fold excess of either pUC-J67 or pUC-H50 caused significant inhibition of HIV LTR activation, 40% inhibition with pUC-J67 and 50% inhibition with pUC-H50; lower concentrations of pUC-H50 seemed to cause greater inhibition (Fig. 2B).
FIG. 2.
Competitive inhibition of Tat-induced transactivation by the truncated Tat proteins. (A) CV-1 cells were cotransfected with 0.05 μg of pHIV-CAT, 0.1 μg of HIV tat plasmid pUC-Htat, and the indicated amount of the competitors (pUC-J67 expresses the truncated JDV Tat protein and pUC-H50 expresses the truncated HIV Tat protein). The indicated values represent the percentages of the amount of CAT in the presence of competitor relative to that in the absence of competitor (defined as 100%). (B) CV-1 cells were cotransfected with 0.05 μg of pHIV-CAT plasmid, 0.25 μg of JDV tat plasmid pUC-Jtat, and the indicated amounts of competitors. (C) CV-1 cells were cotransfected with 0.5 μg of pJU3R-CAT plasmid, 0.25 μg of JDV tat plasmid pUC-Jtat, and the indicated amounts of competitors. (D) CV-1 cells were cotransfected with 0.15 μg of pHTLV-CAT plasmid, 0.1 μg of pCMV-Tax plasmid, and the indicated amounts of competitors. The total amount of DNA used in each transfection was equalized with a control plasmid. Control in the figure represents no competitor addition.
We next examined the effect of the truncated Tat proteins on transactivation of the JDV LTR by JDV Tat (Fig. 2C). Cotransfection of CV-1 cells with pJU3R-CAT, pUC-Jtat, and varying concentrations of competitor plasmids (either pUC-H50 or pUC-J67) inhibited JDV Tat transactivation to levels similar to those observed with the HIV LTR. A 10-fold excess of either of the Tat inhibitors HTat50 and JTat67 inhibited activation by about 50%. At lower concentrations of the inhibitors, the HIV Tat truncated protein also seemed to be more effective than the JDV truncated Tat protein. These results demonstrated that both truncated JDV and HIV Tat may inhibit transactivation by the intact Tat, possibly by competing for common cellular factors used by both JDV and HIV Tat.
To confirm that the inhibition by the JDV and HIV Tat activation domains was specific to Tat-mediated activation, we also examined the effects of these activation domains on the activation of human T-cell leukemia virus (HTLV) LTR by its cognate transactivator Tax. The presence of either HIV or JDV Tat transactivation domain did not dramatically affect the activation of the HTLV LTR by Tax. However, a slight inhibition by a fivefold or more excess of HIV Tat transactivation domain expression plasmid pUC-H50 and a 10-fold or more excess of JDV Tat transactivation domain expression plasmid pUC-J67 was observed. These results suggested that the inhibition effects of the JDV and HIV Tat activation domain were Tat specific but may also have involved some nonspecific transcription factors (Fig. 2D).
JDV Tat can complement HIV tat(−) provirus expression.
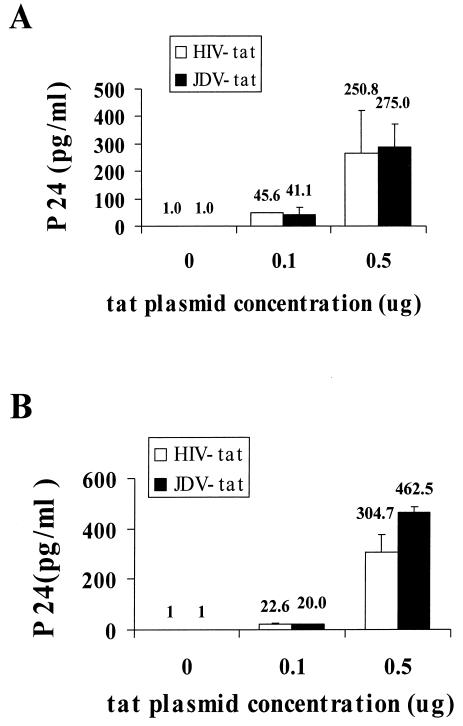
Tat is a potent and essential transcriptional transactivator of HIV LTR, and HIV-1 proviruses mutated in tat are nonviable (18, 49). However, this defect can be complemented by coexpression of the HIV Tat in trans. To determine whether JDV Tat can also complement the HIV tat(−) provirus expression in trans, two tat mutant HIV proviral plasmids, pMtat(−) and pMtat30, were cotransfected into CV-1 cells with a JDV Tat expression plasmid. Both tat mutants were demonstrated previously to be defective due to the mutations at the initiation codon of the Tat protein or at the cysteine-rich transactivation domain (49, 50). At 48 h posttransfection, supernatant was collected and assayed for the expression of HIV by p24 analysis. It was determined that transfection of the two tat mutant cDNA plasmids alone into CV-1 cells did not produce detectable levels of HIV as measured by p24 assays. However, in the presence of either the JDV tat or the HIV tat plasmid, HIV p24 was detected (Fig. 3). An increase in the amount of the JDV or HIV Tat plasmid transfected resulted in higher levels of HIV production. Our results demonstrated that both tat HIV mutants were complemented equally well by the presence of JDV or HIV Tat. This complementation assay demonstrated that the JDV Tat could functionally complement the defect in the tat gene in the HIV provirus to enable the replication of HIV in the proviral DNA-transfected cell.
FIG. 3.
Complementation of the tat mutant HIV proviruses by JDV Tat and HIV Tat. A 0.1-μg amount of tat mutant HIV proviral DNA was cotransfected into CV-1 cells with varying concentrations of the JDV and HIV tat plasmids, as indicated. The culture supernatant was collected at 72 h posttransfection, and the amount of HIV produced was determined by p24 ELISA. (A) Complementation of the HIV tat mutant clone pMtat(−) by JDV Tat and HIV Tat. This mutant carries a termination codon in place of the ATG (methionine) initiator codon in the Tat coding region (49). (B) Complementation of the HIV tat mutant clone pMtat30 by JDV and HIV Tat. This mutant has a Cys→Gly substitution within the Tat cysteine cluster and produces a missense Tat (50).
Construction and characterization of chimeric HIV carrying the JDV tat gene.
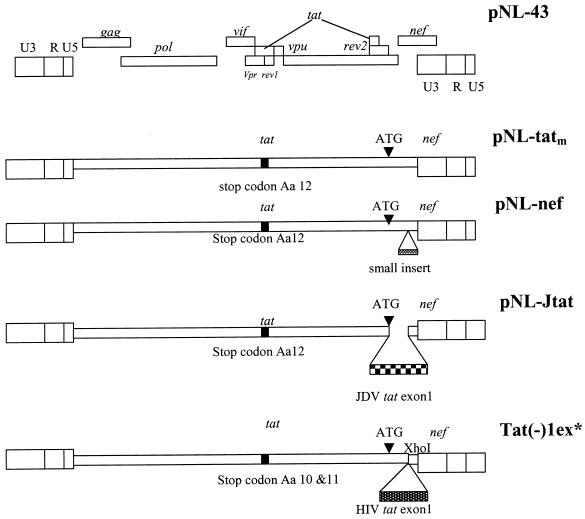
To further investigate whether JDV tat could functionally substitute for the HIV tat gene, we constructed a chimeric proviral clone by inserting the JDV tat gene into HIV proviral backbone, pNL43 (Fig. 4). To avoid affecting the function of other genes which overlap with the tat gene, we introduced a stop codon into the tat region of the HIV proviral plasmid pNL43 to inactivate tat, and an EcoRI site was generated in the mutated tat region. The resulting plasmid, pNL-tatm, with a tat mutation, otherwise isogenic to pNL43, was wild type in all other genes. A chimeric pNL-Jtat virus was also constructed, wherein a small fragment of nef was replaced with cDNA coding for JDV Tat exon 1 but still using the native nef ATG translation initiation site. The pNL43 nef could be effectively replaced since such nef mutations do not significantly affect the in vitro replication and infectivity of the mutant HIV in T-cell lines (45). As a positive control, a chimeric virus, Tat(−)1ex, containing the HIV exon 1 tat, was constructed using a similar strategy (45). A pNL-nef plasmid, in which random sequences were inserted into the nef region of pNL-tatm to further inactivate nef, was used as a negative control.
FIG. 4.
Schematic representation of the wild-type pNL43 and the various modified virus genomes. The pNL-tatm was generated by introducing a stop codon at amino acid 12 of the tat gene to render it inactive. This clone was then used to construct the other modified HIVs. pNL-Jtat express the JDV Tat exon 1 using the ATG of the nef gene. pNL-nef contains a random sequence insertion in the nef gene. Tat(−)1ex was used and described previously (45).
We first characterized the ability of these modified proviral clones to replicate and generate viruses in transfected cells. When 1 μg of the above plasmids was transfected into nonpermissive cells such as CV-1, HeLa, and 293T cells, the tat mutant proviral clones pNL-tatm and pNL-nef did not produce detectable levels of HIV in HeLa cells and produced much lower levels of HIV in 293T and CV-1 cells than did wild-type virus pNL43 (Table 1). In contrast, Tat(−)1ex and pNL-Jtat, which contain the HIV tat and JDV tat, respectively, produced wild-type levels of expression of HIV in all the tested cell lines.
TABLE 1.
p24 assay of supernatant from chimeric proviral DNA-transfected cellsa
| Plasmid(s) | Level of p24 in cell line:
|
||
|---|---|---|---|
| HeLa (pg/ml) | 293T (ng/ml) | CV-1 (pg/ml) | |
| pNL43 | 636.5 | 21.8 | >1,000.0 |
| pNL43 + pUC-Jtat | >600.0 | 16.3 | 1,063.9 |
| pNL43 + pUC-Htat | 625.3 | 19.9 | 1,064.8 |
| pNL43 + pΔCys1-4 | ND | 20.6 | ND |
| pNL-tatm | 5.8 | 1.3 | 267.8 |
| pNL-tatm + pUC-Jtat | >600.0 | 15.8 | >1,000.0 |
| pNL-tatm + pUC-Htat | >600.0 | 20.6 | 1,105.6 |
| pNL-tatm + pΔCys1-4 | ND | 0.7 | ND |
| pNL-nef | 1.2 | 0.6 | 77.4 |
| pNL-nef + pUC-Jtat | 177.7 | 18.2 | 1,005.6 |
| pNL-nef + pUC-Htat | 638.2 | 15.7 | 885.2 |
| pNL-nef + pΔCys1-4 | ND | 0.05 | ND |
| pNL-Jtat | 605.7 | 12.6 | 1,122.1 |
| pNL-Jtat + pUC-Jtat | >600.0 | 11.3 | 1,132.1 |
| pNL-Jtat + pUC-Htat | 645.7 | 13.3 | 1,137.8 |
| pNL-Jtat + pΔCys1-4 | ND | 8.5 | ND |
| Tat(−)1ex | 650.2 | 13.4 | 1,058.6 |
| Tat(−)1ex + pUC-Jtat | >600.0 | 8.9 | 1,160.4 |
| Tat(−)1ex + pUC-Htat | >600.0 | 8.4 | >1,000.0 |
| Tat(−)1ex + pΔCys1-4 | ND | 3.4 | ND |
One microgram of proviral DNA was transfected into either HeLa, 293T, or CV-1 cells with or without tat plasmids. At 48 h posttransfection, the supernatants were collected and assayed for p24 production. ND, not determined.
To rule out the possibility that the failure to express virus in cells transfected with pNL-tatm or pNL-nef was due to other unexpected defects besides the tat mutation in the genome, we cotransfected these cells with tat mutant proviral DNAs and either JDV or HIV Tat-expressing plasmids. For both pNL-tatm and pNL-nef, in the presence of HIV or JDV tat, the replication of the viruses in the transfected cells was restored and high levels of p24 were detected in the culture supernatant. Cotransfection of Tat-expressing plasmids with Tat(−)1ex, pNL-Jtat, or pNL43 proviral DNA only slightly increased or did not change the level of HIV expression. Cotransfection of a mutant HIV Tat-expressing plasmid, pΔCys1-4, which carries four Cys→Ser amino acid substitutions in the Cys-rich region and has lost its transactivation ability, failed to rescue the tat-defective virus (Table 1).
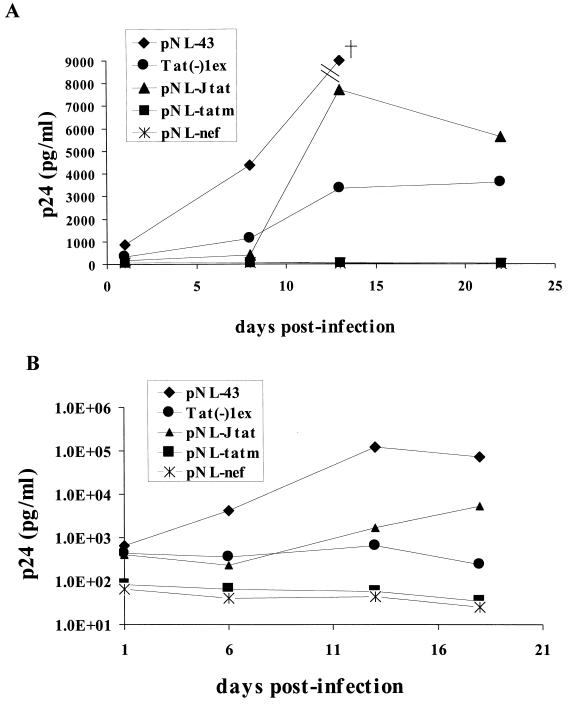
To further determine whether the presence of JDV tat in the chimeric virus could fully complement the HIV tat functions and enable the virus to productively infect CD4+ T cells, the viruses generated by transfection were used to infect both primary PBMC and a T-cell line, C8166 (45). The amount of virus produced from the transfected 293T cells was determined by p24 antigen levels, a similar amount of virus from each transfection experiment was then used to infect both fresh PBMC and C8166 cells, and the infected cells were monitored for virus growth by p24 assay at various time points after infection (Fig. 5A). The control chimeric virus Tat(−)1ex, containing the HIV tat gene, replicated in the T-cell line as demonstrated previously (45). The NL-Jtat virus, containing JDV tat, also replicated efficiently in this T-cell line, and substantial amounts of p24 were detected 14 days after infection. Almost twice as much p24 was detected in NL-Jtat-infected cells as in those infected by the control Tat(−)1ex virus. Our results thus showed that the chimeric virus with the HIV tat replaced by the JDV tat was fully competent in its ability to infect and replicate in C8166 T cells. As expected, the negative control viruses, NL-tatm and NL-nef, did not replicate.
FIG. 5.
Growth and replication kinetics of the various recombinant HIV-1 viruses in the C8166 T-cell line and PBMC. (A) Different viral stocks generated by transfection of 293T cells were used to infect the C8166 T-cell line. The 293T cells were transfected with the recombinant proviral DNA using Lipofectamine. At 48 h posttransfection, supernatants were collected, and the p24 levels were determined. C8166 cells (2 × 105) were infected by 10 ng of the different viruses as determined by the p24 assay. The amount of virus produced was monitored at the indicated time points by p24 analysis. The cross in the figure represents a p24 concentration of 103 ng/ml, which is beyond the y-axis scale. (B) PBMC (2 × 106) were infected with 10 ng of virus as determined by the p24 assay, and the amount of virus produced was monitored at the indicated time points as described above.
To further characterize the chimeric viruses, their ability to infect and replicate in primary T cells was tested. It was previously shown that the HIV chimera with the HIV nef gene replaced by HIV tat was unable to replicate in PBMC (45), suggesting that either the HIV tat exon 1 alone or the destruction of the HIV nef affected the ability of HIV to replicate in primary cells. Indeed, when we tested our various chimeric viruses in PBMC (Fig. 5B), the Tat(−)1ex virus containing the HIV tat exon 1 did not replicate in PBMC even though it replicated in C8166 cells, similar to results obtained previously with this virus (45). In contrast, the chimeric NL-Jtat which contained the JDV tat infected and replicated effectively in the primary PBMC, even though the levels seemed to be lower than those of the wild-type virus NL43. It produced lower levels of virus expression initially, but by 14 days the levels seemed to have increased and by 18 days were much higher. Our results suggested that the presence of JDV exon 1 tat complemented both HIV tat and nef to render the virus fully replication competent in both PBMC and T-cell lines. As expected, both tat mutant virus NL-tatm and NL-nef failed to replicate in PBMC.
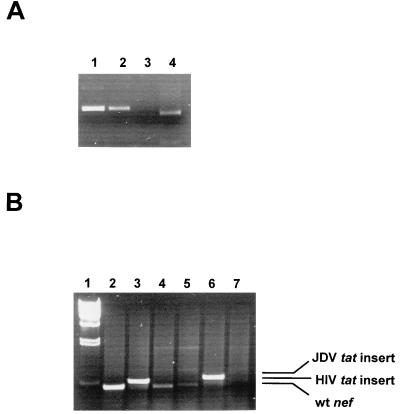
To rule out the possibility that infection of PBMC and C8166 cells by chimeric virus NL-Jtat was due to the back mutation of the HIV tat mutation to wild type, the HIV tat gene of the infected cells was analyzed by PCR and restriction analysis of the infected PBMC viral DNA (Fig. 6). Since the mutated tat gene was engineered to have an EcoRI site, the presence of the restriction site would indicate persistence of the mutation. Indeed, the amplified tat gene product from the chimeric virus NL-Jtat-infected cellular DNA, but not the wild-type virus-infected cellular DNA, was cut completely by EcoRI to generate a smaller tat gene fragment of the expected size (Fig. 6A, compare lanes 1 and 4). This result suggested that the mutation in the tat gene for NL-Jtat persisted and that reversion to the wild type was not responsible for the ability of this virus to replicate in PBMC. To further confirm that the JDV and HIV tat gene insertion in the nef region had not been deleted, the nef region of the viruses from the infected PBMC was analyzed by PCR. As was expected, the PCR-amplified fragments from NL-Jtat and Tat(−)1ex viruses were relatively larger than the wild-type nef fragment, indicating that the tat insert was retained (Fig. 6B).
FIG. 6.
PCR analysis of the viruses generated from the infected PBMC cultures. The figure shows PCR amplification of tat and nef gene sequences from the infected PBMC cellular DNA. At 18 days after infection, PBMC cellular DNA was extracted from cells infected with different viruses, and PCR amplification with the appropriate primers was performed. (A) Verification of tat mutation in the recombinant virus by EcoRI digestion. PCR products of the tat region amplified from infected PBMC DNA were purified from 1% agarose gels and then cut with EcoRI. To avoid amplification of the inserted tat sequences in the recombinant virus, the primers NL-tat1 (nt 5762 to 5781, 5′ GTT TAT CCA TTT CAG AAT TG 3′) and NL-tat2 (nt 6428 to 6409, 5′ CCA AAC ATT ATG TAC CTC TG 3′) used were located outside the tat sequences (nt 5830 to 6044) in the NL43 genome (GenBank accession no. M19921). Lane 1, NL43 virus tat DNA; lane 2, Tat(−)1ex virus tat DNA; lane 3, NL-tatm virus tat DNA; lane 4, NL-Jtat virus tat DNA. (B) PCR amplification of nef sequences from the infected PBMC DNA to verify the presence of the tat insert in the nef region of the recombinant virus genome. PCR was performed with primers NL-nef1 (nt 8447 to 8465, 5′ TCC ATT CGA TTA GTG AAC G3′) and NL-nef2 (nt 8904 to 8886, 5′ CTA CTT GTG ATT GCT CCA T3′). Lane 1, lambda/HindIII; lane 2, NL43; lane 3, Tat(−)1ex; lane 4, NL-tatm; lane 5, NL-nef, lane 6, NL-Jtat; lane 7, mock-infected control. wt, wild type.
Phylogenetic analysis of lentivirus Tat proteins.
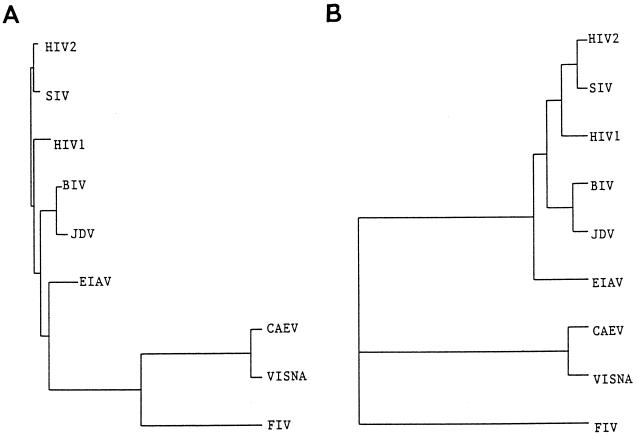
The cross-transactivation by JDV Tat of the HIV LTR suggested a close evolutionary relationship between JDV and other primate lentiviruses. To analyze the evolutionary genetic relationship between JDV and other lentiviruses, a neighbor-joining phylogenetic tree based on the published sequences of lentiviral Tat peptides which are competent in transactivation was constructed (3, 11, 17, 20, 21, 36, 41, 51, 52, 55) (Fig. 7A). With the constructed tree, two major subgroups of Tat were formed (primate versus nonprimate lentivirus Tat proteins). Within the primate lentivirus subgroup, the simian immunodeficiency virus Tat was more closely related to HIV-2 Tat than to HIV-1 Tat. Within the nonprimate lentivirus Tat group, the JDV and BIV Tat proteins were closely related phylogenetically. Interestingly, the two bovine lentivirus Tat proteins were more closely related to the primate lentivirus Tat proteins than they were to the nonprimate lentivirus Tat proteins. Another phylogenetic tree created by using UPGMA also showed a similar phylogenetic relationship, and although no distinctive nonprimate lentiviral Tat subgroup was formed with this method of analysis (Fig. 7B), JDV and BIV Tat proteins were within a subgroup which mainly contained the primate lentivirus Tat proteins.
FIG. 7.
Phylogenetic analysis of the various lentivirus Tat proteins. The Tat amino acid sequences were deduced from the tat nucleotide sequences: JDV genome (GenBank accession no. U21603), BIV (GenBank accession no. M32690), HIV-1 (GenBank locus hivhxb2cg), HIV-2 (GenBank accession no. M38432), simian immunodeficiency virus (SIV) (GenBank accession no. M19499), equine infectious anemia virus (EIAV) (GenBank accession no. M30138), caprine arthritis-encephalitis virus (CAEV) (GenBank accession no. M34092), feline immunodeficiency virus (FIV) (GenBank accession no. M36968), and visna virus tat sequence (accession no. J04359). The trees were reconstructed using two separate programs as described in Materials and Methods. Phylogenetic analysis used the Evolution program in GCG. (A) Analysis using the neighbor-joining algorithm; (B) analysis using the UPGMA algorithm.
DISCUSSION
A comparison of the amino acid sequences of the JDV and HIV Tat showed a striking homology in the cysteine-rich and the core activation domain, but very little similarity in the RNA binding basic domain (11). Indeed, competitors containing the HIV-1 and JDV Tat activation domain could inhibit the HIV and JDV Tat transactivation of the HIV LTR or the JDV Tat transactivation of the JDV LTR. Since both truncated proteins did not contain the basic RNA binding domain that is responsible for the TAR binding, the inhibition was unlikely to be due to competition for binding to TAR and more likely was due to the effects of the transactivating domain itself. These observations support the notion that the JDV and HIV Tat activation domains may involve a similar transactivation mechanism and both may involve interaction with cellular cofactors. The cross-inhibition of Tat functions may be due to the sequestering of the cellular factors involved.
In this study, we have demonstrated that the transactivation effect of JDV Tat on HIV LTR is at least partially TAR dependent. Mutations of TAR in the HIV LTR, except for mutations in the loop region as represented by mutant Mod3CAT, resulted in significant loss of responsiveness to JDV Tat transactivation. However, differences in the Tat of JDV and HIV were noted. First, as the HIV LTR mutant Mod3CAT was responsive to transactivation by JDV Tat but not to that by HIV Tat, this suggested that a sequence in the loop region of HIV TAR was not critical for transactivation by JDV Tat but was essential for transactivation by HIV Tat. Second, most HIV LTR mutants retained some responsiveness to the JDV Tat even though they were not activated by HIV Tat. These differences could be due to the JDV Tat transactivating the HIV LTR using other mechanisms in addition to the TAR-dependent pathway, or the JDV Tat-TAR interaction may be different from the HIV Tat-TAR interaction. We favor the latter explanation, since our results suggest that the loop sequence in the TAR is not critical for JDV transactivation, which is different from that by HIV. Studies of HIV have shown that the loop region is the binding site for cyclin T, and the binding of cyclin T to the loop can significantly stabilize the HIV Tat-TAR interaction (56). Recent data have convincingly demonstrated that the binding of the HIV Tat activation domain to the cyclin T1 subunit of the human TAK/P-TEFb transcription elongation complex is a critical first step in TAR RNA recognition and Tat-mediated transactivation (32). It is possible that JDV Tat can directly interact with the TAR elements independent of cyclin T complex binding, and thus the mutations of the cyclin target in the loop region would not affect the transactivation function of the JDV Tat. However, this does not exclude the possibility that the cyclin T complex is still involved in the JDV Tat-induced transactivation of the HIV LTR. Further studies are necessary in order to decipher the role of cyclin T in JDV Tat transactivation.
In addition to a transactivation function, Tat is essential for viral replication and pathogenesis (12, 18, 45, 62). HIV Tat was reported to activate quiescent T lymphocytes and induce apoptosis (38, 40) and may contribute to the selective depletion of CD4+ lymphocytes in vivo. Tat is also vital for viral replication and infection; mutations in tat led to viruses that were defective (3, 4). Relocation of tat in the HIV genome or the substitution of HIV tat for the nef gene still led to viruses that were defective; they were able to replicate in T-cell lines but not in PBMC (45). Substitution for the HIV tat by alternative transactivators, such as HTLV Tax and Vp16, which were cloned into HIV tat(−) has resulted in viruses that replicate very poorly in PBMC and other cell types (45). Our chimeric virus that used JDV tat exon 1 is therefore unique, as JDV tat exon 1 was able to substitute for HIV tat and enable successful replication in both a T-cell line and PBMC, even though resulting virus replication was less than that with the wild-type virus. Our results indicated that JDV Tat can functionally replace HIV Tat and suggest that JDV Tat should play a role in viral replication and pathogenesis of infection similar to that of HIV Tat in HIV. In addition, the JDV Tat could rescue a mutant HIV when it was mutated not only in the tat but also in the nef gene, suggesting that JDV Tat can substitute for both HIV Tat and Nef to enable the virus to replicate effectively in vitro. However, it is possible that this HIV-JDV chimeric virus may be nonpathogenic and attenuated in vivo. Further characterization of this virus and its pathogenicity will be important not only for elucidation of the functions of Tat but also for vaccine development if the HIV-JDV chimeric virus is found to be attenuated.
ACKNOWLEDGMENTS
This study was supported in part by the University of Nebraska Center of Biotechnology Area of Concentration grant to C.W.
We thank K. T. Jeang for kindly providing the plasmid Tat(−)1ex, R. Gaynor for plasmid pΔCys1-4, and F. Kashanchi for plasmids pHTLV-CAT and pCMV-Tax. The plasmids pMtat(−) and pMtat30 were obtained through the AIDS Research and Reference Reagent Program from Reza Sadaie. We also thank Malcolm Martin for providing the pNL43 plasmid through the AIDS Research and Reference Reagent Program.
REFERENCES
- 1.Adachi A, Koenig S, Gendelman H E, Daugherty D, Gattoni-Celli S, Fauci A S, Martin M A. Productive, persistent infection of human colorectal cell lines with human immunodeficiency virus. J Virol. 1987;61:209–213. doi: 10.1128/jvi.61.1.209-213.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albini A, Barillari G, Benelli R, Gallo R C, Ensoli B. Angiogenic properties of human immunodeficiency virus type 1 Tat protein. Proc Natl Acad Sci USA. 1995;92:4838–4842. doi: 10.1073/pnas.92.11.4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arya S K, Beaver B, Jagodzinski L, Ensoli B, Kanki P J, Albert J, Fenyo E M, Biberfeld G, Zagury J F, Laure F, Essex M, Norrby E, Wong-Staal F, Gallo R C. New human and simian HIV-related retroviruses possess functional transactivator (tat) gene. Nature. 1987;328:548–550. doi: 10.1038/328548a0. [DOI] [PubMed] [Google Scholar]
- 4.Arya S K, Guo C, Josephs S F, Wong-Staal F. Trans-activator gene of human T-lymphotropic virus type III (HTLV-III) Science. 1985;229:69–73. doi: 10.1126/science.2990040. [DOI] [PubMed] [Google Scholar]
- 5.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates; 1989. [Google Scholar]
- 6.Barillari G, Gendelman R, Gallo R C, Ensoli B. The Tat protein of human immunodeficiency virus type 1, a growth factor for AIDS Kaposi sarcoma and cytokine-activated vascular cells, induces adhesion of the same cell types by using integrin receptors recognizing the RGD amino acid sequence. Proc Natl Acad Sci USA. 1993;90:7941–7945. doi: 10.1073/pnas.90.17.7941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Budiarso I T, Hardjosworo S. Jembrana disease in Bali cattle. Aust Vet J. 1976;52:97. doi: 10.1111/j.1751-0813.1976.tb13867.x. [DOI] [PubMed] [Google Scholar]
- 8.Buonaguro L, Barillari G, Chang H K, Bohan C A, Kao V, Morgan R, Gallo R C, Ensoli B. Effects of the human immunodeficiency virus type 1 Tat protein on the expression of inflammatory cytokines. J Virol. 1992;66:7159–7167. doi: 10.1128/jvi.66.12.7159-7167.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chadwick B J, Coelen R J, Sammels L M, Kertayadnya G, Wilcox G E. Genomic sequence analysis identifies Jembrana disease virus as a new bovine lentivirus. J Gen Virol. 1995;76:189–192. doi: 10.1099/0022-1317-76-1-189. [DOI] [PubMed] [Google Scholar]
- 10.Chadwick B J, Coelen R J, Wilcox G E, Sammels L M, Kertayadnya G. Nucleotide sequence analysis of Jembrana disease virus: a bovine lentivirus associated with an acute disease syndrome. J Gen Virol. 1995;76:1637–1650. doi: 10.1099/0022-1317-76-7-1637. [DOI] [PubMed] [Google Scholar]
- 11.Chen H, Wilcox G E, Kertayadnya G, Wood C. Characterization of the Jembrana disease virus tat gene and the cis- and trans-regulatory elements in its long terminal repeats. J Virol. 1999;73:658–666. doi: 10.1128/jvi.73.1.658-666.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng-Mayer C, Shioda T, Levy J A. Host range, replicative, and cytopathic properties of human immunodeficiency virus type 1 are determined by very few amino acid changes in Tat and gp120. J Virol. 1991;65:6931–6941. doi: 10.1128/jvi.65.12.6931-6941.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clements J E, Zink M C. Molecular biology and pathogenesis of animal lentivirus infections. Clin Microbiol Rev. 1996;9:100–117. doi: 10.1128/cmr.9.1.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coffin J M, Hughes S H, Varmus H E. Retroviruses. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 475–636. [PubMed] [Google Scholar]
- 15.Cullen B R. HIV-1 auxiliary proteins: making connections in a dying cell. Cell. 1998;93:685–692. doi: 10.1016/s0092-8674(00)81431-2. [DOI] [PubMed] [Google Scholar]
- 16.Cullen B R. Regulation of HIV gene expression. AIDS. 1995;9:S19–32. [PubMed] [Google Scholar]
- 17.Davis J L, Clements J E. Characterization of a cDNA clone encoding the visna virus transactivating protein. Proc Natl Acad Sci USA. 1989;86:414–418. doi: 10.1073/pnas.86.2.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dayton A I, Sodroski J G, Rosen C A, Goh W C, Haseltine W A. The trans-activator gene of the human T cell lymphotropic virus type III is required for replication. Cell. 1986;44:941–947. doi: 10.1016/0092-8674(86)90017-6. [DOI] [PubMed] [Google Scholar]
- 19.Deacon N J, Tsykin A, Solomon A, Smith K, Ludford-Menting M, Hooker D J, McPhee D A, Greenway A L, Ellett A, Chatfield C, Lawson V A, Crowe S, Dowton D, Mills J. Genomic structure of an attenuated quasi species of HIV-1 from a blood transfusion donor and recipients. Science. 1995;270:988–991. doi: 10.1126/science.270.5238.988. [DOI] [PubMed] [Google Scholar]
- 20.de Parseval A, Elder J H. Demonstration that orf2 encodes the feline immunodeficiency virus transactivating (Tat) protein and characterization of a unique gene product with partial Rev activity. J Virol. 1999;73:608–617. doi: 10.1128/jvi.73.1.608-617.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dorn P, DaSilva L, Martarano L, Derse D. Equine infectious anemia virus tat: insights into the structure, function, and evolution of lentivirus trans-activator proteins. J Virol. 1990;64:1616–1624. doi: 10.1128/jvi.64.4.1616-1624.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ensoli B, Gendelman R, Markham P, Fiorelli V, Colombini S, Raffeld M, Cafaro A, Chang H K, Brady J N, Gallo R C. Synergy between basic fibroblast growth factor and HIV-1 Tat protein in induction of Kaposi's sarcoma. Nature. 1994;371:674–680. doi: 10.1038/371674a0. [DOI] [PubMed] [Google Scholar]
- 23.Ensoli B, Barillari G, Salahuddin S Z, Gallo R C, Wong-Staal F. Tat protein of HIV-1 stimulates growth of cells derived from Kaposi's sarcoma lesions of AIDS patients. Nature. 1990;345:84–86. doi: 10.1038/345084a0. [DOI] [PubMed] [Google Scholar]
- 24.Ensoli B, Buonaguro L, Barillari G, Fiorelli V, Gendelman R, Morgan R A, Wingfield P, Gallo R C. Release, uptake, and effects of extracellular human immunodeficiency virus type 1 Tat protein on cell growth and viral transactivation. J Virol. 1993;67:277–287. doi: 10.1128/jvi.67.1.277-287.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fong S E, Smanik P, Thais T, Smith M C, Jaskunas S R. Detection of specific human immunodeficiency virus type 1 Tat-TAR complexes in the presence of mild denaturing conditions. J Virol Methods. 1997;66:91–101. doi: 10.1016/s0166-0934(97)00039-6. [DOI] [PubMed] [Google Scholar]
- 26.Garcia J A, Harrich D, Pearson L, Mitsuyasu R, Gaynor R B. Functional domains required for tat-induced transcriptional activation of the HIV-1 long terminal repeat. EMBO J. 1988;7:3143–3147. doi: 10.1002/j.1460-2075.1988.tb03181.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harrich D, Ulich C, Garcia-Martinez L F, Gaynor R B. Tat is required for efficient HIV-1 reverse transcription. EMBO J. 1997;16:1224–1235. doi: 10.1093/emboj/16.6.1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harrich D, Ulich C, Gaynor R B. A critical role for the TAR element in promoting efficient human immunodeficiency virus type 1 reverse transcription. J Virol. 1996;70:4017–4027. doi: 10.1128/jvi.70.6.4017-4027.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harrich D, Mavankal G, Mette-Snider A, Gaynor R B. Human immunodeficiency virus type 1 TAR element revertant viruses define RNA structures required for efficient viral gene expression and replication. J Virol. 1995;69:4906–4913. doi: 10.1128/jvi.69.8.4906-4913.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Howcroft T K, Strebel K, Martin M A, Singer D S. Repression of MHC class I gene promoter activity by two-exon Tat of HIV. Science. 1993;260:1320–1322. doi: 10.1126/science.8493575. [DOI] [PubMed] [Google Scholar]
- 31.Huang L M, Joshi A, Willey R, Orenstein J, Jeang K T. Human immunodeficiency viruses regulated by alternative trans-activators: genetic evidence for a novel non-transcriptional function of Tat in virion infectivity. EMBO J. 1994;13:2886–2896. doi: 10.1002/j.1460-2075.1994.tb06583.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ivanov D, Kwak Y T, Nee E, Guo J, Garcia-Martinez L F, Gaynor R B. Cyclin T1 domains involved in complex formation with Tat and TAR RNA are critical for tat-activation. J Mol Biol. 1999;288:41–56. doi: 10.1006/jmbi.1999.2663. [DOI] [PubMed] [Google Scholar]
- 33.Iversen A K, Shpaer E G, Rodrigo A G, Hirsch M S, Walker B D, Sheppard H W, Merigan T C, Mullins J I. Persistence of attenuated rev genes in a human immunodeficiency virus type 1-infected asymptomatic individual. J Virol. 1995;69:5743–5753. doi: 10.1128/jvi.69.9.5743-5753.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jeang K T. Tat, Tat-associated kinase, and transcription. J Biomed Sci. 1998;5:24–27. doi: 10.1007/BF02253352. [DOI] [PubMed] [Google Scholar]
- 35.Jung M, Wood C. Activation of retroviral promoters by trans-acting factors. Virology (Life Sci Adv) 1991;10:77–88. [Google Scholar]
- 36.Kalinski H, Mashiah P, Rotem D, Orzech Y, Sherman L, Miki T, Yaniv A, Gazit A, Tronick S R. Characterization of cDNA species encoding the Tat protein of caprine arthritis encephalitis virus. Virology. 1994;204:828–834. doi: 10.1006/viro.1994.1602. [DOI] [PubMed] [Google Scholar]
- 37.Kashanchi F, Duvall J F, Lindholm P F, Radonovich M F, Brady J N. Sequences downstream of the RNA initiation site regulate human T-cell lymphotropic virus type I basal gene expression. J Virol. 1993;67:2894–2902. doi: 10.1128/jvi.67.5.2894-2902.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Katsikis P D, Garcia-Ojeda M E, Torres-Roca J F, Greenwald D R, Herzenberg L A. HIV type 1 Tat protein enhances activation—but not Fas (CD95)-induced peripheral blood T cell apoptosis in healthy individuals. Int Immunol. 1997;9:835–841. doi: 10.1093/intimm/9.6.835. [DOI] [PubMed] [Google Scholar]
- 39.Kirchhoff F, Easterbrook P J, Douglas N, Troop M, Greenough T C, Weber J, Carl S, Sullivan J L, Daniels R S. Sequence variations in human immunodeficiency virus type 1 nef are associated with different stages of disease. J Virol. 1999;73:5497–5508. doi: 10.1128/jvi.73.7.5497-5508.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li C J, Friedman D J, Wang C, Metelev V, Pardee A B. Induction of apoptosis in uninfected lymphocytes by HIV-1 Tat protein. Science. 1995;268:429–431. doi: 10.1126/science.7716549. [DOI] [PubMed] [Google Scholar]
- 41.Liu Z Q, Sheridan D, Wood C. Identification and characterization of the bovine immunodeficiency-like virus tat gene. J Virol. 1992;66:5137–5140. doi: 10.1128/jvi.66.8.5137-5140.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mariani R, Kirchhoff F, Greenough T C, Sullivan J L, Desrosiers R C, Skowronski J. High frequency of defective nef alleles in a long-term survivor with nonprogressive human immunodeficiency virus type 1 infection. J Virol. 1996;70:7752–7764. doi: 10.1128/jvi.70.11.7752-7764.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McCloskey T W, Ott M, Tribble E, Khan S A, Teichberg S, Paul M O, Pahwa S, Verdin E, Chirmule N. Dual role of HIV Tat in regulation of apoptosis in T cells. J Immunol. 1997;158:1014–1019. [PubMed] [Google Scholar]
- 44.Michael N L, Chang G, d'Arcy L A, Ehrenberg P K, Mariani R, Busch M P, Birx D L, Schwartz D H. Defective accessory genes in a human immunodeficiency virus type 1-infected long-term survivor lacking recoverable virus. J Virol. 1995;69:4228–4236. doi: 10.1128/jvi.69.7.4228-4236.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Neuveut C, Jeang K T. Recombinant human immunodeficiency virus type 1 genomes with tat unconstrained by overlapping reading frames reveal residues in Tat important for replication in tissue culture. J Virol. 1996;70:5572–5581. doi: 10.1128/jvi.70.8.5572-5581.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nowak M A, Anderson R M, Boerlijst M C, Bonhoeffer S, May R M, McMichael A J. HIV-1 evolution and disease progression. Science. 1996;274:1008–1011. doi: 10.1126/science.274.5289.1008. [DOI] [PubMed] [Google Scholar]
- 47.Parada C A, Roeder R G. Enhanced processivity of RNA polymerase II triggered by Tat-induced phosphorylation of its carboxyl-terminal domain. Nature. 1996;384:375–378. doi: 10.1038/384375a0. [DOI] [PubMed] [Google Scholar]
- 48.Pocsik E, Higuchi M, Aggarwal B B. Down-modulation of cell surface expression of p80 form of the tumor necrosis factor receptor by human immunodeficiency virus-1 Tat gene. Lymphokine Cytokine Res. 1992;11:317–325. [PubMed] [Google Scholar]
- 49.Sadaie M R, Benter T, Wong-Staal F. Site-directed mutagenesis of two trans-regulatory genes (tat-III, trs) of HIV-1. Science. 1988;239:910–913. doi: 10.1126/science.3277284. [DOI] [PubMed] [Google Scholar]
- 50.Sadaie M R, Rappaport J, Benter T, Josephs S F, Willis R, Wong-Staal F. Missense mutations in an infectious human immunodeficiency viral genome: functional mapping of tat and identification of the rev splice acceptor. Proc Natl Acad Sci USA. 1988;85:9224–9228. doi: 10.1073/pnas.85.23.9224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sakuragi J, Sakuragi S, Ueda S, Adachi A. Functional analysis of simian immunodeficiency virus SIVAGM long terminal repeat. Virus Genes. 1996;12:21–25. doi: 10.1007/BF00369997. [DOI] [PubMed] [Google Scholar]
- 52.Saltarelli M J, Schoborg R, Gdovin S L, Clements J E. The CAEV tat gene trans-activates the viral LTR and is necessary for efficient viral replication. Virology. 1993;197:35–44. doi: 10.1006/viro.1993.1564. [DOI] [PubMed] [Google Scholar]
- 53.Scala G, Ruocco M R, Ambrosino C, Mallardo M, Giordano V, Baldassarre F, Dragonetti E, Quinto I, Venuta S. The expression of the interleukin 6 gene is induced by the human immunodeficiency virus 1 Tat protein. J Exp Med. 1994;179:961–971. doi: 10.1084/jem.179.3.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Taylor J P, Cupp C, Diaz A, Chowdhury M, Khalili K, Jimenez S A, Amini S. Activation of expression of genes coding for extracellular matrix proteins in Tat-producing glioblastoma cells. Proc Natl Acad Sci USA. 1992;89:9617–9621. doi: 10.1073/pnas.89.20.9617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Viglianti G A, Mullins J I. Functional comparison of transactivation by simian immunodeficiency virus from rhesus macaques and human immunodeficiency virus type 1. J Virol. 1988;62:4523–4532. doi: 10.1128/jvi.62.12.4523-4532.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wei P, Garber M E, Fang S M, Fischer W H, Jones K A. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell. 1998;92:451–462. doi: 10.1016/s0092-8674(00)80939-3. [DOI] [PubMed] [Google Scholar]
- 57.Weiss R A. How does HIV cause AIDS? Science. 1993;260:1273–1279. doi: 10.1126/science.8493571. [DOI] [PubMed] [Google Scholar]
- 58.Westendorp M O, Frank R, Ochsenbauer C, Stricker K, Dhein J, Walczak H, Debatin K M, Krammer P H. Sensitization of T cells to CD95-mediated apoptosis by HIV-1 Tat and gp120. Nature. 1995;375:497–500. doi: 10.1038/375497a0. [DOI] [PubMed] [Google Scholar]
- 59.Westendorp M O, Li-Weber M, Frank R W, Krammer P H. Human immunodeficiency virus type 1 Tat upregulates interleukin-2 secretion in activated T cells. J Virol. 1994;68:4177–4185. doi: 10.1128/jvi.68.7.4177-4185.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wilcox G E, Chadwick B J, Kertayadnya G. Recent advances in the understanding of Jembrana disease. Vet Microbiol. 1995;46:249–255. doi: 10.1016/0378-1135(95)00089-s. [DOI] [PubMed] [Google Scholar]
- 61.Wilcox G E, Soeharsono S, Dharma D M N, Copland J W. Jembrana disease and the bovine lentiviruses. ACIAR Proc. 1997;75:10–75. [Google Scholar]
- 62.Zagury J F, Sill A, Blattner W, Lachgar A, Le Buanec H, Richardson M, Rappaport J, Hendel H, Bizzini B, Gringeri A, Carcagno M, Criscuolo M, Burny A, Gallo R C, Zagury D. Antibodies to the HIV-1 Tat protein correlated with nonprogression to AIDS: a rationale for the use of Tat toxoid as an HIV-1 vaccine. J Hum Virol. 1998;1:282–292. [PubMed] [Google Scholar]