Abstract
Background
Cytokine storm is known to impact the prognosis of coronavirus disease 2019 (COVID-19), since pro-inflammatory cytokine variants are associated with cytokine storm. It is tempting to speculate that pro-inflammatory cytokines variants may impact COVID-19 outcomes by modulating cytokine storm. Here, we verified this hypothesis via a comprehensive analysis.
Methods
PubMed, Cochrane Library, Central, CINAHL, and ClinicalTrials.gov were searched until December 15, 2023. Case–control or cohort studies that investigated the impacts of rs1800795 or rs1800629 on COVID-19 susceptibility, severity, mortality, IL-6, TNF-α, or CRP levels were included after an anonymous review by two independent reviewers and consultations of disagreement by a third independent reviewer.
Results
47 studies (8305 COVID-19 individuals and 17,846 non-COVID-19 individuals) were analyzed. The rs1800629 A allele (adenine at the −308 position of the promoter was encoded by the A allele) was associated with higher levels of tumor necrosis factor-α (TNF-α) and C-reactive protein (CRP). In contrast, the rs1800795 C allele (cytosine at the −174 position of the promoter was encoded by the C allele) was linked to higher levels of interleukin-6 (IL-6) and CRP. In addition, the A allele of rs1800629 increased the severity and mortality of COVID-19. However, the C allele of rs1800795 only increased COVID-19 susceptibility.
Conclusions
rs1800629 and rs1800795 variants of pro-inflammatory cytokines have significant impacts on systemic inflammatory profile and COVID-19 clinical outcomes. rs1800629 may serve as a genetic marker for severe COVID-19.
Supplementary Information
The online version contains supplementary material available at 10.1007/s44197-024-00204-w.
Keywords: IL-6, Cytokine storm, Coronavirus disease 2019, Severity
Introduction
The COVID-19 pandemic is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). It first appeared in December 2019 with the characteristics of a highly contagious and high mortality rate [1]. According to the World Health Organization (WHO) report, COVID-19 infected about 701 million individuals and caused more than 6.97 million deaths [2].
The clinical manifestations of COVID-19 differ substantially in severity, varying from asymptomatic or mildly symptomatic to severe or critical illness [3, 4]. Among the infected individuals with symptoms, the majority presented with mild illness [5–7], while approximately 10% progressed to a severe or critical stage requiring intensive care or mechanical ventilation support [8, 9]. The variation in symptoms or severity of COVID-19 might be attributed to some known risk factors, including males [10], older age [11], alcohol consumption [12], menopause [13], smoking [14], and underlying comorbidities (e.g., hypertension [14, 15], diabetes [14–16], cardiovascular disease [14–16], chronic pulmonary disease [15–17], obesity [15, 18], cancer [15, 19], and immunodeficiencies [15]). Although older age [11], menopause [13] and comorbidities [14–19] were associated with illness severity, these risk factors (i.e., older age, menopause and comorbidities) alone did not explain why some young [20], healthy individuals [21] suffered a severe or life-threatening illness. Interestingly, this aberrant phenomenon might be partly attributed to genetic underliers imparting inter-individual differences in susceptibility to COVID-19 infection and illness severity [22–25].
The procedure for COVID-19 infection is hierarchical. First, the spike protein S binds the angiotensin-converting enzyme 2 (ACE2) receptor to enter the host cell [26]. Then, the host immune response, such as the innate immune response, is initiated against virus infection [25]. Subsequently, some critical signaling pathways are activated, including Toll-like receptor (TLR) [24, 27–29], C-lectin type receptors (CLR) [24, 28], neuropilin-1 (NPR1) [28], and inflammasome (cytokine storm) [24, 27, 29]. Notably, the genetic variants of viral entry and innate immunity (eg, ACE1 rs4343/rs4646994/rs1799752 [30, 31], ACE2 rs2285666 [30], and IFITM3 rs12252 [32]) may influence susceptibility to COVID-19 infection [25] and confer altered clinical outcomes [25].
The IL-6 gene is located on the short arm of human chromosome 7 (7p21–24), including five exons. rs1800795 (also known as -174 G > C), is located in the promoter at position -174, formed by a transversion from guanine (G) to cytosine (C) and is known to increase the transcriptional activity of IL-6 [33]. The TNF-α gene contains four exons on human chromosome 6 (6p21.31). rs1800629 (also known as -308 G > A), is located in the promoter at position -308, formed by substitution from guanine (G) to adenine (A), resulting in a 2–3 time increase in the transcriptional activity of TNF-α [34]. In addition to the rs1800795 C allele and the rs1800629 A allele may elevate plasma levels of C-reactive protein (CRP) [35, 36], it indicates that pro-inflammatory cytokines variants (i.e., 1800795 and rs1800629) may impact systemic inflammatory profile (i.e., IL-6, TNF-α, and CRP). Here, we conducted this study to investigate this hypothesis.
Cytokine storm (an aberrant systemic hyperinflammatory state characterized by high plasma levels of cytokines, including IL-1β, IL-2, IL-6, IL-7, IL-8, IL-10, IL-15, TNF-α, CRP, and MCP-1) [37–39] is closely linked to acute respiratory distress syndrome (ARDS) [40–42] and COVID-19 outcomes [43–48]. For instance, plasma levels of TNF-α, IFN-γ, IL-2, IL-4, IL-6, IL-10, and CRP were higher in COVID-19 patients compared with healthy individuals [43], indicating that cytokine storm may be related to COVID-19 infection. In contrast, plasma levels of IL-6, IL-8, IL-10, and TNF-α were higher in patients with severe COVID-19 compared with those without severe COVID-19 [44, 45], indicating that cytokine storm may be associated with COVID-19 severity. In addition, plasma levels of IL-6, IL-10, TNF-α, and CRP were higher in COVID-19 death cases than in non-death patients [46–48], suggesting that cytokine storm may be correlated with COVID-19 mortality.
IL-6 [37, 49–54] and TNF-α [37, 51, 53–55] are two critical components of cytokine storm. Since plasma levels of TNF-α [34, 56, 57] and IL-6 [33, 58, 59] are at least partly determined by variants of rs1800629 [34, 56, 57] and rs1800795 [33, 58, 59], it indicates that variants of rs1800629 and rs1800795 may impact COVID-19 outcomes by modulating cytokine storm. To verify this hypothesis, this study is required to investigate the impacts of rs1800795 and rs1800629 on systemic inflammatory profile (i.e., IL-6, TNF-α, and CRP) and COVID-19 clinical outcomes (i.e., susceptibility, severity, and mortality).
Materials and Methods
Study Selection
Studies that meet the following PICOS principle are preliminary selected: (1) P (population): Caucasians, Asians, Indians, and Mexicans, etc.; (2) I (intervention): no particular intervention; (3) C (comparison): the studies compare inflammatory parameters (e.g., IL-6, TNF-α, or CRP) and/or COVID-19 outcomes (e.g., susceptibility, severity, or mortality) between carriers of the rs1800795 G allele (or rs1800629 G allele) and carriers of the rs1800795 C allele (or rs1800629 A allele); (4) O (outcome): inflammatory parameters are expressed as mean with standard deviation (SD), or the number of genotype in case group, and control group is provided, to facilitate the subsequent calculation of standardized mean difference (SMD) and 95% confidence intervals (CI), or odds ratio (OR) and corresponding 95% CI; (5) S (study design): case–control studies or cohort studies, published in English, and funded by a funding body or institution.
Literature Search
PubMed, Cochrane Library, Central, CINAHL, and ClinicalTrials.gov were searched from August 05, 2022 to December 15, 2023. The following keywords were used in the search: (“cytokines,” “inflammatory cytokines,” “pro-inflammatory cytokines”) OR (“interleukin 6,” “tumor necrosis factor-α,” “IL-6,” “TNF-α”) AND (“rs1800795,” “rs1800629,” “-174 G > C,” “-308 G > A”) AND (“variant,” “variation,” “mutant,” “mutation,” “polymorphism,” “SNP”) OR (“single nucleotide polymorphism”) AND (“IL-6,” “TNF-α,” “CRP,” “interleukin 6,” “tumour necrosis factor-α,” “C-reactive protein”) OR (“COVID-19,” “SAR-CoV-2,” “coronavirus disease 2019”) OR (“severe acute respiratory syndrome coronavirus 2”) AND/OR (“clinical outcomes,” “susceptibility,” “severity,” “mortality”).
Inclusion and Exclusion Criteria
The inclusion criteria for the impacts of inflammatory cytokines variants on inflammatory biomarkers include:
Article type: case–control studies or cohort studies that investigated the effects of rs1800629 or rs1800795 on IL-6, TNF-α, or CRP levels.
Data type: studies that provided mean IL-6, TNF-α, or CRP levels with SD.
Inflammatory biomarkers: studies that at least provided two of three parameters in the inflammatory profiles (i.e., IL-6, TNF-α, and CRP).
Genetic information: studies that provided the genotype frequencies of rs1800795 and rs1800629.
Human subjects: test subjects were limited to humans.
Language: the language of eligible studies was restricted to English.
The inclusion criteria for the impacts of inflammatory cytokines variants on COVID-19 clinical outcomes include:
Article type: case–control studies that investigated the effects of rs1800795 or rs1800629 on COVID-19 susceptibility, severity, or mortality.
Population: COVID-19 patients were confirmed by RT-PCR or PCR–RFLP.
Genetic information: studies that provided case and control genotype frequencies.
Language: studies published in English language only.
Studies were rejected if they met one or more of the following exclusion criteria:
Studies did not relate to rs1800795 or rs1800629.
Studies did not relate to inflammatory biomarkers or COVID-19.
Studies did not present genetic information.
Studies with incomplete data.
Pedigree or overlapping studies.
Abstract/comments/review/case report/animal studies.
Data Extraction
Two authors (XD and KT) independently extracted the data using standardized data extraction sheets (Table S1–S11). The discrepancy in data collected was resolved by consensus or a discussion with the senior author (ZL). The following data were extracted from each eligible study: the first author’s name (Table S1), year (Table S1), country (Table S1), ethnicity (Table S1), gender (Table S1), genotyping methods (Table S1), total sample size (Table S1), mean inflammatory biomarkers levels with SD or SE by genotype (Table S2–S5), and genotype counts (Table S6–S11).
Data Analysis
The SMD and 95% CI were used to evaluate the differences in inflammatory biomarkers between different genotypes. The OR with 95% CI was used to evaluate the impacts of rs1800795 and rs1800629 on COVID-19 susceptibility, severity, and mortality. The pooled OR was performed for the allelic model [(A vs. G) for rs1800629, (C vs. G) for rs1800795], additive model [(AA vs. GG) for rs1800629, (CC vs. GG) for rs1800795], dominant model [(GA + AA) vs. GG for rs1800629, (GC + CC) vs. GG for rs1800795] and recessive model [(GG + GA) vs. AA for rs1800629, (GG + GC) vs. CC for rs1800795]. Since most of the included studies presented inflammatory biomarkers in a dominant model [(GA + AA) vs. GG for rs1800629, (GC + CC) vs. GG for rs1800795], a dominant model was adopted to ensure adequate statistical power. All statistical tests were conducted with the Cochrane Collaboration meta-analysis software, Review Manager 5.4. P < 0.05 was recognized as statistically significant.
Subgroup Analysis
Subgroup analysis was carried out on Caucasians, Asians, Indians, and Mexicans. In some studies, the subjects were divided into more than one subpopulation (e.g., the subjects originated from different ethnicities or genders). Each subpopulation was regarded as an independent comparison in this study.
Heterogeneity Processing
Heterogeneity was tested by I2 statistic and Cochran's χ2-based Q statistic. If heterogeneity was significant (I2 > 50%, P ≤ 0.05), the random-effect model (DerSimonian-Laird method) was used to calculate the results. Otherwise, the fixed-effect model (Mantel–Haenszel method) would be adopted (I2 < 50%, P > 0.05) [60]. In addition, the Galbraith plot was employed to detect the potential sources of heterogeneity. To completely eliminate the impact of heterogeneity on the results, all results were recalculated after excluding the studies with heterogeneity.
Publication Bias Test
The Begg funnel plot and Egger linear test evaluated the probability of publication bias among the included studies [61].
Risk of Bias/Quality Assessment
The risk bias among the included studies was evaluated by the risk-of-bias plot [62], in which different colors represent different levels of risk bias. For instance, green indicates a low risk bias, while red suggests a high risk bias.
Results
Study Selection
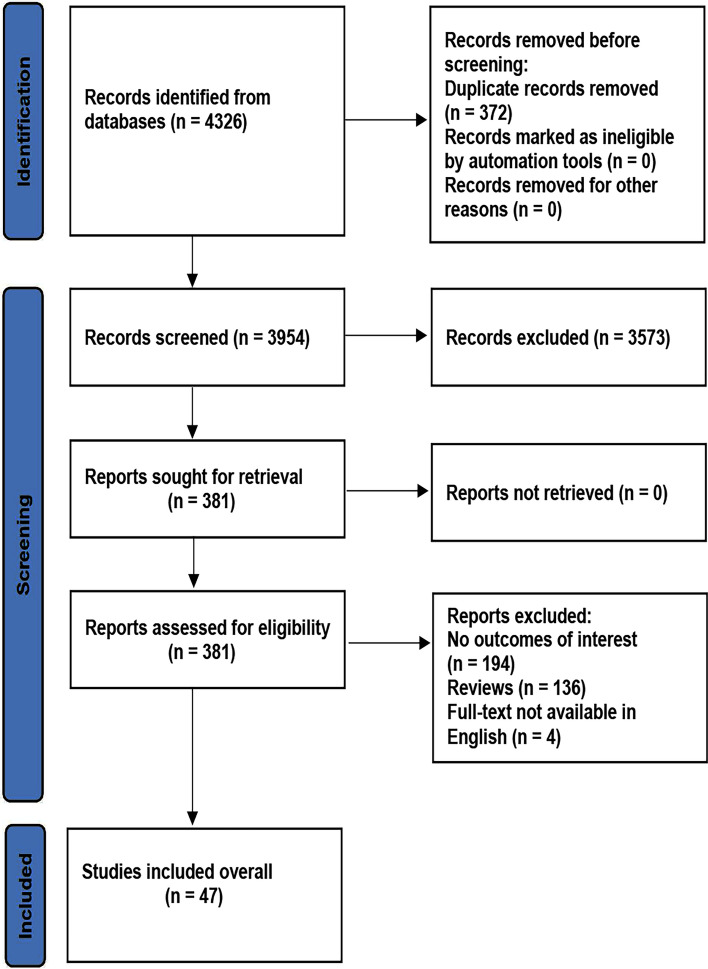
Preferred Reporting Items for Systematic Reviews and Meta-analyses (PRISMA) is an essential reference material and reporting standard for conducting meta-analysis, including seven parts and 27 projects. The present meta-analysis follows the PRISMA Checklist 2020 (Table S12). The initial search of the databases yielded 4326 studies. After screening, 3573 studies were excluded by their title, abstract, and content. The remaining 381 studies were re-estimated by the inclusion criteria. Three hundred and thirty-four studies were further excluded due to the following reasons: 194 studies did not provide outcomes of interest, 136 studies were reviews, and four studies did not provide full-text in English. Finally, 47 studies (26,151 individuals) were included in this study (Fig. 1).
Fig. 1.
Flow diagram of the literature search process
Characteristics of the Included Studies
The characteristics of the included studies are presented in Supplementary Material: Table S1. The plasma TNF-α levels by the genotype of rs1800629 are presented in Supplementary Material: Table S2. The plasma CRP levels by the genotype of rs1800629 are presented in Supplementary Material: Table S3. The plasma IL-6 levels by the genotype of rs1800795 are presented in Supplementary Material: Table S4. The plasma CRP levels by the genotype of rs1800795 are presented in Supplementary Material: Table S5. The genotype distribution frequency of rs1800629 in COVID-19 and non-COVID-19 individuals is presented in Supplementary Material: Table S6. The genotype distribution frequency of rs1800795 in COVID-19 and non-COVID-19 individuals is presented in Supplementary Material: Table S7. The genotype distribution frequency of rs1800629 in severe and non-severe COVID-19 individuals is presented in Supplementary Material: Table S8. The genotype distribution frequency of rs1800795 in severe and non-severe COVID-19 individuals is presented in Supplementary Material: Table S9. The genotype distribution frequency of rs1800629 in COVID-19 dead and non-dead individuals is presented in Supplementary Material: Table S10. The genotype distribution frequency of rs1800795 in COVID-19 dead and non-dead individuals is presented in Supplementary Material: Table S11. The PRISMA Checklist 2020 is presented in Supplementary Material: Table S12. The forest plot of the meta-analysis between rs1800629 and COVID-19 susceptibility is presented in Supplementary Material: Figure S1. The forest plot of the meta-analysis between rs1800795 and COVID-19 severity is presented in Supplementary Material: Figure S2. The forest of the meta-analysis between rs1800795 and COVID-19 mortality is presented in Supplementary Material: Figure S3. The risk bias plot of the meta-analysis between rs1800795 and interleukin-6 levels is presented in Supplementary Material: Figure S4. The risk bias plot of the meta-analysis between rs1800795 and C-reactive protein levels is presented in Supplementary Material: Figure S5. The risk bias plot of the meta-analysis between rs1800629 with COVID-19 severity is presented in Supplementary Material: Figure S6. The risk bias plot of the meta-analysis between rs1800629 with tumor necrosis factor-αis presented in Supplementary Material: Figure S7. The risk bias plot of the meta-analysis between rs1800629 with C-reactive protein levels is presented in Supplementary Material: Figure S8. The risk bias plot of the meta-analysis between rs1800795 with COVID-19 severity is presented in Supplementary Material: Figure S9.
Impacts of rs1800629 and rs1800795 on Inflammatory Biomarkers
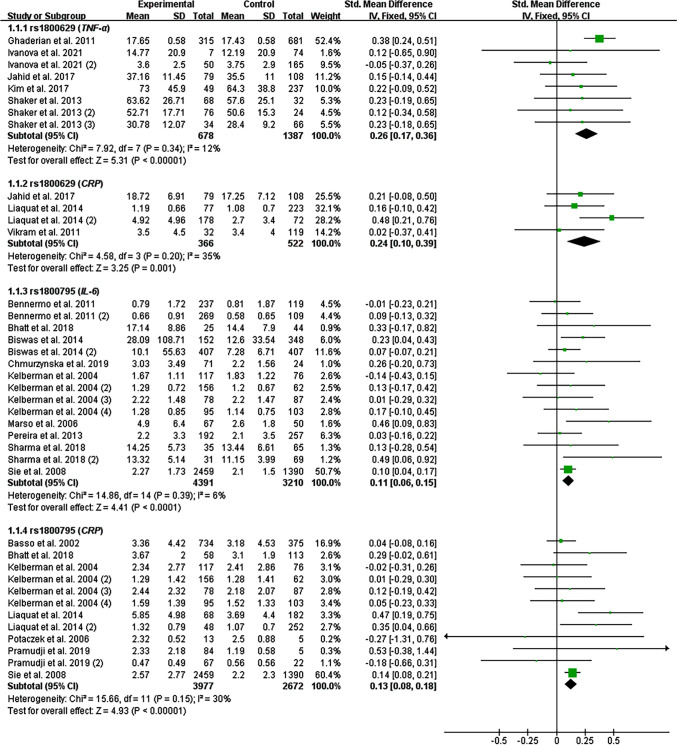
All the results stated below were the data excluded heterogeneity. The consistent finding for the impacts of rs1800629 (Table 1, Fig. 2) and rs1800795 (Table 2, Fig. 2) on inflammatory biomarkers was increased CRP levels. In addition, the rs1800629 A allele (Table 1, Fig. 2) and rs1800795 C allele (Table 2, Fig. 2) elevated TNF-α and IL-6 levels, respectively. Subgroup analysis indicated that the impacts of rs1800629 (Table 1) and rs1800795 (Table 2) on inflammatory biomarkers were significant in Caucasians.
Table 1.
Impacts of TNF-α rs1800629 variant on systemic inflammatory profiles and COVID-19 clinical outcomes
| Groups or subgroups | PH | OR (95% CI) | POR | Groups or subgroups | PH | OR (95% CI) | POR |
|---|---|---|---|---|---|---|---|
| Overall results | Recalculated results | ||||||
| TNF-α | TNF-α | ||||||
| Dominant model (GA + AA vs. GG) | Dominant model (GA + AA vs. GG) | ||||||
| All | < 0.001 | 0.62 (0.27–0.97) | < 0.01 | All | 0.34 | 0.26 (0.17–0.36) | < 0.001 |
| Caucasian | < 0.001 | 0.44 (0.08–0.81) | 0.02 | Caucasian | 0.22 | 0.29 (0.18–0.40) | < 0.001 |
| Indian | < 0.001 | 0.41 (−0.53–1.34) | 0.39 | Indian | – | – | – |
| CRP | CRP | ||||||
| Dominant model (GA + AA vs. GG) | Dominant model (GA + AA vs. GG) | ||||||
| All | < 0.01 | 0.39 (0.17–0.60) | < 0.001 | All | 0.20 | 0.24 (0.10–0.39) | < 0.01 |
| Caucasian | 0.01 | 0.44 (0.15–0.73) | < 0.01 | Caucasian | 0.09 | 0.31 (0.12–0.50) | < 0.01 |
| Indian | 0.02 | 0.31 (−0.06–0.69) | 0.10 | Indian | 0.46 | 0.14 (−0.09–0.38) | 0.23 |
| COVID-19 susceptibility | COVID-19 susceptibility | ||||||
| Allelic model (A vs. G) | Allelic model (A vs. G) | ||||||
| All | < 0.001 | 1.19 (0.81–1.75) | 0.38 | All | 0.06 | 0.85 (0.67–1.07) | 0.16 |
| Caucasian | < 0.001 | 1.25 (0.76–2.06) | 0.39 | Caucasian | 0.77 | 1.59 (1.07–2.37) | 0.02 |
| Asian | 0.04 | 1.02 (0.46–2.25) | 0.97 | Asian | 0.04 | 0.97 (0.72–1.31) | 0.84 |
| Additive model (AA vs. GG) | Additive model (AA vs. GG) | ||||||
| All | < 0.001 | 1.04 (0.41–2.64) | 0.93 | All | 0.33 | 1.01 (0.59–1.73) | 0.97 |
| Caucasian | < 0.001 | 1.07 (0.36–3.19) | 0.91 | Caucasian | 0.33 | 1.06 (0.57–1.95) | 0.86 |
| Asian | 0.16 | 0.96 (0.13–7.2) | 0.97 | Asian | 0.16 | 0.79 (0.25–2.46) | 0.68 |
| Heterozygote model (GA vs. GG) | Heterozygote model (GA vs. GG) | ||||||
| All | < 0.001 | 1.35 (0.89–2.06) | 0.16 | All | 0.15 | 0.78 (0.60–1.02) | 0.07 |
| Caucasian | < 0.001 | 1.42 (0.78–2.59) | 0.25 | Caucasian | 0.38 | 1.88 (1.20–2.97) | 0.01 |
| Asian | 0.36 | 1.01 (0.72–1.43) | 0.94 | Asian | 0.36 | 1.01 (0.72–1.40) | 0.97 |
| Recessive model (AA vs. GG + GA) | Recessive model (AA vs. GG + GA) | ||||||
| All | < 0.01 | 0.98 (0.50–1.92) | 0.96 | All | 0.30 | 1.01 (0.59–1.73) | 0.97 |
| Caucasian | < 0.001 | 0.98 (0.46–2.07) | 0.96 | Caucasian | 0.27 | 1.06 (0.58–1.94) | 0.86 |
| Asian | 0.17 | 0.96 (0.13–7.00) | 0.96 | Asian | 0.17 | 0.78 (0.25–2.45) | 0.67 |
| Dominant model (GA + AA vs. GG) | Dominant model (GA + AA vs. GG) | ||||||
| All | < 0.001 | 1.30 (0.82–2.05) | 0.26 | All | 0.11 | 0.80 (0.62–1.04) | 0.09 |
| Caucasian | < 0.001 | 1.37 (0.73–2.60) | 0.33 | Caucasian | 0.74 | 1.80 (1.16–2.79) | 0.01 |
| Asian | 0.16 | 1.06 (0.57–1.97) | 0.86 | Asian | 0.16 | 0.99 (0.72–1.36) | 0.94 |
| Overdominant model (GA vs. GG + AA) | Overdominant model (GA vs. GG + AA) | ||||||
| All | 0.01 | 1.23 (0.93–1.64) | 0.15 | All | 0.36 | 1.08 (0.93–1.26) | 0.33 |
| Asian | 0.37 | 1.01 (0.73–1.41) | 0.94 | Asian | 0.37 | 1.01 (0.73–1.41) | 0.94 |
| Caucasian | < 0.01 | 1.28 (0.87–1.89) | 0.20 | Caucasian | 0.18 | 0.90 (0.75–1.07) | 0.22 |
| COVID-19 severity | COVID-19 severity | ||||||
| Allelic model (A vs. G) | Allelic model (A vs. G) | ||||||
| All | < 0.001 | 1.71 (0.72–4.06) | 0.22 | All | 0.34 | 1.21 (0.99–1.48) | 0.06 |
| Caucasian | < 0.001 | 1.66 (0.54–5.05) | 0.37 | Caucasian | 0.11 | 1.16 (0.92–1.45) | 0.21 |
| Asian | 0.67 | 1.45 (0.39–5.40) | 0.58 | Asian | 0.67 | 1.47 (0.40–5.39) | 0.56 |
| Mexican | 0.40 | 1.39 (0.86–2.23) | 0.18 | Mexican | 0.40 | 1.41 (0.88–2.27) | 0.15 |
| Additive model (AA vs. GG) | Additive model (AA vs. GG) | ||||||
| All | < 0.001 | 2.44 (0.21–28.16) | 0.47 | All | 0.93 | 1.09 (0.58–2.05) | 0.79 |
| Caucasian | < 0.001 | 3.56 (0.17–74.49) | 0.41 | Caucasian | 0.81 | 1.14 (0.59–2.20) | 0.71 |
| Heterozygote model (GA vs. GG) | Heterozygote model (GA vs. GG) | ||||||
| All | < 0.001 | 1.60 (0.89–2.89) | 0.12 | All | 0.25 | 1.26 (0.99–1.59) | 0.06 |
| Caucasian | < 0.001 | 1.63 (0.69–3.86) | 0.27 | Caucasian | 0.09 | 1.18 (0.89–1.55) | 0.25 |
| Asian | 0.98 | 2.73 (0.57–13.15) | 0.21 | Asian | 0.98 | 2.73 (0.57–13.17) | 0.21 |
| Mexican | 0.37 | 137 (0.84–2.22) | 0.21 | Mexican | 0.37 | 1.40 (0.86–2.26) | 0.18 |
| Recessive model (AA vs. GG + GA) | Recessive model (AA vs. GG + GA) | ||||||
| All | < 0.001 | 1.66 (0.45–6.10) | 0.45 | All | 0.94 | 0.84 (0.46–1.54) | 0.58 |
| Caucasian | < 0.001 | 2.01 (0.48–8.49) | 0.34 | Caucasian | 0.86 | 1.25 (0.67–2.33) | 0.49 |
| Dominant model (GA + AA vs. GG) | Dominant model (GA + AA vs. GG) | ||||||
| All | < 0.001 | 1.83 (0.86–3.93) | 0.12 | All | 0.27 | 1.26 (1.00–1.58) | 0.05 |
| Caucasian | < 0.001 | 2.01 (0.64–6.29) | 0.23 | Caucasian | 0.08 | 1.19 (0.91–1.55) | 0.21 |
| Asian | 0.81 | 1.97 (0.47–8.30) | 0.36 | Asian | 0.81 | 1.99 (0.47–8.32) | 0.35 |
| Mexican | 0.37 | 1.39 (0.85–2.25) | 0.19 | Mexican | 0.37 | 1.41 (0.87–2.29) | 0.16 |
| Overdominant model (GA vs. GG + AA) | Overdominant model (GA vs. GG + AA) | ||||||
| All | < 0.01 | 1.10 (0.75–1.60) | 0.62 | All | 0.21 | 0.81 (0.65–1.02) | 0.08 |
| Caucasian | < 0.01 | 0.98 (0.63–1.52) | 0.91 | Caucasian | 0.08 | 1.15 (0.88–1.50) | 0.32 |
| Asian | 0.99 | 2.81 (0.58–13.51) | 0.20 | Asian | 0.99 | 2.81 (0.58–13.53) | 0.20 |
| Mexican | 0.37 | 1.37 (0.84–2.22) | 0.21 | Mexican | 0.37 | 1.40 (0.86–2.26) | 0.18 |
| COVID-19 mortality | COVID-19 mortality | ||||||
| Allelic model (A vs. G) | Allelic model (A vs. G) | ||||||
| All | < 0.001 | 2.64 (0.54–12.84) | 0.23 | All | 0.76 | 1.07 (0.70–1.65) | 0.74 |
| Caucasian | < 0.001 | 4.43 (0.10–190.76) | 0.44 | Caucasian | 0.96 | 0.95 (0.54–1.65) | 0.84 |
| Asian | 0.44 | 133 (0.69–2.56) | 0.40 | Asian | 0.44 | 1.30 (0.68–2.51) | 0.43 |
| Additive model (AA vs. GG) | Additive model (AA vs. GG) | ||||||
| All | < 0.001 | 2.35 (0.15–36.72) | 0.54 | All | 0.94 | 0.71 (0.20–2.57) | 0.60 |
| Caucasian | < 0.01 | 3.05 (0.08–122.11) | 0.55 | Caucasian | 0.88 | 0.65 (0.16–2.70) | 0.56 |
| Heterozygote model (GA vs. GG) | Heterozygote model (GA vs. GG) | ||||||
| All | 0.92 | 1.44 (0.85–2.43) | 0.18 | All | 0.92 | 1.44 (0.85–2.43) | 0.18 |
| Caucasian | 0.78 | 1.38 (0.62–3.07) | 0.43 | Caucasian | 0.78 | 1.38 (0.62–3.07) | 0.43 |
| Asian | 0.53 | 1.48 (0.74–2.98) | 0.27 | Asian | 0.53 | 1.48 (0.74–2.98) | 0.27 |
| Recessive model (AA vs. GG + GA) | Recessive model (AA vs. GG + GA) | ||||||
| All | < 0.001 | 2.72 (0.09–80.55) | 0.56 | All | 0.92 | 1.65 (0.49–5.61) | 0.42 |
| Caucasian | < 0.001 | 3.77 (0.04–351.71) | 0.57 | Caucasian | 0.86 | 0.56 (0.15–2.13) | 0.39 |
| Dominant model (GA + AA vs. GG) | Dominant model (GA + AA vs. GG) | ||||||
| All | 0.07 | 2.30 (1.45–3.66) | < 0.001 | All | 0.07 | 2.30 (1.45–3.66) | < 0.001 |
| Caucasian | 0.01 | 3.17 (1.62–6.18) | < 0.01 | Caucasian | 0.01 | 3.17 (1.62–6.18) | < 0.01 |
| Asian | 0.51 | 1.41 (0.70–2.83) | 0.33 | Asian | 0.51 | 1.41 (0.70–2.83) | 0.33 |
| Overdominant model (GA vs. GG + AA) | Overdominant model (GA vs. GG + AA) | ||||||
| All | < 0.001 | 0.92 (0.21–3.97) | 0.91 | All | 0.92 | 0.66 (0.40–1.11) | 0.11 |
| Caucasian | < 0.001 | 0.43 (0.02–10.55) | 0.60 | Caucasian | 0.77 | 1.53 (0.72–3.26) | 0.27 |
| Asian | 0.53 | 1.51 (0.75–3.04) | 0.25 | Asian | 0.53 | 1.49 (0.74–3.00) | 0.26 |
TNF-α tumor necrosis factor-α gene; CRP C-reactive protein; PH P for heterogeneity; OR odds ratio
Fig. 2.
Forest plot of the meta-analysis between inflammatory cytokines variants and inflammatory biomarkers
Table 2.
Impacts of IL-6 rs1800795 variant on systemic inflammatory profiles and COVID-19 clinical outcomes
| Groups or subgroups | PH | OR (95% CI) | POR | Groups or subgroups | PH | OR (95% CI) | POR |
|---|---|---|---|---|---|---|---|
| Overall results | Recalculated results | ||||||
| IL-6 | IL-6 | ||||||
| Dominant model (GC + CC vs. GG) | Dominant model (GC + CC vs. GG) | ||||||
| All | < 0.001 | 0.24 (0.14–0.35) | < 0.001 | All | 0.39 | 0.11 (0.06–0.15) | < 0.001 |
| Caucasian | < 0.001 | 0.20 (0.06–0.33) | < 0.01 | Caucasian | 0.40 | 0.10 (0.04–0.15) | < 0.01 |
| Indian | < 0.001 | 0.37 (0.15–0.60) | < 0.01 | Indian | 0.29 | 0.16 (0.15–0.26) | < 0.01 |
| CRP | CRP | ||||||
| Dominant model (GC + CC vs. GG) | Dominant model (GC + CC vs. GG) | ||||||
| All | < 0.001 | 0.23 (0.08–0.37) | < 0.01 | All | 0.15 | 0.13 (0.08–0.18) | < 0.01 |
| Caucasian | < 0.001 | 0.20 (0.04–0.36) | < 0.01 | Caucasian | 0.13 | 0.13 (0.04–0.22) | 0.01 |
| Indian | 0.06 | 0.55 (−0.01–1.11) | 0.06 | Indian | – | – | – |
| COVID-19 susceptibility | COVID-19 susceptibility | ||||||
| Allelic model (C vs. G) | Allelic model (C vs. G) | ||||||
| All | < 0.001 | 0.85 (0.24–2.97) | 0.80 | All | 0.74 | 1.63 (1.22–2.19) | < 0.01 |
| Additive model (CC vs. GG) | Additive model (CC vs. GG) | ||||||
| All | 0.01 | 0.72 (0.06–8.58) | 0.80 | All | 0.69 | 1.72 (1.31–2.27) | < 0.001 |
| Heterozygote model (GC vs. GG) | Heterozygote model (GC vs. GG) | ||||||
| All | < 0.001 | 0.78 (0.21–2.85) | 0.71 | All | 0.21 | 2.72 (1.76–4.22) | < 0.001 |
| Recessive model (CC vs. GG + GC) | Recessive model (CC vs. GG + GC) | ||||||
| All | 0.06 | 0.88 (0.13–5.85) | 0.90 | All | 0.70 | 0.44 (0.19–1.03) | 0.06 |
| Dominant model (GC + CC vs. GG) | Dominant model (GC + CC vs. GG) | ||||||
| All | < 0.001 | 0.78 (0.19–3.26) | 0.73 | All | 0.63 | 1.71 (1.21–2.41) | < 0.01 |
| Overdominant model (GC vs. GG + CC) | Overdominant model (GC vs. GG + CC) | ||||||
| All | 0.01 | 0.85 (0.30–2.44) | 0.76 | All | 0.32 | 2.23 (1.47–3.37) | < 0.001 |
| COVID-19 severity | COVID-19 severity | ||||||
| Allelic model (C vs. G) | Allelic model (C vs. G) | ||||||
| All | 0.01 | 0.96 (0.64–1.45) | 0.85 | All | 0.11 | 0.86 (0.68–1.09) | 0.22 |
| Caucasian | 0.18 | 1.01 (0.62–1.65) | 0.97 | Caucasian | 0.18 | 1.01 (0.74–1.36) | 0.96 |
| Additive model (CC vs. GG) | Additive model (CC vs. GG) | ||||||
| All | 0.08 | 0.83 (0.45–1.52) | 0.54 | All | 0.08 | 0.83 (0.45–1.51) | 0.54 |
| Caucasian | 0.05 | 0.99 (0.49–2.02) | 0.98 | Caucasian | 0.05 | 0.99 (0.49–2.02) | 0.98 |
| Heterozygote model (GC vs. GG) | Heterozygote model (GC vs. GG) | ||||||
| All | 0.01 | 0.92 (0.53–1.59) | 0.76 | All | 0.09 | 0.83 (0.61–1.13) | 0.24 |
| Caucasian | 0.90 | 1.01 (0.68–1.49) | 0.98 | Caucasian | 0.90 | 1.01 (0.68–1.49) | 0.98 |
| Recessive model (CC vs. GG + GC) | Recessive model (CC vs. GG + GC) | ||||||
| All | 0.08 | 0.83 (0.46–1.51) | 0.55 | All | 0.08 | 1.20 (0.66–2.18) | 0.55 |
| Caucasian | 0.05 | 0.99 (0.49–1.98) | 0.97 | Caucasian | 0.05 | 0.99 (0.49–1.98) | 0.97 |
| Dominant model (GC + CC vs. GG) | Dominant model (GC + CC vs. GG) | ||||||
| All | < 0.01 | 0.90 (0.52–1.56) | 0.72 | All | 0.06 | 0.83 (0.62–1.11) | 0.20 |
| Caucasian | 0.55 | 1.01 (0.70–1.45) | 0.98 | Caucasian | 0.55 | 1.01 (0.70–1.46) | 0.97 |
| Overdominant model (GC vs. GG + CC) | Overdominant model (GC vs. GG + CC) | ||||||
| All | 0.01 | 0.93 (0.54–1.59) | 0.79 | All | 0.09 | 1.18 (0.87–1.59) | 0.29 |
| Caucasian | 0.93 | 1.00 (0.68–1.47) | 0.99 | Caucasian | 0.93 | 1.00 (0.68–1.47) | 0.99 |
| COVID-19 mortality | COVID-19 mortality | ||||||
| Allelic model (C vs. G) | Allelic model (C vs. G) | ||||||
| All | 0.08 | 1.01 (0.52–1.96) | 0.98 | All | 0.08 | 1.01 (0.52–1.96) | 0.98 |
| Caucasian | 0.08 | 1.01 (0.52–1.96) | 0.98 | Caucasian | 0.08 | 1.01 (0.52–1.96) | 0.98 |
| Additive model (CC vs. GG) | Additive model (CC vs. GG) | ||||||
| All | 0.25 | 1.09 (0.29–4.10) | 0.90 | All | 0.25 | 1.09 (0.29–4.10) | 0.90 |
| Caucasian | 0.25 | 1.09 (0.29–4.10) | 0.90 | Caucasian | 0.25 | 1.09 (0.29–4.10) | 0.90 |
| Heterozygote model (GC vs. GG) | Heterozygote model (GC vs. GG) | ||||||
| All | 0.21 | 0.90 (0.35–2.31) | 0.83 | All | 0.21 | 0.90 (0.35–2.31) | 0.83 |
| Caucasian | 0.21 | 0.90 (0.35–2.31) | 0.83 | Caucasian | 0.21 | 0.90 (0.35–2.31) | 0.83 |
| Recessive model (CC vs. GG + GC) | Recessive model (CC vs. GG + GC) | ||||||
| All | 0.53 | 0.86 (0.26–2.84) | 0.81 | All | 0.53 | 0.86 (0.26–2.84) | 0.81 |
| Caucasian | 0.53 | 0.86 (0.26–2.84) | 0.81 | Caucasian | 0.53 | 0.86 (0.26–2.84) | 0.81 |
| Dominant model (GC + CC vs. GG) | Dominant model (GC + CC vs. GG) | ||||||
| All | 0.12 | 0.96 (0.41–2.28) | 0.93 | All | 0.12 | 0.96 (0.41–2.28) | 0.93 |
| Caucasian | 0.12 | 0.96 (0.41–2.28) | 0.93 | Caucasian | 0.12 | 0.96 (0.41–2.28) | 0.93 |
| Overdominant model (GC vs. GG + CC) | Overdominant model (GC vs. GG + CC) | ||||||
| All | 0.42 | 1.10 (0.46–2.66) | 0.83 | All | 0.42 | 1.10 (0.46–2.66) | 0.83 |
| Caucasian | 0.42 | 1.10 (0.46–2.66) | 0.83 | Caucasian | 0.42 | 1.10 (0.46–2.66) | 0.83 |
IL-6 interleukin-6 gene; CRP C-reactive protein; PH P for heterogeneity; OR odds ratio
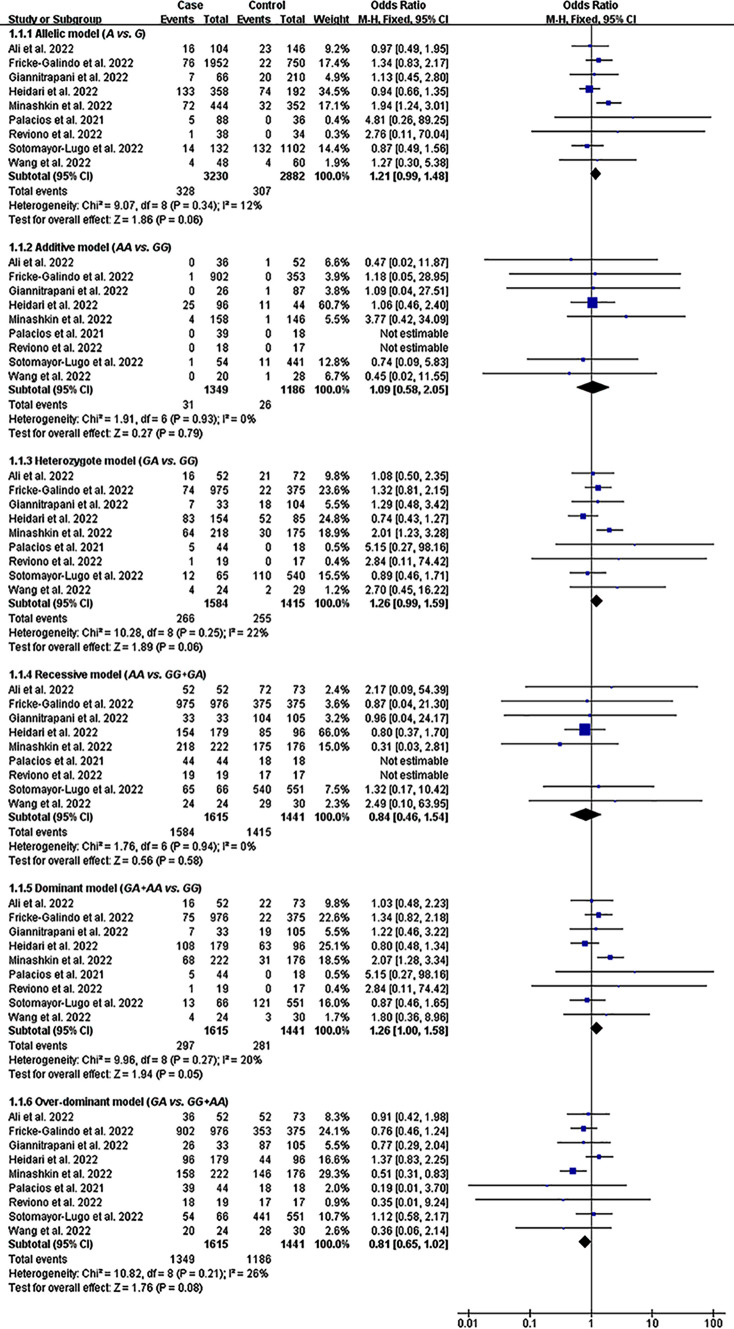
Impacts of rs1800629 and rs1800795 on COVID-19 Susceptibility
The impact of rs1800795 on COVID-19 susceptibility was significant in five genetic models (Table 2, Fig. 3). However, the impact of rs1800629 on COVID-19 susceptibility did not show statistically significant in all genetic models (Table 1, Figure S1). Subgroup analysis indicated that the A allele of rs1800629 significantly increased COVID-19 risk in Caucasians under the allelic, heterozygote, and dominant models (Table 1).
Fig. 3.
Forest plot of the meta-analysis between rs1800795 and COVID-19 susceptibility
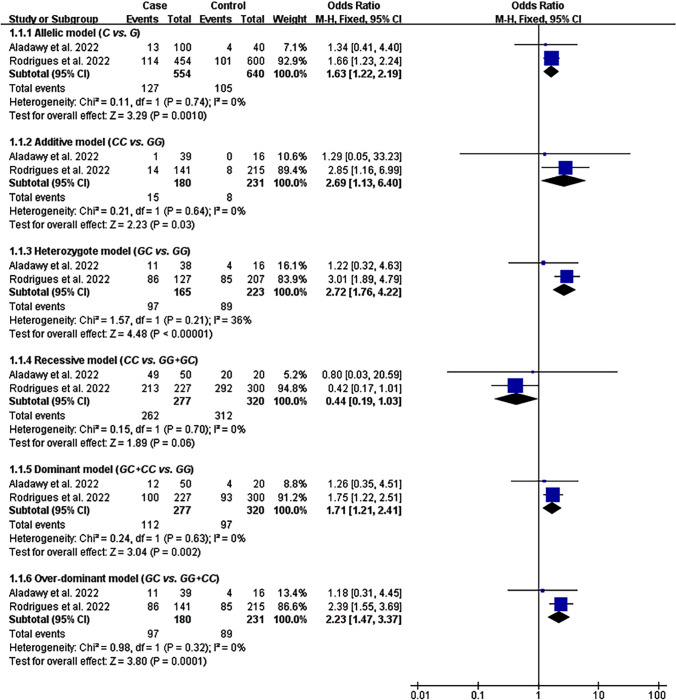
Impacts of rs1800629 and rs1800795 on COVID-19 Severity
The A allele of rs1800629 significantly increased the severity of COVID-19 under the dominant model (Table 1, Fig. 4). In addition, a marginally significant impact was detected between rs1800629 and COVID-19 severity under the allelic, heterozygote, and over-dominant models (Table 1, Fig. 4). In contrast, rs1800795 did not show a statistically significant impact on COVID-19 severity (Table 2, Figure S2).
Fig. 4.
Forest plot of the meta-analysis between rs1800629 and COVID-19 severity
Impacts of rs1800629 and rs1800795 on COVID-2019 Mortality
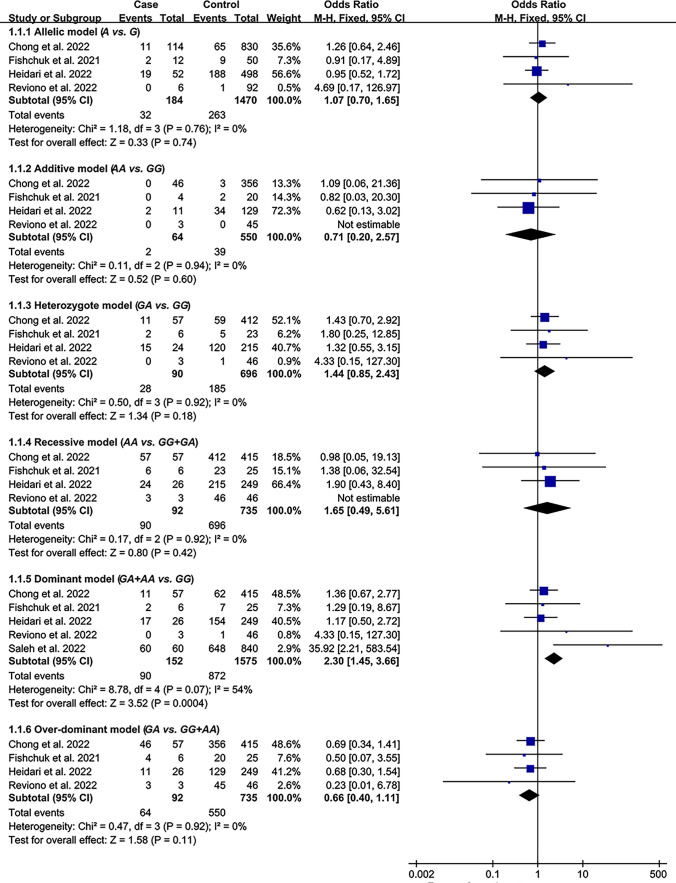
The rs1800629 A allele significantly increased the mortality of COVID-19 under the dominant model (Table 1, Fig. 5). Subgroup analysis indicated that the impact of rs1800629 on COVID-19 mortality was significant in Caucasians (Table 1). However, rs1800795 did not show a statistically significant impact on COVID-19 mortality (Table 2, Figure S3).
Fig. 5.
Forest plot of the meta-analysis between rs1800629 and COVID-19 mortality
Evaluation of Heterogeneity
In analyzing the impacts of rs1800629 on COVID-19 clinical outcomes, two [67] and Heidari et al. (2022), one [67], and one [67] comparison were identified as the main heterogeneity contributor to COVID-19 susceptibility, severity, and mortality. Notably, the recalculated results for susceptibility and severity changed substantially after excluding those comparisons (please see Table 1 for more details).
In analyzing the impacts of rs1800795 on COVID-19 clinical outcomes, one (Balzanelli et al. 2022) and one (Verma et al. 2022) comparison were identified as the main heterogeneity contributor to COVID-19 susceptibility and severity. Notably, the recalculated results for susceptibility changed substantially after excluding those comparisons (please see Table 2 for more details).
Publication Bias Test
Begg's test did not find any publication bias in the present study, which was confirmed by Egger's regression test.
Risk of Bias/Quality Assessment
In an analysis of the risk bias of rs1800795 with IL-6 (Figure S4) and CRP (Figure S5) and rs1800629 with COVID-19 severity (Figure S6), the majority of studies (80–92.4%) presented with green color (Figure S4–S6), indicating a low risk of bias. In addition, no risk of bias was detected for rs1800629 with TNF-α (Figure S7) and CRP (Figure S8) and rs1800795 with COVID-19 severity (Figure S9). In summary, the current literature included is of high quality due to a low risk of bias (Figure S4–S9).
Discussion
The A allele of rs1800629 significantly elevated TNF-α and CRP levels and increased COVID-19 severity and mortality. In contrast, the C allele of rs1800795 elevated IL-6 and CRP levels and increased COVID-19 susceptibility.
The up-regulated inflammatory parameters (Tables 1, 2, Fig. 2) associated with pro-inflammatory cytokines variants may be attributed to the increased transcriptional activity of IL-6 and TNF-α [33, 34]. Moreover, two plausible mechanisms can be proposed to explain the impacts of pro-inflammatory cytokines variants on COVID-19 clinical outcomes. (1) By inducing a cytokine storm. IL-6 and TNF-α are critical components of cytokine storm [37, 49–55]. The elevated TNF-α and IL-6 levels associated with pro-inflammatory cytokines variants (Tables 1, 2) may be helpful to the formation of cytokine storm, thus deteriorating COVID-19 outcomes (Tables 1, 2). (2) By inducing lymphocytopenia. Lymphocytes play a critical role in controlling SARS-CoV-2 infection [63]. A low abundance of CD8+ T and CD4+ T lymphocytes was associated with severe illness and high mortality of COVID-19 [64–66]. Therefore, the lymphocytopenia associated with rs1800629 [67] and rs1800795 [68] may worsen COVID-19 outcomes.
The A allele of rs1800629 vastly increased TNF-α and CRP levels (Table 1, Fig. 2), indicating that individuals with the rs1800629 A allele are at high risk of cytokine storm and may have poor outcomes for COVID-19. Intriguingly, this speculation was verified in the present study, whereas the rs1800629 A allele significantly increased the severity and mortality of COVID-19 (Table 1, Figs. 4, 5). Notably, the increased levels of IL-6 were recognized as a maker of severe COVID-19 [69–71]. However, rs1800795 did not impact the severity of COVID-19 despite elevating IL-6 levels (Table 2, Fig. 2, Figure S2), since only 381 individuals were included for analyzing the impact of rs1800795 on COVID-19 severity, which largely lowered the statistical power and needs to be confirmed by future clinical trials.
The susceptibility of COVID-19 was increased 1.59–1.88 fold in Caucasians with the rs1800629 A allele (Table 1), indicating that Caucasians with the rs1800629 A allele are at high risk of suffering COVID-19. In addition, the mortality of COVID-19 increased 3.17-fold in Caucasians with the rs1800629 A allele (Table 1), suggesting that Caucasians with the rs1800629 A allele are at high risk of death. The specific reason why the impacts of rs1800629 on COVID-19 susceptibility and mortality were significant in Caucasians rather than in Asians was likely that the distribution frequency of the A allele was much higher in Caucasian individuals with COVID-19 (Caucasian individuals with COVID-19 vs. Asian individuals with COVID-19 = [10.7–23.4%] vs. [1.8–7.3%]) [72, 73].
The present study showed that pro-inflammatory cytokines variants remodeled the systemic inflammatory profile and impacted COVID-19 outcomes. Since the cytokine storm was closely linked to COVID-19 outcomes [43–48], it indicated that the impacts of pro-inflammatory cytokines variants on COVID-19 outcomes (Tables 1, 2) were mediated, at least partly, by the impacts of pro-inflammatory cytokines variants on systemic inflammatory profile (Tables 1, 2). Since anti-TNF-α (e.g., infliximab) and anti-IL-6 (e.g., tocilizumab) therapies were effective in individuals with severe illness [74–76], it indicated that targeting TNF-α and IL-6 may help prevent COVID-19 progression in individuals with rs1800629 and rs1800795. Large-scale clinical trials are urgently needed to verify this hypothesis.
Moreover, according to Anastassopoulou et al. [23]. proposals, safe and effective vaccines should be given priority to individuals at high genetic risk of developing COVID-19. Since rs1800629 A allele and rs1800795 C allele significantly increased the risk of COVID-19 (Tables 1, 2, Fig. 3), it indicated that individuals with variants of rs1800795 and rs1800629 should be prioritized for vaccination against COVID-19. Genetic screening of rs1800795 and rs1800629 is necessary for the public to achieve this goal.
Conclusions
The C allele of rs1800795 increased the risk of COVID-19 and plasma levels of IL-6 and CRP. In contrast, the A allele of rs1800629 increased the severity and mortality of COVID-19 and plasma levels of TNF-α and CRP. These results hint that rs1800629 and rs1800795 variants of pro-inflammatory cytokines have significant impacts on COVID-19 clinical outcomes and systemic inflammatory profile. rs1800629 may serve as a genetic marker for severe COVID-19.
Supplementary Information
Below is the link to the electronic supplementary material.
Author Contributions
ZL, ZW, and SH contributed to the study's conception and design. Material preparation, data collection, and formal analysis were performed by KT. The first draft of the manuscript was written by XD and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Funding
The authors did not receive support from any organization for the submitted work.
Data Availability
All data generated or analyzed during this study are included in this published article and its Supplementary Material.
Declarations
Conflict of Interest
The authors have no competing interests to declare that are relevant to the content of this article.
Ethics Approval
This is an observational study. The Suining Central Hospital Research Ethics Committee has confirmed that no ethical approval is required.
Consent
Not applicable.
Footnotes
XueJun Deng and Kai Tang contributed equally to this work.
Contributor Information
Zhiqiang Wang, Email: wzq3344@163.com.
Suyu He, Email: hesuyu2009@163.com.
Zhi Luo, Email: 2020103030002@whu.edu.cn.
References
- 1.Ghazvini K, Karbalaei M, Keikha M. What are the clinical benefits of tocilizumab for COVID-19 patients? Evidence from available case-control studies. Le Pharm Hosp Clin. 2021;56:217–221. [Google Scholar]
- 2.World Health Organization Data. WHO COVID-19 dashboard. https://covid19.who.int/. Accessed Jan 2020.
- 3.Liao Y, Feng Y, Wang B, et al. Clinical characteristics and prognostic factors of COVID-19 patients progression to severe: a retrospective, observational study. Aging (Albany NY) 2020;12:18853–18865. doi: 10.18632/aging.103931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Covino M, De Matteis G, Santoro M, et al. Clinical characteristics and prognostic factors in COVID-19 patients aged ≥80 years. Geriatr Gerontol Int. 2020;20:704–708. doi: 10.1111/ggi.13960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andrade LA, Bagno FF, Sérgio SA, et al. Heterogeneity of humoral response patterns in mildly symptomatic, non-hospitalized COVID-19 patients: a one-year longitudinal study. Exp Biol Med (Maywood) 2023;248:874–882. doi: 10.1177/15353702231157941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Di Pietro GM, Ronzoni L, Meschia LM, et al. SARS-CoV-2 infection in children: a 24 months experience with focus on risk factors in a pediatric tertiary care hospital in Milan, Italy. Front Pediatr. 2023;11:1082083. doi: 10.3389/fped.2023.1082083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang C, Wang Z, Wang G, Lau JY, Zhang K, Li W. COVID-19 in early 2021: current status and looking forward. Signal Transduct Target Ther. 2021;6:114. doi: 10.1038/s41392-021-00527-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Verity R, Okell LC, Dorigatti I, et al. Estimates of the severity of coronavirus disease 2019: a model-based analysis. Lancet Infect Dis. 2020;20:669–677. doi: 10.1016/S1473-3099(20)30243-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.World Health Organization. Report of the WHO-China joint mission on coronavirus disease 2019 (COVID-19). 2020. https://www.who.int/publications/i/item/report-of-the-who-china-joint-mission-oncoronavirus-disease-2019-(covid-19). Accessed 28 Feb 2020.
- 10.Attaway AH, Scheraga RG, Bhimraj A, Biehl M, Hatipoğlu U. Severe covid-19 pneumonia: pathogenesis and clinical management. BMJ. 2021;372:n436. doi: 10.1136/bmj.n436. [DOI] [PubMed] [Google Scholar]
- 11.Zhou F, Yu T, Du R, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wei B, Liu Y, Li H, Peng Y, Luo Z. Impact of alcohol consumption on coronavirus disease 2019 severity: a systematic review and meta-analysis. J Med Virol. 2023;95:e28547. doi: 10.1002/jmv.28547. [DOI] [PubMed] [Google Scholar]
- 13.Liu Y, Li H, Peng Y, et al. Impacts of pregnancy and menopause on COVID-19 severity: a systematic review and meta-analysis of 46 million women. QJM. 2023;116:755–765. doi: 10.1093/qjmed/hcad106. [DOI] [PubMed] [Google Scholar]
- 14.Zheng Z, Peng F, Xu B, et al. Risk factors of critical & mortal COVID-19 cases: a systematic literature review and meta-analysis. J Infect. 2020;81:16–25. doi: 10.1016/j.jinf.2020.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao YD, Ding M, Dong X, et al. Risk factors for severe and critically ill COVID-19 patients: a review. Allergy. 2021;76:428–455. doi: 10.1111/all.14657. [DOI] [PubMed] [Google Scholar]
- 16.Docherty AB, Harrison EM, Green CA, et al. Features of 20,133 UK patients in hospital with covid-19 using the ISARIC WHO clinical characterisation protocol: prospective observational cohort study. BMJ. 2020;369:m1985. doi: 10.1136/bmj.m1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakanishi T, Willett J, Farjoun Y, et al. Alternative splicing in lung influences COVID-19 severity and respiratory diseases. Nat Commun. 2023;14:6198. doi: 10.1038/s41467-023-41912-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang Y, Lu Y, Huang YM, et al. Obesity in patients with COVID-19: a systematic review and meta-analysis. Metabolism. 2020;113:154378. doi: 10.1016/j.metabol.2020.154378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hardy N, Vegivinti CTR, Mehta M, et al. Mortality of COVID-19 in patients with hematological malignancies versus solid tumors: a systematic literature review and meta-analysis. Clin Exp Med. 2023;23:1945–1959. doi: 10.1007/s10238-023-01004-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yonker LM, Swank Z, Bartsch YC, et al. Circulating Spike protein detected in post-COVID-19 mRNA vaccine myocarditis. Circulation. 2023;147:867–876. doi: 10.1161/CIRCULATIONAHA.122.061025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rakanidis Machado N, Fagundes BO, Fernandes IG, Terra De ApoenaReche D, Sato MN, Victor JR. IgG from patients with mild or severe COVID-19 reduces the frequency and modulates the function of peripheral mucosal-associated invariant T cells in PBMCs from healthy individuals. Biomed Rep. 2023;19:95. doi: 10.3892/br.2023.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.COVID-19 Host Genetics Initiative. Mapping the human genetic architecture of COVID-19. Nature. 2021;600:472–7. [DOI] [PMC free article] [PubMed]
- 23.Anastassopoulou C, Gkizarioti Z, Patrinos GP, Tsakris A. Human genetic factors associated with susceptibility to SARS-CoV-2 infection and COVID-19 disease severity. Hum Genomics. 2020;14:40. doi: 10.1186/s40246-020-00290-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elhabyan A, Elyaacoub S, Sanad E, Abukhadra A, Elhabyan A, Dinu V. The role of host genetics in susceptibility to severe viral infections in humans and insights into host genetics of severe COVID-19: a systematic review. Virus Res. 2020;289:198163. doi: 10.1016/j.virusres.2020.198163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grolmusz VK, Bozsik A, Papp J, Patócs A. Germline genetic variants of viral entry and innate immunity may influence susceptibility to SARS-CoV-2 infection: toward a polygenic risk score for risk stratification. Front Immunol. 2021;12:653489. doi: 10.3389/fimmu.2021.653489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jackson CB, Farzan M, Chen B, Choe H. Mechanisms of SARS-CoV-2 entry into cells. Nat Rev Mol Cell Biol. 2022;23:3–20. doi: 10.1038/s41580-021-00418-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Debnath M, Banerjee M, Berk M. Genetic gateways to COVID-19 infection: implications for risk, severity, and outcomes. FASEB J. 2020;34:8787–8795. doi: 10.1096/fj.202001115R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Subbaram K, Ali PSS, Ali S. Enhanced endocytosis elevated virulence and severity of SARS-CoV-2 due to hyperglycemia in type 2 diabetic patients. Gene Rep. 2022;26:101495. doi: 10.1016/j.genrep.2022.101495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wallach T, Raden M, Hinkelmann L, et al. Distinct SARS-CoV-2 RNA fragments activate Toll-like receptors 7 and 8 and induce cytokine release from human macrophages and microglia. Front Immunol. 2023;13:1066456. doi: 10.3389/fimmu.2022.1066456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alimoradi N, Sharqi M, Firouzabadi D, Sadeghi MM, Moezzi MI, Firouzabadi N. SNPs of ACE1 (rs4343) and ACE2 (rs2285666) genes are linked to SARS-CoV-2 infection but not with the severity of disease. Virol J. 2022;19:48. doi: 10.1186/s12985-022-01782-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gupta K, Kaur G, Pathak T, Banerjee I. Systematic review and meta-analysis of human genetic variants contributing to COVID-19 susceptibility and severity. Gene. 2022;844:146790. doi: 10.1016/j.gene.2022.146790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gómez J, Albaiceta GM, Cuesta-Llavona E, et al. The interferon-induced transmembrane protein 3 gene (IFITM3) rs12252 C variant is associated with COVID-19. Cytokine. 2021;137:155354. doi: 10.1016/j.cyto.2020.155354. [DOI] [PubMed] [Google Scholar]
- 33.Totaro F, Cimmino F, Pignataro P, et al. Impact of interleukin-6 -174 G>C gene promoter polymorphism on neuroblastoma. PLoS ONE. 2013;8:e76810. doi: 10.1371/journal.pone.0076810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moscovis SM, Gordon AE, Al Madani OM, et al. Genetic and environmental factors affecting TNF-α responses in relation to sudden infant death syndrome. Front Immunol. 2015;6:374. doi: 10.3389/fimmu.2015.00374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liaquat A, Asifa GZ, Zeenat A, Javed Q. Polymorphisms of tumor necrosis factor-alpha and interleukin-6 gene and C-reactive protein profiles in patients with idiopathic dilated cardiomyopathy. Ann Saudi Med. 2014;34:407–414. doi: 10.5144/0256-4947.2014.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grira N, Lahidheb D, Lamine O, et al. The association of IL-6, TNFα and CRP gene polymorphisms with coronary artery disease in a Tunisian population: a case–control study. Biochem Genet. 2021;59:751–766. doi: 10.1007/s10528-021-10035-0. [DOI] [PubMed] [Google Scholar]
- 37.Frisoni P, Neri M, D'Errico S, et al. Cytokine storm and histopathological findings in 60 cases of COVID-19-related death: from viral load research to immunohistochemical quantification of major players IL-1β, IL-6, IL-15 and TNF-α. Forensic Sci Med Pathol. 2022;18:4–19. doi: 10.1007/s12024-021-00414-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Da BL, Kushner T, El Halabi M, et al. Liver injury in patients hospitalized with coronavirus disease 2019 correlates with hyperinflammatory response and elevated interleukin-6. Hepatol Commun. 2020;5:177–188. doi: 10.1002/hep4.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luo W, Li YX, Jiang LJ, Chen Q, Wang T, Ye DW. Targeting JAK-STAT signaling to control cytokine release syndrome in COVID-19. Trends Pharmacol Sci. 2020;41:531–543. doi: 10.1016/j.tips.2020.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nagarkatti P, Miranda K, Nagarkatti M. Use of cannabinoids to treat acute respiratory distress syndrome and cytokine storm associated with coronavirus disease-2019. Front Pharmacol. 2020;11:589438. doi: 10.3389/fphar.2020.589438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yasuma T, D'Alessandro-Gabazza CN, Kobayashi T, Gabazza EC, Fujimoto H. Microbial burden-associated cytokine storm may explain nonresolving acute respiratory distress syndrome in patients with COVID-19. Am J Respir Crit Care Med. 2022;206:1182–1183. doi: 10.1164/rccm.202207-1266LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lotfi M, Rezaei N. SARS-CoV-2: a comprehensive review from pathogenicity of the virus to clinical consequences. J Med Virol. 2020;92:1864–1874. doi: 10.1002/jmv.26123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Han H, Ma Q, Li C, et al. Profiling serum cytokines in COVID-19 patients reveals IL-6 and IL-10 are disease severity predictors. Emerg Microbes Infect. 2020;9:1123–1130. doi: 10.1080/22221751.2020.1770129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leisman DE, Ronner L, Pinotti R, et al. Cytokine elevation in severe and critical COVID-19: a rapid systematic review, meta-analysis, and comparison with other inflammatory syndromes. Lancet Respir Med. 2020;8:1233–1244. doi: 10.1016/S2213-2600(20)30404-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mulchandani R, Lyngdoh T, Kakkar AK. Deciphering the COVID-19 cytokine storm: systematic review and meta-analysis. Eur J Clin Invest. 2021;51:e13429. doi: 10.1111/eci.13429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tian W, Jiang W, Yao J, et al. Predictors of mortality in hospitalized COVID-19 patients: a systematic review and meta-analysis. J Med Virol. 2020;92:1875–1883. doi: 10.1002/jmv.26050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Melo AKG, Milby KM, Caparroz ALMA, et al. Biomarkers of cytokine storm as red flags for severe and fatal COVID-19 cases: a living systematic review and meta-analysis. PLoS ONE. 2021;16:e0253894. doi: 10.1371/journal.pone.0253894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Udomsinprasert W, Jittikoon J, Sangroongruangsri S, Chaikledkaew U. Circulating levels of interleukin-6 and interleukin-10, but not tumor necrosis factor-alpha, as potential biomarkers of severity and mortality for COVID-19: systematic review with meta-analysis. J Clin Immunol. 2021;41:11–22. doi: 10.1007/s10875-020-00899-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen LYC, Hoiland RL, Stukas S, Wellington CL, Sekhon MS. Confronting the controversy: interleukin-6 and the COVID-19 cytokine storm syndrome. Eur Respir J. 2020;56:2003006. doi: 10.1183/13993003.03006-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Boretti A, Banik B. Modulation of Covid-19 cytokine storm by tocilizumab. J Med Virol. 2022;94:823–828. doi: 10.1002/jmv.27380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Que Y, Hu C, Wan K, et al. Cytokine release syndrome in COVID-19: a major mechanism of morbidity and mortality. Int Rev Immunol. 2022;41:217–230. doi: 10.1080/08830185.2021.1884248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Paniri A, Akhavan-Niaki H. Emerging role of IL-6 and NLRP3 inflammasome as potential therapeutic targets to combat COVID-19: role of lncRNAs in cytokine storm modulation. Life Sci. 2020;257:118114. doi: 10.1016/j.lfs.2020.118114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Donina ZA, Baranova EV, Aleksandrova NP. A comparative assessment of effects of major mediators of acute phase response (IL-1, TNF-α, IL-6) on breathing pattern and survival rate in rats with acute progressive hypoxia. J Evol Biochem Physiol. 2021;57:936–944. doi: 10.1134/S0022093021040177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang P, Zuo Q, Li Y, et al. A vicious cycle: in severe and critically Ill COVID-19 patients. Front Immunol. 2022;13:930673. doi: 10.3389/fimmu.2022.930673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Feldmann M, Maini RN, Woody JN, et al. Trials of anti-tumour necrosis factor therapy for COVID-19 are urgently needed. Lancet. 2020;395:1407–1409. doi: 10.1016/S0140-6736(20)30858-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Umapathy D, Krishnamoorthy E, Mariappanadar V, Viswanathan V, Ramkumar KM. Increased levels of circulating (TNF-α) is associated with (-308G/A) promoter polymorphism of TNF-α gene in diabetic nephropathy. Int J Biol Macromol. 2018;107:2113–2121. doi: 10.1016/j.ijbiomac.2017.10.078. [DOI] [PubMed] [Google Scholar]
- 57.Nascimento H, Vieira E, Coimbra S, et al. Adipokine gene single-nucleotide polymorphisms in Portuguese obese adolescents: associations with plasma concentrations of adiponectin, resistin, IL-6, IL-1β, and TNF-α. Child Obes. 2016;12:300–313. doi: 10.1089/chi.2015.0235. [DOI] [PubMed] [Google Scholar]
- 58.Rana BK, Flatt SW, Health DD, et al. The IL-6 gene promoter SNP and plasma IL-6 in response to diet intervention. Nutrients. 2017;9:552. doi: 10.3390/nu9060552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zakharyan R, Petrek M, Arakelyan A, Mrazek F, Atshemyan S, Boyajyan A. Interleukin-6 promoter polymorphism and plasma levels in patients with schizophrenia. Tissue Antigens. 2012;80:136–142. doi: 10.1111/j.1399-0039.2012.01886.x. [DOI] [PubMed] [Google Scholar]
- 60.DerSimonian R, Kacker R. Random-effects model for meta-analysis of clinical trials: an update. Contemp Clin Trials. 2007;28:105–114. doi: 10.1016/j.cct.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 61.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088–1101. doi: 10.2307/2533446. [DOI] [PubMed] [Google Scholar]
- 62.Savović J, Weeks L, Sterne JA, et al. Evaluation of the Cochrane Collaboration's tool for assessing the risk of bias in randomized trials: focus groups, online survey, proposed recommendations and their implementation. Syst Rev. 2014;3:37. doi: 10.1186/2046-4053-3-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sette A, Crotty S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell. 2021;184:861–880. doi: 10.1016/j.cell.2021.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xu B, Fan CY, Wang AL, et al. Suppressed T cell-mediated immunity in patients with COVID-19: a clinical retrospective study in Wuhan. China J Infect. 2020;81:e51–e60. doi: 10.1016/j.jinf.2020.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Du RH, Liang LR, Yang CQ, et al. Predictors of mortality for patients with COVID-19 pneumonia caused by SARS-CoV-2: a prospective cohort study. Eur Respir J. 2020;55:2000524. doi: 10.1183/13993003.00524-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen J, Qi T, Liu L, et al. Clinical progression of patients with COVID-19 in Shanghai, China. J Infect. 2020;80:e1–e6. doi: 10.1016/j.jinf.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Saleh A, Sultan A, Elashry MA, et al. Association of TNF-αG-308 a promoter polymorphism with the course and outcome of COVID-19 patients. Immunol Invest. 2022;51:546–557. doi: 10.1080/08820139.2020.1851709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ortlepp JR, Metrikat J, Vesper K, et al. The interleukin-6 promoter polymorphism is associated with elevated leukocyte, lymphocyte, and monocyte counts and reduced physical fitness in young healthy smokers. J Mol Med (Berl) 2003;81:578–584. doi: 10.1007/s00109-003-0471-6. [DOI] [PubMed] [Google Scholar]
- 69.Broman N, Rantasärkkä K, Feuth T, et al. IL-6 and other biomarkers as predictors of severity in COVID-19. Ann Med. 2021;53:410–412. doi: 10.1080/07853890.2020.1840621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Henry BM, de Oliveira MHS, Benoit S, Plebani M, Lippi G. Hematologic, biochemical and immune biomarker abnormalities associated with severe illness and mortality in coronavirus disease 2019 (COVID-19): a meta-analysis. Clin Chem Lab Med. 2020;58:1021–1028. doi: 10.1515/cclm-2020-0369. [DOI] [PubMed] [Google Scholar]
- 71.Aziz M, Fatima R, Assaly R. Elevated interleukin-6 and severe COVID-19: a meta-analysis. J Med Virol. 2020;92:2283–2285. doi: 10.1002/jmv.25948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kim YC, Jeong BH. Strong correlation between the case fatality rate of COVID-19 and the rs6598045 single nucleotide polymorphism (SNP) of the interferon-induced transmembrane protein 3 (IFITM3) gene at the population-level. Genes (Basel) 2020;12:42. doi: 10.3390/genes12010042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Leite MM, Gonzalez-Galarza FF, Silva BCCD, Middleton D, Santos EJMD. Predictive immunogenetic markers in COVID-19. Hum Immunol. 2021;82:247–254. doi: 10.1016/j.humimm.2021.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Du P, Geng J, Wang F, Chen X, Huang Z, Wang Y. Role of IL-6 inhibitor in treatment of COVID-19-related cytokine release syndrome. Int J Med Sci. 2021;18:1356–1362. doi: 10.7150/ijms.53564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Soin AS, Kumar K, Choudhary NS, et al. Tocilizumab plus standard care versus standard care in patients in India with moderate to severe COVID-19-associated cytokine release syndrome (COVINTOC): an open-label, multicentre, randomised, controlled, phase 3 trial. Lancet Respir Med. 2021;9:511–521. doi: 10.1016/S2213-2600(21)00081-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Syed MN, Shah M, Shin DB, Wan MT, Winthrop KL, Gelfand JM. Effect of anti-tumor necrosis factor therapy on the risk of respiratory tract infections and related symptoms in patients with psoriasis—a meta-estimate of pivotal phase 3 trials relevant to decision making during the COVID-19 pandemic. J Am Acad Dermatol. 2021;84:161–163. doi: 10.1016/j.jaad.2020.08.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its Supplementary Material.