Abstract
Rhesus monkey rhadinovirus (RRV) is a gamma-2 herpesvirus that exhibits a considerable degree of similarity to the human Kaposi's sarcoma-associated herpesvirus (KSHV). The R1 protein of RRV is distantly related to the K1 protein of KSHV, and R1, like K1, can contribute to cell growth transformation. In this study we analyzed the ability of the cytoplasmic tail of R1 to function as a signal transducer. The cytoplasmic domain of the R1 protein contains several tyrosine residues whose phosphorylation is induced in cells expressing Syk kinase. Expression of a CD8 chimera protein containing the extracellular and transmembrane domains of CD8 fused to the cytoplasmic domain of R1 mobilized intracellular calcium and induced cellular tyrosine phosphorylation in B cells upon stimulation with anti-CD8 antibody. None of the CD8-R1 cytoplasmic deletion mutants tested were able to mobilize intracellular calcium or to induce tyrosine phosphorylation to a significant extent upon addition of anti-CD8 antibody. Expression of wild-type R1 protein activated nuclear factor of activated T lymphocytes (NFAT) eightfold in B cells in the absence of antibody stimulation; expression of the CD8-R1C chimera strongly induced NFAT activity (60-fold) but only upon the addition of anti-CD8 antibody. We conclude that the cytoplasmic domain of R1 is capable of transducing signals that elicit B-lymphocyte activation events. The signal-inducing properties of R1 appear to be similar to those of K1 but differ in that the required sequences are distributed over a much longer stretch of the cytoplasmic domain (>150 amino acids). In addition, the induction of calcium mobilization was considerably longer in duration and stronger with R1 than with K1.
Src homology-2 (SH2) domains are present in a number of signal-transducing proteins including protein kinases, phosphatases, and phospholipases (6, 23, 24). Sequences that bind to SH2 domains are characterized by phosphorylated tyrosine residues present in a variety of cellular signaling proteins such as lymphocyte receptors, adapter proteins, protein kinases, and transcription factors. The sequences containing these phosphorylated tyrosine residues are referred to as SH2 binding motifs and serve to recruit cellular proteins containing SH2 domains into lymphocyte receptor-mediated signaling complexes. SH2 binding motifs consist of a tyrosine residue followed by three amino acids, YXX(L/I/V), whose sequence is specific for binding a particular SH2 domain (23, 28). These motifs can occur in either a singular or tandem fashion. Hematopoietic lymphocyte receptors including immunoglobulin-α (Igα), Igβ, T-cell receptor-ɛ, IgE-β, and IgE-γ contain two such SH2 binding motifs in tandem, and this motif has been termed the immunoreceptor tyrosine-based activation motif (ITAM) (3, 4, 13, 16, 32, 33). The archetypal ITAM consists of a stretch of negatively charged amino acids preceding the two tyrosine-containing sequences: (D/E)X7(D/E)X2YX2LX7–10YX2L/I, where X is any amino acid. In B cells, T cells, and mast cells, the ITAMs have been shown to be critical for interactions with Src family (Src, Lyn, Blk, Lck, and Fyn) and Syk family (Syk and Zap70) kinases (16, 19, 29). The antigen binding site of the B-cell receptor (BCR) is a membrane-bound Ig that is noncovalently attached to a disulfide-linked Igα-Igβ heterodimer that serves as the major signaling component of the BCR complex. In the resting state of B cells, the Src family of protein tyrosine kinases is associated with the Igα subunit of the Igα-Igβ heterodimer. Binding of antigen to the membrane Ig results in a higher affinity of the Src kinase family members for the BCR, leading to the phosphorylation of the tyrosine residues present in the ITAMs of the Igα and Igβ subunits. This in turn recruits the Syk family of protein kinases to the BCR complex in a manner that is SH2 domain dependent. The immediate effect is an activated receptor complex that provides binding sites for signal adapter and transducer proteins that initiate a signal cascade leading to proliferation and differentiation of the B cell.
Kaposi's sarcoma-associated herpesvirus (KSHV), also called Human herpesvirus 8, has been consistently identified in Kaposi's sarcomas, body cavity-based lymphomas, and some forms of multicentric Castleman's disease (7, 14, 25, 26, 30). Recently a new herpesvirus called Rhesus monkey rhadinovirus (RRV), which is closely related to but distinct from KSHV, has been isolated from rhesus monkeys (10). RRV is the closest known relative of KSHV on the basis of sequence similarities and structural organization of the genomes (1, 27). Similar to KSHV, RRV has been shown to be present predominantly in B lymphocytes (22). At a genomic location equivalent to that of the saimiri transforming protein (STP) of herpesvirus saimiri (HVS) and K1 of KSHV, we have identified a novel open reading frame called the R1 open reading frame at the left end of the RRV genome (9). The R1 protein is a glycosylated transmembrane protein of approximately 70 kDa. Expression of R1 in rodent fibroblasts induces transformation, resulting in morphological changes and focus formation (9). Injection of R1-expressing cells into nude mice induced the formation of multifocal, disseminated tumors (9). A recombinant herpesvirus in which the STP oncogene of HVS was replaced with the R1 gene was capable of immortalizing T lymphocytes to interleukin-2-independent growth, further indicating the transforming potential of the R1 protein (9).
R1, like K1, has features of a receptor protein. While the predicted extracellular domain of R1 of RRV exhibits considerable homology and is similar in length to the extracellular domain of K1 of KSHV, the cytoplasmic domain of R1 is markedly different from that of K1. The K1 protein has a short cytoplasmic tail containing only 38 amino acids (20). This short cytoplasmic tail has been shown to contain an ITAM that is capable of eliciting cellular signal transduction, evidenced by the induction of cellular tyrosine phosphorylation and calcium mobilization in B cells (17, 19). In contrast, the R1 protein has a 171-amino-acid cytoplasmic tail. Unlike K1, which contains 2 tyrosine residues in a single ITAM sequence in its cytoplasmic tail, R1 contains a total of 13 tyrosine residues in its cytoplasmic domain. Five of these 13 tyrosines are present in contiguous YXXL ITAM-like sequences and are localized to a defined stretch proximal to the carboxyl terminus.
To investigate the potential signal-transducing activity of the cytoplasmic region of R1, we constructed a chimeric protein in which the cytoplasmic tail of the human CD8α polypeptide was replaced with that of R1. We demonstrate that expression of this chimera in B cells induced cellular tyrosine phosphorylation, intracellular calcium mobilization, and nuclear factor of activated T lymphocytes (NFAT) activity upon stimulation with an anti-CD8 antibody. Our results demonstrate that the cytoplasmic domain of the RRV transforming protein R1 is an efficient transducer of signals to elicit lymphocyte activation events.
MATERIALS AND METHODS
Plasmids.
The pFJ-R1 expression plasmid has been described previously (9). For the CD8-R1 chimera proteins, the coding sequences for the cytoplasmic deletion mutants of R1 were cloned in-frame with those for the CD8 extracellular and transmembrane domains into the EcoRI-XbaI site of the pcDNA3-CD8Δ vector, which contained the coding sequences for the CD8 extracellular and transmembrane domains inserted in the BglII and EcoRI sites. The R1 cytoplasmic deletions were obtained by PCR using EcoRI and XbaI containing primers. To facilitate detection, the CD8-R1 chimeras were tagged with the AU1 epitope (BAbCO) at the C-terminal end. PFJ-K1 has been described previously (19). The NFAT-luciferase construct was obtained from Don Ganem. The Syk expression plasmid was provided by A. Veillette, and the Src expression plasmid was provided by T. M. Roberts.
Cell culture and transfection.
Cos-1 cells and Rat-1 fibroblasts were grown in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum (FCS). Calcium phosphate was used for transient transfection of Cos-1 cells. For Rat-1 fibroblasts stably expressing the R1 constructs, cells were transfected using Lipofectamine (Gibco-BRL) and stable transfectants were selected using 2 mg of geneticin per ml. BJAB cells were cultured in RPMI media containing 10% FCS. Ten micrograms of each CD8-R1 chimera plasmid was electroporated into BJAB cells at 200 V and 960 μF in Optimem (Gibco-BRL) media. Forty-eight hours postelectroporation, cells were pelleted and transferred into RPMI media supplemented with geneticin (2 mg per ml) and cultured for 5 weeks. Cells were sorted by fluorescence-activated cell sorting (FACS).
FACS analysis.
For FACS, 2 × 107 B cells expressing chimera proteins were washed with Optimem and stained with phycoerythrin-conjugated CD8 antibody (Pharmingen) for 30 min at 4°C. Excess antibody was washed away, and cells were sorted based on CD8 expression using a FACSVantage (Becton Dickinson). After being sorted, cells were washed and cultured in complete RPMI media.
Calcium mobilization assay.
Cells (106) were resuspended in 2 ml of complete RPMI medium and incubated with 1 μM indo-1 for 20 min at 37°C. The cells were subsequently placed on ice till ready to be used. Five minutes prior to use, cells were warmed at 37°C and baseline calcium levels were read for 1 min. Cells were then stimulated with 10 μg of anti-CD8 antibody, OKT8 (American Type Culture Collection), or anti-human IgM antibody (Upstate Biotechnology), and data were collected for 4 min for the IgM stimulation and 8 min for the OKT8 stimulation. A FACSVantage (Becton Dickinson) was used for data collection and analysis.
Antibody stimulation.
Cells (106) were stimulated with 10 μg of OKT8 antibody or anti-human IgM antibody, after which cells were flash frozen in liquid nitrogen and subsequently thawed and lysed in cold lysis buffer containing 1 mM Na2VO3 and protease inhibitors. Cell lysates were used for immunoprecipitation and Western analysis.
Immunoprecipitation and immunoblotting.
After transfection and antibody stimulation, cells were harvested and lysed in NP-40 lysis buffer (0.15 M NaCl, 50 mM Tris [pH 8], 1% NP-40) containing 1 mM Na2VO3, and protease inhibitors. Cellular debris was pelleted at 10,000 rpm for 10 min, and the supernatants were used for immunoprecipitation with the appropriate antibody and protein A/G-Sepharose beads (Santa Cruz Biotechnology). Immune complexes were pelleted and washed at least five times with lysis buffer. Complexes were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose. Immunoblot detection was performed using the enhanced chemiluminescence ECL kit (Amersham).
For immunoprecipitations and immunoblotting, anti-AU1 antibody was obtained from BAbCO. Anti-Src and anti-Syk antibodies were obtained from Upstate Biotechnology. Anti-pTyr (4G10) primary antibody and horseradish peroxidase-conjugated antiphosphotyrosine (anti-pTyr) antibody was obtained from Upstate Biotechnology.
In vitro kinase assay.
Coimmunoprecipitations were performed as described above. After being washed, the complexes were incubated with 5 μCi of [γ-32P]ATP in kinase buffer (50 mM Tris, 10 mM MgCl2, 10 mM CaCl2) for 30 min at room temperature. At the end of the incubation, complexes were subjected to SDS-PAGE, and the gel was subsequently exposed to X-ray film.
Luciferase assays.
Luciferase assays were performed by a method similar to the previously described protocol with a few modifications (18). BJAB cells (106) were electroporated with 20 μg of NFAT luciferase plasmid, 40 μg of pFJ-R1 or pFJ-K1 plasmids, and 5 μg of β-galactosidase expression plasmid. Cells were harvested 24 h posttransfection, and luciferase assays were performed using the luciferase assay kit from Promega. For the experiments involving antibody stimulation, BJAB cells were electroporated with the indicated plasmids and 24 h postelectroporation cells were pelleted and washed in serum-free media. Cells were incubated with anti-CD8 antibody for 5 min at 37°C and subsequently resuspended in 10 ml of medium and placed in the incubator. Aliquots of cells were removed at various time points, and luciferase assays were performed as described above.
RESULTS
R1 contains multiple YXXL motifs in its cytoplasmic domain.
The cytoplasmic domain of R1 contains several potential SH2 binding motifs (Fig. 1). Analysis of the tyrosine residues and their surrounding sequences revealed similarities to sequences that are capable of binding the SH2 domains of signal transducer proteins. YXXA, YXXP, YXXT, and YXXV motifs have been shown to bind SH2 domains within proteins such as the Src family of kinases, Shc, Abl, Nck, and phosphatidylinositol 3-kinase (6, 29). Five of the 13 tyrosines in the cytoplasmic domain of R1 have such a sequence (Fig. 1). The five C-terminal tyrosines in R1 are present within YXXL sequences. Similar YXXL sequence elements have been previously shown to be important for the binding of B-cell-specific Syk kinase (16, 19, 29). The third and fourth and fourth and fifth YXXL motifs resemble ITAMs in that they and the surrounding sequences are spaced in a fashion consistent with that of the ITAM consensus sequence, (D/E)X7(D/E)X2YX2LX7–10YX2L/I (Fig. 1).
FIG. 1.
Amino acid sequences of the R1 cytoplasmic tail. The 170 amino acids of the C-terminal cytoplasmic domain of R1 are shown. The cytoplasmic domain of R1 contains several potential SH2 binding motifs (boldface). Dotted underlining, SH2 binding motifs YXXA, YXXV, YXXT, and YXXP; solid underlining, last five YXXL motifs. The third and fourth and fourth and fifth YXXL motifs resemble ITAMs in that they and the surrounding sequences are spaced in a fashion consistent with the consensus sequence, (D/E)X7(D/E)X2YX2LX7–10YX2L/I. These motifs are boxed.
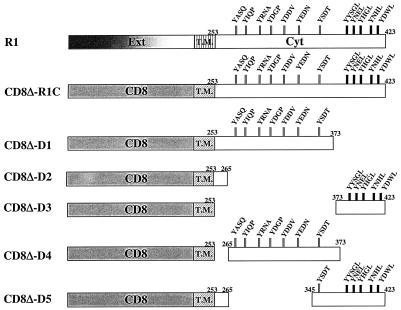
Construction of CD8Δ-R1 chimeras.
The full-length R1 cytoplasmic domain (R1C) and deletion mutants derived from it were fused to the extracellular and transmembrane domains of CD8 in order to analyze the contributions of the various tyrosine-containing elements to B-cell signaling. Six CD8Δ-R1 chimera constructs were made (Fig. 2). The cytoplasmic deletion mutations were selected based on the types of SH2 binding motifs present in the R1 cytoplasmic domain. The last five tyrosine residues in R1 are all present in YXXL motifs, and some or all could potentially function as ITAMs. Hence, this region was separated from the remaining portion which contains a more diverse set of tyrosine-containing motifs. CD8Δ-R1C represents the full-length R1 cytoplasmic tail extending from amino acids 253 to 423 fused to the CD8 extracellular domain and transmembrane region. CD8Δ-D1 contains the first 121 amino acids of the R1 cytoplasmic tail which includes 7 tyrosine residues, 2 of which are in YXXP sequences, 1 of which is in a YXXA sequence, and another of which is in a YXXT sequence. Such sequences have been previously shown to function as SH2 binding motifs and protein tyrosine kinase docking sites in T-cell receptors, BCRs, Epstein-Barr virus (EBV) LMP2A, and KSHV K1 (2, 3, 5, 8, 16, 18, 20, 23, 32, 33). The CD8Δ-D4 construct has a stretch of 106 amino acids extending from the first tyrosine residue (amino acid 265) in the R1 cytoplasmic tail to amino acid 373, which is the amino acid preceding the start of the putative ITAMs in the R1 cytoplasmic domain. Hence, CD8Δ-D4 differs from CD8Δ-D1 in that it is missing amino acids 253 to 264, a region that is immediately adjacent to the R1 transmembrane domain. CD8Δ-D2 contains a short stretch of 13 amino acids in the R1 cytoplasmic tail and is devoid of any tyrosine residues. CD8Δ-D3 contains the five YXXL motifs present at the C-terminal end of R1. CD8Δ-D5 contains the same stretch of amino acids as CD8Δ-D2 in addition to the residues extending from amino acid 345 to amino acid 423. All chimera proteins, including CD8Δ-R1C, were tagged with the AU1 epitope at the C-terminal end.
FIG. 2.
Construction of CD8-R1 chimeras. The cytoplasmic domain (Cyt) of R1 was fused to the extracellular (Ext) and transmembrane (T.M.) domains of CD8. The CD8 chimera containing the full-length cytoplasmic domain of R1 (CD8Δ-R1C) and cytoplasmic domains with various deletions (CD8Δ-D1, CD8Δ-D2, CD8Δ-D3, CD8Δ-D4, CD8Δ-D5) are depicted. YXXX, tyrosine residues and their surrounding sequences.
In order to construct stable cell lines expressing the six CD8Δ-R1 chimera proteins, BJAB (EBV− KSHV−) cells were electroporated with plasmids encoding the individual chimeras. pCDNA3 constructs containing CD8Δ, CD8Δ-R1C, CD8Δ-D1, CD8Δ-D2, CD8Δ-D3, CD8Δ-D4 and CD8Δ-D5 coding sequences were electroporated into BJAB cells, and neomycin-resistant cells were selected for 6 weeks. To further enrich for CD8 chimera-expressing cell lines, an anti-CD8 antibody was used to sort these cell lines by flow cytometry to obtain a population of cells, greater than 95% of which expressed the CD8Δ-R1 chimeric proteins. The CD8Δ-R1C cell line was sequentially sorted four times in order to obtain a >95% pure population of cells (data not shown).
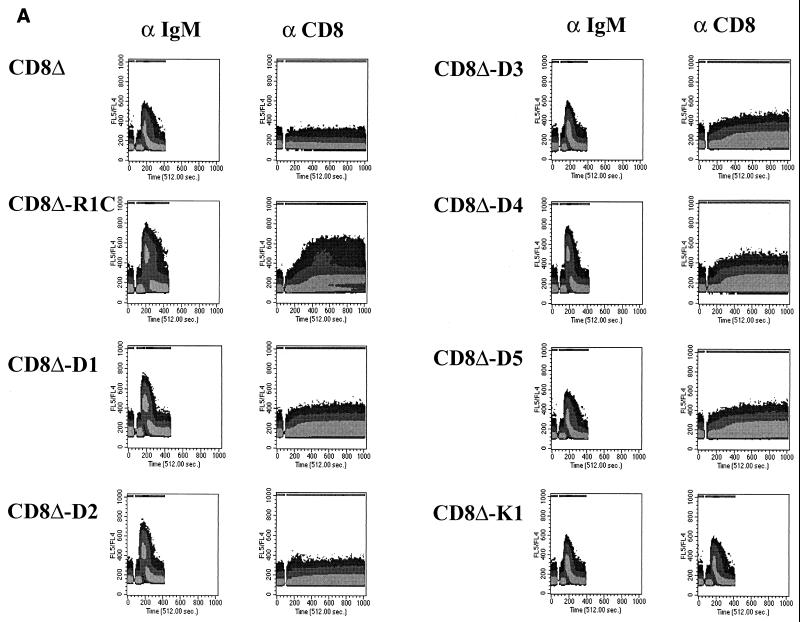
The cytoplasmic tail of R1 can elicit a calcium response in B cells.
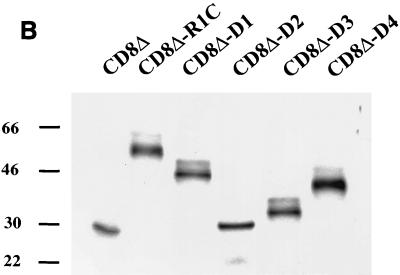
Many B- and T-lymphocyte receptors, when activated by extracellular stimulation, are capable of transducing signals that cause the release of calcium from intracellular stores. Since R1 has several SH2 binding motifs, we tested the ability of the CD8Δ-R1 chimeras to induce intracellular calcium mobilization in B cells. BJAB cells expressing the chimeric proteins were individually stimulated with either anti-IgM antibody or anti-CD8 antibody, and levels of free intracellular calcium were monitored by flow cytometry (Fig. 3A). All seven cell lines, CD8Δ, CD8Δ-R1C, CD8Δ-D1, CD8Δ-D2, CD8Δ-D3, CD8Δ-D4, and CD8Δ-D5 cells, were capable of inducing calcium when stimulated with an anti-IgM antibody, suggesting that these cells were similarly capable of eliciting intracellular calcium mobilization through the B-cell antigen receptor (Fig. 3A). In order to test whether the cytoplasmic domain of R1 and the various deletion mutants could elicit calcium mobilization, we stimulated the BJAB cell lines expressing CD8-R1 chimeras with an anti-CD8 antibody. The CD8Δ, CD8Δ-D1, CD8Δ-D2, CD8Δ-D3, and CD8Δ-D4 cell lines exhibited little or no calcium mobilization upon stimulation with an anti-CD8 antibody as monitored by flow cytometry (Fig. 3A). However, BJAB cells expressing the CD8Δ-R1C chimera did mobilize intracellular calcium, and there was a prolonged period of calcium release from intracellular stores that lasted more than 5 min (Fig. 3A). This is in sharp contrast to the calcium response seen with the anti-IgM antibody, which peaked rapidly and returned to the basal level within a short period of time (Fig. 3A). The calcium response of a CD8Δ-K1cytoplasmic BJAB cell line (20) was also tested using the anti-CD8 antibody (Fig. 3A), and the response was found to be similar in nature to that induced by anti-IgM antibody. The prolonged release of calcium observed in the CD8Δ-R1C cell line in response to anti-CD8 antibody suggests that the full-length cytoplasmic tail of R1 can transduce signals to elicit calcium mobilization. None of the other CD8Δ-R1 deletion mutants could mobilize calcium to a significant extent, including CD8Δ-D3 and CD8Δ-D5, which contain the putative ITAM-like sequences. However, all of the tyrosine-containing chimeras did show a very small increase in intracellular calcium levels, which were only slightly above baseline (Fig. 3A). In order to determine the levels of CD8Δ and CD8Δ-R1 chimeric tagged proteins expressed in the B-cell lines, protein extracts from B cells were separated by SDS-PAGE and Western blotting was performed using an anti-AU1 antibody. The data in Fig. 3B indicate that equivalent levels of CD8Δ and CD8Δ-R1 chimera proteins were expressed in the stable cell lines. In addition, the failure of the deletion mutants to signal could not be explained by differences in the levels of surface expression of these proteins as measured by flow cytometry (data not shown).
FIG. 3.
Mobilization of intracellular free calcium upon antibody stimulation. (A) Intracellular calcium release from the chimera cell lines was monitored by flow cytometry. Either an anti-IgM antibody (left) or an anti-CD8 antibody (right) was used to stimulate cells. Data are presented as histograms of the numbers of cells expressing blue fluorescence (y axis) versus time. The intensities of the responses are depicted as different shades of gray, with the darkest shade representing the highest levels of free calcium in the cell. (B) Western blot analysis of CD8-R1 chimera cell extracts using an anti-AU1 antibody to detect expression levels of the AU1-tagged chimera proteins.
R1 induces NFAT activity in B cells.
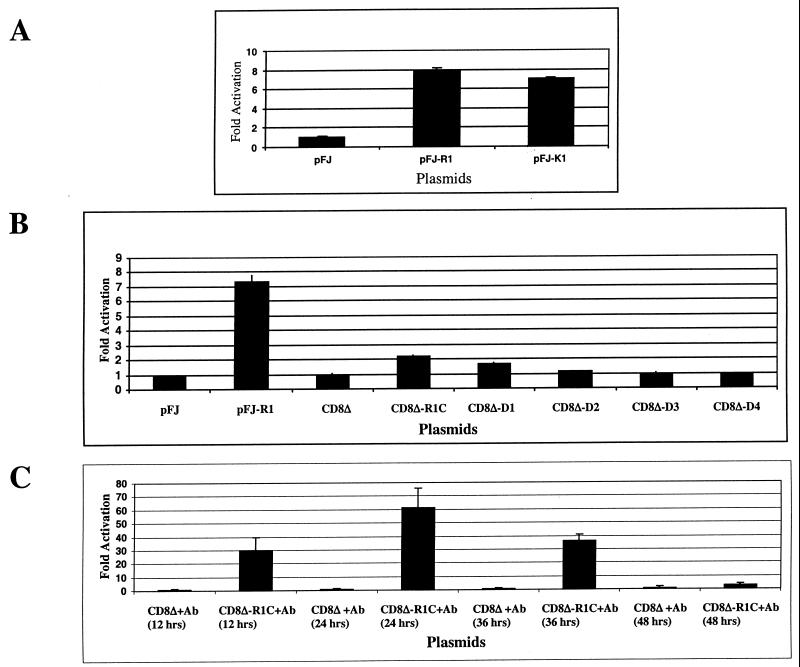
The release of calcium from intracellular stores induces further signaling events that eventually result in the activation of transcription factors in the nucleus. Recently Lagunoff et al. (18) demonstrated that the cytoplasmic domain of the K1 protein, specifically the ITAM, is responsible for the activation of NFAT in BJAB (EBV−) and Raji (EBV+) cells. NFAT is a transcription factor that is present in both T and B lymphocytes (31). We tested the ability of R1 to activate NFAT in BJAB cells using an NFAT-luciferase reporter construct (18). BJAB cells were electroporated with the NFAT-luciferase reporter construct along with pFJ vector alone, pFJ-R1, or pFJ-K1 (Fig. 4A), and cells were harvested 24 h posttransfection. A β-galactosidase expression plasmid was also included as a control for transfection efficiency. Figure 4A graphs the result of these experiments. We observed that R1 could activate NFAT eightfold with respect to the vector control and that K1 could activate NFAT sevenfold.
FIG. 4.
Activation of NFAT activity in BJAB cells by the R1 protein. (A) BJAB cells were transfected with an NFAT luciferase and β-galactosidase reporter plasmid along with either a pFJ vector control plasmid, a pFJ-R1-expressing plasmid, or a pFJ-K1-expressing plasmid. At 48 h posttransfection cells were harvested and tested for luciferase activity. Luciferase counts for each sample were normalized with respect to β-galactosidase activity. Luciferase assays were repeated three times, and the average of these values was graphed as the fold activation of NFAT by the R1 and K1 proteins versus that of control vector alone. (B) Transfections similar to those in panel A except that the CD8Δ-R1 chimera-expressing plasmids indicated in the graph were electroporated into BJAB cells. (C) BJAB cells were transfected with NFAT luciferase and β-galactosidase reporter plasmids along with a CD8Δ-R1C-expressing plasmid. At 24 h posttransfection cells were harvested and incubated with anti-CD8 antibody for 12, 24, 36, and 48 h. Ab, antibody.
We subsequently tested the ability of the CD8Δ-R1 chimeras to induce NFAT activity in BJAB cells. We performed an experiment similar to that described above except that this time each CD8Δ-R1 chimera construct was electroporated into the B cells in lieu of the pFJ-R1 plasmid. Figure 4B demonstrates that none of the CD8Δ-R1 chimeras were able to induce a significant level of NFAT activity in these cells, including the CD8Δ-R1C chimera. We conducted an additional experiment in which each CD8Δ-R1 chimera plasmid was electroporated into BJAB cells and 24 h posttransfection these cells were incubated with an anti-CD8 antibody for different periods of time (for 12, 24, 36, and 48 h following addition of anti-CD8 antibody). The results are graphed in Fig. 4C. We observed that the CD8Δ-R1C chimera, upon stimulation with anti-CD8 antibody, could strongly induce NFAT activity in B cells. The effect of this activation peaked 24 h after the addition of the anti-CD8 antibody, inducing a 60-fold increase in the level of NFAT activity compared to that induced by the CD8Δ chimera alone. Similar experiments conducted with the other CD8Δ-R1 deletion mutants showed them incapable of inducing NFAT activity even when incubated with the anti-CD8 antibody (data not shown). These results demonstrate that the CD8Δ-R1C chimera requires antibody stimulation to induce NFAT activity.
The cytoplasmic tail of R1 induces cellular phosphorylation.
Calcium mobilization and NFAT activation are indicators of lymphocyte activation. The ability of the cytoplasmic tail of R1 to induce calcium mobilization prompted us to determine whether this event involved cellular tyrosine phosphorylation as is the case with BCR activation.
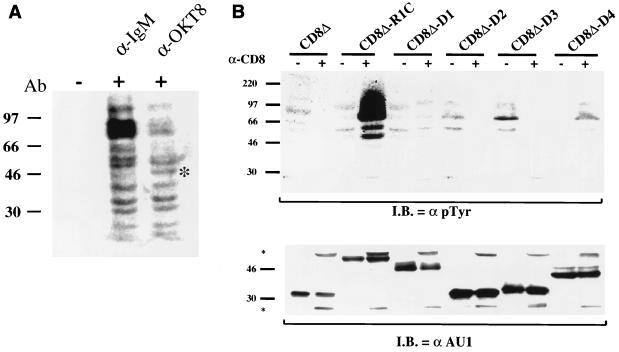
We stimulated the CD8Δ-R1C-expressing cell line with an anti-IgM or an anti-CD8 antibody and performed Western blot analysis with cellular extract using an anti-pTyr antibody. As shown in Fig. 5A, both the anti-IgM and anti-CD8 antibodies were capable of inducing tyrosine phosphorylation in the B-cell line compared to unstimulated cells. Cross-linking with the anti-CD8 antibody induced the phosphorylation of an additional 48-kDa protein, whose molecular mass correlates with that of the CD8Δ-R1C protein (Fig. 5A).
FIG. 5.
Induction of cellular tyrosine phosphorylation upon antibody stimulation. (A) Comparison of anti-IgM- and anti-CD8-stimulated CD8Δ-R1C cells. Cross-linking of the CD8Δ-R1C cell line with an anti-IgM or anti-CD8 (OKT8) antibody (Ab) induced tyrosine phosphorylation of cellular proteins as determined by Western analysis using an anti-pTyr antibody. Asterisk, 50-kDa phosphorylated protein seen in the CD8 antibody-stimulated, but not IgM antibody-stimulated, B cells. (B) Mutational analysis of CD8Δ-R1 chimeras for the induction of tyrosine phosphorylation upon antibody stimulation. Cells were incubated in the absence (−) or presence (+) of anti-CD8 antibody for 1 min at 37°C and immediately lysed. Cell extracts were subjected to gel electrophoresis, transferred to nitrocellulose, and reacted with an anti-pTyr antibody (top). Cell extracts were probed with an anti-AU1 antibody (bottom) to show comparative levels of chimeric proteins in these cells. Asterisks, heavy and light chains of the CD8 antibody used for stimulation. I.B., immunoblotting.
We next tested the abilities of the different R1 cytoplasmic deletion mutants to induce cellular tyrosine phosphorylation (Fig. 5B, top). In contrast to what was seen for cells expressing CD8Δ-R1C, which showed a marked degree of cellular tyrosine phosphorylation upon stimulation with the anti-CD8 antibody, a very slight increase in tyrosine phosphorylation was seen with the CD8Δ-D1 and CD8Δ-D4 deletion mutants (Fig. 5B, top). In order to control for the level of CD8Δ-R1 chimera expression, we reacted extracts from stimulated and unstimulated cells with an anti-AU1 antibody and observed that similar levels of chimeric proteins were expressed in these cells (Fig. 5B, bottom). These results indicate that an intact cytoplasmic tail of the R1 protein is required not only for the calcium mobilization described above but also for the induction of tyrosine phosphorylation in cells, although the deletion mutants may exhibit partial function.
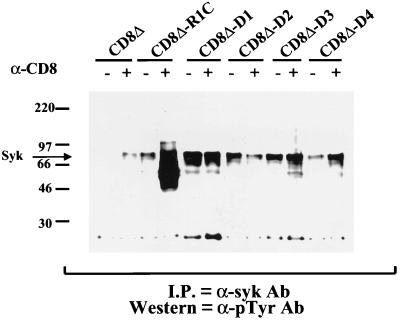
The wild-type CD8Δ-R1C cell line showed the presence of an ∼70 kDa protein whose phosphorylation was increased upon antibody stimulation. The molecular mass of this protein corresponds with that of Syk kinase. In order to determine whether the hyperphosphorylated band represented phosphorylated Syk kinase, we performed immunoprecipitations using an anti-Syk antibody with the same extracts from anti-CD8-stimulated and unstimulated cells. The immunoprecipitated complexes were subjected to Western blot analysis using anti-pTyr antibody. Extracts from stimulated CD8Δ-R1C cells showed the presence of Syk kinase as a 70-kDa protein which was phosphorylated to a greater extent than Syk in unstimulated CD8Δ-R1C cells (Fig. 6). In addition, the stimulated CD8Δ-R1C cells showed a phosphorylated protein of ∼50 kDa coimmunoprecipitating with Syk kinase. This protein corresponds in size to the CD8Δ-R1C chimera protein itself, suggesting that the phosphorylated full-length R1 cytoplasmic domain is complexed with phosphorylated Syk kinase when activated. The CD8Δ-D3 and CD8Δ-D4 immunoprecipitation complexes (Fig. 6) revealed a slight increase in the levels of phosphorylated Syk kinase compared to that of unstimulated cells.
FIG. 6.
Tyrosine phosphorylation of Syk kinase and CD8 chimeras. Shown is the induction of tyrosine phosphorylation by Syk kinase. The CD8-R1 chimera cell lines were incubated with (+) or without (−) anti-CD8 antibody. Extracts were subjected to immunoprecipitation (I.P.) with an anti-Syk antibody (Ab), and immune complexes were resolved by gel electrophoresis and subjected to a Western blot analysis using an anti-pTyr antibody.
Syk kinase can induce phosphorylation of R1.
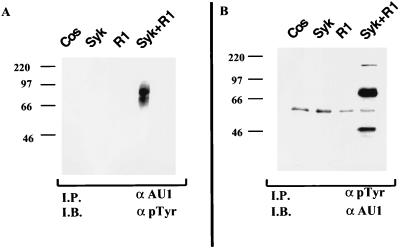
The above results indicated that the cytoplasmic domain of R1 is phosphorylated and raised the possibility that R1 may potentially be phosphorylated by a B-cell kinase. Since Syk is a major B-cell kinase, we conducted additional experiments to determine whether Syk kinase can induce phosphorylation of the R1 protein. Cos-1 cells were cotransfected with a Syk expression plasmid in the presence or absence of a vector expressing a C-terminal, AU1 epitope-tagged R1 protein (Fig. 7). Forty-eight hours after transfection, lysates from transfected cells were reacted with anti-AU1 antibody and immune complexes were analyzed by SDS-PAGE, transferred to nitrocellulose, and reacted with a pTyr antibody. Tyrosine phosphorylation of a 70-kDa protein was detected only in cells transfected with Syk and R1 expression plasmids (Fig. 7A). Since both Syk and R1 have an apparent molecular mass of 70 kDa, it is possible that both of these proteins were represented in the 70-kDa phosphorylated band. We also performed coimmunoprecipitations using an anti-pTyr antibody and a Western analysis with an anti-AU1 antibody to detect the presence of phosphorylated R1 protein. We observed that the R1 protein was tyrosine phosphorylated when Cos-1 cells were cotransfected with Syk expression and R1 expression plasmids (Fig. 7B).
FIG. 7.
Syk kinase induces the phosphorylation of full-length R1. (A) Cotransfection of R1 and Syk expression plasmids in Cos-1 cells. A full-length, C-terminal AU1 epitope-tagged R1-expressing plasmid was transfected into Cos-1 cells either alone or in the presence of Syk kinase. Cells were harvested 48 h posttransfection, and extracts were subjected to immunoprecipitation (I.P.) with an anti-AU1 antibody followed by a Western blot analysis (I.B.) using an anti-pTyr antibody. (B) The same extracts were coimmunoprecipitated with an anti-pTyr antibody followed by Western blotting with an anti-AU1 antibody.
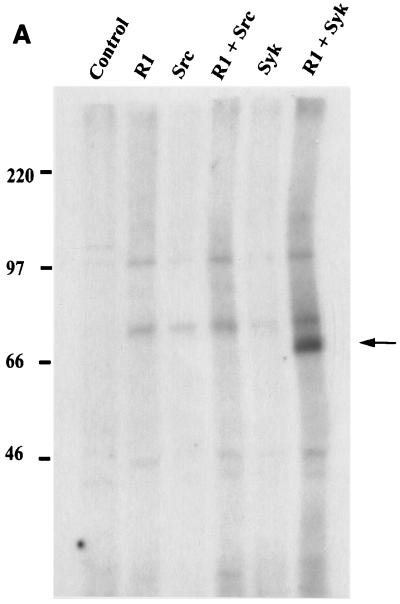
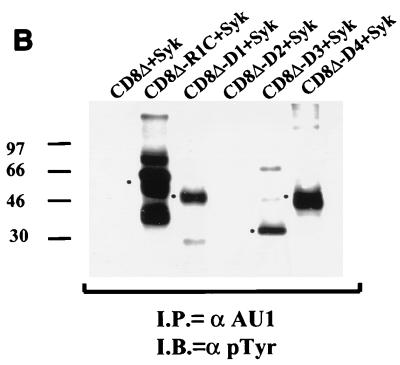
An in vitro kinase assay was performed using extracts from cells cotransfected with an R1 expression plasmid in the absence or presence of Syk kinase- or Src kinase-expressing plasmids (Fig. 8A). Anti-AU1 immune complexes from these cells were washed extensively and incubated with [γ-32P]ATP. The in vitro kinase assay showed that a 70-kDa phosphorylated protein was specifically immunoprecipitated with the AU1 antibody (Fig. 8A). In order to confirm that R1 was indeed phosphorylated in Syk kinase-expressing cells, we cotransfected Cos-1 cells with a Syk kinase expression vector and individual CD8Δ-R1 chimera expression plasmids. Figure 8B shows that Syk kinase induced phosphorylation of the CD8Δ-R1C protein as well as of the three other tyrosine-containing mutants, CD8Δ-D1, CD8Δ-D3, and CD8Δ-D4. Syk kinase could not induce phosphorylation of the CD8Δ-D2 mutant, which does not contain any tyrosine residues. These results show that the cytoplasmic domain of the R1 protein contains several phosphorylation sites whose phosphorylation is induced in cells expressing Syk kinase.
FIG. 8.
Syk-induced phosphorylation of R1 and CD8Δ-R1 chimeras. (A) Phosphorylation of R1 in vitro. An in vitro kinase assay was performed using extracts from Cos-1 cells transfected with R1 alone or in combination with Syk and Src kinase. Immune complexes were washed extensively and subsequently incubated with [γ-32P]ATP. Complexes were then separated by gel electrophoresis and exposed to autoradiography. Arrow, presence of a 70 kDa phosphorylated protein that corresponds to the molecular sizes of both Syk and R1. (B) Tyrosine phosphorylation of CD8Δ-R1 chimeras induced by Syk kinase. Cos-1 cells were cotransfected with a Syk expression plasmid along with the CD8Δ-R1 chimera-expressing plasmids. The chimeras were tagged with an anti-AU1 antibody. Immunoprecipitations (I.P.) were performed using an anti-AU1 antibody and an anti-pTyr antibody was used for a Western blot analysis (I.B.). Dots, positions of mutant proteins.
Syk kinase can interact with R1.
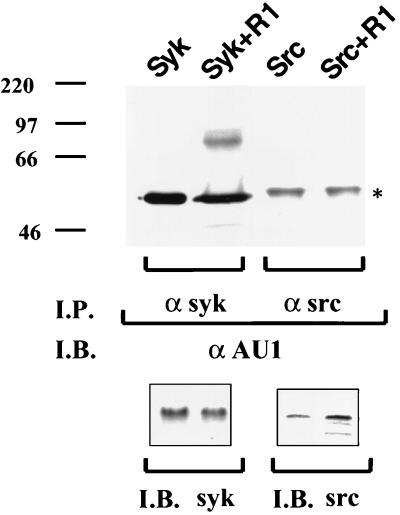
To investigate whether the interaction between Syk and R1 is sufficiently stable to be detected by coimmunoprecipitation, Cos-1 cells were transfected with vectors expressing Src kinase or Syk kinase together with the AU1-tagged R1 gene, and immunoprecipitations were performed using an anti-Src or anti-Syk antibody. Immune complexes were resolved by SDS-PAGE, transferred to nitrocellulose, and reacted with the anti-AU1 epitope antibody. As shown in Fig. 9, R1 was specifically coimmunoprecipitated with Syk kinase but not with Src kinase. The lower panel indicates that there was equal expression of Syk kinase and Src kinase in these cells. Thus, these data indicate that Syk kinase can interact with and induce phosphorylation of R1.
FIG. 9.
R1 can specifically interact with Syk kinase. Coimmunoprecipitations (I.P.) were performed using an anti-Src or anti-Syk kinase antibody on cells transfected with an AU1-tagged R1-expressing plasmid alone or together with Src or Syk kinase-expressing plasmids. Immune complexes were subjected to gel electrophoresis. The gel was transferred to nitrocellulose and subsequently probed with an anti-AU1 antibody. Bottom, levels of Syk and Src kinase in transfected cells. I.B., immunoblotting.
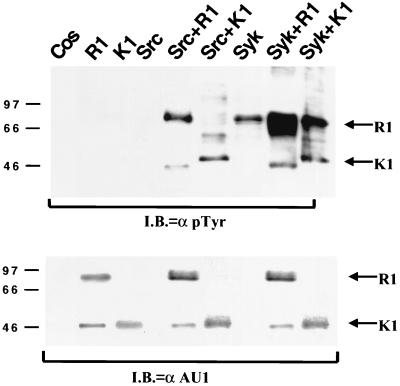
We compared the abilities of both Syk and Src kinases to phosphorylate RRV R1 and KSHV K1 proteins. Cos-1 cells were cotransfected with plasmids expressing R1 or K1 in the presence or absence of Src kinase or Syk kinase expression plasmids (Fig. 10). The top panel represents an immunoblot of the corresponding cell extracts reacted with an anti-pTyr antibody and shows that Syk kinase can phosphorylate R1 to a greater extent than K1, whereas the levels of R1 and K1 phosphorylation induced by Src kinase are similar. The bottom panel represents the same reaction mixtures probed with an AU1 antibody and demonstrates that equivalent levels of K1 and R1 were expressed in these cells.
FIG. 10.
Tyrosine phosphorylation of the R1 and K1 proteins. Comparison of the phosphorylation of R1 and K1 proteins by Src and Syk kinase. Both R1- and K1-expressing plasmids were transfected into Cos-1 cells in the absence or presence of Src and Syk kinases. The immunoblot (I.B.) was reacted with a pTyr antibody (Top). Arrows, R1 and K1 proteins. (Bottom) Western blot analysis of cell extracts used for the immunoprecipitations in the upper panel. The extracts were probed with an AU1 antibody to detect expression levels of R1 and K1 proteins expressed in transfected cells.
DISCUSSION
The R1 open reading frame, the first open reading frame of RRV, encodes a transmembrane protein predicted to contain an extracellular amino-terminal domain and a cytoplasmic carboxy-terminal domain. The extracellular domain of R1 is similar in length and sequence to that of KSHV K1 and shows homology to the Ig receptor superfamily. These properties suggest that R1, like other receptor proteins, may be capable of binding a stimulating ligand, which can result in the transduction of signals inside the cell.
Our results demonstrate that upon cross-linking with an anti-CD8 antibody, B cells expressing a CD8Δ-R1C chimera protein containing the full-length cytoplasmic domain of R1 become fully activated, as indicated by the release of free calcium from intracellular stores. This event differs from that induced by BCR cross-linking by an anti-IgM antibody in that the release of free calcium is prolonged, lasting significantly longer than the BCR-induced response. Importantly, none of the R1 deletion mutants could elicit calcium mobilization to a significant degree. Mutants CD8Δ-D1 and CD8Δ-D4, which contain putative SH2 binding domains, as well as CD8Δ-D3, which contains putative ITAM-like sequences, showed only a marginal increase in calcium mobilization, suggesting that the release of calcium from intracellular stores is dependent on the full-length cytoplasmic tail. The fact that the CD8Δ-D3 mutant could not significantly mobilize calcium was surprising since it contains sequences that match well with the ITAM consensus. One possibility that we considered was that the CD8Δ-D3 mutant requires a linker sequence between the CD8 extracellular and transmembrane domains and the carboxy-terminal SH2 binding motifs in R1 in order to induce calcium mobilization. However, CD8Δ-D5, which contains such a linker sequence, was also incapable of eliciting a significant calcium response. Thus these data are supportive of the need for the full-length R1 cytoplasmic domain for calcium mobilization. The differences in the lengths of the R1 versus K1 cytoplasmic domains required to achieve calcium mobilization could possibly result from additional functions for R1 that remain to be determined or could reflect the fact that the K1 protein is more highly evolved. It seems likely that the amino acids that are required for calcium mobilization reside in two or more distinct domains in the R1 cytoplasmic tail and that proper folding of the cytoplasmic tail is needed to bring together disparate parts of the molecule.
In agreement with our calcium mobilization data, we have also observed that the R1 protein is capable of inducing NFAT activity eightfold in B lymphocytes. Both these events are indicative of B-cell activation. We determined that none of the CD8Δ-R1 chimera proteins could activate NFAT activity to a significant level in B cells by themselves, but upon addition of anti-CD8 antibody we observed a dramatic increase in the activation of NFAT by the CD8Δ-R1C chimera protein. This is suggestive of a role for the extracellular and transmembrane regions of the R1 protein for lymphocyte activation, in addition to that for a full-length cytoplasmic tail. The extracellular domain of the R1 protein may be binding a ligand present at least to some extent in the media or on the surfaces of B cells. Another possibility is that the extracellular and transmembrane domains of R1 may undergo self-oligomerization leading to the phosphorylation of the R1 cytoplasmic domain followed by B-cell activation events.
In agreement with the calcium mobilization assays, we observed that only the chimera with the full-length R1 cytoplasmic domain (CD8Δ-R1C) could induce a state of cellular phosphorylation in B cells. Cross-linking of the CD8Δ-R1C chimeric protein by an anti-CD8 antibody induced a pattern of tyrosine phosphorylation similar to that seen with BCR.
We detected complex formation between R1 and the major B-cell kinase, Syk. Although both Src and Syk kinase were capable of phosphorylating R1, only Syk kinase specifically interacted detectably with R1. The ability of Src kinase to phosphorylate R1 may be indirect. Syk induced the phosphorylation of the CD8Δ-R1C chimera. Syk also induced the phosphorylation of the deletion mutants CD8Δ-D1, CD8Δ-D3, and CD8Δ-D4, and these same mutants showed marginal responses in the calcium mobilization assay. Taken together, these data suggest that these mutants may exhibit partial function to a slight extent. In addition, stimulation of the CD8Δ-R1C cell line with an anti-CD8 antibody resulted in increased phosphorylation of Syk kinase indicative of the need for a full-length R1 cytoplasmic protein to elicit tyrosine phosphorylation in B cells.
The cytoplasmic tail of R1 is 170 amino acids in length and contains 13 tyrosine residues, 5 of which are part of YXXL motifs at the carboxyl-terminal end of the protein and match consensus sequences for ITAMs. Five of the other eight tyrosine residues could also potentially function as SH2 binding motifs since the associated sequences, YXXA, YXXP, YXXT, and YXXV, match sequences previously shown to bind SH2 domains. YXXA can bind Src, Lyn, Fyn, Shc, phosphatidylinositol 3 kinase, and phospholipase Cγ (28, 29). YXXP has been shown to interact with Abl, Crk, and Nck SH2 binding domains (29). Thus the R1 protein has rich potential for interacting with a number of different signal-transducing molecules. Nonetheless, our data have shown that one contiguous stretch containing seven tyrosines and a separate overlapping stretch containing seven different contiguous tyrosines are insufficient for a significant mobilization of calcium, induction of tyrosine phosphorylation, and NFAT activity.
Both RRV R1 and KSHV K1 have been shown to transform rodent fibroblasts, to substitute for STP in HVS-induced immortalization of T cells to interleukin-2-independent growth, and to induce cellular phosphorylation and signal transduction (20, 21). Although these proteins have sequence and structural homology in their external domains, the length of the R1 cytoplasmic domain and the number of potential SH2 binding motifs contained in this domain differ markedly from those of K1, which has a short 38-amino-acid cytoplasmic tail and only two SH2 binding motifs, which constitute a single ITAM (18, 20). One striking difference in the activities of these proteins is the sustained calcium mobilization induced in B cells by R1 versus K1. This prolonged activation event may be a result of the phosphorylation of multiple tyrosine residues in the R1 cytoplasmic tail as these residues are part of SH2 binding motifs that could possibly interact with a variety of cellular kinases.
Cellular receptors, upon ligand stimulation, are capable of undergoing oligomerization and of inducing cellular proliferation. LMP1 protein of EBV mimics an activated CD40 receptor by undergoing multimerization of its transmembrane domains in a ligand-independent manner, which results in the activation of the NF-κB pathway leading to cellular proliferation (11, 12, 15, 24). It remains to be determined whether signal induction via R1 and K1 results from stimulation by an exogenous ligand or self-multimerization. If there is no self-multimerization, a ligand appears to be present, at least to some extent, on the surfaces of BJAB cells, in the culture supernatant, or in the serum. Regardless of the mechanism, both R1 and K1 appear to be able to activate B cells independent of the traditional BCR engagement.
ACKNOWLEDGMENTS
We thank Don Ganem and Mike Lagunoff for the NFAT-luciferase plasmid.
This work was supported by U.S. Public Health Service grants RR00168, CA82057, and AI38131. B. Damania is a fellow of the Cancer Research Institute.
REFERENCES
- 1.Alexander, L., L. Denenkamp, A. Knapp, M. Auerbach, S. Czajak, B. Damania, and R. C. Desrosiers. 1999. The primary sequence of rhesus rhadinovirus isolate 26-95: sequence similarities to Kaposi's sarcoma herpesvirus and rhesus rhadinovirus isolate 17577. J. Virol. [DOI] [PMC free article] [PubMed]
- 2.Beaufils P, Choquet D, Mamoun R Z, Malissen B. The (YXXL/I)2 signaling motif found in the cytoplasmic segments of the bovine leukaemia virus envelope protein and Epstein-Barr virus latent membrane protein 2A can elicit early and late lymphocyte activation events. EMBO J. 1993;12:5105–5112. doi: 10.1002/j.1460-2075.1993.tb06205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cambier J C. Antigen and Fc receptor signaling. The awesome power of the immunoreceptor tyrosine-based activation motif (ITAM) J Immunol. 1995;155:3281–3285. [PubMed] [Google Scholar]
- 4.Cambier J C, Jensen W A. The hetero-oligomeric antigen receptor complex and its coupling to cytoplasmic effectors. Curr Opin Genet Dev. 1994;4:55–63. doi: 10.1016/0959-437x(94)90091-4. [DOI] [PubMed] [Google Scholar]
- 5.Cambier J C, Pleiman C M, Clark M R. Signal transduction by the B cell antigen receptor and its coreceptors. Annu Rev Immunol. 1994;12:457–486. doi: 10.1146/annurev.iy.12.040194.002325. [DOI] [PubMed] [Google Scholar]
- 6.Cantley L C, Songyang Z. Specificity in recognition of phosphopeptides by src-homology 2 domains. J Cell Sci. 1994;18(Suppl.):121–126. doi: 10.1242/jcs.1994.supplement_18.18. [DOI] [PubMed] [Google Scholar]
- 7.Cesarman E, Chang Y, Moore P S, Said J W, Knowles D M. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med. 1995;332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 8.Clark M R, Johnson S A, Cambier J C. Analysis of Ig-alpha-tyrosine kinase interaction reveals two levels of binding specificity and tyrosine phosphorylated Ig-alpha stimulation of Fyn activity. EMBO J. 1994;13:1911–1919. doi: 10.1002/j.1460-2075.1994.tb06460.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Damania B, Li M, Choi J K, Alexander L, Jung J U, Desrosiers R C. Identification of the R1 oncogene and its protein product from the rhadinovirus of rhesus monkeys. J Virol. 1999;73:5123–5131. doi: 10.1128/jvi.73.6.5123-5131.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Desrosiers R C, Sasseville V G, Czajak S, Zhang X, Mansfield K G, Kaur A, Johnson R P, Lackner A A, Jung J U. A herpesvirus of rhesus monkeys related to the human Kaposi's sarcoma-associated herpesvirus. J Virol. 1997;71:9764–9769. doi: 10.1128/jvi.71.12.9764-9769.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Devergne O, Hatzivassiliou E, Izumi K M, Kaye K M, Kleijnen M F, Kieff E, Mosialos G. Association of TRAF1, TRAF2, and TRAF3 with an Epstein-Barr virus LMP1 domain important for B-lymphocyte transformation: role in NF-κB activation. Mol Cell Biol. 1996;16:7098–7108. doi: 10.1128/mcb.16.12.7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Devergne O, McFarland E C, Mosialos G, Izumi K M, Ware C F, Kieff E. Role of the TRAF binding site and NF-κB activation in Epstein-Barr virus latent membrane protein 1-induced cell gene expression. J Virol. 1998;72:7900–7908. doi: 10.1128/jvi.72.10.7900-7908.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flaswinkel H, Reth M. Dual role of the tyrosine activation motif of the Ig-alpha protein during signal transduction via the B cell antigen receptor. EMBO J. 1994;13:83–89. doi: 10.1002/j.1460-2075.1994.tb06237.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gessain A, Sudaka A, Briere J, Fouchard N, Nicola M A, Rio B, Arborio M, Troussard X, Audouin J, Diebold J. Kaposi sarcoma-associated herpes-like virus (human herpesvirus type 8) DNA sequences in multicentric Castleman's disease: is there any relevant association in non-human immunodeficiency virus-infected patients? Blood. 1996;87:414–416. [PubMed] [Google Scholar]
- 15.Izumi K M, Kieff E D. The Epstein-Barr virus oncogene product latent membrane protein 1 engages the tumor necrosis factor receptor-associated death domain protein to mediate B lymphocyte growth transformation and activate NF-κB. Proc Natl Acad Sci USA. 1997;94:12592–12597. doi: 10.1073/pnas.94.23.12592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson S A, Pleiman C M, Pao L, Schneringer J, Hippen K, Cambier J C. Phosphorylated immunoreceptor signaling motifs (ITAMs) exhibit unique abilities to bind and activate Lyn and Syk tyrosine kinases. J Immunol. 1995;155:4596–4603. [PubMed] [Google Scholar]
- 17.Jung J U, Trimble J J, King N W, Biesinger B, Fleckenstein B W, Desrosiers R C. Identification of transforming genes of subgroup A and C strains of Herpesvirus saimiri. Proc Natl Acad Sci USA. 1991;88:7051–7055. doi: 10.1073/pnas.88.16.7051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lagunoff M, Majeti R, Weiss A, Ganem D. Deregulated signal transduction by the K1 gene product of Kaposi's sarcoma-associated herpesvirus. Proc Natl Acad Sci USA. 1999;96:5704–5709. doi: 10.1073/pnas.96.10.5704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Latour S, Fournel M, Veillette A. Regulation of T-cell antigen receptor signaling by Syk tyrosine protein kinase. Mol Cell Biol. 1997;17:4434–4441. doi: 10.1128/mcb.17.8.4434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee H, Guo J, Li M, Choi J K, DeMaria M, Rosenzweig M, Jung J U. Identification of an immunoreceptor tyrosine-based activation motif of K1 transforming protein of Kaposi's sarcoma-associated herpesvirus. Mol Cell Biol. 1998;18:5219–5228. doi: 10.1128/mcb.18.9.5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee H, Veazey R, Williams K, Li M, Guo J, Neipel F, Fleckenstein B, Lackner A, Desrosiers R C, Jung J U. Deregulation of cell growth by the K1 gene of Kaposi's sarcoma-associated herpesvirus. Nat Med. 1998;4:435–440. doi: 10.1038/nm0498-435. [DOI] [PubMed] [Google Scholar]
- 22.Mansfield K, Westmoreland S V, DeBakker C D, Czajak S, Lackner A A, Desrosiers R C. Experimental infection of rhesus and pig-tailed macaques with macaque rhadinoviruses. J Virol. 1999;73:10320–10328. doi: 10.1128/jvi.73.12.10320-10328.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marengere L E, Songyang Z, Gish G D, Schaller M D, Parsons J T, Stern M J, Cantley L C, Pawson T. SH2 domain specificity and activity modified by a single residue. Nature. 1994;369:502–505. doi: 10.1038/369502a0. [DOI] [PubMed] [Google Scholar]
- 24.Mosialos G, Birkenbach M, Yalamanchili R, VanArsdale T, Ware C, Kieff E. The Epstein-Barr virus transforming protein LMP1 engages signaling proteins for the tumor necrosis factor receptor family. Cell. 1995;80:389–399. doi: 10.1016/0092-8674(95)90489-1. [DOI] [PubMed] [Google Scholar]
- 25.Neipel F, Albrecht J C, Fleckenstein B. Human herpesvirus 8—the first human rhadinovirus. J Natl Cancer Inst Monogr. 1998;23:73–77. doi: 10.1093/oxfordjournals.jncimonographs.a024178. [DOI] [PubMed] [Google Scholar]
- 26.Pastore C, Gloghini A, Volpe G, Nomdedeu J, Leonardo E, Mazza U, Saglio G, Carbone A, Gaidano G. Distribution of Kaposi's sarcoma herpesvirus sequences among lymphoid malignancies in Italy and Spain. Br J Haematol. 1995;91:918–920. doi: 10.1111/j.1365-2141.1995.tb05410.x. [DOI] [PubMed] [Google Scholar]
- 27.Searles R P, Bergquam E P, Axthelm M K, Wong S W. Sequence and genomic analysis of a rhesus macaque rhadinovirus with similarity to Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8. J Virol. 1999;73:3040–3053. doi: 10.1128/jvi.73.4.3040-3053.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Songyang Z, Shoelson S E, Chaudhuri M, Gish G, Pawson T, Haser W G, King F, Roberts T, Ratnofsky S, Lechleider R J, Neel B G, Birge R B, Fajardo J E, Chou M M, Hanafusa H, Schaffhausen B, Cantley L C. SH2 domains recognize specific phosphopeptide sequences. Cell. 1993;72:767–778. doi: 10.1016/0092-8674(93)90404-e. [DOI] [PubMed] [Google Scholar]
- 29.Songyang Z, Shoelson S E, McGlade J, Olivier P, Pawson T, Bustelo X R, Barbacid M, Sabe H, Hanafusa H, Yi T, Ren R, Baltimore D, Ratnofsky S, Feldman R A, Cantley L C. Specific motifs recognized by the SH2 domains of Csk, 3BP2, fps/fes, GRB-2, HCP, SHC, Syk, and Vav. Mol Cell Biol. 1994;14:2777–2785. doi: 10.1128/mcb.14.4.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soulier J, Grollet L, Oksenhendler E, Cacoub P, Cazals-Hatem D, Babinet P, d'Agay M F, Clauvel J P, Raphael M, Degos L. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood. 1995;86:1276–1280. [PubMed] [Google Scholar]
- 31.Verweij C L, Guidos C, Crabtree G R. Cell type specificity and activation requirements for NFAT-1 (nuclear factor of activated T-cells) transcriptional activity determined by a new method using transgenic mice to assay transcriptional activity of an individual nuclear factor. J Biol Chem. 1990;265:15788–15795. [PubMed] [Google Scholar]
- 32.Wange R L, Samelson L E. Complex complexes: signaling at the TCR. Immunity. 1996;5:197–205. doi: 10.1016/s1074-7613(00)80315-5. [DOI] [PubMed] [Google Scholar]
- 33.Weiss A, Littman D R. Signal transduction by lymphocyte antigen receptors. Cell. 1994;76:263–274. doi: 10.1016/0092-8674(94)90334-4. [DOI] [PubMed] [Google Scholar]