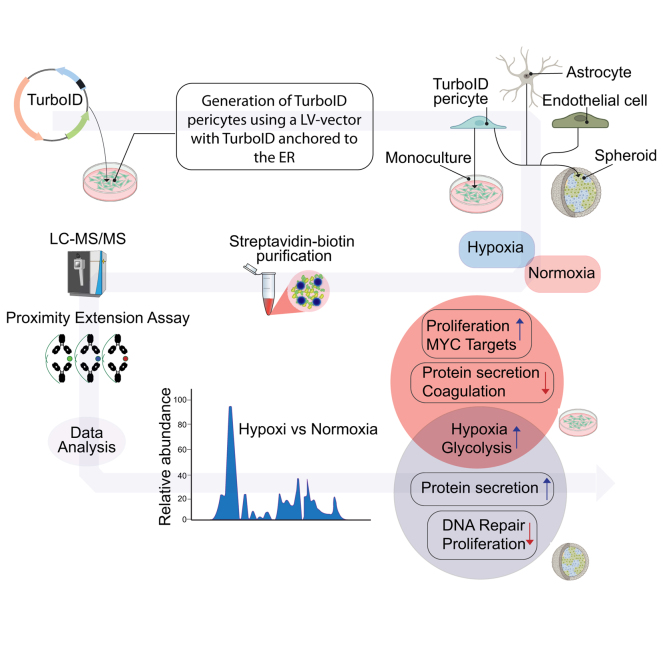
Abstract
Cellular communication within the brain is imperative for maintaining homeostasis and mounting effective responses to pathological triggers like hypoxia. However, a comprehensive understanding of the precise composition and dynamic release of secreted molecules has remained elusive, confined primarily to investigations using isolated monocultures. To overcome these limitations, we utilized the potential of TurboID, a non-toxic biotin ligation enzyme, to capture and enrich secreted proteins specifically originating from human brain pericytes in spheroid cocultures with human endothelial cells and astrocytes. This approach allowed us to characterize the pericyte secretome within a more physiologically relevant multicellular setting encompassing the constituents of the blood-brain barrier. Through a combination of mass spectrometry and multiplex immunoassays, we identified a wide spectrum of different secreted proteins by pericytes. Our findings demonstrate that the pericytes secretome is profoundly shaped by their intercellular communication with other blood-brain barrier–residing cells. Moreover, we identified substantial differences in the secretory profiles between hypoxic and normoxic pericytes. Mass spectrometry analysis showed that hypoxic pericytes in coculture increase their release of signals related to protein secretion, mTOR signaling, and the complement system, while hypoxic pericytes in monocultures showed an upregulation in proliferative pathways including G2M checkpoints, E2F-, and Myc-targets. In addition, hypoxic pericytes show an upregulation of proangiogenic proteins such as VEGFA but display downregulation of canonical proinflammatory cytokines such as CXCL1, MCP-1, and CXCL6. Understanding the specific composition of secreted proteins in the multicellular brain microvasculature is crucial for advancing our knowledge of brain homeostasis and the mechanisms underlying pathology. This study has implications for the identification of targeted therapeutic strategies aimed at modulating microvascular signaling in brain pathologies associated with hypoxia.
Keywords: pericytes, TurboID, biotin-ligase, secretome, hypoxia, neurovascular, blood-brain-barrier, spheroid, monoculture, proteins, secreted, oxygen
Graphical Abstract
Highlights
-
•
TurboID tagged to the ER enables cell selective secretome profiling in co-cultures and hypoxia.
-
•
The secretome from hypoxic pericytes highlights their role as hypoxia sensors.
-
•
Co-culture spheroids reflect the pericyte secretome in hypoxia better than monocultures.
In Brief
This study investigated pericyte-specific signaling in cellular communication within the BBB. We utilized TurboID, a biotin ligase, for proteomic evaluation, enabling the identification of pericyte-secreted proteins in multi-cellular environments under hypoxia. Our findings highlight the differences in the secretome between pericyte monocultures and spheroids, providing insights into microvascular crosstalk and protein release under hypoxic conditions, regulating angiogenesis, inflammation, and cell proliferation. These findings contribute to our understanding of pericyte communication with BBB cells and advance our knowledge of microvascular signaling in hypoxia-related pathologies.
The blood-brain barrier (BBB) is an essential constituent of the neurovascular unit, comprising endothelial cells, astrocytes, and pericytes. Pericytes have many functions at the BBB, including control of endothelial tight- and adherens-junctions, transcytosis, clearing of toxic debris, regulating angiogenesis but are also known to release a large variety of signaling molecules (1, 2, 3, 4).
The release of signaling molecules into the extracellular space is essential for intercellular communication. Evidently, secreted signals are necessary to maintain homeostasis and play an important role in pathology. Notably, recent studies show that beneficial effects from stem cell transplantation are mainly derived from their secreted factors rather than cell engraftment after central nervous system injury (5, 6). Pericytes share several features with mesenchymal stem cells and have become increasingly recognized for their secretory properties, regulating a diverse response to important biological processes as well as disease progression. Because of their unique position at the interface between the circulating blood and brain parenchyma, pericytes are at the frontline to sense and respond to environmental signals. For instance, pericytes in hypoxia are known to secrete different cytokines, chemokines, extracellular matrix components, and growth factors through paracrine signaling but also communicate to distant cells and organs by the release of extracellular vesicles (7, 8, 9, 10, 11, 12).
Most previous studies that investigated the pericyte secretome have relied on the use of in vitro monocultures because they provide controlled conditions where the secreted molecules have a certain association with the cell type of origin (3, 13). Albeit useful for specific purposes, monocultures are poorly resembling in vivo conditions. Thus, the current understanding of the pericyte-specific secretome is limited concerning its dynamic regulation and response to pathological stimuli in the context of multicellular environments as these studies have previously been hampered by the lack of sufficient methodologies.
Novel biotechnological tools have now been developed to enable cell-specific proteomic evaluation (14, 15, 16). One approach is to express a mutated form of tRNA-synthase that incorporates azidonorleucine in place of methionine where proteins can then be isolated based on azidonorleucine affinity probes. However, reliance on a single cognate tRNA target codon potentially restricts proteomic coverage and downstream analysis. Perturbations in protein folding, stability, posttranslational modifications, or other unwanted effects due to unnatural amino acid incorporation could also affect subsequent analysis (17, 18, 19).
Another approach is the use of cell-specific puromycylation of newly synthesized proteins, which can later be detected by puromycin-specific antibodies. Although this is an easy technique to apply, its drawbacks comprise the generation of many aberrant or truncated proteins and short time periods of labeling due to its cellular toxicity (20, 21).
Recently, innovative research has led to the development of TurboID which is a mutant nontoxic biotin ligase with a highly reactive biotin-5′-AMP–binding motif, exhibiting superior biotin-labeling efficiency than previous biolabeling enzymes like APEX, AirID, or BioID (22, 23, 24). The TurboID enzyme has proven highly effective for studying protein–protein interactions as it functions as a promiscuous proximity biotin-labeling enzyme within the radius of ∼20 nm, but its application extend beyond studying just protein–protein interaction. In this study, our approach of choice was to utilize TurboID conjugated to the endoplasmic reticulum (ER) anchoring domain of cytochrome-P450 to establish pericyte-specific secretome profiling in an in vitro coculture model (22). Expressing this construct in pericytes permits the isolation, purification, and subsequent analysis of pericyte-secreted proteins under physiological or pathological conditions in complex multicellular environments.
TurboID biotinylation has previously been verified for intracellular labeling of endogenous proteins but has only recently been evaluated for biotinylation of secreted proteins in cell cultures or animal models of rodents. Similarly, another mutated biotin ligase closely related to TurboID has been used in Drosophila and mouse models (22, 25, 26, 27). However, a detailed characterization of human pericyte-secreted factors in hypoxia has been lacking. Here, we aimed to (I) use TurboID for secretome profiling and optimize the method of protein recovery and then (II) utilize this method to characterize the specific signaling of pericytes at the BBB in response to hypoxic insult, as well as (III) characterize the differences in the secretome between pericyte monocultures and pericyte cocultures. We demonstrated by using mass spectrometry and immunoassays that TurboID expression allows the purification and analysis of the cell-specific secretome in a co-culture environment and that hypoxic pericytes increase their secretion of proteins related to angiogenesis, inflammation, cellular metabolism, and proliferation.
Experimental Procedures
Cell Culture
Human brain vascular pericytes (HBVPs, Sciencell Catalog No.#1200) were cultured in pericyte medium (PM, Sciencell) supplemented with 2% fetal bovine serum (FBS), (100 μg/ml) penicillin/streptomycin (P/S), and growth supplement supplied by the manufacturer. HBVP culture plates were coated with 0.1% gelatine under monoculture conditions. Human brain microvascular endothelial cells (Cell Systems, Catalog No. #ACBRI376) were cultured on 0.1% gelatine-coated plates and endothelial cell medium MV2 with 5% FBS (ECM, PromoCell) was used. Primary human hippocampal astrocytes (NGC-407) were cultured on fibronectin-coated plates (1 μg/ml) and the astrocyte medium composition contained Advanced Dulbecco's Modified Eagle Medium (DMEM F12, Gibco) supplemented with 10% FBS and (100 μg/ml) P/S.
Virus Production and Stable Cell Line Generation
For the preparation of lentiviruses, HEK293 cells were grown in DMEM high glucose, + sodium pyruvate (110 mg/l) with 10% FBS and (100 μg/ml) P/S and transfected at ∼80% confluency in 6-well plates. The lentiviral vector TurboID-V5 pLX304 (1573 ng) together with the viral envelope plasmid pMD2.G (397 ng) and lentiviral packaging plasmid pBR8.91 (849 ng) were diluted in 125 μl of Optimem medium (Gibco). Separately, (Thermo Fischer Scientific) 5 μl of Lipofectamine 2000 was added to 125 μl of Optimem medium before the solutions were mixed and incubated for 5 min at RT for lipid droplet formation, before being applied to HEK293 cells. After 6 h, the medium containing lipofectamine 2000 was replaced with normal PM. After 48 and 72 h, the cell medium containing the lentivirus was harvested and filtered through a 0.45-μm filter. To generate stable TurboID expression in the cell line of interest, HBVP at ∼60% confluency in 6-well plates were transduced with 1.5 ml of the virus titer and incubated for 24 h at 37 °C and 5% CO2. Subsequently, HBVPs were expanded in selection medium (PM + 4 μg/ml blasticidin) for 14 days, replacing the medium every 3 days.
Spheroid Coculture Generation
Spheroids were generated by mixing HBVPs, human brain microvascular endothelial cells, and human hippocampal astrocytes in a 1:1:1 ratio with 10,000 cells of each cell type. The spheroids were cultured in 96-well plates coated with 1% agarose in PBS and cultured in spheroid medium generated from mixing PM, ECM, and astrocyte medium in a 1:1:1 ratio.
Biotin Treatment and Hypoxia Induction
To enable sufficient protein-biotinylation, 500 μM (22) of exogenous biotin in DMSO was added to the cell-culture medium for both mono- and spheroid-cultures as a final concentration unless stated otherwise. For hypoxia treatment, cells were transferred to a humidified gas-tight hypoxia chamber (Electrotek) with a gas composition of 85% N2, 10% H2, and 5% CO2 generating an oxygen supply between ∼0.5 and 1%. In addition, the medium used under hypoxic incubation was pre-bubbled with N2 gas for 15 min to deoxygenate the medium before applying it to the cells. Throughout experiments, an anaerobic indicator solution (Electrotek) was used for monitoring oxygen levels under 1%, containing 2% w/v C6H12O6, 9% w/v NaHCO3, and 1% w/v methylene blue solution in water.
Sample Generation for Proteomic Analysis
Spheroid Cultures
For each independent experimental replicate, 96 spheroids were transferred from 96-well plates and transferred to 6-well plates (16 spheroids/well) corresponding to 960,000 pericytes. Monocultures: The same amount of pericytes was seeded for monocultures (160,000 cells/well in 6-well plates). The same setup was used for normoxic, hypoxic, and negative controls (TurboID with omit biotin or WT omitting TurboID expression). Subsequently, the cells were pre-incubated with 500 μM biotin for 24 h before media exchange and hypoxic or normoxic incubation for 24 h. Supernatant samples were collected and kept on ice before being centrifuged at 700g for 10 min at 4 °C. Total supernatant volume of 12 ml/sample was concentrated to 1 ml using an Amicon centrifuge filter unit with a 3 kDa cut-off (EMD Millipore, UFC900324). The supernatant was then transferred to protein low-binding microcentrifuge tubes and stored at −80 °C until further use. The same samples were used for both liquid chromatography mass spectrometry (LC-MS/MS) analysis and the Olink Proximity Extension Assay (Olink).
Protein-Purification of LC-MS/MS
Streptavidin magnetic beads (Thermo Fisher Scientific, 88817) were made protease resistant through chemical modification according to a previously described protocol (28). In short, lysine residues were di-methylated with 4% paraformaldehyde and sodium cyanoborohydride (Sigma, 25895-60-7) as a reducing agent. In addition, 1,2-cyclohexanedione (Sigma, C101400) was used to condense Arginine residues. For each supernatant sample of 450 μl, we used 30 μl of protease-resistant streptavidin beads (PrS) that were first washed twice with 500 μl RIPA lysis buffer before supernatant samples, and 450 μl of RIPA buffer were incubated with PrS beads O/N at 4 °C. Next, we followed the enrichment protocol previously published (24). Protein-enriched PrS beads were washed twice with RIPA lysis buffer, once with 1 M KCL, once with 0.1 M Na2CO3, once with 2 M urea in 10 mM Tris–HCL, twice with RIPA, once with 50 mM Tris–HCL, and finally two washes with 2 M urea in 50 mM Tris–HCL. Then, 5% of the washed beads were taken out for enrichment analysis. Throughout the purification protocol, the flow-through fraction, the last wash fraction, and the input fraction were also saved for further analysis. Next, the PrS beads were incubated with 80 μl of 2 M urea in 50 mM Tris–HCL containing 1 mM DTT and 0.4 μg sequencing grade trypsin (Promega, V5111) at 37 °C for 1 h with shaking 1000 r.p.m. Next, the supernatant was removed, and the beads were washed twice with 60 μl of 2 M urea in 50 mM Tris–HCL and combined with the on-bead digest supernatant. The disulphide bonds were reduced with a final concentration of 4 mM DTT and incubated at 25 °C for 30 min with shaking at 1000 r.p.m. Next, iodoacetamide at a final concentration of 10 mM was added to the solution and incubated at 25 °C for 45 min in the dark while shaking 1000 r.p.m. Another 0.5 μg of trypsin was added to the samples before O/N incubation at 37 °C with shaking 750 r.p.m. After O/N digestion, a final concentration of TFA ∼1% was added making sure the sample was below pH = 3. The samples were then desalted using C18 Ultra Microspin columns 3 to 30 μg (The Nest Group Inc, SS18V). Briefly, stage tips were conditioned with 100 μl of 100% acetonitrile (ACN), then washed twice with 0.1% TFA before adding the samples, followed by two more washes with 0.1% TFA before eluting with 80% ACN in 0.1% TFA. All centrifuge steps were done at 150g for 1 to 2 min. Next, samples were vacuum centrifuged at 45 °C for 1 h and stored in RT until further use.
LC-MS/MS Analysis
The samples were analyzed on an Exploris 480 mass spectrometer (Thermo Fischer Scientific) coupled with a Vanquish Neo Ultra-high performance liquid chromatography system (Thermo Fischer Scientific). Two-column setup was used on the HPLC system and peptides were loaded into an Acclaim PepMap 100 C18 precolumn (75 μm × 2 cm, Thermo Fisher Scientific) and then separated on an EASY spray column (75 μm × 25 cm, C18, 2 μm, 100 Å, ES902) with the flow rate of 300 nl/min. The column temperature was set at 45 °C. Solvent A (0.1% formic acid in water) and solvent B (0.1% formic acid in 80% ACN) were used to create a 90 min linear gradient from 5 to 38% of solvent B to elute the peptides. The samples were analyzed with a data-dependent acquisition in positive mode. The full MS1 resolution was set to 120,000 at m/z 200 and the normalized automatic gain control target was set to 3 × 106 with the maximum injection time of 45 ms. The full mass range was set at 350 to 1400 m/z. Precursors were isolated with the isolation window of 1.3 m/z and fragmented by HCD with the normalized collision energy of 30. MS2 was detected in the Orbitrap with a resolution of 15,000. The normalized automatic gain control target and the maximum injection time were set to 1 × 105 and auto, respectively. The intensity threshold for precursor selection was set to 1 × 104 and 45 s of dynamic exclusion was applied.
Data Analysis MS/MS
The raw data-dependent acquisition data were analyzed with Proteome Discoverer 2.5 Software (https://knowledge1.thermofisher.com/Software_and_Downloads/Chromatography_and_Mass_Spectrometry_Software/Proteome_Discoverer/Proteome_Discoverer_Operator_Manuals/Proteome_Discoverer_2.5_overview, Thermo Fisher Scientific), and the peptides were identified using SEQUEST HT against UniProtKB Human database (UP000005640_9606) and IgG goat sequences (29). The UniProtKB FASTA file in which the analysis was run against contained 20,528 different sequences with a false discovery rate set at 0.01 for high confidence, 0.05 for medium confidence, and 0.1 for low confidence for peptide score. The search was performed with the following parameters applied: static modification: cysteine carbamidomethylating and dynamic modifications, N-terminal acetylation, and methionine oxidation. The precursor tolerance was set to 10 ppm, and the fragment tolerance was set to 0.02. Up to two missed cleavages were allowed, and Percolator was used for peptide validation at a q-value of a maximum of 0.05 (supplemental Table S1, A and B).
Bioinformatics
MS/MS or Olink values were imported into msImpute from raw text data protein expression values. Imputation was done by setting v2 in the msImpute R-package from filtered data demanding four present values per protein or replicate (msFilter) (30). Olink data was analyzed for significant protein differences in Graph pad PRISM version 10 and a p-value <0.05 with multiple t-tests and Sidak’s multiple comparison test. For LC-MS/MS, the resulting data frame was fed into Limma, and contrasts were made for conditions of interest (31). Subsequently, WT control contaminant proteins were removed by contrasts to WT in Limma by comparing WT monocultures to normoxic monocultures (NM), hypoxic monocultures (HM), or WT spheroids to normoxic spheroids (NS) or hypoxic spheroids (HS) and removing all proteins with a log2 FC >0.5 compared to WT, which generated 974 variables. Where applicable, that is, for heatmaps of multiple contrasts, duplicate proteins were filtered out with base R functions of log2 FC >2, p < 0.05 data. The gplots function heatmap.2 was used for heatmap plotting. An upset plot was constructed from protein lists of log2 FC >1 filtered data for indicated conditions (upsetplot R) (32). Volcano plots were plotted with a log2 FC >1, p < 0.05 stringency selection (https://github.com/kevinblighe/EnhancedVolcano) using a moderate t-statistic. Gene set enrichments were made with fGSEA using the Hallmark database with default setting except for minSize = 3 proteins per pathway (33). fGSEA data were plotted with the dot plot function in R. See session info for R-package information (supplemental Table S2). Preceding Venn diagram analysis, the gene IDs from signalp6.0 (https://doi.org/10.1038/s41587-021-01156-3), the predicted secreted molecules from the Human Protein Atlas v.23 subcellular location dataset (https://www.proteinatlas.org/about/download), and the contrasted 974 protein dataset from this work, were run through the (https://www.syngoportal.org/convert) to ensure that the gene symbol nomenclature was the same.
Protein-Purification Olink
Monomeric avidin-agarose slurry (75 μl, Thermo Fischer Scientific) was washed with 500 μl of RIPA buffer two times. After, 450 μl of the sample of interest and 450 μl of RIPA buffer was added to the slurry and left rolling at 4 °C, O/N. Next, the samples were washed as in the LC-MS/MS purification protocol, with 1 ml of RIPA buffer twice, 1 ml of (1 M KCl) once, 1 ml of Na2CO3 once, 1 ml of (2 M urea in 10 mM Tris pH 8.0) once, and finally with 1 ml of RIPA buffer twice. For each washing step, the supernatant was removed after the avidin-agarose slurry was centrifuged at 600g for 1 min. All flow-through and washing fractions were saved for further analysis. Subsequently, the avidin beads were eluted using 2 mM biotin diluted in PBS, making the samples compatible for the Olink Proximity extension assay (Olink). All samples were randomized in a 96-well plate and sent to be further processed for proximity extension assay using a 96-target inflammation panel (SciLifeLab).
Western Blot and Silver Staining
For quantifying biotinylated proteins from supernatants without protein purification, samples were mixed with Laemmli buffer (Bio-Rad) and 0.1 M DTT and denatured at 95 °C for 5 min for further analysis. For the Western blot, the proteins were separated using 4 to 15% gradient SDS-PAGE precast gels (Bio-Rad). Subsequently, gels were transferred onto nitrocellulose membranes (Bio-Rad) and blocked with 5% milk in TBST with 0.1% Tween-20 for 1 h. For detection of biotinylated proteins, the membranes were subsequently incubated with streptavidin conjugated to horseradish peroxidase (Southern Biotech) for 1 h in 1% bovine serum albumin diluted in TBST. After, the membrane was washed 3 × 5 min with TBST before being developed using ECL substrate (Clarity Max, Bio-Rad). For detection of TurboID, cell lysates were lysed directly in Laemmli buffer with 0.1 M DTT. The primary anti-V5 antibody (Invitrogen) was incubated for 1 h at RT in 5% milk. Next, membranes were washed 3 × 5 min with TBST before the secondary antibody goat-anti-mouse-HRP-conjugated (1:5000, Dako) in 5% bovine serum albumin was incubated with the membranes for 1 h at RT. After washing with TBST 3 × 5 min, membranes were developed with ECL (Clarity, Bio-Rad). All western blots were developed using chemiluminescence (Chemidoc, Bio-Rad).
The silver staining (Thermo Fischer Scientific) was performed according to the manufacturer’s protocol. Briefly, the gel was washed with MilliQ water before being fixed with (30% ethanol and 10% acetic acid). After, the gel was washed with 10% ethanol solution and MilliQ water. The gels were then submerged in sensitizer solution and washed with MiliQ water. Then the gels were stained with working solution for 30 min at RT before being washed and developed for 2 to 3 min before adding the stop solution. The gels were then imaged using a Chemidoc.
Immunocytochemistry and Image Acquisition
For monocultures, HBVPs were seeded on fibronectin (1 μg/ml)-coated coverslips. The cells were then fixed with PFA and added directly to the medium with a final concentration of 2% and incubated for 20 min. The fixed cells were then washed 3 × 5 min with PBS and blocked for 1 h with normal donkey serum 5% in PBS supplemented with 0.3% Triton x-100 (PBST) at RT. Primary antibodies were suspended in blocking solution and left O/N at 4 °C. After washing 3 × 5 min with PBS, secondary antibodies in blocking solution were added to the cells and incubated for 1 h at RT and washed 3 × 5 min with PBS. DAPI (1:10,000, Thermo fischer Scientific) was added and incubated for 5 min at RT before washing 3 × 5 min. Finally, the coverslips were mounted on glass slides using PVA-DABCO (Sigma-Aldrich). For spheroids, the cells were fixed in 2% PFA for 30 min, then washed 3 × 5 min before being transferred to optimal cutting temperature solution, and embedded and frozen on dry ice before sectioning. The spheroids were cryo-sectioned at 16 μm and mounted on cryo-slides for imaging. For visualization of fluorescent immunostainings, images were obtained using a Leica DMi8 confocal microscope with the oil immersion 40× objective and 0.75 zoom. A full list of antibodies used for immunocytochemistry and western blots is found in Table 1.
Table 1.
List of antibodies used in this study
| Antibody | Species | Dilution | Company |
|---|---|---|---|
| Anti-V5 (R96025) | Mouse | 1:400 | Thermofischer Scientific |
| PDGFRβ (28E1) | Rabbit | 1:500 | Cell Signaling Technology |
| CD31 (550389) | Mouse | 1:50 | BD Pharmingen |
| GFAP (ab4674) | Chicken | 1:1000 | Abcam |
| Streptavidin-488 (s1223) | - | 1:500 | Thermofischer Scientific |
| Streptavidin-HRP (7100-05) | - | 1:50,000 | Southern Biotech |
| Calnexin (PA5-34754) | Rabbit | 1:500 | Thermofischer Scientific |
| Alexa-488 | Donkey | 1:500 | Jackson ImmunoResearch |
| Alexa-647 | Donkey | 1:500 | Jackson ImmunoResearch |
| Cy3 | Donkey | 1:500 | Jackson ImmunoResearch |
Flow Cytometry
Single cells were prepared from spheroids exposed to 24 h normoxia or hypoxia in DMEM/F12 with 15 mM Hepes according to https://doi.org/10.1038/nm.4309. The spheres were subjected to 30 min of enzymatic digestion before satisfactory single cell dissociation was achieved. The suspension cells were stained for LSR II (Benton Dickinson) flow cytometry in AnnexinV buffer (BioLegend) with 1 μg/ml Annexin V-Pacific blue (for apoptotic cells) and/or 1:25 v/v 7AAD (BioLegend) 1:25 (for plasma membrane disruption in dying cells). The cells were acquired in an LSR II flow cytometry and analyzed with FlowJo 10, including all events in the forward and side scatter in for the live/dead analysis in order not to exclude dead or dying cells in the forward and side scatter-gate. A 2:1 mix of freshly dissociated NM cells and NM cells treated in 60 °C for 10 min to induce cell death were used as positive controls for the AnnexinV and 7AAD stains. Fluorescence minus one stainings were used to set the gates for positive stainings.
Experimental Design and Statistical Rationale
The samples for this study were prepared using TurboID-expressing pericytes and WT-pericytes as controls, either in monoculture or in spheroid cocultures together with endothelial cells and astrocytes. The samples were then exposed to hypoxic (<1% O2) or atmospheric oxygen pressure in standard culturing conditions. For each of our samples including controls, there were three independent replicates. The number of replicates was chosen based on budgeted allowance and an a priori power analysis was not performed. The samples were run blinded for mass spectrometry and Olink data acquisition. Samples for Olink were randomized using a random number generator. For statistical analysis and the filtering of the proteomic data, see the section Bioinformatics.
Results
TurboID Activity and Biotinylation of Secreted Proteins
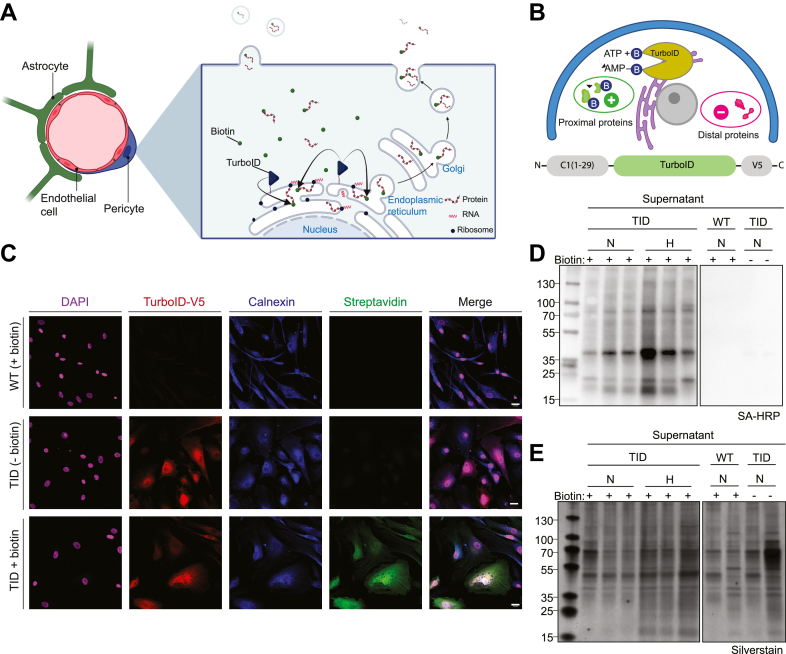
Since conventional transport of secretory proteins occurs via ER–Golgi trafficking before secretion into the extracellular space, we predicted the genetic fusion of the ER-anchoring domain of cytochrome-P450 to TurboID would capture most of the secreted proteins, as shown previously to have a 92% specificity to proteins of the secretory pathway (22, 27) (Fig. 1, A and B). First, we characterized intracellular biotinylation for functional assessment of TurboID-pericytes in monoculture. Exogenous biotin (500 μM) was added for 24 h to the medium and we observed robust biotin-labeling of proteins, evident from the streptavidin immunolabeling in TurboID-pericytes compared to WT with omitted TurboID ligase or TurboID-pericytes with omitted biotin (Fig. 1C). We verified TurboID expression in pericytes by immunocytochemistry of the V5-tag epitope of the fusion construct where most pericytes were expressing TurboID which also colocalized to the ER-marker calnexin (Fig. 1C). Although immunofluorescence assays can verify intracellular TurboID activity, they cannot easily differentiate between secreted or non-secreted proteins, nor ligase self-biotinylation. Thus, for verification, we performed streptavidin-blotting from the cell-culture supernatant to evaluate secreted proteins in environments of normoxia or hypoxia. TurboID-pericytes induced biotin labeling of a variety of secreted proteins in both conditions, where most of the biotinylated proteins corresponded to the total protein analysis, seen from the streptavidin Western blot compared to the silver stain (Fig. 1, D and E). Importantly, control conditions of WT treated with biotin or TurboID-pericytes with omitted biotin showed no or only minimal signal of biotinylated proteins in the supernatant evident from the Western blot, while a strong signal was observed from total protein analysis in the silver stain (Fig. 1, D and E), verifying that protein labeling only occurs when the TurboID ligase is expressed in the presence of exogenous biotin.
Fig. 1.
TurboID biotinylates intracellular and secreted proteins in pericyte monocultures.A, illustration of TurboID biotin-labeling of translated proteins at the ER and the conventional secretory pathways with ER-Golgi trafficking and/or release of extracellular vesicles. B, illustration of the TurboID construct with the CYP-450 – C1(1–29) ER-anchoring domain at the N-terminal and the V5-tag at the C-terminal and its functionality as a biotin labeling enzyme within ∼20 nm of proximal proteins. C, TurboID-pericytes treated with 500 μM biotin or negative controls including TurboID-pericytes with omitted biotin or WT-pericytes with omitted TurboID ligase were stained for anti-V5 verifying TurboID expression, calnexin marking the ER, and streptavidin-488 for biotin-labeling. D, Western blot of supernatant samples using SA-HRP after 24 h of either normoxic (N) or hypoxic (H) incubation. E, total protein content from the supernatant samples were analyzed with silver stain. Scale bar represents 25 μm. CYP-450, cytochrome P450; ER, endoplasmic reticulum; SA-HRP, streptavidin-horse radish peroxidase; TID, TurboID.
Next, we verified that TurboID expression was confined to our cells of interest (pericytes) and not secreted into the extracellular space, which could potentially lead to transmigration into other cell types confounding its feasibility to be used in spheroid coculture settings. We did not detect any secreted TurboID in the supernatant although as expected, we did observe biotinylated soluble biotinylated proteins from the TurboID-pericytes while no signal was detected for the WT or TurboID-pericytes with omitted biotin. In contrast, TurboID was clearly expressed when blotting the cell lysates where biotinylation was especially enriched at ∼37 kDa, the molecular weight corresponding to TurboID. This indicates TurboID self-biotinylation to some degree, which has previously been observed but shown not to affect ligase activity (22) (supplemental Fig. S1).
Since TurboID has much faster labeling kinetics than any previously generated proximity ligation enzyme such as BioID (22), we tested different labeling times and biotin concentrations to evaluate labeling yields and feasibility of different experimental designs. First, we analyzed the yields by streptavidin-blotting for six different time-points of duration of biotin administration illustrated in (supplemental Fig. S2A). Pre-treatment of biotin for 10 min (referred to as condition A) conducted 24 h before media exchange effectively capture a large portion of biotinylated proteins that are subsequently secreted in response to environmental stimuli such as hypoxia. Alternative strategies of biotin addition for 3 h, 1 h, and 10 min before the experiment (conditions B–E) were evaluated for efficiency in labeling pericyte-secreted proteins. Among these conditions, the most robust signal was obtained from (condition E) where the cells were pre-treated with exogenous biotin for 24 h (supplemental Fig. S2B). These results highlight the rapid detectability of TurboID-initiated biotinylation within 10 min of labeling, although the abundance of biotinylated proteins increases with longer labeling durations. Different concentrations of biotin had less of an effect on the number of secreted biotinylated proteins but were slightly higher at 500 μM than lower concentrations of either 50 μM or 250 μM of biotin (supplemental Fig. S2C). For optimal biotin labeling of the pericyte-secretome, we decided to adopt condition E of 24 h pre-labeling with 500 μM biotin for our experimental design.
TurboID-Pericytes in a Spheroid Coculture Model
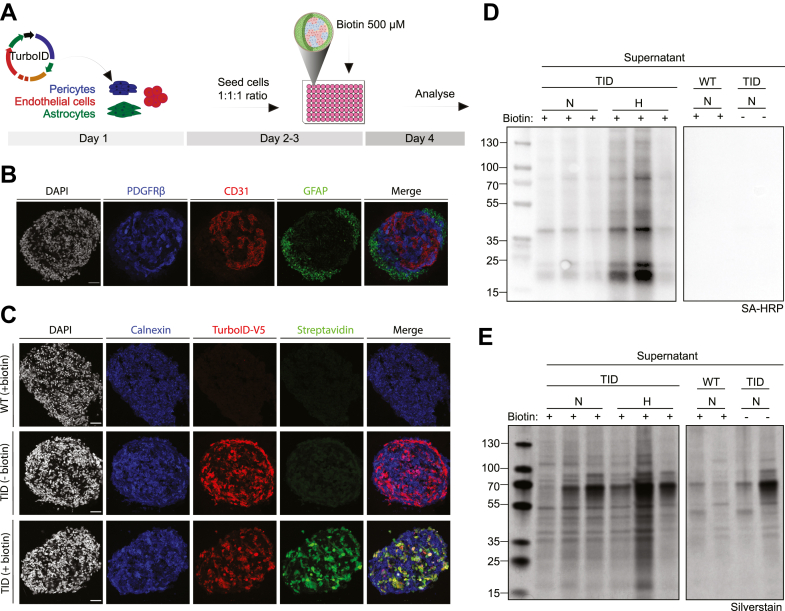
To determine if pericyte-specific labeling of secreted proteins was feasible in coculture settings, we set up spheroids of human TurboID-pericytes with human endothelial cells and astrocytes to facilitate the cell–cell interactions of the BBB (Fig. 2A). The spheroids showed an even distribution of CD31+ endothelial cells and PDGFRβ+ pericytes, whereas GFAP+ astrocytes were located more towards the spheroid border (Fig. 2B). We also evaluated pericyte intracellular protein biotinylation in spheroid settings by administering 500 μM biotin for 24 h. Biotinylated proteins were clearly detected using streptavidin-immunoassays and colocalized to the TurboID V5-tag and calnexin in TurboID-pericytes treated with biotin. The WT and TurboID-pericytes with omitted biotin controls had no to very low signal of streptavidin labeling (Fig. 2C). We next verified the biotinylation of secreted proteins in spheroid cocultures. We observed a strong signal from streptavidin blotting in both normoxic and hypoxic environments. Notably, our control conditions (WT and TurboID-pericytes with omitted biotin) had undetectable levels of biotinylated proteins in the supernatant while exhibiting a clear signal from the total protein content in the silver stain (Fig. 2, D and E).
Fig. 2.
TurboID biotinylates intracellular and secreted proteins in pericyte spheroid cultures.A, illustration of the experimental workflow and cells used for spheroid cultures. B, cryosection of spheroid immune-stained for PDGFRβ (pericytes), CD31 (endothelial cells), and GFAP (astrocytes). C, spheroids with TurboID-pericytes treated with 500 μM biotin for 24 h or negative controls including TurboID-pericytes with omitted biotin or WT-pericytes with omitted TurboID ligase were stained for anti-V5 verifying TurboID expression, calnexin marking the ER, and streptavidin-488 for biotin-labeling. D, Western blot of supernatant samples from spheroids using SA-HRP after 24 h of either normoxic (N) or hypoxic (H) incubation. E, total protein content from the supernatant samples from spheroids were analyzed with silver stain. Scale bar represents 50 μm. ER, endoplasmic reticulum; SA-HRP, streptavidin-horse radish peroxidase; TID, TurboID.
In addition, we investigated cellular viability in the spheroids by live/dead stainings of 7AAD and Annexin V using flow cytometry. Most cells were alive with only 5.97% and 8.66% cell death after 24 h of normoxia versus hypoxia, respectively (supplemental Fig. S3, A and B). Furthermore, we did not detect any overt changes in cellular populations in normoxia compared to hypoxia when examining the different cell types using immunocytochemistry (supplemental Fig. S3C).
Proteomics of Pericyte-Secreted Proteins in Hypoxia
Cells are constantly exchanging biological signals as a means of adaptation and maintaining homeostasis, and the communication between neighboring cells in the BBB vascular niche is direct and complex (34, 35).
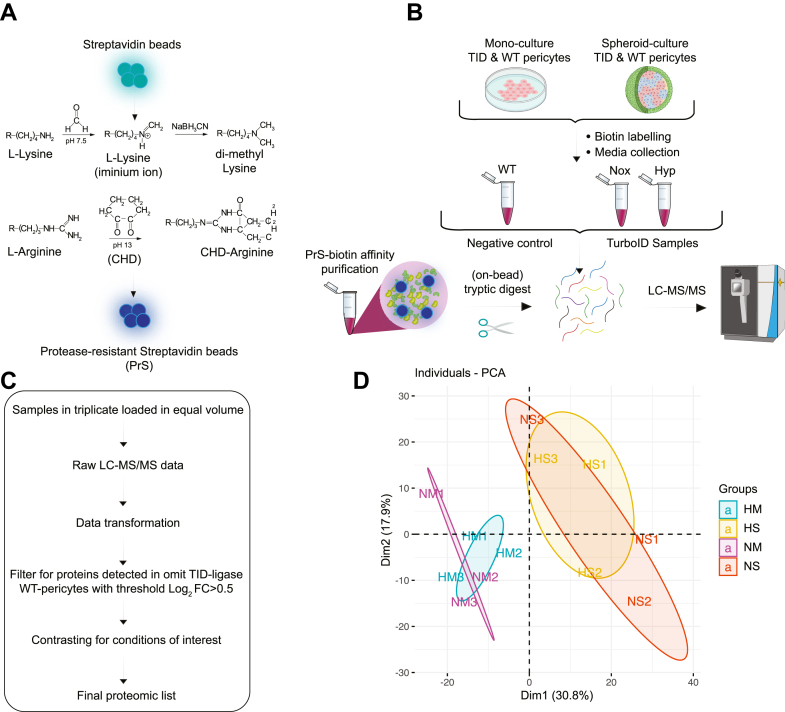
Thus, we performed a shotgun proteomics experiment to determine the protein identities secreted from pericytes in normoxic or hypoxic conditions. In addition, we analyzed the differences in secretion between mono- and spheroid-culture settings for each condition. Towards this end, biotinylated proteins were purified from media supernatant using PrS beads generated according to a previously published protocol (28). Subsequently, captured proteins underwent on-bead digestion and analyzed with LC-MS/MS (Fig. 3, A and B). To enable ratio metric and statistical testing, triplicates of each condition were included. From the supernatant samples, we detected 14,935 unique peptides corresponding to 2112 different proteins (supplemental Table S1, A and B). Missing values were imputed using msImpute with a filtering requirement of four values per protein or replicate (supplemental Fig. S4) Next, we employed a filtering regime to select for peptides originating from biotinylated proteins with a ratio against WT-pericytes with omit TurboID ligase, with a cut-off at log2 FC >0.5 generating 974 different proteins for the final dataset (Fig. 3C). This step is necessary to determine pericyte-specific proteins in spheroid cocultures as the ratio would exclude any endogenously biotinylated proteins originating from other cell types and the possibility of unspecific protein binding to the PrS beads. For the data to be comparable between spheroid cocultures and monocultures, the same filtering regime was applied on the monoculture data. Before proteomic analysis by LC-MS/MS, we validated the purity and sufficient enrichment of pericyte-derived biotinylated proteins from our PrS-biotin affinity purifications. From the streptavidin-blotting, we detected a clear signal of biotinylated proteins eluted from 5% of the enriched PrS beads before on-bead digestion, while the flow-through and washing step only elicited a weak to undetectable signal, ensuring sufficient protein capture and purity of our samples. In addition, we only detected a weak signal from 10% of the digested beads after on-bead digestion indicating successful tryptic digestion. This was further verified as the same samples were analyzed by silver stain, showing the elimination of unspecific or non-pericyte–secreted factors in the flow through while only detecting a weak signal in the washing fraction (supplemental Fig. S5, A and B).
Fig. 3.
Pericyte-specific protein purification and LC-MS/MS proteomics.A, streptavidin beads were made protease resistant (PrS) through chemical modification of lysine and arginine residues, inhibiting tryptic digestion. B, experimental design to identify pericyte-specific proteins in spheroid cultures in response to hypoxia using PrS-biotin affinity purification and mass spectrometry. C, filtering scheme for mass spectrometric data. Raw LC-MS/MS data was transformed, and global protein abundance was normalized. Biotinylated proteins were filtered for with a cut-off at log2 FC >0.5 compared to WT-pericytes with omitted biotin, and results were contrasted between the conditions of interest. D, principal component analysis of PrS-purified secreted proteins after 24 h of normoxic or hypoxic incubation both in mono- and spheroid-cultures, n = 3. HM, hypoxia–monoculture; HS, hypoxia–spheroids; NM, normoxia–monoculture; NS, normoxia–spheroids.
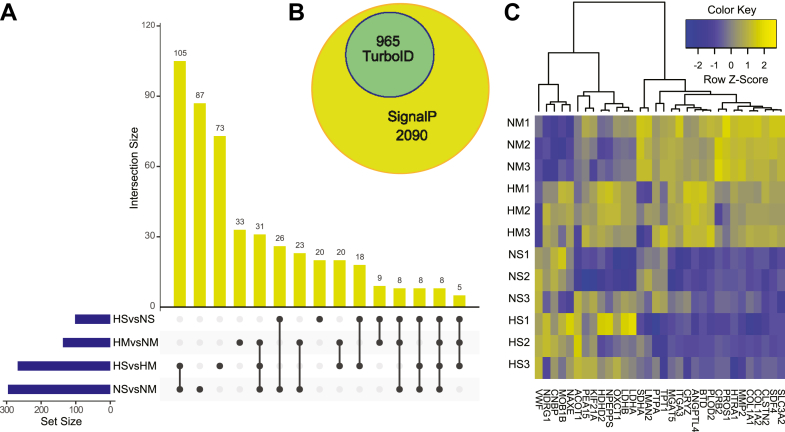
Although we ran LC-MS/MS containing the different samples in a single batch, abundance values are still subject to other factors such as sample handling during the protein purification or other potential biases. Thus, we performed data median-normalization on our global protein abundance where each sample showed similar percentiles and reduced variability, indicating successful normalization (supplemental Fig. S6). Our principal component analysis revealed some separation of biotinylated proteins between normoxic and hypoxic environments in monocultures denoted NM and HM, respectively. Notably, we also observed some variation between experimental replicates, especially in the spheroid cocultures denoted NS and HS for normoxia and hypoxia, respectively. Interestingly, spheroid cultures and monocultures showed a larger separation between each respective condition than the hypoxic to normoxic comparison (Fig. 3D). Next, we generated an UpSet plot of differentially expressed proteins to give additional insight into the secretome landscape of pericytes. We identified 33 and 20 specific proteins that were significantly different in hypoxia than normoxia, in the monocultures and spheroid cultures, respectively (Fig. 4A). However, the largest differences were observed between mono- and spheroid-cultures. For instance, 73 significantly different proteins were specific in HS compared to HM, and 87 proteins were specific for NS compared to NM, where a total of 105 specific proteins were shared between the conditions of HS versus HM and NS versus NM (Fig. 4A). This data further demonstrates that the experimental condition regarding cell culture conditions of either mono- or spheroid-cultures indicates that cell–cell interactions at the BBB have a large impact on the pericyte-derived secretome. In the conventional secretory pathway, signal peptides (SignalP) are short N-terminal sequences that control ER trafficking and protein secretion in eukaryotes. Our analysis revealed that all proteins in our dataset were predicted to have SignalP, indicating high confidence in secretion (Fig. 4B).
Fig. 4.
Proteomic differences of pericyte-secreted proteins in response to environmental cues.A, upset plot demonstrating shared or specific differentially secreted proteins between the different experimental conditions. The blue bars on the y-axis represent total number of proteins for comparison in each set size. The yellow bars on the x-axis represent the number of significantly differentially secreted proteins specific to a singular comparison or shared across comparisons illustrated by the black dots. B, Venn diagram of our TurboID dataset with predicted SignalP peptides. C, heatmap of hierarchical clustering by z-score intensities to identify the top 10 (maximum) FC >2 with p < 0.05 upregulated genes after 24 h of hypoxia for mono- or spheroid-cultures, respectively. The numbering indicate independent replicates, n = 3. HM, hypoxia–monoculture; HS, hypoxia–spheroids; NM, normoxia–monoculture; NS, normoxia–spheroids.
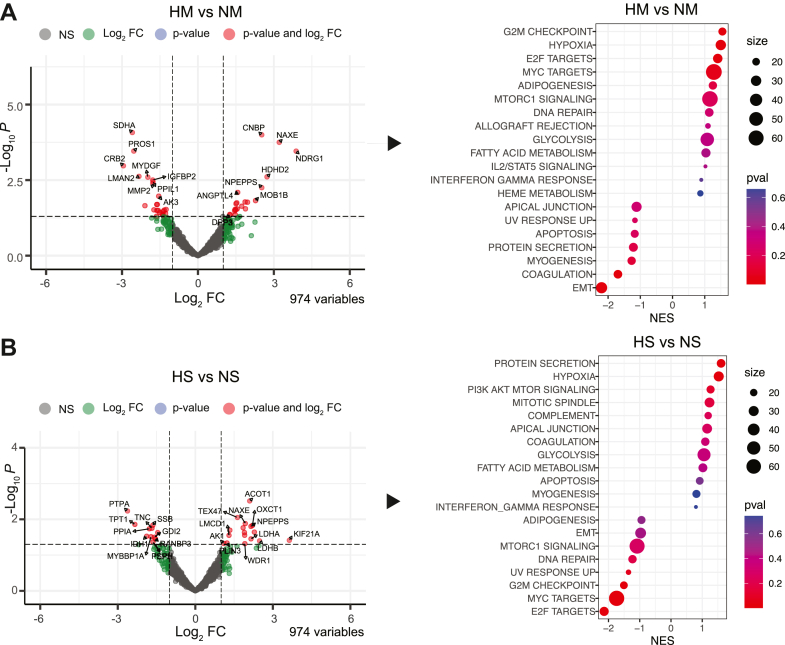
To further understand and illustrate the quantitative differences between the set of proteins of the different experimental conditions, we used a hierarchical clustering heatmap. Proteins with a log2 fold change greater than 2 and p < 0.05 between normoxia and hypoxia were plotted for all studied conditions and the WT controls (Fig. 4C). A maximum of ten proteins per condition were plotted in the heatmap, illustrating differences in the detection of secreted proteins between hypoxia and normoxia from monocultured pericytes or pericytes in spheroids (Fig. 4C). To focus on the significant differences in the pericyte secretome profiles, volcano plots of the binary comparisons between hypoxia and normoxia for monocultures and spheroid cultures are reported in (Fig. 5, A and B), respectively. In the monoculture comparison of HM versus NM, hypoxia-enriched proteins included cellular nucleic acid binding protein, NAD(P)HX epimerase (NAXE), N-Myc downstream-regulated gene 1 angiopoietin-like 4 (ANGPTL4) and others, while we observed depletion in succinate dehydrogenase complex subunit A, protein S (PROS1), crumbs cell polarity complex component 2 (CRB2), insulin-like growth factor binding protein 2, as well as others (Fig. 5A). Functional enrichment analysis of the differentially secreted proteins in hypoxic compared to normoxic monocultures correlated to an upregulation in pathways related to G2M checkpoint, hypoxia, E2F targets, Myc targets, adipogenesis, MTORC1 signaling, and others. Instead, the downregulated pathways correlated to apical junction, UV response, apoptosis, protein secretion, myogenesis, coagulation, and epithelial-mesenchymal transition (Fig. 5A). In contrast, the spheroid-culture of HS versus NS comparison showed a different pericyte secretome profile. Hypoxia-enriched proteins from pericytes in spheroids included acyl-CoA thioesterase 1 (ACOT1), NAXE, 3-oxoacid CoA-transferase 1 (OXCT1), alanine aminopeptidase (NPEPPS), adenylate kinase-1 (AK1), and others, while we observed depletion in protein phosphatase 2A, tumor protein-translationally-controlled 1, peptidylprolyl isomerase A, and others (Fig. 5B). In spheroid cultures, the functional enrichment analysis of differentially expressed proteins in hypoxia revealed an upregulation in pathways correlating to protein secretion, hypoxia, PI3K–AKT–MTOR signaling, mitotic spindle, complement, coagulation, glycolysis, and others. The downregulated pathways correlated to adipogenesis, epithelial-mesenchymal transition, MTORC1 signaling, DNA repair, G2M checkpoints, Myc targets, and E2F targets (Fig. 5B).
Fig. 5.
Proteomics of pericyte-specific secreted proteins in response to hypoxia.A, volcano plot showing the differentially expressed proteins between HM compared to NM (left panel) and the significantly regulated signaling pathways from the GSEA hallmark database (right panel), after 24 h of hypoxia, n = 3. B, volcano plot showing the differentially expressed proteins between HS compared to NS (left panel) and the significantly regulated signaling pathways from the GSEA hallmark database (right panel), after 24 h of hypoxia, n = 3. EMT, epithelial mesenchymal transition; HM, hypoxia–monoculture; HS, hypoxia–spheroids; NM, normoxia–monoculture; NS, normoxia–spheroids.
Changes in the Pericyte Secretome in Spheroids Compared to Monocultures
Given that the pericyte secretome in response to hypoxia was considerably different between spheroid cultures and monocultures, we wanted to assess the differences between cell-culture conditions in more detail. Volcano plot of spheroids compared to monocultures in normoxia showed large differences in pericyte-derived secreted factors, indicating a majority of downregulated factors in the spheroid cultures including calsyntenin 2 (CLSTN2), matrix metalloproteinase-2, integrin subunit Alpha 3, serine protease (HTRA1), and others, while a minority of secreted factors were upregulated in spheroid cultures, where those with the highest fold-change included, von Willebrand factor (vWF), zinc finger nucleic acid binding protein (CNBP), and tenascin C (supplemental Fig. S6A). In the normoxic spheroids, the enriched pathways with the largest fold change were all downregulated compared to monocultures including hypoxia, coagulation, glycolysis, complement, apoptosis, and epithelial-mesenchymal transition (Supplemental Fig. S7A). Likewise, most of the differentially secreted factors in hypoxia were downregulated in spheroids compared to monocultures. Downregulated proteins with the largest fold change included ANGPTL4, CLSTN2, Procollagen-lysine-oxoglutarate 5dioxygenase 2, biotinidase, matrix metalloproteinase-2, collagen type 1 alpha 1 chain (COL1A1), collagen type 1 alpha 2 chain, and others, while we observed an enrichment in vWF, regulator of microtubule dynamics 3, AK3, ACOT1, exosome component 1, and others (supplemental Fig. S6B). When subjected to functional enrichment analysis, the top differentially regulated pathways were all downregulated in spheroids compared to monocultures, including MTORC1-signaling, complement, xenobiotic metabolism, IL2 and STAT5-signaling, glycolysis, hypoxia, epithelial-mesenchymal transition, and apoptosis (supplemental Fig. S7B).
Proximity Extension Assay for Characterization of the Pericyte Secretome in Hypoxia
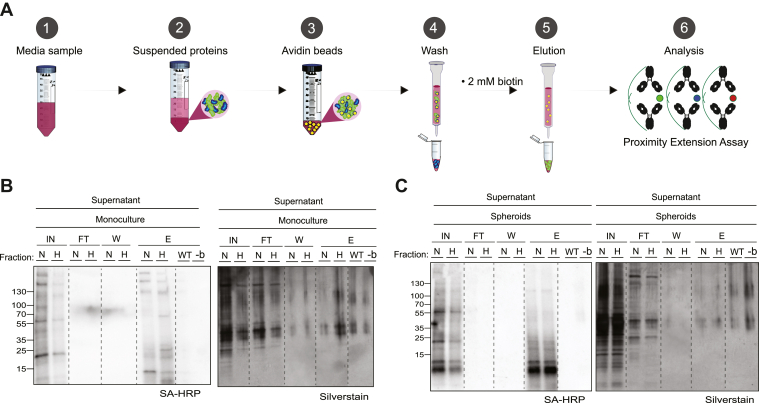
Although LC-MS/MS achieves high throughput, accuracy, and reproducibility in proteomic analysis, its limitation in sensitivity is still an issue when characterizing proteomic data (36). Thus, to be able to detect small and low abundant proteins such as cytokines, chemokines, and growth factors, we decided to incorporate an additional targeted approach using a proximity extension assay to further analyze the pericyte-specific secretome in spheroid cultures. However, since this assay requires amplification of DNA reporter sequences by real-time qPCR, we modified our protein purification protocol to eliminate proteolytic or denaturing conditions of protein recovery, for compatibility.
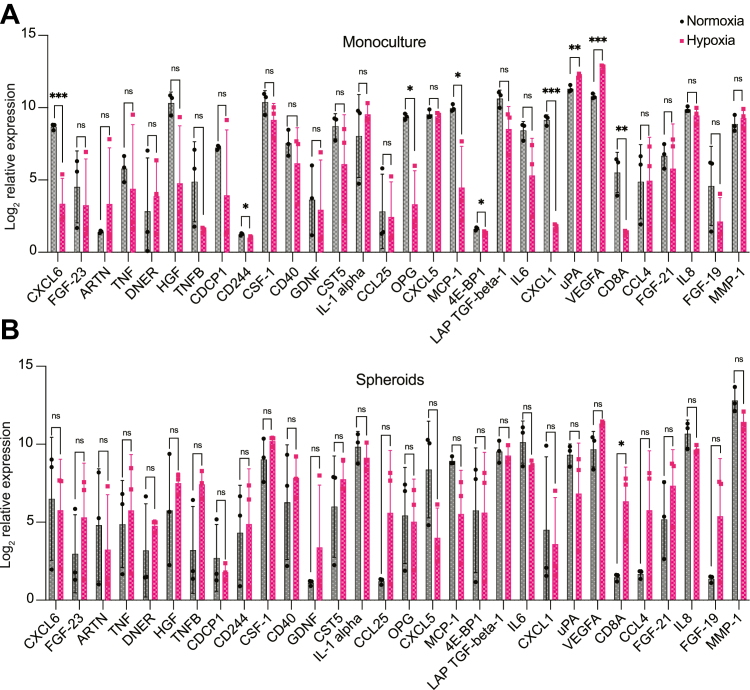
The high affinity between biotin–avidin complexes, having the strongest known noncovalent bond formation, allows easy isolation of biotinylated proteins but also introduces limitations in protein recovery. Therefore, we used monomeric avidin beads with a moderate affinity of Kd = 10−8 M compared to conventional streptavidin having a biotin binding-affinity of Kd = 10−15 M, to allow a more efficient elution of biotinylated proteins (37, 38). For protein recovery of captured biotinylated proteins, we used 2 mM of biotin in PBS as a competitive agonist for protein elution (Fig. 6A). Before proteomic analysis, we evaluated the different fractions for protein purification efficiency including, input (IN), flow-through (FT) washing (W), and biotin elution (E) by streptavidin-blotting and silver staining to confirm successful enrichment and minimal contamination. We observed a clear signal of biotinylated proteins in the IN fraction from the streptavidin blotting, confirming the efficient binding of the avidin beads. Furthermore, we detected no signal of biotinylated proteins in the FT, while we observed many nonbiotinylated or nonspecific–binding proteins in the silver stain FT. The W-fraction showed no signal from biotinylated proteins in the streptavidin blotting and minimal signal from the silver stain. The E-fraction had a similar band pattern to the IN-fraction from the streptavidin blotting, suggesting the efficient release of biotinylated proteins from the avidin beads. Importantly, we detected no signal of biotinylated proteins in the E-fraction from our negative controls of WT-pericytes or TurboID-pericytes with omitted biotin (Fig. 6, B and C). From the proximity extension assay, we were able to analyze 30 different target proteins presented in (Fig. 7, A and B). Missing values were imputed using the same filtering scheme as for the mass spectrometry data (supplemental Fig. S8). In pericyte-monocultures exposed to 24 h of hypoxia, we detected a significant increase in urokinase-type plasminogen activator (uPA) and vascular endothelial growth factor (VEGFA) compared to normoxic pericytes, while we detected a significant downregulation of chemokine C-X-C motif ligand 6 (CXCL6), cluster of differentiation 244, osteoprotegerin, monocyte chemoattractant protein-1, eukaryotic translation initiation factor 4E-binding protein 1, chemokine C-X-C motif ligand, and cluster of differentiation 8A (CD8A) (Fig. 7A). In spheroid cocultures, the only significant difference was the upregulation of CD8A in hypoxic pericytes compared to normoxia (Fig. 7B).
Fig. 6.
Pericyte-specific protein purification and sample evaluation.A, illustration of protein-purification workflow using monomeric avidin beads to capture biotinylated proteins with subsequent elution with first 2 mM biotin solution as a competitive agonist. B, Western blotting with SA-HRP or silver stain were used to evaluate the purification efficiency of supernatant samples after 24 h of hypoxia, originating from monocultures and (C) spheroids of TurboID-pericytes and negative controls before proteomic analysis. E, elution; FT, flow-through; IN, input; W, wash.
Fig. 7.
Pericyte-specific secretome analysis using a proximity extension assay.A, bar plots of pericyte-secreted proteins after 24 h of hypoxia compared to normoxic monocultures and (B) spheroids, n = 3. Statistical testing was performed using unpaired multiple t-tests with correction for multiple comparisons using the Holm–Sidak method, and the data is displayed as mean ± SD.
Discussion
The proteins that are secreted from human cells, collectively known as the secretome, are an important aspect to understand basic human biology to identify cellular activation in pathogenesis and for future development of diagnostic and therapeutic interventions (39, 40). Our understanding of cell-specific signaling by secreted molecules without compromising physiological relevance has previously been hampered by the lack of sufficient biological tools. Here, we use a novel approach of cell type–selective secretome profiling in more complex coculture settings in vitro, by utilizing TurboID tagged to the ER-lumen, to allow bio-labeling of proteins within the secretory pathway that can be purified and analyzed after being secreted into the extracellular space. Given the low cellular toxicity of TurboID-dependent proteome biotinylation in different cellular compartments (22), alterations or inhibition of protein function is likely kept to a minimum, an important feature when studying communication between different cell types in hypoxia. In summary, this method allows rapid and broad mapping of the dynamic pericyte secretome under pathophysiological conditions such as hypoxia.
Pericytes are known to respond to different stimuli by secreting various proteins that affect the function of BBB resident cells (13). Increasing evidence supports pericytes as early sensors of hypoxia in the brain, likely due to their unique position between the circulation and the brain parenchyma (41, 42, 43). However, most of the previous studies on pericyte-secreted factors in hypoxia have been analyzed from monocultures (9, 12, 13), which may not reflect the pericyte secretome in vivo under similar pathological conditions, and a full characterization of the pericyte secretome in a hypoxic environment has been lacking.
The pericyte secretome data from this study further contextualize previous findings and provide new insights into pericyte signaling after a hypoxic insult. Although we observed large differences in secreted factors depending on monoculture or spheroid-culture conditions, some factors were consistently increased in both conditions of hypoxia. Two of the secreted proteins with the highest increase in fold change in hypoxic conditions and conserved in both monocultures and spheroids were NAXE and NPEPPS. NAXE is a highly evolutionary conserved enzyme catalyzing the repair of damaged forms of NAD(P)H, to protect several different metabolic pathways (44, 45). Interestingly, recent studies have identified NAXE (also known as APOA1 binding protein), as a secreted protein regulating angiogenesis and inflammation. NAXE has been shown to accelerate the cholesterol efflux in endothelial cells, leading to depletion in membrane lipid rafts that inhibits VEGFR2 dimerization and signaling which results in reduced angiogenesis (46). In addition, low NAXE expression in hypoxic tumor environments showed reduced NADPH levels, leading to an increase in reactive oxygen species and activation of hypoxia-inducible factor 1-alpha (47). NAXE expression has also been reported to be increased in ischemia, where NAXE KO mice showed elevated angiogenic activity correlated to inhibition of Notch signaling (48). Our data indicates a strong induction of NAXE being secreted from hypoxic pericytes, both in monocultures and spheroid cultures. Interestingly, hypoxia is associated with increased angiogenesis as a compensatory mechanism to an oxygen deficit, but the hypoxic state may also lack sufficient reducing agents for controlled vascular formation during angiogenesis (49). It is possible that the release of NAXE is a necessary response to prevent an imbalanced angiogenic response in hypoxia leading to the excessive immature vascular formation and increased risk of haemorrhagic transformation.
Furthermore, we identified NPEPPS as one of the most significantly hypoxia-induced secreted protein in pericytes from both monocultures and spheroids. A previous study identified NPEPPS secretion detected in extracellular vesicles taken from cerebrospinal fluid (50). NPEPPS is known to regulate the angiotensin-aldosterone system modulating immune response as well as regulating proteolytic events in cell cycle progression (51). In another study, NPEPPS was identified as one of the top upregulated genes in hypoxic tumors compared to normoxic controls (52). Notably, NPEPPS has also been shown to be involved in cholesterol biosynthesis and a previous study identified NPEPPS as 1 out of 18 common disease-specific genes between pathologies including Alzheimer's disease, multiple sclerosis, and ischemic stroke (53). While there is some evidence of NPEPPS having neuroprotective functions in Alzheimer’s disease by direct proteolytic cleavage of TAU proteins (54), its physiological function in the hypoxic/ischemic brain is still somewhat elusive.
We identified ACOT1 to be one of the top upregulated proteins of the pericyte secretome in hypoxia that was specific for spheroid cultures. ACOT1 is an important protein regulating lipid metabolism by catalyzing fatty acid oxidation of acyl-CoAs to CoA-SH and free fatty acids (55). Notably, human ACOT1 has been suggested to be expressed both in the cytosol and in the peroxisomes (56). A recent study performed a genome-wide CRISPR knock-out screen examining cell fitness in normoxic and hypoxic environments. Interestingly, the authors identified many proteins related to lipid metabolism and peroxisome metabolism/biogenesis to be essential for cell survival in hypoxia (57). Reduced cell viability was correlated to decreased availability of unsaturated fatty acids in hypoxia leading to impaired cell growth, likely due to decreased membrane fluidity. In addition, another study showed that ACOT1 downregulation is associated with reduced biosynthesis of unsaturated fatty acids both in vivo and in vitro (58). Considering these previous findings, the pericyte release of ACOT1 upon hypoxic insult may regulate cell survival in BBB resident cells.
Interestingly, the hallmark pathway analysis indicated that both pericyte monocultures and spheroids had similar adaptive responses to hypoxia regarding energy metabolism-related pathways, known to play a major role in cellular fitness under hypoxia. However, there were also substantial differences in other enriched pathways related to protein secretion, coagulation, and cellular proliferation between the two culturing conditions under hypoxia. Signature proteins related to protein secretion and coagulation were upregulated in hypoxic pericyte-spheroids while they were some of the top downregulated pathways in pericyte monocultures. Likewise, pathways related to cellular proliferation, for example, G2M checkpoint, E2F-targets, and Myc-targets were highly upregulated in monocultures whereas they were the top downregulated in pericyte spheroid cultures. These findings can hopefully increase our understanding of how pericytes respond to hypoxic stress and further indicates the large differences between culturing conditions.
In affinity pull-downs of biotinylated proteins, protein recovery is a crucial concern. On-bead tryptic digestion provided sufficient peptide yields (∼0.5 μg/μl) for mass spectrometry analysis, yet we were unable to detect small and low abundant proteins like cytokines and growth factors. Therefore, we adopted a targeted approach relying on antigen recognition and genetic sequence amplification that requires an elution protocol where we generated protein concentrations in the (pg/ml) range. While we had significantly lower protein recovery yields for our elution approach, we were still able to detect 30 different proteins from the proximity extension assay where nine were significantly differentially expressed in hypoxic pericyte monocultures. For instance, we noticed a significant increase in VEGFA secretion from hypoxic pericytes, which is a key initiator of angiogenesis (59). This finding is also supported by previous studies showing VEGFA increase in hypoxic retinal pericytes (12) but also in hypoxic human brain pericytes (60). Furthermore, we observed a significant increase in pericyte secretion of uPA in hypoxic conditions. One of the major functions of uPA is to facilitate the conversion of inactive plasminogen to its active form plasmin, which can dissolve blood clots by fibrinolysis (61). While the release of uPA in response to hypoxia-induced by, for example, ischemic stroke may have beneficial effects in restoring brain perfusion, recent studies have demonstrated additional functions of uPA such as neuronal protection and regulation of angiogenesis (62). We also observed a clear downregulation of CXCL6 in hypoxic monocultures. CXCL6 knockdown in an ischemia-reperfusion model has previously been reported to promote cell proliferation and decrease apoptosis (63). Likewise, cluster of differentiation 244, osteoprotegerin, monocyte chemoattractant protein-1, 4E-binding protein 1, and chemokine C-X-C motif ligand were also significantly downregulated in hypoxia suggesting an anti-inflammatory response by pericytes after 24 h of hypoxic exposure in monocultures. When examining the spheroid cultures, we only detected a significant difference showing a strong upregulation of CD8A—a pro-inflammatory marker. However, we did observe some strong trends towards an upregulation of trophic and growth factors such as FGF-23, GDNF, FGF-21, FGF-19, VEGF, and other proinflammatory markers including, TNFB, CCL4, and CCL25.
Another important aspect of this study was the characterization of the pericyte-secretome differences between monocultures and spheroids. While previous studies have shown that maintaining vascular homeostasis at the BBB requires close interplay between pericytes, endothelial cells, and astrocytes (35, 64), a full understanding of the microvascular crosstalk and its effect on cellular specific secretomes is still elusive. For instance, a recent study utilizing RNA-seq when comparing the human pericyte transcriptome cultured in the absence or presence of human endothelial cells identified a total of 6704 genes that were differentially expressed in pericytes in coculture environments (65). One of the main findings from that study was that pericytes in coculture with endothelial cells suppressed overall expression of ECM proteins including a variety of different collagens (65). Interestingly, we identified COL1A1 as one of the top downregulated proteins in normoxic spheroids compared to monocultures and both COL1A1 and collagen type 1 alpha 2 chain as two of the top downregulated proteins in spheroids compared to monocultures under hypoxic conditions. Notably, recent studies into the existence of various vascular cell types and zonation markers have led to the categorization of pericytes into three distinct subtypes (66, 67). Interestingly, our mass spectrometry data suggests potential features of matrix-pericytes secreting different types of collagens, while we did not detect any specific markers for transporter-pericytes expressing various solute carrier membrane transporter proteins. However, further research is needed to conclusively address the diversity of pericyte subtypes within spheroids or organoids and their potential implications on the secretome signaling. Another interesting observation from our proteomic dataset was the strong induction of the pericyte-derived release of vWF in spheroid cultures specifically, observed both in normoxic and hypoxic conditions, which has been previously identified as a pericyte-secreted protein in vivo by Wei et al. (27) who used PDGFRβ-CreERT2 mice to drive TurboID expression in pericytes. While there was a limited overlap between our proteomic dataset, we identified several common secreted proteins although using different models and species. As mentioned, our analysis identified vWF but also other proteins overlapping with those found in mouse plasma, including transglutaminase 2. Notably we were unable to detect A1AT4 and KV5A6, which were previously indicated to be released by pericytes in the plasma of mice. The limited overlap between these two datasets may arise from variances in experimental models.
This is another indication that pericytes may regulate thrombosis and angiogenic processes through vWF release at the microvasculature. Interestingly, many of the differentially secreted proteins between spheroids and monocultures were similar in both hypoxic and normoxic conditions also evident from the downregulated pathway analysis, suggesting that most differentially secreted proteins between culturing conditions were independent of hypoxia.
As with all studies, our results also need to be considering potential limitations. We only used a single human pericyte cell line for this study. Although all replicates in this study were performed independently, the fact remains that it is hard to procure human biological replicate cell lines and the conclusions presented here are limited to the cell line used in this study and may not reflect the pericyte population in general. Moreover, a recent study using bioinformatic predictions of human secreted proteins based on sequence analysis of signal peptides with no transmembrane spanning regions identified 2641 proteins out of the human proteome to be secreted factors (40). From this study, we detected on average 2112 different pericyte-secreted proteins and were able to analyze 974 variables after our filtering regime suggesting that pericytes are highly secretory cells. While we controlled for cell viability during hypoxic or normoxic incubation and removed any cellular debris from the supernatant samples before analysis, the possibility of proteins detected from cell lysis remains. However, we believe contamination of intracellular proteins to have had a minimal effect on our final analysis, also evident from the fact that all the proteins in our analyzed dataset were predicted to have a signal peptide.
Although the human secretome is being increasingly explored, much information regarding the spectrum and functional annotation of secreted factors remains to be elucidated. Further studies in vivo and in humans are necessary to shed light on secreted factors under healthy and pathological conditions. We believe that this study can provide an additional resource in the methodological aspects of cell-specific secretome profiling and contribute to the understanding of the pericytes' secretory response at the BBB under pathological hypoxia.
Data Availability
All data generated in this study are available and will be shared by the corresponding author upon reasonable request. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD044689 (68). For accessing the data, dataset: https://www.ebi.ac.uk/pride/archive?keyword=PXD044689 (supplemental Table S1, A and B). The number code for each sample used for mass spectrometry analysis is presented in (supplemental Table S3) and the number code for annotated spectra regarding protein identification from single unique peptides is presented in (supplemental Table S4).
Supplemental Data
This article contains supplemental data.
Conflict of interest
The authors declare no competing or interests.
Acknowledgments
Author contributions
A. E., R. C., and G. P. conceptualization; A. E., C. B., and M. L. data curation; A. E. and R. C. formal analysis; A. E. and G. P. funding acquisition; A. E., R. C., C. B., M. L., and G. P. investigation; A. E., R. C., C. B., and M. L. methodology; A. E. and R. C. software; A. E., R. C., C. B., and M. L. validation; A. E., R. C., and C. B. visualization; A. E. writing–original draft; A. E., R. C., C. B., and G. P. writing–review and editing; R. C. and G. P. supervision; G. P. project administration; G. P. resources.
Funding and additional information
This work is supported by the Swedish Research Council, the Swedish Stroke Foundation, Gunvor and Josef Anérs Foundation, the Swedish Brain Foundation, Fang Foundation, and The Royal Physiographic Society of Lund.
Supplemental data
References
- 1.Armulik A., Genové G., Mäe M., Nisancioglu M.H., Wallgard E., Niaudet C., et al. Pericytes regulate the blood-brain barrier. Nature. 2010;468:557–561. doi: 10.1038/nature09522. [DOI] [PubMed] [Google Scholar]
- 2.Keaney J., Campbell M. The dynamic blood-brain barrier. FEBS J. 2015;282:4067–4079. doi: 10.1111/febs.13412. [DOI] [PubMed] [Google Scholar]
- 3.Gaceb A., Ozen I., Padel T., Barbariga M., Paul G. Pericytes secrete pro-regenerative molecules in response to platelet-derived growth factor-BB. J. Cereb. Blood Flow Metab. 2018;38:45–57. doi: 10.1177/0271678X17719645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kovac A., Erickson M.A., Banks W.A. Brain microvascular pericytes are immunoactive in culture: cytokine, chemokine, nitric oxide, and LRP-1 expression in response to lipopolysaccharide. J. Neuroinflammation. 2011;8:139. doi: 10.1186/1742-2094-8-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gomes E.D., Mendes S.S., Assunção-Silva R.C., Teixeira F.G., Pires A.O., Anjo S.I., et al. Co-transplantation of adipose tissue-derived stromal cells and olfactory ensheathing cells for spinal cord injury repair. Stem Cells. 2018;36:696–708. doi: 10.1002/stem.2785. [DOI] [PubMed] [Google Scholar]
- 6.Teixeira F.G., Carvalho M.M., Panchalingam K.M., Rodrigues A.J., Mendes-Pinheiro B., Anjo S., et al. Impact of the secretome of human mesenchymal stem cells on brain structure and animal behavior in a rat model of Parkinson's disease. Stem Cells Transl. Med. 2017;6:634–646. doi: 10.5966/sctm.2016-0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen M.P., Frank R.N., Khalifa A.A. Collagen production by cultured retinal capillary pericytes. Invest. Ophthalmol. Vis. Sci. 1980;19:90–94. [PubMed] [Google Scholar]
- 8.Geranmayeh M.H., Rahbarghazi R., Farhoudi M. Targeting pericytes for neurovascular regeneration. Cell Commun. Signal. 2019;17:26. doi: 10.1186/s12964-019-0340-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ishitsuka K., Ago T., Arimura K., Nakamura K., Tokami H., Makihara N., et al. Neurotrophin production in brain pericytes during hypoxia: a role of pericytes for neuroprotection. Microvasc. Res. 2012;83:352–359. doi: 10.1016/j.mvr.2012.02.009. [DOI] [PubMed] [Google Scholar]
- 10.Makridakis M., Vlahou A. Secretome proteomics for discovery of cancer biomarkers. J. Proteomics. 2010;73:2291–2305. doi: 10.1016/j.jprot.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 11.Rustenhoven J., Jansson D., Smyth L.C., Dragunow M. Brain pericytes as mediators of neuroinflammation. Trends Pharmacol. Sci. 2017;38:291–304. doi: 10.1016/j.tips.2016.12.001. [DOI] [PubMed] [Google Scholar]
- 12.Takagi H., King G.L., Robinson G.S., Ferrara N., Aiello L.P. Adenosine mediates hypoxic induction of vascular endothelial growth factor in retinal pericytes and endothelial cells. Invest. Ophthalmol. Vis. Sci. 1996;37:2165–2176. [PubMed] [Google Scholar]
- 13.Gaceb A., Paul G. Pericyte secretome. Adv. Exp. Med. Biol. 2018;1109:139–163. doi: 10.1007/978-3-030-02601-1_11. [DOI] [PubMed] [Google Scholar]
- 14.Kalocsay M. APEX peroxidase-catalyzed proximity labeling and multiplexed quantitative proteomics. Methods Mol. Biol. 2019;2008:41–55. doi: 10.1007/978-1-4939-9537-0_4. [DOI] [PubMed] [Google Scholar]
- 15.Ngo J.T., Champion J.A., Mahdavi A., Tanrikulu I.C., Beatty K.E., Connor R.E., et al. Cell-selective metabolic labeling of proteins. Nat. Chem. Biol. 2009;5:715–717. doi: 10.1038/nchembio.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sears R.M., May D.G., Roux K.J. BioID as a tool for protein-proximity labeling in living cells. Methods Mol. Biol. 2019;2012:299–313. doi: 10.1007/978-1-4939-9546-2_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eichelbaum K., Winter M., Berriel Diaz M., Herzig S., Krijgsveld J. Selective enrichment of newly synthesized proteins for quantitative secretome analysis. Nat. Biotechnol. 2012;30:984–990. doi: 10.1038/nbt.2356. [DOI] [PubMed] [Google Scholar]
- 18.Witzke K.E., Rosowski K., Müller C., Ahrens M., Eisenacher M., Megger D.A., et al. Quantitative secretome analysis of activated Jurkat cells using click chemistry-based enrichment of secreted glycoproteins. J. Proteome Res. 2017;16:137–146. doi: 10.1021/acs.jproteome.6b00575. [DOI] [PubMed] [Google Scholar]
- 19.Yang A.C., du Bois H., Olsson N., Gate D., Lehallier B., Berdnik D., et al. Multiple click-selective tRNA synthetases expand mammalian cell-specific proteomics. J. Am. Chem. Soc. 2018;140:7046–7051. doi: 10.1021/jacs.8b03074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barrett R.M., Liu H.W., Jin H., Goodman R.H., Cohen M.S. Cell-specific profiling of nascent proteomes using orthogonal enzyme-mediated puromycin incorporation. ACS Chem. Biol. 2016;11:1532–1536. doi: 10.1021/acschembio.5b01076. [DOI] [PubMed] [Google Scholar]
- 21.Du S., Wang D., Lee J.S., Peng B., Ge J., Yao S.Q. Cell type-selective imaging and profiling of newly synthesized proteomes by using puromycin analogues. Chem. Commun. (Camb.) 2017;53:8443–8446. doi: 10.1039/c7cc04536k. [DOI] [PubMed] [Google Scholar]
- 22.Branon T.C., Bosch J.A., Sanchez A.D., Udeshi N.D., Svinkina T., Carr S.A., et al. Efficient proximity labeling in living cells and organisms with TurboID. Nat. Biotechnol. 2018;36:880–887. doi: 10.1038/nbt.4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cho K.F., Branon T.C., Rajeev S., Svinkina T., Udeshi N.D., Thoudam T., et al. Split-TurboID enables contact-dependent proximity labeling in cells. Proc. Natl. Acad. Sci. U. S. A. 2020;117:12143–12154. doi: 10.1073/pnas.1919528117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cho K.F., Branon T.C., Udeshi N.D., Myers S.A., Carr S.A., Ting A.Y. Proximity labeling in mammalian cells with TurboID and split-TurboID. Nat. Protoc. 2020;15:3971–3999. doi: 10.1038/s41596-020-0399-0. [DOI] [PubMed] [Google Scholar]
- 25.Droujinine I.A., Meyer A.S., Wang D., Udeshi N.D., Hu Y., Rocco D., et al. Proteomics of protein trafficking by in vivo tissue-specific labeling. Nat. Commun. 2021;12:2382. doi: 10.1038/s41467-021-22599-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim K.E., Park I., Kim J., Kang M.G., Choi W.G., Shin H., et al. Dynamic tracking and identification of tissue-specific secretory proteins in the circulation of live mice. Nat. Commun. 2021;12:5204. doi: 10.1038/s41467-021-25546-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wei W., Riley N.M., Yang A.C., Kim J.T., Terrell S.M., Li V.L., et al. Cell type-selective secretome profiling in vivo. Nat. Chem. Biol. 2021;17:326–334. doi: 10.1038/s41589-020-00698-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rafiee M.R., Sigismondo G., Kalxdorf M., Förster L., Brügger B., Béthune J., et al. Protease-resistant streptavidin for interaction proteomics. Mol. Syst. Biol. 2020;16 doi: 10.15252/msb.20199370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwartz J.C., Philp R.L., Bickhart D.M., Smith T.P.L., Hammond J.A. The antibody loci of the domestic goat (Capra hircus) Immunogenetics. 2018;70:317–326. doi: 10.1007/s00251-017-1033-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hediyeh-zadeh S., Webb A.I., Davis M.J. MSImpute: imputation of label-free mass spectrometry peptides by low-rank approximation. bioRxiv. 2020 doi: 10.1101/2020.08.12.248963. [preprint] [DOI] [Google Scholar]
- 31.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43 doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Conway J.R., Lex A., Gehlenborg N. UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics. 2017;33:2938–2940. doi: 10.1093/bioinformatics/btx364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Korotkevich G., Sukhov V., Budin N., Shpak B., Artyomov M.N., Sergushichev A. Fast gene set enrichment analysis. bioRxiv. 2021 doi: 10.1101/060012. [preprint] [DOI] [Google Scholar]
- 34.Heymans M., Figueiredo R., Dehouck L., Francisco D., Sano Y., Shimizu F., et al. Contribution of brain pericytes in blood-brain barrier formation and maintenance: a transcriptomic study of cocultured human endothelial cells derived from hematopoietic stem cells. Fluids Barriers CNS. 2020;17:48. doi: 10.1186/s12987-020-00208-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sweeney M.D., Ayyadurai S., Zlokovic B.V. Pericytes of the neurovascular unit: key functions and signaling pathways. Nat. Neurosci. 2016;19:771–783. doi: 10.1038/nn.4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Catherman A.D., Skinner O.S., Kelleher N.L. Top down proteomics: facts and perspectives. Biochem. Biophys. Res. Commun. 2014;445:683–693. doi: 10.1016/j.bbrc.2014.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheah J.S., Yamada S. A simple elution strategy for biotinylated proteins bound to streptavidin conjugated beads using excess biotin and heat. Biochem. Biophys. Res. Commun. 2017;493:1522–1527. doi: 10.1016/j.bbrc.2017.09.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scheurer S.B., Roesli C., Neri D., Elia G. A comparison of different biotinylation reagents, tryptic digestion procedures, and mass spectrometric techniques for 2-D peptide mapping of membrane proteins. Proteomics. 2005;5:3035–3039. doi: 10.1002/pmic.200402069. [DOI] [PubMed] [Google Scholar]
- 39.Stastna M., Van Eyk J.E. Secreted proteins as a fundamental source for biomarker discovery. Proteomics. 2012;12:722–735. doi: 10.1002/pmic.201100346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Uhlen M., Karlsson M.J., Hober A., Svensson A.S., Scheffel J., Kotol D., et al. The human secretome. Sci. Signal. 2019;12 doi: 10.1126/scisignal.aaz0274. [DOI] [PubMed] [Google Scholar]
- 41.Duz B., Oztas E., Erginay T., Erdogan E., Gonul E. The effect of moderate hypothermia in acute ischemic stroke on pericyte migration: an ultrastructural study. Cryobiology. 2007;55:279–284. doi: 10.1016/j.cryobiol.2007.08.009. [DOI] [PubMed] [Google Scholar]
- 42.Enstrom A., Carlsson R., Ozen I., Paul G. RGS5: a novel role as a hypoxia-responsive protein that suppresses chemokinetic and chemotactic migration in brain pericytes. Biol. Open. 2022;11 doi: 10.1242/bio.059371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gonul E., Duz B., Kahraman S., Kayali H., Kubar A., Timurkaynak E. Early pericyte response to brain hypoxia in cats: an ultrastructural study. Microvasc. Res. 2002;64:116–119. doi: 10.1006/mvre.2002.2413. [DOI] [PubMed] [Google Scholar]
- 44.Becker-Kettern J., Paczia N., Conrotte J.F., Zhu C., Fiehn O., Jung P.P., et al. NAD(P)HX repair deficiency causes central metabolic perturbations in yeast and human cells. FEBS J. 2018;285:3376–3401. doi: 10.1111/febs.14631. [DOI] [PubMed] [Google Scholar]
- 45.Bommer G.T., Van Schaftingen E., Veiga-da-Cunha M. Metabolite repair enzymes control metabolic damage in glycolysis. Trends Biochem. Sci. 2020;45:228–243. doi: 10.1016/j.tibs.2019.07.004. [DOI] [PubMed] [Google Scholar]
- 46.Fang L., Choi S.H., Baek J.S., Liu C., Almazan F., Ulrich F., et al. Control of angiogenesis by AIBP-mediated cholesterol efflux. Nature. 2013;498:118–122. doi: 10.1038/nature12166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun B., Yu L., Xu C., Li Y.M., Zhao Y.R., Cao M.M., et al. NAD(P)HX epimerase downregulation promotes tumor progression through ROS/HIF-1alpha signaling in hepatocellular carcinoma. Cancer Sci. 2021;112:2753–2769. doi: 10.1111/cas.14925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mao R., Meng S., Gu Q., Araujo-Gutierrez R., Kumar S., Yan Q., et al. AIBP limits angiogenesis through gamma-secretase-mediated upregulation of Notch signaling. Circ. Res. 2017;120:1727–1739. doi: 10.1161/CIRCRESAHA.116.309754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schito L., Rey S. Hypoxia: turning vessels into vassals of cancer immunotolerance. Cancer Lett. 2020;487:74–84. doi: 10.1016/j.canlet.2020.05.015. [DOI] [PubMed] [Google Scholar]
- 50.Muraoka S., Jedrychowski M.P., Yanamandra K., Ikezu S., Gygi S.P., Ikezu T. Proteomic profiling of extracellular vesicles derived from cerebrospinal fluid of Alzheimer's disease patients: a pilot study. Cells. 2020;9:1959. doi: 10.3390/cells9091959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pan Y., Sun X., Mi X., Huang Z., Hsu Y., Hixson J.E., et al. Whole-exome sequencing study identifies four novel gene loci associated with diabetic kidney disease. Hum. Mol. Genet. 2023;32:1048–1060. doi: 10.1093/hmg/ddac290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Song Z., Pearce M.C., Jiang Y., Yang L., Goodall C., Miranda C.L., et al. Delineation of hypoxia-induced proteome shifts in osteosarcoma cells with different metastatic propensities. Sci. Rep. 2020;10:727. doi: 10.1038/s41598-019-56878-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tseveleki V., Rubio R., Vamvakas S.S., White J., Taoufik E., Petit E., et al. Comparative gene expression analysis in mouse models for multiple sclerosis, Alzheimer's disease and stroke for identifying commonly regulated and disease-specific gene changes. Genomics. 2010;96:82–91. doi: 10.1016/j.ygeno.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kudo L.C., Parfenova L., Ren G., Vi N., Hui M., Ma Z., et al. Puromycin-sensitive aminopeptidase (PSA/NPEPPS) impedes development of neuropathology in hPSA/TAU(P301L) double-transgenic mice. Hum. Mol. Genet. 2011;20:1820–1833. doi: 10.1093/hmg/ddr065. [DOI] [PubMed] [Google Scholar]
- 55.Wang F., Wu J., Qiu Z., Ge X., Liu X., Zhang C., et al. ACOT1 expression is associated with poor prognosis in gastric adenocarcinoma. Hum. Pathol. 2018;77:35–44. doi: 10.1016/j.humpath.2018.03.013. [DOI] [PubMed] [Google Scholar]
- 56.Hunt M.C., Siponen M.I., Alexson S.E. The emerging role of acyl-CoA thioesterases and acyltransferases in regulating peroxisomal lipid metabolism. Biochim. Biophys. Acta. 2012;1822:1397–1410. doi: 10.1016/j.bbadis.2012.03.009. [DOI] [PubMed] [Google Scholar]
- 57.Jain I.H., Calvo S.E., Markhard A.L., Skinner O.S., To T.L., Ast T., et al. Genetic screen for cell fitness in high or low oxygen highlights mitochondrial and lipid metabolism. Cell. 2020;181:716–727.e1. doi: 10.1016/j.cell.2020.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu Y., Zeng L., Yang Y., Chen C., Wang D., Wang H. Acyl-CoA thioesterase 1 prevents cardiomyocytes from doxorubicin-induced ferroptosis via shaping the lipid composition. Cell Death Dis. 2020;11:756. doi: 10.1038/s41419-020-02948-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zechariah A., ElAli A., Doeppner T.R., Jin F., Hasan M.R., Helfrich I., et al. Vascular endothelial growth factor promotes pericyte coverage of brain capillaries, improves cerebral blood flow during subsequent focal cerebral ischemia, and preserves the metabolic penumbra. Stroke. 2013;44:1690–1697. doi: 10.1161/STROKEAHA.111.000240. [DOI] [PubMed] [Google Scholar]
- 60.Sharma K., Zhang Y., Paudel K.R., Kachelmeier A., Hansbro P.M., Shi X. The emerging role of pericyte-derived extracellular vesicles in vascular and neurological health. Cells. 2022;11:3108. doi: 10.3390/cells11193108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pannell R., Li S., Gurewich V. Fibrin-specific and effective clot lysis requires both plasminogen activators and for them to be in a sequential rather than simultaneous combination. J. Thromb. Thrombolysis. 2017;44:210–215. doi: 10.1007/s11239-017-1514-0. [DOI] [PubMed] [Google Scholar]
- 62.Merino P., Diaz A., Manrique L.G., Cheng L., Yepes M. Urokinase-type plasminogen activator (uPA) promotes ezrin-mediated reorganization of the synaptic cytoskeleton in the ischemic brain. J. Biol. Chem. 2018;293:9234–9247. doi: 10.1074/jbc.RA118.002534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang X., Dai Y., Zhang X., Pan K., Deng Y., Wang J., et al. CXCL6 regulates cell permeability, proliferation, and apoptosis after ischemia-reperfusion injury by modulating Sirt3 expression via AKT/FOXO3a activation. Cancer Biol. Ther. 2021;22:30–39. doi: 10.1080/15384047.2020.1842705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hellstrom M., Gerhardt H., Kalén M., Li X., Eriksson U., Wolburg H., et al. Lack of pericytes leads to endothelial hyperplasia and abnormal vascular morphogenesis. J. Cell Biol. 2001;153:543–553. doi: 10.1083/jcb.153.3.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brandt M.M., van Dijk C.G.M., Maringanti R., Chrifi I., Kramann R., Verhaar M.C., et al. Transcriptome analysis reveals microvascular endothelial cell-dependent pericyte differentiation. Sci. Rep. 2019;9 doi: 10.1038/s41598-019-51838-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Garcia F.J., Sun N., Lee H., Godlewski B., Mathys H., Galani K., et al. Single-cell dissection of the human brain vasculature. Nature. 2022;603:893–899. doi: 10.1038/s41586-022-04521-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang A.C., Vest R.T., Kern F., Lee D.P., Agam M., Maat C.A., et al. A human brain vascular atlas reveals diverse mediators of Alzheimer's risk. Nature. 2022;603:885–892. doi: 10.1038/s41586-021-04369-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Perez-Riverol Y., Bai J., Bandla C., García-Seisdedos D., Hewapathirana S., Kamatchinathan S., et al. The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 2022;50:D543–D552. doi: 10.1093/nar/gkab1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated in this study are available and will be shared by the corresponding author upon reasonable request. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD044689 (68). For accessing the data, dataset: https://www.ebi.ac.uk/pride/archive?keyword=PXD044689 (supplemental Table S1, A and B). The number code for each sample used for mass spectrometry analysis is presented in (supplemental Table S3) and the number code for annotated spectra regarding protein identification from single unique peptides is presented in (supplemental Table S4).