Highlights
-
•
miRNAs from DLK1-DIO3 are globally down regulated in papillary thyroid carcinomas.
-
•
Only twelve, out of 100 mature miRNAs, are responsible for most of the impact of DLK1-DIO3 region on the post transcriptional regulation in PTC.
-
•
DLK1-DIO3-derived miRNAs act predominantly in PTC tumor progression.
-
•
In vitro functional analises show influence of miR-485–5p on cell migration, invasion and EMT.
-
•
miR-485–5p action is dependent on the tumor oncogenic background, with effect only in BRAF-mutant papillary thyroid carcinoma cell line.
Keywords: MicroRNA, Papillary thyroid cancer, DLK1-DIO3 region, miR-485–5p
Abstract
Background
In previous studies, we demonstrated the downregulation of several miRNAs from the DLK1-DIO3 genomic region in papillary thyroid carcinoma (PTC). Due to the large number of miRNAs within this region, the individual contribution of these molecules to PTC development and progression remains unclear.
Objective
In this study, we aimed to clarify the contribution of DLK1-DIO3-derived miRNAs to PTC.
Methods
We used different computational approaches and in vitro resources to assess the biological processes and signaling pathways potentially modulated by these miRNAs.
Results
Our analysis suggests that, out of more than 100 mature miRNAs originated from the DLK1-DIO3 region, a set of 12 miRNAs accounts for most of the impact on PTC development and progression, cooperating to modulate distinct cancer-relevant biological processes, such as cell migration, extracellular matrix remodeling, and signal transduction. The restoration of the expression of one of these miRNAs (miR-485–5p) in a BRAFT199A-positive PTC cell line impaired proliferation and migration, suppressing the expression of GAB2 and RAC1, validated miR-485–5p targets.
Conclusions
Overall, our results shed light on the role of the DLK1-DIO3 region, which harbors promising tumor suppressor miRNAs in thyroid cancer, and open prospects for the functional exploration of these miRNAs as therapeutic targets for PTC.
Introduction
In the United States, 43,720 new cases of thyroid cancer are estimated in 2023. Of these, 2120 deaths are anticipated [1]. The genetic landscape of papillary thyroid carcinoma has been well characterized [2], [3], [4], [5]. Most of the genetic abnormalities observed in PTC are aligned in the MAPK signaling cascade, with the prevalence of RET fusions, BRAF, and RAS point mutations [5]. The BRAFT1799A oncogene is currently known to be associated with invasion of extrathyroidal tissues and increased risk of lymph node and distant metastases [6].
MiRNAs are small non-coding RNA molecules involved in the post-transcriptional regulation of gene expression in plants and animals [7]. The relevance of these molecules to the thyroid development and function has been extensively explored [8], as miRNAs participate in the regulation of thyroid-specific transcription factors and iodine-metabolism genes [8], [9], [10]. Importantly, the aberrant expression of miRNAs has been widely described in thyroid cancer [11] and these molecules are currently being explored as promising molecular markers for diagnosis and recurrence [12].
A previous study by our group described the global deregulation of the expression of miRNAs located in the genomic region DLK1-DIO313. This region, located on human chromosome 14, is present in placental mammals and originated due to a tandem amplification of the locus [14]. It is controlled by genomic imprinting, with the protein-coding genes DLK1, RTL1 and DIO3 being expressed from the paternal allele and the noncoding genes MEG3, MEG8 and several miRNAs being expressed from the maternal allele [15]. DLK1 is involved in Notch signaling in different tissues and DIO3 is a type 3 deiodinase, involved in the metabolism of thyroid hormone in different organs [16]. MEG3 has been described as a regulator of physiological and pathological processes such as cell proliferation, differentiation, and survival [17,18]. Regarding the miRNAs, the DLK1-DIO3 region hosts the largest known miRNA cluster in the human genome [15]. Although miRNAs derived from the DLK1-DIO3 region have been described as tumor suppressors in several types of cancer, their role in thyroid cancer remains to be explored [19], [20], [21]. Moreover, in Temple Syndrome patients, in which partial or total deletions of the DLK1-DIO3 region are observed, an increased risk of developing thyroid cancer prematurely has been detected [22]. This suggests a close relationship between the DLK1-DIO3 locus and the biology of the thyroid follicular cell. Due to the large number of mature miRNAs derived from the cluster, the functional investigation of the individual contribution of each miRNA to the oncogenic process becomes challenging. Thus, we have developed a computational approach based on the target prediction, filtering against thyroid cancer gene expression datasets and gene set enrichment analysis to better understand how these miRNAs might act as tumor suppressors in PTC. In this study we aimed to shed light on the potential of the DLK1-DIO3 region as a source of miRNAs that could act as tumor suppressors in PTC.
Materials and methods
Computational analysis of DLK1-DIO3 region miRNAs
The list of miRNA genes from the DLK1-DIO3 region was downloaded using the Table Browser tool (https://genome.ucsc.edu/cgi-bin/hgTables). The miRWalk v.2.0 platform was used for the computational prediction of targets of each mature miRNA from the DLK1-DIO3 region [23]. The miRWalk v2.0 allowed the prediction of miRNA targets by 12 different algorithms, including TargetScan [24], PicTar [25], and the miRWalk native algorithm. The lists of targets of all miRNAs from the DLK1-DIO3 region predicted by the 12 algorithms were downloaded. Due to the high stringency of the TargetScan algorithm, based on conservation between different species and canonical/non-canonical seed pairing to predict miRNA:target interactions, only interactions predicted by the TargetScan platform and at least 7 more of the 12 algorithms used were included. The resulting lists were then combined and processed into a matrix and filtered using “PTC versus Normal” gene expression data from the Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) and The Cancer Genome Atlas (TCGA, http://cancergenome.nih.gov/). The mean fold change expression values between normal and tumor thyroid tissue were obtained from 5 studies from the GEO platform, comprising a total of 101 PTC samples and 69 normal tissues using the GEO2R algorithm, as described previously [26]. Each gene was considered differentially expressed when adjusted P-value < 0.01 and when an agreement between at least 3 of the 5 studies was observed. Similarly, level 3 mRNA expression data from 437 human PTC samples and 59 normal thyroid tissue samples from the TCGA project were downloaded as described previously[13]. Since miRNAs of the DLK1-DIO3 region are downregulated in PTC, every target with mean fold change < 1.25 (25 %) was excluded. Importantly, the use of a higher threshold (>1.5 or 2.0) would significantly impair the target identification. Similarly, gene expression data and its relationship with clinical attributes were downloaded from the cBioPortal platform (cbioportal.org) and statistically analyzed as described below. To assess the expression status of DLK1-DIO3 miRNAs in different types of cancer, miRNA expression data were downloaded from the ENCORI database [27] (Sun Yat-sen University, http://www.sysu.edu.cn), which compiles data from the TCGA project.
We used the DAVID online tool [28] (https://david.ncifcrf.gov/) for the identification of signaling pathways and biological processes enriched among DLK1-DIO3-derived miRNAs. To strengthen our strategy to identify miRNAs involved in PTC, we directed the search to biological processes relevant to cancer, such as “cell cycle”, “cell adhesion”, “regulation of cell migration”, “extracellular matrix”, “positive and negative regulation of apoptosis” and “cell-cell adhesion”. In addition, the list of potential targets was compared with 98 oncogenes characterized in the literature. Venn diagrams were generated using the InteractiVenn online tool [29]. A network of predicted target genes was constructed, according to the same criteria described above, using Cytoscape 3.7.1 with the STRING protein query (StringApp), which annotates protein-protein interactions [30,31]. We used a confidence score of 0.8 and no additional interactions were allowed.
Cell culture
A panel of three thyroid cancer cell lines was used to evaluate the tumor suppressor potential of miR-485–5p in thyroid cancer. N-Thy-ORI is a normal follicular immortalized cell line [32], TPC-1 [33] and BCPAP [34] cell lines are derived from human papillary carcinoma and harbor the RET/PTC1 chromosomal rearrangements and the BRAFT1799A mutation, respectively. N-Thy-ORI was kindly donated by Dr. Edna Kimura, from the University of Sao Paulo, Brazil. The TPC-1 cell line was kindly provided by Dr. James A Fagin (Human Oncology and Pathogenesis Program, Memorial Sloan-Kettering Cancer Center, New York, NY, USA). The BCPAP cells were gently donated by Dr. Massimo Santoro (Medical School, University ‘Federico II’ of Naples, Naples, Italy). N-Thy-ORI cells were maintained in RPMI medium supplemented with 10 % fetal bovine serum (SFB, Thermo Fisher, Waltham, MA, USA). TPC-1 and BCPAP were maintained in DMEM medium supplemented with 5 % and 10 % fetal bovine serum, respectively. Importantly, the restoration of miR-485–5p in the thyroid cancer cells did not cause apparent morphological changes in the cancer cell lines.
Overexpression of miR-485–5p in thyroid cancer cell lines
Primers flanking the genomic region of the MIR485 gene were used for amplification and cloning of the miR-485–5p genomic region in the pGEM-T Easy Vector System (Promega) (Supplementary Table 1). The insert was removed by double digestion with XhoI and EcoRI restriction enzymes and cloned in the MSCV-puro vector, generating a pMSCV-485 vector. Sanger sequencing confirmed the identity of the miR-485–5p genomic region in the construct. Transfection of the plasmid constructs (pMSCV-485 or the empty vector) was performed using Lipofectamine 2000 (Thermo Fisher) according to the manufacturer's instructions. Puromycin-resistant (5 μg/mL) clones were selected for further experiments. Overexpression of miR-485–5p was confirmed by RT-qPCR.
Quantitative real-time PCR
One hundred thousand cells were seeded in 60 mm plates and collected after 72 h in TRIzol reagent. Total RNA was extracted based on Chomczynski and Sacchi [35]. Specific complementary DNA of miR-485–5p and RNU6B was synthesized using TaqMan MicroRNA Reverse Transcription Kit (ThermoFisher) and qPCR reactions were performed using Taqman MicroRNA Assay (miR-485–5p: Assay ID 001,036; RNU6B: Assay ID 001093, ThermoFisher), using the TaqMan Universal Master Mix II, No UNG kit in the ABI 7300 thermal cycler. The quantification of mRNA target genes was performed using SYBR Green Dye reagent (Applied Biosystems). Reactions were performed in a final volume of 20 % μL, using 5 % μL of cDNA diluted 10 times from the final volume of cDNA synthesis (∼ 5 ng), 10 μL of 2x Master Mix SYBR reagent Green (Applied Biosystems), and 5 μL of specific primers (Supplementary Table 1). The 7300 SDS Software program was used to analyze the data obtained. Relative expression was calculated as described elsewhere[36].
Cell proliferation
The cell lines overexpressing miR-485–5p and those carrying the empty vector were seeded in a density of 2.5 × 104 cells per well in 12-well plates. At periods of 48 and 72 h, the cells were trypsinized, fixed at 3.7 % formaldehyde, and maintained at 4 °C until counting, which was performed using a Neubauer chamber.
Migration and invasion
The miR-485–5p-overexpressing cell lines were submitted to the wound healing assay, as previously described[37]. After total confluence, a vertical lesion line was introduced on the cell monolayer in the center of each well using a sterile 200 μL tip. The healing process was assessed soon after the lesion was performed (0 h) and 16 and 24 h later. The size of each lesion was measured at different time points using ImageJ® software[38]. The cell migration and invasion assays were executed in a modified Boyden chamber using 8.0 μm pore membrane inserts (Millipore, MA). 2 × 104 cells were resuspended in medium containing 0.5 % fetal bovine serum and plated in the upper chamber compartment. The lower compartment was filled with a medium containing 10 % fetal bovine serum. After 12 h, the culture medium was removed, and the chamber was washed twice with PBS. The cells in the upper compartment were removed and the cells in the lower compartment were fixed, stained with 0.5 % Violet Crystal, and photographed under a Nikon Eclipse E600 microscope. For cell invasion assays, extracellular matrix (ECM Gel from Engelbreth-Holm-Swarm murine sarcoma - liquid, BioReagent 8.42 μg), diluted in 22.5 μL of DMEM, and 30 μL of the diluted matrix were plated on top of each insert.
Statistical analysis
The results were submitted to statistical analysis using GraphPad Prism version 9.0 for Windows (GraphPad Software, San Diego, California, USA). Values were expressed as mean ± standard deviation. The Student's t-test and the Mann-Whitney test were used for comparisons between two data populations with Gaussian and non-Gaussian distributions, respectively. The Two-Way ANOVA test was used for comparisons between three or more data groups and the Log-rank test was used for Kaplan-Meier survival analysis. Statistical significance was considered when p < 0.05, unless otherwise specified.
Data access statement: The results shown here are in part based upon public data generated by the TCGA Research Network: https://www.cancer.gov/tcga.
Ethics statement: This study solely relied on publicly available data and did not include any materials requiring ethical approval.
Results
In silico characterization of the DLK1-DIO3-derived miRNAs in PTC
Expression data from the TCGA project revealed that miRNAs from the DLK1-DIO3 region were downregulated in kidney papillary and chromophobe renal cell carcinoma, as well as breast invasive carcinoma and head and neck squamous cell carcinoma (Fig. 1a). However, a global upregulation was observed in other types of cancer, such as gastric and lung carcinoma, suggesting that, depending on the tumor of origin, the DLK1-DIO3-derived miRNAs may act as oncogenes or tumor suppressor genes.
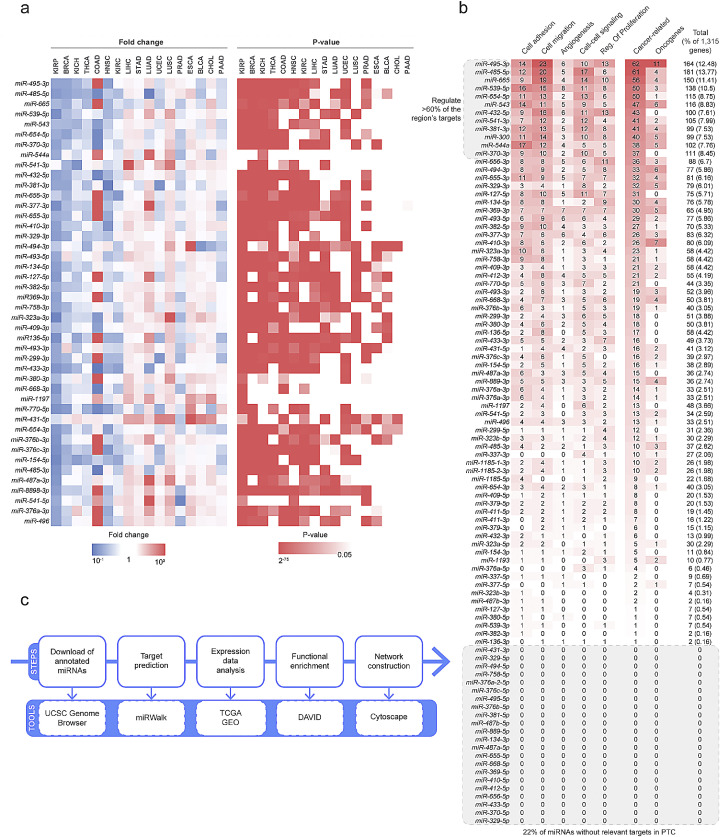
Fig. 1.
Abnormal expression of DLK1-DIO3-derived miRNAs in cancer and target prediction in papillary thyroid carcinoma. (a) The miRNAs from the DLK1-DIO3 region are deregulated in different types of cancer. The mean fold change between tumor and non-tumor samples for each miRNA in each cancer type is represented in the heatmap on the left. The statistical significance is represented in the heatmap on the right. The miRNA expression data were downloaded from the ENCORI database, which compiles data from the TCGA project. (b) The DLK1-DIO3-derived miRNAs were ranked according to the total number of predicted targets involved in cancer-related processes. (c) Scheme of the computational strategy adopted for identification of DLK1-DIO3-derived miRNAs with tumor suppressor potential.
Over 100 mature miRNAs are transcribed from the 53 miRNA genes present in the DLK1-DIO3 region. Thus, the functional characterization of individual miRNAs may be time- and labor-consuming. To improve efficiency in identifying the most relevant DLK1-DIO3-derived miRNAs for PTC we combined different bioinformatics resources, as shown in Fig. 1b. Firstly, the miRWalk v.2.0 platform was used to identify the list of predicted targets for each of the mature miRNAs from the DLK1-DIO3 region. As described in the Methods section, the miRWalk v2.0 platform provides the list of predicted targets of any given miRNA according to 12 different algorithms. In this study, only interactions predicted by at least 8 out of the 12 algorithms were considered valid, resulting in a list of 6231 targets. Once the miRNAs from DLK1-DIO3 are downregulated in PTC, the list of targets was compared with mRNA expression data downloaded from GEO and TCGA to select targets whose expression was increased by at least 25 % (fold change> 1.25; p < 0.01). The resulting 1315 targets were then submitted to Gene Set Enrichment Analysis on the DAVID online tool. Then, the miRNAs were ranked according to the number of targets in cancer-related ontological categories (Fig. 1c).
Interestingly, although approximately a hundred mature miRNAs may originate from the DLK1-DIO3 region, 22 (23 %) have no valid interactions in our analysis. On the other hand, we observed that more than 63 % of the genes are targeted by the 12 top-ranked miRNAs on the list (Fig. 1c). The enrichment analysis of the list of predicted targets of all DLK1-DIO3-derived miRNAs combined revealed similar enriched GO categories with the list of the top-12 miRNAs combined (Fig. 2a). These findings suggest that the top-12 miRNAs may have a larger contribution to the regulation of biological processes in PTC than the remaining miRNAs from the DLK1-DIO3 region. Interestingly, although the combined top-12 miRNAs mimic the enrichment observed in targets of all miRNAs from the DLK1-DIO3, each miRNA displays particular enriched processes. The reason for that may lie in the large fraction of genes targeted by only one miRNA in our model: 523 (39.77 %) of the total of the 1315 genes submitted to the enrichment analysis. For example, for the top-12 miRNAs, 35.51 % of the genes were targeted exclusively by a single miRNA.
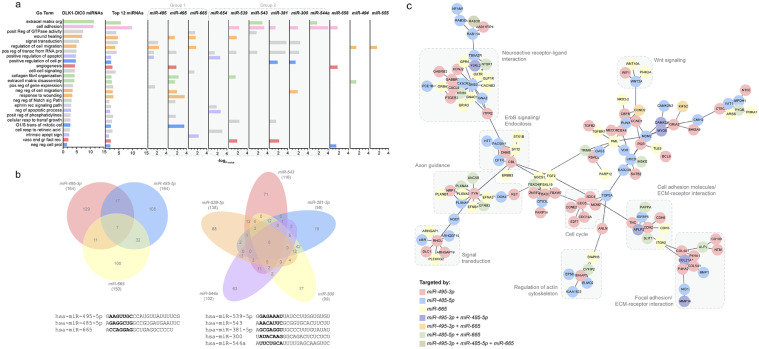
Fig. 2.
DLK1-DIO3-derived miRNAs cooperate to modulate cancer-related processes in PTC. (a) Gene set enrichment analysis of the list of the predicted targets of all miRNAs in the DLK1-DIO3 region compared to the list of predicted targets of the top 12 miRNAs combined and individually. miRNAs with similar enrichment patterns were marked as group 1 (miR-495–3p, miR-485-p, miR-665) and 2 (miR-539–5p, miR-543, miR-381–3p, miR-300, miR-544a, miR-432–5p). (b) Venn diagrams show a small number of common targets between miRNAs in groups 1 and 2. The mature sequences of the miRNAs in the diagrams are shown at the bottom, with seed sequences in bold. (c) Network of protein-protein interactions between predicted targets of miRNAs from group 1. The color code represents the targeting of each node by one or more miRNAs. The network was built using STRING protein in Cytoscape 3.7.1. Confidence score = 0.8 and no additional interactions were allowed.
As an example, the group of miRNAs miR-495–3p, miR-485–5p, and miR-665 (named group 1 for comparison purposes) may have a major impact on cell migration while the group containing miR-539–5p, miR-543, miR-381–3p, miR-300, miR-544a and miR-432–5p (named group 2 for comparison purpose) may contribute to deregulate cell adhesion, extracellular matrix remodeling and gene expression (Fig. 2a). Thus, each small group of miRNAs could contribute to PTC development and progression through the deregulation of different biological processes. Except for the miRNAs miR-665, miR-370–3p, and miR-432–5p, most of the top-12 miRNAs genes are located at a 39.8 kb region (chr14:101,028,292–101,068,115) upstream from MEG9 gene with no apparent relationship regarding their genomic location, sequence conservation and biological function (data not shown).
As mentioned above, each of the top-12 miRNAs modulates a small percentage of targets in our model: miR-485–5p (13.76 %), miR-495–3p (12.47 %), miR-665 (11.40 %), miR-539–5p (10.49 %), miR-543 (8.82 %), miR-654–5p (8.75 %), miR-370–3p, miR-541–3p (7.99 %), miR-544a (7.76 %), miR-432–5p (7.61 %), miR-300 (7.53 %) and miR-381–3p (7.53 %). Moreover, even miRNAs with similar enriched categories within each group have few or no common targets. As an example, miRNAs from group 1 (miR-495–3p, miR-485–5p, miR-665) have only 7 common targets, while group 2 (miR-539–5p, miR-543, miR-381–3p, miR-300, miR-544a) have none (Fig. 2b). This suggests that these miRNAs cooperate by targeting distinct members within each biological process. The differential targeting may be explained by the differences in the seed sequences of each miRNA (Fig. 2b), which is the sequence segment from the second to the seventh nucleotides of each miRNA, critical for the recognition and stabilization with the target mRNA[4]. An example is shown in Fig. 2c, where we observed that miRNAs from group 1 (miR-495–3p, miR-485–5p, miR-665) cooperate to modulate different cancer-related processes, such as cell cycle, signal transduction, and extracellular matrix remodeling, where simultaneous targeting was less frequent.
miR-485–5p as a tumor suppressor candidate in PTC
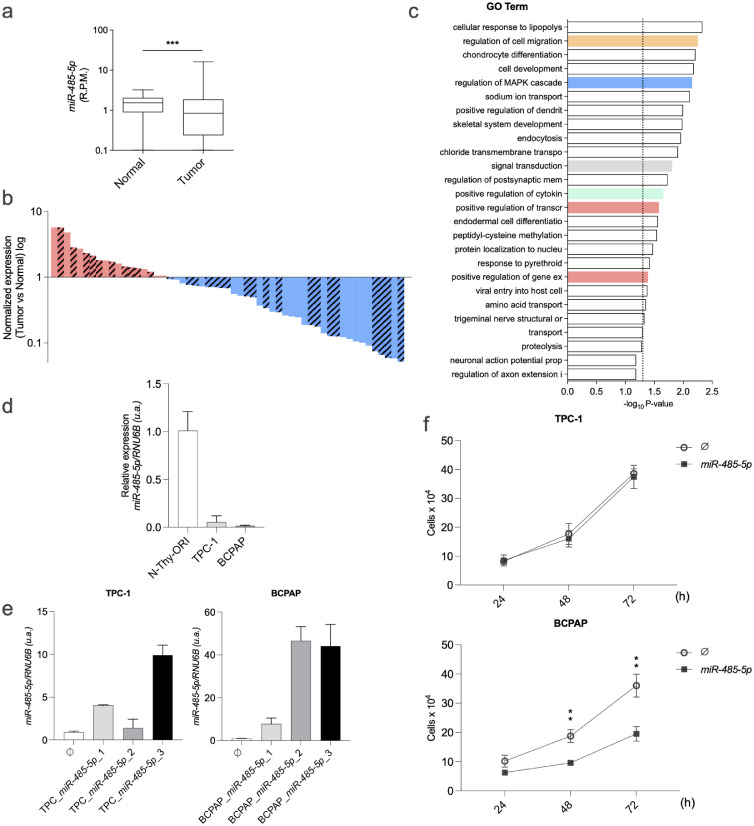
As a proof of concept, we aimed to test whether the identified miRNAs within the network could act as tumor suppressors in PTC. One of the top-ranked miRNAs in our in silico analysis, miR-485–5p, has been described as a tumor suppressor in different types of cancer [24], [25], [26], [27], [28] and is downregulated in PTC in comparison with normal thyroid tissue, according to TCGA data (Fig. 4a,b). The enriched biological processes and signaling pathways among miR-485–5p targets include relevant processes for PTC, such as “regulation of cell migration”, “regulation of MAPK cascade” and “signal transduction”, targeting several key modulators of distinct signaling pathways relevant for cancer progression (Fig. 3c, Supplementary Table 2). miR-485–5p was downregulated in two papillary thyroid cancer cell lines (TPC-1 and BCPAP) in comparison with the human immortalized normal thyroid cell line N-Thy-ORI (Fig. 3d). Interestingly the overexpression of miR-485–5p abolished cell proliferation in the BRAF-mutated cell line BCPAP (Fig. 3e,f). In contrast, no significant changes were observed in the RET/PTC1-positive TPC-1 cells.
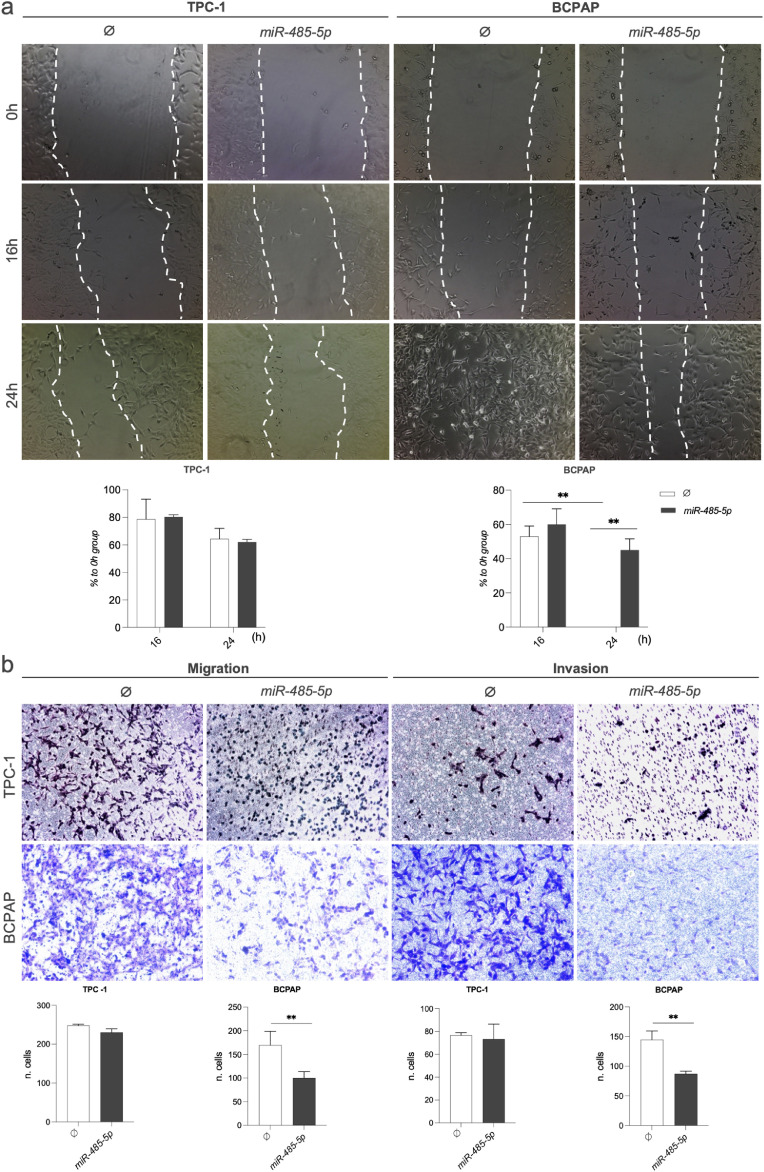
Fig. 4.
miR-485–5p suppresses migration and invasion of PTC cells in vitro. (a) TPC-1 and BCPAP cells, overexpressing miR-485–5p or not, were cultured until near total confluence and a lesion was performed on the monolayer using a pipette tip. Photographs were taken at times 0, 16, and 24 h (N = 3). The BCPAP 24 h time point on the graph has no column due to the complete wound closure. (b) For the migration assay, TPC-1 and BCPAP cells, overexpressing miR-485–5p or not, were seeded in the upper compartment of modified Boyden chambers. For the invasion assay, the chambers were covered with a commercial reconstitutable extracellular matrix. Migrated/invaded cells at the lower chamber compartment were fixed, stained, and photographed after 12 h. Data is presented as one representative experiment (from three independent experiments) and bars represent the standard deviation. P ≤ 0,01(**). 40x magnification.
Fig. 3.
miR-485–5p as a candidate tumor suppressor in PTC. (a) miR-485–5p downregulation in human thyroid cancer samples from miRNA-seq data (TCGA project). Mean expression of miR-485–5p in 437 PTC samples versus 59 normal thyroid tissues. (b) “Waterfall-plot” graph of miR-485–5p expression in 59 PTC samples compared to matched normal thyroid samples from miRNAseq data (TCGA project). Pink bars represent upregulation and blue bars represent downregulation. Hatched bars represent BRAFT1799A-positive samples. (c) Functional enrichment of miR-485–3p predicted targets. GO categories relevant to this study are shown in colors. Dashed line marks P-value = 0.05. (d) miR-485–5p is downregulated in PTC-derived cell lines compared to a normal thyroid follicular immortalized cell line. For each cell line 1 × 105 cells were cultivated for 72 h, total RNA was extracted and cDNA was synthesized. (e) Two PTC cell lines (TPC-1 and BCPAP) were transfected with miR-485–5p-expressing plasmid or transfected with the empty vector. For each cell line, the transfectants with higher miR-485–5p expression were selected for further analysis. (f) TPC-1 and BCPAP cells, transfected with pMSCV-485 plasmid or the empty vector, were collected 24, 48, and 72 h after seeding, fixed, and counted. In (d) and (e)RNU6B gene was used as endogenous control. The y-axis shows values in arbitrary units (a.u.). Data is presented as the average of three independent experiments and bars represent the standard deviation. P ≤ 0,01(**). R.P.M: reads per million.
We then evaluated the influence of miR-485–5p on the regulation of cell migration, the main enriched GO term among miR-485–5p predicted targets. In the wound healing assay, as observed for cell proliferation, the restoration of miR-485–5p levels inhibited cell migration only in the BRAF-mutated cell line BCPAP (Fig. 4a), without changes in TPC-1 cells. The role of miR-485–5p in the suppression of migration of the BRAF-mutated BCPAP cell line was confirmed in the modified Boyden chamber assay (Fig. 4b). Importantly, miR-485–5p also inhibited cell invasion of BCPAP cells seeded on the top of inserts coated with commercial extracellular matrix.
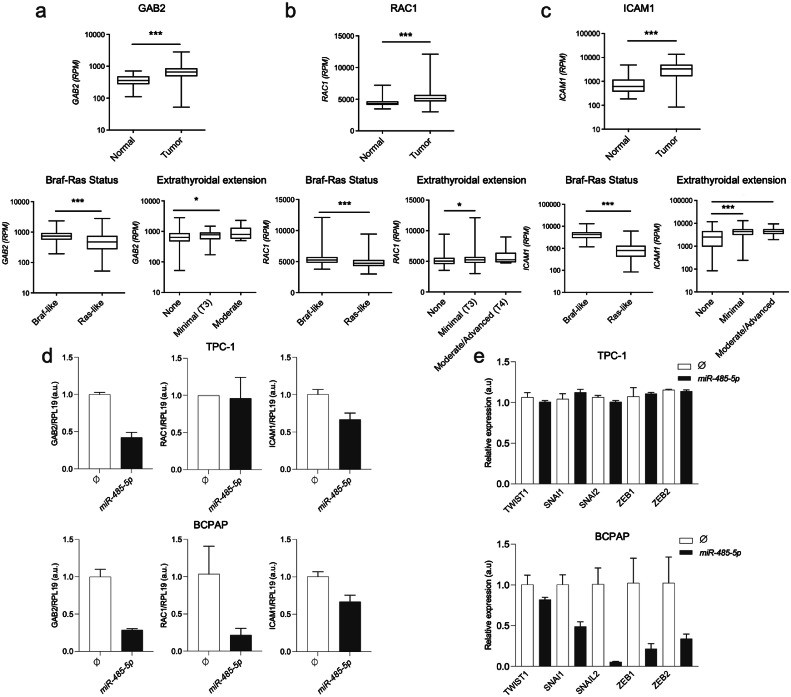
To test our hypothesis, we further examined the ability of miR-485–5p to regulate the expression levels of three candidate genes predicted by our bioinformatic approach, GAB2, RAC1, and ICAM1 (Supplementary Table 2). GAB2 and RAC1 are regulators of cell migration in different types of cancers and have been experimentally validated as direct targets of miR-485–5p[24,29]. In addition, ICAM1 is positively correlated with aggressiveness in thyroid cancer samples and is a promising target for advanced thyroid cancer therapy [30,31]. According to TCGA data, these genes are upregulated in PTC samples in comparison with normal tissue, in PTCs with a "BRAF-like" rather than a "RAS-like" phenotype, and in patients with extrathyroidal extension by the time of diagnosis (Fig. 5a,b,c). Interestingly, GAB2 and ICAM1 were downregulated by miR-485–5p overexpression in both cell lines, whereas RAC1 levels were diminished only in the BRAF-mutated cell line BCPAP (Fig. 5d), which could, at least in part, explain the differential migratory behavior between TPC-1 and BCPAP cell lines. Importantly, the overexpression of miR-485–5p was capable of inhibiting the expression of key genes in Epithelial-to-Mesenchymal transition (EMT) in the BRAF-mutated cell line BCPAP, such as TWIST1, SNAI1, SNAI2, ZEB1 and ZEB2 (Fig. 5e). In contrast, no significant changes were observed in the RET/PTC-bearing cell line TPC-1. No changes were observed in the expression levels of thyroid differentiation genes SLC5A5, TPO, TG, and TSHR upon restoration of miR-485–5p in both cell lines (data not shown).
Fig. 5.
GAB2, RAC1, and ICAM1 as targets of miR-485–5p in PTC. (a-c, Top) Mean expression of GAB2, RAC1, or ICAM1 in 437 PTC samples versus 59 normal thyroid samples from RNA-seq (TCGA project) and respective correlation with clinical attributes according to the TCGA project. (N = 388); (R.P.M) reads per million; (d, e) Total RNA was extracted from TPC-1 and BCPAP cells transfected with pMSCV-485 plasmid or the empty vector and used for cDNA synthesis. Expression of GAB2, RAC1, ICAM1, TWIST1, SNAI1, SNAI2, ZEB1 and ZEB2 was quantified by qPCR. RPL19 gene was used as endogenous control. The y-axis shows values in arbitrary units (a.u). Data is presented as the average of three independent experiments and bars represent the standard deviation. P ≤ 0,05 (*), P ≤ 0,001 (***).
Discussion
In a prior study, we reported the global downregulation of miRNAs from DLK1-DIO3 region in PTC cell lines, human PTC samples, and a murine model of BRAF-mutated PTC [13]. The Cancer Genome Atlas project data show the abnormal expression of several DLK1-DIO3-derived miRNAs in various cancer types (Fig. 1a), and a number of these miRNAs have validated oncogenic or tumor suppressor roles in a variety of neoplastic malignancies [[19], [20], [21],37,[39], [40], [41], [42], [43], [44], [45]]. In this study, we used computational tools and online-available gene expression data to shed light on the contribution of DLK1-DIO3-derived miRNAs to PTC.
Due to the large number of potential targets predicted by miRNA-target prediction algorithms, the functional analysis of miRNAs can be challenging. Also, the coordinated aberrant expression of a large number of miRNAs may hinder the identification of those with higher oncogenic impact on PTC. Therefore, bioinformatics tools may help to provide a more detailed scenario of post-transcriptional regulation by miRNAs in PTC [26,46]. Here we used different computational resources to generate a rank of DLK1-DIO3-derived miRNAs according to the targeting of cancer-related genes in PTC. Our analysis suggests that the top-12 miRNAs on the rank are responsible for most of the impact of DLK1-DIO3-derived miRNAs in PTC.
Glazov and colleagues have shown that processes related to neurogenesis and embryonic development are enriched among the targets of DLK1-DIO3-derived miRNAs [14]. Interestingly, despite the use of PTC-derived gene expression from TCGA to construct the miRNA:target matrix, we observed enriched GO categories related to axon guidance, nervous system development, and neuro-active ligand-receptor interaction, indicating that deregulation of these processes in the follicular cell could participate in thyroid oncogenesis.
Several miRNAs derived from the DLK1-DIO3 region have been implicated in the tumorigenesis and progression of different types of cancer. miR-485–5p, for example, has been shown to inhibit pathways related to cell growth and motility in colorectal cancer and to affect the regulation of oral squamous cell carcinoma stemness and drug resistance [40,47]. miR-495–3p is shown to be involved with tumor growth in gastric cancer and a recent study of our group has also identified the involvement of miR-495–3p in cell migration and invasion in PTC [37,39]. A set of seven DLK1-DIO3 miRNAs (miR-300, −382, −494, −495, −539, −543, and −544) have been shown to act coordinately to control epithelial-to-mesenchymal transition in breast cancer cell lines [48]. Accordingly, our analysis suggests that five out of the above-mentioned miRNAs (miR-494, −495, −539, −543, and −544a) modulate processes related to extracellular matrix organization. On the other hand, it is known that miRNAs derived from the DLK1-DIO3 region may act as oncogenes depending on the tumor type, for example, metastatic prostate cancer and hepatocellular carcinoma [49], [50], [51].
In our study, the restoration of miR-485–5p affected cell migration, invasion and proliferation in the BCPAP cell line. The expression of miR-485–5p in thyroid cancer cells have been investigated by others. The downregulation of this molecule has been observed in the anaplastic thyroid carcinoma cell lines CAL-62 and FRO in comparison with N-Thy-ORI cells, regulating cell growth and migration in the circ_0023990-miR-485–5p-FOXM1 axis [52]. Moreover, Li & Kong (2019) have shown the expression of this miRNA in TPC-1 and BCPAP cell lines, however, without comparison with normal cells [53].
Interestingly, we only observed a significant impact of miR-485–5p overexpression on cell migration and invasion in the BRAFT1799A positive cell line BCPAP. This incidental finding suggests that the oncogenic background may influence the miRNA-mediated post-transcriptional regulation. Although the role of BRAFT1799A mutation in PTC is well-documented[54], its relationship with the DLK1-DIO3 region is unclear. Our bioinformatics analyses show that miR-485–5p may target several genes involved in the transduction of MAPK and PI3K pathways, such as EREG, EGFR, MET, INSR, and GAB2. Li and colleagues have shown the direct targeting of GAB2 by miR-485–5p in colorectal cancer [40]. Indeed, apart from any other mutation in PTC, BRAFT1799A-positive tumors present a unique transcriptional profile and are often diagnosed as less differentiated and more aggressive carcinomas [5]. Indeed, the BCPAP cell line, commonly described as derived from papillary thyroid carcinoma, was derived from a PTC with local and lymph node metastasis, characteristics of a poorly differentiated thyroid carcinoma [34]. In this context, miR-485–5p could be participating in the repression of crucial BRAFT1799A partners and subsequently impacting cell migration and invasion. Given the role of GAB2 and RAC1 in the modulation of the cytoskeletal organization [55], [56], [57], [58], [59], [60], their downregulation could explain, at least in part, the suppression of the migratory potential in miR-485–5p-transfected BCPAP and not in TPC-1 cells. Additionally, different studies have shown the relevance of ICAM1 in aggressive thyroid cancer, which highlights it as a promising therapeutic target [61], [62], [63]. Therefore, the restoration of miR-485–5p levels emerges as a potential strategy for adjuvant therapy of BRAFT1799A-positive aggressive PTC.
In conclusion, the combination of computation analysis and data mining allowed us to shed light on the biological role of DLK1-DIO3-derived miRNAs in PTC. The major impact of DLK1-DIO3 miRNAs on thyroid oncogenesis and progression may derive from the top-12 miRNAs, which cooperate to modulate important cancer-related biological processes. Taken together, our results provide a new strategy to identify tumor suppressor candidates for PTC, opening prospects for a new approach to the treatment of aggressive thyroid cancer.
Funding
This work was supported by São Paulo Research Foundation (FAPESP, grants MVG: 2017/03635-8; LAM: 2016/09107-0 and LFA: 2017/21660-0), Coordination for the Improvement of Higher Education Personnel (CAPES, grant 88887.372858/2019-00) and Fund for Support to Teaching, Research and Outreach Activities – FAEPEX (3124/16, 2587/19 and 2607/21).
CRediT authorship contribution statement
Letícia Ferreira Alves: Conceptualization, Methodology, Software, Visualization, Writing – original draft, Writing – review & editing. Leonardo Augusto Marson: Conceptualization, Methodology, Investigation, Writing – original draft. Micheli Severo Sielski: Investigation, Writing – review & editing. Cristina Pontes Vicente: Conceptualization, Writing – review & editing. Edna Teruko Kimura: Conceptualization, Writing – review & editing. Murilo Vieira Geraldo: Conceptualization, Project administration, Supervision, Writing – review & editing.
Declaration of competing interest
All the following authors declare that there is no financial/personal interest or belief that could affect their objectivity
Acknowledgments
The authors thank Espaço da Escrita – Pró-Reitoria de Pesquisa – UNICAMP – for the language services provided.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2023.101849.
Appendix. Supplementary materials
References
- 1.Siegel R.L., Miller K.D., Wagle N.S., et al. Cancer statistics, 2023. CA Cancer J. Clin. 2023;73(1):17–48. doi: 10.3322/caac.21763. [DOI] [PubMed] [Google Scholar]
- 2.Fusco A., Grieco M., Santoro M., et al. A new oncogene in human thyroid papillary carcinomas and their lymph-nodal metastases. Nature. 1988;328(6126) doi: 10.1038/328170a0. [DOI] [PubMed] [Google Scholar]
- 3.Suarez H.G., Du Villard J.A., Caillou B., et al. Detection of activated ras oncogenes in human thyroid carcinomas. Oncogene. 1988;2(4) [PubMed] [Google Scholar]
- 4.Nikiforova M.N., Kimura E.T., Gandhi M., et al. BRAF mutations in thyroid tumors are restricted to papillary carcinomas and anaplastic or poorly differentiated carcinomas arising from papillary carcinomas. J. Clin. Endocrinol. Metab. 2003;88(11):5399–5404. doi: 10.1210/jc.2003-030838. [DOI] [PubMed] [Google Scholar]
- 5.Agrawal N., Akbani R., Aksoy B.A., et al. Integrated genomic characterization of papillary thyroid carcinoma. Cell. 2014 doi: 10.1016/j.cell.2014.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hall R.D., Kudchadkar R.R. Braf mutations: signaling, epidemiology, and clinical experience in multiple malignancies. Cancer Control. 2014;21(3) doi: 10.1177/107327481402100307. [DOI] [PubMed] [Google Scholar]
- 7.Bartel D.P. Metazoan MicroRNAs. Cell. 2018;173(1) doi: 10.1016/j.cell.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lakshmanan A., Wojcicka A., Kotlarek M., et al. microRNA-339-5p modulates Na+/I- symporter-mediated radioiodide uptake. Endocr. Relat. Cancer. 2015;22(1) doi: 10.1530/ERC-14-0439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li L., Lv B., Chen B., et al. Inhibition of miR-146b expression increases radioiodine-sensitivity in poorly differential thyroid carcinoma via positively regulating NIS expression. Biochem. Biophys. Res. Commun. 2015;462(4) doi: 10.1016/j.bbrc.2015.04.134. [DOI] [PubMed] [Google Scholar]
- 10.Riesco-Eizaguirre G., Wert-Lamas L., Perales-Paton J., et al. The miR-146b-3p/PAX8/NIS regulatory circuit modulates the differentiation phenotype and function of thyroid cells during carcinogenesis. Cancer Res. 2015;75(19) doi: 10.1158/0008-5472.CAN-14-3547. [DOI] [PubMed] [Google Scholar]
- 11.Fuziwara C.S., Kimura E.T. MicroRNAs in thyroid development, function and tumorigenesis. Mol. Cell Endocrinol. 2017:456. doi: 10.1016/j.mce.2016.12.017. [DOI] [PubMed] [Google Scholar]
- 12.Ulisse S., Baldini E., Lauro A., et al. Papillary thyroid cancer prognosis: an Evolving Field. Cancers (Basel) 2021;13(21) doi: 10.3390/cancers13215567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geraldo M.V., Nakaya H.I., Kimura E. Down-regulation of 14q32-encoded miRNAs and tumor suppressor role for miR-654-3p in papillary thyroid cancer. Oncotarget. 2017;8(6):9597–9607. doi: 10.18632/oncotarget.14162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glazov E.A., McWilliam S., Barris W.C., et al. Origin, evolution, and biological role of miRNA cluster in DLK-DIO3 genomic region in placental mammals. Mol. Biol. Evol. 2008;25(5):939–948. doi: 10.1093/molbev/msn045. [DOI] [PubMed] [Google Scholar]
- 15.Benetatos L., Hatzimichael E., Londin E., et al. The MicroRNAs within the DLK1-DIO3 genomic region: involvement in disease pathogenesis. Cell Mol. Life Sci. 2013;70(5) doi: 10.1007/s00018-012-1080-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.da Rocha ST, CA Edwards, Ito M., et al. Genomic imprinting at the mammalian Dlk1-Dio3 domain. Trends Genet. 2008;24(6):306–316. doi: 10.1016/j.tig.2008.03.011. [DOI] [PubMed] [Google Scholar]
- 17.Zhou Y., Zhang X., Klibanski A. MEG3 noncoding RNA: a tumor suppressor. J. Mol. Endocrinol. 2012;48(3) doi: 10.1530/JME-12-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Benetatos L., Vartholomatos G., Hatzimichael E. MEG3 imprinted gene contribution in tumorigenesis. Int. J. Cancer. 2011;129(4) doi: 10.1002/ijc.26052. [DOI] [PubMed] [Google Scholar]
- 19.Zhao G., Zhang L., Qian D., et al. MiR-495-3p inhibits the cell proliferation, invasion and migration of osteosarcoma by targeting C1q/TNF-related protein 3. Onco. Targets Ther. 2019:12. doi: 10.2147/OTT.S193937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao X.G., Hu J.Y., Tang J., et al. miR-665 expression predicts poor survival and promotes tumor metastasis by targeting NR4A3 in breast cancer. Cell Death Dis. 2019;10(7) doi: 10.1038/s41419-019-1705-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gong Y.B., Fan X.H. MiR-539-3p promotes the progression of epithelial ovarian cancer by targeting SPARCL1. Eur. Rev. Med. Pharmacol .Sci. 2019;23(6) doi: 10.26355/eurrev_201903_17381. [DOI] [PubMed] [Google Scholar]
- 22.Severi G., Bernardini L., Briuglia S., et al. New patients with Temple syndrome caused by 14q32 deletion: genotype-phenotype correlations and risk of thyroid cancer. Am. J. Med. Genet Part A. 2016;170(1):162–169. doi: 10.1002/ajmg.a.37346. [DOI] [PubMed] [Google Scholar]
- 23.Dweep H., Gretz N. MiRWalk2.0: a comprehensive atlas of MicroRNA-Target interactions. Nat. Methods. 2015;12(8) doi: 10.1038/nmeth.3485. [DOI] [PubMed] [Google Scholar]
- 24.Agarwal V., Bell G.W., Nam J.W., et al. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4(AUGUST2015) doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krek A., Grün D., Poy M.N., et al. Combinatorial microRNA target predictions. Nat. Genet. 2005;37(5) doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 26.Geraldo M.V., Kimura E.T. Integrated analysis of thyroid cancer public datasets reveals role of post-transcriptional regulation on tumor progression by targeting of immune system mediators. PLoS ONE. 2015;10(11) doi: 10.1371/journal.pone.0141726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li J.H., Liu S., Zhou H., et al. StarBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic. Acids Res. 2014;42(D1) doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang D.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1) doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 29.Heberle H., Meirelles V.G., da Silva F.R., et al. InteractiVenn: a web-based tool for the analysis of sets through Venn diagrams. BMC Bioinformatics. 2015;16(1) doi: 10.1186/s12859-015-0611-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shannon P., Markiel A., Ozier O., et al. Cytoscape: a software Environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11) doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Doncheva N.T., Morris J.H., Gorodkin J., et al. cytoscape stringapp: network analysis and visualization of proteomics data. J. Proteome Res. 2019;18(2):623–632. doi: 10.1021/acs.jproteome.8b00702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lemoine N.R., Mayall E.S., Jones T., et al. Characterisation of human thyroid epithelial cells immortalised in vitro by simian virus 40 DNA transfection. Br. J. Cancer. 1989;60(6) doi: 10.1038/bjc.1989.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tanaka J., Ogura T., Sato H., et al. Establishment and biological characterization of an in vitro human cytomegalovirus latency model. Virology. 1987;161(1) doi: 10.1016/0042-6822(87)90171-1. [DOI] [PubMed] [Google Scholar]
- 34.Fabien N., Fusco A., Santoro M., et al. Description of a human papillary thyroid carcinoma cell line. Morphologic study and expression of tumoral markers. Cancer. 1994;73(8) doi: 10.1002/1097-0142(19940415)73:8<2206::AID-CNCR2820730828>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 35.Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987;162(1):156–159. doi: 10.1016/0003-2697(87)90021-2. [DOI] [PubMed] [Google Scholar]
- 36.Pfaffl M.W. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic. Acids Res. 2001;29(9):e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alves L.F., Geraldo M.V. MiR-495-3p regulates cell migration and invasion in papillary thyroid carcinoma. Front. Oncol. 2023:13. doi: 10.3389/fonc.2023.1039654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abràmoff M.D., Magalhães P.J., Ram S.J. Image Processing with Image. J.. Biophotonics Int. 2004;11(7) doi: 10.1201/9781420005615.ax4. [DOI] [Google Scholar]
- 39.Eun J.W., Kim H.S., Shen Q., et al. MicroRNA-495-3p functions as a tumor suppressor by regulating multiple epigenetic modifiers in gastric carcinogenesis. J. Pathol. 2018;244(1) doi: 10.1002/path.4994. [DOI] [PubMed] [Google Scholar]
- 40.Li J., Xu J., Yan X., et al. MicroRNA-485 plays tumour-suppressive roles in colorectal cancer by directly targeting GAB2. Oncol. Rep. 2018;40(1) doi: 10.3892/or.2018.6449. [DOI] [PubMed] [Google Scholar]
- 41.Lou C., Xiao M., Cheng S., et al. MiR-485-3p and miR-485-5p suppress breast cancer cell metastasis by inhibiting PGC-1α expression. Cell Death Dis. 2016;7:e2159. doi: 10.1038/cddis.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu X.X., Xu X.N., He B.S., et al. microRNA-485-5p functions as a tumor suppressor in colorectal cancer cells by targeting CD147. J. Cancer. 2018;9(15) doi: 10.7150/jca.24918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang X., Ruan H., Hu X., et al. miR-381-3p suppresses the proliferation of oral squamous cell carcinoma cells by directly targeting FGFR2. Am. J. Cancer Res. 2017;7(4) [PMC free article] [PubMed] [Google Scholar]
- 44.Yang Y.B., Tan H., Wang Q. MiRNA-300 suppresses proliferation, migration and invasion of non-small cell lung cancer via targeting ETS1. Eur. Rev. Med. Pharmacol. Sci. 2019;23(24) doi: 10.26355/eurrev_201912_19786. [DOI] [PubMed] [Google Scholar]
- 45.Duan J., Zhang H., Li S., et al. The role of miR-485-5p/NUDT1 axis in gastric cancer. Cancer Cell Int. 2017;17(1) doi: 10.1186/s12935-017-0462-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang Z., Yuan Z., Fan Y., Deng X.Z.Q. Integrated analyses of microRNA and mRNA expression profiles in aggressive papillary thyroid carcinoma. Mol. Med. Rep. 2013 doi: 10.3892/mmr.2013.1699. [DOI] [PubMed] [Google Scholar]
- 47.Jang T.H., Huang W.C., Tung S.L., et al. MicroRNA-485-5p targets keratin 17 to regulate oral cancer stemness and chemoresistance via the integrin/FAK/Src/ERK/β-catenin pathway. J. Biomed. Sci. 2022;29(1) doi: 10.1186/s12929-022-00824-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Haga C.L., Phinney D.G. MicroRNAs in the imprinted DLK1-DIO3 region repress the epithelial-to-mesenchymal transition by targeting the TWIST1 protein signaling network. J. Biol. Chem. 2012;287(51) doi: 10.1074/jbc.M112.387761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Luk J.M., Burchard J., Zhang C., et al. DLK1-DIO3 genomic imprinted microRNA cluster at 14q32.2 defines a stemlike subtype of hepatocellular carcinoma associated with poor survival. J. Biol. Chem. 2011;286(35) doi: 10.1074/jbc.M111.229831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gururajan M., Josson S., Chu G.C.Y., et al. MiR-154∗ and miR-379 in the DLK1-DIO3 MicroRNA mega-cluster regulate epithelial to mesenchymal transition and bone metastasis of prostate cancer. Clin. Cancer Res. 2014;20(24) doi: 10.1158/1078-0432.CCR-14-1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jin A.L., Ding L., Yang W.J., et al. Exosomal microRNAs in the DLK1-DIO3 imprinted region derived from cancer-associated fibroblasts promote progression of hepatocellular carcinoma by targeting hedgehog interacting protein. BMC Gastroenterol. 2022;22(1) doi: 10.1186/s12876-022-02594-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang Q., Wu L., Liu S.Z., et al. Hsa_circ_0023990 Promotes Tumor Growth and Glycolysis in Dedifferentiated TC via Targeting miR-485-5p/FOXM1 Axis. Endocrinol. (United States) 2021;162(12) doi: 10.1210/endocr/bqab172. [DOI] [PubMed] [Google Scholar]
- 53.Li G., Kong Q. LncRNA LINC00460 promotes the papillary thyroid cancer progression by regulating the LINC00460/miR-485-5p/Raf1 axis. Biol. Res. 2019;52(1) doi: 10.1186/s40659-019-0269-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fagin J.A., Wells S.A. Biologic and clinical perspectives on thyroid cancer. N. Engl. J. Med. 2016;375(11):1054–1067. doi: 10.1056/NEJMra1501993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Abreu M.T.H., Hughes W.E., Mele K., et al. Gab2 regulates cytoskeletal organization and migration of mammary epithelial cells by modulating RhoA activation. Mol. Biol. Cell. 2011;22(1) doi: 10.1091/mbc.E10-03-0185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang Y., Sheng Q., Spillman M.A., et al. Gab2 regulates the migratory behaviors and E-cadherin expression via activation of the PI3K pathway in ovarian cancer cells. Oncogene. 2012;31(20) doi: 10.1038/onc.2011.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ding C., Luo J., Li L., et al. Gab2 facilitates epithelial-to-mesenchymal transition via the MEK/ERK/MMP signaling in colorectal cancer. J. Exp. Clin. Cancer Res. 2016;35(1) doi: 10.1186/s13046-015-0280-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ding C.B., Yu W.N., Feng J.H., et al. Structure and function of gab2 and its role in cancer (Review) Mol. Med. Rep. 2015;12(3) doi: 10.3892/mmr.2015.3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Katoh H., Hiramoto K., Negishi M. Activation of Rac1 by RhoG regulates cell migration. J. Cell Sci. 2006;119(1) doi: 10.1242/jcs.02720. [DOI] [PubMed] [Google Scholar]
- 60.Liang J., Oyang L., Rao S., et al. Rac1, a potential target for tumor therapy. Front. Oncol. 2021:11. doi: 10.3389/fonc.2021.674426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Buitrago D., Keutgen X.M., Crowley M., et al. Intercellular adhesion molecule-1 (ICAM-1) is upregulated in aggressive papillary thyroid carcinoma. Ann. Surg. Oncol. 2012;19(3) doi: 10.1245/s10434-011-2029-0. [DOI] [PubMed] [Google Scholar]
- 62.Min I.M., Shevlin E., Vedvyas Y., et al. CAR T therapy targeting ICAM-1 eliminates advanced human thyroid tumors. Clin. Cancer Res. 2017;23(24) doi: 10.1158/1078-0432.CCR-17-2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gray K.D., McCloskey J.E., Vedvyas Y., et al. PD1 blockade enhances ICAM1-directed CAR T therapeutic efficacy in advanced thyroid cancer. Clin. Cancer Res. 2020;26(22) doi: 10.1158/1078-0432.CCR-20-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.