Summary
Adult humans cannot regenerate the enamel-forming cell type, ameloblasts. Hence, human induced pluripotent stem cell (hiPSC)-derived ameloblasts are valuable for investigating tooth development and regeneration. Here, we present a protocol for generating three-dimensional induced early ameloblasts (ieAMs) utilizing serum-free media and growth factors. We describe steps for directing hiPSCs toward oral epithelium and then toward ameloblast fate. These cells can form suspended early ameloblast organoids. This approach is critical for understanding, treating, and promoting regeneration in diseases like amelogenesis imperfecta.
For complete details on the use and execution of this protocol, please refer to Alghadeer et al.1
Subject areas: Developmental biology, Stem Cells, Cell Differentiation, Organoids
Graphical abstract
Highlights
-
•
Generate hiPSC-derived ameloblasts
-
•
Step-by-step hiPSCs to oral epithelium to early ameloblast differentiation
-
•
Cultivate serum-free 3D ameloblast organoids
-
•
Gain insight into future enamel regeneration and disease
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Adult humans cannot regenerate the enamel-forming cell type, ameloblasts. Hence, human induced pluripotent stem cell (hiPSC)-derived ameloblasts are valuable for investigating tooth development and regeneration. Here, we present a protocol for generating three-dimensional induced early ameloblasts (ieAMs) utilizing serum-free media and growth factors. We describe steps for directing hiPSCs toward oral epithelium and then toward ameloblast fate. These cells can form suspended early ameloblast organoids. This approach is critical for understanding, treating, and promoting regeneration in diseases like amelogenesis imperfecta.
Before you begin
The following protocol outlines the precise procedures for using human induced pluripotent stem cells (hiPSCs) to produce early ameloblasts. In this procedure, hiPSCs and their derivatives are kept in a humidified incubator at 37°C and 5% CO2. Before commencing the differentiation, preparing cell culture medium and matrix-coated culture plates is necessary. It is recommended that small molecules be made as stock aliquots and that required quantities of them be freshly introduced to the basal media shortly before usage. It is recommended to preheat the culture media to a temperature of 37°C before use. All cell culture work should be done aseptically in a Class II biosafety cabinet.
Institutional permissions
Investigators must obtain an approved Biological Use Authorization (BUA) from their Institutional Biosafety Committee (IBC) before beginning this protocol. Experiments on pluripotent stem cells need to be performed in accordance with relevant institutional and national guidelines. Depending on the institution where this protocol is to be executed, it may be necessary to obtain prior approval for the utilization of hiPSCs, such as from a Stem Cell Research Oversight committee.
Preparation of matrigel-coated plates
Timing: 30 min
-
1.
Thaw a bottle of growth factor-reduced Corning Matrigel matrix (Matrigel) for 12–16 h at 4°C.
-
2.
Set out 10 cm and 35 mm tissue culture dishes, and 12-well or 24-well cell culture plates in a biosafety cabinet.
-
3.
Place a 500 mL bottle of sterile cold (4°C) 1X phosphate buffered saline (PBS) in biosafety cabinet.
-
4.
Prepare two Matrigel dilutions in cold 1X PBS, 1:100 for 10 cm and 35 mm tissue culture dishes to be used for maintaining hiPSCs, and 1:20 for the 12-well cell culture plates to be used for differentiation.
-
5.
To coat the plate, add 1 mL of Matrigel diluted solution into the 35 mm tissue culture dishes, and add 0.5 mL of Matrigel diluted solution into each well of the 12-well cell culture plates. Add 5 mL to the 10 cm plates.
-
6.
Wrap the Matrigel-coated plates with Parafilm and store them at 4°C to prevent evaporation for up to two weeks.
-
7.
If the coated plates need to be used immediately, incubate at 20°C–24°C for at least 1 h.
Preparation of cell culture medium
Timing: 30 min
-
8.
Prepare Wash media.
-
9.
Prepare Base iAM media.
Note: Both media should be stored at 4°C for a maximum of 4 weeks.
Culture of human induced pluripotent stem cells (hiPSCs)
Timing: 30 min
Revival of induced pluripotent stem cells
-
10.
To thaw hiPSCs, prepare 5 mL of wash media for each straw to be thawed in a 15 mL centrifuge tube, and warm to 37°C .
-
11.
Get stored frozen straws with hiPSCs (WTC-11 human induced pluripotent stem cells) (Coriell, #GM25256) from liquid nitrogen tank and thaw in a water container at 37°C for 5 s.
-
12.
Quickly disinfect the straws and transfer them inside the biosafety cabinet.
-
13.
Hold the straws vertically (cotton side up) and using a sterile pair of scissors cut the lower end of the straw.
-
14.
Then insert the exposed end of the straw inside a prepared 15 mL centrifuge tube containing wash media, then cut the top end of the straw to allow the thawed cells to flow inside the 15 mL tube.
-
15.
Close the caps and mix by inverting the tube gently. Avoid breaking them into single cells.
-
16.
Spin at 300 × g for 3 min.
-
17.
Discard the supernatant.
-
18.
Aspirate and discard all excess liquid in the precoated Matrigel plates.
-
19.
Resuspend and plate the hiPSC cells on a 10 cm tissue culture dishes in the fresh 10 mL mTeSR media (STEMCELL Technologies) containing 10 μM of Rock inhibitor (Ri) (Y-27632, Selleckchem, #S1049), and incubate at 37°C and 5% CO2.
-
20.
Change the media on the next day with fresh mTeSR media without Ri and observe the number of cells successfully plated.
-
21.
Continue changing media every 24 h and observe the morphology of the hiPSC colonies (Figure 1).
Note: hiPSC colonies will initially have rough edges in the presence of Ri. After changing the media to a fresh mTeSR media without Ri, the colonies will have smooth edges as they become more mature in 2–3 days.
-
22.
When the cells reach 70% confluency and are ready for passaging, warm up Versene (Thermo Fisher Scientific, #15040066) in a water bath at 37°C.
-
23.
Aspirate the old media and wash with sterile 1X PBS at 20°C–24°C.
-
24.
Aspirate the 1X PBS and add 3 mL of the preheated Versene, and incubate at 37°C for 5 min.
-
25.
Gently aspirate the excess Versene. Add fresh mTESR media supplemented with Ri, then resuspend the cells by gently pipetting and flushing all areas of the plate with the media.
-
26.
Plate the cells in a new Matrigel coated plate at a ratio of 1:7 or 50,000 cells per well.
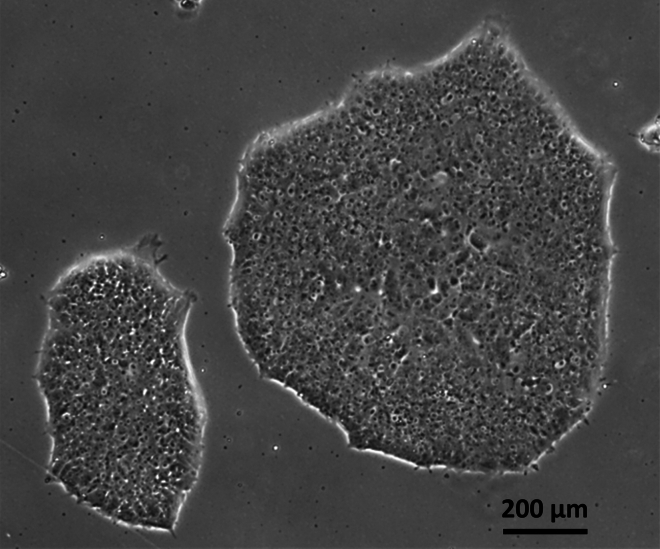
Figure 1.
Bright-field image of hiPSC colonies
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies (suggested dilution/concentration) | ||
| AMBN (1:50) | Santa Cruz | sc-271012; RRID: AB_10613795 |
| DSPP (1:50) | Santa Cruz | sc-73632; RRID: AB_2230660 |
| KRT14 (1:100) | Thermo Fisher Scientific | LL002; RRID: AB_306091 |
| KRT5 (1:100) | Sigma-Aldrich | HPA059479; RRID: AB_2684034 |
| ZO-1 (1:200) | Thermo Fisher Scientific | 33-9100; RRID: AB_87181 |
| ZO-1 (1:200) | Thermo Fisher Scientific | 40-2200; RRID: AB_2533456 |
| SP6 (1:200) | Atlas Antibodies | HPA024516; RRID: AB_10960551 |
| DAPI (1 μg/mL) | Thermo Fisher Scientific | D1306, RRID: AB_2629482 |
| Goat anti-mouse IgG, Alexa Fluor 488 (1:400) | Thermo Fisher Scientific | A11001; RRID: AB_2534069 |
| Goat anti-rabbit IgG, Alexa Fluor 488 (1:400) | Thermo Fisher Scientific | A32731; RRID: AB_2633280 |
| Goat anti-mouse IgG, Alexa Fluor 568 (1:400) | Thermo Fisher Scientific | A11004; RRID: AB_2534072 |
| Goat anti-rabbit IgG, Alexa Fluor 568 (1:400) | Thermo Fisher Scientific | A11036, RRID: AB_10563566 |
| Goat anti-mouse IgG, Alexa Fluor 647 (1:400) | Thermo Fisher Scientific | A32728; RRID: AB_2633277 |
| Goat anti-rabbit IgG, Alexa Fluor 647 (1:400) | Thermo Fisher Scientific | A32733; RRID: AB_2633282 |
| Alexa Fluor 488 Phalloidin (1:500) | Thermo Fisher Scientific | A12379; RRID: AB_3096417 |
| Alexa Fluor 568 Phalloidin (1:500) | Thermo Fisher Scientific | A12380; RRID: AB_3096418 |
| Chemicals, peptides, and recombinant proteins | ||
| Matrigel matrix | Corning | #356231 |
| β-Mercaptoethanol | Sigma | #M7522 |
| Smoothened agonist (SAG) | Selleckchem | #S7779 |
| Bone morphogenic protein-4 (BMP4) | R&D Systems | #314-BP-010 |
| BMP inhibitor (LDN-193189) | Cayman Chemical | #11802 |
| GSK3 inhibitor (CHIR99021) | Cayman Chemical | #13122 |
| Epidermal growth factor (EGF) | R&D Systems | #236-EG |
| N-2 supplement (100X) | Gibco | #17502048 |
| Neurotrophin-4 (NT4) | R&D Systems | #268-N4 |
| Transforming growth factor beta 1(TGFβ1) | R&D Systems | #7754-BH |
| ROCKi (Y-27632) | Selleckchem | #S1049 |
| Knockout serum replacer | Thermo Fisher Scientific | #10828-028 |
| Nonessential amino acids (NEAA) | Gibco | #11140050 |
| Hydrocortisone stock solution (96 μg/mL) | STEMCELL Technologies | #07925 |
| GlutaMAX | Gibco | #35050038 |
| Sodium pyruvate | Gibco | #11360070 |
| Penicillin-Streptomycin | Gibco | #15140122 |
| Paraformaldehyde | EMS | #15710 |
| EpiCult-C Human medium kit | STEMCELL Technologies | #05630 |
| mTeSR1 stem cell medium | STEMCELL Technologies | #85850 |
| DMEM/F-12 | Thermo Fisher Scientific | #11320074 |
| Versene | Thermo Fisher Scientific | #15040066 |
| TrypLE Express | Gibco | #12604013 |
| Experimental models: Cell lines | ||
| WTC-11 human induced pluripotent stem cells | Coriell | #GM25256 |
| Software and algorithms | ||
| Fiji (ImageJ2 v2.14.0) | Schindelin et al., 20122; Schindelin et al., 20153 | https://imagej.net/software/fiji/ |
| BioRender | BioRender | https://www.biorender.com/ |
| NIS-Elements | Nikon | RRID: SCR_014329 |
Materials and equipment
Wash media
| Reagent | Final concentration | Amount |
|---|---|---|
| DMEM/F-12 (Gibco) | N/A | 38.5 mL |
| Knock-out serum replacer (Fisher Scientific) | 20% | 10 mL |
| 100X Nonessential amino acids (NEAA) (Gibco) | 1X | 500 μL |
| 100 mM sodium pyruvate (Gibco) | 1 mM | 500 μL |
| 100X Penicillin-Streptomycin (Gibco) | 1X | 500 μL |
| Total | N/A | 50 mL |
Base iAM media
| Reagent | Final concentration | Amount |
|---|---|---|
| EpiCult-C Basal Medium (STEMCELL Technologies) | N/A | 48.5 mL |
| 100X EpiCult-C Proliferation Supplement (STEMCELL Technologies) | 1X | 500 μL |
| 100X Penicillin-Streptomycin (Gibco) | 1X | 500 μL |
| 200X Hydrocortisone Stock Solution (96 μg/mL) | 1X (0.48 μg/mL) | 250 μL |
| 100X GlutaMAX (Gibco) | 0.05X | 25 μL |
| 100X NEAA (Gibco) | 0.4X | 200 μL |
| β-mercaptoethanol 0.1 M (Sigma-Aldrich) | 80 μM | 40 μL |
| Total | N/A | 50 mL |
Note: Both the wash media and the base iAM media should be stored at 4°C for a maximum of 4 weeks.
Step-by-step method details
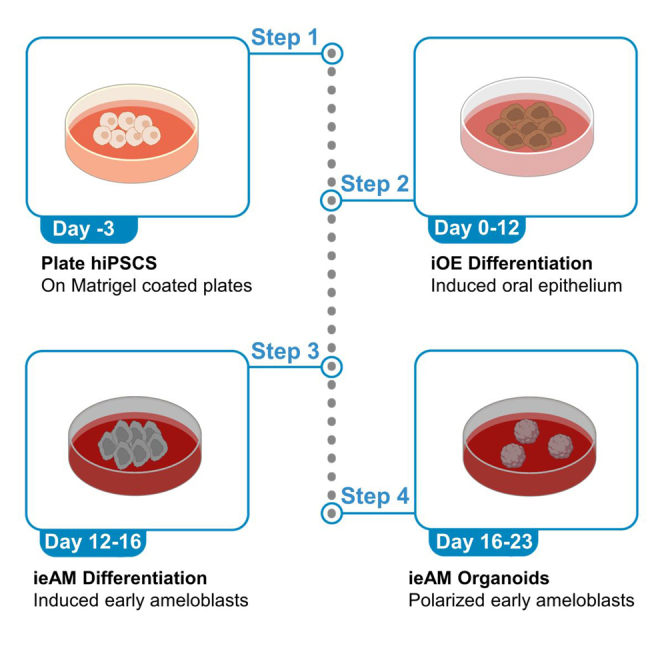
Plating and growing hiPSCs
Timing: 3 days
This step prepares the cells for the initiation of the differentiation.
-
1.
Passage and plate the hiPSCs (Figure 1) (50,000 cells per well) into a 12-well plate in fresh mTeSR media containing 10 μM of Ri at 37°C and 5% CO2.
-
2.
The day of plating the hiPSCs is deemed as day −3.
-
3.
Change the media on day −2 with fresh mTeSR media without Ri and initiate the differentiation after the cells reach 70%–80% confluency on day −1.
Differentiation into oral epithelium
Timing: 10 days
This step generates oral epithelium cells positive for specific markers (PITX1, PITX2, TBX1, KRT14, TP63) that can be used not only to generate ameloblasts but also to generate salivary gland cells (Figure 2).
-
4.Induction at day 0.
-
a.Aspirate the mTeSR media from the 12-well plate.
-
b.Replace the media with base iAM media with no growth factors for at least 6 h to acclimate the cells to the new media.
-
c.Replace the media with base media containing 400 nM of Smoothened agonist (SAG; Selleckchem, S7779) for two days.
-
a.
-
5.On day 3, replace the media with fresh base media containing 400 nM of SAG and 150 pM of monomeric Bone morphogenic protein-4 (BMP4; R&D Systems, #314-BP).
-
a.Refresh the media every day for four more days with base media containing 400 nM of SAG and 150 pM of BMP4.
-
a.
-
6.On day 8, replace the media with fresh base media containing 400 nM of SAG, 5 μM of GSK3-Inhibitor (GSK-I) (CHIR99021) (Cayman Chemical, #13122), 500 pM of monomeric Epidermal growth factor (EGF) (R&D Systems, #236-EG), 1 μM of BMP inhibitor (LDN-193189) (Cayman Chemical, #11802) and 70 pM of monomeric Neurotrophin-4 (NT4) (R&D Systems, #268-N4).
-
a.No media refresh is needed on day 9.
-
a.
-
7.
On day 10, three wells should be harvested for analysis of oral epithelial markers, and the rest can be differentiated into preameloblasts/early ameloblasts.
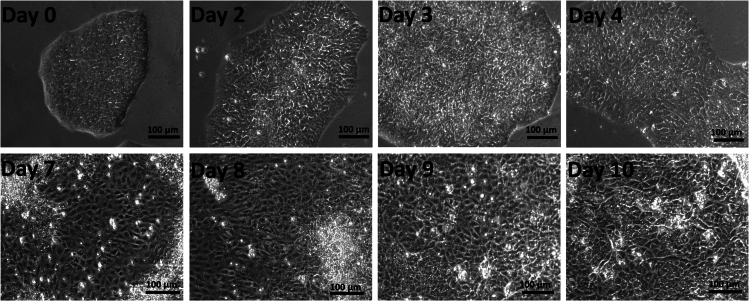
Figure 2.
Bright-field images of day 0 to day 10 iOE of in vitro differentiation
A change in morphology is seen, starting from the periphery moving inwards. The cells’ shape changes from circular to polygonal and they increase in size. The nuclear-cytoplasmic ratio decreases, and the nuclei become darker. The cells become flatter, spread out, and assume a cobblestone morphology by day 10.
Differentiation of oral epithelium into pre-ameloblast/early ameloblasts
Timing: 6 days
This step generates preameloblasts and early ameloblasts (Figure 3) positive for specific markers (AMBN, SP6) that can be used to generate early ameloblast organoids.
-
8.
On day 0 (or day 10 of oral epithelium differentiation), we continue with the same factors used at day 8.
-
9.
On day 2 (or day 12 of oral epithelium differentiation), add fresh base media containing 400 nM of SAG, 5 μM of GSK-I, 500 pM of EGF, 300 pM of BMP4 and 2 nM of TGF-β1 (R&D Systems, #7754-BH).
-
10.
Replace with fresh base media containing the same growth factors (mentioned above) every other day.
-
11.
On day 6 (or continuous day 16), three wells should be harvested for analysis of preameloblasts/early ameloblasts, and the rest can be used to generate early ameloblasts (ieAM) organoids.
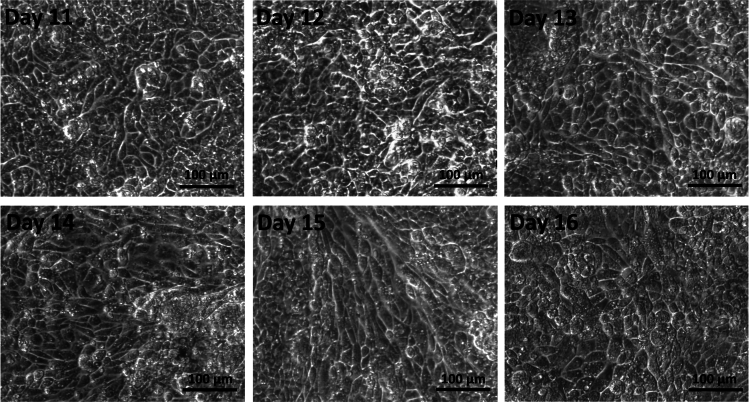
Figure 3.
Bright-field images of day 11 to day 16 of iAM in vitro differentiation
Here, the cells continue to change in morphology. The cells become elongated. The nuclei contract in size and become denser. Cytoplasmic granules become evident.
Early ameloblasts (ieAM) organoid
Timing: 7 days
This step generates polarized early ameloblast organoids in suspension that can be used to study early ameloblasts in vitro (Figure 4).
-
12.Detach the cells.
-
a.Aspirate the old media.
-
b.Wash the cells with sterile 1X PBS.
-
c.Warm an aliquot of TrypLE Express (TrypLE) (Gibco, #12604013) at 37°C.
-
d.Incubate the early ameloblast (day 16 ieAM) cells in TrypLE (500 μL per well) for 5–7 min at 37°C.
-
e.Gently aspirate the TrypLE. Add 1 mL of fresh iAM organoid media (iAM base media with 1X N-2 supplement (Gibco, #17502048) + 10 μM Ri) to each well. Resuspend the cells by gently pipetting and flushing all areas of the plate with media.
-
a.
Note: It is important to keep the cells in clumps.
-
13.
Transfer the cells into 24-well ultra-low attachment plates. Split 1 well of a 12-well plate of day 16 ieAM into 2 wells of a 24-well plate, 500 μL volume each.
-
14.
Every other day: Move the organoids to a 1.5 mL tube (Eppendorf) and let the organoids settle to the bottom of the tube. Aspirate ⅓ of the old media and add fresh media (Figure 4A).
-
15.
On day 7, the organoids can be fixed in 4% paraformaldehyde and prepared for staining and confocal microscopy (Methods video S1).
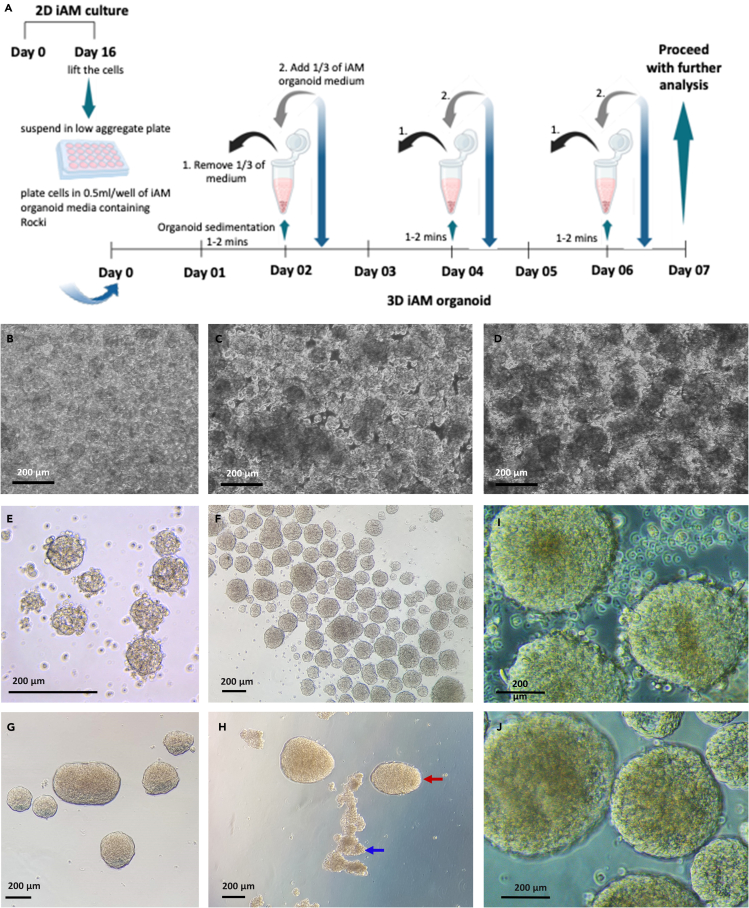
Figure 4.
Formation of ieAM organoids
(A) A schematic of the culture procedure for organoid formation and maintenance.
(B‒D) Bright-field images of ieAM organoids on day 0 to day 2.
(E‒G) Bright-field image of ieAM organoids on day 2 after washing (E), day 4 (F), and day 7 (G).
(H) Bright-field image showing examples of healthy organoids (red arrows), and irregular organoids (blue arrows).
(I and J) Bright-field images of day 7 ieAM organoids before wash (I) and after wash (J).
Polarized early ameloblasts can be seen at different z stacks as indicated by the presence of multiple apically generated lumens (arrowheads). Phalloidin (green), DAPI (blue).
Expected outcomes
Markers of oral epithelium
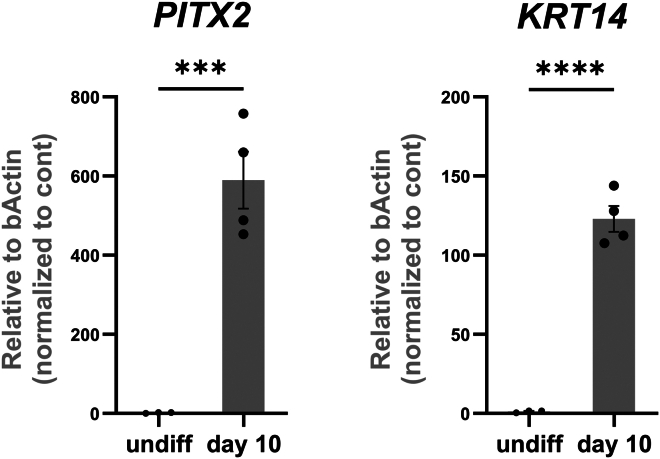
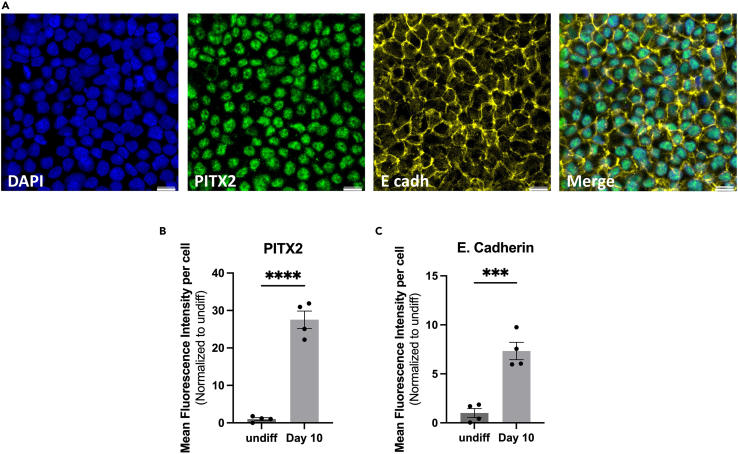
To assess the success of the differentiation into oral epithelium at day 10 of the differentiation, certain criteria must be achieved. First, epithelial cells must have a cobblestone morphology when viewed under a light microscope (Figure 2). Second, the expression of E-cadherin (E-CADH) and PITX2 should be validated with immunostaining, and PITX1, PITX2, TBX1, and KRT14 with qRT-PCR (Figures 5 and 6). Around 94% of the cells should successfully express PITX2.
List of qRT-PCR primers
| Primer name | Sequence |
|---|---|
| PITX1_F | TCCACCAAGAGCTTCACCTT |
| PITX1_R | CGGTGAGGTTGTTGATGTTG |
| PITX2_F | GTGTGGACCAACCTTACGGAAG |
| PITX2_R | CGAAGCCATTCTTGCATAGCTCG |
| KRT14_F | CATGAGTGTGGAAGCCGACAT |
| KRT14_R | GCCTCTCAGGGCATTCATCTC |
| bActin_F | TCCCTGGAGAAGAGCTACG |
| bActin_R | GTAGTTTCGTGGATGCCACA |
| TP63_F | TTCTTAGCGAGGTTGGGCTG |
| TP63_R | GATCGCATGTCGAAATTGCTC |
| TBX1_F | CGCAGTGGATGAAGCAAATCGTG |
| TBX1_R | TTTGCGTGGGTCCACATAGACC |
| AMBN_F | TTGAGCCTTGAGACAATGAGAC |
| AMBN_R | AGACCGTGCATCCACAAAGAA |
| SP6_F | AGCCTCTCCAAACTTACCAGGG |
| SP6_R | TCATAGCCCTGCGAGAAGTCCA |
Figure 5.
qRT-PCR of oral epithelium markers
The cells at Day 10 of differentiation (iOE) show upregulated expression of oral epithelium markers PITX2 and KRT14 as assessed by qRT-PCR. ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001; Graph error bars are the means ± SEM.
Figure 6.
Immunofluorescence staining of day 10 iOE
(A) The staining shows PITX2-positive nuclei (green) and E-cadherin-positive (yellow) membranes. Scale bars: 50 μm.
(B and C) Quantification of the mean fluorescence intensity of day 10 iOE markers PITX2 (B) and E-cadherin (C) in comparison to undifferentiated hiPSCs. Quantification was done using Image J (Image J2 v2.14.0). For PITX2, nuclear signal intensity was quantified and normalized to DAPI signal intensity per cell. For E-cadherin, the overall signal intensity per field was normalized to DAPI overall signal intensity per field (n = 4). 329 cells out of 350 cells showed PITX2+ expression, hence 94% of cells successfully expressed PITX2. Graph error bars are the means ± SEM. Statistical significance was determined using student T-test; ∗∗∗p ≤ 0.001; ∗∗∗∗p ≤ 0.0001.
Markers of early ameloblasts
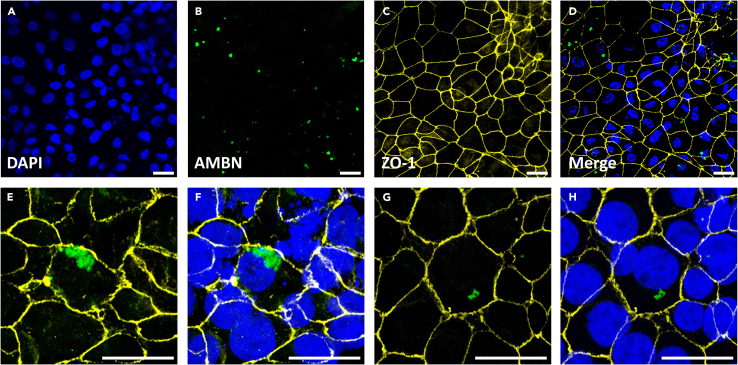
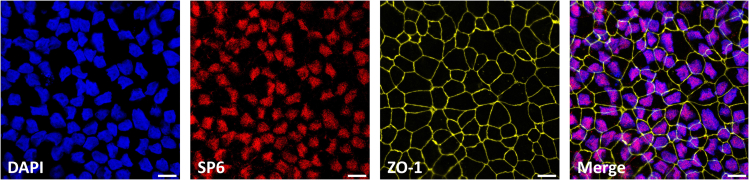
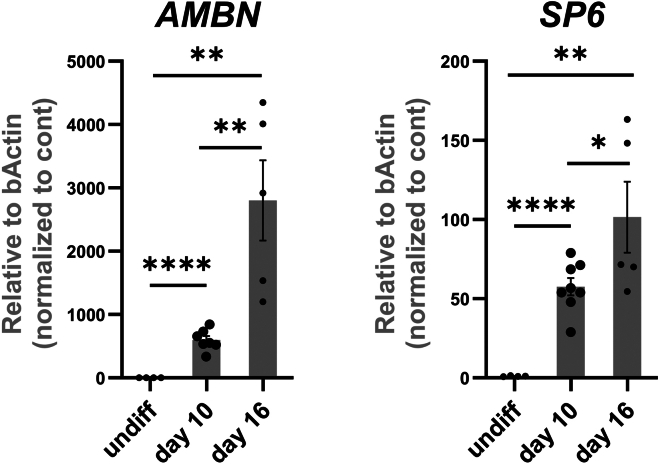
To assess the success of the differentiation into early ameloblasts at day 16 of the differentiation, the cells must express AMBN and SP6 which could be assessed by Western blotting, qRT-PCR1 or Immunofluorescence staining (Figures 7, 8, and 9). Around 88% of the cells should express SP6 and approximately 30%–50% of cells should successfully express AMBN.
Figure 7.
Immunofluorescence staining of AMBN at day 16 ieAM
(A‒H) The staining shows AMBN-positive cells (green) and the tight junction marker ZO-1 (yellow) at low power magnification (A‒D), and high-power magnification (E‒F) and (G‒H). Scale bar: 20 μm.
Figure 8.
Immunofluorescence staining of SP6 at day 16 ieAM
The staining shows SP6-positive nuclei (red) and the tight junction marker ZO-1 (yellow). Scale bar: 20 μm.
Figure 9.
QRT-PCR of early ameloblasts markers
The cells at Day 16 of differentiation (iAM) show upregulated expression of early ameloblast markers AMBN and SP6 as assessed by QRT-PCR. ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001; Graph error bars are the means ± SEM.
Polarized early ameloblasts
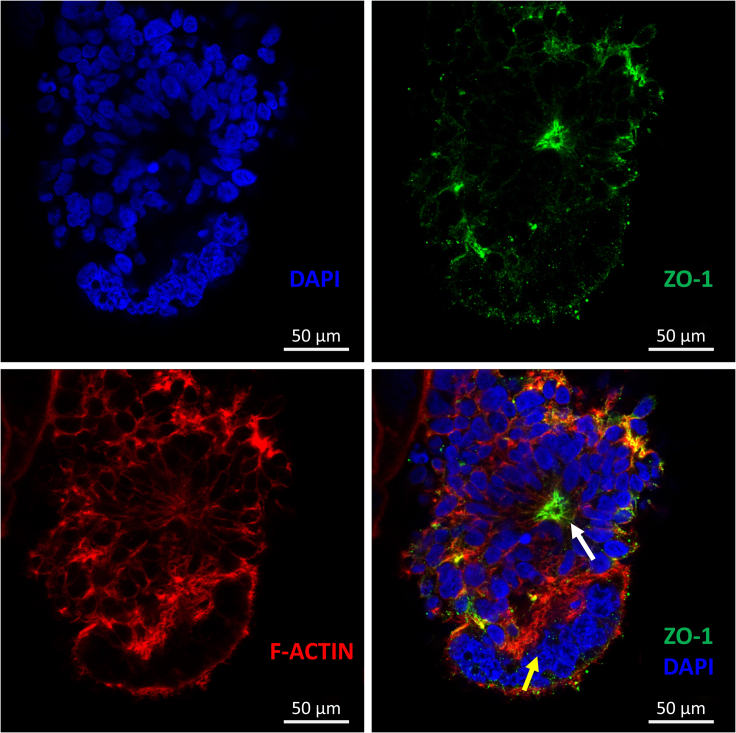
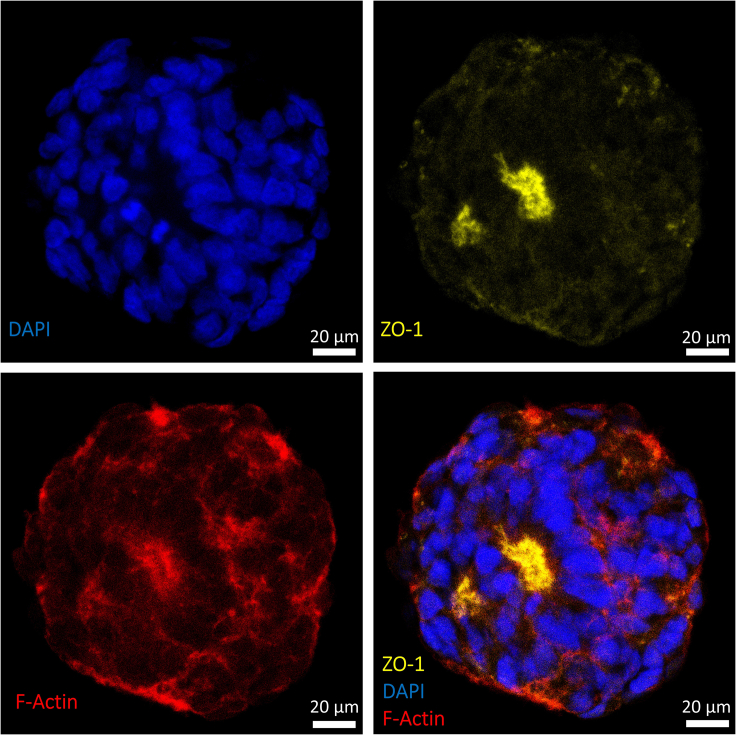
After the formation of the ieAM organoid in suspension, the morphology should be assessed. Stable ieAM organoids should have smooth borders and dark centers. Unhealthy organoids will have blurry and ill-defined boundaries. Polarized cells should be confirmed by immunofluorescence staining and confocal microscopy, where the luminal/apical side will densely stain with ZO-1 (Figures 10 and 11, Methods video S1).
Figure 10.
Immunofluorescence staining of medium-sized day 7 ieAM organoid showing polarized early ameloblasts
Apical sides of these polarized epithelial cells (marked by ZO-1, green) produce a lumen. The lumen is indicated by a white arrow, and the dying cells are indicated by a yellow arrow.
Figure 11.
Immunofluorescence staining of smaller-sized day 7 ieAM organoid showing polarized early ameloblasts toward a lumen marked by high expression of ZO-1 (yellow)
Limitations
This protocol has been tested with the WTC-11 hiPSCs cell line (Coriell, #GM25256) and may require further optimization for the application to other PSC lines. While designed for the generation of early ameloblasts, it is important to note that, for the production of secretory ameloblasts, co-culturing the organoid with dental pulp stem cells (DPSCs) is essential.1 We are still working to optimize this co-culturing process in future publications.
Troubleshooting
Problem 1
In step 1 of plating and growing hiPSCs, cells sometimes do not grow evenly across the plate.
Potential solution
-
•
Confirm the expiry date of Matrigel. Dilute Matrigel to achieve a final concentration of 5 μg/mL (0.5 μg/cm2, 1:20 dilution). Ensure uniform coverage of the plate wells and incubate for a minimum of 1 h at 20°C–24°C.
-
•
To ensure that the plate surface is evenly coated with cold Matrigel, after adding Matrigel solution, always keep the plate horizontal during transfer and storage.
-
•
To ensure that cells are plated evenly, before plating, transfer the cell suspension to a 15 mL tube and gently invert it to mix the cells evenly. Use a pipette to transfer the correct volume of cell suspension to several areas of each well and by gently moving plates backward and forward, then right to left to right, repeat same motions x 5 to distribute the cells evenly across the plate.
Problem 2
In Step 5 of the oral epithelium differentiation, the differentiation efficiency is lower than expected.
Potential solution
-
•
Make fresh media with small molecules and reagents and initiate the differentiation again.
Problem 3
In step 10 of the differentiation into ameloblasts stage, cells may undergo apoptosis due to lack of nutrients.
Potential solution
-
•
Perform a cell count before plating hiPSCs on the differentiation plate. Do not exceed plating 50,000 cells per well.
-
•
Ensure that media change takes place every other day.
-
•
Use 1 mL media for each well of the 12-well plate.
Problem 4
In Step 14 of the early ameloblast organoid formation process, irregular organoids lacking cell polarity frequently emerge. Optimizing the protocol is recommended to minimize the occurrence of these aberrant or unhealthy organoids.
Potential solution
-
•
Optimize the timing for detaching the cells with TrypLE. Never exceed 10 min.
-
•
Avoid vigorously pipetting of the cells while mixing and plating.
-
•
Ensure that 10 μM Ri is added while plating the cells into the suspension plate.
-
•
Evenly distribute the cells by gently moving plates backward and forward, then right to left to right.
-
•
Do not disturb the plate for the next 24 h after plating.
-
•
Keeping the cell in clumps increases the number of healthy organoids.
Problem 5
Differentiation efficiency is lower than expected.
Potential solution
-
•
Check that all reagents are correctly prepared and stored.
-
•
Confirm that the cells are not over-confluent or stressed, as this can impact their ability to differentiate efficiently.
-
•
Review the differentiation protocol to ensure all steps are being followed correctly and in the proper order. Assess the timing and duration of the differentiation process. Ensure that cells are exposed to differentiation-inducing conditions for the appropriate length of time.
-
•
Optimize the timing of media changes and treatments during the differentiation process.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Hannele Ruohola-Baker (hannele@uw.edu).
Technical contact
Technical questions regarding the protocol should be directed to the technical contact, Ammar Alghadeer (aaalghadeer@iau.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
This protocol does not include any datasets.
Acknowledgments
This work was supported by grants from the National Institutes of Health 1P01GM081619, R01GM097372, R01GM97372-03S1, and R01GM083867 and the NHLBI Progenitor Cell Biology Consortium (U01HL099997; UO1HL099993) for H.R.-B. and R01DE033016 for J.M. and H.R.-B. This work was supported by the Brotman Baty Institute (BBI) grants for J.M. and H.R.-B. This work was also supported by Dr. Douglass L. Mourell Research Fund for H.R.-B. and A.A. The Birth Defects Research Laboratory (BDRL) was supported by NIH award number 5R24HD000836 from the Eunice Kennedy Shriver National Institute of Child Health and Human Development. A.A. was supported by Imam Abdulrahman bin Faisal University and Saudi Arabian Cultural Mission (SACM) to the USA. We thank the Ruohola-Baker lab members for their helpful discussions. We thank Chris Cavanaugh, Fatima Al-Shimmary, Christopher Kelley, Aaron Liu, Infencia Xavier, Dibyo Maiti, Mickey Kim, Gwen L. Tilmes, Yan Ting Zhao, Sesha Hanson-Drury, and Anoushka Amath for their technical assistance. We are grateful to Profs. Carol Ware and Irma Thesleff for their inspiring discussions and guidance. We acknowledge Kimberly A. Aldinger, Ian G. Phelps, Jennifer C. Dempsey, Kevin Lee, and Lucy Cort from BDRL for their support in sample processing and troubleshooting nuclei isolation. We appreciate the technical support and guidance provided by the BBI in sequencing project design.
Author contributions
Conceptualization, A.A., J.M., and H.R.-B.; methodology, A.A., A.P.P., Y.C.L., Z.L., J.M., and H.R.-B.; investigation, A.A., A.P.P., Y.C.L., and Z.L.; funding acquisition, H.R.-B. and J.M.; resources, H.R.-B.; supervision, H.R.-B. and J.M.; visualization, A.A., Z.L., A.P.P., and Y.C.L.; formal analysis, A.A., A.P.P., and Y.C.L.; data curation, A.A., A.P.P., and Y.C.L.; project administration, H.R.-B.; validation, A.A. and A.P.P.; writing – original draft, A.A., A.P.P., J.M., and H.R.-B.; writing – review and editing, A.A., A.P.P., Z.L., Y.C.L., J.M., and H.R.-B.
Declaration of interests
A.A., A.P.P., J.M., and H.R.-B. are co-inventors on a patent application entitled “A Method to Direct the Differentiation of Human Induced Pluripotent Stem Cells into Early Ameloblasts” (PCT/US2022/053517 filed 12/20/2022).
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xpro.2024.103100.
References
- 1.Alghadeer A., Hanson-Drury S., Patni A.P., Ehnes D.D., Zhao Y.T., Li Z., Phal A., Vincent T., Lim Y.C., O’Day D., et al. Single-cell census of human tooth development enables generation of human enamel. Dev. Cell. 2023;58:2163–2180.e9. doi: 10.1016/j.devcel.2023.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schindelin J., Rueden C.T., Hiner M.C., Eliceiri K.W. The ImageJ ecosystem: An open platform for biomedical image analysis. Mol. Reprod. Dev. 2015;82:518–529. doi: 10.1002/mrd.22489. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Polarized early ameloblasts can be seen at different z stacks as indicated by the presence of multiple apically generated lumens (arrowheads). Phalloidin (green), DAPI (blue).
Data Availability Statement
This protocol does not include any datasets.