Abstract
Background & Aims
Irritable bowel syndrome (IBS) shows genetic predisposition, and large-scale genome-wide association studies (GWAS) are emerging, based on heterogeneous disease definitions. We investigated the genetic architecture of IBS defined according to gold standard Rome Criteria.
Methods
We conducted GWAS meta-analyses of Rome III IBS and its subtypes in 24,735 IBS cases and 77,149 asymptomatic control subjects from 2 independent European cohorts (UK Biobank and Lifelines). Single-nucleotide polymorphism (SNP)-based heritability (h2SNP) and genetic correlations (rg) with other traits were calculated. IBS risk loci were functionally annotated to identify candidate genes. Sensitivity and conditional analyses were conducted to assess impact of confounders. Polygenic risk scores were computed and tested in independent datasets.
Results
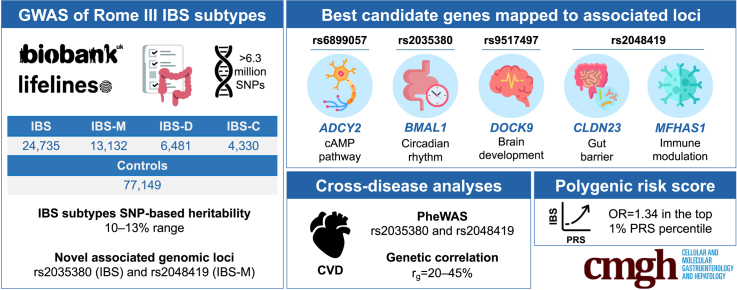
Rome III IBS showed significant SNP-heritability (up to 13%) and similar genetic architecture across subtypes, including those with manifestations at the opposite ends of the symptom spectrum (rg = 0.48 between IBS-D and IBS-C). Genetic correlations with other traits highlighted commonalities with family history of heart disease and hypertension, coronary artery disease, and angina pectoris (rg = 0.20–0.45), among others. Four independent GWAS signals (P < 5×10-8) were detected, including 2 novel loci for IBS (rs2035380) and IBS-mixed (rs2048419) that had been previously associated with hypertension and coronary artery disease. Functional annotation of GWAS risk loci revealed genes implicated in circadian rhythm (BMAL1), intestinal barrier (CLDN23), immunomodulation (MFHAS1), and the cyclic adenosine monophosphate pathway (ADCY2). Polygenic risk scores allowed the identification of individuals at increased risk of IBS (odds ratio, 1.34; P = 1.1×10-3).
Conclusions
Rome III Criteria capture higher SNP-heritability than previously estimated for IBS. The identified link between IBS and cardiovascular traits may contribute to the delineation of alternative therapeutic strategies, warranting further investigation.
Keywords: Genome-Wide Association Study, IBS, Genetic Correlation, CVD
Graphical abstract
Summary.
Genome-wide association studies on irritable bowel syndrome defined according to consensus Rome criteria shows higher single-nucleotide polymorphism–based heritability than previously reported, and novel risk loci with attractive druggable targets. Genetic profiling shows overlap with cardiovascular diseases and may contribute to the stratification of patients with irritable bowel syndrome.
Irritable bowel syndrome (IBS) is a common disorder of gut–brain interaction.1 It affects women more often than men and has a prevalence of 3%–11% in European populations, based on the consensus Rome criteria from the Rome Foundation.2 These criteria define IBS as a recurrent abdominal pain or discomfort associated with abnormal stool frequency and consistency, and classify IBS subtypes as constipation (IBS-C), diarrhea (IBS-D), or mixed (IBS-M) based on the predominant stool type.3 The latest Rome IV criteria are the most stringent, focusing only on abdominal pain (rather than also discomfort, as in Rome III) and an increased frequency of symptoms (1 day/week vs 3 days/month in Rome III), thus possibly capturing a more severe phenotype compared with earlier versions.4,5
Existing treatments for IBS are mostly directed toward the amelioration of symptoms, but are not effective in all patients.6 The contribution of dietary and psychological factors, gut dysbiosis, prior gastrointestinal (GI) infection, and genetic predisposition is recognized.1 A heritable component of IBS has been demonstrated in twin and family studies, although with varying degrees of estimates depending on the adopted IBS definition.7 The highest heritability was estimated in the Australian Twin study, with 57.9% (95% confidence interval, 40.6–75.9) of the variance in the reporting of IBS symptoms being attributed to additive genetic effects.8 Whether these genetic factors differently predispose to various IBS forms and/or subtypes is unclear, which hampers personalized approaches to treat patients with distinct clinical presentations.6
Interest in the genetic architecture of IBS has been growing, because it holds potential to reveal actionable pathways and biomarkers.9 This can be sought, for instance, by analogy with other diseases showing similar pathophysiologic mechanisms, including those accounted for by shared genetic factors. Despite demonstrated heritability, however, previous IBS candidate-gene studies lacked power and replication, except for serotonin, sucrase-isomaltase and ion channel genes.10, 11, 12, 13 Recently, well-powered genome-wide association studies (GWAS) of IBS and endophenotypes (eg, gut motility) have emerged, which made use of genotypic and health-related data from large cohorts and population-based biobanks.9,14, 15, 16, 17 However, these studies used heterogeneous IBS definitions that show poor overlap and inconsistent findings across definitions. For instance, the largest GWAS meta-analysis included results from 53,400 IBS cases, collectively identified through hospital-inpatient records, self-reported diagnoses, and Rome criteria: among those included from the UK Biobank (UKBB), less than 1% (n = 340/40,548) satisfied all IBS definitions simultaneously.15 The applicability and clinical relevance of genetic findings obtained using pooled IBS definitions are yet to be determined, nor have different IBS manifestations been adequately explored based on stratification of patients in different Rome subtype groups.
Here, we investigate the genetic underpinnings of IBS and its subtypes, based on available genotype and Rome III questionnaire data from 2 large European population-based biobanks (UKBB and Lifelines [LL]), totaling 24,735 cases and 77,149 asymptomatic control subjects.
Results
Characteristics of Rome III IBS Patients
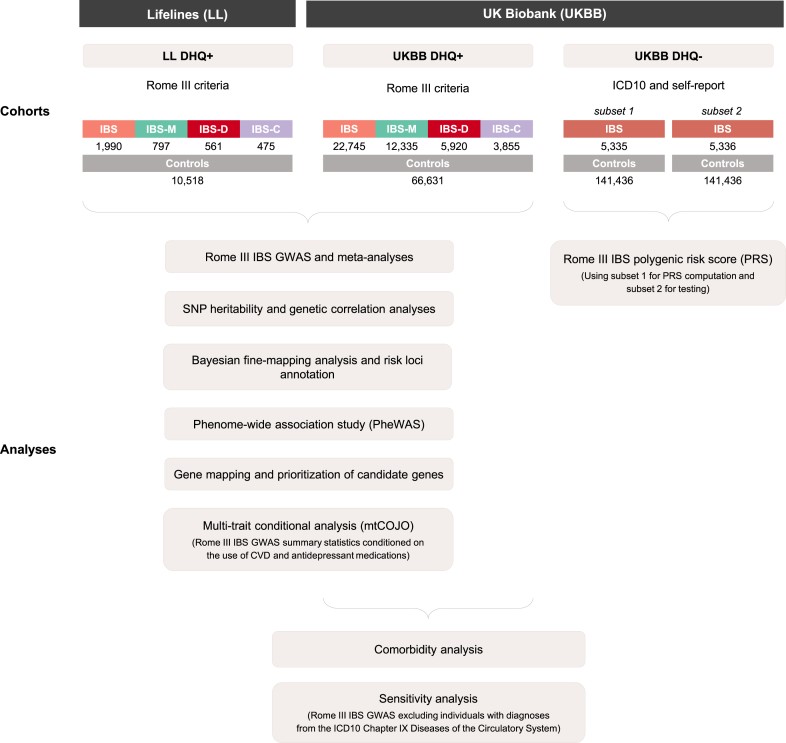
Rome III IBS patients and asymptomatic control subjects were defined based on digestive questionnaire data available for UKBB and LL participants (see Methods, Figure 1). In total, we identified 24,735 Rome III IBS patients (22,745 UKBB patients and 1990 LL patients) and 77,149 asymptomatic control subjects (66,631 from UKBB and 10,518 from LL). The demographics of study participants are summarized in Table 1. In line with previous studies, IBS-M was the most common subtype both in UKBB (54% of patients with IBS) and LL (40%). Patients with IBS were predominantly women, with the IBS-C group showing the most pronounced female predominance (84.0% of IBS-C cases were female in both UKBB and LL).18,19
Figure 1.
Infographic summarizing the derivation of study cohorts (IBS cases and control subjects) and their inclusion in different analyses. Digestive health questionnaire (DHQ) responders are described as “DHQ+” and nonresponders as “DHQ-.”
Table 1.
Demographics of Study Cohorts Included in Rome III IBS GWAS Meta-analyses
| Cohort | N | Age, mean (SD) | Sex, female (%) | |
|---|---|---|---|---|
| UKBB | IBS | 22,745 | 54.4 (7.7) | 73.0 |
| IBS-M | 12,335 | 56.6 (7.6) | 72.4 | |
| IBS-D | 5,920 | 54.2 (7.7) | 67.4 | |
| IBS-C | 3,855 | 55.0 (7.8) | 83.9 | |
| Control subjects | 66,631 | 56.6 (7.6) | 46.9 | |
| LL | IBS | 1,990 | 42.5 (14.0) | 76.2 |
| IBS-M | 797 | 40.4 (14.2) | 78.9 | |
| IBS-D | 561 | 43.1 (13.4) | 67.9 | |
| IBS-C | 475 | 44.0 (14.1) | 84.0 | |
| Control subjects | 10,518 | 47.2 (13.8) | 48.0 | |
GWAS, genome-wide association study; IBS, irritable bowel syndrome; IBS-C, irritable bowel syndrome constipation-predominant subtype; IBS-D, irritable bowel syndrome diarrhea-predominant subtype; IBS-M, irritable bowel syndrome mixed subtype; LL, Lifelines; SD, standard deviation; UKBB, UK Biobank.
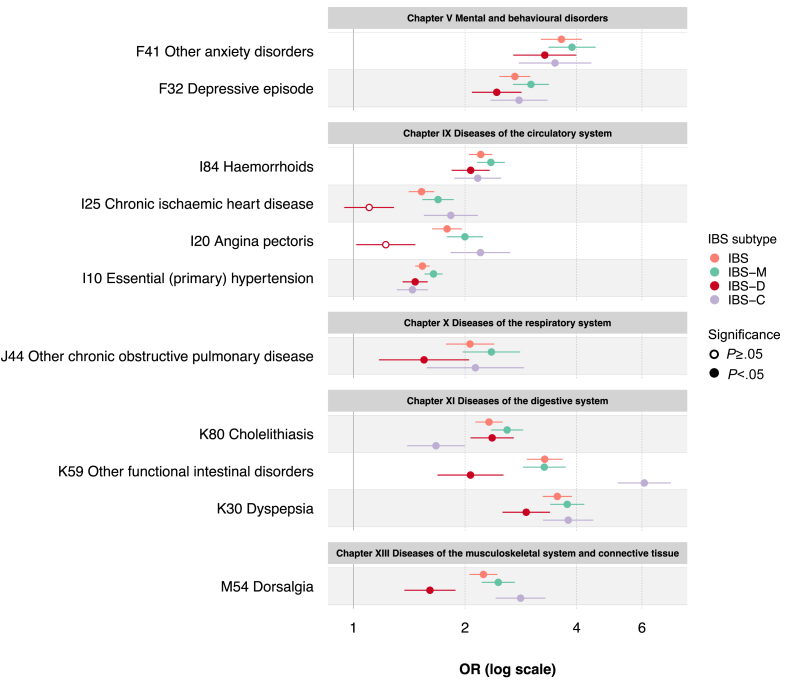
Health-related data available in UKBB allowed other diseases and conditions to be studied in IBS cases and control subjects, based on hospital-inpatient records in relation to diseases (International Classification of Diseases, 10th version [ICD10], Chapters I-XIV) and laboratory findings (ICD10 Chapter XVIII), across 22,344,000 data-points (250 data-fields covering 89,376 individuals) (Methods). Age- and sex-adjusted regression analysis revealed patients with IBS (all subtypes) to suffer from a significantly higher number of comorbidities compared with control subjects (P ≤ 5.1×10-161) (Table 2). At both the chapter (domain) and 3-digit (trait-specific) ICD10 levels, strongest associations were detected for GI (chapter XI diseases of the digestive system) and psychiatric (chapter V mental and behavioral disorders) disorders, whose risk was more than doubled in Rome III IBS versus control subjects (odds ratio [OR], 2.0–2.7; P ≤ 7.7×10-39) (Table 3), particularly for F41 other anxiety disorders (OR, 3.3–3.9; P ≤ 4.5×10-26) and K30 dyspepsia (OR, 2.9–3.8; P ≤ 1.0×10-44) (Figure 2, Supplementary Table 1). As a novel observation, traits from the chapter IX diseases of the circulatory system showed increased prevalence among patients with IBS (Figure 2, Supplementary Table 1). This included I10 essential (primary) hypertension, which was associated with all IBS subtypes (OR, 1.4–1.6; P ≤ 1.3×10-12), more pronouncedly IBS-M (Figure 2). Other differences, also related to diseases of the circulatory system, were observed across subtypes including I20 angina pectoris and I25 chronic ischemic heart disease, which were more common in IBS-C patients (OR, 2.2 [P = 1.2×10-15] and OR, 1.8 [P = 2.1×10-11], respectively) (Figure 2, Supplementary Table 1).
Table 2.
Number of ICD10 Comorbidities in UKBB Subjects
| Cohort | Number of ICD10 comorbidities |
||
|---|---|---|---|
| Mean (SD) | Beta (SE) | P (Beta)a | |
| IBS | 4.5 (5.1) | 1.81 (0.03) | <1.0E-300 |
| IBS-M | 4.7 (5.3) | 2.02 (0.04) | <1.0E-300 |
| IBS-D | 4.1 (4.6) | 1.42 (0.05) | 5.1E-161 |
| IBS-C | 4.6 (5.3) | 1.93 (0.06) | 1.9E-194 |
| Control subjects | 3.0 (3.9) | — | — |
IBS, irritable bowel syndrome; IBS-C, irritable bowel syndrome constipation-predominant subtype; IBS-D, irritable bowel syndrome diarrhea-predominant subtype; IBS-M, irritable bowel syndrome mixed subtype; ICD10, International Classification of Diseases, 10th revision; SD, standard deviation; SE, standard error; UKBB, UK Biobank.
From sex- and age-adjusted linear regression, testing the effect of IBS on the number of ICD10 comorbidities. Comorbidities included ICD10 codes for diseases (Chapters I-XIV) and laboratory findings (Chapter XVIII), excluding K58 for IBS and confounding gastrointestinal diagnoses (see Methods).
Table 3.
Chapter-Level ICD10 Comorbidities in UKBB Subjects
| ICD10 Chapter | OR (95% CI) |
P value (FDR)a |
||||||
|---|---|---|---|---|---|---|---|---|
| IBS | IBS-M | IBS-D | IBS-C | IBS | IBS-M | IBS-D | IBS-C | |
| I Certain infectious and parasitic diseases | 2.1 (1.9–2.2) | 2.2 (2.1–2.4) | 2.0 (1.8–2.2) | 1.9 (1.6–2.1) | 5.0E-104 | 1.4E-87 | 6.5E-35 | 4.2E-19 |
| II Neoplasms | 1.2 (1.2–1.3) | 1.2 (1.2–1.3) | 1.3 (1.2–1.3) | 1.3 (1.2–1.4) | 3.5E-28 | 3.2E-17 | 3.0E-10 | 4.4E-10 |
| III Diseases of the blood and blood-forming organs and certain disorders involving the immune mechanism | 1.5 (1.4–1.7) | 1.6 (1.5–1.8) | 1.4 (1.2–1.6) | 1.6 (1.4–1.9) | 3.1E-30 | 3.8E-27 | 9.5E-06 | 6.0E-10 |
| IV Endocrine, nutritional and metabolic diseases | 1.6 (1.5–1.7) | 1.8 (1.7–1.9) | 1.5 (1.4–1.6) | 1.5 (1.4–1.7) | 2.0E-100 | 3.2E-98 | 3.3E-22 | 9.6E-18 |
| V Mental and behavioral disorders | 2.2 (2.1–2.4) | 2.4 (2.3–2.6) | 2.0 (1.8–2.2) | 2.3 (2.0–2.6) | 4.1E-140 | 2.3E-120 | 7.7E-39 | 3.3E-39 |
| VI Diseases of the nervous system | 1.7 (1.6–1.8) | 1.8 (1.7–1.9) | 1.5 (1.4–1.7) | 1.8 (1.6–2.0) | 4.4E-80 | 1.2E-63 | 3.3E-18 | 2.3E-27 |
| VII Diseases of the eye and adnexa | 1.2 (1.1–1.3) | 1.2 (1.1–1.3) | 1.2 (1.0–1.3) | 1.2 (1.0–1.3) | 8.3E-10 | 4.7E-08 | 1.4E-02 | 3.8E-02 |
| VIII Diseases of the ear and mastoid process | 1.5 (1.4–1.7) | 1.6 (1.4–1.8) | 1.5 (1.3–1.8) | 1.6 (1.3–2.0) | 8.2E-15 | 2.8E-11 | 1.8E-05 | 3.0E-05 |
| IX Diseases of the circulatory system | 1.6 (1.6–1.7) | 1.7 (1.7–1.8) | 1.5 (1.4–1.6) | 1.6 (1.5–1.7) | 1.3E-146 | 7.5E-125 | 1.3E-33 | 7.5E-33 |
| X Diseases of the respiratory system | 1.7 (1.6–1.8) | 1.8 (1.7–1.9) | 1.5 (1.4–1.6) | 1.7 (1.6–1.9) | 5.2E-106 | 2.8E-90 | 2.4E-23 | 1.9E-28 |
| XI Diseases of the digestive system | 2.5 (2.4–2.6) | 2.7 (2.5–2.8) | 2.3 (2.2–2.4) | 2.6 (2.4–2.8) | <1.0E-300 | <1.0E-300 | 1.5E-178 | 1.8E-157 |
| XII Diseases of the skin and subcutaneous tissue | 1.3 (1.2–1.4) | 1.3 (1.2–1.4) | 1.3 (1.2–1.4) | 1.3 (1.1–1.4) | 1.3E-19 | 1.8E-16 | 3.0E-06 | 1.2E-04 |
| XIII Diseases of the musculoskeletal system and connective tissue | 1.5 (1.4–1.5) | 1.6 (1.5–1.6) | 1.3 (1.3–1.4) | 1.5 (1.4–1.7) | 1.2E-102 | 1.0E-85 | 2.0E-20 | 3.4E-30 |
| XIV Diseases of the genitourinary system | 1.5 (1.5–1.6) | 1.6 (1.6–1.7) | 1.4 (1.3–1.5) | 1.7 (1.5–1.8) | 1.0E-123 | 3.3E-101 | 3.6E-28 | 6.4E-41 |
| XVIII Symptoms, signs and abnormal clinical and laboratory findings, not elsewhere classified | 2.1 (2.1–2.2) | 2.3 (2.2–2.4) | 2.0 (1.9–2.1) | 2.2 (2.0–2.4) | <1.0E-300 | <1.0E-300 | 1.4E-119 | 1.8E-108 |
CI, confidence interval; FDR, false discovery rate; IBS, irritable bowel syndrome; IBS-C, irritable bowel syndrome constipation-predominant subtype; IBS-D, irritable bowel syndrome diarrhea-predominant subtype; IBS-M, irritable bowel syndrome mixed subtype; ICD10, International Classification of Diseases, 10th revision; OR, odds ratio; SD, standard deviation; SE, standard error; UKBB, UK Biobank.
From sex- and age-adjusted logistic regressions, comparing the risk of ICD10 diagnoses (at the chapter level) in IBS cases versus control subjects. Nonsignificant results (FDR >0.05) are omitted (indicated by a dash). Only ICD10 chapters for diseases (Chapters I-XIV) and laboratory findings (Chapter XVIII) are included, excluding K58 for IBS and confounding gastrointestinal diagnoses (see Methods).
Figure 2.
Analysis of Rome III IBS comorbidities in UKBB. Selected ICD10 diagnoses at the 3-digit code (trait-specific) level are reported, with corresponding OR and 95% confidence interval versus asymptomatic control subjects, derived from logistic regression adjusted for sex and age. Nonsignificant OR (pFDR ≥ .05) are shown as empty circles. Full results are reported in Supplementary Table 1.
Rome III IBS GWAS Meta-Analyses: Heritability and Genetic Correlation with Other Traits
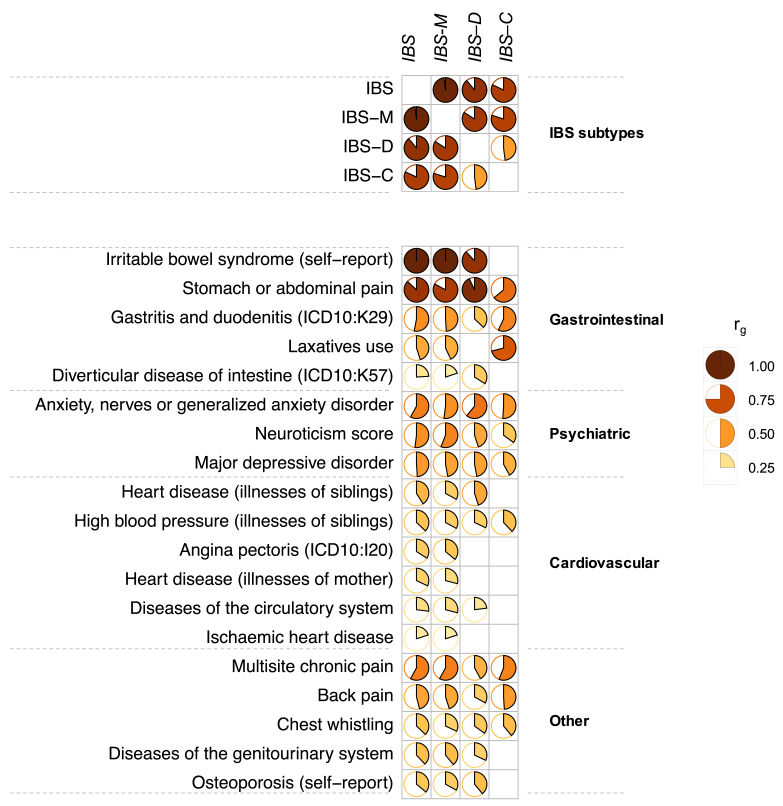
Individual Rome III IBS GWAS analyses were carried out in UKBB and LL, adopting a common analytic pipeline for quality control (per-sample and per-marker), imputation, and association testing adjusting for age, sex, and principal components (PCs) (Methods). UKBB and LL GWAS summary statistics for 6,311,313 high-quality single-nucleotide polymorphism (SNP) markers were included in GWAS meta-analyses spanning, respectively, 24,735 IBS, 13,132 IBS-M, 6481 IBS-D, and 4330 IBS-C cases and a common set of 77,149 asymptomatic control subjects (Table 1). GWAS meta-analysis results showed no population stratification (λ=1.03–1.09, linkage disequilibrium score regression [LDSC], intercept=0.97–1.00). Based on LDSC analyses (Methods), all Rome III IBS subtypes showed detectable and significant SNP-based heritability (h2SNP), estimated at 10.3% (P = 7.1×10-25) for IBS, 13.4% (P = 3.2×10-15) for IBS-M, 12.4% (P = 1.7×10-8) for IBS-D, and 11.7% (P = 1.6×10-4) for IBS-C. Considerable genetic overlap was observed across all traits, with almost complete correlation between IBS and IBS-M (rg=0.98; P < 1.0×10-300) (Figure 3, Supplementary Table 2). Of note, although weaker than with other subtypes, positive genetic correlation was also observed between traits at the opposite ends of the stool consistency spectrum, such as IBS-C and IBS-D (rg=0.48; P = 5.6×10-3).
Figure 3.
Genetic correlations (rg) across Rome III IBS subtypes and other traits. IBS, its subtypes, and other traits are reported based on respective significant (pFDR < .05) genetic correlations, divided into different domains. Empty cells correspond to nonsignificant results or rg between the same traits. Full results are reported in Supplementary Table 2.
The genetic architecture of IBS was then compared with that of other traits and diseases via LDSC analyses implemented in the Complex Traits Genetics Virtual Lab (CTG-VL) platform (Methods), which revealed significant correlations (pFDR < .05) between IBS types and 278 other traits across the GI, psychiatric, musculoskeletal, respiratory, cardiovascular, and metabolic domains (full results in Supplementary Table 2). Strongest correlations were observed for self-reported IBS and stomach or abdominal pain (Figure 3). Indeed, among different IBS types, IBS-C is the one that most often departed from others in its genetic similarities with other traits, illustrated for instance by reduced or augmented genetic correlations with stomach or abdominal pain, laxatives, and back pain (Figure 3). Among non-GI and non-pain-related traits, the conditions most closely correlating with IBS were psychiatric conditions, including generalized anxiety disorder (rg=0.51–0.61; P = ≤ 2.2×10-2), major depressive disorder (rg=0.42–0.49; p≤1.9×10-5), and neuroticism score (rg=0.35–0.56; P ≤ 4.1×10-4), all 3 showing stronger mean correlations with IBS-M and IBS-D than IBS-C. As an interesting and new observation, significant genetic correlations with IBS (particularly the IBS-M subtype) were noted for cardiovascular diseases (CVD), including family history of heart disease (measured as illness of mother and of siblings; rg=0.29–0.45; P ≤ 1.3×10-3) and high blood pressure (BP, measured as illness of siblings; rg=0.32–0.38; P ≤ 2.2×10-2), among others (Figure 3, Supplementary Table 2).
Annotation of Genomic Risk Loci and Fine Mapping
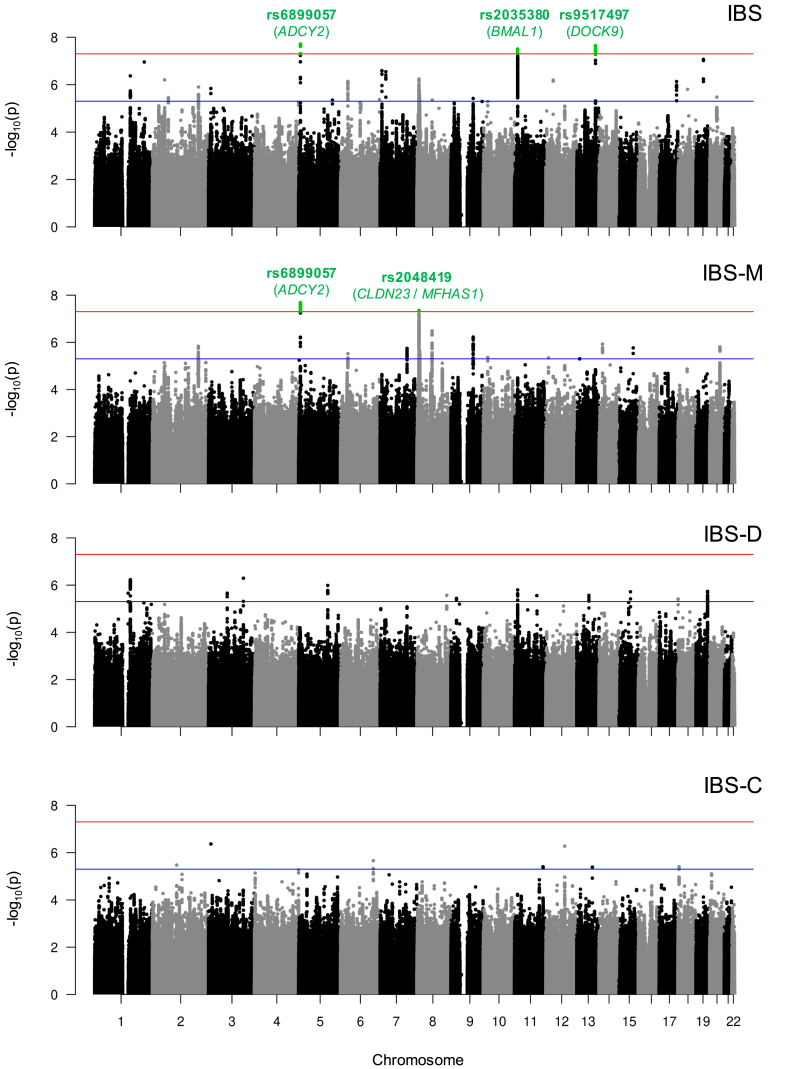
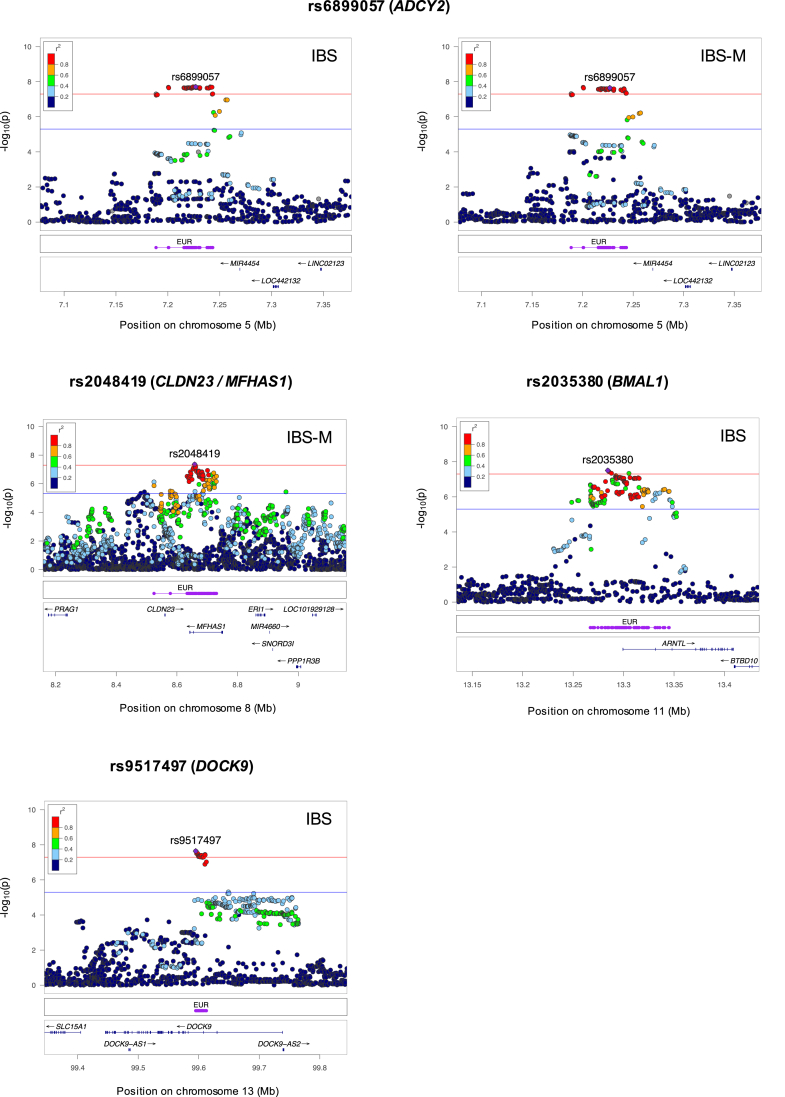
Sixty-four genome-wide significant SNP associations (P ≤ 5×10-8) were detected in Rome III IBS GWAS meta-analyses, corresponding to 4 independent loci: 2 in IBS (tagged by rs2035380 and rs9517497 on chromosome 11 and 13), 1 in IBS-M (tagged by rs2048419 on chromosome 8), and 1 common to IBS and IBS-M (tagged by the lead SNP rs6899057 on chromosome 5), as shown in Table 4 and Figure 4. No genome-wide significant associations were detected for IBS-D or IBS-C (Figure 4). Association signals from the 4 Rome IBS risk loci (regional plots in Figure 5) showed no heterogeneity (Cochran P-Het>0.05) and consistent genetic risk effects (OR direction) across GWAS performed individually in UKBB and LL (Table 4). Notably, rs2035380 (candidate gene BMAL1) and rs2048419 (CLDN23/MFHAS1) represent novel associations identified in this study, respectively, for IBS and the IBS-M subtype. In addition, although genome-wide associations for rs6899057 (ADCY2) and rs9517497 (DOCK9) had been previously reported for IBS,10 the former is also linked here to IBS-M for the first time.
Table 4.
Rome III IBS Risk Loci Identified Via GWAS Meta-analyses
| CHR | Lead SNP | Start–end (BP) | EA | OA | EAF | GWAS |
Credible Variants, n |
Gene mapping | Novel risk locus | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IBS subtype | OR (95% CI) | P value | OR in individual cohorts (UKBB/LL) | P-Het | |||||||||
| 5 | rs6899057 | 7188657–7257239 | T | C | 0.36 | IBS | 1.07 (1.04–1.09) | 2.0E-08 | 1.07/1.03 | 0.40 | 41 | ADCY2e | x |
| IBS-M | 1.09 (1.05–1.12) | 2.3E-08 | 1.08/1.09 | 0.97 | |||||||||
| 8 | rs2048419 | 8524474–8730488 | A | G | 0.53 | IBS-M | 1.08 (1.05–1.11) | 4.4E-08 | 1.08/1.03 | 0.34 | 77 |
CLDN23e ERI1e MFHAS1p,e PPP1R3Be TNKSe |
✓ |
| 11 | rs2035380 | 13267867–13346294 | T | C | 0.30 | IBS | 1.07 (1.04–1.09) | 3.1E-08 | 1.07/1.04 | 0.46 | 99 | BMAL1p,e | ✓ |
| 13 | rs9517497 | 99594682–99612588 | T | C | 0.63 | IBS | 1.07 (1.04–1.09) | 2.3E-08 | 1.06/1.09 | 0.48 | 20 | DOCK9p,e | x |
BP, base-pair position (genome build hg19); CHR, chromosome; CI, confidence interval; EA, effect allele; EAF, effect allele frequency; GWAS, genome-wide association study; IBS, irritable bowel syndrome; IBS-M, irritable bowel syndrome mixed subtype; LL, Lifelines; OA, other allele; OR, odds ratio; SNP, single-nucleotide polymorphism; UKBB, UK Biobank.
OR in individual cohorts: magnitude of mean effects described to the risk allele in UKBB and LL individual GWAS; P-Het: Cochran's Q statistics P value for heterogeneity across individual GWAS; Number of credible variants: number of markers in each fine-mapped credible set per risk locus; Gene mapping: genes mapped to the locus, based on FUMA positional (p) or cis-eQTL (e) analyses. The best candidate genes at each locus are highlighted in bold.
Figure 4.
Manhattan plots of subtype-specific IBS ΩGWAS meta-analyses. Each circle denotes a marker with regard to its physical location (based on the Genome Reference Consortium Human Build 37) and associated -log10P value. SNPs reaching the genome-wide significance threshold (P = 5×10-8, indicated with a horizontal red line) are colored in green. Suggestive significance threshold (P = 5×10-6) is indicated with a horizontal blue line. The lead SNP rs ID and its respective best candidate protein-coding gene symbol are annotated in green.
Figure 5.
Regional plots of associated IBS and IBS-M risk loci. SNPs (dots) are reported with their physical location (based on the Genome Reference Consortium Human Build 37), associated -log10P value, and degree of linkage disequilibrium (r2) with the lead SNP (purple diamond labelled with the rs ID). The red line indicates the genome-wide threshold at P = 5×10-8 and the blue line indicates the suggestive threshold at P = 5×10-6. Purple lines labelled as “EUR” indicate the genomic location of fine-mapped credible variants.
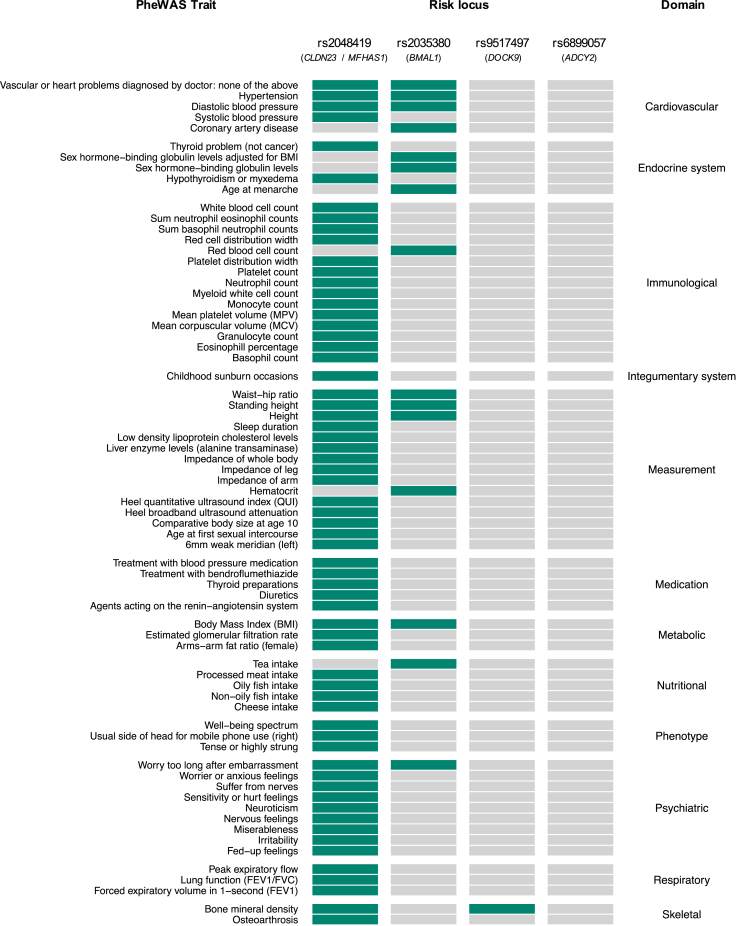
To gain initial biologic insight, we adopted a phenome-wide association study approach to screen Rome III IBS risk loci for known associations (P ≤ 5×10-8) reported in previous GWAS studies (Methods). Similar to the results obtained from genetic correlations, these analyses revealed the 2 novel risk loci, tagged by rs2048419 (CLDN23/MFHAS1) and rs2035380 (BMAL1), had both been previously associated with multiple traits including cardiovascular (hypertension, coronary artery disease), psychiatric (anxiety-like), and metabolic (body mass index) traits (Figure 6).
Figure 6.
Phenome-wide association (PheWAS) results for IBS and IBS-M risk loci. Known GWAS associations (P ≤ 5×10-8) for lead SNPs and/or their LD proxies (R2 >0.8) were extracted from OpenTargets, PhenoScanner v2, and GWAS ATLAS (see Supplementary Methods). Domains were harmonized according to GWAS ATLAS classifications and are sorted alphabetically.
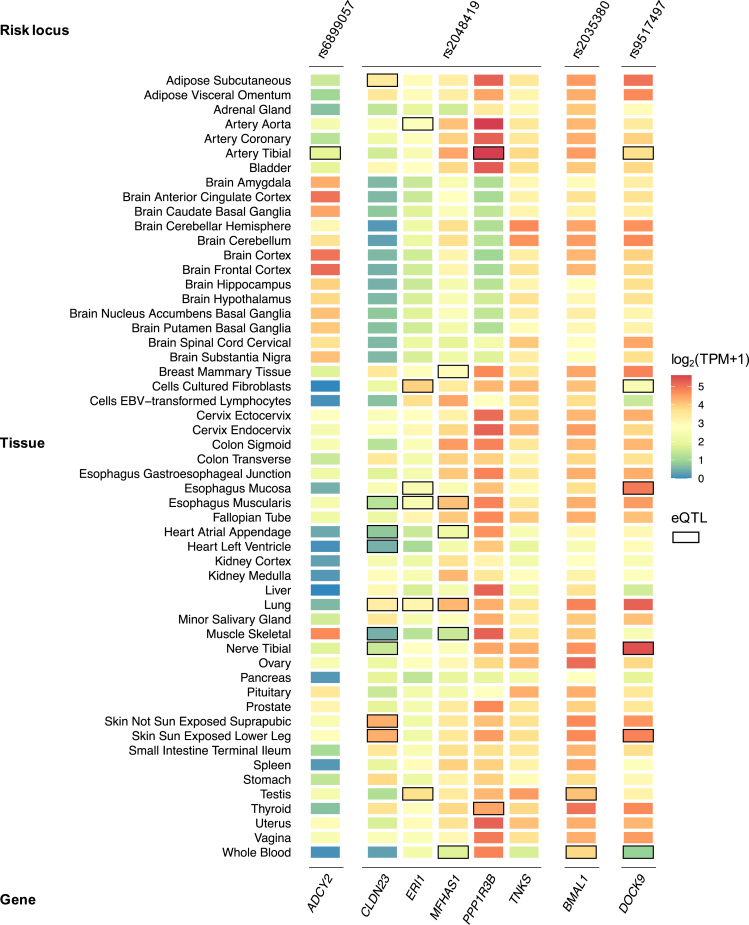
A Bayesian approach to fine-mapping was used to try and identify the most likely causative variants from each locus (Methods). This returned 4 independent credible sets of SNPs (range, 20–99 markers each) with high posterior probabilities to include causative variants (posterior probability, 82.0%–91.4%). However, no individual variant was estimated to be causative with high confidence (all variants were lower than 8.0% probability), and the identity of the individual causal variant from each locus remained elusive. Although there was no coding variant among SNPs from the credible set of each locus, several mapped near regulatory regions and transcription factors binding sites (Table 5). Furthermore, many were associated with differential expression (expression quantitative trait loci [eQTL]) of multiple genes in various tissues from the Genotype-tissue Expression (GTEx) Portal (Figure 7; see Methods), including eQTLs in tissues of the GI tract (for rs2048419 and rs9517497 loci) and heart (for rs2048419).
Table 5.
List of Fine-Mapped Credible Variants That Are Near Regulatory Sites Based on Ensembl Variation Annotation
| Lead SNP | Credible SNP | Biotype | CHR | BP | EA | OA | EAF | GWAS |
||
|---|---|---|---|---|---|---|---|---|---|---|
| IBS subtype | OR (95% CI) | P | ||||||||
|
rs6899057 ADCY2 |
rs4286635 | Enhancer | 5 | 7242559 | A | C | 0.60 | IBS | 0.94 (0.92–0.96) | 2.0E-08 |
| rs4286635 | Enhancer | 5 | 7242559 | A | C | 0.60 | IBS-M | 0.92 (0.90–0.94) | 4.1E-08 | |
| rs7718889 | CTCF binding site | 5 | 7200526 | A | T | 0.63 | IBS | 0.94 (0.92–0.96) | 2.1E-08 | |
| rs7718889 | CTCF binding site | 5 | 7200526 | A | T | 0.63 | IBS-M | 0.92 (0.90–0.94) | 2.1E-08 | |
|
rs2048419 CLDN23 / MFHAS1 |
rs1039915 | Enhancer | 8 | 8679614 | T | C | 0.53 | IBS-M | 1.07 (1.05–1.09) | 2.3E-07 |
| rs1039916 | Enhancer | 8 | 8685854 | A | G | 0.52 | IBS-M | 1.07 (1.05–1.09) | 2.7E-07 | |
| rs1039917 | Enhancer | 8 | 8718850 | A | G | 0.37 | IBS-M | 0.93 (0.91–0.95) | 9.6E-07 | |
| rs11775523 | Enhancer | 8 | 8679176 | A | G | 0.53 | IBS-M | 1.07 (1.05–1.09) | 2.7E-07 | |
| rs11992186 | CTCF binding site | 8 | 8524474 | C | G | 0.49 | IBS-M | 1.07 (1.05–1.09) | 1.4E-06 | |
| rs13270070 | Enhancer | 8 | 8691622 | A | T | 0.44 | IBS-M | 0.93 (0.91–0.95) | 1.4E-06 | |
| rs13282015 | Enhancer | 8 | 8659675 | T | G | 0.76 | IBS-M | 0.92 (0.88–0.96) | 3.4E-06 | |
| rs2409092 | Enhancer | 8 | 8682192 | A | T | 0.53 | IBS-M | 1.07 (1.05–1.09) | 1.5E-07 | |
| rs332039 | Enhancer | 8 | 8723651 | C | G | 0.57 | IBS-M | 1.07 (1.05–1.09) | 1.8E-06 | |
| rs35039922 | Enhancer | 8 | 8675325 | A | T | 0.53 | IBS-M | 1.07 (1.05–1.09) | 2.4E-07 | |
| rs35900578 | Enhancer | 8 | 8719513 | A | G | 0.40 | IBS-M | 0.93 (0.91–0.95) | 4.7E-07 | |
| rs3789843 | Enhancer | 8 | 8724257 | T | C | 0.41 | IBS-M | 0.93 (0.91–0.95) | 3.7E-07 | |
| rs3789845 | Enhancer | 8 | 8723918 | T | G | 0.34 | IBS-M | 0.93 (0.91–0.95) | 1.0E-06 | |
| rs3789849 | Enhancer | 8 | 8687054 | C | G | 0.43 | IBS-M | 0.93 (0.91–0.95) | 3.0E-07 | |
| rs3827806 | Enhancer | 8 | 8724276 | T | C | 0.41 | IBS-M | 0.93 (0.91–0.95) | 3.8E-07 | |
| rs4840362 | Enhancer | 8 | 8670082 | C | G | 0.46 | IBS-M | 0.93 (0.91–0.95) | 1.4E-07 | |
| rs4841040 | Promoter | 8 | 8654527 | T | C | 0.53 | IBS-M | 1.08 (1.06–1.10) | 7.8E-08 | |
| rs4841042 | Enhancer | 8 | 8664622 | A | G | 0.46 | IBS-M | 0.93 (0.91–0.95) | 1.4E-07 | |
| rs4841051 | Enhancer | 8 | 8685646 | T | C | 0.52 | IBS-M | 1.07 (1.05–1.09) | 2.2E-07 | |
| rs560544 | CTCF binding site | 8 | 8637429 | A | G | 0.52 | IBS-M | 1.07 (1.05–1.09) | 3.7E-07 | |
| rs56367294 | Enhancer | 8 | 8722527 | A | G | 0.32 | IBS-M | 0.93 (0.89–0.97) | 1.4E-06 | |
| rs60315134 | Enhancer | 8 | 8670599 | A | G | 0.54 | IBS-M | 1.07 (1.05–1.09) | 1.9E-07 | |
| rs7820146 | Enhancer | 8 | 8699757 | A | T | 0.44 | IBS-M | 0.93 (0.91–0.95) | 6.1E-07 | |
| rs7823757 | Enhancer | 8 | 8670177 | A | T | 0.46 | IBS-M | 0.93 (0.91–0.95) | 1.6E-07 | |
| rs7833171 | Enhancer | 8 | 8699761 | T | C | 0.44 | IBS-M | 0.93 (0.91–0.95) | 5.8E-07 | |
| rs882462 | Enhancer | 8 | 8678530 | A | G | 0.47 | IBS-M | 0.93 (0.91–0.95) | 3.5E-07 | |
|
rs2035380 BMAL1 |
rs10766064 | Open chromatin region | 11 | 13277932 | A | G | 0.74 | IBS | 0.94 (0.92–0.96) | 5.7E-07 |
| rs10766065 | Open chromatin region | 11 | 13277961 | T | C | 0.72 | IBS | 0.94 (0.92–0.96) | 9.4E-07 | |
| rs10766066 | Open chromatin region | 11 | 13278027 | A | G | 0.72 | IBS | 0.94 (0.92–0.96) | 9.4E-07 | |
| rs10832018 | Enhancer | 11 | 13312424 | A | G | 0.71 | IBS | 0.94 (0.92–0.96) | 8.9E-08 | |
| rs10832021 | Enhancer | 11 | 13324530 | A | G | 0.71 | IBS | 0.94 (0.92–0.96) | 5.2E-07 | |
| rs11022733 | Enhancer | 11 | 13281557 | A | C | 0.71 | IBS | 0.94 (0.92–0.96) | 8.3E-07 | |
| rs11022734 | Enhancer | 11 | 13281580 | A | G | 0.29 | IBS | 1.06 (1.04–1.08) | 8.3E-07 | |
| rs11022735 | Enhancer | 11 | 13292727 | A | C | 0.30 | IBS | 1.06 (1.04–1.08) | 7.8E-08 | |
| rs11022743 | Promoter | 11 | 13297800 | A | G | 0.27 | IBS | 1.06 (1.04–1.08) | 5.7E-07 | |
| rs11022753 | CTCF binding site | 11 | 13310854 | T | C | 0.73 | IBS | 0.94 (0.92–0.96) | 7.8E-07 | |
| rs11022754 | Enhancer | 11 | 13313243 | A | G | 0.27 | IBS | 1.06 (1.04–1.08) | 7.3E-07 | |
| rs11022756 | Enhancer | 11 | 13315439 | A | C | 0.29 | IBS | 1.06 (1.04–1.08) | 8.9E-08 | |
| rs11022757 | Enhancer | 11 | 13322580 | A | G | 0.29 | IBS | 1.06 (1.04–1.08) | 5.5E-07 | |
| rs11605776 | Enhancer | 11 | 13318524 | A | C | 0.71 | IBS | 0.94 (0.92–0.96) | 4.1E-07 | |
| rs12290622 | Enhancer | 11 | 13314307 | A | G | 0.71 | IBS | 0.94 (0.92–0.96) | 8.5E-08 | |
| rs12361893 | Promoter | 11 | 13301114 | C | G | 0.30 | IBS | 1.06 (1.04–1.08) | 1.5E-07 | |
| rs1351525 | Promoter | 11 | 13301548 | A | T | 0.29 | IBS | 1.06 (1.04–1.08) | 1.5E-07 | |
| rs1384030 | Enhancer | 11 | 13292909 | T | C | 0.30 | IBS | 1.06 (1.04–1.08) | 8.3E-08 | |
| rs1481891 | CTCF binding site | 11 | 13310718 | T | C | 0.29 | IBS | 1.06 (1.04–1.08) | 8.7E-08 | |
| rs1481892 | Promoter | 11 | 13301921 | C | G | 0.71 | IBS | 0.94 (0.92–0.96) | 1.5E-07 | |
| rs2035380 | Enhancer | 11 | 13284345 | T | C | 0.30 | IBS | 1.07 (1.04–1.09) | 3.1E-08 | |
| rs2219998 | Enhancer | 11 | 13292864 | A | G | 0.57 | IBS | 0.94 (0.92–0.96) | 1.8E-07 | |
| rs2279284 | Promoter | 11 | 13298750 | T | C | 0.27 | IBS | 1.06 (1.04–1.08) | 5.3E-07 | |
| rs2279285 | Promoter | 11 | 13298687 | T | G | 0.70 | IBS | 0.94 (0.92–0.96) | 9.4E-08 | |
| rs2279286 | Promoter | 11 | 13298519 | A | G | 0.70 | IBS | 0.94 (0.92–0.96) | 8.4E-08 | |
| rs2279287 | Promoter | 11 | 13298485 | T | C | 0.30 | IBS | 1.06 (1.04–1.08) | 8.4E-08 | |
| rs2403661 | Enhancer | 11 | 13285281 | T | G | 0.30 | IBS | 1.07 (1.05–1.09) | 3.6E-08 | |
| rs34148132 | Enhancer | 11 | 13314475 | T | C | 0.29 | IBS | 1.06 (1.04–1.08) | 8.2E-08 | |
| rs34796300 | Enhancer | 11 | 13315205 | T | C | 0.43 | IBS | 1.06 (1.04–1.08) | 2.9E-07 | |
| rs4603287 | Promoter | 11 | 13301335 | T | G | 0.29 | IBS | 1.06 (1.04–1.08) | 1.5E-07 | |
| rs4757138 | Promoter | 11 | 13297925 | A | G | 0.30 | IBS | 1.06 (1.04–1.08) | 8.8E-08 | |
| rs4757139 | Promoter | 11 | 13300456 | T | C | 0.70 | IBS | 0.94 (0.92–0.96) | 1.3E-07 | |
| rs4757140 | Promoter | 11 | 13300540 | T | G | 0.70 | IBS | 0.94 (0.92–0.96) | 1.4E-07 | |
| rs55769038 | Enhancer | 11 | 13331808 | A | G | 0.59 | IBS | 0.94 (0.92–0.96) | 6.1E-07 | |
| rs61882122 | Promoter | 11 | 13299895 | A | G | 0.30 | IBS | 1.06 (1.04–1.08) | 9.7E-08 | |
| rs6486116 | CTCF binding site | 11 | 13319838 | A | C | 0.29 | IBS | 1.06 (1.04–1.08) | 6.3E-07 | |
| rs6486118 | Enhancer | 11 | 13323786 | A | G | 0.71 | IBS | 0.94 (0.92–0.96) | 4.0E-07 | |
| rs6486119 | Enhancer | 11 | 13323789 | T | C | 0.29 | IBS | 1.06 (1.04–1.08) | 4.3E-07 | |
| rs6486120 | Enhancer | 11 | 13324142 | T | G | 0.29 | IBS | 1.06 (1.04–1.08) | 5.1E-07 | |
| rs7114573 | Enhancer | 11 | 13323498 | A | C | 0.29 | IBS | 1.06 (1.04–1.08) | 5.2E-07 | |
| rs7125487 | Enhancer | 11 | 13282058 | C | G | 0.28 | IBS | 1.07 (1.05–1.09) | 1.3E-07 | |
| rs72867447 | Promoter | 11 | 13301875 | C | G | 0.43 | IBS | 1.06 (1.04–1.08) | 3.3E-07 | |
| rs7928655 | Promoter | 11 | 13300252 | C | G | 0.30 | IBS | 1.06 (1.04–1.08) | 8.7E-08 | |
| rs7937432 | CTCF binding site | 11 | 13320525 | A | G | 0.71 | IBS | 0.94 (0.92–0.96) | 4.1E-07 | |
| rs7938307 | CTCF binding site | 11 | 13320526 | A | C | 0.71 | IBS | 0.94 (0.92–0.96) | 3.7E-07 | |
| rs7949336 | CTCF binding site | 11 | 13319894 | A | G | 0.74 | IBS | 0.94 (0.92–0.96) | 7.6E-07 | |
| rs7951393 | Enhancer | 11 | 13318431 | T | C | 0.71 | IBS | 0.94 (0.92–0.96) | 4.7E-07 | |
| rs998089 | Enhancer | 11 | 13284111 | T | C | 0.27 | IBS | 1.06 (1.04–1.08) | 4.5E-07 | |
|
rs9517497 DOCK9 |
rs6491472 | Enhancer | 13 | 99598367 | A | G | 0.37 | IBS | 0.94 (0.92–0.96) | 3.1E-08 |
| rs7337807 | Enhancer | 13 | 99610581 | A | C | 0.63 | IBS | 1.06 (1.04–1.08) | 3.8E-08 | |
| rs7338982 | Enhancer | 13 | 99610373 | T | C | 0.63 | IBS | 1.06 (1.04–1.08) | 3.6E-08 | |
| rs7339076 | Enhancer | 13 | 99599128 | C | G | 0.63 | IBS | 1.06 (1.04–1.08) | 4.1E-08 | |
| rs7987680 | Enhancer | 13 | 99601519 | T | G | 0.63 | IBS | 1.06 (1.04–1.08) | 4.7E-08 | |
| rs9513513 | Open chromatin region | 13 | 99595622 | T | C | 0.63 | IBS | 1.06 (1.04–1.08) | 2.4E-08 | |
| rs9517497 | Open chromatin region | 13 | 99594953 | T | C | 0.63 | IBS | 1.07 (1.04–1.09) | 2.3E-08 | |
| rs9517502 | Enhancer | 13 | 99599727 | A | C | 0.37 | IBS | 0.94 (0.92–0.96) | 4.5E-08 | |
| rs9517503 | Enhancer | 13 | 99600658 | A | G | 0.63 | IBS | 1.06 (1.04–1.08) | 4.6E-08 | |
| rs9517508 | Enhancer | 13 | 99606192 | T | G | 0.37 | IBS | 0.94 (0.92–0.96) | 5.2E-08 | |
| rs9557097 | Enhancer | 13 | 99612588 | T | C | 0.62 | IBS | 1.06 (1.04–1.08) | 9.4E-08 | |
NOTE. Functional consequence of variants was performed using the Ensembl Variant Effect Predictor tool (https://grch37.ensembl.org/info/docs/tools/vep). OR: mean effects described to the risk allele in the meta-analysis.
BP, base-pair position (genome build hg19); CHR, chromosome; CI, confidence interval; EA, effect allele; EAF, effect allele frequency; GWAS, genome-wide association study; IBS, irritable bowel syndrome; IBS-M, irritable bowel syndrome mixed subtype; LL, Lifelines; OA, other allele; OR, odds ratio; SNP, single-nucleotide polymorphism; UKBB, UK Biobank.
Figure 7.
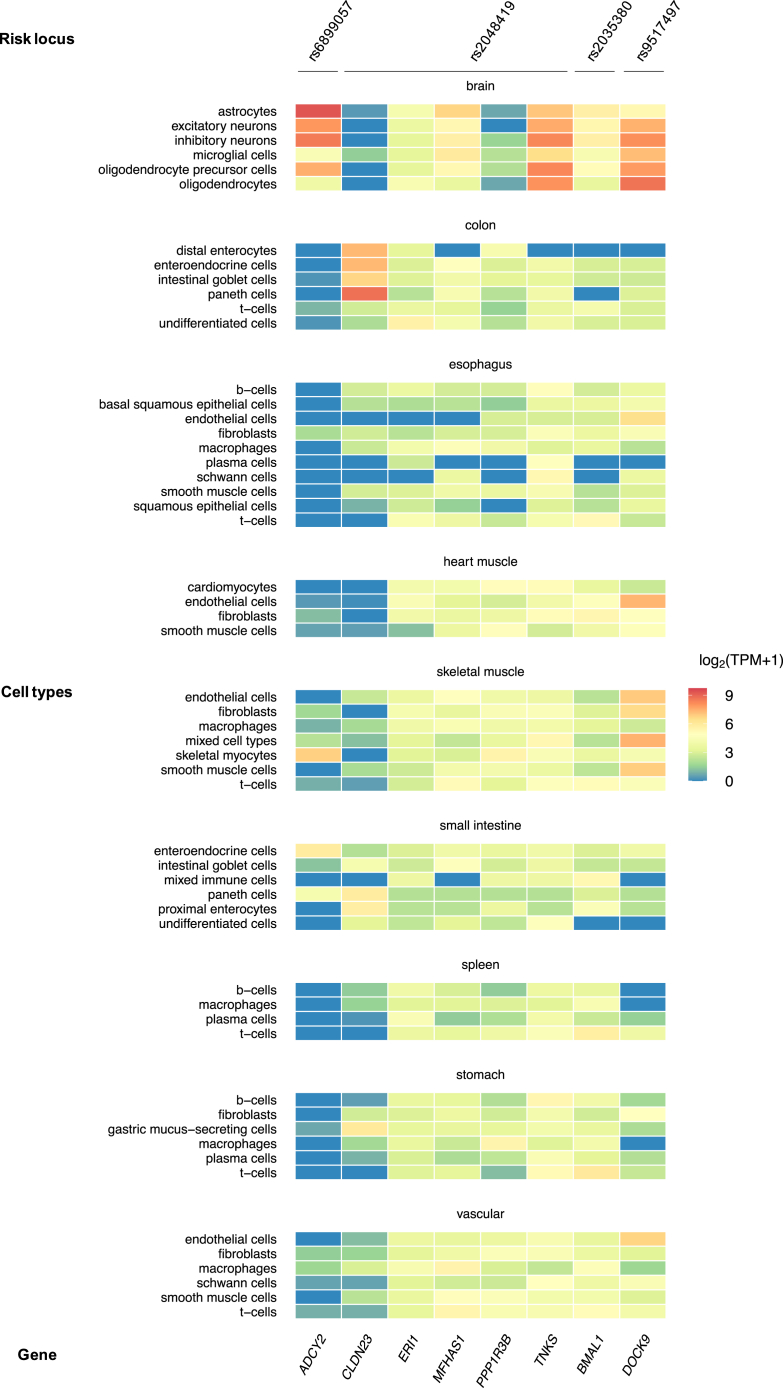
Heat map of gene expression from Rome III IBS risk loci, based on GTEx data. IBS candidate genes from IBS risk loci (locus membership annotated at the top) are reported with their level of expression (mRNA) in 54 human tissues using data from GTEx v8 (https://gtexportal.org). The log2(TPM+1), where TPM (transcripts per million) is the averaged expression per tissue and per gene, is indicated on a color-coded scale. eQTLs (pFDR < .05) linked to lead SNP or variants in high LD (r2 >0.8) are boxed.
Gene Mapping and Prioritization of Candidate Genes
Positional and cis-eQTL functional mapping (FUMA) and annotation of GWAS (Methods) resulted in a total of 8 protein-coding genes as plausible causative candidates at the 4 IBS and IBS-M risk loci (Table 4). Their expression in different human tissues and cell types are shown in Figures 7 and 8. Previous GWAS associations of these genes with various traits are listed in Supplementary Table 3.
Figure 8.
Heat map of gene expression from Rome III IBS and IBS-M risk loci, based on single-cell transcriptomic data. IBS candidate genes from IBS risk loci (locus membership annotated at the top) are reported with their level of expression (mRNA) in cell types of human tissues using single-cell transcriptomic data from the Human Protein Atlas (www.proteinatlas.org). The log2(TPM+1), where TPM (transcripts per million) is the averaged expression per tissue and per gene, is indicated on a color-coded scale.
Single genes were mapped to 3 risk loci, hence these represent bona fide candidates to be causative of the observed associations at respective loci: BMAL1 (basic helix-loop-helix ARNT Like 1) at the new rs2035380 locus for IBS, ADCY2 (adenylate cyclase 2) at the rs6899057 locus in common for IBS and IBS-M, and DOCK9 (dedicator of cytokinesis 9) at the rs9517497 locus for IBS (Table 1). BMAL1, mapped positionally and via eQTL, codes for a key clock protein that controls circadian rhythms and is highly expressed in the brain and the GI tract (colon, esophagus, small intestine, and stomach) (Figure 7), and in multiple cell types (Figure 8). This gene has been implicated via GWAS in several other traits, including CVDs and psychiatric disorders (Supplementary Table 3). DrugBank search revealed BMAL1 regulators are targeted by antibacterial drugs with indications for IBS-D (rifaximin, DrugBank ID: DB01220) and traveler’s diarrhea (norfloxacin and ofloxacin, DrugBank ID: DB01059 and DB01165). For ADCY2, mapped to the rs6899057 locus via eQTLs, considerable expression is observed in brain tissues and cells (Figure 7, Figure 8) and in myocytes (Figure 8). Previous GWAS studies linked this gene to IBS and other traits, including GI diseases and CVDs, possibly via cyclic adenosine monophosphate signaling pathway (Supplementary Table 3). The encoded enzyme (AC2) is targeted by the drug colforsin indicated for smooth muscle relaxation and vasodilation (DrugBank ID: DB02587). Finally, at the rs9517497 locus, DOCK9 is involved in GTPase activity regulation and brain development, showing high expression in brain and GI tissues (colon, esophagus, small intestine, and stomach) (Figure 7), and muscle and endothelial cell types (Figure 8). It is associated with eQTLs in the esophagus mucosa (Figure 7) and has been previously linked to IBS and many other traits via GWAS (Supplementary Table 3).
Five candidate genes were mapped to the rs2048419 locus for IBS-M, the second novel locus identified in this study (Table 4). We aimed to prioritize the best candidates at this locus based on functional evidence at multiple levels, including eQTL data for GI and brain tissues, gene expression, previous GWAS findings, mechanistic links to IBS, relevant phenotypes from knockout mice, and DrugBank data (Methods). Multiple functional annotations used for gene prioritization in the rs2048419 locus are reported in Table 6. By these means, CLDN23 (claudin 23), which encodes a tight-junction protein that plays a role in gut barrier function, and MFHAS1 (multifunctional ROCO family signaling regulator 1), which modulates immune responses, were deemed the most likely causative genes. Both show expression in the GI tract (Figure 7; including expression in specific colonic cell types, such as enteroendocrine cells, Figure 8), are associated with eQTLs in GI tissues via fine-mapped credible variants (Table 6), and are linked via GWAS data to other traits, such as neuroticism and hypertension (Supplementary Table 3).
Table 6.
Functional Evidence Used for Prioritization of rs2048419 Candidate Genes
| Evidence level | Candidate genes (rs2048419 locus) |
||||
|---|---|---|---|---|---|
|
CLDN23 (claudin 23) |
MFHAS1 (multifunctional ROCO family signaling regulator 1) |
ERI1 (exoribonuclease 1) |
PPP1R3B (protein phosphatase 1 regulatory subunit 3B) |
TNKS (tankyrase) |
|
| (1) GI/brain expression | Colon, small intestine, stomach | Colon, esophagus, small intestine, stomach, brain | Colon, esophagus, small intestine, stomach | Colon, esophagus, small intestine, brain | |
| (2) GI/brain eQTL | Esophagus | Esophagus | Esophagus | ||
| (3) Gene-trait association | Yes (others) | Yes (others) | Yes (others) | Yes (others) | Yes (others) |
| (4) Mechanistic link | Tight-junction intestinal barrier function (PMID: 37798277) | Immune response modulation (PMID:28609714, 26599367, 27783989) | Lipid and glucose metabolism (PMID: 28473467) | Intestinal epithelium homeostasis (PMID: 30260955, 27190037) Immune response modulation (PMID: 35362478) |
|
| (5) Phenotype from KO model or human studies | Intestinal barrier function alterations (PMID: 37798277) Intestinal-type gastric cancer (PMID: 12736707) Immune-induced atopic dermatitis following tight-junction barrier disruption (PMID: 21163515) |
Mitigation of colorectal tumors associated with inhibition of M2 macrophage polarization (PMID: 27783989) | Human intellectual disability (PMID: 36208065) | Reduced hepatic glycogen, glucose-intolerant and insulin-resistant (PMID: 28473467) | Small intestinal crypts and decreased intestinal stem cells (PMID: 30260955) Systemic inflammation (PMID: 35362478) |
NOTE. Functional evidence used for gene prioritization included: (1) GI/brain expression: high expression (defined as log2TPM≥3) in GI and/or brain tissues based on GTEx data; (2) GI/brain eQTL: colocalization between a credible variant and an eQTL in a GI and/or brain tissue; (3) Gene-trait association: association with IBS or other traits in previous GWAS studies via credible variants in the gene region; (4) Mechanistic link: plausible mechanistic link between the gene and IBS; (5) Phenotype from KO model or human studies: relevant phenotype from experimental knockout/knockdown models or human studies. Obs: No genes in this locus were directly targeted by current drugs with IBS-relevant indications based on DrugBank search.
eQTL, expression quantitative trait loci; GI, gastrointestinal; GTEx, genotype-tissue expression; GWAS, genome-wide association study; IBS, inflammatory bowel syndrome.
Genetic Link to Heart-Related Traits: Sensitivity and Conditional Analyses
Given the observed associations between IBS and heart-related traits, we sought to gain further insight from 2 additional, alternative analyses of their relationship. First, using UKBB data, we repeated our IBS Rome III GWAS analyses excluding all participants with diseases of the circulatory system (ICD10 chapter IX; diagnoses I00-I99), leaving 5993 IBS cases and 15,041 control subjects (Methods). This analysis revealed the genetic architecture of Rome III IBS to be virtually identical to that estimated devoid of heart-related conditions (rg>0.98 and P ≤ .4×10-144, across all Rome III IBS subtypes based on LDSC analysis of GWAS data). At the same time, the genetic risk effect estimates (ORs) associated with the 4 GWAS signals remained largely unchanged (despite the reduction in sample size and, expectedly, the strength of the associations) (Table 7). Next, we performed a proper conditional analysis to assess again, based on a different approach, whether the genetic underpinnings of Rome III IBS are independent of heart-disease predisposing factors. For this purpose, using mtCOJO20 (Methods), we conditioned our Rome III IBS GWAS meta-analysis results on the use of CVD medications (as proxies for their indications), using GWAS summary-level data from Wu et al21 (Methods). As shown in Table 8, Rome III IBS GWAS signals were not attenuated on conditioning, and the genetic architecture of IBS subtypes remained substantially unaltered, as assessed via LDSC analyses of conditioned and unconditioned GWAS data (rg>0.92 and P ≤ 7.4×10-263, across all Rome III IBS subtypes). Similarly, conditioning on antidepressants as proxies for anxiety and mood disorders (conditions known to share underlying genetics with IBS) did not result in substantial changes of the genetic risk effects (Table 8; and rg>0.94 and P ≤ 1.0×10-300 across all Rome III IBS subtypes). Overall, these results suggest that the genetic predisposition to Rome III IBS is not dependent on co-occurring heart-related diseases (and/or anxiety and mood disorders). Instead, the detected GWAS risk signals are intrinsic to the genetic architecture of IBS.
Table 7.
Sensitivity GWAS Analysis After Removing Individuals with CVD Diagnoses in UKBB
| Lead SNP | IBS subtype | Discoverya IBS: 24,735 / IBS-M: 13,132 Control subjects: 77,149 |
UKBB ALL IBS: 22,745 / IBS-M: 12,335 Control subjects: 66,631 |
UKBB CVD- IBS: 16,752 / IBS-M: 8,955 Control subjects: 51,590 |
|||
|---|---|---|---|---|---|---|---|
| OR (95% CI) | P value | OR (95% CI) | P value | OR (95% CI) | P value | ||
| rs6899057 ADCY2 |
IBS | 1.07 (1.04–1.09) | 2.0E-08 | 1.08 (1.06–1.11) | 6.6E-08 | 1.08 (1.05–1.11) | 9.1E-06 |
| IBS-M | 1.09 (1.05–1.12) | 2.3E-08 | 1.07 (1.05–1.09) | 2.0E-08 | 1.06 (1.04–1.09) | 5.9E-06 | |
| rs2048419 CLDN23 / MFHAS1 |
IBS-M | 1.08 (1.05–1.11) | 4.4E-08 | 1.08 (1.06–1.11) | 3.2E-08 | 1.09 (1.06–1.12) | 3.2E-07 |
| rs2035380 BMAL1 |
IBS | 1.07 (1.04–1.09) | 3.1E-08 | 1.07 (1.05–1.10) | 4.0E-08 | 1.07 (1.04–1.10) | 1.1E-06 |
| rs9517497 DOCK9 |
IBS | 1.07 (1.04–1.09) | 2.3E-08 | 1.06 (1.04–1.09) | 3.1E-07 | 1.06 (1.04–1.09) | 8.1E-06 |
CI, confidence interval; CVD, cardiovascular diseases; GWAS, genome-wide association study; IBS, irritable bowel syndrome; IBS-C, irritable bowel syndrome constipation-predominant subtype; IBS-D, irritable bowel syndrome diarrhea-predominant subtype; IBS-M, irritable bowel syndrome mixed subtype; LL, Lifelines; OR, odds ratio; SD, standard deviation; SNP, single-nucleotide polymorphism; UKBB, UK Biobank.
Estimates from Rome III IBS GWAS meta-analyses (UKBB and LL).
Table 8.
Conditional GWAS Analysis on the Use of CVD and Antidepressant Medications
| Risk locus | Conditional medication |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| C01D Vasodilators used in cardiac diseases | C02 Antihypertensives | C03 Diuretics | C07 Beta blocking agents | C08 Calcium channel blockers | C09 Agents acting on the renin-angiotensin system | C10AA HMG CoA reductase inhibitors | N06A Antidepressants | ||
| rs6899057 (IBS) ADCY2 |
OR | 1.07 (1.04 - 1.09) | |||||||
| P | 2.0E-08 | ||||||||
| OR-cond | 1.07 (1.04–1.09) | 1.07 (1.04–1.09) | 1.07 (1.04–1.09) | 1.07 (1.04–1.09) | 1.07 (1.04–1.09) | 1.07 (1.04–1.09) | 1.07 (1.04–1.09) | 1.06 (1.04–1.09) | |
| P-cond | 1.2E-07 | 2.4E-08 | 9.1E-09 | 8.0E-09 | 1.2E-08 | 1.3E-08 | 1.4E-08 | 2.6E-07 | |
| rs6899057 (IBS-M) ADCY2 |
OR | 1.09 (1.05–1.12) | |||||||
| P | 2.3E-08 | ||||||||
| OR-cond | 1.09 (1.05–1.12) | 1.09 (1.05–1.12) | 1.09 (1.05–1.12) | 1.09 (1.06–1.12) | 1.09 (1.05–1.12) | 1.09 (1.05–1.12) | 1.09 (1.05–1.12) | 1.08 (1.05–1.11) | |
| P-cond | 8.3E-08 | 2.5E-08 | 1.9E-08 | 1.4E-08 | 2.2E-08 | 2.2E-08 | 2.0E-08 | 1.1E-07 | |
| rs2048419 (IBS-M) CLDN23 / MFHAS1 |
OR | 1.08 (1.05–1.11) | |||||||
| P | 4.4E-08 | ||||||||
| OR-cond | 1.08 (1.05–1.11) | 1.08 (1.05–1.11) | 1.08 (1.05–1.11) | 1.08 (1.05–1.11) | 1.08 (1.05–1.11) | 1.08 (1.05–1.11) | 1.08 (1.05–1.11) | 1.07 (1.04–1.10) | |
| P-cond | 1.4E-07 | 1.5E-08 | 2.7E-08 | 1.8E-08 | 4.0E-08 | 3.2E-08 | 3.5E-08 | 1.0E-06 | |
| rs2035380 (IBS) BMAL1 |
OR | 1.07 (1.04–1.09) | |||||||
| P | 3.1E-08 | ||||||||
| OR-cond | 1.07 (1.04–1.10) | 1.07 (1.05–1.10) | 1.07 (1.05–1.10) | 1.07 (1.05–1.10) | 1.07 (1.04–1.10) | 1.07 (1.05–1.10) | 1.07 (1.05–1.10) | 1.07 (1.04–1.10) | |
| P-cond | 1.8E-07 | 2.8E-08 | 1.5E-08 | 3.0E-09 | 2.5E-08 | 1.4E-08 | 1.6E-08 | 1.5E-07 | |
| rs9517497 (IBS) DOCK9 |
OR | 1.07 (1.04–1.09) | |||||||
| P | 2.3E-08 | ||||||||
| OR-cond | 1.06 (1.04–1.09) | 1.07 (1.04–1.09) | 1.07 (1.04–1.09) | 1.07 (1.04–1.09) | 1.07 (1.04–1.09) | 1.07 (1.04–1.09) | 1.06 (1.04–1.09) | 1.07 (1.05–1.10) | |
| P-cond | 5.9E-07 | 2.7E-08 | 1.4E-08 | 3.6E-08 | 2.5E-08 | 2.6E-08 | 3.8E-08 | 9.5E-09 | |
NOTE. Conditional analyses used GWAS summary-level data on the use of CVD and antidepressant medications, obtained from Wu et al21 (PMID:31015401).
CoA, coenzyme A; CVD, cardiovascular diseases; GWAS, genome-wide association study; HMG, 3-hydroxy-3-methylglutaryl; IBS, irritable bowel syndrome; IBS-M, irritable bowel syndrome mixed subtype; OR, odds ratio (with 95% confidence interval) of the associated lead SNP for the discovery IBS and IBS-M GWAS; OR-cond, odds ratio (with 95% confidence interval) of the associated lead SNP after conditioning IBS/IBS-M GWAS results on medication use.
Rome III IBS Polygenic Risk Scores
Finally, we computed and tested Rome III IBS polygenic risk score (PRS), to assess the relevance and translational potential of our GWAS results in predicting IBS risk. For this purpose, in the absence of other large cohorts with Rome III and genotype data available, we used phenotype and genotype data from the independent set of UKBB participants who did not fill the digestive health questionnaire (DHQ nonresponders), and who were therefore not included in the Rome III IBS GWAS (Figure 1). In this dataset, IBS cases were identified based on hospital-inpatient records and self-reported diagnoses, whereas control subjects were the remainder of DHQ nonresponders (Methods). Two similarly sized nonoverlapping fractions (n = 146,771 and 146,772) of the DHQ nonresponder dataset were then used, respectively, for computation and testing of Rome III IBS PRS (see Table 9 for demographics).
Table 9.
Demographics of UKBB Study Cohorts Included in PRS Analyses
| Cohort | N | Age, mean (SD) | Sex, female (%) | |
|---|---|---|---|---|
| Subset 1 (PRS computation) | IBS | 5,335 | 57.0 (8.1) | 73.9 |
| Control subjects | 141,436 | 57.2 (8.2) | 52.2 | |
| Subset 2 (PRS testing) | IBS | 5,336 | 56.9 (8.1) | 74.8 |
| Control subjects | 141,436 | 57.2 (8.2) | 52.1 | |
NOTE. IBS cases and control subjects for PRS analyses were identified based on ICD10 K58 (code for IBS) and self-reported IBS diagnosis from touchscreen questionnaire (see Methods).
IBS, irritable bowel syndrome; ICD10, International Classification of Diseases, 10th revision; PRS, polygenic risk score; SD, standard deviation; UKBB, UK Biobank.
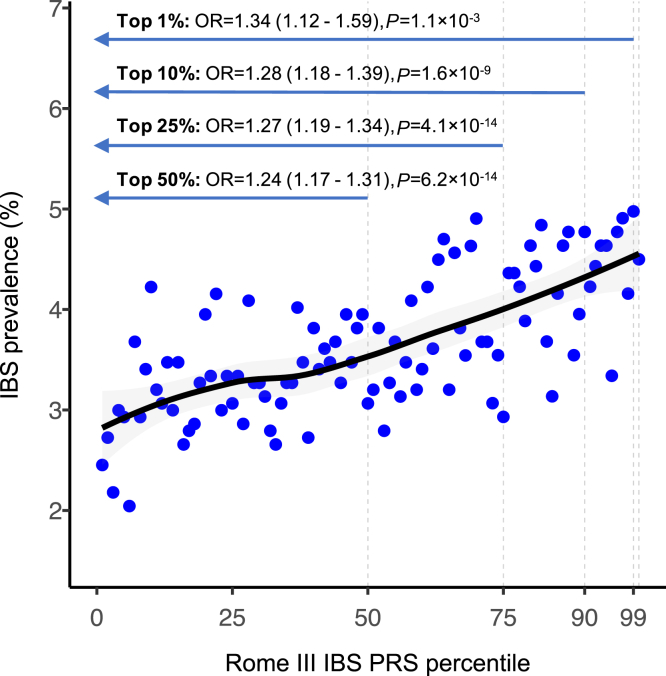
Using the PRSice-222 pipeline (Methods), we computed a PRS model based on the inclusion of 51,509 SNP markers that were best able to differentiate IBS cases (n = 5335) from control subjects (n = 141,436) in the first dataset (P = 6.0×10-13). Using this marker set in the testing group (5336 IBS cases and 141,436 control subjects), we detected significantly higher PRS values in IBS cases compared with control subjects (P = 7.9×10-22). Notably, IBS prevalence progressively increased across the PRS distribution, reaching 4.5% in the top 1% percentile (OR, 1.34; P = 1.1×10-3 vs the rest of the population) (Figure 9).
Figure 9.
Rome III IBS PRS testing analysis. Prevalence of IBS across PRS percentiles (polynomial regression fitting line in black), and OR relative to various upper percentiles compared with the rest of the population (estimated from logistic regression adjusted for sex, 10 top principal components, and genotyping array). The testing cohort consisted of IBS cases (n = 5336) and control subjects (n = 141,436) identified based on ICD10 K58 (for IBS) and self-reported IBS diagnosis.
Discussion
We report a large-scale, population-based survey of clinical and genetic data in relation to IBS and its subtypes according to the gold standard Rome III criteria. Based on GWAS meta-analyses of 2 independent cohorts comprising 101,884 individuals of European ancestry, we show that Rome III IBS and its subtypes are heritable traits, with SNP-based heritability ranging between 10% and 13%. This is higher than 5.8% SNP heritability reported in previous larger IBS GWAS meta-analyses of multiple heterogeneous IBS definitions,15 suggesting that standardized Rome III criteria might better capture the contribution of core common genetic risk factors to IBS. The incomplete overlap of GWAS associations signals detected here and in previous studies may also indicate that different definitions of IBS (ICD10 diagnoses from electronic medical records, self-reported doctor diagnoses, Rome criteria, or all combined) may capture slightly different genetic underpinnings.9 At the same time, the genetic architecture of Rome III IBS is largely shared across subtypes, including those at the opposite end of the stool consistency spectrum (as shown in genetic correlations of IBS-D and IBS-C). Rome III IBS also shows considerable genetic overlap with other GI, psychiatric, and extraintestinal pain-related conditions that, as shown in previous studies,23,24 are often comorbid as in our UKBB ICD10 analyses. Novel from this study is the observation of strong genetic correlation between IBS and heart-related conditions and traits, including family history of heart disease and hypertension, ischemic (coronary) heart disease, and angina pectoris, ranging between 20% and 45%.
This novel link between IBS and cardiovascular traits was further consolidated using 2 independent, different analytical approaches: GWAS devoid of individuals with heart diseases, and GWAS conditioned on CVD medications (as proxy for CVD) both provided evidence that the genetic risk effects observed for Rome III IBS are not dependent on heart-related comorbid conditions, nor on their genetic underpinnings. Showing correlation with all Rome III subtypes, the genetic (sibling history of) hypertension was most pronouncedly similar to that of IBS-C. Of note, similar observations, albeit lacking a genetic basis, have been recently made in large epidemiologic surveys showing that patients with IBS-C are exposed to a 2-fold increased risk of poor cardiovascular outcomes compared with healthy individuals.25, 26, 27 This was evident across a series of heart-related conditions including hypertension, venous thromboembolism, angina, peripheral artery disease, myocardial infarction, stroke, and arrhythmias. In relation to the latter, it is worth noting that genetic commonalities with IBS have also been described for the SCN5A gene (coding for the NaV1.5 sodium channel with a pacemaker role in myocytes and gut interstitial cells of Cajal), whose rare defective variants play a role both in Brugada syndrome and IBS-C.11,28 In addition, other cross-mechanisms may link IBS to heart-related traits, including alterations of the autonomic/sympathetic control of BP and cardiovascular activity, as observed in patients with IBS more severe symptoms.29 Therefore, similar to what has been previously shown for anxiety and mood disorders, pleiotropic mechanisms seem to be important also for the predisposition to IBS and CVDs, something that is highlighted here for the first time.
Evidence linking Rome III IBS to cardiovascular traits also came from the analysis of individual GWAS signals. The 2 novel IBS risk loci identified here, led by rs2035380 on chromosome 11 (for IBS) and rs2048419 on chromosome 8 (for IBS-M), had been previously linked to hypertension, diastolic/systolic BP, and coronary artery disease (as per results publicly available in PhenoScanner and GWAS Atlas).30, 31, 32, 33 Although multiple genes were mapped to the rs2048419 locus, only 1, BMAL1, is associated with rs2035380. BMAL1 (or ARNTL) codes for a core transcriptional factor that controls circadian rhythm and chronophysiologic body mechanisms, including digestive and cardiac functions.34,35 Of pharmacologic interest, BMAL1 regulators are targeted by antibiotic drugs used to treat IBS-D and traveler’s diarrhea (eg, rifaximin, norfloxacin, and ofloxacin).6,36 BMAL1-deficient mice show altered diurnal oscillations in gut microbiota composition associated with changes in the colonic concentration of short-chain fatty acids and bile acids, microbial metabolites known to be relevant to both GI motility and BP regulation.37, 38, 39 In line with this, functional studies demonstrated GI clock disruption is associated with abnormal colonic motility.40 Altogether, these findings suggest that similar pathogenetic mechanisms may contribute to Rome III IBS and heart disease via genetic disruption of the circadian system, warranting further investigations.
Interesting observations also came from the functional annotation of the other 3 GWAS risk loci. Association signals at rs6899057 on chromosome 5 and rs9517497 on chromosome 13 had been previously reported,15 and correspond to single gene annotations (respectively ADCY2 and DOCK9). In the context of findings from this study, it is noteworthy that ADCY2 (adenylyl cyclase isoform II) is the target of colforsin (NKH477), a compound that has smooth muscle relaxant and vasodilation properties with indication for acute heart failure.41,42 Moreover, ADCY2 mutations have been associated with congenital heart disease and abnormal calcium signaling.43 Of relevance to IBS, this gene acts downstream of G-protein coupled receptors, such as the muscarinic acetylcholine44 and serotonin (5-hydroxytryptamine)45 receptors, which play key roles in the autonomic control of GI motility46,47 and cardiovascular function,48,49 which are pathways targeted by current drugs used to treat IBS.6 Hence, AC2 may hold potential for IBS therapeutic exploitation.
At the novel rs2048419 IBS-M locus on chromosome 8 CLDN23 and MFHAS1 were prioritized among 5 candidate genes mapped to the region. CLDN23 encodes the nonclassical tight-junction claudin-23 protein that has been recently demonstrated, both in vitro and in vivo, to modulate the intestinal epithelial barrier.50 This occurs via interaction of claudin-23 with other claudin isoforms (particularly claudin-3 and -4) at the luminal surface of intestinal epithelial cells, leading to regulation of paracellular permeability to ions and macromolecules and strengthening of barrier function.50 Changes in the expression of other claudins have been documented in patients with IBS (especially IBS-D and IBS-M), and linked to severity of GI symptoms.51,52 Hence, it is possible that similar (still uncharacterized) genotype-drive mechanisms are responsible for the putative involvement of claudin-23 in IBS. MFHAS1, however, plays a role in the modulation of inflammatory responses via macrophage polarization and toll-like receptor signaling (particularly toll-like receptor 2 and toll-like receptor 4).53, 54, 55 The latter drives inflammation on recognition of microbial components (eg, lipopolysaccharide), and multiple studies report increased toll-like receptor expression in the intestinal mucosa of patients with IBS.56,57 The interplay between gut barrier disruption and immune overactivation is hypothesized to be linked to IBS progression via sensory afferent overstimulation, resulting in pain dysfunctions.51 Interestingly, CLDN23 and MFHAS1 are expressed in intestinal enteroendocrine cells, which are key sensors of the intestinal environment and play important roles in modulating gut-brain sensory and immune responses.58 Additional genetic and functional studies are needed to unequivocally establish weather CLDN23 or MFHAS1 or both contribute to IBS pathophysiology.
Rome III IBS GWAS data were also tested for their potential to contribute to IBS risk prediction, via PRS computation and testing. Despite the lack of accurate Rome III classifications in the independent UKBB dataset used for this purpose, our PRS model still allowed the identification of individuals at higher risk of IBS (self-reported or from hospital admissions), with those in the top 1% of PRS distribution being exposed to more than 1.3 times higher risk of disease. Similar results were recently obtained using stool frequency PRS to capture IBS in UKBB,14 which collectively indicates that, once refined and integrated with other predictive markers, IBS PRS may find translational scope for clinical utility: they may contribute to algorithms designed for differential diagnosing GI diseases (eg, IBS vs inflammatory bowel disease) and/or the decision-making process associated with costly and/or invasive procedures (endoscopy), with positive repercussions on the health care system and the patient.
Finally, although subtype-specific analyses did not reveal major differences, we observed IBS-C was the trait that most often departed from other subtypes in its genetic correlations with other conditions, suggesting specific genetic risk factors and mechanisms may be associated with constipation. Nevertheless, no GWAS significant signals were detected for Rome III IBS-C and IBS-D subtypes: it cannot be excluded that this is caused, for instance, by the current sample size that is insufficient (ie, smaller than IBS-M) to capture weak individual risk effects. Indeed, the measured heritability of these traits seems to be slightly lower than that of IBS-M. This issue can be better addressed in follow-up, possibly larger GWAS meta-analyses in independent cohorts, or via alternative gene-hunting strategies based on the analysis of endophenotypes, such as stool frequency and consistency.14
Important study limitations need to be acknowledged, including: (1) having studied IBS defined according to the older version of the Rome III Criteria, which warrants evaluation of current findings in patients defined according to stricter Rome IV (or possibly upcoming Rome V) criteria when suitable large cohorts with these data become available; (2) GWAS analyses not performed for ancestries other than European; (3) minimal understanding of the pleiotropic mechanisms linking IBS to cardiovascular traits; (4) sample size still likely inadequate to fully capture genetic liability for IBS subtypes; and (5) PRS testing performed on suboptimal IBS classifications because of the lack of other suitable Rome III IBS cohorts with genetic data available.
In summary, we show that IBS types defined according to Rome III criteria are heritable traits whose genetic architecture is shared with other polygenic traits, including CVDs highlighted here for the first time. We also identify novel candidate genes and plausible mechanisms that represent potential actionable targets warranting further investigation in follow-up studies.
Materials and Methods
The study design is summarized in Figure 1. Detailed methodologic description is provided below.
Study Cohorts and Definition of Rome III IBS Cases and Control Subjects
UK Biobank
The UKBB is a population-based longitudinal cohort of >500,000 UK individuals (aged 40–69 years) with available genotype, demographic, and health-related data.59 On exclusion of individuals of non-White ethnic background (data field 21000), sex mismatches (data fields 31 and 22001), and low-quality genotypes (data field 22010), we focused on data from the DHQ (which includes a Rome III module for IBS), available for a subset of UKBB participants who responded to the invitation to participate in this follow-up. After exclusion of all individuals with other potentially confounding GI diagnoses that could result in IBS-like symptoms, including celiac disease, inflammatory bowel disease (and others listed in Table 10), patients with IBS (N = 22,745 total; 12,335 IBS-M; 5920 IBS-D; 3855 IBS-C) were identified using Rome III DHQ questions (data fields 21025, 21027, 21028, 21031–21034). Asymptomatic control subjects (n = 66,631) were defined as the remainder of the population without abdominal pain/discomfort in the previous 3 months (data field 21025) and without IBS, based on IBS diagnoses from hospital inpatients records (ICD10 K58 code; data fields 41202 and 41204) and self-reported IBS from questionnaire data (data fields 21024 and 20002). The study was approved by the Monash University Institute Research Ethics Committee with protocol number 20326 and performed using UKBB data accessed under application number 17435.
Table 10.
Exclusion Criteria Applied for UKBB IBS Cases and Control Subjects
| Data-field | Trait | |
|---|---|---|
| ICD10 | 41202 and 41204 | K50 (Crohn's disease) |
| K51 (Ulcerative colitis) | ||
| K52 (Other and unspecified noninfective gastroenteritis and colitis) | ||
| K86 (Other diseases of pancreas) | ||
| K90 (Intestinal malabsorption) | ||
| K550 (Acute vascular disorders of intestine) | ||
| K551 (Chronic vascular disorders of intestine) | ||
| K627 (Radiation proctitis) | ||
| C15 (Malignant neoplasm of esophagus) | ||
| C16 (Malignant neoplasm of stomach) | ||
| C17 (Malignant neoplasm of small intestine) | ||
| C18 (Malignant neoplasm of colon) | ||
| C19 (Malignant neoplasm of rectosigmoid junction) | ||
| C20 (Malignant neoplasm of rectum) | ||
| C21 (Malignant neoplasm of anus and anal canal) | ||
| C22 (Malignant neoplasm of liver and intrahepatic bile ducts) | ||
| C23 (Malignant neoplasm of gallbladder) | ||
| C24 (Malignant neoplasm of other and unspecified parts of biliary tract) | ||
| C25 (Malignant neoplasm of pancreas) | ||
| C26 (Malignant neoplasm of other and ill-defined digestive organs) | ||
| K57 (Diverticular disease of intestine) | ||
| K58 (Irritable bowel syndrome)a | ||
| Self-report | 20002 | 1462 (Crohn’s disease) |
| 1463 (Ulcerative colitis) | ||
| 1459 (Colitis / not Crohn’s or ulcerative colitis) | ||
| 1461 (Inflammatory bowel disease) | ||
| 1164 (Pancreatic disease) | ||
| 1165 (Pancreatitis) | ||
| 1456 (Malabsorption/coeliac disease) | ||
| 1191 (Gastrointestinal bleeding) | ||
| 1509 (Gastroenteritis/dysentery) | ||
| 1600 (Bowel / intestinal perforation) | ||
| 1601 (Bowel / intestinal infarction) | ||
| 1602 (Bowel / intestinal obstruction) | ||
| 1135 (Stomach disorder) | ||
| 1458 (Diverticular disease / diverticulitis) | ||
| 1562 (Food intolerance) | ||
| 1154 (Irritable bowel syndrome)a | ||
| DHQ | 21068 | Positive answer for "have you been diagnosed with coeliac disease/gluten sensitivity?" |
| 21024 | Positive answer for "have you ever been diagnosed with IBS?" | |
| 21025 | Different answer than 'never' for "in the last 3 months, how often did you have discomfort or pain anywhere in your abdomen?"a | |
| 21025–21034 | Fulfilling Rome III criteria for irritable bowel syndromea | |
DHQ, Digestive Health Questionnaire; IBS, irritable bowel syndrome; ICD10, International Classification of Diseases,10th Edition; UKBB, UK Biobank.
Exclusion criterion applied to control subjects only.
Lifelines
LL is a multidisciplinary prospective population-based cohort study examining in a unique 3-generation design the health and health-related behaviors of 167,729 persons living in the North of the Netherlands.60 It uses a broad range of investigative procedures in assessing the biomedical, sociodemographic, behavioral, physical, and psychological factors that contribute to the health and disease of the general population, with a special focus on multimorbidity and complex genetics. In this study, we used health-related and genotype data for IBS GWAS analyses in an LL subset with ethical approval for genetic studies. Similar to UKBB, 1990 IBS cases (797 IBS-M, 561 IBS-D, and 475 IBS-C) and 10,518 asymptomatic control subjects were identified according to the Rome III criteria from digestive questionnaire data. The LL protocol was approved by the UMCG Medical ethical committee with protocol number 2007/152.
Comorbidity Analysis in UKBB
For comorbidity analysis, we analyzed 250 UKBB ICD10-related data-fields (from 41202-0.0/0.65 to 41204-0.0/0.183) across 89,376 individuals, totaling 2,234,400 data-points. We tested 1116 ICD10 traits, coded for diseases (chapters I to XIV) and laboratory findings (chapter XVIII), for their differential risk across Rome III IBS cases and asymptomatic control subjects. Associations for the number of recorded ICD10 comorbidities (excluding K58 for IBS) were tested in a linear regression model, adjusting for sex and age in R v3.6.2 (https://www.r-project.org/). ORs were calculated based on logistic regression models (also adjusted for age and sex) at 2 ICD10 levels: chapters (hierarchically subdivided into domains; see https://icd.who.int/browse10/2019/en#); and 3-digit code level (including only 96 conditions with ≥1% of prevalence in the DHQ subset of UKBB). To control for type I error, the Benjamini-Hochberg false discovery rate (FDR) method was applied in the calculation of statistical significance for multiple tests correction (corrected α=0.05).
Genotyping, Quality Control, and Genotype Imputation
Similar quality control pipelines and imputation methods were applied to both UKBB and LL cohorts, and are described as follows.
UK Biobank
Genome-wide genotyping of UKBB participants was carried out using custom Axiom (Affymetrix UK Biobank and UK BiLEVE) arrays. Nongenotyped variants were imputed centrally by UKBB researchers, using Haplotype Reference Consortium, UK10K, and 1000 Genomes (1KG) as reference panels. Genetic quality control procedures and PC analysis performed by UKBB researchers are thoroughly described by Bycroft et al.59 Quality control included checking for deviations from Hardy-Weinberg equilibrium, batch and plate effects, sex effects, and array effects across control replicates. Only SNPs with a call rate >0.99, imputation quality score INFO ≥0.9, minor allele frequency ≥0.01, nonindels, nonmultiallelic, and in autosomal chromosomes were tested in association analyses, adjusted for the top 10 PCs (data field 22009). Only individuals with White European ethnic background (categorized as “British,” “Irish,” or “Any other White background”; data field 21000) and without poor heterozygosity and missing rates (data field 22010) were included, leaving 22,745 Rome III IBS cases and 66,631 asymptomatic control subjects for GWAS analyses.
Lifelines
Genome-wide genotyping of LL participants was carried out within the UMCG Genotyping Lifelines Initiative. A total of 38,020 participants were genotyped using Infinium Global Screening Array Multi-ethnic Disease Version 1.0 (Illumina).60 Quality controls were based on checking Hardy-Weinberg equilibrium and call rate >0.95 using PLINK 1.9, sex and expected relationships from pedigree, and PC analysis to check for population outliers (as described in http://wiki-lifelines.web.rug.nl/doku.php?id=ugli). Before imputation, genotypes were prephased using SHAPEIT261 and aligned to the reference panels using Genotype Harmonizer62 to resolve strand issues. Imputation was performed with Beagle 3.1.063 and Minimac,64 based on whole-genome sequencing data from the Haplotype Reference Consortium reference panel. Only SNPs with INFO ≥0.9, minor allele frequency ≥0.01, nonindels, nonmultiallelic, and in autosomal chromosomes were tested in 1990 samples identified as IBS cases and 10,518 asymptomatic control subjects.
Rome III IBS GWAS and Meta-Analyses
Rome III IBS (and subtypes) GWAS analyses were performed with a logistic mixed model using SAIGE v0.42.165 adjusted for age, sex, 10 top PCs, and array (only for UKBB analyses). Individual IBS subtype GWAS summary statistics were inspected with the R package EasyQC v9.0,66 to check for data integrity, remove invalid or unmapped markers, and harmonize SNP identifiers (rs IDs) and allele strand coding across datasets. Markers with allele mismatches or with allele frequency deviating >0.2 compared with the Haplotype Reference Consortium reference panel were excluded. IBS-subtype specific meta-analyses were performed using a fixed-effect inverse-variance-based analysis with METAL v2011-03-25,67 including 6,311,313 markers in common between UKBB and LL individual GWAS. A Cochran Q test was performed to assess heterogeneity of effect-size estimates between individual GWAS, and markers with a P-Het <0.05 were excluded from subsequent subtype-specific analyses.
Bayesian Fine-Mapping Analysis
A Bayesian fine-mapping analysis was carried out using FINEMAP v1.468, to determine a minimum set of variants (credible set) containing likely causal variants for each associated risk locus. Posterior inclusion probability was estimated for each credible variant. As input, all genetic variants located within each risk locus were extracted, and the local linkage disequilibrium (LD) structure was calculated using genotypes from European UKBB individuals as reference.
Annotation of GWAS Risk Loci and Gene Mapping
For annotation of GWAS risk loci and gene mapping, we used the online tool FUMA v1.3.569 (https://fuma.ctglab.nl/tutorial). All loci at genome-wide significance (P ≤ 5×10-8) were annotated by FUMA, with lead SNPs being defined as those with the lowest P values in each genomic risk locus. Manhattan plots were produced using the R package qqman70 and Locuszoom71 was used for regional visualization of risk loci. Independent significant SNPs were identified based on LD with the lead SNPs (R2>0.6, using 1KG-EUR as reference panel) and P value (P ≤ 5×10-8). The maximum distance between LD blocks to merge into a locus was set to 250 kb. Gene mapping at each risk locus was based on position (distance <10 kb) and cis eQTL (pFDR < .05) analyses, excluding the major histocompatibility complex region, in view of its LD complexity. The analyzed eQTL database included 54 tissues from GTEx v872 (https://gtexportal.org/).
SNP Heritability and Genetic Correlation Analyses Via LDSC
SNP-based heritability (h2SNP) and genetic correlations (rg) with other 1400 traits were estimated using LDSC,73 as implemented in the CTG-VL74 platform. Traits with <1000 cases, <5000 overall participants, and duplicated were removed. P values were adjusted for multiple comparisons using the FDR method (corrected α=0.05).
Phenome-Wide Association Study
For each IBS risk locus, lead SNPs and/or their LD proxies (R2 >0.8) were used to screen for previously reported GWAS signals (P ≤ 5×10-8), inspecting data from OpenTargets,75 PhenoScanner v2,32 and GWAS ATLAS.33 Domains associated with individual GWAS findings were harmonized according to GWAS ATLAS classifications.
Prioritization of Candidate Genes
For the novel risk locus with multiple candidate genes (rs2048419), we attempted to identify those most likely to play a causative role based on functional evidence from fine-mapping (FINEMAP68), eQTL and gene expression (GTEx72 and Human Protein Atlas76), GWAS databases (OpenTargets,75 PhenoScanner v2,32 and GWAS ATLAS33), DrugBank (https://go.drugbank.com/), and literature mining. Genes were prioritized based on the following criteria: (1) considerable expression (set as log2 transcript per million ≥3) in GTEx tissues of interest (GI and/or brain), (2) credible SNP-eQTL colocalization in tissues of disorder of gut–brain interaction interest (GI and/or brain), (3) known association with relevant traits from previous GWAS studies, (4) plausible mechanistic link to IBS pathophysiology; and (5) relevant phenotype from knockout/knockdown models or human studies.
Rome III IBS Risk Loci Sensitivity Analysis of CVD Traits
Reevaluation of Rome III IBS loci association signals in UKBB devoid of heart-related conditions was obtained by removing from the analyses individuals with diagnoses from the ICD10 chapter IX diseases of the circulatory system. These included: I10 essential (primary) hypertension, I20 angina pectoris, I25 chronic ischemic heart disease, I44 atrioventricular and left bundle-branch block, I48 atrial fibrillation and flutter, I73 other peripheral vascular diseases, I83 varicose veins of lower extremities, I84 hemorrhoids, and I95 hypotension. As previously, association testing was carried out with SAIGE v0.42.1,65 adjusted for age, sex, 10 top PCs, and genotyping array. In total, 15,041 individuals with chapter IX diagnoses were removed from asymptomatic control subjects, whereas 5993 were removed from Rome III IBS cases (5993 from IBS, 3380 from IBS-M, 1475 from IBS-D, and 998 from IBS-C).
Multitrait Conditional Analysis
Multitrait-based Conditional and Joint analysis (mtCOJO 1.93.2 beta20) was used to generate Rome III IBS GWAS summary statistics (outcome) conditioned on GWAS data for the use of CVD and antidepressant medications (exposures). mtCOJO estimates the effect of the exposure on the outcome either by generalized summary-data-based mendelian randomization or from genetic correlation (rg) analysis, when there are not enough LD independent (r2 <0.05) genome-wide significant SNPs for the exposure or outcome traits (at least 10 required by default). mtCOJO analyses were carried out on IBS and IBS-M GWAS meta-analyses data versus GWAS of CVD medication traits derived from Wu et al,21 including: C01D vasodilators used in cardiac diseases, C02 antihypertensives, C03 diuretics, C07 beta blocking agents, C08 calcium channel blockers, C09 agents acting on the renin-angiotensin system, C10AA 3-hydroxy-3-methylglutaryl coenzyme A (HMG CoA) reductase inhibitors, and N06A antidepressants, according to the drug coding classification of the World Health Organization Anatomical Therapeutic Chemical Classification System.
Polygenic Risk Score Analysis
We adopted a 2-step approach to PRS computation and testing in independent subsets of UKBB, only studying participants who did not fill the DHQ questionnaire (DHQ nonresponders) and were, thus, not included in the original Rome III GWAS meta-analyses. In the absence of Rome III data for these individuals (Rome III modules are part of the DHQ questionnaire), IBS cases were identified based on hospital-inpatient records (ICD10 diagnosis K58; data fields 41202 and 41204) and self-reported doctors’ IBS diagnosis (1154 code; data field 20002), whereas control subjects were the remainder of QCed DHQ nonresponders. Cases and control subjects were randomly assigned to 2 independent and similarly sized groups for PRS computation (5335 cases and 141,436 control subjects) and testing (5336 cases and 141,436 control subjects).
For PRS computation, according to the standard LD pruning and P value thresholding of PRSice-2 v2.2.11.5022 pipeline, varying numbers of SNPs with GWAS P values ranging from 5×10-8 to 1 were tested using the Rome III IBS GWAS summary statistics as the base file. Using default PRSice-2 pruning parameters (clump-kb=250, clump-p=1, clump-r2= 0.1) and 1KG-EUR LD as the reference panel, a total of 214,641 LD pruned variants entered the analyses, after exclusion of poor-quality markers (genotype missingness geno <0.1 and imputation quality INFO<0.9) and rare markers (minor allele frequency <0.01). The best Rome IBS PRS model was derived in the computation dataset, by selecting the largest and most significant Nagelkerke R2 value obtained in the P value thresholding strategy (P(R2) = 6.0×10-13; GWAS P threshold = .1056), which included 51,509 SNPs.
For the testing group, PRSs were then calculated for IBS and control subjects as the sum of effect sizes of the 51,509 markers that fitted the best Rome IBS PRS model. PRS distributions were scaled (mean, 0; standard deviation, 1) using the “scale”’ R function. A multivariate linear regression (adjusted for sex, the first 10 top genetic PCs, and UKBB genotyping array) was used to determine the significance of the difference between the mean Rome IBS PRS in IBS cases and control subjects. Individuals within a given magnitude of increased PRS in top percentiles (binned using the “ntile” dplyr R function) were compared in relation to the rest of the population for their risk to IBS in a logistic regression including previous covariates, and FDR was applied in the calculation of statistical significance for multiple tests correction.
Acknowledgements
This research has been conducted using the UK Biobank Resource under application number 17435. The generation and management of genotype data for the Lifelines Cohort Study was supported by the UMCG Genetics Lifelines Initiative. We thank the UMCG Genomics Coordination Center and the UG Center for Information Technology and their sponsors (BBMRI-NL and TarGet) for storage and computational infrastructure for analyses of Lifelines data (project number: OV20_00051). We also thank the Monash University M3 High Performance Computing facility for the computational infrastructure. Esteban Alexander Lopera-Maya and Ferdinando Bonfiglio contributed equally.
CRediT Authorship Contributions
Leticia Camargo Tavares, MSc, PhD (Conceptualization: Supporting; Data curation: Lead; Formal analysis: Lead; Investigation: Lead; Methodology: Lead; Project administration: Lead; Resources: Supporting; Visualization: Lead; Writing – original draft: Lead; Writing – review & editing: Lead)
Esteban Alexander Lopera-Maya, PhD candidate (Data curation: Supporting; Formal analysis: Supporting; Methodology: Supporting; Writing – review & editing: Supporting)
Ferdinando Bonfiglio, PhD (Data curation: Supporting; Formal analysis: Supporting; Methodology: Supporting; Writing – review & editing: Supporting)
Tenghao Zheng, MD, PhD (Data curation: Supporting; Formal analysis: Supporting; Methodology: Supporting; Writing – review & editing: Supporting)
Trishla Sinha, MD (Writing – review & editing: Supporting)
Francine Marques, PhD (Conceptualization: Supporting; Writing – review & editing: Supporting)
Alexandra Zhernakova, PhD (Conceptualization: Supporting; Resources: Supporting; Writing – review & editing: Supporting)
Serena Sanna, PhD (Conceptualization: Supporting; Resources: Supporting; Writing – review & editing: Supporting)
Mauro D'Amato, PhD (Conceptualization: Lead; Funding acquisition: Lead; Investigation: Lead; Methodology: Supporting; Project administration: Lead; Resources: Lead; Supervision: Lead; Visualization: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting)
Footnotes
Conflicts of interest This author discloses the following: Mauro D’Amato received unrestricted research grants and consulting fees from QOL Medical. The remaining authors disclose no conflicts.
Funding Leticia Camargo Tavares is supported by an Australian Government Research Training (RTP) Scholarship. Esteban Alexander Lopera-Maya is supported by the Colombian Ministry of Sciences External Fellowship (786). Francine Zanchetta Marques is supported by a Senior Medical Research Fellowship from the Sylvia and Charles Viertel Charitable Foundation Fellowship, a National Heart Foundation Future Leader Fellowship (105663), and a National Health and Medical Research Council Emerging Leader Fellowship (GNT2017382). Alexandra Zhernakova is supported by the ERC Starting Grant (715772), Netherlands Organization for Scientific Research (NWO-VIDI) Grant (016.178.056), the Netherlands Heart Foundation CVON Grant (2018-27), and the NWO Gravitation Grant Exposome (NL 024.004.017). Serena Sanna is supported by ERC Starting Grant (101075624), and by the National Recovery and Resilience Plan PNRR grant (PE00000015/Age-it), funded by the Next-Generation EU program. Mauro D’Amato and Serena Sanna are supported by funds from the Italian Ministry of University and Research PRIN 2022 (2022PMZKEC; CUP E53D23004910008 to Mauro D’Amato and CUPB53D23008300006 to Serena Sanna), funded by the Next-Generation EU program. The Lifelines initiative has been made possible by subsidy from the Dutch Ministry of Health, Welfare and Sport, the Dutch Ministry of Economic Affairs, the University Medical Center Groningen, Groningen University and the Provinces in the North of the Netherlands (Drenthe, Friesland, Groningen).
Note: To access the supplementary material accompanying this article, go to the full text version at http://doi.org/10.1016/j.jcmgh.2024.04.002
Supplementary Material
References
- 1.Ford A.C., Sperber A.D., Corsetti M., et al. Irritable bowel syndrome. Lancet. 2020;396:1675–1688. doi: 10.1016/S0140-6736(20)31548-8. [DOI] [PubMed] [Google Scholar]
- 2.Sperber A.D., Bangdiwala S.I., Drossman D.A., et al. Worldwide prevalence and burden of functional gastrointestinal disorders, results of Rome Foundation Global Study. Gastroenterology. 2021;160:99–114.e3. doi: 10.1053/j.gastro.2020.04.014. [DOI] [PubMed] [Google Scholar]
- 3.Lacy B.E., Patel N.K. Rome criteria and a diagnostic approach to irritable bowel syndrome. J Clin Med. 2017;6:99. doi: 10.3390/jcm6110099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aziz I., Törnblom H., Palsson O.S., et al. How the change in IBS criteria from Rome III to Rome IV impacts on clinical characteristics and key pathophysiological factors. Am J Gastroenterol. 2018;113:1017–1025. doi: 10.1038/s41395-018-0074-z. [DOI] [PubMed] [Google Scholar]
- 5.Vork L., Weerts Z.Z.R.M., Mujagic Z., et al. Rome III vs Rome IV criteria for irritable bowel syndrome: a comparison of clinical characteristics in a large cohort study. Neurogastroenterol Motil. 2018;30 doi: 10.1111/nmo.13189. [DOI] [PubMed] [Google Scholar]
- 6.Camilleri M., Boeckxstaens G. Irritable bowel syndrome: treatment based on pathophysiology and biomarkers. Gut. 2023;72:590–599. doi: 10.1136/gutjnl-2022-328515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saito Y.A. The role of genetics in IBS. Gastroenterol Clin North Am. 2011;40:45–67. doi: 10.1016/j.gtc.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morris-Yates A., Talley N.J., Boyce P.M., et al. Evidence of a genetic contribution to functional bowel disorder. Am J Gastroenterol. 1998;93:1311–1317. doi: 10.1111/j.1572-0241.1998.440_j.x. [DOI] [PubMed] [Google Scholar]
- 9.Camilleri M., Zhernakova A., Bozzarelli I., et al. Genetics of irritable bowel syndrome: shifting gear via biobank-scale studies. Nat Rev Gastroenterol Hepatol. 2022;19:689–702. doi: 10.1038/s41575-022-00662-2. [DOI] [PubMed] [Google Scholar]
- 10.Kapeller J., Houghton L.A., Mönnikes H., et al. First evidence for an association of a functional variant in the microRNA-510 target site of the serotonin receptor-type 3E gene with diarrhea predominant irritable bowel syndrome. Hum Mol Genet. 2008;17:2967–2977. doi: 10.1093/hmg/ddn195. [DOI] [PubMed] [Google Scholar]
- 11.Beyder A., Mazzone A., Strege P.R., et al. Loss-of-function of the voltage-gated sodium channel NaV1.5 (channelopathies) in patients with irritable bowel syndrome. Gastroenterology. 2014;146:1659–1668. doi: 10.1053/j.gastro.2014.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henström M., Hadizadeh F., Beyder A., et al. Polymorphisms associated with increased risk of IBS-C and IBS-M. Gut. 2017;66:1725–1727. doi: 10.1136/gutjnl-2016-313346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng T., Camargo-Tavares L., Bonfiglio F., et al. Rare hypomorphic sucrase isomaltase variants in relation to irritable bowel syndrome risk in UK Biobank. Gastroenterology. 2021;161:1712–1714. doi: 10.1053/j.gastro.2021.06.063. [DOI] [PubMed] [Google Scholar]
- 14.Bonfiglio F., Liu X., Smillie C., et al. GWAS of stool frequency provides insights into gastrointestinal motility and irritable bowel syndrome. Cell Genom. 2021;1 doi: 10.1016/j.xgen.2021.100069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eijsbouts C., Zheng T., Kennedy N.A., et al. Genome-wide analysis of 53,400 people with irritable bowel syndrome highlights shared genetic pathways with mood and anxiety disorders. Nat Genet. 2021;53:1543–1552. doi: 10.1038/s41588-021-00950-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bonfiglio F., Zheng T., Garcia-Etxebarria K., et al. Female-specific association between variants on chromosome 9 and self-reported diagnosis of irritable bowel syndrome. Gastroenterology. 2018;155:168–179. doi: 10.1053/j.gastro.2018.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu Y., Murray G.K., Byrne E.M., et al. GWAS of peptic ulcer disease implicates Helicobacter pylori infection, other gastrointestinal disorders and depression. Nat Commun. 2021;12:1146. doi: 10.1038/s41467-021-21280-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oka P., Parr H., Barberio B., et al. Global prevalence of irritable bowel syndrome according to Rome III or IV criteria: a systematic review and meta-analysis. Lancet Gastroenterol Hepatol. 2020;5:908–917. doi: 10.1016/S2468-1253(20)30217-X. [DOI] [PubMed] [Google Scholar]
- 19.Johansen S.G., Ness-Jensen E. The changes in prevalence and risk of irritable bowel syndrome over time in a population-based cohort, the HUNT study, Norway. Scand J Gastroenterol. 2022:1–7. doi: 10.1080/00365521.2022.2028005. [DOI] [PubMed] [Google Scholar]
- 20.Zhu Z., Zheng Z., Zhang F., et al. Causal associations between risk factors and common diseases inferred from GWAS summary data. Nat Commun. 2018;9:224. doi: 10.1038/s41467-017-02317-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu Y., Byrne E.M., Zheng Z., et al. Genome-wide association study of medication-use and associated disease in the UK Biobank. Nat Commun. 2019;10:1891. doi: 10.1038/s41467-019-09572-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choi S.W., O'Reilly P.F. PRSice-2: Polygenic Risk Score software for biobank-scale data. Gigascience. 2019;8 doi: 10.1093/gigascience/giz082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Whitehead W.E., Palsson O., Jones K.R. Systematic review of the comorbidity of irritable bowel syndrome with other disorders: what are the causes and implications? Gastroenterology. 2002;122:1140–1156. doi: 10.1053/gast.2002.32392. [DOI] [PubMed] [Google Scholar]
- 24.Seeling K.S., Hehl L., Vell M.S., et al. Comorbidities, biomarkers and cause specific mortality in patients with irritable bowel syndrome: a phenome-wide association study. United European Gastroenterol J. 2023;11:458–470. doi: 10.1002/ueg2.12397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Judkins C.P., Wang Y., Jelinic M., et al. Association of constipation with increased risk of hypertension and cardiovascular events in elderly Australian patients. Sci Rep. 2023;13 doi: 10.1038/s41598-023-38068-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sundbøll J., Szépligeti S.K., Adelborg K., et al. Constipation and risk of cardiovascular diseases: a Danish population-based matched cohort study. BMJ Open. 2020;10 doi: 10.1136/bmjopen-2020-037080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen J., Sun Y., Fu T., et al. Risk of incident cardiovascular disease among patients with gastrointestinal disorder: prospective cohort study of 340,862 individuals. medRxiv. 2023 doi: 10.1093/ehjqcco/qcad059. 2023.04.09.23288329. [DOI] [PubMed] [Google Scholar]
- 28.Strege P.R., Mazzone A., Bernard C.E., et al. Irritable bowel syndrome patients have SCN5A channelopathies that lead to decreased NaV1.5 current and mechanosensitivity. Am J Physiol Gastrointest Liver Physiol. 2018;314:G494–G503. doi: 10.1152/ajpgi.00016.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davydov D.M., Naliboff B., Shahabi L., et al. Asymmetries in reciprocal baroreflex mechanisms and chronic pain severity: focusing on irritable bowel syndrome. Neurogastroenterol Motil. 2018;30 doi: 10.1111/nmo.13186. [DOI] [PubMed] [Google Scholar]
- 30.Zhu Z., Wang X., Li X., et al. Genetic overlap of chronic obstructive pulmonary disease and cardiovascular disease-related traits: a large-scale genome-wide cross-trait analysis. Respir Res. 2019;20:64. doi: 10.1186/s12931-019-1036-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van der Harst P., Verweij N. Identification of 64 novel genetic loci provides an expanded view on the genetic architecture of coronary artery disease. Circ Res. 2018;122:433–443. doi: 10.1161/CIRCRESAHA.117.312086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kamat M.A., Blackshaw J.A., Young R., et al. PhenoScanner V2: an expanded tool for searching human genotype-phenotype associations. Bioinformatics. 2019;35:4851–4853. doi: 10.1093/bioinformatics/btz469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Watanabe K., Stringer S., Frei O., et al. A global overview of pleiotropy and genetic architecture in complex traits. Nat Genet. 2019;51:1339–1348. doi: 10.1038/s41588-019-0481-0. [DOI] [PubMed] [Google Scholar]
- 34.Segers A., Depoortere I. Circadian clocks in the digestive system. Nat Rev Gastroenterol Hepatol. 2021;18:239–251. doi: 10.1038/s41575-020-00401-5. [DOI] [PubMed] [Google Scholar]
- 35.Hayter E.A., Wehrens S.M.T., Van Dongen H.P.A., et al. Distinct circadian mechanisms govern cardiac rhythms and susceptibility to arrhythmia. Nat Commun. 2021;12:2472. doi: 10.1038/s41467-021-22788-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Diemert D.J. Prevention and self-treatment of traveler's diarrhea. Clin Microbiol Rev. 2006;19:583–594. doi: 10.1128/CMR.00052-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heddes M., Altaha B., Niu Y., et al. The intestinal clock drives the microbiome to maintain gastrointestinal homeostasis. Nat Commun. 2022;13:6068. doi: 10.1038/s41467-022-33609-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O'Donnell J.A., Zheng T., Meric G., et al. The gut microbiome and hypertension. Nat Rev Nephrol. 2023;19:153–167. doi: 10.1038/s41581-022-00654-0. [DOI] [PubMed] [Google Scholar]
- 39.Zheng Z., Tang J., Hu Y., et al. Role of gut microbiota-derived signals in the regulation of gastrointestinal motility. Front Med (Lausanne) 2022;9 doi: 10.3389/fmed.2022.961703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hibberd T.J., Ramsay S., Spencer-Merris P., et al. Circadian rhythms in colonic function. Front Physiol. 2023;14 doi: 10.3389/fphys.2023.1239278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hosono M. Cardiovascular effects of colforsin daropate hydrochloride, a novel drug for the treatment of acute heart failure. Nihon Yakurigaku Zasshi. 1999;114:83–88. doi: 10.1254/fpj.114.83. [DOI] [PubMed] [Google Scholar]
- 42.Shafiq J., Suzuki S., Itoh T., et al. Mechanisms of vasodilation induced by NKH477, a water-soluble forskolin derivative, in smooth muscle of the porcine coronary artery. Circ Res. 1992;71:70–81. doi: 10.1161/01.res.71.1.70. [DOI] [PubMed] [Google Scholar]
- 43.Izarzugaza J.M.G., Ellesøe S.G., Doganli C., et al. Systems genetics analysis identifies calcium-signaling defects as novel cause of congenital heart disease. Genome Med. 2020;12:76. doi: 10.1186/s13073-020-00772-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shen J.X., Wachten S., Halls M.L., et al. Muscarinic receptors stimulate AC2 by novel phosphorylation sites, whereas Gβγ subunits exert opposing effects depending on the G-protein source. Biochem J. 2012;447:393–405. doi: 10.1042/BJ20120279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nawoschik S.P., Olsen M., Smith D.L., et al. Stable expression of adenylyl cyclase 2 leads to the functional rescue of human 5-HT6 receptor in a CHODUKX cell line. J Pharmacol Toxicol Methods. 2007;55:323–331. doi: 10.1016/j.vascn.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 46.Tanahashi Y., Komori S., Matsuyama H., et al. Functions of muscarinic receptor subtypes in gastrointestinal smooth muscle: a review of studies with receptor-knockout mice. Int J Mol Sci. 2021;22:926. doi: 10.3390/ijms22020926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mawe G.M., Hoffman J.M. Serotonin signalling in the gut: functions, dysfunctions and therapeutic targets. Nat Rev Gastroenterol Hepatol. 2013;10:473–486. doi: 10.1038/nrgastro.2013.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harvey R.D. Muscarinic receptor agonists and antagonists: effects on cardiovascular function. Handb Exp Pharmacol. 2012:299–316. doi: 10.1007/978-3-642-23274-9_13. [DOI] [PubMed] [Google Scholar]
- 49.Côté F., Fligny C., Fromes Y., et al. Recent advances in understanding serotonin regulation of cardiovascular function. Trends Mol Med. 2004;10:232–238. doi: 10.1016/j.molmed.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 50.Raya-Sandino A., Lozada-Soto K.M., Rajagopal N., et al. Claudin-23 reshapes epithelial tight junction architecture to regulate barrier function. Nat Commun. 2023;14:6214. doi: 10.1038/s41467-023-41999-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barbara G., Barbaro M.R., Fuschi D., et al. Inflammatory and microbiota-related regulation of the intestinal epithelial barrier. Front Nutr. 2021;8 doi: 10.3389/fnut.2021.718356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hanning N., Edwinson A.L., Ceuleers H., et al. Intestinal barrier dysfunction in irritable bowel syndrome: a systematic review. Therap Adv Gastroenterol. 2021;14 doi: 10.1177/1756284821993586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shi Q., Xiong B., Zhong J., et al. MFHAS1 suppresses TLR4 signaling pathway via induction of PP2A C subunit cytoplasm translocation and inhibition of c-Jun dephosphorylation at Thr239. Mol Immunol. 2017;88:79–88. doi: 10.1016/j.molimm.2017.06.017. [DOI] [PubMed] [Google Scholar]
- 54.Zhong J., Shi Q.Q., Zhu M.M., et al. MFHAS1 is associated with sepsis and stimulates TLR2/NF-κB signaling pathway following negative regulation. PLoS One. 2015;10 doi: 10.1371/journal.pone.0143662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen W., Xu Y., Zhong J., et al. MFHAS1 promotes colorectal cancer progress by regulating polarization of tumor-associated macrophages via STAT6 signaling pathway. Oncotarget. 2016;7:78726–78735. doi: 10.18632/oncotarget.12807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dlugosz A., Zakikhany K., Acevedo N., et al. Increased expression of toll-like receptors 4, 5, and 9 in small bowel mucosa from patients with irritable bowel syndrome. Biomed Res Int. 2017;2017 doi: 10.1155/2017/9624702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Belmonte L., Beutheu Youmba S., et al. Role of toll like receptors in irritable bowel syndrome: differential mucosal immune activation according to the disease subtype. PLoS One. 2012;7 doi: 10.1371/journal.pone.0042777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Worthington J.J., Reimann F., Gribble F.M. Enteroendocrine cells-sensory sentinels of the intestinal environment and orchestrators of mucosal immunity. Mucosal Immunol. 2018;11:3–20. doi: 10.1038/mi.2017.73. [DOI] [PubMed] [Google Scholar]
- 59.Bycroft C., Freeman C., Petkova D., et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sijtsma A., Rienks J., van der Harst P., et al. Cohort profile update: Lifelines, a three-generation cohort study and biobank. Int J Epidemiol. 2022;51:e295–e302. doi: 10.1093/ije/dyab257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Delaneau O., Marchini J., Zagury J.F. A linear complexity phasing method for thousands of genomes. Nat Methods. 2011;9:179–181. doi: 10.1038/nmeth.1785. [DOI] [PubMed] [Google Scholar]
- 62.Deelen P., Bonder M.J., van der Velde K.J., et al. Genotype harmonizer: automatic strand alignment and format conversion for genotype data integration. BMC Res Notes. 2014;7:901. doi: 10.1186/1756-0500-7-901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Browning B.L., Browning S.R. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am J Hum Genet. 2009;84:210–223. doi: 10.1016/j.ajhg.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Howie B., Fuchsberger C., Stephens M., et al. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44:955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhou W., Nielsen J.B., Fritsche L.G., et al. Efficiently controlling for case-control imbalance and sample relatedness in large-scale genetic association studies. Nat Genet. 2018;50:1335–1341. doi: 10.1038/s41588-018-0184-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Winkler T.W., Day F.R., Croteau-Chonka D.C., et al. Quality control and conduct of genome-wide association meta-analyses. Nat Protoc. 2014;9:1192–1212. doi: 10.1038/nprot.2014.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Willer C.J., Li Y., Abecasis G.R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Benner C., Spencer C.C., Havulinna A.S., et al. FINEMAP: efficient variable selection using summary data from genome-wide association studies. Bioinformatics. 2016;32:1493–1501. doi: 10.1093/bioinformatics/btw018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Watanabe K., Taskesen E., van Bochoven A., et al. Functional mapping and annotation of genetic associations with FUMA. Nat Commun. 2017;8:1826. doi: 10.1038/s41467-017-01261-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Turner S.D. qqman: an R package for visualizing GWAS results using Q-Q and manhattan plots. bioRxiv. 2014 [Google Scholar]
- 71.Pruim R.J., Welch R.P., Sanna S., et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Consortium G. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science. 2020;369:1318–1330. doi: 10.1126/science.aaz1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bulik-Sullivan B.K., Loh P.R., Finucane H.K., et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47:291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cuéllar-Partida G., Lundberg M., Kho P.F., et al. Complex-Traits Genetics Virtual Lab: a community-driven web platform for post-GWAS analyses. bioRxiv. 2019 [Google Scholar]
- 75.Carvalho-Silva D., Pierleoni A., Pignatelli M., et al. Open Targets Platform: new developments and updates two years on. Nucleic Acids Res. 2019;47:D1056–D1065. doi: 10.1093/nar/gky1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Karlsson M., Zhang C., Méar L., et al. A single-cell type transcriptomics map of human tissues. Sci Adv. 2021;7 doi: 10.1126/sciadv.abh2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.