Abstract
The transcription of two early “leftwardly” expressed genes carrying repetitive sequences, IR2 and IR4, has been studied for Epstein-Barr virus-associated tumors, and for established B-cell lines, using sequence-specific probes generated for this purpose. Whereas the IR4 transcript was identified in every tumor and cell line assessed (except B95-8, with a deletion that removes the gene), expression of the IR2 gene was restricted to B lymphocytes. Though the promoters for both transcripts lie within homologous regions (DL and DR) in the viral genome, the IR2 promoter appears more tightly regulated. Detailed characterization of the IR4 transcript from a nasopharyngeal carcinoma tumor, C15, identifies a sequence variant of this gene that differs from those reported for B cells; in situ hybridization methods show transcription to be restricted to a subset of cells, with the strongest signals seen adjacent to host stroma. As with B cells in culture (Y. Gao, P. R. Smith, L. Karran, Q. L. Lu, and B. E. Griffin, J. Virol. 71:84–94, 1997), chemical induction enhanced transcriptional expression of the IR4 gene in the C15 tumor, although staining for both the IR4 antigen and that of the virus lytic switch, Zta, gave negative results. In a Burkitt's lymphoma biopsy specimen, however, both proteins were found expressed, notably in the same subset of cells. The data here and elsewhere (Gao et al., J. Virol., 1997) are consistent with a block to intracellular transport of the transcript(s) and suggest nuclear roles for it in tumors, possibly in RNA processing and viral lytic replication. Both roles could be fulfilled in the absence of translation.
The human herpesvirus Epstein-Barr virus (EBV), the etiologic agent of infectious mononucleosis, is associated in high frequency with several human malignancies, including the fast-growing B-cell malignancy Burkitt's lymphoma (BL) and the undifferentiated form of the epithelial tumor nasopharyngeal carcinoma (NPC). In more recent years, an EBV association has been identified with other hematological malignancies, including Hodgkin's disease and T-cell lymphoma, as well as with numerous lymphoepitheliomas, including gastric carcinoma (as reviewed in reference 1), and also with some cases of breast cancer (4, 24). The viral genome is a double-stranded DNA molecule ranging from 172 kbp in B95-8 cells (3) to even larger sizes in other B-cell lines (22). It contains several major internal repeats, designated IR1 to IR4, interspersed throughout the genome and a terminal repeat located at the ends of virion DNA or internally in episomal forms of the genome. The size of the genome is largely determined by copy numbers of these repeats (Fig. 1). In some BL-derived lines that have not been continuously passaged in culture, the viral DNA does not appear to be uniform in size (22), whereas in established and frequently passaged lines, a single-sized molecule appears to predominate (28). The same may be true for NPC (36).
FIG. 1.
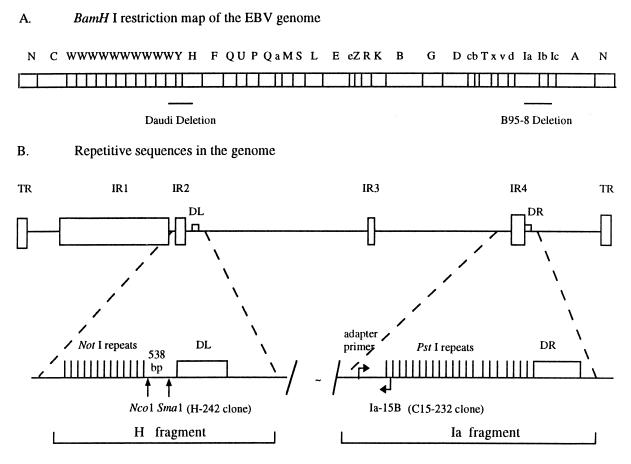
Schematic diagrams showing the BamHI restriction map of the EBV genome and locations of the deletions in Daudi and B95-8 cells (A) and the IR2 (NotI) and IR4 (PstI) and direct repeats, DL and DR, relative to other repetitive sequences in the EBV genome (B). Most viral strains contain genes associated with both IR2 and IR4, but Daudi cells have a deletion that removes the former (19), whereas the B95-8 genome lacks the latter (3), as noted. These two genes are separated by about 100 kbp of DNA. The locations of the clones generated specifically to recognize either IR2 (H-242) or IR4 (C15-232) transcripts are shown. TR, terminal repeat.
All the repetitive sequences are expressed as polyadenylated mRNAs and, under appropriate conditions, as proteins. The gene for the only nuclear antigen, EBNA1, consistently expressed in many tumors encompasses IR3, and the repetitive sequence has been identified as at least partially responsible for the ability of its protein to escape immune surveillance in the host (26). The protein products expressed from genes containing IR1, IR3, and the terminal repeat have been designated as “latent” functions, whereas IR2 and IR4 are thought to be components of early genes associated with the viral life cycle (29–33, 37). The latter are adjacent, but not identically so, to the duplicated sequences, DL and DR (Fig. 1), both of which are present in most but not all viral genomes. The IR2 (125-bp) repeat sequence can be excised from the BamHI H EBV DNA fragment (2) by digestion with the NotI restriction enzyme, and the IR4 (102-bp) repeat can be excised from BamHI Ia using PstI (13). Both regions appear to be associated with lytic viral replication (16, 17).
Our interest, particularly in IR4, was stimulated by finding its transcript in an NPC (18). The major transcription products in NPC are a family of related, highly spliced polyadenylated RNAs, transcribed over a 25-kbp region of the viral genome (39). They are expressed from the DNA strand complementary to that encoding a number of previously recognized genes associated with the viral life cycle (3). A quantitative picture of EBV-containing RNAs in a comprehensive cDNA library made from an NPC xenograft, C15 (6), shows these so-called complementary-strand transcripts (CSTs) to account for >90% of all viral transcripts in this tumor (18). The promoter and splice sites in these RNAs were mapped within the BamHI I and A regions on the conventional map of EBV DNA (20, 39). Notably, the major intron within the CST coding region (about 10 kbp in size), from the BamHI Ia region, contains the IR4 repeat (18). In several NPCs, BLs, and BL-derived cell lines, an IR4-containing RNA was previously identified (7, 15). In the cDNA library from the C15 NPC tumor, the CST major intron region (39) also appeared to be transcriptionally active (18). We thus set out to characterize this transcript, being particularly interested in determining whether, in addition to genes associated with latency (37), an early gene transcript might generally be expressed in EBV-associated tumors and, if so, in the longer term, seeking its function.
In this study, the expression patterns of the IR4 gene have been extensively analyzed in NPCs of North African and Chinese origins and in African BL biopsy specimens, and results have been compared with data from BL-derived cell lines passaged in culture. Since this gene shares considerable sequence homology with IR2 and the promoters for both genes contain viral lytic origins (oriLyt) of replication, the study was partially extended to cover IR2. Differences in patterns were observed, with expression from the epithelial cell-derived tumors being more restricted than that from B cells. These data and their possible significance are discussed, alongside data from in situ hybridization experiments and immunostaining for IR4 proteins.
MATERIALS AND METHODS
Tumor materials.
Three North African NPC tumors, C15, C17, and C18 (6), were gifts from P. Busson and T. Tursz (Institute Gustav Roissy, Villejuif, France) and were maintained by propagation in nude mice. Primary Hong Kong NPC biopsy materials were provided by M. H. Ng (Department of Microbiology) and J. Nichols (Department of Pathology), Hong Kong University, Queen Mary Hospital, Hong Kong. They are designated NPC 1 to 7. Five primary biopsy specimens (SD, IB, LC, TJ, and GE), clinically identified as BLs, obtained from R. Broadhead, E. Molyneux, and E. Borgstein, Blantyre, Malawi, were further characterized by histopathology and shown by us to be EBV positive by Southern blot analysis. A sixth biopsy specimen (CC), not a BL, was EBV negative and was used as a control.
Cell lines.
B95-8 is the prototype EBV-immortalized marmoset cell line whose primary DNA sequence has been determined elsewhere (3). M-ABA is a marmoset lymphoblastoid line established with virus from an NPC patient. Raji, Daudi, and P3HR-1 are all EBV-positive BL-derived cell lines. Ramos and HEp-2C are EBV-negative BLs and epithelial cell lines, respectively. B cells were cultured in suspension in RPMI medium under standard conditions. Epithelial monolayer cell lines were maintained in Dulbecco's minimal essential medium (Gibco-BRL). B-cell lines were induced to activate gene expression in complete medium by adding 12-O-tetradecanoylphorbol-13-acetate (TPA; 20 ng/ml) and sodium n-butyrate (3 mM). Actively dividing cells were treated at densities of 5 × 105 cells per ml and harvested 3 days later.
Induction in the C15 xenograft.
TPA and sodium n-butyrate were dissolved in phosphate-buffered saline (PBS) to final concentrations of 30 μg/ml and 1.5 M, respectively. Induction was carried out by injecting 150 μl of this solution on two occasions (over a week) into C15 tumors, about 10 by 10 mm in size, in nude mice. Animals were sacrificed a week later. As a control, a xenograft was injected with PBS only.
cDNA isolation and analysis.
The construction of an oligo(dT)-primed λgt10 library from twice-selected C15 mRNA has been described elsewhere (18). Probes used for hybridizing the library were from B95-8 BamHI H and Raji BamHI Ia fragments of the EBV genome. DNA isolated from recombinant phage was subcloned into an EcoRI-digested Bluescribe cloning vector (Stratagene) and subjected to restriction enzyme analysis and DNA sequencing.
RNA isolation and Northern blotting.
Total RNA was isolated from tumors, cell lines, and fresh biopsy specimens by the guanidinium-cesium chloride method. Polyadenylated RNA was selected on oligo(dT) mRNA purification columns (Pharmacia). In general, mRNA (6 μg, as measured by A260) was electrophoretically separated on 1% agarose gels and transferred to membranes, as described earlier (18). Northern blots were hybridized with 32P-labeled DNA fragments as described elsewhere (20), using probes given below.
Reverse transcription-PCR (RT-PCR) analysis. (i) DNase treatment.
Polyadenylated [poly(A)+] RNA (1 μg) or total RNA (5 μg) in 10 mM Tris-HCl (pH 7.5)–10 mM MgCl2–50 mM NaCl–1 mM dithiothreitol was treated with 10 U of DNase I (Boehringer). After being mixed well, the reaction mixture was incubated at 37°C for 30 min and then extracted twice with phenol-chloroform, and products were precipitated with ethanol in the presence of 0.3 M NaOAc. To control for enzymatic digestion, 0.1 μg of DNA was similarly treated and subjected to PCR amplification (see below).
(ii) cDNA synthesis.
DNase I-treated RNAs [1 μg of poly(A)+ RNA or 5 μg of total RNA] were used to synthesize the first-strand cDNA using either gene-specific or random primers, as described previously (12). The synthesized cDNA was diluted in 10 mM Tris-HCl (pH 8.0)–0.1 mM EDTA (100 μl).
(iii) PCR.
Amplification was performed in a reaction mixture consisting of 75 mM Tris-HCl (pH 8.8), 20 mM (NH4)2SO4, 0.01% (vol/vol) Tween 20, 1.5 mM MgCl2, 0.16 mM (each) deoxynucleoside triphosphate, and 30 pmol of each primer; 2 U of Vent DNA polymerase (New England Biolabs) and 5 μl of the first-strand cDNA were added to the PCR mix, and 50 μl of mineral oil was layered above the reaction mixture (50 μl). For the reaction, the initial denaturing time was 5 min, and a program of denaturation at 95°C for 40 s, annealing at 62°C for 30 s, and extension at 72°C for 1 min was followed through 39 cycles. PCR products were separated by electrophoresis on 1% agarose gels containing ethidium bromide, and their identities were verified by Southern blot hybridization.
RACE analysis.
To confirm the position of the 3′ end of the IR4 transcript in C15 cells, the protocol for rapid amplification of cDNA ends (RACE) (12) was used. mRNA (1 μg) was primed with an oligo(dT)17 primer and reverse transcribed with Moloney murine leukemia virus reverse transcriptase (Gibco-BRL). The cDNA was amplified by PCR with a gene-specific primer, Ia-15B, and an adapter primer, as given in Table 1. Positive bands, identified by Southern blotting, were subcloned and then sequenced using the ABI PRISM Dye Terminator cycle sequencing kit as specified by the manufacturer (Applied Biosystems, Inc.).
TABLE 1.
Oligonucleotides for RT-PCRa
| Oligonucleotide | Sequence (5′→3′) | Coordinates (bp) |
|---|---|---|
| Ia-1T | CTGCAGCCGGGTCCGGGGTT | 3470–3489 |
| Ia-2T | GGTCCGGGGTTCCGGCCCTG | 3479–3498 |
| Ia-3T | CTCCGGCGGGGATGGGGGTGC | 3552–3572 |
| C15-3T | CTCCGGCGGGGGGTGGGGATG | 3552–3572 |
| H-1T | GGTCCCCATGGCACAGGCCTAG | 3702–3723 |
| H-2T | GCCCAGCGCGCCCCGTTCA | 3744–3762 |
| Ia-3B | CGCTCGGGGGGTGCACACCT | 3684–3665 |
| Ia-13B | CTGGGTCCTGAGACCCAA | 1046–1029 |
| Ia-14B | GGCAGCGGACCCAGCGGA | 1100–1083 |
| Ia-15B | CCCGAGCTCCAGGGCCGGAA | 1161–1142 |
| dT-17 primer | GACTCGAGTCGACATCGA(T)17 | |
| Adapter primer | GACTCGAGTCGACATCG |
Oligonucleotide numbers for Raji and C15 BamHI Ia fragments are taken from the work of Parker et al. (32) and those for B95-8 BamHI H are from the work of Jones et al. (19). T designates sequence from the top strand and B designates that from the bottom strand, on the conventional map of EBV DNA (3).
Specific probes for IR2 and IR4 transcripts.
To create a specific probe for the IR2 gene, the BamHI H fragment from B95-8 DNA was cleaved with NcoI and SmaI, generating a 242-bp fragment from the unique region of the gene, between IR2 and DL. The product, following separation by gel electrophoresis, was eluted and religated into the pBIISK vector (Stratagene). For generating an IR4 gene-specific probe, a 3′-end RACE clone from C15 mRNA, designated C15-232, was used as a template and was amplified by PCR using oligonucleotide Ia-14B as a gene-specific primer combined with the adapter primer (Table 1). A 160-bp PCR product in the unique 3′ end of the gene was obtained and purified.
In situ hybridization.
Tissues were formalin fixed and paraffin embedded, using conventional methods. Cultured cells were chemically induced as described elsewhere (13) and washed with PBS, and a single drop was applied to a silanized microscope slide. Specific localization of IR4 RNA was accomplished by in situ hybridization using an antisense riboprobe containing multiple copies of the EBV IR4 PstI repeat, generated from a C15 clone isolated from a cDNA library (18). This probe was chosen to allow hybridization to the repeat sequence in IR4. The DNA cloned into pBIISK was linearized with SalI and transcribed with T3 RNA polymerase in the presence of 35S-UTP (∼800 Ci/mmol; Amersham, Little Chalfont, United Kingdom). The antisense probe (SalI 14A, containing about 700 bases of IR4 sequence plus 96 bases of vector) was used without hydrolysis. Methods for pretreatment, hybridization, and washing and dipping of slides in Ilford K5 for autoradiography have been described previously (34). The presence of hybridizable mRNA throughout the tissues was established in adjacent serial sections using an antisense human β-actin probe (42). Autoradiography was carried out at 4°C, using two exposures per section (8 and 18 days, respectively) for the EBV transcript and 8 days for β-actin mRNA before development in Kodak D19 and counterstaining by the Giemsa method. Sections were examined under conventional or reflected-light dark-field conditions (Olympus BH2 with epi-illumination) that allowed individual autoradiographic silver grains to be seen as bright objects on a dark background.
Western blotting and immunoprobing.
Fresh C15 tumor was disaggregated and solubilized by homogenization in Laemmli sodium dodecyl sulfate (SDS) sample buffer (0.1 M Tris-HCl [pH 6.8], 2% [wt/vol] SDS, 250 mM β-mercaptoethanol, 20% [vol/vol] glycerol, 0.01% [wt/vol] bromophenol blue). The lysate was sonicated at an amplitude of 15 μm (MSE Soniprep 150) for 30 s and spun for 10 min at 4°C, and the supernatant was stored at −70°C. Total protein was separated on an SDS–9% polyacrylamide gel and electrotransferred to an Immobilon-P membrane (Millipore), followed by probing the blot with an antibody against the IR4 protein, as described previously (31).
Double marker staining of cells.
Slides containing BL smears and sections (6 μm) cut from frozen blocks of the C15 xenograft were used. Cells and sections were fixed with 4% paraformaldehyde in PBS (10 min), blocked with 3% bovine serum albumin–PBS (15 min), and stained with primary polyclonal rabbit anti-IR4 serum (1:50 dilution [31]) alone or in combination with mouse monoclonal antibodies to Zta (1:200 dilution; DAKO) for 1 h at room temperature. Primary antibodies were detected simultaneously by Texas Red conjugated donkey anti-rabbit immunoglobulins (Amersham Life Science; 1:200 dilution) and fluorescein-conjugated goat anti-mouse immunoglobulins (DAKO; 1:200) for 45 min. Two washes were carried out with PBS (5 min) between each step. Cells and sections were counterstained with DAPI (4′,6′-diamidino-2-phenylindole) and mounted with fluorescence mount medium (DAKO). Fluorescent signals were viewed by Zeiss Axiophot microscopy and imaged by MetaMorph 3.5 (Princeton Instrument Ltd.).
RESULTS
Isolation and analysis of clones from the C15 cDNA library.
The transcriptional map profile of C15 in a cDNA library (18) suggested that transcripts coming from the IR4 (but not IR2) region of the genome might be expressed in the tumor. From the library, 1.2 × 106 PFU was plated out and screened (in duplicate) with DNA fragments from BamHI Ia (2, 14) and BamHI H (2), the latter showing strong partial sequence homology to Ia (11). Phage plaques that hybridized to either of the probes were selected and rescreened. Six were identified. Three were preliminarily characterized by hybridization as being derived from BamHI Ia, and three were characterized as being derived from BamHI H. Recloned DNAs from all six plaques, digested with NotI and PstI restriction enzymes, respectively, proved resistant to NotI but cleavable by PstI. Because there is about 70% sequence homology between the two repeat regions, and both are very GC rich (>80%) and flanked by duplicate sequences, DL and DR, the BamHI H probe must have recognized the insert from the Ia region. The data show that the IR4 transcript is expressed in the C15 xenograft, but if the IR2 repeat region is transcribed, the level is much lower than that for IR4.
Sequence of the C15 IR4 transcript.
To examine whether elements in the viral genome could be found that would allow for specific identification of transcripts relating either to the IR2 or to the IR4 genes, sequence analyses were carried out on the six cDNA clones described above. As none of them contained a complete cDNA, a RACE experiment (12) was used to identify 3′ ends of the transcripts. Clones analyzed contained a poly(A) sequence initiating at position 974 (using numbering taken from reference 32), 12 bases downstream of a polyadenylation signal (AAUAAA) used by the IR4 transcript both in M-ABA (25) and in Daudi (13) cells. RT-PCR experiments were carried out to obtain the sequence of the 5′ end of the transcripts. It was not possible using any protocol to determine the sequence across all the repeats because of their GC-rich nature. The repeat copy numbers determined by digestion with DdeI and BanI (data not given) showed C15 to have 22.7 copies (one copy is incomplete) of IR4, that is, two copies fewer than reported for the viral genome in Raji cells (32).
The entire C15 gene sequence over the IR4 transcriptional unit, as determined, is given in Fig. 2. One C-rich region within the open reading frame differs in a significant manner from that reported for Raji (32) and Daudi (13). These in turn differed from that reported for M-ABA cells (25) at this site. The importance of this difference is that it changes the reading frame used for the IR4 repeats.
FIG. 2.
Sequence of the IR4 gene and possible translation product in the C15 NPC tumor. The polarity of the IR4 transcript was that previously described for Daudi cells (13). Of the leftward open reading frames within IR4, only that designated LF3 (32) contains an AUG initiation codon. The 5′ end of the transcript (arrow, as also seen in M-ABA cells [25]) and PstI (CTGCAG) sites in the gene (underlined) are indicated. In the gene, the CAAT and GATA boxes in the promoter, the ATG initiation codon, the TGA stop codon, and the polyadenylation signal, AATAAA, used by C15 are in boldface and underlined. The polyadenylation sequence found in the cDNA of C15 is given, and the C-rich sites where sequence variations between Raji (or Daudi), M-ABA, and C15 have been found are noted (dashed underlines). The oligonucleotide primers used in RT-PCR and RACE experiments are shown.
RACE analysis on EBV strains from other cells.
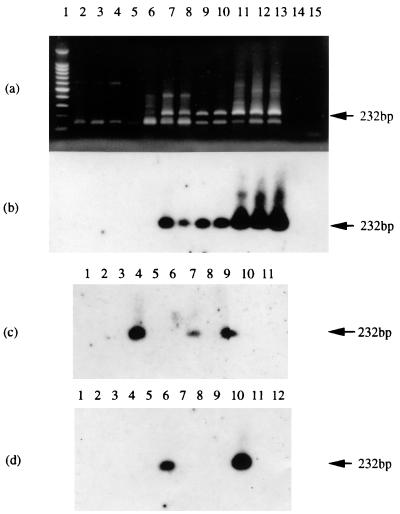
RACE experiments were subsequently carried out to compare the 3′ end of the C15 message with those of other EBV strains. Raji cells—for which Parker et al. (32) had proposed the use of a termination codon which is 141 nucleotides 3′ of the polyadenylation signal at position 992 in the LF3 open reading frame—were included in this study. The results (Fig. 3a and b) showed that the IR4 transcripts in all EBV strains investigated (including Raji) gave the same-sized RACE products, suggesting that the 3′ ends of the IR4 transcripts in all EBV strains terminate at the same position, with a poly(A) sequence at nucleotide 974, as shown previously (13, 25). This finding was further confirmed in tumors by sequencing RACE products from an NPC (NPC 1 [Fig. 3c]) and a BL (IB) biopsy specimen (Fig. 3d), both of which also utilized the first internal polyadenylation AAUAAA signal within the open reading frame. Subsequent work (data not shown) was consistent with transcripts in other tumors and cell lines using this polyadenylation signal—as given (Fig. 2) for the C15 product.
FIG. 3.
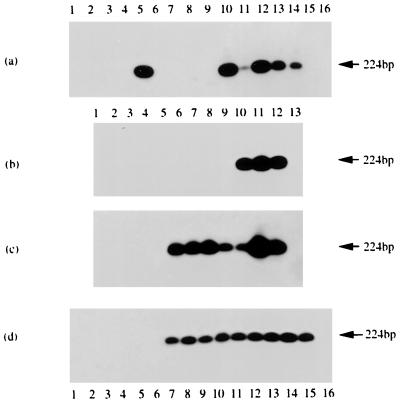
Southern blot analysis of 3′-end RACE products from IR4-containing gene transcripts in different cell lines, as observed by ethidium bromide staining (a) and hybridization (b to d). The probe used in each case was radiolabeled Ia-13B (Table 1). Tracks 1 contain size marker DNAs, and the last tracks in each case contain material from RACE experiments where only primers were used. (a) Products from uninduced Ramos, B95-8, M-ABA, Raji, Daudi, and P3HR-1 cells (tracks 2, 4, 6, 8, 10, and 12, respectively) or from the corresponding cells treated with chemical inducing agents (TPA and n-butyrate; tracks 3, 5, 7, 9, 11, and 13). Track 14 contains a PCR sample from Daudi DNA. (b) Panel a blot hybridized with the radiolabeled Ia-13B probe. (c) Hybridization of RACE products from NPCs. The tracks contain, respectively, material from C15, C17, C18, NPC 1, and NPC 2 (tracks 4 to 8); a Daudi control (track 9); HEp-2C and B95-8 cells (tracks 2 and 3); and a PCR product from C15 DNA (track 10). (d) Hybridization of RACE products from BLs. Tracks 5 to 9 contain materials from SD, IB, LC, TJ, and GE, and other tracks contain controls from Daudi (track 10), Ramos (track 2), and B95-8 (track 3) cells and from a non-BL tumor, CC (track 4). Track 11 contains a PCR sample from Daudi DNA. Whereas the 3′ ends of all cell lines examined gave products similar to those in panel a and all products hybridized with the probe (as shown [b]), among the tumors, only C15 and NPC 1 (panel c, tracks 4 and 7, respectively) and IB (panel d, track 6) gave detectable RACE products.
Production of gene-specific probes for IR4 and IR2. (i) IR4 probe.
Due to the apparent cross-hybridization that can occur between sequences within the IR2 and IR4 transcriptional units, as identified above (Fig. 1), in order to prove gene expression from either of these two regions by Northern blotting, it was necessary to develop gene-specific probes. The only region in IR4 which could serve this aim is a 70-bp unique region that corresponds to the 3′ end of the IR4 RNA, lying between the last repetitive sequence and the polyadenylation site. It contains no useful restriction enzyme sites for cloning purposes. Thus, to produce a specific probe, the 3′-end RACE clone (C15-232 [Fig. 1]) was used to generate a 160-bp PCR product (designated C15-160) within this region (see Materials and Methods). Northern blot hybridization data (see below) demonstrated that it is specific for the IR4 transcript.
(ii) IR2 probe.
A 538-bp unique sequence lies in the middle of the BHLF1 gene, between the NotI repeats and DL in the BamHI H region (Fig. 1). By cleaving the BamHI H fragment of B95-8 DNA with NcoI and SmaI to produce a 242-bp fragment from this unique sequence and religating it into the pBIISK vector (Stratagene), a clone (H-242) was obtained whose DNA functions as a BHLF1 unique probe (see below).
Northern blot analysis of RNAs from IR4 and IR2 regions of the viral genome.
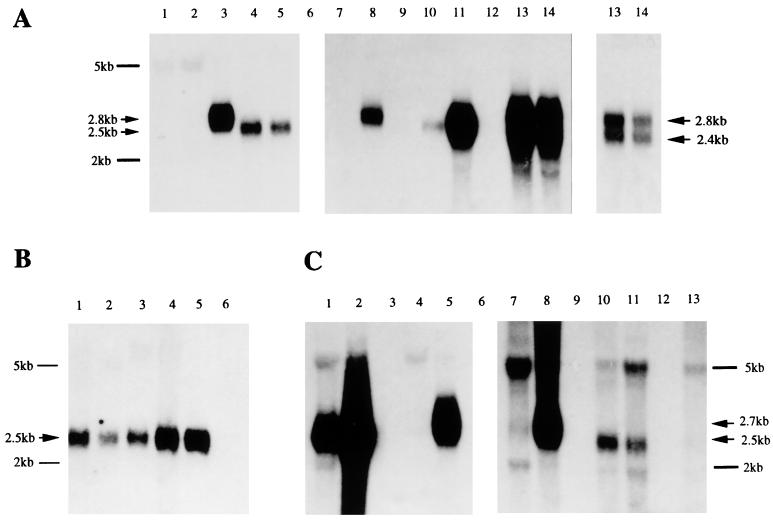
Data given in Fig. 4A show products obtained when the IR4 gene-specific probe, C15-160, was used to hybridize poly(A)-selected RNAs: a positive band about 2.5 kb in size was clearly seen in tracks containing transcripts from C15 tumors (tracks 4 and 5), whereas a slot equally loaded with B95-8 RNA (track 1) gave negative results, confirming that C15 RNA contains reasonable levels of a transcript derived from the IR4 region. On this gel, IR4 transcripts from uninduced (track 2) and induced (track 3) M-ABA cells are shown. As reported elsewhere (25), induction is required to generate this transcript, which is described as 2.8 kb in size (11). Studies with Raji, Daudi, and P3HR-1 mRNAs were also carried out: with Raji (compare tracks 7 and 8), a positive band of about 2.8 kb of mRNA is observed, but only upon induction, as reported elsewhere (32); uninduced Daudi cells showed relatively low-level expression of a 2.5-kb transcript (track 10) which was considerably amplified (track 11) upon chemical induction, as predicted elsewhere (13); P3HR-1 was unique in giving two abundant transcripts, the levels of which were little altered by induction (compare tracks 13 and 14). Differences in RNA sizes probably reflect different copy numbers of the PstI repeat. The uncloned P3HR-1 line that we used gave two bands by Southern blot analysis, consistent with it being a mixed cell population.
FIG. 4.
Northern blot analyses. (A) Hybridization of polyadenylated RNAs with a 32P-labeled IR4 gene-specific probe, C15-160. Tracks 1 to 14 contain poly(A)+ RNAs from B95-8 (track 1), M-ABA uninduced (track 2) and induced with TPA and n-butyrate (track 3), C15 (passaged subcutaneously or propagated under the kidney capsule [tracks 4 and 5, respectively]), Raji uninduced (track 7) and induced (track 8), Daudi uninduced (track 10) and induced (track 11), and P3HR-1 uninduced (track 13) and induced (14), with sizes as indicated. Materials were run on a single gel, blotted, and probed. Products in tracks 1 to 5 were visualized by long-term (about 3 days) exposure of the gel, whereas those in 7 to 14 (center panel) were from a short-term (1 day) exposure. At the right, a 1-h exposure of materials from P3HR-1 cells is shown. Tracks 6, 9, and 12 are blanks. Size markers are indicated on the left. (B) Poly(A)+ RNA (6 μg) from C15 tumors, probed with C15-160. Tracks 1 to 4 contain C15 tumors propagated subcutaneously and under the kidney capsule (tracks 1 and 2, respectively) and subcutaneously propagated tumor treated with PBS (track 3) or with TPA and n-butyrate (track 4). Tracks 5 and 6 contain poly(A)+ Daudi RNA (1 μg) and poly(A)− RNA from C15 cells, respectively. Here the same exposure time was used throughout. (C) Poly(A)+ RNAs from B-cell lines and the C15 tumor, probed with the IR2-specific probe, H-242. Tracks 3, 6, 9, and 12 are blanks, and the others contain RNAs from B95-8 cells uninduced (track 1) and induced (track 2), Raji cells uninduced (track 4) and induced (track 5), M-ABA cells uninduced (track 7) and induced (track 8), C15 (subcutaneous [track 10] and kidney capsule [track 11]), and Daudi uninduced (track 13). The left panel (tracks 1 to 5) shows data from a short-term (1-day) exposure, whereas the right panel (tracks 7 to 13) shows a longer exposure (5 days) needed for visualizing products in the C15 tracks.
We asked whether nude mouse xenografts could be chemically induced so as to alter the levels of the IR4 transcripts. C15 xenografts were injected with TPA and n-butyrate, as described in Materials and Methods, and equal amounts of RNA from induced and uninduced tumors were loaded onto gels. Representative data are given in Fig. 4B. Comparing materials from C15 tumors propagated by two routes (tracks 1 and 2) and a tumor that had been induced (track 4) or treated with PBS (track 3), induction was observed to enhance the level of the IR4 transcript severalfold, relative to that in a noninduced tumor, although not to the same extent as seen for most B cells (Fig. 4A). For example, see the comparison here between poly(A)+ RNA from uninduced Daudi cells (1 μg; track 5) and induced C15 cells (6 μg). The data imply nonetheless that the transcript in epithelial cells, and in a tumor setting, is also subject to induction, like that in most B cells.
The IR2-specific probe, H-242, was used to evaluate transcription from its gene in BamHI H; the data are given in Fig. 4C. B95-8 cells gave an abundant 2.5-kb transcript, especially after induction (compare tracks 2 and 1, respectively). Induced Raji and M-ABA cells also gave high levels of expression of a 2.7-kb transcript (tracks 5 and 8, respectively), whereas the corresponding uninduced cells (tracks 4 and 7) expressed no detectable transcript. We also observed a low-level 2.5-kb band from C15, on a long exposure of the gel, using two different tumors (tracks 10 and 11), suggesting that there could be a very low level of IR2 product in this tumor. Because the transcript has the same size as that observed using the IR4-specific probe, the possibility exists that this is a cross-hybridizing species. However, Daudi mRNA—with a deletion that removes IR2 (19)—gave a negative result (track 13) with the probe at this same exposure. A more likely explanation for the tumor is that long-term passage of this tumor in vivo has resulted in expression alterations, including up-regulation of genes that break the latent expression pattern. There is indeed evidence in the literature that EB virus can be isolated from NPC xenografts (40). However, cross-hybridization cannot be totally excluded in that long exposures with the H-242 probe also showed some hybridization to the 5-kb rRNA.
RT-PCR analysis of gene expression from IR4 and IR2 regions in NPC and BL biopsy specimens.
To explore this topic further and to determine whether one or both of these two genes are transcriptionally expressed in bona fide human biopsy specimens, RT-PCR studies on small tumor aspirate samples from NPC and BL biopsy specimens were carried out. Three sample series were examined. One contained NPC samples, C15, C17, and C18, from North African sources, passaged as xenografts in nude mice, and seven biopsy specimens (NPC 1 to 7) were from Chinese NPCs. The second series consisted of five surgically removed EBV-positive BL biopsy specimens (SD, IB, LC, TJ, and GE) from sub-Saharan African patients; a sixth biopsy specimen (CC), not a BL, was used as a control. The third series involved materials from B-cell lines, including the LCL line, M-ABA, and BL-derived lines Raji, Daudi, and P3HR-1 (with or without induction), since RNAs in these cells had previously been analyzed on Northern blots (Fig. 4). In all three sets of experiments, materials from B95-8 and Daudi cells served as negative and positive controls, respectively, against cross-hybridizing artifacts. As a further control, an EBV-negative epithelial cell line, HEp-2C, was used in the NPC experiments and the EBV negative B-cell line, Ramos, was used with B cells. Neither the IR4 nor IR2 transcript is spliced (3, 25, 32). To eliminate false-positive data from contaminating DNA, all RNA samples were treated with DNase I before cDNA synthesis, and controls to confirm enzymic degradation were included. All of the oligonucleotides used in RT-PCR are described in Table 1 (see Materials and Methods).
(i) IR4 transcriptional expression.
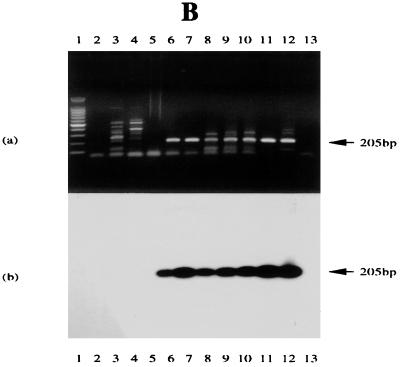
The first-strand cDNA was synthesized using the oligonucleotide Ia-1T as primer. A combination of Ia-2T and Ia-3B was used to carry out subsequent PCR amplifications. For hybridizing Southern blots of RT-PCR products from these three series of samples, to allow for sequence variation (Fig. 2), either an oligonucleotide (Ia-3T) from the Raji IR4 gene sequence (in the junction region between the PstI repeat and the DR region) or a corresponding sequence (C15-3T) from C15 was used as probe (Fig. 2 and Table 1). As shown in Fig. 5, all tumor samples used in this experiment gave a positive band of the anticipated size (205 bp), regardless of whether the source materials were taken from NPCs (Fig. 5A) or BLs (Fig. 5B), and these were identical with products expressed in the EBV-positive B-cell lines examined (Fig. 5C). This transcript was not seen in EBV-negative or other control cells, including B95-8 with a genome deletion that removes IR4. Sequence variation among different EBV strains was apparent, however. When the RT-PCR products from different tumor cell lines and biopsy specimens were hybridized with the radiolabeled Ia-3T probe (based on Raji sequence), hybridization was observed with products from Chinese NPC biopsy specimens, NPC 1 and NPC 2 (Fig. 5A, panel b), and BLs (Fig 5B, panel b), as well as with B-cell lines M-ABA, Raji, and Daudi (with or without induction) (Fig. 5C, panel b), but not with the North African NPC tumors C15, C17, and C18 (Fig. 5A, panel b), nor with P3HR-1 (Fig. 5C, panel b). The same blot, however, hybridized with probe C15-3T (from the C15 sequence) and recognized products from C15, C17, and C18 (Fig. 5A, panel c) and P3HR-1 (Fig. 5C, panel c), but not the products that hybridized to probe Ia-3T. Interestingly, although P3HR-1 is a BL-derived line, its sequence in this junction region of IR4 appears to be more similar to that from the North African NPC tumors than to that from the other B cells. Chinese NPC biopsy specimens were more similar to B-cell lines than to the African NPCs. Mixtures of Ia-3T and C15-3T produced positive bands in all EBV-positive tumor products, as illustrated in Fig. 5A, panels d and e, and 5C, panel d.
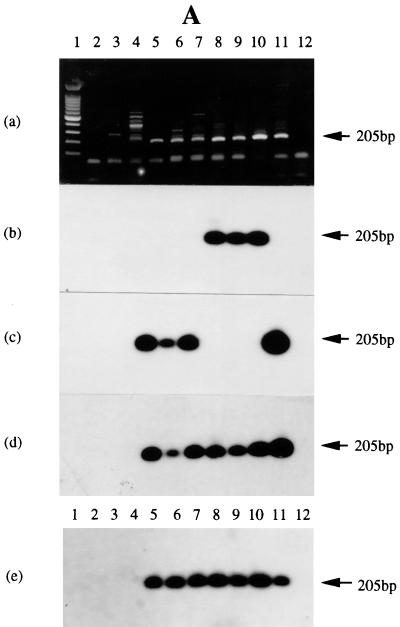
FIG. 5.
RT-PCR assessment of IR4 transcripts in NPC tumors, BLs, and BL-derived B-cell lines. (A) In panels a to d, the tracks contain PCR products from C15 DNA treated with DNase I (tracks 2), untreated C15 DNA (tracks 11), or primers only (tracks 12) or RT-PCR products from HEp-2C cells (tracks 3), B95-8 cells (tracks 4), C15 (tracks 5), C17 (tracks 6), C18 (tracks 7), NPC 1 (tracks 8), NPC 2 (tracks 9), and Daudi (tracks 10). In panel e, tracks 1 to 4 and 10 to 12 contain the same materials as in panels a to d; tracks 5 to 9 contain products from NPC tumors 3 to 7. (a) Ethidium bromide-stained gel; (b) blot probed with 32P-radiolabeled oligonucleotide Ia-3T; (c) same blot probed with oligonucleotide C15-3T; (d) same blot probed with a mixture of the two probes; (e) analysis of NPC tumors 3 to 7, as noted above, using a mixture of the two probes. (B) Tracks contain PCR products from Daudi DNA treated with DNase I (tracks 2), Daudi DNA (tracks 12) or primers only (tracks 13), or RT-PCR products from Ramos cells (tracks 3); B95-8 cells (tracks 4); tumor CC (tracks 5); BL tumors SD, IB, LC, TJ, and GE (tracks 6 to 10, respectively); and Daudi cells (tracks 11). (a) Ethidium bromide-stained gel; (b) gel blot probed with the radiolabeled oligonucleotide Ia-3T. (C) Tracks contain PCR products from C15 DNA treated with DNase I (tracks 2), untreated C15 DNA (tracks 15) or primers only (tracks 16), or RT-PCR products from Ramos cells uninduced (tracks 3) and induced (tracks 4), B95-9 cells uninduced (tracks 5) and induced (tracks 6), M-ABA cells uninduced (tracks 7) and induced (tracks 8), Raji cells uninduced (tracks 9) and induced (tracks 10), Daudi uninduced (tracks 11) and induced (tracks 12), and P3HR-1 uninduced (tracks 13) and induced (tracks 14). (a) Ethidium bromide-stained gel. (b to d) Blot probed with radiolabeled oligonucleotides: Ia-3T (b), C15-3T (c), or a mixture of the two probes (d). The location of the 205-bp product is noted on all gels. In each gel, tracks 1 contain molecular size markers differing by 100 bp (Promega).
(ii) IR2 transcriptional expression.
The same three series of RNA samples were used to explore IR2 gene expression. Daudi cells, which cannot express this gene, were used to control for possible cross-hybridization; other controls were the same as those employed to analyze IR4 gene transcriptional expression. A combination of oligonucleotides H-1T and Ia-3B (from the duplicated region [Table 1]) were used in the RT-PCR experiment. The reverse transcription was carried out using random hexanucleotide primers for the poly(A)+ mRNA or total RNA, and hybridization of the RT-PCR blot was carried out using the radiolabeled H-2T oligonucleotide. Hybridization data (Fig. 6) showed that the product from C15 (Fig. 6b), as well as all the BL biopsy specimens (Fig. 6c), gave positive bands with sizes of 224 bp, as expected from the sequence of the gene. Most of the B-cell lines (whether induced or uninduced) gave positive results (Fig. 6d). The Daudi control was negative, whereas the smaller deletion in P3HR-1, compared with Daudi cells (19), still allows its gene to be expressed. Notably, however, apart from C15 (Fig. 6a, track 5) all other NPC tumors, including C17, C18, and seven Chinese NPC biopsy specimens (NPC 1 to 7), gave negative results (Fig. 6a and b). To confirm that this finding was not a consequence of sequence alterations that led to false-negative results, DNAs from the samples were also assessed by the PCR-hybridization protocol. EBV DNA could be detected (Fig. 6a, tracks 10 to 14) using the same oligonucleotide probe, demonstrating the presence of the corresponding DNA sequence in these tumors and arguing that the transcriptional results are genuine and that expression is restricted in the NPCs.
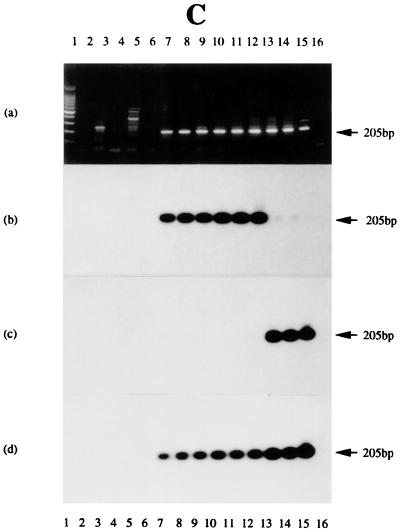
FIG. 6.
RT-PCR and DNA PCR experiments to assess IR2 transcripts and gene presence in NPC tumors, BLs, and BL-derived cell lines. The blots were probed with 32P-radiolabeled oligonucleotide H-2T, and bands were visualized by autoradiography. (a) RT-PCR products from HEp-2C (track 3) and Daudi (track 4) cells and C15 (track 5), C17 (track 6), C18 (track 7), NPC 1 (track 8), and NPC 2 (track 9) tumors. Other tracks contain PCR products from C15 DNA treated with DNase I (track 2) and untreated (track 10); from C17 (track 11), C18 (track 12), NPC 1 (track 13), NPC 2 (track 14), and Daudi (track 15) DNAs; or from primers only (track 16). (b) RT-PCR products from HEp-2C cells (track 3), Daudi cells (track 4), NPCs 3 to 7 (tracks 5 to 9, respectively), C15 (track 10), or B95-8 cells (track 11). Other tracks contain PCR products from C15 DNA treated with DNase I (track 2) or untreated (track 12) or from primers only (track 13). (c) RT-PCR products from Ramos (track 3), Daudi (track 4), and B95-8 (track 11) cells; CC tumor (track 5); and BL tumors SD, IB, LC, TJ, and GE (tracks 6 to 10, respectively). Other tracks contain PCR products from B95-8 DNA treated with DNase I (track 2) or untreated (track 12) or from primers only (track 13). (d) RT-PCR products from Ramos uninduced (track 3) and induced (track 4) cells, Daudi uninduced (track 5) and induced (track 6) cells, M-ABA uninduced (track 7) and induced (track 8) cells, Raji uninduced (track 9) and induced (track 10) cells, P3HR-1 uninduced (track 11) and induced (track 12) cells, and B95-8 uninduced (track 13) and induced (track 14) cells. Other tracks contain PCR products from B95-8 DNA treated with DNase I (track 2) or untreated (track 15) or from primers only (track 16). In each panel, track 1 contains molecular size markers, as noted for Fig. 5.
In situ hybridization.
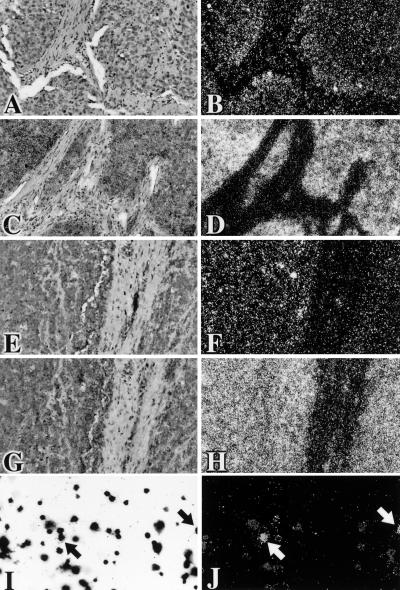
Thin sections from the C15 xenograft or a Chinese NPC tumor were examined for the presence of IR4 and β-actin RNAs, using radiolabeled antisense riboprobes to either the EBV IR4 PstI repetitive RNA sequence (Fig. 7A and B and 7E and F) or the human β-actin transcript (Fig. 7C and D and 7G and H). For comparison, induced Daudi cells were examined for the presence of the IR4 transcript (Fig. 7J). All materials were counterstained with Giemsa stain (Fig. 7A, C, E, G, and I, respectively). Specific patterns of hybridization to IR4 transcripts were observed for the C15 xenograft (Fig. 7A and B) and NPC tumors (Fig. 7E and F). Positive signals in C15 cells were strongest where the tumor was adjacent to host stromal elements, clearly visualized in a dark field (Fig. 7B) as clusters of reflective silver grains. In the NPC, whereas a subset of tumor cells had strong signals, there were no detectable IR4 transcripts in other cells. In contrast, using a probe that does not cross-hybridize to mouse β-actin, human β-actin mRNA was abundant throughout the C15 xenograft (Fig. 7D) and NPC (Fig. 7H) epithelia. In control induced Daudi cells, strong expression of IR4 transcripts was restricted to a small subpopulation, as evidenced by clusters of silver grains (arrows in Fig. 7I and J) over certain cells, without labeling of adjacent cells with similar morphologies.
FIG. 7.
Paired conventional and reflected-light dark-field autoradiographs following hybridization in situ to 35S-labeled riboprobes. (A to D) Histological sections of C15 cells from a tumor xenograft in a mouse (6). (A and B) Sites of hybridization of the IR4 PstI antisense probe (14A SalI) are revealed as clusters of discrete white grains on a dark field accumulated over a subset of tumor cells, especially those close to host stroma. The nuclei of unlabeled cells are just visible. (C and D) Intense hybridization signals for human β-actin mRNA are apparent over the majority of tumor cells. Cross-hybridization to murine β-actin mRNA in host cells would not be expected. (E to H) Sections of a Chinese NPC. Hybridization to the IR4 PstI antisense probe is confined to a subset of carcinoma cells (E and F), whereas β-actin mRNA is abundant throughout the carcinoma and detectable also in some stromal cells (G and H). (I and J) Chemically induced Daudi cells (Giemsa stained in panel I). A small subset (panel J, arrows) show strong hybridization signals for the EBV probe. Adjacent cells with similar morphologies either did not express the target sequence or expressed it at low levels.
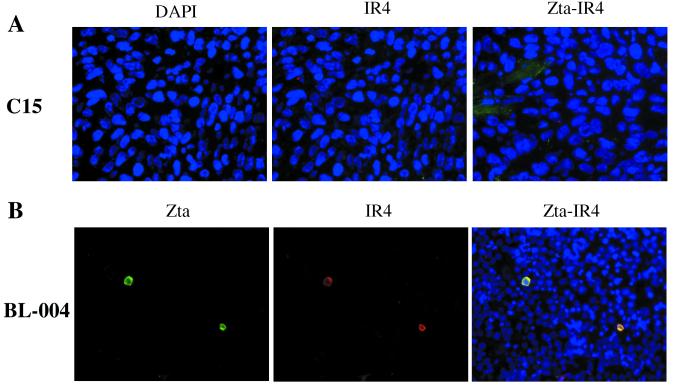
IR4 protein expression.
By Western blotting, a protein was detected in induced P3HR-1 cells, as reported elsewhere (31), but no protein consistent with the size expected for the IR4 gene product from either C15 or Raji cells was detected using the same antibody; sequence analysis for C15, although interestingly not that of Raji (Fig. 2), suggests that a protein could be made from the former. To look more closely into this topic, an immunostaining assay protocol to assess protein expression at the individual cell level was employed. Here, although C15 again gave negative results (Fig. 8A), a BL biopsy specimen (BL-004) examined in the same manner showed positive staining in a small proportion of the cells (Fig. 8B). Since the IR2 promoter—which is identical to the IR4 promoter—is highly responsive to the lytic switch gene product Zta (27) and both promoters encompass origins of EBV lytic DNA replication (oriLyt), we asked if the IR4 gene product might be directly associated with the Zta-induced lytic cycle cascade. Using a double marker staining technique on BL-004 cells, and anti-IR4 and anti-Zta antibodies, not only were both IR4 and Zta proteins detected (Fig. 7B) but, notably, their expression was found to be confined to the same cell population.
FIG. 8.
Double marker staining with anti-IR4 and anti-Zta antibodies using the C15 NPC tumor and a BL biopsy specimen. Frozen sections of C15 and a formalin-fixed BL biopsy specimen (BL-004) slide were stained for either IR4 or Zta or for both antigens (Zta-IR4), as indicated (see Materials and Methods). DAPI shows the nuclear staining of the cells. The samples used here were not chemically induced. The data show that C15 expresses neither IR4 nor Zta antigen. The BL biopsy specimen, on the other hand, expresses both antigens, with expression confined to the same cell populations, suggesting that some BL biopsy specimens may undergo an abortive lytic cycle.
DISCUSSION
We have identified and in part solved some of the problems associated with studying two apparently related EBV genes, those encoding the internal repetitive sequences IR2 and IR4, whose transcripts use promoters in the duplicated homologous sequences, DL and DR, in the viral genome. To distinguish between transcriptional expression of these two genes, specific probes have been generated and used to examine RNAs (by Northern blotting, RT-PCR, and in situ hybridization protocols) in NPC and BL biopsy specimens and in B-cell lines. The IR2 and IR4 transcripts were previously identified as products of early viral genes and were observed mainly in chemically induced cells (11, 13, 25, 30–32). Recently, endogenous expression of the IR4 transcript has been observed in some noninduced B-cell lines (13, 14), and earlier data suggested that it might also be expressed in an NPC tumor (18). In this study, we found these genes to be transcriptionally expressed in primary tumors, with patterns that differ among cells from different sources (as shown in Fig. 4 to 8). After compensating for sequence variation (Fig. 2) (13, 25, 32), the IR4 gene transcripts were found in every cell population examined, regardless of tissue type. The IR2 gene, on the other hand, while being detected in BL biopsy specimens and BL-derived cell lines, was not generally expressed in NPCs from either North African or Chinese sources, although the corresponding DNA sequences are intact and recognizable by the probes used. These data are notable in that both IR4 and IR2 transcripts initiate within the near-perfect homologs, DL and DR. Thus, control over expression probably comes from the cell itself, dependent on either cell-specific functions or epigenetic (for example, methylation) events. Our data suggest expression of the IR4 promoter to be less tightly regulated than that of the corresponding IR2 promoter and also more susceptible to inducing agents. If regulation is controlled by cellular functions—and in IR2 transcripts the promoter has been shown to contain a binding site for cellular proteins (16)—it is not surprising to find preference for one promoter in B cells and for another in epithelial cells. Work elsewhere which shows that EBV viral latency is disrupted by immediate-early gene products in a cell-type-specific manner (41, 43) would support this argument.
Transcription of these genes in tumor cells raises several interesting questions. One relates to their function(s) per se, and a second relates to explaining whether and how, if they act strictly as early genes, expression of an early gene might be advantageous to a tumor. With regard to the second point, the possibility must be considered that endogenous and induced transcripts (11, 13, 14) might serve different functions, with only the latter giving rise to a protein which acts as an early gene. It seems reasonable to explore a role for the RNAs themselves, at least in tumor settings, since for IR4 in situ hybridization shows the transcript to localize mainly to the nucleus (13). Its polarity and mapping position on the viral genome (13) suggest that it could act as a control for the expression and processing of gene products on the opposite strand. For example, in the case of the CSTs, large unspliced nuclear transcripts (18, 39) would contain sequence complementary to that found in IR4 RNA. Temporal coexpression of primary CST and IR4 transcripts could lead to viral double-stranded RNAs being generated, which should be degraded in the cell or, if not, interfere with the splicing process that produces mature CSTs. Similarly, the IR2 transcript (35) could play a controlling role in the maturation of the EBNA genes.
Equally compelling is a second possible role for these transcripts in the virus life cycle (10, 21). The two lytic origins of replication identified within the EBV genome, one within DL and the other in DR (17), have been linked to transcription (38). Replication origins in general contain core elements that determine the initiation site of replication and auxiliary enhancer components that control replication efficiency (23). With EBV, the duplicated elements themselves have organizational structures similar to those seen for simian virus 40, BK virus, and polyomavirus (8, 9) and could represent the ori core for EBV lytic replication, with the IR2-IR4 repetitive sequences providing the auxiliary enhancer elements. Most strains of EBV carry both copies of the repeats, exceptions being Daudi and P3HR-1 (with no DL) and B95-8 (with no DR). Notably, no viral isolate that lacks both copies has been identified. It thus seems reasonable to postulate that these elements are essential for the virus. It follows also that, under the appropriate conditions, every virally infected cell should be able to undergo lytic replication. This is not the case, however, and only a few EBV-infected cells produce virus to any significant extent (44). This would be readily explained were replication to depend, at least in part, upon expression of the IR2 or IR4 gene (or both), which under normal conditions is present at a low level in cells (13).
To look further at the function, particularly of IR4, since it is observed to be expressed in both epithelial and B cells, in situ hybridization was carried out using a riboprobe that should recognize the PstI repetitive region of the gene. Here (Fig. 7), as shown for two NPCs, whereas many cells appeared to be transcriptionally active, the levels of transcripts were fairly low, compared for example with β-actin. Interestingly, in the C15 xenograft, transcription was enhanced in the region of the cellular stroma (Fig. 7B), suggesting some participation by this cellular component in the induction of transcription. This was not, however, observed in the case of the Chinese NPC (Fig. 7F), where strongly expressing cells were scattered throughout the tumor. Overall, there is no apparent absolute block to transcriptional expression of IR4 in these tumors. With Daudi cells, upon chemical treatment, a subpopulation of the cells (Fig. 7J) showed the presence of the IR4 transcript. Earlier work indicated that transcripts were confined mainly to the nucleus (13) and that protein products were only very occasionally expressed in uninduced cells (31). We thus asked whether there was a block to translation, possibly dependent upon a defective mechanism of transport of the messenger from the nucleus. Our data (Fig. 8) suggest that such a block might exist. In the case of the C15 NPC, neither Zta nor IR4 antigen was observed by immunostaining protocols (Fig. 8A; compare with the data in Fig. 7), although when the same technique was used on a BL biopsy specimen, BL-004, even without induction some expression of both Zta and IR4 was observed, and notably expression of both was confined to the same subpopulation of cells (Fig. 8B). It is clear that here, unlike in C15, some cells must at least be undergoing abortive expression of the virus. At this stage in our knowledge, it is far from clear what advantage, if any, this expression might confer on a tumor. A topic which might be addressed in the future and shed light on this subject would involve a comparison of expression of nonlatent viral functions in the rapidly proliferating BLs with that in the much slower epithelial cell malignancies. The data presented here raise very important questions about promoter control, as examined earlier for the IR2 transcript (27), and intracellular transport as a possible route for maintaining viral latency.
ACKNOWLEDGMENTS
We thank Irvin Lampert for histopathology analyses on BLs and Rosemary Jeffery and Jan Longcroft for skilled technical assistance with the in situ hybridization studies.
We also thank the European Community (contract no. IC18CT96-0132), the Leverhulme Trust, and the Imperial Cancer Research Fund for support of this work.
REFERENCES
- 1.Anagnostopoulos I, Hummel M. Epstein-Barr virus in tumours. Histopathology. 1996;29:297–315. doi: 10.1111/j.1365-2559.1996.tb01414.x. [DOI] [PubMed] [Google Scholar]
- 2.Arrand J R, Rymo L, Walsh J E, Björck E, Lindahl T, Griffin B E. Molecular cloning of the complete Epstein-Barr virus genome as a set of overlapping restriction endonuclease fragments. Nucleic Acids Res. 1981;9:2999–3014. doi: 10.1093/nar/9.13.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baer R, Bankier A T, Biggin M D, Deininger P L, Farrell P J, Gibson T H, Hatfull G, Hudson G S, Satchwell S C, Seguin C, Tufnell P S, Barrell B G. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature. 1984;310:207–211. doi: 10.1038/310207a0. [DOI] [PubMed] [Google Scholar]
- 4.Bonnet M, Guinebretiere J M, Kremmer E, Grunewald V, Benhamou E, Contesso G, Joab I. Detection of Epstein-Barr virus in invasive breast cancers. J Natl Cancer Inst. 1999;91:1376–1381. doi: 10.1093/jnci/91.16.1376. [DOI] [PubMed] [Google Scholar]
- 5.Brousset P, Knecht H, Rubin B, Drouet E, Chittal S, Meggetto F, Saati T A, Bachmann E, Denoyel G, Sergeant A, Delsol G. Demonstration of Epstein-Barr virus replication in Reed-Sternberg cells of Hodgkin's disease. Blood. 1993;82:872–876. [PubMed] [Google Scholar]
- 6.Busson P, Ganem G, Flores P, Mugneret F, Clauss B, Caillou B, Braham K, Wakasugi H, Lipinski M, Tursz T. Establishment and characterisation of three transplantable EBV-containing nasopharyngeal carcinomas. Int J Cancer. 1988;42:599–606. doi: 10.1002/ijc.2910420422. [DOI] [PubMed] [Google Scholar]
- 7.Dambaugh T, Nkrumah F K, Biggar R J, Kieff E. Epstein-Barr virus RNA in Burkitt tumor tissue. Cell. 1979;16:313–322. doi: 10.1016/0092-8674(79)90008-4. [DOI] [PubMed] [Google Scholar]
- 8.DePamphilis M L. Transcriptional elements as components of eukaryotic origins of replication. Cell. 1988;52:635–638. doi: 10.1016/0092-8674(88)90398-4. [DOI] [PubMed] [Google Scholar]
- 9.DePamphilis M L. Eukaryotic DNA replication: anatomy of an origin. Annu Rev Biochem. 1993;62:29–63. doi: 10.1146/annurev.bi.62.070193.000333. [DOI] [PubMed] [Google Scholar]
- 10.Fixman E D, Hayward G S, Hayward S D. Replication of Epstein-Barr virus oriLyt: lack of a dedicated virally encoded origin-binding protein and dependence on Zta in cotransfection assays. J Virol. 1995;69:2998–3006. doi: 10.1128/jvi.69.5.2998-3006.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freese U K, Laux G, Hudewentz J, Schwarz E, Bornkamm G W. Two distant clusters of partially homologous small repeats of Epstein-Barr virus are transcribed upon induction of an abortive or lytic cycle of the virus. J Virol. 1983;48:731–743. doi: 10.1128/jvi.48.3.731-743.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frohman M A, Dush M K, Martin G R. Rapid production of full length cDNAs from rare transcripts. Amplification using a single gene specific oligonucleotide primer. Proc Natl Acad Sci USA. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao Y, Smith P R, Karran L, Lu Q L, Griffin B E. Induction of an exceptionally high-level, nontranslated, Epstein-Barr virus-encoded polyadenylated transcript in the Burkitt's lymphoma line Daudi. J Virol. 1997;71:84–94. doi: 10.1128/jvi.71.1.84-94.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao Y, Xue S-A, Griffin B E. Sensitivity of an Epstein-Barr virus-positive tumor line, Daudi, to alpha interferon correlates with expression of a GC-rich viral transcript. Mol Cell Biol. 1999;19:7305–7313. doi: 10.1128/mcb.19.11.7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Griffin B E, Xue S-A. Epstein-Barr virus infections and their association with human malignancies: some key questions. Ann Med. 1998;30:249–259. doi: 10.3109/07853899809005852. [DOI] [PubMed] [Google Scholar]
- 16.Gruffat H, Renner O, Pich D, Hammerschmidt W. Cellular proteins bind to the downstream component of the lytic origin of DNA replication of Epstein-Barr virus. J Virol. 1995;69:1878–1886. doi: 10.1128/jvi.69.3.1878-1886.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hammerschmidt W, Sugden B. Identification and characterization of oriLyt, a lytic origin of DNA replication of Epstein-Barr virus. Cell. 1988;55:427–433. doi: 10.1016/0092-8674(88)90028-1. [DOI] [PubMed] [Google Scholar]
- 18.Hitt M M, Allday M J, Hara T, Karran L, Jones M D, Busson P, Tursz T, Ernberg I, Griffin B E. EBV gene expression in an NPC-related tumour. EMBO J. 1989;8:2639–2651. doi: 10.1002/j.1460-2075.1989.tb08404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones M D, Foster L, Sheedy T, Griffin B E. The EB virus genome in Daudi Burkitt's lymphoma cells has a deletion similar to that observed in a non-transforming strain (P3HR-1) of the virus. EMBO J. 1984;3:813–821. doi: 10.1002/j.1460-2075.1984.tb01890.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karran L, Gao Y, Smith P R, Griffin B E. Expression of a family of complementary-strand transcripts in Epstein-Barr virus-infected cells. Proc Natl Acad Sci USA. 1992;89:8058–8062. doi: 10.1073/pnas.89.17.8058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kieff E. Epstein-Barr virus and its replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2343–2396. [Google Scholar]
- 22.Kinchington D, Griffin B E. Size heterogeneity of EBV and mitochondrial DNAs in Burkitt's lymphoma lines. Nucleic Acids Res. 1987;15:10345–10354. doi: 10.1093/nar/15.24.10345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kornberg A, Baker T A. DNA replication. 2nd ed. New York, N.Y: W. H. Freeman and Co.; 1992. pp. 641–647. [Google Scholar]
- 24.Labrecque L G, Barnes D M, Fentiman I S, Griffin B E. Epstein-Barr virus in epithelial cell tumors: a breast cancer study. Cancer Res. 1995;55:39–45. [PubMed] [Google Scholar]
- 25.Laux G, Freese U K, Bornkamm G W. Structure and evolution of two related transcription units of Epstein-Barr virus carrying small tandem repeats. J Virol. 1985;56:987–995. doi: 10.1128/jvi.56.3.987-995.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levitskaya J, Coram M, Levitsky V, Imreh S, Steigerwald-Mullen P M, Klein G, Kurilla M G, Masucci M G. Inhibition of antigen processing by the internal repeat region of the Epstein-Barr virus nuclear antigen-1. Nature. 1995;375:685–688. doi: 10.1038/375685a0. [DOI] [PubMed] [Google Scholar]
- 27.Lieberman P M, Hardwick J M, Hayward S D. Responsiveness of the Epstein-Barr virus NotI repeat promoter to the Z transactivator is mediated in a cell-type-specific manner by two independent signal regions. J Virol. 1989;63:3040–3050. doi: 10.1128/jvi.63.7.3040-3050.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lindahl T, Adams A, Bjursell G, Bornkamm G W, Kaschka-Dierich C, Jehn U. Covalently closed circular duplex DNA of Epstein-Barr virus in a human lymphoid cell line. J Mol Biol. 1976;102:511–530. doi: 10.1016/0022-2836(76)90331-4. [DOI] [PubMed] [Google Scholar]
- 29.Miller G. The switch between latency and replication of Epstein-Barr virus. J Infect Dis. 1990;161:833–844. doi: 10.1093/infdis/161.5.833. [DOI] [PubMed] [Google Scholar]
- 30.Nuebling C M, Mueller-Lantzsch N. Identification and characterization of an Epstein-Barr virus early antigen that is encoded by the NotI repeats. J Virol. 1989;63:4609–4615. doi: 10.1128/jvi.63.11.4609-4615.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nuebling C M, Mueller-Lantzsch N. Identification of the gene product encoded by the PstI repeats (IR4) of the Epstein-Barr virus genome. Virology. 1991;185:519–523. doi: 10.1016/0042-6822(91)90812-p. [DOI] [PubMed] [Google Scholar]
- 32.Parker B D, Bankier A, Satchwell S, Barrell B, Farrell P J. Sequence and transcription of Raji Epstein-Barr virus DNA spanning the B95-8 deletion region. Virology. 1990;179:339–346. doi: 10.1016/0042-6822(90)90302-8. [DOI] [PubMed] [Google Scholar]
- 33.Pearson G R, Luka J, Petti L, Sample J, Birkenbach M, Braun D, Kieff E. Identification of an Epstein-Barr virus early gene encoding a second component of the restricted early antigen complex. Virology. 1987;160:151–161. doi: 10.1016/0042-6822(87)90055-9. [DOI] [PubMed] [Google Scholar]
- 34.Poulsom R, Longcroft J M, Jeffery R E, Rogers L, Steel J H. A robust method for isotopic riboprobe in situ hybridisation to localise mRNAs in routine pathology specimens. Eur J Histochem. 1998;42:121–131. [PubMed] [Google Scholar]
- 35.Prang N, Wolf H, Schwarzmann F. Latency of Epstein-Barr virus is stabilized by antisense-mediated control of the viral immediate-early gene BZLF1. J Med Virol. 1999;59:512–519. doi: 10.1002/(sici)1096-9071(199912)59:4<512::aid-jmv15>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 36.Raab-Traub N, Flynn K. The structure of the termini of the Epstein-Barr virus as a marker of clonal cellular proliferation. Cell. 1986;47:883–889. doi: 10.1016/0092-8674(86)90803-2. [DOI] [PubMed] [Google Scholar]
- 37.Rickinson A B, Kieff E. Epstein-Barr virus. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2397–2446. [Google Scholar]
- 38.Schepers A, Pich D, Hammerschmidt W. A transcription factor with homology to the AP-1 family links RNA transcription and DNA replication in the lytic cycle of Epstein-Barr virus. EMBO J. 1993;12:3921–3929. doi: 10.1002/j.1460-2075.1993.tb06070.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith P R, Gao Y, Karran L, Jones M D, Snudden D, Griffin B E. Complex nature of the major viral polyadenylated transcripts in Epstein-Barr virus-associated tumors. J Virol. 1993;67:3217–3225. doi: 10.1128/jvi.67.6.3217-3225.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trumper P A, Epstein M A, Giovanella B G, Finerty S. Isolation of infectious EB virus from the epithelial tumour cells of nasopharyngeal carcinoma. Int J Cancer. 1977;20:655–662. doi: 10.1002/ijc.2910200503. [DOI] [PubMed] [Google Scholar]
- 41.Zalani S, Holley-Guthrie E, Kenney S. Epstein-Barr viral latency is disrupted by the immediate-early BRLF1 protein through a cell-specific mechanism. Proc Natl Acad Sci USA. 1996;93:9194–9199. doi: 10.1073/pnas.93.17.9194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zandvliet D W J, Hanby A M, Austin C A, Marsh K L, Clark I B N, Wright N A, Poulsom R. Analysis of foetal expression sites of human type II DNA topoisomerase α and β mRNAs by in situ hybridisation. Biochim Biophys Acta. 1996;1307:239–247. doi: 10.1016/0167-4781(96)00063-2. [DOI] [PubMed] [Google Scholar]
- 43.Zhang Q, Holley-Guthrie E, Ge J Q, Dorsky D, Kenney S. The Epstein-Barr virus (EBV) DNA polymerase accessory protein, BMRF1, activates the essential downstream component of the EBV oriLyt. Virology. 1997;230:22–34. doi: 10.1006/viro.1997.8470. [DOI] [PubMed] [Google Scholar]
- 44.Zur Hausen H, Bornkamm G W, Schmidt R, Hecker E. Tumor initiators and promoters in the induction of Epstein-Barr virus. Proc Natl Acad Sci USA. 1979;76:782–785. doi: 10.1073/pnas.76.2.782. [DOI] [PMC free article] [PubMed] [Google Scholar]