Abstract
The binding of adenovirus (Ad) fiber knob to its cellular receptor, the coxsackievirus and Ad receptor (CAR), promotes virus attachment to cells and is a major determinant of Ad tropism. Analysis of the kinetics of binding of Ad type 5 (Ad5) fiber knob to the soluble extracellular domains of CAR together (sCAR) and each immunoglobulin (Ig) domain (IgV and IgC2) independently by surface plasmon resonance demonstrated that the IgV domain is necessary and sufficient for binding, and no additional membrane components are required to confer high-affinity binding to Ad5 fiber knob. Four Ad5 fiber knob mutations, Ser408Glu and Pro409Lys in the AB loop, Tyr477Ala in the DG loop, and Leu485Lys in β strand F, effectively abolished high-affinity binding to CAR, while Ala406Lys and Arg412Asp in the AB loop and Arg481Glu in β strand E significantly reduced the level of binding. Circular dichroism spectroscopy showed that these mutations do not disorder the secondary structure of the protein, implicating Ser408, Pro409, Tyr477, and Leu485 as contact residues, with Ala406, Arg412, and Arg481 being peripherally or indirectly involved in CAR binding. The critical residues have exposed side chains that form a patch on the surface, which thus defines the high-affinity interface for CAR. Additional site-directed mutagenesis of Ad5 fiber knob suggests that the binding site does not extend to the adjacent subunit or toward the edge of the R sheet. These findings have implications for our understanding of the biology of Ad infection, the development of novel Ad vectors for targeted gene therapy, and the construction of peptide inhibitors of Ad infection.
Adenovirus (Ad) is a group of nonenveloped double-stranded DNA viruses associated with a range of respiratory, ocular, and gastrointestinal infections (15). There is a tendency for virus types within the same subgroups to be associated with similar host tissue tropism and pathogenicity (15, 28). Entry of human Ad into human cells is a stepwise process (10). The primary event in this sequence is attachment that involves an interaction between the Ad fiber protein and its high-affinity cellular receptor (4, 12, 26). Internalization of virus particles as well as cell membrane permeabilization is subsequently mediated through a specific interaction of the viral penton base protein with cell surface integrins (1, 2, 5, 17, 18, 31–33).
The coxsackievirus and Ad receptor (CAR) is a 46-kDa transmembrane protein that was initially identified as a cellular receptor for coxsackie B viruses, Ad type 2 (Ad2), and Ad5 (3, 27). In addition to subgroup C Ad fibers, CAR was also shown to bind to subgroup A, D, E, and F Ad fibers but not to subgroup B Ad fibers like serotype 3 or to the short fiber of subgroup F Ad (24). The CAR sequence predicts a structure related to the immunoglobulin (Ig) superfamily with the extracellular domain consisting of two Ig-related regions, the amino-terminal IgV and IgC2 domains (3). The IgV domain appears sufficient for virus attachment and infection (8), while the transmembrane and intracellular regions appear dispensable for these functions (30). Expression and cellular localization of CAR correlate with Ad5 infection and is therefore an important determinant of Ad tropism (3, 5, 20, 22, 27, 29). The major histocompatibility complex class I α2 domain was also proposed as a high-affinity receptor for Ad2 and Ad5 fibers (14). Its role, however, remains uncertain since the major histocompatibility complex class I allele HLA-A∗0201 permanently expressed in hamster cells failed to bind to Ad5 fiber knob with high affinity and there was no evidence of cooperativity between the two proteins (6).
The Ad5 fiber is a homotrimer with each subunit consisting of three domains: the amino-terminal tail that associates with the penton base protein (7, 21); the shaft, which consists of a motif of approximately 15 residues that is repeated 22 times (11); and the knob, which interacts with the cellular receptor. The structure of Ad5 fiber knob monomer is an eight-stranded antiparallel β sandwich composed of two β sheets, with loops and turns connecting the β strands (13, 34).
We recently showed that β strands E and F in Ad5 fiber knob may contain binding residues for CAR (16, 25) and that deletion of two consecutive amino acids in the DG loop of the protein abolished binding to CAR through disruption of local interactions (16). In the present study, we demonstrate that Ser408 and Pro409 in the AB loop, Tyr477 in the DG loop, and Leu485 in β strand F are in direct contact with CAR and that they constitute at least part of the binding site which is located at the periphery of the protein.
MATERIALS AND METHODS
Site-directed mutagenesis of Ad5 fiber knob.
Ad5 fiber knob and 15 residues of the terminal repeating unit of the shaft (nucleotides 32197 to 32783) were cloned into pKK233-3, an isopropyl-β-d-thiogalactopyranoside-inducible procaryotic expression vector (Pharmacia), to generate plasmid pF5knob, as previously described (16, 25). Plasmid pF5knob was used as the basis for the mutagenesis of Ad5 fiber knob. Single amino acid substitutions were introduced in pF5knob using a QuikChange site-directed mutagenesis kit (Stratagene). All mutations and the integrity of the remaining sequence were confirmed by DNA sequencing. The following pairs of complementary oligonucleotide primers were used to define each mutation in Ad5 fiber: Ala406Lys, 5′-ttgtggaccacaccaaaaccatctcctaactgt and 5′-acagttaggagatggttttggtgtggtccacaa; Ser408Glu, 5′-accacaccagctccagaacctaactgtagacta and 5′-tagtctacagttaggttctggagctggtgtggt; Pro409Lys, 5′-acaccagctccatctaagaactgtagactaaat and 5′-atttagtctacagttcttagatggagctggtgt; Arg412Asp, 5′-ccatctcctaactgtgacctaaatgcagagaaa and 5′-tttctctgcatttaggtcacagttaggagatgg; Asn414Lys, 5′-cctaactggagactaaaggcagagaaagatgct and 5′-agcatctttctctgcctttagtctccagttagg; His456Glu, 5′-acagttcaaagtgctgagcttattataagattt and 5′-aaatcttataataagctcagcactttgaactgt; Ile458Ala, 5′-caaagtgctcatcttgcgataagatttgacgaa and 5′-ttcgtcaaatcttatcgcaagatgagcactttg; Ile458Glu, 5′-caaagtgctcatcttgagataagatttgacgaa and 5′-ttcgtcaaatcttatctcaagatgagcactttg; Asn470Glu, 5′-ggagtgctactaaacgaatccttcctggaccca and 5′-tgggtccaggaaggattcgtttagtagcactcc; Phe472Ala, 5′-ctactaaacaattccgcactggacccagaatat and 5′-atattctgggtccagtgcggaaatgtttagtag; Phe472Lys, 5′-ctactaaacaattcaagactggacccagaatat and 5′-atattctgggtccagcttggaaatgtttagtag; Asp474Arg, 5′-aacaattccttcctgcggccagaatattggaac and 5′-gttccaatattctggccgcaggaaggaattgtt; Tyr477Ala, 5′-ttcctggacccagaagcttggaactttagaaat and 5′-atttctaaagttccaagcttctgggtccaggaa; Arg481Glu, 5′-gaatattggaactttgagaatggagatcttact and 5′-agtaagatctccattctcaaagttccaatattc; Asn482Glu, 5′-attggaactttagagagggagatcttactgaa and 5′-ttcagtaagatctccctctctaaagttccaata; Gly483Glu, 5′-tggaactttagaaatgaagatcttactgaaggc and 5′-gccttcagtaagatcttcatttctaaagttcca; Asp484Ala, 5′-aactttagaaatggagctcttactgaaggcaca and 5′-tgtgccttcagtaagagctccatttctaaagtt; Leu485Lys, 5′-tttagaaatggagataaaactgaaggcacagcc and 5′-ggctgtgccttcagttttatctccatttctaaa; Lys506Ala, 5′-ctatcagcatatccagcatctcacggtaaaact and 5′-agttttaccgtgagatgctggatatgctgatag; Ser507Ala, 5′-tcagcatatccaaaagctcacggtaaaactgcc and 5′-ggcagttttaccgtgagcttttggatatgctga; and Lys510Glu+Lys506Ala, 5′-ctatcagcatatccagcatctcacggtgaaactgccaaaagtaac and 5′-gttacttttggcagtttcaccgtgagatgctgctggatatgctgatag.
Expression and purification recombinant proteins.
Expression and purification of wild-type and mutated Ad5 fiber knobs in Escherichia coli were performed as previously described (13, 16, 25, 34).
The baculovirus expression vector system was used to express the soluble extracellular domains of CAR (sCAR) and each domain (IgV and IgC2) alone. The FLAG-tagged CAR sequences (24) were cloned into the pAcSG2 baculovirus transfer vector downstream of the promoter in a XhoI/KpnI-restricted manner to generate transfer vectors pAcSG2-sCAR, pAcSG2-IgV, and pAcSG2-IgC2. These were cotransfected with BaculoGold (Pharmingen) DNA vector in Sf9 cells to generate recombinant baculovirus vectors. Recombinant baculoviruses were amplified three to four times to obtain a high-titer stock for protein purification.
CAR sequences corresponding to IgV, IgC2, and both domains together (sCAR) (3) were synthesized by PCR on CAR DNA using the following oligonucleotide primer pairs: IgV, 5′-ACTCTCGAGATGGTATCACTACTCCTGAAGAGATGATTG and 5′-TGAGGTACCCTACTTGTCGTCGTCGTCCTTATAGTCCGCACCTGAAGGCTT; IgC2, 5′-ACTCTCGAGATGAGATGTTACGTTGATGGATCTGAAGAAATT and 5′-TGAGGTACCCTACTTGTCGTCGTCGTCCTTATAGACTGAAGGAGGGACAACG; and sCAR, 5′-ACTCTCGAGATGGTATCACTACTCCTGAAGAGATGATTG and 5′-TGAGGTACCCTACTTGTCGTCGTCGTCCTTATAGACTGAAGGAGGGACAACG.
Surface plasmon resonance (SPR) analysis of interactions between wild-type and mutant Ad5 fibers and soluble CAR. (i) Preparation of SPR sensor surface.
Purified recombinant sCAR, IgV, and IgC2 domains were coupled to the CM5 sensor chip, using the amine coupling reaction according to the manufacturer's instructions. Briefly, the CM5 sensor chip was activated using a solution of 50 mM N-hydroxysuccinimide and 200 mM N-ethyl-N′-(dimethylaminopropyl)carbonide to activate the esters on the chip surface. sCAR, IgV, and IgC2 were then injected over the activated surface until a sufficient quantity had bound to the activated esters on the chip surface. A wide range of resonance units (RU) were tested, with immobilization densities of 600 to 800 RU used in all final experiments. Residual esters were inactivated by an injection of 1 M ethanolamine hydrochloride (pH 8.5). Purified soluble Ad5 fiber knob was also immobilized on the chip, using the same approach as described above. Biacore AB supplied the BIAcore system, CM5 sensor chip, and amine coupling kit.
(ii) SPR analysis of wild-type and mutant Ad5 fibers.
SPR analysis of immobilized CAR proteins with recombinant wild-type and mutant Ad5 fiber knobs was performed as follows. All interactions were carried out at 25°C, using HBS (10 mM HEPES [pH 7.4], 150 mM NaCl, 3 mM EDTA, 0.005% P-20 surfactant) as the continuous flow buffer at different flow rates (5, 10, 20, and 40 μl/min). A wide range of concentrations (10 nM to 5 μM) of each protein was used as analyte, with the analyte diluted in HBS and injected for a 120-s association phase followed by HBS for approximately 60 s to monitor dissociation of the bound analyte. The chip was then regenerated with two 2-μl pulses of 25 mM HCl to remove analyte bound to the chip surface. Sample injections onto a sensor chip with immobilized bovine serum albumin as an irrelevant protein assessed nonspecific binding of the analyte; injection onto an uncoated sensor chip assessed nonspecific binding to the sensor chip. Nonspecific binding (<1%) was subtracted from total binding for each case.
(iii) Kinetic analysis of SPR data.
The association (ka) and dissociation (kd) rate constants for a monophasic model of binding were obtained using the BIAevaluation analysis package (version 2.1). The ability of the models to describe the experimental data was determined by examination of the residual plots, which were calculated by subtracting the experimental data points from the fitted curve. Residual was small and randomly distributed around zero (range between 0.5 to −0.5). Nonspecific binding (<1%) was subtracted from specific binding prior to kinetic analysis.
Cell binding competition experiments.
The efficiency and affinity of wild-type and mutant recombinant Ad5 mutant fibers to functional receptors on CHO-CAR cells (3) was determined from cell binding competition assays between Ad5Luc3 (Ad5Luc3, obtained from P. Boulanger, is a replication-competent virus which contains the luciferase gene under the control of the simian virus 40 early promoter inserted in the E3 region of the Ad5 genome) and recombinant Ad5 fiber knobs, using the level of luciferase gene expression as the endpoint assay.
Confluent monolayers CHO-CAR cells were preincubated with increasing amounts of recombinant proteins (0 to 100 μg/ml [final concentration] in serum-free media) at 4°C for 10 min prior to the addition of Ad5Luc3 at a multiplicity of infection (MOI) of 10. After incubation for 1 h at 4°C, the monolayers were washed to remove unabsorbed virus; the cell monolayers were covered with prewarmed complete medium, transferred to 37°C, incubated at that temperature for 18 h, and then processed for luciferase assay. Luciferase activity, expressed in relative light units, was assayed in cell lysates by using the Promega luciferase assay system. The efficiency with which mutant Ad5 fiber bound to CHO-CAR cell receptors was assessed by measuring the reduction in luciferase activity in the presence of various concentrations of mutant fiber protein. The relative affinities of wild-type and mutant fiber binding to CAR were assessed by measuring the amount of each protein required to achieve 50% of maximal inhibition of luciferase activity (IC50).
The capacity of recombinant sCAR, IgV, and IgC2 domains to attach to Ad5 binding domains was assessed in cell binding competition experiments. Ad5Luc3 (MOI of 50) were preincubated in the presence or absence of recombinant CAR proteins (0.1 to 100 μg/ml [final concentration]) prior to application to CHO-CAR cells for 1 h at 4°C. Cells were washed to remove unattached viral particles. Luciferase activity was assayed as described above.
Phenotypic analysis of recombinant mutated fibers.
Circular dichroism (CD) measurements were performed on Jobin-Yvon (Longjumeau, France) CD6 spectrophotometer using cylindrical quartz cells of 0.2-cm path lengths. The spectrophotometer was calibrated for wavelength and ellipticity using d-10-camphorsulfonic acid. Samples were measured in the concentration range 0.9 to 1.4 mg/ml in 50 mM sodium perchlorate (pH 6.5) at constant temperature in thermostated cell holder. The CD data were analyzed for percentage α helix and β sheet as previously described (23).
Cells and viruses.
CHO cells and CHO-CAR cells (3) were maintained in alpha medium (Gibco) supplemented with 10% fetal calf serum. Ad5Luc3 was plaque purified twice and propagated in 293 cells in accordance with established methods (9).
RESULTS
Selection of Ad5 fiber knob mutations.
Ad5 fiber knob mutants deleted of two consecutive amino acids between residues 485 and 493 in the DG loop of the protein abolished high-affinity binding to CAR (16). These five deletion mutant fibers accumulated as stable trimers, but their CD spectra indicated that although there was no disruption of the overall folding and secondary structure of the protein, local conformational changes had occurred (16). This implied the involvement of residues in neighboring loop regions. One candidate region was the segment containing the β strands E and F and the turn between them (residues 479 to 486) because substitution of this sequence with the corresponding sequence in Ad3 resulted in reduced receptor binding (16).
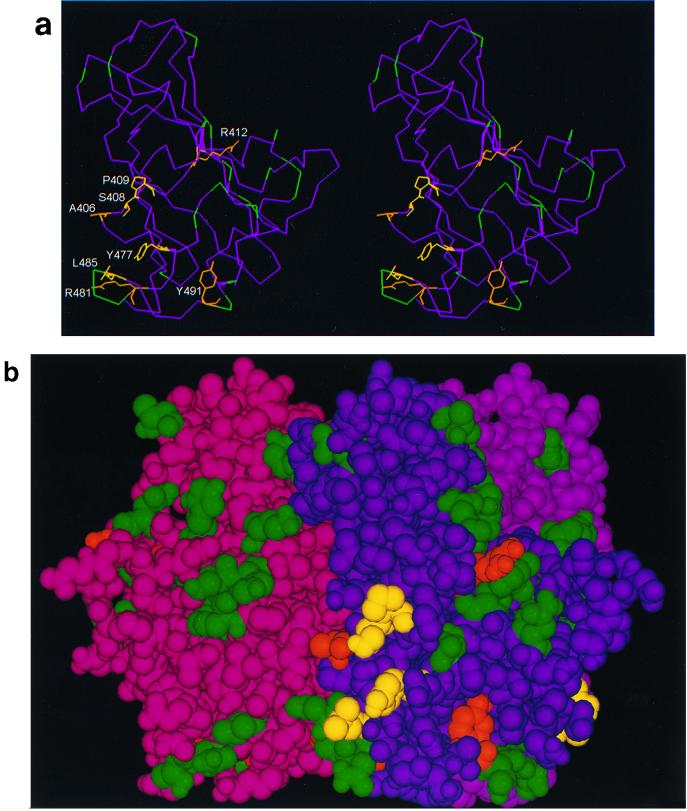
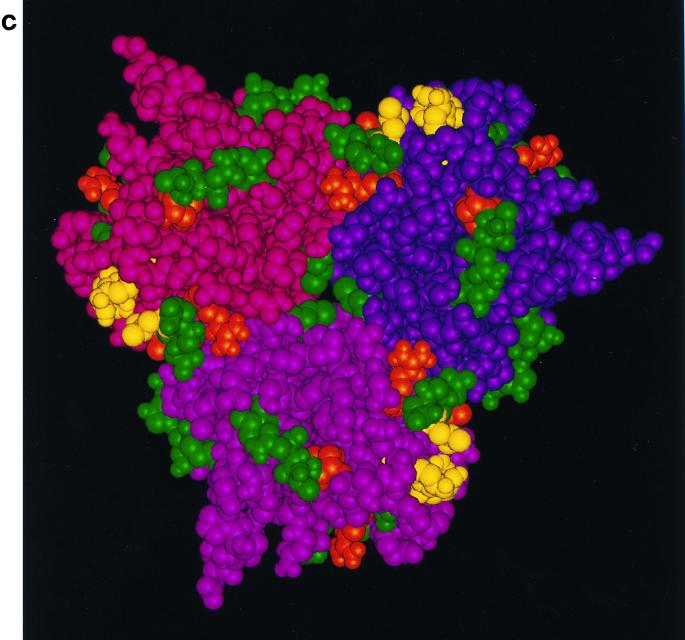
In this study, the region between β strands E and F and the connecting loop was analyzed further. All amino acid residues with surface-exposed side chains were mutated to generate mutant Ad5 fiber knobs Arg481Glu, Asn482Glu, Gly483Glu, Asp484Ala, and Leu485Lys. We also mutated residues Asn414 in the AB loop, His456 and Iso458 in β strand D, and Asn470, Phe472, and Asp474 in the DG loop because we hypothesized that the region defined by these residues may be maintained in its correct structural conformation by β strands E and F and the adjoining part of the DG loop. Subsequent mutagenesis was based on information obtained from functional analysis of the above mutants. Residues Ala406, Ser408, Pro409, Arg412, and Tyr477 have exposed side chains in regions adjacent to β strands E and F (Fig. 1a and b). In addition, mutations were introduced in a region between residues Lys506 and Lys510 in the DG loop (Lys506Ala, Ser507Ala, His508Glu, and the double mutant Lys506Ala+Lys510Glu) since residues in this region are only 2 nm from the EF region on the adjacent subunit (Fig. 1b).
FIG. 1.
(a) Stereo image showing the course of the polypeptide chain (Ca atoms only) within a subunit of the fiber knob protein, and the residues implicated in CAR binding in this study. In yellow are residues which, when mutated, profoundly affect the affinity for CAR (Ser408 and Pro409 in the AB loop, Tyr477 in the DG loop, and Leu485 in strand F) and implicated as contact residues. In orange are residues that may be only peripherally or indirectly involved in CAR binding (Ala406 and Arg412 in the AB loop, Arg481 in strand E [this report], and Tyr491 in the DG loop [16]). In green are locations of residues (Ca atoms only) which, when mutated, have no effect on CAR binding (Asn414, His456, Ile458, Asn470, Phe472, Asp474, Asn482, Gly483, Asp484, Lys506, Ser507, His508, and Lys510 [this report]; Thr492, Asn493, and Val495 [16]; and Cys428, Thr451, Lys526, Gly538, and Glu566 [data not shown]). The four-stranded β sheet known as the R sheet (see text) can be seen edge-on at the top right. (Coordinates were taken from the crystal structure [13, 34], and the images were produced by INSIGHTII [Molecular Simulations Inc., Cambridge, United Kingdom]). (b) Space-filling image (showing all nonhydrogen atoms) of the fiber knob subunit in the same orientation as in panel a, together with the two other subunits that form the trimer. The three subunits are distinguished by color (purple, magenta, and maroon), but the residues investigated by mutagenesis are colored in the same way as in panel a, in each subunit. Only the CAR binding site in the purple subunit is visible in this orientation, in which the trimer is viewed at 90° to the trimer axis with the shaft connection at the bottom. (c) The trimeric fiber knob protein as shown in panel b, rotated 90° and viewed along the threefold axis, from the side connected to the shaft. All three CAR-binding sites are visible in this orientation.
The nucleotide sequences of all mutant fibers were determined; each contained the appropriate mutation without any additional sequence alterations. All mutant fiber knobs were expressed as soluble proteins at high levels in bacteria (∼10 mg/liter).
The first extracellular domain of CAR is responsible for binding to Ad5 fiber knob.
It was recently shown that IgV domain of CAR formed complexes with Ad2 fiber knob in vitro and was sufficient to inhibit Ad2 infection of HeLa cells (8). The kinetics of binding for this interaction and for the interaction between the IgC2 domain IgC2 and CAR were, however, not assessed. Recombinant CAR proteins expressed in bacteria are produced as mainly insoluble and aggregated proteins (8); we therefore expressed the two extracellular domains of CAR together (sCAR) and independently (IgV and IgC2) in insect cells. The biological activity of purified recombinant sCAR, IgV, and IgC2 proteins was assessed first in cell binding competition experiments. We found that sCAR and IgV domains inhibited Ad5 infection of CHO-CAR cells with similar efficiencies and that the IgC2 domain had no effect on Ad5 infection of these cells (data not shown).
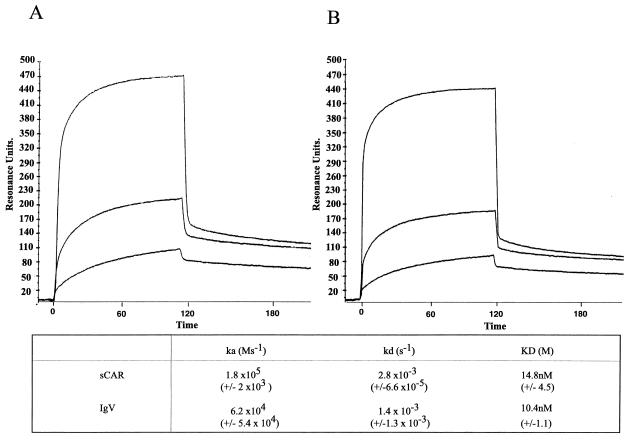
The kinetics of the interaction between wild-type Ad5 fiber knob and CAR was assessed by SPR. Assays were performed with sCAR and the IgV and IgC2 domains alone immobilized on the sensor surface, and both the association and dissociation phases of the response curve were well fitted to monophasic models (residual values ranged between 0.06 and −0.06).
The SPR analyses indicate that binding of sCAR and IgV domains to Ad5 fiber knob was specific, saturable, and dose dependent (Fig. 2). There was no specific binding between Ad5 fiber knob and the IgC2 domain at all concentrations used (data not shown). Figure 2 shows sensograms for the interaction of Ad5 fiber knob with sCAR and IgV. The ka and kd values for Ad5 fiber knob binding to sCAR were similar to the ka and kd values of Ad5 fiber knob binding to the IgV domain (Fig. 2), which indicates that sequence determinants of binding to Ad5 fiber knob are localized within the first extracellular domain of CAR. The dissociation constant for Ad5 fiber knob binding to sCAR (14.8 × 10−9 M) and IgV (10.4 × 10−9 M), as shown in Fig. 2, was also comparable to that previously determined by Scatchard analysis for the interaction between Ad5 fiber knob and full-length CAR expressed on the cell surface of hamster cells (CHO-CAR cells) (4.75 × 10−9 M) (6). These values are also in keeping with the binding constants (Kd ∼ 10−9 M) reported for Ad5 fiber obtained from infected cells and KB and HeLa cell plasma membranes (19). This suggests that CAR binding to Ad5 fiber knob requires no additional membrane cofactors. Assays were also performed with wild-type Ad5 fiber knob immobilized on the chip and sCAR, IgV, and IgC2 domains used as the analyte, with similar results (data not shown).
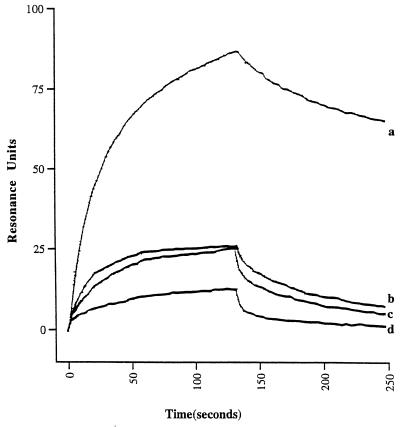
FIG. 2.
SPR sensograms for wild-type Ad5 fiber knob interacting with sCAR (A) and IgV domain alone (B). Ad5 fiber knob was injected over the sensor surface at multiple concentrations in the range of 10 nM to 5 μM. A 90- to 120-s association phase was followed by a 60-s dissociation phase with HBS buffer flowing over the sensor surface at 10, 20, 30, and 40 μl/min. Representative sensograms of Ad5 fiber knob as analyte at concentrations of 200 nM, 400 nM, and 5 μM are shown.
Leu485 in β strand F is critical for Ad5 fiber knob binding to CAR.
We mutated all residues with surface-exposed side chains within short β strands E and F and the turn between them (residues 479 to 486) in Ad5 fiber knob and assessed their effect on fiber knob binding to CAR.
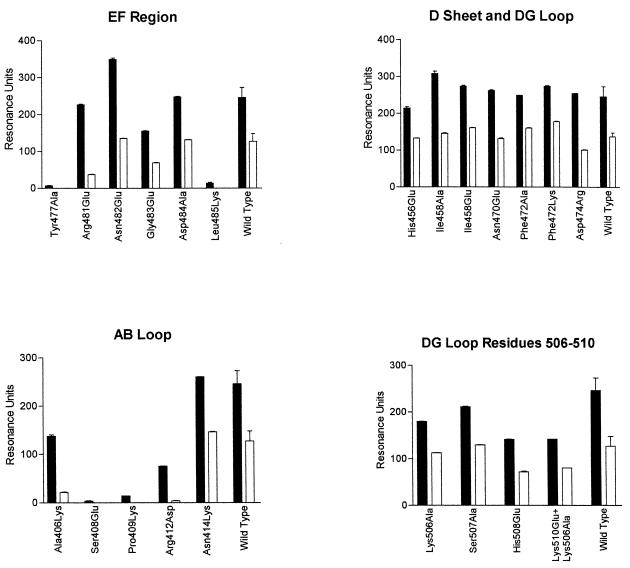
Mutant Ad5 fiber knob binding to sCAR was determined by analysis of the sensograms generated for the interaction between each of the mutants and immobilized sCAR (Fig. 3 and 4). First, the maximal association responses at equilibrium (which was attained in all cases) and the dissociation kinetics at a fixed time point (in RU) for each mutant were compared to the response obtained for wild-type Ad5 fiber knob bound to sCAR (Fig. 3). Second, the kinetics of binding of mutant Ad5 fiber knob proteins to immobilized sCAR was also determined from the sensograms (Fig. 4). Finally, the capacity for cell attachment of recombinant mutant fibers to functional receptors on CHO-CAR cells was estimated from cell binding competition assays between Ad5Luc3 and recombinant fibers, using the level of luciferase gene expression as the endpoint assay (Fig. 5). This assay was used extensively in our previous studies and was found to correlate well with binding data obtained by SPR and direct binding studies using 125I-labeled Ad5 fiber proteins (16, 25).
FIG. 3.
SPR analysis of interactions between sCAR and wild-type and mutant Ad5 fiber knobs. sCAR was immobilized on the CM5 chip. The filled bars represent the maximal association response at equilibrium in RU, and the open bars represent the response at a fixed time point (60 s) into the dissociation phase. This permits comparison to be made between wild-type and mutant fiber knob proteins in relation to their association and dissociation kinetics.
FIG. 4.
SPR sensograms of wild-type Ad5 fiber knob (a) and of mutant Ad5 fiber knobs Ala406Lys (b), Arg481Glu (c), and Arg412Asp (d) binding to immobilized sCAR. The various Ad5 fiber ligands were injected over the sensor surface at three different concentrations (10, 50, and 200 nM). A 2-min association phase was followed by a 1-min dissociation phase with HBS buffer flowing over the sensor surface at 10, 20, 30, and 40 μl/min. Representative sensograms with wild-type and mutant Ad5 fiber knob proteins at a concentration of 200 nM used as analyte are shown.
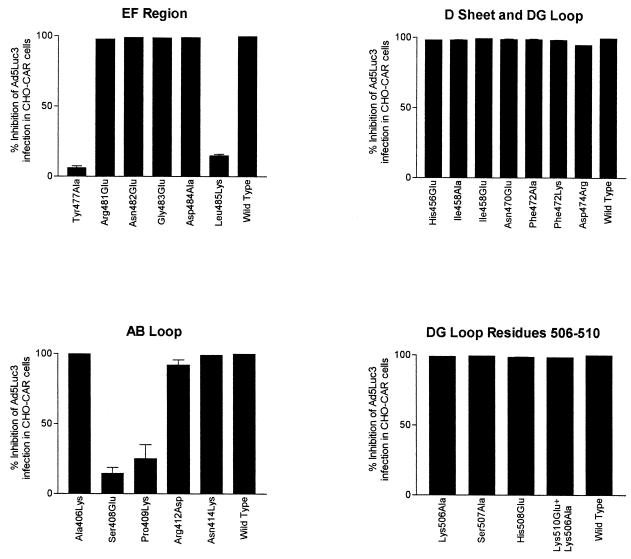
FIG. 5.
CHO-CAR cell binding competition experiments between wild-type or mutant Ad5 fiber knobs and Ad5Luc3. Cells were infected with Ad5Luc3 (MOI of 10) in the presence of large excess of recombinant full-length Ad5 fiber proteins (100 μg/ml). Ad5Luc3 was preincubated with recombinant proteins at room temperature, and the mixture was added to CHO-CAR cells precooled on ice. After incubation for 1 h at 0°C, unabsorbed virus was rinsed off; the cell monolayers were covered with prewarmed medium, transferred to 37°C, further incubated at that temperature for 18 h, and then processed for luciferase assay. Luciferase activity, expressed in relative light units, was assayed in cell lysates using luciferase substrate solution. Results were expressed as percentage reduction in luciferase activity compared to the control cells (i.e., no recombinant fiber = 0%). The data presented are means and standard errors of the means (n = 3) of three representative experiments.
We found that Ad5 fiber mutant Leu485Lys failed to associate with sCAR at all concentrations used (Fig. 3). Leu485Lys also failed to compete efficiently with Ad at concentrations up to 100 μg/ml (Fig. 5), and its IC50 (10 μg/105 cells) was 100-fold higher than the IC50 of wild-type Ad5 fiber knob (0.095 μg/105 cells) for membrane-bound CAR. These findings demonstrate that Leu485Lys dramatically reduced Ad5 fiber knob binding to CAR.
Four additional exposed residues in β strands E and F were also mutated. Arg481Glu bound to sCAR with approximately 10-fold-lower affinity than wild-type Ad5 fiber knob (Fig. 4 and Table 1), predominantly due to an effect on the dissociation rate (1.2 × 10−2, compared to 2.8 × 10−3 of the wild-type protein), as shown in Table 1. Although Arg481Glu competed efficiently with Ad5Luc3 for CHO-CAR cell receptors (Fig. 5), its IC50 (1 μg/105 cells) was 10-fold higher than the IC50 of Ad5 fiber knob (0.095 μg/105 cells), indicating that it binds to CHO-CAR cell receptors with reduced affinity. Asn482Glu, Gly483Glu, and Asp484Ala mutant proteins bound to sCAR with similar affinity as the wild-type protein (Fig. 3 and Table 1) and competed for CHO-CAR cell receptors with similar efficiency (Fig. 5) and affinity (IC50s of 0.1, 0.09, and 0.35 μg/105 cells, respectively) as wild-type Ad5 fiber knob. This would indicate that Asn482, Gly483, and Asp484 are not in direct contact with CAR. This and the fact that residues with exposed side chains in the immediate vicinity of Arg481 interacted with CAR with the same affinity as the wild-type protein (16) indicate that Arg481 may not itself be in direct contact with CAR, but that it may have an indirect effect on the conformation of nearby contact residues.
TABLE 1.
Summary of kinetic data for the interaction of wild-type and mutant Ad5 fiber knob proteins with sCAR, as determined by SPRa
| Protein assayed | ka (Ms−1) (% WT) | kd (s−1) (% WT) | Kd (nM) |
|---|---|---|---|
| Wild type | (1.8 ± 0.02) × 105 | (2.8 ± 0.06) × 10−3 | 14.8 ± 4.5 |
| Ala406Lys | (2.4 ± 0.20) × 105 (133) | (1.5 ± 0.20) × 10−2 (535) | 73.7 ± 9.8 |
| Arg412Asp | (5.0 ± 0.11) × 104 (27) | (1.5 ± 0.40) × 10−2 (535) | 227 ± 77 |
| Asn414Lys | (2.0 ± 0.07) × 105 (111) | (3.7 ± 0.50) × 10−3 (132) | 19.2 ± 2.8 |
| His456Glu | (1.4 ± 0.03) × 105 (78) | (2.6 ± 0.10) × 10−3 (93) | 17.8 ± 0.9 |
| Ile458Glu | (1.2 ± 0.01) × 105 (67) | (1.7 ± 0.06) × 10−3 (60) | 14.3 ± 5.5 |
| Asn470Glu | (1.6 ± 0.03) × 105 (89) | (3.6 ± 0.04) × 10−3 (129) | 21.5 ± 2.4 |
| Phe472Lys | (2.1 ± 0.17) × 105 (117) | (2.7 ± 0.07) × 10−3 (96) | 12.7 ± 1.2 |
| Arg481Glu | (8.9 ± 1.80) × 104 (49) | (1.2 ± 0.12) × 10−2 (429) | 137 ± 4.5 |
| Asn482Glu | (1.3 ± 0.01) × 105 (72) | (2.3 ± 0.04) × 10−3 (82) | 17.9 ± 1.2 |
| Gly483Glu | (1.4 ± 0.01) × 105 (78) | (5.4 ± 0.02) × 10−3 (192) | 37.8 ± 1.9 |
| Asp484Ala | (1.6 ± 0.04) × 105 (89) | (3.6 ± 0.01) × 10−3 (128) | 21.7 ± 3.8 |
| Lys506Ala | (1.4 ± 0.02) × 105 (78) | (2.9 ± 0.02) × 10−3 (103) | 20.7 ± 1.4 |
| Ser507Ala | (1.8 ± 0.02) × 105 (100) | (2.9 ± 0.01) × 10−3 (105) | 16.1 ± 2.1 |
| His508Glu | (1.6 ± 0.05) × 105 (89) | (4.2 ± 0.18) × 10−3 (150) | 27.2 ± 1.5 |
| Lys510Glu+Lys506Ala | (1.2 ± 0.01) × 105 (67) | (3.2 ± 0.26) × 10−3 (114) | 26.1 ± 2.6 |
Sensorgrams were analyzed using a monophasic model for interaction. Results are expressed as mean ± sd for six separate determinations.
Identification of further candidate contact residues in the CAR-binding site of Ad5 fiber knob.
Following the identification of Leu485 as a critical residue for the interaction of Ad5 fiber knob with CAR, we obtained additional mutants in order to characterize the CAR-binding site. Four residues, Ala406, Ser408, and Pro409 in the AB loop and Tyr477 in the DG loop, have exposed side chains in the same region as Leu485 (Fig. 1a and b). We found that Ser408, Pro409, and Tyr477 failed to bind to sCAR at all concentrations used (Fig. 3) and also failed to compete efficiently with Ad5Luc3 for CHO-CAR cell receptors (Fig. 5). In addition, their IC50s were strikingly higher than that of Ad5 fiber knob (for Tyr477Ala, 100 μg/105 cells; for Ser408Glu, >100 μg/105 cells; for Ser408Glu, >100 μg/105 cells). These results demonstrate that Ser408, Pro409, and Tyr477 are critical for Ad5 fiber knob binding to CAR. Two other mutants, Ala406Lys and Arg412Asp, competed as efficiently as the wild-type protein for CHO-CAR cell receptors (Fig. 5), but their IC50s (0.5 and 2.5 μg/105 cells) were 5- and 25-fold higher, respectively, than the IC50 of Ad5 fiber knob. Ala406Lys and Arg412Asp both exhibited significantly lower levels of binding to sCAR, principally due to a higher dissociation rate in the case of Arg412Asp at least (Table 1 and Fig. 4). These finding suggest that Ala406 and Arg412 are likely to be peripheral to the binding site (Fig. 1b).
Residues in β strand D and the DG loop (residues Asn470 to Asn474 and Lys506 to Lys510) do not interact with CAR.
Our previous findings indicated that deletions in the DG loop dramatically reduced Ad5 fiber knob binding to CAR due to local conformational changes (16). One possibility was that these conformational changes had disrupted the interaction between neighboring residues in the R sheet of the protein and CAR. We therefore mutated residues Asn414 in the AB loop, His456 and Ile458 in β strand D, and Asn470, Phe472, and Asn474 in the DG loop. All of these residues have exposed and accessible side chains at the edge of the R sheet and in the region between this sheet and those residues already identified as contact residues (Fig. 1a).
No significant differences were observed between the sensograms of wild-type Ad5 fiber knob and each of eight mutant fiber knobs (Fig. 3). The numbers of RU recorded during the association and dissociation phases of the interaction were similar to those obtained with the wild-type protein (Fig. 3), and analysis of the kinetics showed that the ka and kd of mutant fiber knobs Asn414Lys, His456Glu, Ile458Glu, Asn470Glu, Phe472Lys, and Asp474Arg were, within error, the same as those of wild-type Ad5 fiber knob (Table 1). In addition, all eight mutant proteins competed with the same efficiency (Fig. 5) and affinity for CHO-CAR cell receptors (the IC50 of each of these mutant fiber knobs was <0.1 μg/105 cells) as the wild-type protein. These findings demonstrate that these residues are unlikely to be in direct contact with CAR.
We also investigated the possibility that the CAR-binding region in Ad5 fiber knob may involve residues on adjacent subunits. We therefore mutated four surface-exposed residues in the DG loop, between Asn500 and Lys510. These residues are approximately 2 nm away from β strands E and F across the subunit interface (Fig. 1b). We found that mutant fiber knobs Lys506Ala, Ser507Ala, and His508Glu and the double mutant Lys510Glu+Lys506Ala bound to sCAR with comparable affinities as the wild-type protein (Fig. 3 and Table 1). Also, all four mutants competed with similar efficiency (Fig. 5) and affinity with Ad5Luc3 for CHO-CAR cell receptors (their IC50s were <0.1 μg/105 cells). Based on these findings, we propose that the region of interaction between Ad5 fiber knob and CAR does not span adjacent subunits.
Five other mutations, Cys428Ser in β strand B at the bottom of the central depression, Lys526Tyr in the rim of the central depression, Thr451Arg in the CD loop, Gly538Cys in the HI loop, and Glu566Lys in the IJ loop, were analyzed and found to bind to sCAR and membrane-bound CAR with similar affinities as their wild-type counterpart (data not shown).
Analysis of wild-type mutant Ad5 fiber knob proteins by CD spectroscopy.
The consequence each mutation on the folding and secondary structure of the Ad5 fiber knob was analyzed by CD spectroscopy and by the ability to form stable trimers.
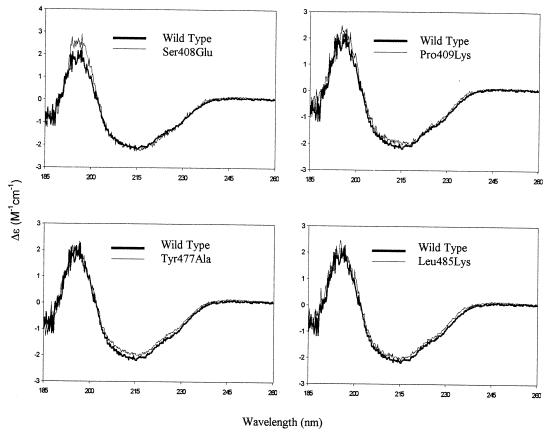
The secondary structure of mutant fiber knobs was assessed by CD spectroscopy. In this and a recent study (16), we show that the positive signal at 203 nm and the negative signal at 215 nm observed in the spectrum of the wild-type Ad5 fiber knob (Fig. 6) are in full agreement with the known β-sheet structure of the Ad5 fiber knob monomer (11, 34). The CD spectra of all Ad5 fiber mutants were identical to that of the wild-type protein over the entire recorded spectrum (Fig. 6 and data not shown). All mutants formed stable trimers as determined by gel filtration chromatography and native gel electrophoresis at all concentrations used in the various assays (data not shown).
FIG. 6.
Comparisons of the CD spectrum of wild-type Ad5 fiber knob with those of mutant fiber knob proteins. All samples were measured in the concentration range of 0.9 to 1.4 mg/ml in 50 mM sodium perchlorate (pH 6.5) in a 0.2-cm path length cell at constant temperature.
DISCUSSION
In this study, we set out to characterize the CAR-binding site in Ad5 fiber knob and to identify specific residues that may be in direct contact with CAR. This work built on our previous studies, which showed that the segment containing β strands E and F and the turn between them (residues 479 to 486) in Ad5 fiber knob might contain residues that are critical for receptor recognition (16).
Before discussing individual mutations, the interaction between wild-type Ad5 fiber knob and CAR needs to be addressed. We found that the dissociation constant determined by SPR for wild-type Ad5 fiber knob binding to sCAR was similar to that derived using conventional binding assays on CAR-transfected hamster cells (6) and KB and HeLa cells (19) and almost identical to the dissociation constant for Ad5 fiber knob binding to IgV alone. Since the IgC2 domain failed to bind to Ad5 fiber knob at all concentrations used, it is clear that the IgV domain of CAR is both necessary and sufficient for binding. It is also clear that there is no requirement for additional membrane components to confer high-affinity binding to Ad5 fiber knob. These observations support and extend those of Freimuth et al. (8).
Four mutations, Ser408Glu, Pro409Lys, Tyr477Ala, and Leu485Lys, abolished specific binding to sCAR as assessed by SPR and cell binding competition experiments. These findings indicate that Ser408, Pro409, Tyr477, and Leu485 are critical residues in the Ad5 fiber knob-CAR binding interaction. Two other mutations, Ala406Lys and Arg412Asp, had significant effects on binding kinetics, with dissociation constants 5 and 16 times lower, respectively, than those for their wild-type counterpart. The side chain of Ala406 lies very close to those of Ser408, Pro409, Tyr477, and Leu485, whereas that of Arg412 is further away (Fig. 1a). However, the long side chain of arginine may allow sufficient flexibility of movement to bring it closer to the other exposed side chains (Fig. 1a). All six mutant proteins were analyzed by CD spectroscopy and found to be indistinguishable from the wild-type Ad5 fiber knob. This, and the fact that these proteins accumulated as stable trimers, would indicate that these mutations did not alter the secondary or tertiary structure of the protein. This leads us to propose that Ser408, Pro409, Tyr477, and Leu485 are critical contact residues and together with Ala406 and Arg412, which may be only peripherally involved, constitute at least part of the high-affinity binding site for CAR. Determination of the crystal structure of Ad5 fiber-CAR complex will resolve the precise contribution of these residues in the binding interaction.
The only other residue in β strands E and F that, when mutated, altered the binding kinetics significantly was Arg481. Replacement of this residue with glutamic acid reduced the dissociation constant mainly through an increase in the rate of dissociation. In contrast, adjacent residues Asn482, Glu483, and Asp484 bound to CAR with similar kinetics and overall affinity as wild-type protein. This and the fact that the side chain of Arg481 is partially buried at the subunit interface, with the head group exposed at a distance from the other contact residues (Fig. 1), suggests that Arg481 is unlikely to be a contact residue. However, it may well exert an indirect effect on binding by causing a subtle local structural rearrangement.
Residues Ala406, Ser408, and Pro409 lie close to the interface between adjacent subunits, raising the possibility that the binding site may span two subunits. However, this was not supported by our findings, which showed that Lys506Ala, Ser507Ala, His508Glu, and Lys510Glu+Lys506Ala mutant fiber knobs bound to CAR with almost the same affinity as their wild-type counterpart. These residues lie close to residues Ala406, Ser408, and Pro409 across the subunit interface (Fig. 1b and c). Another adjacent region that might include contact residues for receptor binding lies toward and encompasses the edge of the R sheet. The conformation of this could be affected by β strands E and F and the adjoining part of the DG loop, which, when rearranged, reduced Ad5 fiber binding to CAR. We therefore mutated Asn414 in the AB loop, His456 and Iso458 in β strand D, as well as Asn470, Phe472, and Asp474 in the DG loop, since all of these residues lie within this region and have surface-exposed side chains. All fiber knob proteins with mutations in this region of the structure bound to CAR with the same affinity as the wild-type protein, demonstrating that none of the above residues is involved in the binding interaction and that the boundaries of the binding patch do not extend toward this part of the R sheet.
Our findings are consistent with the known pattern of reactivity of different Ad serotypes with CAR. Two of the proposed contact residues, Ser408 and Pro409, are identically conserved in Ad2 and Ad9 fibers as well as the long fibers of Ad40 and Ad41 that bind to CAR but are not conserved in Ad3 fiber and the short fiber of Ad41 which do not bind to CAR. Residues Tyr477 and Leu485 are either nonconservatively substituted or have no counterpart at all in Ad3 fiber or the short fiber of Ad41. Epitope mapping of two monoclonal antibodies that neutralized Ad5 infection of HeLa cells by blocking the Ad5 fiber-binding site characterized two contiguous regions in Ad5 fiber knob spanning residues 438 to 486 (14). The fact that critical residues Arg477 and Leu485 are within a region spanned by one of the two epitopes (residues 473 to 486) provides a molecular explanation for those observations.
The three binding sites for CAR are located around the periphery of the trimer (Fig. 1c). They are, however, disposed toward the underside of the protein near the shaft domain and not, as had been speculated, on the upper more exposed surface (Fig. 1a and b) (34). The definition of the CAR binding site and the identification of individual contact residues in Ad5 fiber knob have important implications for our understanding of the molecular determinants of Ad tropism, will permit modulation of the Ad-host cell interaction for the development of novel target-specific Ad vectors for human gene therapy, and will allow the development of peptide inhibitors of Ad infection.
ACKNOWLEDGMENTS
We thank J. M. Bergelson and R. W. Finberg for CHO-CAR cells, and we thank S. S. Hong and P. Boulanger for Ad5Luc3.
This work was funded by grants from the Wellcome Trust and the Special Trustees for Guy's and St. Thomas' Hospitals to G. Santis. B. J. Sutton and A. J. Beavil also thank the Wellcome Trust for support.
REFERENCES
- 1.Bai M, Campisi L, Freimuth P. Vitronectin receptor antibodies inhibit infection of HeLa and A549 cells by adenovirus type 12 but not by adenovirus type 2. J Virol. 1994;68:5925–5932. doi: 10.1128/jvi.68.9.5925-5932.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belin M, Boulanger P. Involvement of cellular adhesion sequences in the attachment of adenovirus to the HeLa cell surface. J Gen Virol. 1994;74:1485–1497. doi: 10.1099/0022-1317-74-8-1485. [DOI] [PubMed] [Google Scholar]
- 3.Bergelson J M, Cunningham J A, Droguett G, Kurt-Jones E A, Krithivas A, Hong J, Horwitz M S, Crowell R L, Finberg R W. Isolation of a common receptor for coxsackie B viruses and adenoviruses 2 and 5. Science. 1997;275:1320–1323. doi: 10.1126/science.275.5304.1320. [DOI] [PubMed] [Google Scholar]
- 4.Chroboczek J, Ruigrok R W, Cusack S. Adenovirus fiber. Curr Top Microbiol Immunol. 1995;199:163–200. doi: 10.1007/978-3-642-79496-4_10. [DOI] [PubMed] [Google Scholar]
- 5.Davison E, Diaz R M, Hart I R, Santis G, Marshall J F. Integrin α5β1-mediated adenovirus infection is enhanced by the integrin-activating antibody TS2/16. J Virol. 1997;71:6204–6207. doi: 10.1128/jvi.71.8.6204-6207.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davison E, Kirby I, Elliot T, Santis G. The human HLA-A0201 allele, when expressed in hamster cells, is not a high-affinity receptor for adenovirus type 5 fiber. J Virol. 1999;73:4513–4517. doi: 10.1128/jvi.73.5.4513-4517.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Devaux C, Adrian M, Berthet C C, Cusack S, Jacrot B. Structure of adenovirus fibre. I. Analysis of crystals of fibre from adenovirus serotypes 2 and 5 by electron microscopy and X-ray crystallography. J Mol Biol. 1990;215:567–588. doi: 10.1016/S0022-2836(05)80169-X. [DOI] [PubMed] [Google Scholar]
- 8.Freimuth P, Springer K, Berard C, Hainfeld J, Bewley M, Flanagan J. Coxsackievirus and adenovirus receptor amino-terminal immunoglobulin V-related domain binds adenovirus type 2 and fiber knob from adenovirus type 12. J Virol. 1999;73:1392–1398. doi: 10.1128/jvi.73.2.1392-1398.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Graham F, Prevec L. Adenovirus based expression vectors and recombinant veccines. In: Ellis R, editor. Vaccine: new approaches to immunological problems. Oxford, England: Butterworth-Heinemann; 1992. pp. 363–390. [DOI] [PubMed] [Google Scholar]
- 10.Greber U F, Willets M, Webster P, Helenius A. Stepwise dismantling of adenovirus 2 entry into cells. Cell. 1993;75:477–486. doi: 10.1016/0092-8674(93)90382-z. [DOI] [PubMed] [Google Scholar]
- 11.Green N M, Wringley N G, Russell W C, Martin S R, MacLachlan A D. Evidence of a repeating cross-∫ sheet structure in the adenovirus fibre. EMBO J. 1983;2:1357–1365. doi: 10.1002/j.1460-2075.1983.tb01592.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hennache B, Boulanger P. Biochemical study of KB-cell receptor for adenovirus. Biochem J. 1977;166:237–247. doi: 10.1042/bj1660237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henry L J, Xia D, Wilke M E, Deisenhofer J, Gerard R D. Characterization of the knob domain of the adenovirus type 5 fiber protein expressed in Escherichia coli. J Virol. 1994;68:5239–5246. doi: 10.1128/jvi.68.8.5239-5246.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hong S S, Karayan L, Tournier J, Curiel D T, Boulanger P A. Adenovirus type 5 fiber knob binds to MHC class I alpha 2 domain at the surface of human epithelial and B lymphoblastoid cells. EMBO J. 1997;16:2294–2306. doi: 10.1093/emboj/16.9.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horwitz M. Adenoviruses. Vol. 2. New York, N.Y: Raven Press; 1990. [Google Scholar]
- 16.Kirby I, Davison E, Beavil A J, Soh C P C, Wickham T J, Roelvink P W, Kovesdi I, Sutton B J, Santis G. Mutations in the DG loop of adenovirus 5 fiber protein abolish high-affinity binding to its cellular receptor CAR. J Virol. 1999;73:9508–9514. doi: 10.1128/jvi.73.11.9508-9514.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mathias P, Galleno M, Nemerow G R. Interactions of soluble recombinant integrins αvβ5 with human adenoviruses. J Virol. 1998;72:8669–8675. doi: 10.1128/jvi.72.11.8669-8675.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mathias P, Wickham T, Moore M, Nemerow G. Multiple adenovirus serotypes use αV integrins for infection. J Virol. 1994;68:6811–6814. doi: 10.1128/jvi.68.10.6811-6814.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meager A, Butters T D, Mautner V, Hughes R C. Interaction of KB cell glycoproteins with an adenovirus capsid protein. Eur J Biochem. 1976;61:345–355. doi: 10.1111/j.1432-1033.1976.tb10028.x. [DOI] [PubMed] [Google Scholar]
- 20.Miller C R, Buchsbaum D J, Reynolds P N, Douglas J T, Gillespie G Y, Mayo M S, Raben D, Curiel D T. Differential susceptibility of primary and established human glioma cells to adenovirus infection: targeting via the epidermal growth factor receptor achieves fiber receptor-independent gene transfer. Cancer Res. 1998;58:5738–5748. [PubMed] [Google Scholar]
- 21.Novelli A, Boulanger P A. Deletion analysis of functional domains in baculovirus-expressed adenovirus type 2 fiber. Virology. 1991;185:365–376. doi: 10.1016/0042-6822(91)90784-9. [DOI] [PubMed] [Google Scholar]
- 22.Pickles R J, McCarty D, Matsui H, Hart P J, Randell S H, Boucher R C. Limited entry of adenovirus vectors into well-differentiated airway epithelium is responsible for inefficient gene transfer. J Virol. 1998;72:6014–6023. doi: 10.1128/jvi.72.7.6014-6023.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Provencher S W, Glockner J. Estimation of globular protein secondary structure from circular dichroism. Biochemistry. 1981;20:33–37. doi: 10.1021/bi00504a006. [DOI] [PubMed] [Google Scholar]
- 24.Roelvink P W, Lizonova A, Lee J G M, Li Y, Bergelson J M, Finberg R W, Brough D E, Kovesdi I, Wickham T J. The coxsackie-adenovirus receptor protein can function as a cellular attachment protein for adenovirus serotypes from subgroups A, C, D, E, and F. J Virol. 1998;72:7909–7915. doi: 10.1128/jvi.72.10.7909-7915.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Santis G, Legrand V, Hong S, Davison E, Kirby I, Chartier C, Pavirani A, Bergelson J, Mehtali M, Boulanger P. Molecular determinants and serotype specificity of Ad5 fiber binding to its high affinity receptor CAR. J Gen Virol. 1999;80:1519–1527. doi: 10.1099/0022-1317-80-6-1519. [DOI] [PubMed] [Google Scholar]
- 26.Svensson U, Persson R, Everitt E. Virus-receptor interaction in the adenovirus system: identification of virion attachment proteins of the HeLa cell plasma membrane. J Virol. 1981;38:70–81. doi: 10.1128/jvi.38.1.70-81.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tomko R, Xu R, Philipson L. HCAR and MCAR: the human and mouse cellular receptors for subgroup C adenoviruses and group B coxsackieviruses. Proc Natl Acad Sci USA. 1997;94:3352–3356. doi: 10.1073/pnas.94.7.3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wadell G. Adenoviruses. In: Zuckerman A J, Banatvala J E, Pattison J R, editors. Principles and practice of clinical virology. Chichester, England: John Wiley & Sons; 1990. pp. 267–287. [Google Scholar]
- 29.Walters R, Grunst T, Bergelson J, Finberg R, Welsh M, Zabner J. Basolateral localization of fiber receptors limits adenovirus infection from the apical surface of airway epithelia. J Biol Chem. 1999;274:10219–10226. doi: 10.1074/jbc.274.15.10219. [DOI] [PubMed] [Google Scholar]
- 30.Wang X, Bergelson J M. Coxsackievirus and adenovirus receptor cytoplasmic and transmembrane domains are not essential for coxsackievirus and adenovirus infection. J Virol. 1999;73:2559–2562. doi: 10.1128/jvi.73.3.2559-2562.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wickham T J, Carrion M E, Kovesdi I. Targeting of adenovirus penton base to new receptors through the replacement of its RGD motif with other receptor-specific peptide motifs. Gene Ther. 1995;2:750–758. [PubMed] [Google Scholar]
- 32.Wickham T J, Filardo E J, Cheresh D A, Nemerow G R. Integrin αvβ5 selectively promotes adenovirus cell membrane permeability. J Cell Biol. 1994;127:257–264. doi: 10.1083/jcb.127.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wickham T J, Mathias P, Cheresh D A, Nemerow G R. Integrins alpha v beta 3 and alpha v beta 5 promote adenovirus internalization but not virus attachment. Cell. 1993;73:309–319. doi: 10.1016/0092-8674(93)90231-e. [DOI] [PubMed] [Google Scholar]
- 34.Xia D, Henry L, Gerard R, Desenhofer J. Crystal structure of the receptor-binding domain of adenovirus type 5 fiber protein at 1.7 A resolution. Structure. 1994;2:1259–1270. doi: 10.1016/s0969-2126(94)00126-x. [DOI] [PubMed] [Google Scholar]