Abstract
Human immunodeficiency virus type 1 (HIV-1) particle formation and the subsequent initiation of protease-mediated maturation occur predominantly on the plasma membrane. However, the mechanism by which HIV-1 assembly is targeted specifically to the plasma membrane versus intracellular membranes is largely unknown. Previously, we observed that mutations between residues 84 and 88 of the matrix (MA) domain of HIV-1 Gag cause a retargeting of virus particle formation to an intracellular site. In this study, we demonstrate that the mutant virus assembly occurs in the Golgi or in post-Golgi vesicles. These particles undergo core condensation in a protease-dependent manner, indicating that virus maturation can occur not only on the plasma membrane but also in the Golgi or post-Golgi vesicles. The intracellular assembly of mutant particles is dependent on Gag myristylation but is not influenced by p6Gag or envelope glycoprotein expression. Previous characterization of viral revertants suggested a functional relationship between the highly basic domain of MA (amino acids 17 to 31) and residues 84 to 88. We now demonstrate that mutations in the highly basic domain also retarget virus particle formation to the Golgi or post-Golgi vesicles. Although the basic domain has been implicated in Gag membrane binding, no correlation was observed between the impact of mutations on membrane binding and Gag targeting, indicating that these two functions of MA are genetically separable. Plasma membrane targeting of Gag proteins with mutations in either the basic domain or between residues 84 and 88 was rescued by coexpression with wild-type Gag; however, the two groups of MA mutants could not rescue each other. We propose that the highly basic domain of MA contains a major determinant of HIV-1 Gag plasma membrane targeting and that mutations between residues 84 and 88 disrupt plasma membrane targeting through an effect on the basic domain.
Assembly of type C retroviruses and lentiviruses takes place predominantly at the plasma membrane of infected cells. This process involves multiple steps mediated by the viral Gag proteins, which are both necessary and sufficient for the assembly and release of noninfectious, immature virus-like particles (VLPs). Retroviral Gag proteins are synthesized as polyprotein precursors; in the case of human immunodeficiency virus type 1 (HIV-1), the Gag precursor, Pr55Gag, is composed of matrix (MA), capsid (CA), nucleocapsid (NC), and p6 domains as well as p2 and p1 spacer peptides (reviewed in reference 12). During or immediately after virus particle release, Pr55Gag is cleaved by the viral protease (PR) to generate the mature Gag proteins p17 (MA), p24 (CA), p7 (NC), and p6. The processing of Pr55Gag causes a major transformation in virion morphology; this process, known as maturation, gives rise to virions with condensed, conical cores characteristic of infectious HIV-1 virions.
MA plays a key role in several steps in virus replication, including the binding of Pr55Gag to membrane, the incorporation of Env glycoproteins into budding virions, and early, postentry events. The covalent attachment of myristate to the N terminus of the MA domain of Pr55Gag is crucial for the binding of Gag to membrane and is thus required for virus assembly (4, 18, 23, 51). A highly basic region spanning MA residues 17 to 31 has also been implicated in Gag membrane binding. Structural studies of HIV-1 MA (28, 42) and MA of other retroviruses (for a review, see reference 8) have suggested that basic amino acids in the highly basic domain and at more C-terminal positions form a positively charged surface that may facilitate binding of Gag to membrane by promoting an electrostatic interaction with acidic phospholipids in the inner leaflet of the membrane (42, 70). In support of this hypothesis, in vitro membrane binding assays demonstrated that the N-terminal 31 amino acids of MA could confer membrane binding ability upon otherwise soluble proteins (70). We observed recently that mutation of a nonbasic residue within the basic domain increased the binding of Gag to membrane (33, 34, 45). However, an 11-amino-acid deletion in the MA basic domain was shown to have no impact on virus particle production (68) or the binding of MA to membrane (61). Moreover, deletion of large portions of MA, including the basic domain, did not significantly impair virus assembly and release (36, 54, 63, 64).
In addition to membrane binding, MA has been implicated in the targeting of virus assembly. Large deletions in HIV-1 MA cause either promiscuous virus assembly both on the plasma membrane and at intracellular sites (36, 54, 63) or a redirection of assembly to intracellular locations (11, 17). Small deletions and amino acid substitutions in MA can also cause defects in virus production by inducing intracellular accumulation of Gag (69) or retargeted VLP assembly (5, 16). Interestingly, some of these mutations involved basic amino acids within (69) or C terminal to (5) the highly basic domain. The relationship between Gag targeting and Gag membrane binding for these mutants was not determined. Intracellular VLPs observed in cells expressing large HIV-1 MA deletion mutants displayed an immature morphology by electron microscopy (EM) (11, 17, 54). Conversely, redirection of intracisternal A-type particles from the endoplasmic reticulum (ER) to the plasma membrane reportedly induced both Gag processing and particle maturation (65). These observations led to the hypothesis that retroviral Gag processing and/or virus particle maturation can occur only at the plasma membrane. A role for MA in the targeting of virus assembly is also suggested by studies in other retroviral systems. Redirection of murine leukemia virus Gag proteins from the plasma membrane to intracellular compartments was observed for several mutants (27, 31, 60), including some with amino acid substitutions in the highly basic domain of murine leukemia virus MA (60). In the case of the type D Mason-Pfizer monkey virus, it was demonstrated that a single amino acid substitution in a cytoplasmic targeting/retention signal redirects virus assembly from the cytosol to the plasma membrane (7, 56).
In an effort to characterize the role of MA in HIV-1 replication and to map functional domains within this protein, we have previously used site-directed mutagenesis to introduce over 80 single and double amino acid substitutions in MA and have analyzed the effects on a variety of aspects of the virus life cycle (for a review, see reference 12). Most of the mutants showed wild-type (WT) levels of virus assembly and release, but substantial defects in virus production were caused by mutations in several domains. For example, mutations between MA amino acids 84 and 88 caused a reduction in extracellular virus release due to retargeting of virus assembly to an intracellular compartment (16). MA amino acids 84 to 88 are located within a central buried helix within the globular core of the protein (42). In clear contrast to the phenotype observed for large MA deletion mutants (11, 17, 54), the VLPs which assembled intracellularly contained condensed cores (16). The defective phenotype of one of these mutants was reversed by second-site compensatory changes in MA which arose naturally in infected T-cell line cultures (46). Interestingly, one such second-site mutation was located within the highly basic domain, suggesting a functional relationship between the basic domain and amino acids 84 to 88. Consistent with this interpretation, we have observed that single and double amino acid substitutions in the basic domain caused reductions in extracellular virus production (13).
To understand further the mechanism by which virus formation is targeted to the plasma membrane, we performed a detailed study of MA mutants with amino acid changes in the highly basic domain and between residues 84 and 88. We focused on three aspects of targeting: (i) the connection between virus maturation and the site of assembly, (ii) the nature of the targeting signal in MA, and (iii) the relationship between Gag targeting and membrane binding. EM and confocal microscopy data demonstrate that Gag processing and virus maturation can occur not only on the plasma membrane but also in the Golgi or Golgi-derived vesicles. We provide evidence that the basic domain functions as a plasma membrane targeting signal independent of its potential involvement in Gag membrane binding. Furthermore, we show that the plasma membrane targeting signal can be provided to mutant Gag proteins in trans by coexpression with WT Gag.
MATERIALS AND METHODS
Cell culture and transfection.
HeLa cells were maintained as previously described (14). Transfection of HeLa cells was performed by the calcium phosphate precipitation method as reported previously (16).
Plasmids, mutagenesis, and DNA cloning.
Construction of derivatives of the HIV-1 proviral molecular clone pNL4-3 (1) containing MA amino acid changes 1GA, 85YG, and 87VE, as well as the double mutation 29KT/31KT, has been described previously (13, 16). The pNL4-3 derivatives pNL4-3/PR−, which contains a protease (PR) active site mutation (Asp→Asn change at PR amino acid 25), and pNL4-3/p6−, which contains a stop codon at p6 amino acid 1, have been described previously (30). The pNL4-3 derivative pNL4-3/KFS has been reported previously (15); this clone contains a frameshift mutation at nucleotide (nt) 6343, which eliminates Env expression.
Double mutations 1GA/85YG, 18IK/20LK, 18IE/20LE, and 29KE/31KE were introduced into pNL4-3 by oligonucleotide-directed mutagenesis using methods detailed previously (16). Molecular clones expressing Pr55Gag tagged with hemagglutinin (HA) or FLAG epitopes (pNL4-3-55HA and pNL4-3-55FLAG, respectively) were constructed as follows. (i) A molecular clone (pNL4-3/55X) was made by substituting the Pr55Gag stop codon with a sequence encoding the XbaI restriction site. This was accomplished by oligonucleotide-directed mutagenesis with an M13mp18 subclone harboring the 1.4-kbp SphI-PstI fragment from pNL4-3 (nt 1443 to 2839) as a template and oligonucleotide 5′-GTCACAATCTAGATAGGG-3′. (ii) Short oligonucleotide duplexes (for HA, 5′-CTAGCTACCCATACGATGTTCCAGATTACGCTTAG-3′ hybridized to 5′-CTAGCTAAGCGTAATCTGGAACATCGTATGGGTAG-3′; for FLAG, 5′-CTAGCGATTATAAAGACGATGACGACAAGTAG-3′ hybridized to 5′-CTAGCTACTTGTCGTCATCGTCTTTATAATCG-3′) encoding the HA (YPYDVPDYA) and FLAG (DYKDDDDK) epitope sequences, respectively, followed by stop codons, were inserted into the XbaI site of pNL4-3/55X. These insertions also introduce stop codons in the pol coding region so that no pol-encoded viral enzymes are expressed.
PR−, p6−, 55HA, and 55FLAG versions of MA mutant molecular clones were constructed by introducing the SphI-EcoRI fragments (nt 1443 to 5743) from pNL4-3/PR−, pNL4-3/p6−, pNL4-3-55HA, and pNL4-3-55FLAG, respectively, into MA mutant pNL4-3 derivatives.
Antibodies and reagents.
The following reagents were obtained from the indicated sources: mouse monoclonal antibody which recognizes p17 (MA) but not Pr55Gag, Advanced Biotechnologies (Columbia, Md.); mouse monoclonal antibodies which recognize both Pr55Gag and p17 (MA), Cellular Products (Buffalo, N.Y.) and Capricorn Products (Scarborough, Maine); rabbit polyclonal anti-calreticulin antibody, Affinity Bioreagents (Golden, Colo.); rabbit polyclonal anti-Rab1B and anti-HA antibodies, Zymed Laboratories (South San Francisco, Calif.); rabbit anti-FLAG antibody (OctA-Probe), Santa Cruz Biotechnology (Santa Cruz, Calif.); AIDS patient sera, the National Institutes of Health AIDS Research and Reference Reagent Program (catalog no. 1983 and 1984); anti-human immunoglobulin and anti-rabbit immunoglobulin antibodies conjugated with horseradish peroxidase, Amersham; Texas red-conjugated anti-mouse immunoglobulin G antibody and fluorescein isothiocyanate (FITC)-conjugated anti-rabbit immunoglobulin G antibody, Jackson Immunoresearch Laboratories (West Glove, Pa.); FITC-labeled Ricinus communis agglutinin 60 (RCA60), Sigma; and fluorescein-conjugated wheat germ agglutinin (WGA), Molecular Probes (Eugene, Oreg.).
Metabolic labeling, immunoprecipitation, and Western blotting.
Metabolic labeling of transfected HeLa cells with [35S]Cys was performed as previously described (16). Preparation of cell lysates, pelleting of virions in the ultracentrifuge, and immunoprecipitation of cell- and virion-associated proteins with AIDS patient sera have been detailed previously (16, 66). Cell and virion lysates prepared from nonlabeled, transfected HeLa cells were analyzed by Western blotting as previously described (33).
Membrane binding assay.
Equilibrium flotation centrifugation was performed as detailed previously (45). Briefly, HeLa cells were collected in phosphate-buffered saline (PBS), washed once with 10 mM Tris-HCl (pH 7.5) containing 1 mM EDTA and 1 mM EGTA, and resuspended in 10 mM Tris-HCl containing 1 mM EDTA, 6% (wt/vol) sucrose, and Complete protease inhibitor cocktail. Postnuclear supernatants obtained after sonication of cell suspensions were mixed with 85.5% (wt/vol) sucrose in Tris-EDTA (TE) and placed on the bottom of a centrifuge tube. On top of this postnuclear supernatant-containing 73% (wt/vol) sucrose mixture was layered TE containing 65% (wt/vol) sucrose and 10% (wt/vol) sucrose, respectively. The gradients were centrifuged at 100,000 × g for 18 h at 4°C in a Beckman SW41 rotor. Ten fractions were collected from the top of the centrifuge tube. Fractionated samples were analyzed by Western blotting as previously described (33). Quantitation of Western blotting data was performed by densitometry scanning.
EM.
Fixation of cells, preparation of samples and EM were performed as described previously (16).
Fluorescent staining and confocal microscopy.
Fluorescent staining of transfected HeLa cells with antibodies and lectins was performed as described previously (47), with some modifications. Briefly, cells were grown in chamber slides (Nunc) and transfected by the calcium phosphate precipitation method as described above but without glycerol shock. Twenty-four hours later, cells were washed once with Dulbecco modified Eagle medium supplemented with 5% fetal bovine serum. After another 24 h, cells were rinsed once with PBS and fixed with 3.7% formaldehyde in 100 mM sodium phosphate buffer (pH 7.2) for 20 min at room temperature (r.t.) (all the procedures were carried out at r.t. unless otherwise noted). After being washed four times with PBS, cells were permeabilized either with PBS containing 0.1% Triton X-100 for 2 min at r.t. followed by washing with PBS three times or with methanol for 4 min at −20°C followed by air drying. Subsequently, cells were incubated with 0.1 M glycine in PBS for 10 min and blocked with 3% bovine serum albumin in PBS (BSA-PBS) for 30 min. Cells were then incubated with primary antibodies diluted appropriately in BSA-PBS for 1 h, washed with PBS three times, and incubated with secondary antibodies appropriately diluted in BSA-PBS for 30 min. For double staining with FITC-conjugated lectins, after removal of antibody solution by washing with PBS three times, cells were incubated with respective lectins diluted appropriately in PBS for 30 min. After being washed with PBS three times, cells were mounted with Fluoromount G (Virotech International, Rockville, Md.) and examined with a Zeiss LSM410 laser scanning microscope.
RESULTS
Mutations between HIV-1 MA amino acids 84 and 88 redirect assembly and PR-mediated maturation to intracellular vesicles.
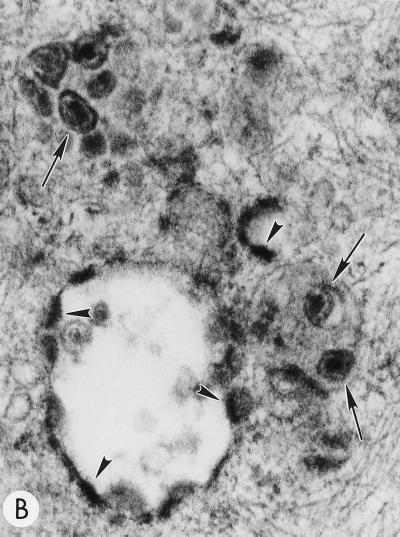
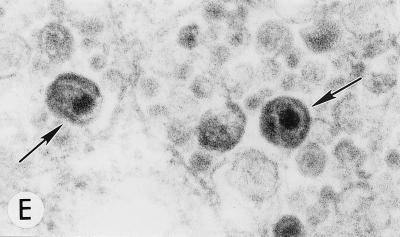
To determine whether the core condensation observed within intracellularly assembled VLPs produced by Gag mutants containing substitutions between MA amino acids 84 and 88 (16) was the result of HIV-1 PR activity, we constructed double mutants containing either the 85YG or the 87VE substitution and an inactivating mutation in the PR active site (PR−). As determined previously with this PR mutation (30, 33, 45), immunoprecipitation analysis confirmed that the processing of Gag expressed by the pNL4-3/85YG/PR− and pNL4-3/87VE/PR− clones was blocked (data not shown). HeLa cells transfected with pNL4-3, pNL4-3/85YG, pNL4-3/87VE, pNL4-3/85YG/PR−, and pNL4-3/87VE/PR− were examined by EM. pNL4-3-transfected cells showed particles budding exclusively from the plasma membrane; many of these particles contained condensed cores (Fig. 1A). Cells expressing the 85YG and 87VE mutants contained numerous intracytoplasmic VLPs, many of which contained condensed cores (Fig. 1B); similar data were obtained for both 85YG and 87VE mutants (16; data not shown). In the absence of an active PR (i.e., in cells transfected with pNL4-3/85YG/PR− and pNL4-3/87VE/PR−) we detected budding structures and immature VLPs but no particles with condensed cores (Fig. 1C; data not shown). These results indicate that core condensation resulting from PR activity can occur at an intracellular site.
FIG. 1.
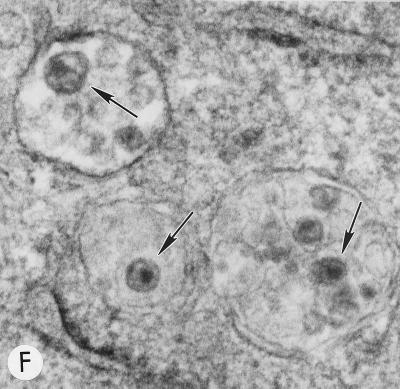
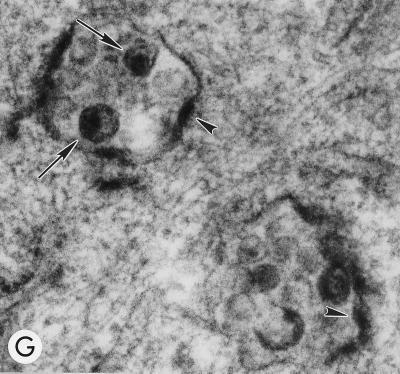
Evaluation of mutant virus targeting and maturation by EM. HeLa cells transfected with the following molecular clones were fixed and analyzed by EM 2 days posttransfection. Arrows and arrowheads indicate VLPs with condensed cores and budding structures, respectively. (A) WT pNL4-3, showing a cluster of typical mature particles on the cell surface. Magnification, ×83,520. (B) pNL4-3/85YG. Magnification, ×72,210. (C) pNL4-3/87VE/PR−. Magnification, ×95,700. (D) pNL4-3KFS/85YG. Magnification, ×89,250. (E) pNL4-3/85YG/p6−. Magnification, ×89,250. (F) pNL4-3/29KE/31KE. Magnification, ×94,600. (G) pNL4-3/29KT/31KT. Magnification, ×93,500.
Intracellular assembly, budding, and maturation of mutant virions require Gag myristylation but are independent of Env and p6.
A mutation in Mason-Pfizer monkey virus Gag that blocks myristylation (and hence membrane binding) does not affect assembly itself, but rather prevents assembled capsids from migrating from the cytosol to the plasma membrane (55). The HIV-1 MA mutants with changes between amino acids 84 and 88 analyzed here show clear evidence of membrane association; however, it is uncertain whether membrane binding is critical to the observed intracellular assembly. To examine this issue, we constructed a double mutant which has, in addition to the 85YG mutation, a Gly→Ala change at MA amino acid 1 (1GA) (16). This mutation abolishes Pr55Gag myristylation and severely impairs Gag membrane binding (4, 45, 61, 71). In the presence of the 1GA mutation, no discernible particles were observed, suggesting that assembly of the MA mutants with changes between amino acids 84 and 88 is dependent upon Gag membrane binding.
Interaction between Env and Gag proteins is thought to play a role in the ER-directed assembly and budding of the spumaretroviruses (19, 20), and in several other retroviral systems, including HIV-1, Env directs virus budding to the basolateral surface of polarized epithelial cells (39, 40, 49). To explore the possibility that Env may play a role in the intracellular budding of the MA mutants under study here, we introduced the 85YG mutation into the env-minus HIV-1 molecular clone pNL4-3KFS (15). EM analysis of HeLa cells transfected with this molecular clone showed that the absence of Env did not affect the intracellular assembly, budding, or subsequent core condensation observed with the Env-expressing pNL4-3/85YG clone (compare Fig. 1B and D). Thus, in this system, the Env glycoprotein is not involved in the retargeting of virus assembly.
The HIV-1 p6Gag protein functions late in the assembly process by stimulating particle release from virus-expressing cells (22, 30); in some systems, introduction of a stop codon at p6 amino acid 1 (p6−) causes an accumulation of immature particles at the plasma membrane and a nearly complete block of virus release (22, 30). To investigate whether p6 deletion might likewise result in the accumulation of immature VLPs at intracellular membranes, we constructed an 85YG/p6− double mutant and examined its assembly, budding, and maturation properties by EM. Interestingly, p6 mutation did not affect the 85YG phenotype (Fig. 1E), suggesting that neither the apparent release of 85YG particles into intracellular vesicles nor the maturation of these particles requires a functional p6.
The 85YG MA mutation redirects assembly to the Golgi complex and/or Golgi-derived vesicles but not to the ER.
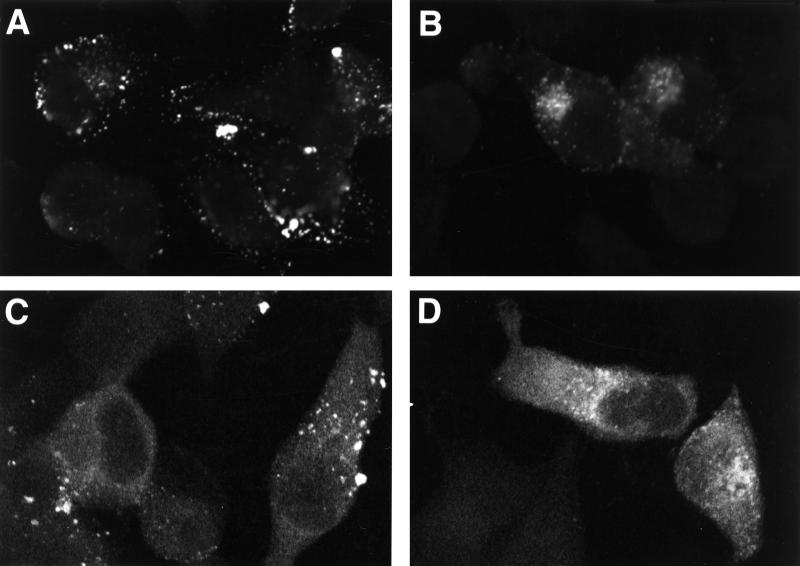
To identify the intracellular site to which VLP assembly is redirected, we used confocal microscopy to examine pNL4-3- and pNL4-3/85YG-transfected HeLa cells stained with several organelle markers as well as anti-Gag antibodies. Gag was detected with two anti-MA monoclonal antibodies. One recognizes the mature p17 (MA) but not the MA domain of the unprocessed Gag precursor Pr55Gag (anti-p17) (71; A. Ono and E. O. Freed, unpublished results); this antibody allowed us to specifically detect mature MA generated upon PR-mediated Gag cleavage. The second antibody recognizes both mature p17 and the MA domain of Pr55Gag (anti-p17/55). When singly stained with anti-p17 antibody, WT-transfected HeLa cells showed a punctate staining pattern on the cell surface, whereas in 85YG-transfected cells we observed an accumulation of punctate signal in the perinuclear region (Fig. 2A and B). As expected, cells transfected with pNL4-3/PR− showed no signal (data not shown), since the anti-p17 antibody does not bind unprocessed Gag. When stained with the anti-p17/55 antibody, WT Gag showed a diffuse cytoplasmic signal in addition to the surface punctate pattern; 85YG also showed a diffuse cytoplasmic staining superimposed on the perinuclear localization pattern (Fig. 2C and D). The hazy staining may represent cytosolic Gag not associated with membrane. Using the anti-p17/55 antibody, cells transfected with the PR− and 85YG/PR− clones displayed a staining pattern similar to those of WT pNL4-3 and pNL4-3/85YG, respectively (data not shown).
FIG. 2.
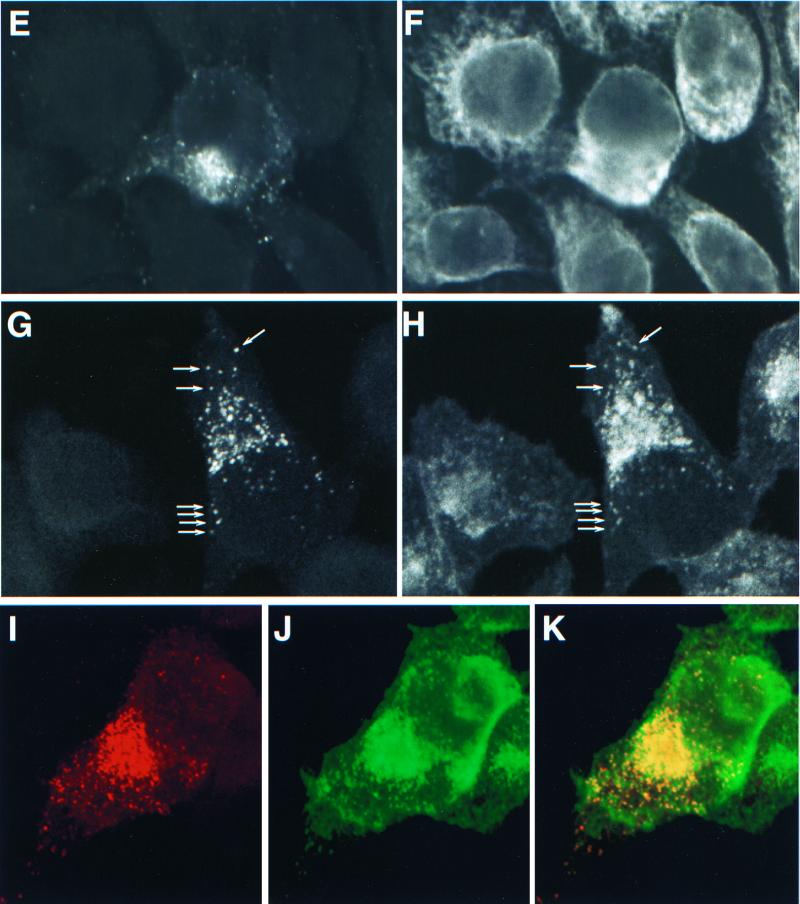
Accumulation of processed mutant Gag in the Golgi and/or post-Golgi vesicles. HeLa cells were transfected with WT pNL4-3 (A and C) or with pNL4-3/85YG (B, D, and E through J) and fixed 2 days posttransfection. After permeabilization, cells were stained with either mouse monoclonal anti-p17 antibody (A and B) or mouse monoclonal anti-p17/55 antibody (C and D) or costained with mouse monoclonal anti-p17 antibody (E, G, and I) and organelle marker rabbit anti-calreticulin antibody (F), RCA60 (H), or WGA (J) and analyzed by confocal microscopy. The same fields are shown for panels E and F, G and H, and I and J. In panels G and H, some vesicles which show double staining for both p17 (MA) and RCA60 are indicated with arrows. Panel I shows 85YG p17 (MA) staining in red and panel J shows WGA staining in green. An overlay of the p17 (MA) and Golgi staining is presented in Panel K; red (MA) and green (Golgi) overlap is visualized as yellow.
The perinuclear staining of 85YG Gag evident with the anti-p17 antibody colocalized neither with calreticulin (Fig. 2E and F), which is an ER luminal protein (35, 44), nor with Rab1B (data not shown), which localizes to the cytoplasmic surface of the ER/cis-Golgi intermediate compartment and/or cis-Golgi (25, 52, 58). In contrast, 85YG p17 (MA) staining substantially colocalized with the region stained by fluoroscein-labeled RCA60 (Fig. 2G and H) and WGA (Fig. 2I to K). RCA60 and WGA have been used as medial/trans- and trans-Golgi markers, respectively (9, 21, 24, 26, 62, 67). These results suggest that VLP assembly of the 85YG mutant is retargeted to the medial Golgi, trans-Golgi, and/or post-Golgi vesicles but not to the ER. Together with the EM results, these observations indicate that core condensation triggered by PR-mediated Gag processing can occur in the Golgi as well as at the plasma membrane.
Highly basic region of MA is functionally related to the domain of MA spanning amino acids 84 to 88.
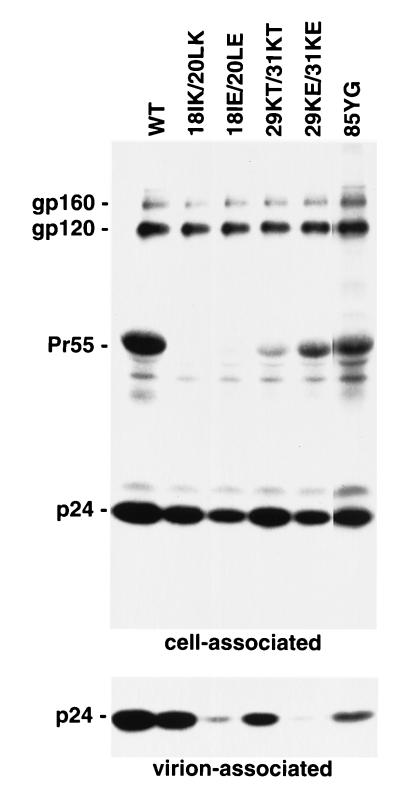
In a previous report, we observed that one of the mutants with a single amino acid change between MA amino acids 84 and 88 (86CS) reverted in culture (46). A second-site, compensatory mutation (27QK) was identified in the highly basic domain of MA. These findings prompted us to examine the possibility that the highly basic domain and the MA domain spanning amino acids 84 to 88 might be functionally related. We characterized several basic domain mutants for virus production, Gag localization, and membrane binding. To assess the impact of changes in the basic domain, we introduced double amino acid mutations that altered the charge of this region. Thus, neutral-to-basic (18IK/20LK), neutral-to-acidic (18IE/20LE), basic-to-neutral (29KT/31KT), and basic-to-acidic (29KE/31KE) substitutions were introduced. One of these mutants (29KT/31KT) was previously shown to cause a threefold reduction in virus particle production (13). HeLa cells were transfected with pNL4-3 or the basic domain mutants and were metabolically labeled. Cell- and virion-associated material was immunoprecipitated with AIDS patient sera and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis followed by autoradiography (Fig. 3). The basic domain changes all caused a decrease in virus particle production, although the severity of this defect varied widely among the mutants. When normalized for cell-associated gp120, the amount of virion-associated p24 released from cells transfected with 18IK/20LK, 18IE/20LE, 29KT/31KT, and 29KE/31KE mutants was 72, 14, 37, and 7% of the WT amount, respectively. Similar effects on virus release were also observed by measuring the production of virion-associated gp120, p66 (RT), or p32 (IN) (data not shown). Levels of cell-associated Pr55Gag detected by immunoprecipitation were significantly reduced by several of the basic domain mutations. This latter observation could be the result of several factors: (i) reduced immunoreactivity previously described for amino acid 20 substitutions (33) and for other MA mutants (6, 10), (ii) Pr55Gag instability, and (iii) increased rate of Pr55Gag processing, also observed previously for residue 20 mutants (33, 34). All of these factors may be operative, since Western blotting revealed less of a reduction in Pr55Gag levels than did the immunoprecipitation analysis; these differences were particularly small but were still present in the absence of a functional PR (data not shown).
FIG. 3.
WT and mutant virus production. HeLa cells transfected with WT pNL4-3 (WT) or its derivatives containing the indicated MA mutations were metabolically labeled with [35S]Cys. Virions were pelleted by ultracentrifugation. Cell- and virion-associated material was immunoprecipitated with AIDS patient sera and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis followed by fluorography. The positions of the Env precursor gp160, the mature surface Env glycoprotein gp120, the Gag precursor Pr55Gag, and p24 (CA) are shown.
HeLa cells transfected with the basic domain mutants were also analyzed by confocal microscopy following immunostaining with the anti-p17 antibody. All four double mutants exhibited substantial amounts of perinuclear staining (Fig. 4); the extent of Gag relocalization correlated with the severity of the defect in assembly or release (Fig. 3). Consistent with these confocal microscopy data, EM analysis of HeLa cells transfected with these mutants showed virus budding into intracellular vesicles (Fig. 1F and G). These results demonstrate that, as observed for mutations between MA residues 84 and 88, alteration of the highly basic domain causes retargeting of Gag to intracellular compartments. Costaining for Gag and cellular markers was performed by confocal microscopy, as described above for 85YG. Again, mutant Gag colocalized with Golgi and post-Golgi markers, but not with the ER-specific calreticulin (data not shown).
FIG. 4.
Accumulation of processed Gag in the Golgi and/or post-Golgi vesicles caused by mutations in the MA basic domain. HeLa cells were transfected with WT pNL4-3 (A), pNL4-3/85YG (B), pNL4-3/18IK/20LK (C), pNL4-3/18IE/20LE (D), pNL4-3/29KT/31KT (E), or pNL4-3/29KE/31KE (F). Two days posttransfection, cells were fixed, permeabilized, stained with monoclonal anti-p17 antibody, and analyzed by confocal microscopy.
Retargeting of virus assembly is not caused by altered Gag membrane binding.
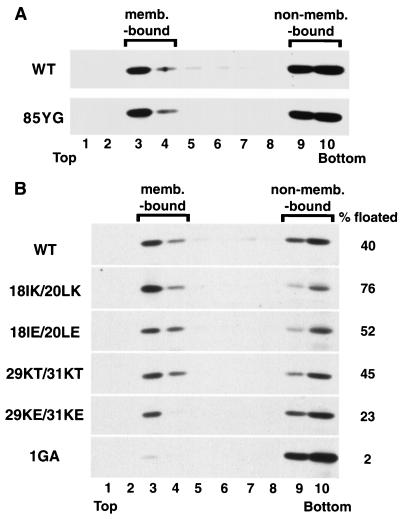
It is possible that an increased affinity for membrane might induce a promiscuous binding of Gag to intracellular membranes, and, as a consequence, a retargeting of virus assembly. To investigate this possibility, we measured the membrane binding ability of 85YG Gag by equilibrium flotation centrifugation. This technique separates membrane-bound Gag from non-membrane-bound Gag complexes, and thus represents a significant improvement over traditional cell fractionation approaches. We focused on Pr55Gag membrane binding by utilizing PR− constructs. HeLa cells transfected with pNL4-3/PR− and pNL4-3/85YG/PR− were sonicated, and postnuclear supernatants of cell homogenates were subjected to equilibrium flotation centrifugation. Membrane-bound material was recovered in fractions 3 and 4, whereas non-membrane-bound Gag remained in the bottom two fractions (45). Consistent with previous studies performed with HeLa cells (45, 50), approximately 40% of WT Pr55Gag was recovered in the membrane-bound fraction (Fig. 5A). The nonmyristylated, 1GA mutant displayed an almost complete block in membrane binding (Fig. 5B). 85YG mutant Pr55Gag showed a behavior essentially identical to that of the WT (Fig. 5A), suggesting that the 85YG mutation does not alter Gag binding to membrane. We emphasize that this assay does not distinguish between plasma and intracellular membrane binding.
FIG. 5.
Effects of MA mutations on Gag membrane binding. HeLa cells were transfected with pNL4-3/PR− (WT) or its derivatives containing the indicated MA mutations. Postnuclear supernatants were prepared and subjected to equilibrium flotation centrifugation (see Materials and Methods), during which membrane-bound (memb.-bound) materials float to the interface between 10 and 65% sucrose (fractions 3 and 4). Pr55Gag detected by Western blotting is shown. In panel B, the percentage of total Pr55Gag that is membrane bound is indicated on the right.
We performed similar equilibrium flotation centrifugation assays to determine the membrane binding properties of the basic domain mutants (Fig. 5B). The two mutants with changes at residues 18 and 20 showed an increased percentage of membrane-bound Pr55Gag relative to WT (18IK/20LK, 76%; 18IE/20LE, 56%). In contrast, the two mutants with changes at residues 29 and 31 showed either unchanged (29KT/31KT, 45%) or decreased (29KE/31KE, 23%) membrane binding relative to that of the WT. These results suggest that, as with the 85YG mutation, the retargeting of assembly observed with the basic domain mutants does not correlate with altered membrane binding.
Targeting defect imposed by 29KE/31KE and 85YG mutants can be reversed by coexpression with WT but not by coexpression with each other.
To understand further the nature of the targeting signal affected by the 85YG and basic domain mutants, we asked whether WT Pr55Gag could rescue mutant Gag into extracellular particles by providing a plasma membrane targeting signal in trans. To distinguish WT from mutant Pr55Gag, we constructed molecular clones which express Pr55Gag containing either HA or FLAG C-terminal epitope tags. HeLa cells were cotransfected with various combinations of clones expressing tagged versions of WT and MA mutant Gag. Cell and virion lysates were analyzed by Western blotting with rabbit anti-HA or anti-FLAG antiserum. Consistent with our observations using untagged Pr55Gag, 85YG Gag tagged with the HA or FLAG epitope produced virion-associated material from transfected HeLa cells with an efficiency that was markedly reduced relative to that of HA- or FLAG-tagged WT Gag (Fig. 6, compare lanes 13 and 14 or lanes 16 and 17). When HA-tagged 85YG Gag was coexpressed with FLAG-tagged WT Gag, the amount of released 85YG Gag increased substantially (Fig. 6, compare lanes 22 and 14). Similar results were obtained when FLAG-tagged 85YG and HA-tagged WT Gag were coexpressed (Fig. 6, compare lanes 19 and 17). Similarly, the release of 29KE/31KE Gag was also increased when it was coexpressed with WT Gag (Fig. 6, compare lanes 15 and 23 or lanes 18 and 20). These results indicate that 85YG and 29KE/31KE mutant Pr55Gag proteins can be rescued into virions by coexpression with WT Gag and imply that a plasma membrane targeting signal can be provided to 85YG and 29KE/31KE Gag by oligomerization with WT Gag.
FIG. 6.
Coexpression of WT and mutant Gag proteins. HeLa cells were transfected at a ratio of 1:1 with various combinations of molecular clones expressing WT, 85YG, or 29KE/31KE Gag tagged with either the HA or the FLAG epitope. Virions were pelleted by ultracentrifugation. Cell and virion lysates were analyzed by Western blotting with anti-HA or anti-FLAG antibodies. Epitope-tagged Pr55Gag is shown.
The ability of a basic domain substitution (27QK) to reverse the phenotype of a mutation between MA residues 84 and 88 (86CS) (46) suggests that these two domains are in the same functional complementation group. To test this hypothesis, we coexpressed 85YG and 29KE/31KE Gag proteins tagged with either the HA or the FLAG epitope and the release of both Gag species was monitored as above. The results indicated that cells coexpressing 85YG and 29KE/31KE Gag showed no increase in virus-associated Gag release (Fig. 6, lanes 21 and 24) compared with singly expressing cells (Fig. 6, lanes 14, 15, 17, and 18). Thus, both 85YG and 29KE/31KE changes may alter the same targeting signal.
DISCUSSION
We previously demonstrated that HIV-1 particle assembly can be retargeted from the plasma membrane to an intracellular site by introducing single amino acid changes between MA amino acids 84 and 88 (16) (Fig. 1). Interestingly, these particles appeared to have a mature-particle-like morphology with a condensed core. In this report, we further characterize the intracellular particles by demonstrating that the core condensation in intracellular mutant virus particles is driven by the viral PR (Fig. 1). Confocal microscopy utilizing a monoclonal antibody which specifically recognizes the p17 (MA) Gag cleavage product indicates that the mutant MA colocalizes with Golgi or post-Golgi markers but not with the ER marker calreticulin (Fig. 2). Since the majority of HIV-1 PR activity is associated with Pr55Gag assembly and particle release (32), these results suggest that these MA mutants undergo VLP formation and subsequent PR-mediated maturation in the Golgi or post-Golgi vesicles but not in the ER. It was reported previously that deletion of MA amino acids 16 to 99 caused intracellular budding of mutant virus in the ER; in contrast to that of our mutants (Fig. 1), the morphology of these virus particles was exclusively immature (11). Collectively, these results suggest that PR-mediated maturation of HIV-1 can occur not only at the plasma membrane but also in Golgi or post-Golgi compartments but that perhaps it cannot take place in the ER. However, we cannot exclude the possibility that the differences in maturation observed between these studies might relate to the use of different mutants and cell types.
It has been demonstrated that elimination of p6 or mutation of the Pro-Thr-Ala-Pro-Pro sequence near the N terminus of p6 causes a defect late in the budding process (22, 30). As a consequence of such mutations, large numbers of immature particles are observed to be tethered to the plasma membrane and virus release is markedly impaired (22, 30). In the case of the retargeted mutants analyzed here, however, the lack of p6 affected neither the apparent release of VLPs into intracellular vesicles nor subsequent core condensation (Fig. 1E). These results indicate that virus assembly and maturation at intracellular vesicles does not require p6. Likewise, mutational inactivation of the env gene did not alter intracellular assembly (Fig. 1D), indicating that this phenomenon is not influenced by Env glycoprotein expression. This is in contrast to the situation observed with the intracellular assembly carried out by the spumaretroviruses; in this system an ER retention signal in the cytoplasmic tail of the transmembrane Env protein reportedly promotes ER-directed virus budding (19, 20).
Relationship between Gag membrane binding and targeting.
The observation that the replication-defective phenotype of a mutant containing a substitution at MA residue 86 (86CS) could be reversed by a second-site change in the MA highly basic domain (46) suggested the existence of a functional relationship between the highly basic domain and amino acids 84 to 88 (see below). Consistent with this hypothesis, we demonstrate that mutations in the highly basic domain and between residues 84 and 88 induce similar phenotypes, i.e., reduced extracellular virus production due to retargeting of virus assembly to intracellular vesicles (Fig. 3 and 4). Based on the crystal and nuclear magnetic resonance structures of the nonmyristylated HIV-1 MA (28, 42) and in vitro membrane binding assays (70) it was proposed that the highly basic domain, along with basic amino acids in other regions of MA, forms a positively charged patch that promotes membrane binding of Gag through electrostatic interaction with acidic phospholipids in the inner leaflet of membranes (70). Thus, it was of interest to determine whether membrane binding and plasma membrane targeting of Gag are interrelated functions of MA. For the mutants analyzed here, this is not the case; the 85YG mutant displays WT membrane binding properties despite the fact that its targeting is markedly affected. Moreover, the basic domain mutations all induce Gag retargeting, yet membrane binding is variably affected: the 18IK/20LK and 18IE/20LE mutations increase membrane binding, the 29KT/31KT mutant behaves like the WT Gag in flotation assays, and the 29KE/31KE changes decrease membrane binding. The 20LK mutation described previously (33) increases membrane binding but does not induce Gag retargeting (A. Ono and E. O. Freed, unpublished results). Taken together, these results indicate that the roles of MA in membrane binding and Gag targeting are genetically separable, although both are influenced by the basic domain.
It has been suggested that Gag mutants containing large MA deletions might display an enhanced membrane binding ability as a result of increased exposure of the N-terminal myristate moiety (54). This increase in membrane binding could potentially enable Gag to override its selectivity for the plasma membrane and cause a promiscuous binding of Gag to both intracellular and plasma membranes. This model clearly does not apply to the mutants analyzed here, since, as discussed above, we observed no direct correlation between effects on Gag membrane binding and targeting. In addition, the intracellular localization of mutant Pr55Gag and the site of retargeted assembly were specific for the Golgi or post-Golgi vesicles. Localization of Gag to the ER was not detected, even though the ER membrane is the most abundant intracellular membrane (41). These results are most consistent with the idea that the intracellular virus assembly observed with the MA mutants studied here is due not to promiscuous assembly but rather to a specific retargeting.
Possible mechanisms for targeting of virus assembly.
Several mechanisms could explain the retargeting of MA mutant virus formation. (i) The substitutions reported here could disrupt a plasma membrane-targeting signal and reveal a weaker but preexisting Golgi-targeting signal. This model could be operative if WT Pr55Gag is directly targeted to the plasma membrane after its synthesis or if it first binds an intracellular membrane prior to plasma membrane transport. It is unclear why HIV-1 Gag would have evolved a cryptic Golgi-targeting signal; however, it has been reported that in certain cell types (e.g., primary human macrophages) the Golgi or post-Golgi vesicles are the primary site of virus assembly (48). Thus, a cryptic Golgi-targeting signal, revealed by these MA mutations, might be dominant in some cases of natural HIV-1 infection. (ii) The MA mutations described here could create a Golgi-targeting signal. This seems somewhat unlikely given the diversity of changes that lead to the retargeting phenotype. (iii) WT Pr55Gag may bind a variety of intracellular membranes during its transport, but assembly may be suppressed until Gag reaches the plasma membrane. In this case, the MA mutations may alter the regulation of virus assembly, leading to premature assembly of VLPs in the Golgi.
Coexpression of WT Gag rescued both 29KE/31KE and 85YG mutant Gag into extracellular virus particles (Fig. 6), indicating that the plasma membrane-targeting signal can be provided in trans. This result, which confirms a similar finding obtained with a MA basic domain deletion mutant (69), suggests that the WT and mutant Gag proteins share at least part of their transport pathways, allowing them to interact. In cells coexpressing WT and mutant Pr55Gag, both Gag proteins may bind an intracellular membrane, oligomerize, and then traffic to the plasma membrane. Alternatively, these two species may oligomerize with each other in the cytosol and then traffic together to the plasma membrane. This latter possibility is consistent with reports indicating that HIV-1 Gag can form large, non-membrane-bound oligomeric complexes in virus-expressing cells (37, 38).
Although WT Gag could efficiently rescue the retargeted mutants into virus particles, 85YG and 29KE/31KE could not complement each other (Fig. 6). It has been reported that two Gag mutants which belong to different phenotypic groups can complement each other when coexpressed (29). The lack of complementation observed here is consistent with mutations in the MA basic domain and between amino acid 84 and 88 affecting the same targeting signal. However, we cannot rule out the possibility that the 85YG and 29KE/31KE mutants may be unable to interact with each other, or that one may exert a trans-dominant-negative effect upon the other.
Structural data obtained for both simian immunodeficiency virus and HIV-1 MA (28, 42, 43, 53) suggest that MA amino acids 84 to 88 are in an α-helical domain located in the center of the tightly packed globular core of MA, whereas the highly basic domain is exposed on the surface of the molecule. Although it is not yet established that the currently available MA structures reflect the folding of the myristylated MA domain of Pr55Gag, it is likely that the basic domain, rather than amino acids 84 to 88, is directly involved in plasma membrane targeting. Mutations between amino acids 84 and 88 may cause a conformational change which, in turn, may affect the function of the basic domain. Alternatively, mutations in the basic domain and MA amino acids 84 to 88 may affect a third domain which directly promotes Gag trafficking to the plasma membrane.
To understand the mechanism by which WT HIV-1 assembly occurs selectively at the plasma membrane, it is important to identify not only the targeting signal in Gag but also the element of the plasma membrane to which Gag is transported. It has been suggested that the basic residues of the highly basic domain may interact with acidic phospholipids on the inner leaflet of the plasma membrane (70). However, we observed some retargeting even for a mutation which increased the positive charge of the basic domain (18IK/20LK) (Fig. 4). Thus, although the putative interaction between basic amino acids and acidic phospholipids may promote membrane binding, it is unlikely to be responsible for the specificity of Gag targeting. Still, it is possible that a certain kind of lipid which specifically resides on the plasma membrane may interact with Gag to direct plasma membrane targeting. Plasma membrane microdomains, which consist of specific sets of lipids and proteins, have been the focus of much interest in recent years. One such microdomain, or raft, is rich in sphingolipids and cholesterol (3, 59). Interestingly, the lipid bilayer of the HIV-1 envelope is reportedly enriched for cholesterol and sphingolipids relative to the host cell plasma membrane (2), raising the possibility that rafts or similar lipid microdomains might be involved in HIV-1 assembly. A proteinaceous Gag receptor could also provide a site on the plasma membrane to which Gag is targeted.
We and others have observed that WT retroviral Gag proteins display a punctate staining pattern on the surface of Gag-expressing cells (Fig. 2 and 4) (27, 31, 57). These sites of intense Gag localization presumably represent active centers of virus assembly. Characterization of such sites will provide additional insights into the mechanism of Gag trafficking and virus assembly.
ACKNOWLEDGMENTS
We thank D. Demirov, T. Murakami, and R. Willey for helpful suggestions and critical review of the manuscript. The following reagent was obtained through the NIH AIDS Research and Reference Reagent Program: HIV-1 patient (neutralizing) serum (from L. Vujcic).
REFERENCES
- 1.Adachi A, Gendelman H E, Koenig S, Folks T, Willey R, Rabson A, Martin M A. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aloia R C, Tian H, Jensen F C. Lipid composition and fluidity of the human immunodeficiency virus envelope and host cell plasma membranes. Proc Natl Acad Sci USA. 1993;90:5181–5185. doi: 10.1073/pnas.90.11.5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown D A, London E. Structure of detergent-resistant membrane domains: does phase separation occur in biological membranes? Biochem Biophys Res Commun. 1997;240:1–7. doi: 10.1006/bbrc.1997.7575. [DOI] [PubMed] [Google Scholar]
- 4.Bryant M, Ratner L. Myristoylation-dependent replication and assembly of human immunodeficiency virus 1. Proc Natl Acad Sci USA. 1990;87:523–527. doi: 10.1073/pnas.87.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cannon P M, Matthews S, Clark N, Byles E D, Iourin O, Hockley D J, Kingsman S M, Kingsman A J. Structure-function studies of the human immunodeficiency virus type 1 matrix protein, p17. J Virol. 1997;71:3474–3483. doi: 10.1128/jvi.71.5.3474-3483.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casella C R, Raffini L J, Panganiban A T. Pleiotropic mutations in the HIV-1 matrix protein that affect diverse steps in replication. Virology. 1997;228:294–306. doi: 10.1006/viro.1996.8355. [DOI] [PubMed] [Google Scholar]
- 7.Choi G, Park S, Choi B, Hong S, Lee J, Hunter E, Rhee S S. Identification of a cytoplasmic targeting/retention signal in a retroviral Gag polyprotein. J Virol. 1999;73:5431–5437. doi: 10.1128/jvi.73.7.5431-5437.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conte M R, Matthews S. Retroviral matrix proteins: a structural perspective. Virology. 1998;246:191–198. doi: 10.1006/viro.1998.9206. [DOI] [PubMed] [Google Scholar]
- 9.Davis G L, Hunter E. A charged amino acid substitution within the transmembrane anchor of the Rous sarcoma virus envelope glycoprotein affects surface expression but not intracellular transport. J Cell Biol. 1987;105:1191–1203. doi: 10.1083/jcb.105.3.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dorfman T, Mammano F, Haseltine W A, Gottlinger H G. Role of the matrix protein in the virion association of the human immunodeficiency virus type 1 envelope glycoprotein. J Virol. 1994;68:1689–1696. doi: 10.1128/jvi.68.3.1689-1696.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fäcke M, Janetzko A, Shoeman R L, Kräusslich H G. A large deletion in the matrix domain of the human immunodeficiency virus gag gene redirects virus particle assembly from the plasma membrane to the endoplasmic reticulum. J Virol. 1993;67:4972–4980. doi: 10.1128/jvi.67.8.4972-4980.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freed E O. HIV-1 Gag proteins: diverse functions in the virus life cycle. Virology. 1998;251:1–15. doi: 10.1006/viro.1998.9398. [DOI] [PubMed] [Google Scholar]
- 13.Freed E O, Englund G, Martin M A. Role of the basic domain of human immunodeficiency virus type 1 matrix in macrophage infection. J Virol. 1995;69:3949–3954. doi: 10.1128/jvi.69.6.3949-3954.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Freed E O, Martin M A. Evidence for a functional interaction between the V1/V2 and C4 domains of human immunodeficiency virus type 1 envelope glycoprotein gp120. J Virol. 1994;68:2503–2512. doi: 10.1128/jvi.68.4.2503-2512.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Freed E O, Martin M A. Virion incorporation of envelope glycoproteins with long but not short cytoplasmic tails is blocked by specific, single amino acid substitutions in the human immunodeficiency virus type 1 matrix. J Virol. 1995;69:1984–1989. doi: 10.1128/jvi.69.3.1984-1989.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Freed E O, Orenstein J M, Buckler-White A J, Martin M A. Single amino acid changes in the human immunodeficiency virus type 1 matrix protein block virus particle production. J Virol. 1994;68:5311–5320. doi: 10.1128/jvi.68.8.5311-5320.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gallina A, Mantoan G, Rindi G, Milanesi G. Influence of MA internal sequences, but not of the myristylated N-terminus sequence, on the budding site of HIV-1 Gag protein. Biochem Biophys Res Commun. 1994;204:1031–1038. doi: 10.1006/bbrc.1994.2566. [DOI] [PubMed] [Google Scholar]
- 18.Gheysen D, Jacobs E, de Foresta F, Thiriart C, Francotte M, Thines D, De Wilde M. Assembly and release of HIV-1 precursor Pr55gag virus-like particles from recombinant baculovirus-infected insect cells. Cell. 1989;59:103–112. doi: 10.1016/0092-8674(89)90873-8. [DOI] [PubMed] [Google Scholar]
- 19.Goepfert P A, Shaw K, Wang G, Bansal A, Edwards B H, Mulligan M J. An endoplasmic reticulum retrieval signal partitions human foamy virus maturation to intracytoplasmic membranes. J Virol. 1999;73:7210–7217. doi: 10.1128/jvi.73.9.7210-7217.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goepfert P A, Shaw K L, Ritter G D, Jr, Mulligan M J. A sorting motif localizes the foamy virus glycoprotein to the endoplasmic reticulum. J Virol. 1997;71:778–784. doi: 10.1128/jvi.71.1.778-784.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gorelick F S, Sarras M P, Jr, Jamieson J D. Regional differences in lectin binding to colonic epithelium by fluorescent and electron microscopy. J Histochem Cytochem. 1982;30:1097–1108. doi: 10.1177/30.11.6897257. [DOI] [PubMed] [Google Scholar]
- 22.Göttlinger H G, Dorfman T, Sodroski J G, Haseltine W A. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc Natl Acad Sci USA. 1991;88:3195–3199. doi: 10.1073/pnas.88.8.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Göttlinger H G, Sodroski J G, Haseltine W A. Role of capsid precursor processing and myristoylation in morphogenesis and infectivity of human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1989;86:5781–5785. doi: 10.1073/pnas.86.15.5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Griffiths G, Brands R, Burke B, Louvard D, Warren G. Viral membrane proteins acquire galactose in trans Golgi cisternae during intracellular transport. J Cell Biol. 1982;95:781–792. doi: 10.1083/jcb.95.3.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Griffiths G, Ericsson M, Krijnse-Locker J, Nilsson T, Goud B, Soling H D, Tang B L, Wong S H, Hong W. Localization of the Lys, Asp, Glu, Leu tetrapeptide receptor to the Golgi complex and the intermediate compartment in mammalian cells. J Cell Biol. 1994;127:1557–1574. doi: 10.1083/jcb.127.6.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Griffiths G, Quinn P, Warren G. Dissection of the Golgi complex. I. Monensin inhibits the transport of viral membrane proteins from medial to trans Golgi cisternae in baby hamster kidney cells infected with Semliki Forest virus. J Cell Biol. 1983;96:835–850. doi: 10.1083/jcb.96.3.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hansen M, Jelinek L, Whiting S, Barklis E. Transport and assembly of gag proteins into Moloney murine leukemia virus. J Virol. 1990;64:5306–5316. doi: 10.1128/jvi.64.11.5306-5316.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hill C P, Worthylake D, Bancroft D P, Christensen A M, Sundquist W I. Crystal structures of the trimeric human immunodeficiency virus type 1 matrix protein: implications for membrane association and assembly. Proc Natl Acad Sci USA. 1996;93:3099–3104. doi: 10.1073/pnas.93.7.3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hong S S, Boulanger P. Assembly-defective point mutants of the human immunodeficiency virus type 1 Gag precursor phenotypically expressed in recombinant baculovirus-infected cells. J Virol. 1993;67:2787–2798. doi: 10.1128/jvi.67.5.2787-2798.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang M, Orenstein J M, Martin M A, Freed E O. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J Virol. 1995;69:6810–6818. doi: 10.1128/jvi.69.11.6810-6818.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones T A, Blaug G, Hansen M, Barklis E. Assembly of gag-β-galactosidase proteins into retrovirus particles. J Virol. 1990;64:2265–2279. doi: 10.1128/jvi.64.5.2265-2279.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaplan A H, Manchester M, Swanstrom R. The activity of the protease of human immunodeficiency virus type 1 is initiated at the membrane of infected cells before the release of viral proteins and is required for release to occur with maximum efficiency. J Virol. 1994;68:6782–6786. doi: 10.1128/jvi.68.10.6782-6786.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kiernan R E, Ono A, Englund G, Freed E O. Role of matrix in an early postentry step in the human immunodeficiency virus type 1 life cycle. J Virol. 1998;72:4116–4126. doi: 10.1128/jvi.72.5.4116-4126.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kiernan R E, Ono A, Freed E O. Reversion of a human immunodeficiency virus type 1 matrix mutation affecting Gag membrane binding, endogenous reverse transcriptase activity, and virus infectivity. J Virol. 1999;73:4728–4737. doi: 10.1128/jvi.73.6.4728-4737.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krause K H, Michalak M. Calreticulin. Cell. 1997;88:439–443. doi: 10.1016/s0092-8674(00)81884-x. [DOI] [PubMed] [Google Scholar]
- 36.Lee P P, Linial M L. Efficient particle formation can occur if the matrix domain of human immunodeficiency virus type 1 Gag is substituted by a myristylation signal. J Virol. 1994;68:6644–6654. doi: 10.1128/jvi.68.10.6644-6654.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee Y-M, Liu B, Yu X-F. Formation of virus assembly intermediate complexes in the cytoplasm by wild-type and assembly-defective mutant human immunodeficiency virus type 1 and their association with membranes. J Virol. 1999;73:5654–5662. doi: 10.1128/jvi.73.7.5654-5662.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee Y M, Yu X F. Identification and characterization of virus assembly intermediate complexes in HIV-1-infected CD4+ T cells. Virology. 1998;243:78–93. doi: 10.1006/viro.1998.9064. [DOI] [PubMed] [Google Scholar]
- 39.Lodge R, Delamarre L, Lalonde J P, Alvarado J, Sanders D A, Dokhelar M C, Cohen E A, Lemay G. Two distinct oncornaviruses harbor an intracytoplasmic tyrosine-based basolateral targeting signal in their viral envelope glycoprotein. J Virol. 1997;71:5696–5702. doi: 10.1128/jvi.71.7.5696-5702.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lodge R, Göttlinger H, Gabuzda D, Cohen E A, Lemay G. The intracytoplasmic domain of gp41 mediates polarized budding of human immunodeficiency virus type 1 in MDCK cells. J Virol. 1994;68:4857–4861. doi: 10.1128/jvi.68.8.4857-4861.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lodish H, Baltimore D, Berk A, Zipursky S L, Matsudaira P, Darnell J. Molecular Cell Biology. 3rd ed. New York, N.Y: Scientific American Books, Inc.; 1995. Cell organization, subcellular structure, and cell division. [Google Scholar]
- 42.Massiah M A, Starich M R, Paschall C, Summers M F, Christensen A M, Sundquist W I. Three-dimensional structure of the human immunodeficiency virus type 1 matrix protein. J Mol Biol. 1994;244:198–223. doi: 10.1006/jmbi.1994.1719. [DOI] [PubMed] [Google Scholar]
- 43.Matthews S, Barlow P, Boyd J, Barton G, Russell R, Mills H, Cunningham M, Meyers N, Burns N, Clark N, et al. Structural similarity between the p17 matrix protein of HIV-1 and interferon-gamma. Nature. 1994;370:666–668. doi: 10.1038/370666a0. [DOI] [PubMed] [Google Scholar]
- 44.Michalak M, Baksh S, Opas M. Identification and immunolocalization of calreticulin in pancreatic cells: no evidence for “calciosomes”. Exp Cell Res. 1991;197:91–99. doi: 10.1016/0014-4827(91)90484-c. [DOI] [PubMed] [Google Scholar]
- 45.Ono A, Freed E O. Binding of human immunodeficiency virus type 1 Gag to membrane: role of the matrix amino terminus. J Virol. 1999;73:4136–4144. doi: 10.1128/jvi.73.5.4136-4144.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ono A, Huang M, Freed E O. Characterization of human immunodeficiency virus type 1 matrix revertants: effects on virus assembly, Gag processing, and Env incorporation into virions. J Virol. 1997;71:4409–4418. doi: 10.1128/jvi.71.6.4409-4418.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ono A, Kawakita M. Transport of envelope proteins of Sendai virus, HN and F0, is blocked at different steps by thapsigargin and other perturbants to intracellular Ca2+ J Biochem (Tokyo) 1994;116:649–656. doi: 10.1093/oxfordjournals.jbchem.a124575. [DOI] [PubMed] [Google Scholar]
- 48.Orenstein J M, Meltzer M S, Phipps T, Gendelman H E. Cytoplasmic assembly and accumulation of human immunodeficiency virus types 1 and 2 in recombinant human colony-stimulating factor-1-treated human monocytes: an ultrastructural study. J Virol. 1988;62:2578–2586. doi: 10.1128/jvi.62.8.2578-2586.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Owens R J, Dubay J W, Hunter E, Compans R W. Human immunodeficiency virus envelope protein determines the site of virus release in polarized epithelial cells. Proc Natl Acad Sci USA. 1991;88:3987–3991. doi: 10.1073/pnas.88.9.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Paillart J-C, Göttlinger H G. Opposing effects of human immunodeficiency virus type 1 matrix mutations support a myristyl switch model of Gag membrane targeting. J Virol. 1999;73:2604–2612. doi: 10.1128/jvi.73.4.2604-2612.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pal, R., M. S. Reitz, Jr., E. Tschachler, R. C. Gallo, M. G. Sarngadharan, and F. D. Veronese. 1990. Myristoylation of gag proteins of HIV-1 plays an important role in virus assembly. AIDS Res. Hum. Retrovir. 6:721–730. [DOI] [PubMed]
- 52.Plutner H, Cox A D, Pind S, Khosravi-Far R, Bourne J R, Schwaninger R, Der C J, Balch W E. Rab1b regulates vesicular transport between the endoplasmic reticulum and successive Golgi compartments. J Cell Biol. 1991;115:31–43. doi: 10.1083/jcb.115.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rao Z, Belyaev A S, Fry E, Roy P, Jones I M, Stuart D I. Crystal structure of SIV matrix antigen and implications for virus assembly. Nature. 1995;378:743–747. doi: 10.1038/378743a0. [DOI] [PubMed] [Google Scholar]
- 54.Reil H, Bukovsky A A, Gelderblom H R, Göttlinger H G. Efficient HIV-1 replication can occur in the absence of the viral matrix protein. EMBO J. 1998;17:2699–2708. doi: 10.1093/emboj/17.9.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rhee S S, Hunter E. Myristylation is required for intracellular transport but not for assembly of D-type retrovirus capsids. J Virol. 1987;61:1045–1053. doi: 10.1128/jvi.61.4.1045-1053.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rhee S S, Hunter E. A single amino acid substitution within the matrix protein of a type D retrovirus converts its morphogenesis to that of a type C retrovirus. Cell. 1990;63:77–86. doi: 10.1016/0092-8674(90)90289-q. [DOI] [PubMed] [Google Scholar]
- 57.Sandefur S, Varthakavi V, Spearman P. The I domain is required for efficient plasma membrane binding of human immunodeficiency virus type 1 Pr55Gag. J Virol. 1998;72:2723–2732. doi: 10.1128/jvi.72.4.2723-2732.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schwaninger R, Plutner H, Bokoch G M, Balch W E. Multiple GTP-binding proteins regulate vesicular transport from the ER to Golgi membranes. J Cell Biol. 1992;119:1077–1096. doi: 10.1083/jcb.119.5.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Simons K, Ikonen E. Functional rafts in cell membranes. Nature. 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 60.Soneoka Y, Kingsman S M, Kingsman A J. Mutagenesis analysis of the murine leukemia virus matrix protein: identification of regions important for membrane localization and intracellular transport. J Virol. 1997;71:5549–5559. doi: 10.1128/jvi.71.7.5549-5559.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Spearman P, Horton R, Ratner L, Kuli-Zade I. Membrane binding of human immunodeficiency virus type 1 matrix protein in vivo supports a conformational myristyl switch mechanism. J Virol. 1997;71:6582–6592. doi: 10.1128/jvi.71.9.6582-6592.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Virtanen I, Ekblom P, Laurila P. Subcellular compartmentalization of saccharide moieties in cultured normal and malignant cells. J Cell Biol. 1980;85:429–434. doi: 10.1083/jcb.85.2.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang C T, Lai H Y, Li J J. Analysis of minimal human immunodeficiency virus type 1 gag coding sequences capable of virus-like particle assembly and release. J Virol. 1998;72:7950–7959. doi: 10.1128/jvi.72.10.7950-7959.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang C T, Zhang Y, McDermott J, Barklis E. Conditional infectivity of a human immunodeficiency virus matrix domain deletion mutant. J Virol. 1993;67:7067–7076. doi: 10.1128/jvi.67.12.7067-7076.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Welker R, Janetzko A, Kräusslich H G. Plasma membrane targeting of chimeric intracisternal A-type particle polyproteins leads to particle release and specific activation of the viral proteinase. J Virol. 1997;71:5209–5217. doi: 10.1128/jvi.71.7.5209-5217.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Willey R L, Bonifacino J S, Potts B J, Martin M A, Klausner R D. Biosynthesis, cleavage, and degradation of the human immunodeficiency virus 1 envelope glycoprotein gp160. Proc Natl Acad Sci USA. 1988;85:9580–9584. doi: 10.1073/pnas.85.24.9580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wills J W, Srinivas R V, Hunter E. Mutations of the Rous sarcoma virus env gene that affect the transport and subcellular location of the glycoprotein products. J Cell Biol. 1984;99:2011–2023. doi: 10.1083/jcb.99.6.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yu X, Yuan X, Matsuda Z, Lee T H, Essex M. The matrix protein of human immunodeficiency virus type 1 is required for incorporation of viral envelope protein into mature virions. J Virol. 1992;66:4966–4971. doi: 10.1128/jvi.66.8.4966-4971.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yuan X, Yu X, Lee T H, Essex M. Mutations in the N-terminal region of human immunodeficiency virus type 1 matrix protein block intracellular transport of the Gag precursor. J Virol. 1993;67:6387–6394. doi: 10.1128/jvi.67.11.6387-6394.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhou W, Parent L J, Wills J W, Resh M D. Identification of a membrane-binding domain within the amino-terminal region of human immunodeficiency virus type 1 Gag protein which interacts with acidic phospholipids. J Virol. 1994;68:2556–2269. doi: 10.1128/jvi.68.4.2556-2569.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhou W, Resh M D. Differential membrane binding of the human immunodeficiency virus type 1 matrix protein. J Virol. 1996;70:8540–8548. doi: 10.1128/jvi.70.12.8540-8548.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]