Abstract
Kaposi's sarcoma-associated herpesvirus (KSHV), or human herpesvirus 8 (HHV-8), belongs to the gammaherpesvirus subfamily and encodes ∼80 open reading frames (ORFs). Among them are a few candidates for immediate-early genes (e.g., K5). We developed a monoclonal antibody (MAb), 328C7, against the K5 antigen. This MAb reacted with the K5 gene product by immunoscreening of a cDNA library from BCBL-1 cells, and this result was confirmed by transfection of the K5 ORF into Cos-7 cells. After induction of lytic infection by treatment with 12-O-tetradecanoylphorbol-13-acetate, MAb 328C7 reacted with an antigen in the cytoplasm of BCBL-1 and BC-3 cells as early as after 4 h of induction. Immunoelectron microscopy showed that the K5 antigen was situated mainly in the endoplasmic reticulum but was not present on the virion or in the nucleus. Northern blotting with a K5-specific probe revealed a single transcript of 1.2 kb, while Western blotting showed the antigen to be a 36-kDa polypeptide. The 5′ and 3′ ends were then determined by rapid amplification of cDNA, followed by sequencing of RACE products, and a splice was revealed upstream of the K5 ORF. K5 expression was unaffected by the respective DNA and protein synthesis inhibitors phosphonoformic acid and cycloheximide plus actinomycin D, confirming its immediate-early nature. Transient-transfection assays showed that the K5 promoter was transactivated by ORF 50 (KSHV Rta), a homolog of Epstein-Barr virus Rta, but the K5 gene product exhibited no transregulation of its own promoter or those of DNA polymerase and the human immunodeficiency virus type 1 long terminal repeat. This is the first such analysis of an immediate-early gene product; determination of its specific biological function requires further investigation.
Kaposi's sarcoma-associated herpesvirus (KSHV), or human herpesvirus 8 (HHV-8), was recently discovered in tissues isolated from varied forms of Kaposi's sarcoma (7, 18, 38), Castleman's disease (39), and body cavity-based lymphoma (BCBL; primary effusion lymphoma) (5). The etiology and pathogenic mechanisms of HHV-8 are as yet unknown, but its origin and kindred strongly suggest that HHV-8 promotes a particular type of cell proliferation (28). All of the cell lines derived from BCBL harbor the HHV-8 genome in a latent state; the virus is transformed to a lytic state by chemical agents such as 12-O-tetradecanoylphorbol-13-acetate (TPA) and n-butyrate (1, 5, 24, 25, 34, 37). The first such cell line reported, BC-1, was also infected with Epstein-Barr virus (EBV) (6, 13), but later BCBL lines were EBV negative (1, 34). Although HHV-8 is difficult to transmit, it can be maintained in some cell lines and virus production can be induced by treatment with TPA or n-butyrate (12, 27).
HHV-8 is a new member of the gammaherpesvirus subfamily and has genetic similarity to herpesvirus saimiri (HVS) and EBV (27, 35). Its genome, which has been completely sequenced, consists of a double-stranded long unique DNA sequence of 140.5 kbp flanked by GC-rich terminal repeat sequences (30, 35). It encodes about 80 complete open reading frames (ORFs), some of which have similarity to those of HVS (35). Among them, 15 ORFs, designated K1 to K15, are unique to HHV-8. In addition, HHV-8 encodes a number of human gene homologs related to immune function and cell cycle regulation, including ORFs 16 (viral Bcl-2) (8, 36), 72 (viral cyclin D) (14, 21, 33), 74 (viral interleukin-8 receptor) (2, 16), K2 (viral interleukin-6) (4, 26, 30, 31), K4 and K6 (viral macrophage inflammatory proteins I and II, respectively) (3, 20, 31), K9 (viral interferon regulatory factor) (19, 22, 47), and K13 (vFLIP/vFLICE [caspase-8]-inhibitory protease) (42). Earlier reports suggested that some of these factors do, indeed, initiate cell proliferation, thereby promoting tumor progression.
Herpesvirus genes are classified as latent, immediate-early (IE), early (E), and late (L) (17, 41, 46). Expression of the IE and E genes is independent of viral DNA replication, and some are involved in gene regulation and DNA replication (17, 23, 40). The L genes, by contrast, are dependent on viral DNA replication and mainly encode structural proteins (17). Regulation of gene expression in cells infected with herpesviruses is generally ordered in a cascade fashion: IE genes are transcribed first following cell penetration of the virus, after which the E and L genes are expressed. Recent analysis of the complete DNA sequence suggests that HHV-8 possesses several IE candidate genes, including K3, K5, K8, ORF 45, ORF 50, and ORF 57 (35). Of these, the K3 and K5 genes have homology to the IE-1 gene from bovine herpesvirus 4 (BHV-4) (35), which is also a gammaherpesvirus (9).
To analyze HHV-8 gene function more precisely, we developed monoclonal antibodies (MAbs) against viral proteins and attempted to identify their antigens using the λgt11 cDNA library of HHV-8-infected BCBL-1 cells. Among our MAbs, six specifically recognized the K5 gene product and we have used one of them (MAb 328C7) as a probe to characterize the K5 gene and its product. To our knowledge, this is the first such analysis of a candidate IE gene product.
MATERIALS AND METHODS
Cell lines and cell culture.
BCBL-1 (34) or BC-3 (1) cells, which harbor HHV-8, and BC-1 cells (6), which harbor both EBV and HHV-8, were grown in RPMI 1640 medium supplemented with 10 or 20% heat-inactivated fetal calf serum (FCS), penicillin, and streptomycin at 37°C under an atmosphere of 95% air and 5% CO2. B95-8 and Raji cells, which harbor EBV, and uninfected Ramos cells, a human B-cell line, were cultured in RPMI 1640 medium supplemented with 10% FCS, penicillin, and streptomycin. The latter three cell lines served as controls throughout the study. When necessary, lytic gene expression was induced in HHV-8-infected and control cells by treatment with TPA (Sigma) at 20 ng/μl. Cos-7 and 293T cells were maintained in Dulbecco modified Eagle medium supplemented with 10% FCS, penicillin, and streptomycin and were used for transient-transfection and cotransfection assays, respectively.
Establishment and characterization of MAbs.
BALB/c mice were immunized with lysates from 107 TPA-induced BCBL-1 cells. The first immunization was carried out with a mixture of cell lysates in complete Freund's adjuvant. That was followed by two or three boosters, 3 to 4 weeks apart, in incomplete Freund's adjuvant. Three to 4 days prior to cell fusion, 107 TPA-induced BCBL-1 cells in 500 μl of phosphate-buffered saline (PBS) were administered intraperitoneally.
Hybridomas were established by fusing splenocytes from the hyperimmune mice with a nonproducing myeloma cell line, Sp-2/0-Ag14. After selection in medium containing hypoxanthine-aminopterin-thymidine, cells secreting MAbs were screened by indirect immunofluorescence assays (IFA; see below for details). TPA-induced and uninduced BCBL-1 cells were fixed in acetone and exposed to supernatants of the hybrid cells. Clones secreting antibodies reactive with TPA-induced BCBL-1 cells were expanded and cloned twice more by limiting dilutions. Ascites fluids with high antibody titers were then accumulated by injecting cloned hybrid cells intraperitoneally into Pristane (Sigma)-treated mice.
IFA.
BCBL-1, BC-3, and BC-1 cells were kept uninduced or induced with TPA. At selected times after induction, cells were harvested, washed twice with PBS, spotted onto slide glass in 24-well plates, air dried, and fixed with acetone for 20 min at −20°C. The fixed cells were incubated with hybridoma supernatants for 30 min at room temperature in a humid chamber. After washing with PBS containing 0.05% Tween 20 for 10 min, the slides were air dried and incubated for another 30 min with appropriately diluted fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse immunoglobulin G (IgG; DAKO, Copenhagen, Denmark). After being washed once again as described above, the slides were mounted with 50% glycerol in PBS and signals were detected by fluorescence microscopy.
Confocal immunohistochemistry.
BCBL-1 cells were induced with TPA for 24 h, washed in PBS, spotted onto slide glass, air dried, and fixed with acetone as described for the IFA. They were then incubated with a primary anti-K5 MAb for 30 min, followed by a 1:100 dilution of FITC-conjugated goat anti-mouse IgG and Hoechst 33342 for another 30 min at 37°C. Specific immunofluorescence was observed under a confocal laser scanning microscope (Carl Zeiss Co. Ltd.).
Immunoelectron microscopy.
BCBL-1 cells were induced with TPA for 24 h, washed with PBS, and fixed in 0.1% glutaraldehyde and 4% paraformaldehyde buffered in 0.1 M phosphate buffer (pH 7.4) for 10 min, after which they were fixed for an additional 12 h in 4% paraformaldehyde. To determine the exact localization of the antigen, we used the cryo-thin-section immunogold method (15). The specimens were immersed in 1.89 M sucrose with 20% polyvinylpyrrolidone and frozen with liquid nitrogen. Thin frozen sections were then cut with a microtome (ULTRACUTS; Reichert-Nissei, Tokyo, Japan) and mounted on Formvar carbon-coated nickel grids. The sections were rinsed with PBS and then Tris-buffered saline containing 1% bovine serum albumin and incubated overnight with MAb 328C7 in Tris-buffered saline. The following day, the cells were incubated for 1 h with anti-mouse IgG conjugated with 5-nm colloidal gold particles, fixed again in 2% glutaraldehyde in PBS, postfixed with 1% OsO4, and stained with uranyl acetate. After dehydration in ethanol, the sections were embedded in LR white (London Resin.) and observed using a Hitachi H-7100 electron microscope.
Western blot analysis.
Cell lysates in Laemmli's buffer were prepared from TPA-induced and uninduced BCBL-1, BC-3, and Raji cells 24 h after induction. Samples were subjected to sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis (SDS-PAGE) under reducing conditions and then electrophoretically transferred to polyvinylidene difluoride membranes (Bio-Rad, Hercules, Calif.) using standard procedures (19). The membranes were then blocked for 1 h while shaking at room temperature in PBS containing 0.05% Tween 20 and 5% skim milk, after which they were incubated with the primary antibody (1:1,000 dilution of ascitic fluid in PBS–0.05% Tween 20–2% skim milk) for an additional 1 h while again shaking at room temperature. The membranes were then washed three times for 10 min each in 0.05% Tween 20 in PBS and finally incubated for 1 h as described above with an appropriate dilution of alkaline phosphatase-conjugated goat anti-mouse IgG (Bio-Rad). After washing, bound enzyme-labeled MAb was detected with 5-bromo-4-chloro-3-indolylphosphate (BCIP) and nitroblue tetrazolium substrate (Wako Chemicals, Osaka, Japan).
Construction of a cDNA library and immunoscreening with anti-HHV-8 MAb.
Construction of a cDNA library from BCBL-1 cells was described previously (19). This library was screened with MAb 328C7 using a picoBlue Immunoscreening Kit (Stratagene) in accordance with the manufacturer's instructions. Briefly, 5 × 104 PFU of recombinant phages per 150-mm-diameter agar plate were mixed with Escherichia coli Y1090 and plated. After incubation at 37°C for 6 to 7 h, expression of fusion proteins was induced by overlaying 10 mM IPTG-treated nitrocellulose membranes. The membranes were blocked for 1 h in buffer containing 20 mM Tris-HCl (pH 7.5)–150 mM NaCl–1% bovine serum albumin and then reacted with the MAb. Following incubation with an appropriate dilution of alkaline phosphatase-conjugated goat anti-mouse IgG, reactivity was detected with BCIP and nitroblue tetrazolium as described for Western blotting. Immunoreactive phages were collected, diluted, replated, and isolated by subsequent screening until they were clonally pure. Four immunoreactive phages were then selected and rescreened through three cycles of cloning. Purified recombinant phages were transferred to SM buffer, and cDNA inserts were amplified by PCR and cloned into TA cloning vector pCR2.1 (Invitrogen).
Analysis of the DNA sequence.
The cDNA subcloned into pCR2.1 was sequenced using SequiTherm EXCEL long-read DNA sequencing kit-LC (Epicentre Technologies). DNA sequence data were compiled, and homology was analyzed by searching data banks using FASTA and BLAST software.
Construction of plasmids.
The viral DNA from HHV-8-harboring BCBL-1 cells was used as a template for all PCR amplifications. The HHV-8 K5 gene fragment was amplified by PCR using primers with NheI and XhoI restriction sites at the 5′ and 3′ ends, respectively. The amplified products were digested with NheI and XhoI and inserted into pcDNA3.1(+)/Zeo (Invitrogen) to create plasmid pcDNA3-K5. To construct plasmid pcDNA3-KSHV/Rta, a gene fragment encoding KSHV Rta was amplified by PCR, digested with XbaI, and inserted into the XbaI site of pcDNA3.1. Luciferase reporter plasmids pk5-Luc and p8pol-Luc were constructed by PCR amplification of fragments extending 1 kbp upstream of the respective genes relative to their translation start (ATG) codons. The amplified products were digested with SacI and NcoI and cloned into pGL3-Basic vectors (Promega) previously digested with the same enzymes. Construction of a heterologous reporter plasmid containing the human immunodeficiency virus type 1 (HIV-1) long terminal repeat (LTR) (PLTR-Luc) was described previously (30).
Transient expression in Cos-7 cells.
Prior to transfection, 4 × 105 Cos-7 cells were seeded onto 60-mm-diameter dishes and incubated for 16 h at 37°C in a 5% CO2 incubator. They were then transfected with expression plasmid pcDNA3-K5 using SuperFect Transfection Reagent (Qiagen) in accordance with the manufacturer's instructions. Twenty-four hours after transfection, cells were washed in PBS, collected by trypsinization, again washed in PBS, and fixed for 20 min in cold acetone at −20°C. Expression of K5 was then detected as described for confocal immunohistochemistry of BCBL-1 cells.
RT-PCR and nested PCR.
Reverse transcription (RT)-PCR was carried out on poly(A)+ RNA extracted from TPA-induced and uninduced BCBL-1 cells using Superscript II reverse transcriptase (GIBCO Bethesda Research Laboratories [BRL]), essentially as indicated in the manufacturer's instructions. Briefly, RT of 0.5 μg of poly(A)+ RNA was performed at 42°C for 50 min in a 20-μl reaction mixture containing 1 μl of Superscript II reverse transcriptase and 0.5 μM primer AP (5′-GGCCA CGCGT CGACT AGTAC TTTTT TTTTT TTTTT TT-3′) (provided in the kit). Thereafter, 2 μl of the reaction mixture was amplified by PCR using primers K5-F (5′-TGGAC GGGCA CGAAG CGTTG AT-3′) and AUAP (5′-GGCCA CGCGT CGACT AG-TAC-3′) (provided in the kit). Nested PCR was carried out using 5 μl of 1:100 diluted RT-PCR mixture and primers K5-F and K5R3 (5′-AGAAT GTTCC CGCAG GAGCA GTTAG GATGA-3′). Control reactions were carried out using genomic HHV-8 DNA as a template.
5′ and 3′ RACE.
All primers used for cDNA synthesis and PCR were designed on the basis of cDNA sequence data from the HHV-8 cDNA library. 3′ and 5′ rapid amplification of cDNA ends (RACE) was performed using a cDNA ends amplification kit (GIBCO BRL) and poly(A)+ RNA extracted from uninduced and TPA-induced BCBL-1 cells. The first-strand cDNA of 3′ RACE was synthesized using Superscript II reverse transcriptase and an oligo(dT) primer provided with the kit. The cDNA was amplified by PCR using an adapter-specific primer also provided in the kit and an HHV-8-specific primer (K5F1) corresponding to the ORF K5 3′ end (5′-GACTT AGGGG GACAG CGCGT ACCGA CCTAT-3′). The first-strand cDNA of 5′ RACE was synthesized using HHV-8-specific primer K5R1 (5′-GTCAC CCGGG TGTTAT TTGCC GCGTA TAAT-3′) and tailed with dCTP. The cDNA was amplified by PCR using an adapter-specific primer provided in the kit and a second HHV-8-specific primer (K5R2) corresponding to the ORF K5 5′ end (5′-CACTT ACGCG GCATA TGCCG CCAAA TGCAA-3′). Both 3′ and 5′ RACE cDNA PCR products were cloned into the pCR2.1 vector (Invitrogen) in accordance with the manufacturer's instructions and sequenced. The TA clones of the 3′ and 5′ RACE PCR products were sequenced using a SequiTherm EXCEL sequencing kit (EPICENTRE TECHNOLOGIES) based on the dideoxynucleotide termination method, and sequence analyses were carried out by comparison with the published HHV-8 sequence data (35).
Primer extension.
Oligonucleotide primer K5R4 (5′-TACGT CCTTG GACGC CATCT-3′) was 5′ end labeled with IRD41 (Aloka), creating a dye primer. Ten micrograms each of poly(A)+ RNA extracted from TPA-induced and uninduced BCBL-1 cells was then mixed with 5 μl of 1 μM K5R4 dye primer, heated to 70°C for 10 min, and then immediately transferred to a 50°C chamber, where the extension reaction was carried out for 50 min in a total volume of 50 μl, which included 200 U of Superscript II reverse transcriptase (GIBCO BRL). After digestion with an RNase mixture for 30 min at 37°C, the samples were precipitated with ethanol and analyzed on 6% polyacrylamide sequencing gels containing 8 M urea and on a Li-Cor DNA sequencer (model 4000; Aloka). Sequence reactions using the same labeled primer were run in parallel experiments using a cDNA clone as a template to verify the size of the primer.
Time course of K5 antigen expression.
The kinetics of HHV-8 gene expression were examined in uninduced and TPA-induced BC-3 and BCBL-1 cells. To analyze K5 expression, cells were harvested and washed in PBS 2, 4, 5, 6, 8, and 12 h after induction, spotted onto antigen plates, and permeabilized with acetone for IFA using MAb 328C7 as a probe. For comparison, the early gene K9 (19) was similarly analyzed using MAb B291.
Northern blotting.
BCBL-1 cells (1 × 107 to 2 × 107) were induced for 12 h with TPA alone, TPA plus the DNA synthesis inhibitor phosphonoformic acid (PFA; 200 μg/ml), or TPA plus cycloheximide (CHX), a protein synthesis inhibitor, at 10 to 100 μg/ml. Total RNA was then isolated from the cells using a total RNA purification kit (Qiagen) in accordance with the manufacturer's instructions. After ethanol precipitation, the RNA was stored at −70°C for further use. For Northern blot hybridization, 20 μg of total RNA was subjected to electrophoresis on 1% agarose-formaldehyde gels, blotted onto Hybond-N nylon membranes (Amersham), and hybridized to an oligonucleotide probe specific for the K5 gene (5′-CCCGC AGGAG CAGTT AGGAT GACCA GGAGC-3′) at 42°C in a 25 mM sodium phosphate buffer containing 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and 5× Denhardt's solution. After hybridization, the membranes were washed at 42°C in solutions of decreasing ionic strength (2× SSC–0.1% SDS and 1× SSC–0.1% SDS) for 30 min each time and then exposed to Kodak XAR film with an intensifying screen. A β-actin probe (5′-AGCATTTGCGGTGGACGATGGAG-3′) was used as a control. An antisense oligonucleotide probe specific for the K9 gene (5′-TTGGCCTGGGTCCATTGTCC-3′) was also used to detect the K9 transcript on the same filter. Antisense oligonucleotides were 5′ labeled with T4 polynucleotide kinase and [γ-32P]ATP using a Mega label kit (Boehringer, Mannheim, Germany).
Metabolic labeling and immunoprecipitation.
BCBL-1 cells (5 × 106) were washed twice with PBS and resuspended in Dulbecco's modified Eagle medium (DMEM; GIBCO BRL) supplemented with 5% dialyzed FCS without methionine. To differentiate HHV-8 IE and E gene products, BCBL-1 cells were induced for 12 h with TPA alone or with TPA plus PFA (300 μg/ml). The cells were labeled with [35S]methionine at 220 μCi/ml during the final 1 h. As a control, uninduced BCBL-1 cells were labeled for the same period.
Sequential application of metabolic inhibitors enabled us to demonstrate the true IE nature of the K5 gene product. A sample of BCBL-1 cells (5 × 107) was divided into two aliquots that were resuspended in DMEM, induced for 4 h with TPA in the presence of CHX, an inhibitor of protein synthesis, at 100 μg/ml, and cultured. One aliquot was then washed three times with PBS to remove the CHX, resuspended for an additional 6 h in labeling medium containing TPA plus actinomycin D (AcD), an mRNA synthesis inhibitor, at 10 μg/ml, and cultured. CHX- and AcD-treated cells were each labeled with [35S]methionine as described above, after which they were harvested, washed with PBS, lysed for 30 min on ice in RIPA buffer (0.05 M Tris-HCl [pH 8.0], 0.15 M NaCl, 0.5% sodium deoxycholate, 1% Triton X-100, 0.1% SDS), and centrifuged at 13,000 × g for 15 min. The resultant supernatant was then used for immunoprecipitation assays as follows.
Two microliters of MAb 328C7 containing ascites fluid was mixed with 200 μl of protein G-Sepharose 4 FF (Pharmacia Biotech) beads and incubated at 4°C for 3 h while rotating. The beads were then washed six times with RIPA buffer to remove unbound protein and isotope. The radiolabeled cell lysate was mixed with protein G-Sepharose antibody complex and rotated overnight as described above. After extensive washing with RIPA buffer, the immunoprecipitate was eluted by boiling in Laemmli's buffer, separated by SDS–10% denaturing PAGE, and analyzed by fluorography.
Transregulation assay.
Prior to transfection, 293T cells were plated to a density of 5 × 105 cells per 60-mm-diameter tissue culture dish in DMEM supplemented with 10% FCS and allowed to grow overnight at 37°C in a 5% CO2 incubator. Transient cotransfection was accomplished by using SuperFect Transfection Reagent (Qiagen). A 4-μg sample of total DNA was used for each 60-mm dish with 1 μg of reporter and 2 μg of effector plasmid, and the total quantity of transfected DNA was kept constant by adding an appropriate amount of pcDNA3.1. Cells were harvested 24 h after transfection, and luciferase activity was assayed. Values were normalized to the protein concentration using a Bio-Rad protein assay with bovine serum albumin as the standard.
Nucleotide sequence accession number.
The nucleotide sequence reported here has been deposited in the GenBank database and assigned accession no. AF117253.
RESULTS
A protein specific for HHV-8-infected cells is recognized by MAb 328C7.
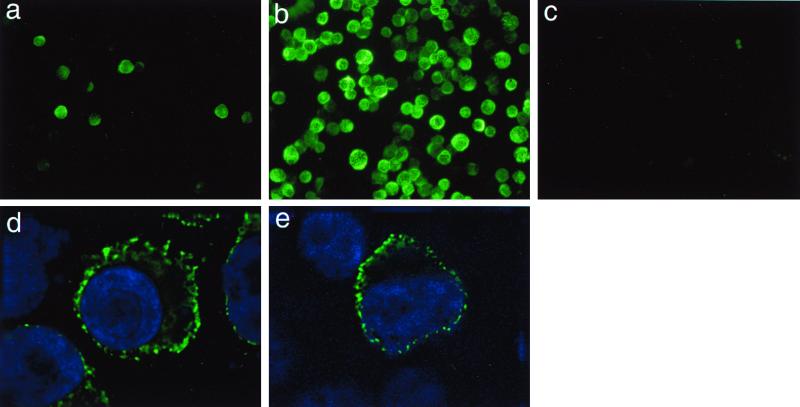
In order to identify individual HHV-8 proteins, we raised several MAbs against whole-cell lysates of TPA-induced BCBL-1 cells. From a list of six MAbs specifically recognizing the K5 gene, in this study we used one of them, named MAb 328C7, to analyze the K5 gene. Using IFA, MAb 328C7 was observed to react with an antigen apparently present in the cytoplasm of BCBL-1 cells (Fig. 1a and b). Although only 4 to 5% of uninduced cells exhibited specific staining, the percentage of positive cells increased to 80 to 90% after TPA treatment. The staining was specific for HHV-8-infected cells: Raji (Fig. 1c), B95-8, and Ramos cells (not shown) were all not stained specifically by MAb 328C7.
FIG. 1.
Fluorescence (FITC) photomicrographs showing MAb 328C7 immunoreactivity in HHV-8-infected, uninfected, and transfected cells after selected periods of induction or after transfection. BCBL-1 and Raji cells were fixed and labeled after 12 and 24 h of exposure to TPA, respectively; pcDNA3-K5-transfected Cos-7 cells were fixed and labeled 24 h posttransfection. a, untreated BCBL-1 cells; b, TPA-treated BCBL-1 cells; c, TPA-treated Raji cells; d, TPA-treated BCBL-1 cells; e, Cos-7 cells transfected with pcDNA3-K5 also stained with Hoechst 33342.
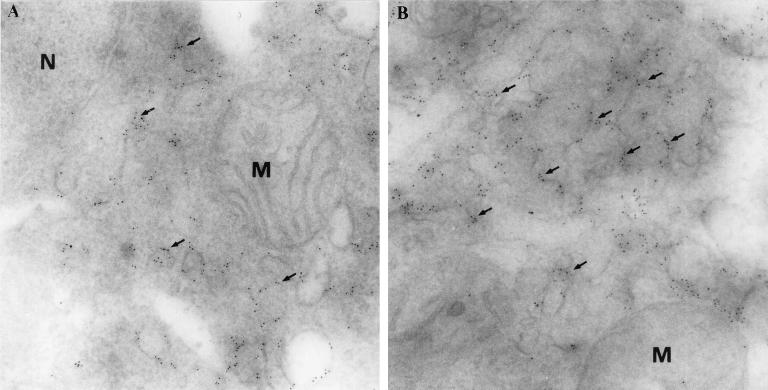
To more accurately localize K5 protein, TPA-induced BCBL-1 cells and K5-transfected Cos-7 cells were double labeled with MAb 328C7 and Hoechst 33342 and examined under a confocal imaging microscope. Figure 1d and e shows that antigen was, indeed, distributed in the cytoplasm of induced and transfected cells, appearing as dots. The greater resolution achieved with immunoelectron microscopy using MAb 328C7 revealed that the antigen was localized particularly in the endoplasmic reticulum (Fig. 2A and B) but not in the nucleus and mitochondria, suggesting that the K5 protein is not a structural protein.
FIG. 2.
Electron micrographs demonstrating the presence of K5 antigen in BCBL-1 cells. Immunogold labeling (arrows) is detected only on membranes of endoplasmic reticulum-like structures (A and B) but not in a nucleus (N) and mitochondria (M). Immunogold labeling was performed using cryo-thin sections (gold particles, 5 nm in diameter). Original magnifications: A, ×57,000; B, ×60,000.
MAb 328C7 recognizes the K5 gene product in HHV-8-infected cells.
To identify a coding gene for the protein recognized by MAb 328C7, the cDNA library constructed from TPA-induced BCBL-1 cells was immunoscreened with the MAb. Proteins from four cDNA clones reacted with MAb 328C7, and in each clone, the size of the inserted cDNA was approximately 0.8 to 1.0 kbp. The clone with the longest cDNA insert was sequenced and compared with the previously published HHV-8 sequence (35); the sequence was found to be identical to that part of the HHV-8 sequence encoding ORF K5 (data not shown). The sequences of the 5′ and 3′ ends of the other three cDNA inserts overlapped those of the K5 clone.
To confirm that the antigen recognized by MAb 328C7 and the K5 protein was the same K5 ORF was amplified by PCR, inserted into pcDNA, and transfected into Cos-7 cells. MAb 328C7 specifically recognized the expressed K5 protein located in the cytoplasm of the transfectants (Fig. 1e).
MAb 328C7 recognizes a 36-kDa protein in Western blots.
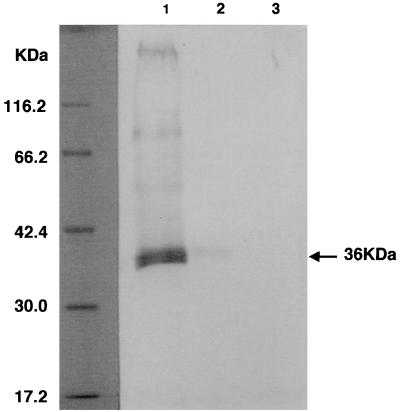
To identify the viral protein recognized by the anti-K5 MAb, we performed Western blot analysis. Figure 3 shows that MAb 328C7 recognized a 36-kDa polypeptide in BCBL-1 cells, and this band was more intensively recognized in cells treated with TPA than in untreated ones (Fig. 3, lanes 1 and 2). It has no reactivity with any polypeptides obtained from EBV-infected Raji cells (Fig. 3, lane 3) or from uninfected cell lines (data not shown). Thus, synthesis of this antigen appears to be highly dependent on induction of HHV-8 infection.
FIG. 3.
Western blot analysis of cell lysates obtained from TPA-treated (lane 1) and untreated (lane 2) BCBL-1 cells and untreated Raji cells (lane 3) using MAb 328C7 as a probe. Polypeptides were separated by SDS–10% PAGE under reducing conditions, transferred to a polyvinylidene difluoride membrane, and reacted with MAb 328C7. Molecular mass markers are shown on the left. The arrow indicates the HHV-8-specific polypeptide recognized by MAb 328C7.
Analysis of K5 gene transcription.
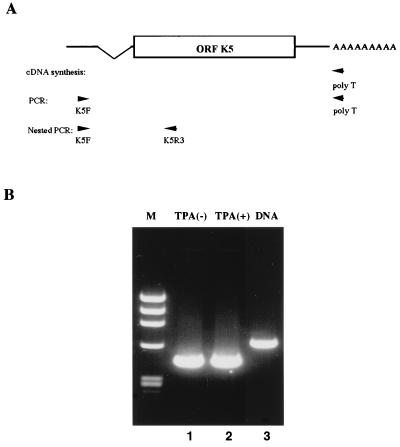
When sequencing of the 5′ RACE products was carried out, based on the sequence data from the cDNA library, it was found that an mRNA with a splice occurring in the 5′ noncoding region was transcribed from the K5 locus of HHV-8 (see Fig. 6). To confirm the 5′ RACE results, RT-PCR and subsequent nested PCR were carried out with RNA extracted from BCBL-1 cells; extracted DNA served as a control against detection of genomic DNA using the K5F and K5R3 primers (Fig. 4A). Amplification of HHV-8 genomic DNA yielded a 610-nucleotide fragment, as predicted by HHV-8 sequence data (Fig. 4, lane 3); however, a fragment of only about 440 nucleotides was amplified by RT-PCR and nested PCR from RNA extracted from TPA-treated or untreated BCBL-1 cells (Fig. 4, lanes 1 and 2, respectively).
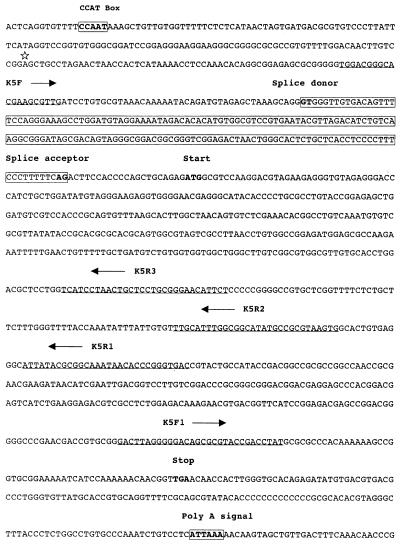
FIG. 6.
Map of the K5 ORF. A putative CCAT box and the poly(A) signal are boxed and in boldface; note that they lack a distinguishable TATA box. The asterisk indicates the initiation site of the K5 transcript. The nucleotide sequence corresponding to the intron is boxed and shaded. The splice donor (GT…) and splice acceptor (…AG) sites are in boldface. The primers (K5F, K5F1, K5R1, K5R2, and K5R3) used for 3′ and 5′ RACE and RT-PCR are underlined. The arrows indicate the orientations of the primers.
FIG. 4.
RT-PCR and nested PCR of K5. Using DNase I-digested poly(A) RNA extracted from untreated and TPA-treated BCBL-1 cells, a spliced fragment encoding K5 was amplified by RT-PCR and subsequent nested PCR. (A) Schematic representation of the primers used in the assays. (B) Nested PCR results obtained with primers K5F and K5R3. Lanes: M, molecular weight markers (φX174 HaeIII digest); 1, untreated cells; 2, TPA-treated cells; 3, control PCR with HHV-8 genomic DNA.
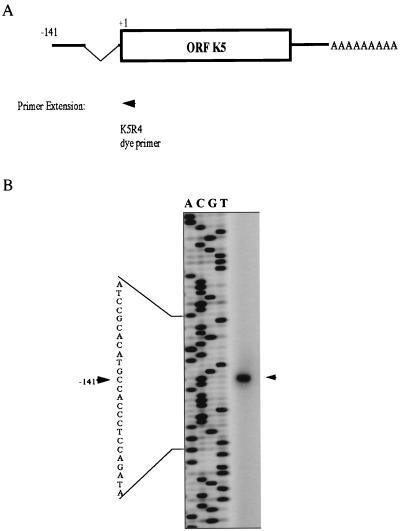
To map precisely the 5′ end of the K5 transcript, a primer extension assay was carried out with K5R4 dye primer (Fig. 5). One product was situated 141 bp upstream of the initiation codon, with a splice occurring at the 5′ noncoding region; a typical splice donor-acceptor site (GT…AG) was present in the intron of the K5 locus (Fig. 5); there was a putative CCAT box-like sequence at a position 126 to 129 bp upstream of the initiation site; but no obvious TATA box-like sequence was found within 1.0 kb upstream of the starting codon using the transcription factor database (32).
FIG. 5.
Map showing the K5 transcription pattern. (A) Schematic representation of the primer used in the primer extension assay and summary of the maps obtained using primer extension and 5′ and 3′ RACE. (B) Initiation site of the K5 transcript identified by primer extension. The size of the primer extension product is indicated by a sequencing ladder initiated with the same primer. The arrowhead indicates the position of the cDNA product, and the number indicates its position with respect to the initiation codon (ATG).
To determine the location of the 3′ end of the K5 transcript, a 3′ RACE assay was performed and only a single product was generated. Sequencing showed the 3′ end to be situated 24 nucleotides downstream of the putative poly(A) signal (Fig. 6).
K5 is expressed as an IE protein.
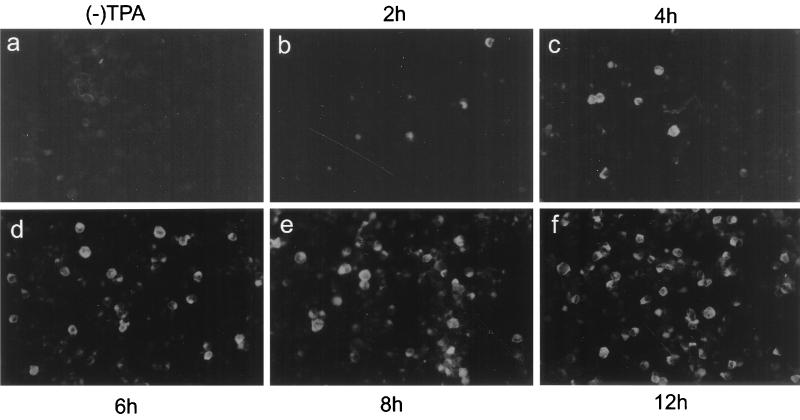
The time course of K5 gene expression was determined using antigen plates prepared after selected periods of TPA treatment. BC-3 cells were used in this experiment instead of BCBL-1 cells because less than 1% of uninduced BC-3 cells expressed the K5 protein (Fig. 7a). This provided a lower background against which to detect increases in K5 expression than would be obtained with BCBL-1 cells (4 to 5%; Fig. 1a). We found that approximately 20% of BC-3 cells were K5 positive after 4 h of TPA induction (Fig. 7c) and that number increased to a maximum (80%) within 12 h after induction (Fig. 7f). By contrast, the K9 gene product, which is an early gene product (19), was first expressed 12 h after treatment (data not shown).
FIG. 7.
Fluorescence (FITC) photomicrographs showing the time course of K5 protein expression. BC-3 cells were left uninduced or induced with TPA. After 2, 4, 6, 8, and 12 h of induction, cells were labeled with MAb 328C7.
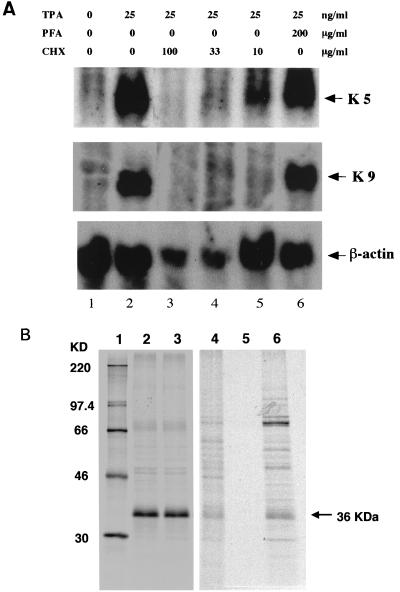
Northern blot analysis of total RNA extracted from BCBL-1 cells carried out using an ORF K5-specific probe showed a single transcript of approximately 1.2 kb (Fig. 8A, lane 2). The same transcript was clearly detected in cells treated with TPA plus PFA for 12 h (Fig. 8A, lane 6), indicating its independence of viral DNA synthesis, and in cells treated with TPA plus CHX at 10 μg/ml (Fig. 8A, lanes 5), although the transcript was barely detectable in cells exposed to CHX at 33 μg/ml (Fig. 8A, lane 4). Diminished β-actin in the presence of either CHX concentration was indicative of inhibited protein synthesis. On the other hand, K9, which is supposed to be an E gene, was repressed in the presence of CHX at any concentration. This result confirmed that K5 was one of the IE genes.
FIG. 8.
(A) Northern blot analysis demonstrating the resistance of K5 transcription to the presence of inhibitors of DNA and protein synthesis (PFA and CHX, respectively). Total cellular RNA was prepared from TPA-stimulated BCBL-1 cells after 12 h of exposure to PFA and CHX, size fractionated on a 1% agarose-formaldehyde gel, transferred to nylon membrane, and probed with K5- and K9-specific antisense DNA probes. From top to bottom, the autoradiograms shown are of blots hybridized with probes for the K5, K9, and β-actin (used as a loading control) genes, respectively. (B) Immunoprecipitation of K5 protein expressed in BCBL-1 cells following metabolic labeling with PFA and CHX. For detection of HHV-8 IE and E proteins, cells were induced with TPA and incubated for 12 h with PFA at 300 μg/ml. For detection of HHV-8 IE genes, cells were induced with TPA and incubated for 4 h with CHX at 100 μg/ml and then for 6 h with AcD at 10 μg/ml. Labeling with [35S]methionine was performed 1 h before cells were harvested. Lysates were prepared and immunoprecipitated with MAb 328C7. Samples were separated by SDS–10% denaturing PAGE. Molecular mass markers are shown on the left (lane 1). The position of the 36-kDa protein is indicated. Samples in lanes 2, 3, 4, 5, and 6 are TPA treated, TPA-PFA treated, untreated, CHX treated, and CHX-AcD treated, respectively.
Immunoprecipitation assays yielded a 36-kDa band in samples treated with TPA alone or with TPA plus PFA (Fig. 8B, lanes 2 and 3). No K5 protein expression was detected in cells treated with CHX (Fig. 8B, lane 5), but replacement of CHX with AcD restored expression of the 36-kDa protein. Thus, synthesis of the K5 gene product is independent of de novo viral gene expression and protein synthesis, which is consistent with its being an IE gene product. In addition, a second prominent band was observed at 70 kDa (Fig. 8B, lane 6). Although its origin is unknown, this band may indicate that under some conditions, the K5 protein exists as a dimer.
Transregulation of the K5 gene.
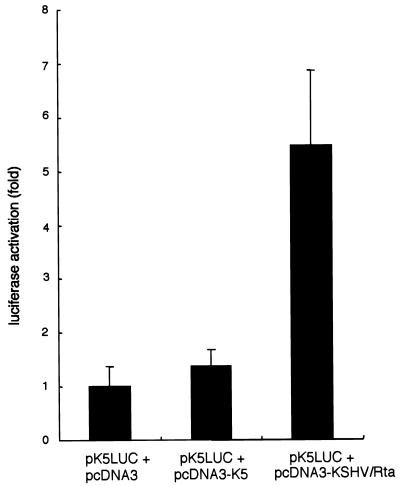
Because the majority of IE genes exert a regulatory effect on E and L gene expression, transregulation of the K5 gene was examined in two types of experiment. In the first, the responsiveness of the K5 gene promoter was assessed in cells transiently cotransfected with expression vectors pcDNA3-K5 and pcDNA3-KSHV/Rta and with reporter plasmid pk5-Luc. After 24 h of transfection, the cells were collected by trypsinization, washed twice with PBS, and assayed for luciferase activity. As shown in Fig. 9, luciferase activity was increased about fivefold when pcDNA3-KSHV/Rta was cotransfected with pk5-Luc. In contrast, when pcDNA3-K5 and pk5-Luc were cotransfected, there was no significant stimulation of luciferase activity compared with cotransfection of an empty vector. In a second experiment, the transregulatory effect of K5 on homologous and heterologous promoters of the HHV-8 polymerase (p8pol-Luc) and the HIV-1 LTR (pLTR-Luc) was tested and in neither case did K5 affect luciferase activity driven by these promoters (data not shown).
FIG. 9.
Responsiveness of the K5 gene promoter to the K5 gene and KSHV Rta. Transfected cells were harvested 24 h after transfection, and luciferase activity was assayed. Shown are the means ± standard deviations of triplicate transfections.
DISCUSSION
The HHV-8 K5 gene is a homologue of BHV-4 IE-1. BHV-4 possesses two IE genes, IE-1 and IE-2 (43), the latter being a homologue of EBV R and HVS R which acts as a transactivator and interacts with other proteins (44, 45). On the other hand, BHV-4 IE-1 has no homology with the EBV genome; rather, it is a positional homologue of HVS gene 14 (9). Its putative product contains a cysteine-rich region that may form zinc fingers characteristic of some DNA-binding proteins (44), but no function has yet been ascribed to BHV-4 IE-1.
Using Western blots probed with an anti-K5 MAb, we identified a 36-kDa polypeptide in BCBL-1 (Fig. 3) and BC-3 cells infected with HHV-8, but not in those infected with EBV, and cDNA screening showed the protein to be encoded by ORF K5. This finding was confirmed by the ability of MAb 328C7 to react with a eukaryotically expressed fusion protein containing ORF K5 (Fig. 1e). Both cell types were strongly positive for virus-specific antigen only after induction with TPA, suggesting that the K5 gene was not expressed during the latent stage of infection but was reactivated as part of a cascade leading to replication. The low level of expression observed in uninduced cells (Fig. 1 and 7) probably indicates that a certain amount of spontaneous virus replication occurs in BCBL-1 and BC-3 cells, as is seen in some cell lines harboring EBV (6, 25).
K5 protein synthesis in the presence of PFA, CHX, and AcD—DNA, protein, and mRNA synthesis inhibitors, respectively—indicates the independence of K5 gene expression from de novo viral protein synthesis, confirming its IE nature. Indeed, IE genes are known to code for regulatory proteins and are expressed by host cell transcription and translation systems prior to viral DNA replication. Other HHV-8 IE and E gene candidates have been reported based on levels of mRNA expression in BCBL-1 and BC-1 cells (23, 40, 41, 46), but the method of their analysis was different from ours. They investigated the mRNA expression level using BCBL-1 and BC-1 cells. K5 protein was detected as early as 4 h after TPA induction in BCBL-1 and BC-3 cells, and expression was maximal within 12 h. In contrast, when BC-1 cells were induced with TPA and reacted with MAb 328C7, antigen expression was not detected until 12 h after induction (data not shown). The reason for the delayed expression in BC-1 cells is unclear, but the presence of EBV in BC-1 cells may inhibit expression of the K5 gene. Therefore, to our knowledge, this is the first account of an HHV-8 IE gene product.
The K5 antigen was present in the cytoplasm—particularly the ER—of both HHV-8-infected cells and ORF K5 transfectants (Fig. 1 and 2) but was absent from the virion and from the nucleus. Almost all other known herpesvirus IE antigens are found in the nucleus and are involved in regulation of the expression of other genes (e.g., E and L genes). One exception is the varicella-zoster virus IE-4 antigen, which, like K5, is localized in the cytoplasm (10, 11). On the other hand, the varicella-zoster virus IE-4 antigen has transactivational activity toward homologous and heterologous promoters (10, 11) and during very early stages of infection, it is detectable in the nucleus (11). We observed no apparent transactivational activity toward homologous and heterologous promoters on the part of K5; nevertheless, by analogy, K5 may exert a regulatory effect on gene expression at times earlier than those at which we were first able to detect the protein in the present study.
Recently, KSHV Rta was shown to encode an IE gene that acts as a transactivator (23, 40). KSHV Rta is a homolog of EBV Rta; its mRNA is partially resistant to treatment with CHX (40, 41); and it transactivates the K5 promoter (Fig. 9). As the K5 protein did not transregulate its own promoter or a promoter driving the expression of DNA polymerase or the HIV-1 LTR, it appears that K5 expression occurs downstream of KSHV Rta expression. Further analysis is required to more precisely evaluate the activity of K5 at very early times and to gain a more complete understanding of the biological function of K5.
ACKNOWLEDGMENT
This study was supported in part by a Grant-in-Aid from the Ministry of Education, Science and Culture of Japan.
REFERENCES
- 1.Arvanitakis L, Mesri E A, Nador R G, Said J W, Asch A S, Knowles D M, Cesarman E. Establishment and characterization of a primary effusion (body cavity-based) lymphoma cell line (BC-3) harboring Kaposi's sarcoma-associated herpesvirus (KHSV/HHV-8) in the absence of Epstein-Barr virus. Blood. 1996;88:2648–2654. [PubMed] [Google Scholar]
- 2.Bais C, Santomasso B, Coso O, Arvanitakis L, Raakar E G, Gutkind J S, Asch A S, Cesarmans E, Gerhengorn M C, Mesri E A. G-protein-coupled receptor of Kaposi's sarcoma-associated herpesvirus is a viral oncogene and angiogenesis activator. Nature. 1998;391:86–89. doi: 10.1038/34193. [DOI] [PubMed] [Google Scholar]
- 3.Boshoff C, Endo Y, Collins P D, Takeuchi Y, Reeves J D, Schweickart V L, Siani M A, Sasaki T, Williams T J, Gray P W, Moore P S, Chang Y, Weiss R A. Angiogenic and HIV-inhibitory functions of KSHV-encoded chemokines. Science. 1997;278:290–294. doi: 10.1126/science.278.5336.290. [DOI] [PubMed] [Google Scholar]
- 4.Burger R, Neipel F, Fleckenstein B, Savino R, Ciliberto G, Kalden J R, Gramatzki M. Human herpesvirus type 8 interleukin-6 homologue is functionally active on human myeloma cells. Blood. 1998;91:1858–1863. [PubMed] [Google Scholar]
- 5.Cesarman E, Chang Y, Moore P S, Said J W, Knowles D M. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med. 1995;332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 6.Cesarman E, Moore P S, Rao P H, Inghirami G, Knowles D M, Chang Y. In vitro establishment and characterization of two AIDS-related lymphoma cell lines (BC-1 and BC-2) containing Kaposi's sarcoma-associated herpesvirus-like (KHSV) DNA sequences. Blood. 1995;86:2708–2714. [PubMed] [Google Scholar]
- 7.Chang Y, Cesarman E, Pessin M S, Lee F, Culpepper J, Knowels D M, Moore P S. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 8.Cheng E H, Nicholas J, Bellows D S, Hayward G S, Guo H G, Reitz M S, Hardwick J M. A Bcl-2 homolog encoded by Kaposi sarcoma-associated virus, human herpesvirus 8, inhibits apoptosis but does not heterodimerize with Bax or Bak. Proc Natl Acad Sci USA. 1997;94:690–694. doi: 10.1073/pnas.94.2.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cruz B R, Zhang L, van Santen V L. Characterization of a bovine herpesvirus 4 (BHV-4) 1.1-kb RNA and its transactivation by immediate-early gene product. Arch Virol. 1998;143:2391–2412. doi: 10.1007/s007050050469. [DOI] [PubMed] [Google Scholar]
- 10.Defechereux P, Debrus S, Baudoux L, Schoonbroodt S, Merville M-P, Rentier B, Piette J. Intracellular distribution of the ORF4 gene product of varicella-zoster virus is influenced by the IE62 protein. J Gen Virol. 1996;77:1505–1513. doi: 10.1099/0022-1317-77-7-1505. [DOI] [PubMed] [Google Scholar]
- 11.Defechereux P, Debrus S, Baudoux L, Rentier B, Piette J. Varicella-zoster virus open reading frame 4 encodes an immediate-early protein with posttranscriptional regulatory properties. J Virol. 1997;71:7073–7079. doi: 10.1128/jvi.71.9.7073-7079.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foreman K E, Friborg J J, Kong W P, Woffendin C, Polverini P J, Nickoloff B J, Nabel G J. Propagation of a human herpesvirus from AIDS-associated Kaposi's sarcoma. N Engl J Med. 1997;336:163–171. doi: 10.1056/NEJM199701163360302. [DOI] [PubMed] [Google Scholar]
- 13.Gaidano G, Cechova K, Chang Y, Moore P S, Knowles D M, Dalla-Favera R. Establishment of AIDS-related lymphoma cell lines from lymphomatous effusions. Leukemia. 1996;10:1237–1240. [PubMed] [Google Scholar]
- 14.Godden-Kent D, Talbot S J, Boshoff C, Chang Y, Moore P, Weiss R A, Mittnacht S. The cyclin encoded by Kaposi's sarcoma-associated herpesvirus stimulates cdk6 to phosphorylate the retinoblastoma protein and histone H1. J Virol. 1997;71:4193–4198. doi: 10.1128/jvi.71.6.4193-4198.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gotow T, Sakata M, Funakoshi T, Uchiyama Y. Preferential localization of annexin V to the axon terminal. Neuroscience. 1996;75:507–521. doi: 10.1016/0306-4522(96)00295-3. [DOI] [PubMed] [Google Scholar]
- 16.Guo H G, Browning P, Nicholas J, Hayward G S, Tschachler E, Jiang Y W, Sadowska M, Raffeld M, Colombini S, Gallo R C, Reitz M S., Jr Characterization of a chemokine receptor-related gene in human herpesvirus 8 and its expression in Kaposi's sarcoma. Virology. 1997;228:371–378. doi: 10.1006/viro.1996.8386. [DOI] [PubMed] [Google Scholar]
- 17.Honess R W, Roizman B. Regulation of herpesvirus macromolecular synthesis. I. Cascade regulation of the synthesis of three groups of viral proteins. J Virol. 1974;14:8–19. doi: 10.1128/jvi.14.1.8-19.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang Y Q, Li J J, Kaplan M H, Poiesz B, Katabira E, Zhang W C, Feiner D, Friedman K A. Human herpesvirus-like nucleic acid in various forms of Kaposi's sarcoma. Lancet. 1995;345:759–761. doi: 10.1016/s0140-6736(95)90641-x. [DOI] [PubMed] [Google Scholar]
- 19.Inagi R, Okuno T, Ito M, Chen J, Mori Y, Haque M, Zou P, Yagi H, Kiniwa S, Saida T, Ueyama Y, Hayashi K, Yamanishi K. Identification and characterization of human herpesvirus 8 open reading frame K9 viral interferon regulatory factor by a monoclonal antibody. J Hum Virol. 1999;2:63–71. [PubMed] [Google Scholar]
- 20.Kledal T N, Rosenkilde M M, Coulin F, Simmons G, Johnsen A H, Alovani S, Power C A, Luttichau H R, Gerstoft J, Clapham P R, Lewis I-C, Wells T N C, Schwartz T W. A broad-spectrum chemokine antagonist encoded by Kaposi's sarcoma-associated herpesvirus. Science. 1997;277:1656–1659. doi: 10.1126/science.277.5332.1656. [DOI] [PubMed] [Google Scholar]
- 21.Li M, Lee H, Yoon D W, Albrecht J C, Fleckenstein B, Neipel F, Jung J U. Kaposi's sarcoma-associated herpesvirus encodes a functional cyclin. J Virol. 1997;71:1984–1991. doi: 10.1128/jvi.71.3.1984-1991.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li M, Lee H, Guo J, Neipel F, Fleckenstein B, Ozato K, Jung J U. Kaposi's sarcoma-associated herpesvirus viral interferon regulatory factor. J Virol. 1998;72:5433–5440. doi: 10.1128/jvi.72.7.5433-5440.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lukac D M, Renne R, Kirshner J R, Ganem D. Reactivation of Kaposi's sarcoma-associated herpesvirus infection from latency by expression of the ORF 50 transactivator, a homolog of the EBV R protein. Virology. 1998;252:304–312. doi: 10.1006/viro.1998.9486. [DOI] [PubMed] [Google Scholar]
- 24.Miller G, Rigsby M O, Heston L, Grogan E, Sun R, Metroka C, Levy J A, Gao S J, Chang Y, Moore P. Antibodies to butyrate-inducible antigens of Kaposi's sarcoma-associated herpesvirus in patients with HIV-1 infection. N Engl J Med. 1996;334:1292–1297. doi: 10.1056/NEJM199605163342003. [DOI] [PubMed] [Google Scholar]
- 25.Miller G, Heston L, Grogan E, Gradoville L, Rigsby M, Sun R, Shedd D, Kushnaryov V M, Grossberg S, Chang Y. Selective switch between latency and lytic replication of Kaposi's sarcoma herpesvirus and Epstein-Barr virus in dually infected body cavity lymphoma cells. J Virol. 1997;71:314–324. doi: 10.1128/jvi.71.1.314-324.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Molden J, Chang Y, You Y, Moore P S, Goldsmith M A. A Kaposi's sarcoma-associated herpesvirus-encoded cytokine homolog (vIL-6) activates signaling through the shared gp130 receptor subunit. J Biol Chem. 1997;272:19625–19631. doi: 10.1074/jbc.272.31.19625. [DOI] [PubMed] [Google Scholar]
- 27.Moore P S, Gao S J, Dominguez G, Cesarman E, Lungu O, Knowles D M, Garber R, McGeoch D J, Pellet P, Chang Y. Primary characterization of a herpesvirus-like agent associated with Kaposi's sarcoma. J Virol. 1996;70:549–558. doi: 10.1128/jvi.70.1.549-558.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moore P S, Chang Y. Antiviral activity of tumor-suppressor pathways: clues from molecular piracy by KSHV. Trends Genet. 1998;14:144–150. doi: 10.1016/s0168-9525(98)01408-5. [DOI] [PubMed] [Google Scholar]
- 29.Mori Y, Yagi H, Shimamoto T, Isegawa Y, Sunagawa T, Inagi R, Kondo K, Tano Y, Yamanishi K. Analysis of human herpesvirus 6 U3 gene, which is a positional homolog of human cytomegalovirus UL24 gene. Virology. 1998;249:129–139. doi: 10.1006/viro.1998.9305. [DOI] [PubMed] [Google Scholar]
- 30.Neipel F, Albrecht J C, Ensser A, Huang Y Q, Li J J, Friedmankien A E, Fleckenstein B. Human herpesvirus 8 encodes a homolog of interleukin-6. J Virol. 1997;71:839–842. doi: 10.1128/jvi.71.1.839-842.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nicholas J, Ruvolo V R, Burns W H, Sandford G, Wan X, Ciufo D, Hendrickson S B, Guo H G, Hayward G S, Reitz M S. Kaposi's sarcoma-associated human herpesvirus-8 encodes homologues of macrophage inflammatory protein-1 and interleukin-6. Nat Med. 1997;3:287–292. doi: 10.1038/nm0397-287. [DOI] [PubMed] [Google Scholar]
- 32.Quandt K, Frech K, Karas H, Wingender E, Werner T. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 1995;23:4878–4884. doi: 10.1093/nar/23.23.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reed J A, Nador R G, Spaulding D, Tani Y, Cesarman E, Knowles D M. Demonstration of Kaposi's sarcoma-associated herpes virus cyclin D homolog in cutaneous Kaposi's sarcoma by colorimetric in situ hybridization using a catalyzed signal amplification system. Blood. 1998;91:3825–3832. [PubMed] [Google Scholar]
- 34.Renne R, Zhong W, Herndier B, McGrath M, Abbey N, Kedes D, Ganem D. Lytic growth of Kaposi's sarcoma associated herpesvirus (human herpesvirus 8) in culture. Nat Med. 1996;2:342–346. doi: 10.1038/nm0396-342. [DOI] [PubMed] [Google Scholar]
- 35.Russo J J, Bohenzky R A, Chien M C, Chen J, Yan M, Maddalena D, Parry J P, Peruzzi D, Edelman I S, Chang Y, Moore P S. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8) Proc Natl Acad Sci USA. 1996;93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sarid R, Sato T, Bohenzky R A, Russo J J, Chang Y. Kaposi's sarcoma-associated herpesvirus encodes a functional bcl-2 homologue. Nat Med. 1997;3:293–298. doi: 10.1038/nm0397-293. [DOI] [PubMed] [Google Scholar]
- 37.Sarid W, Chien K, Takeuchi S, Tasaka T, Asou H, Cho S K, de Vos S, Cesarman E, Knowles D M, Koeffler H P. Kaposi's sarcoma-associated herpesvirus (KSHV or HHV-8) in primary effusion lymphoma: ultrastructural demonstration of herpesvirus in lymphoma cells. Blood. 1996;87:4937–4943. [PubMed] [Google Scholar]
- 38.Schalling M, Ekman M, Kaaya E E, Linde A, Bieberfeld P. A role for a new herpes virus (KSHV) in different forms of Kaposi's sarcoma. Nat Med. 1995;1:707–708. doi: 10.1038/nm0795-707. [DOI] [PubMed] [Google Scholar]
- 39.Soulier J, Grollet, Oskenhendler E, Cacoub P, Cazalas-Hatem D, Babinet P, d'Agay M F, Clauvel J P, Raphael M, Degos L, Sigaux F. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood. 1995;86:1276–1280. [PubMed] [Google Scholar]
- 40.Sun R, Lin S F, Gradoville L, Yuan Y, Zhu F, Miller G. A viral gene that activates lytic cycle expression of Kaposi's sarcoma-associated herpesvirus. Proc Natl Acad Sci USA. 1998;95:10866–10871. doi: 10.1073/pnas.95.18.10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun R, Lin S F, Staskus K, Gradoville L, Grogan E, Haase A, Miller G. Kinetics of Kaposi's sarcoma-associated herpesvirus gene expression. J Virol. 1999;73:2232–2242. doi: 10.1128/jvi.73.3.2232-2242.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thome M, Schneider P, Hofmann K, Fickenscher H, Meinl E, Neipel F, Mattmann C, Burns K, Bodmer J L, Schroter M, Scaffidl C, Krammer P H, Peter M E, Tschopp J. Viral FLICE-inhibitory proteins (FLIPs) prevent apoptosis induced by death receptors. Nature. 1997;386:517–521. doi: 10.1038/386517a0. [DOI] [PubMed] [Google Scholar]
- 43.Van Santen V L. Characterization of the bovine herpesvirus 4 major immediately-early transcript. J Virol. 1991;65:5211–5224. doi: 10.1128/jvi.65.10.5211-5224.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Van Santen V L. Characterization of a bovine herpesvirus 4 immediate-early RNA encoding a homolog of the Epstein-Barr virus R transactivator. J Virol. 1993;67:773–784. doi: 10.1128/jvi.67.2.773-784.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang L, Van Santen V L. Interaction of bovine herpesvirus 4 (BHV-4) immediate early 2 gene product with BHV-4 thymidine kinase promoter-regulatory region. J Gen Virol. 1995;76:2433–2445. doi: 10.1099/0022-1317-76-10-2433. [DOI] [PubMed] [Google Scholar]
- 46.Zhu F X, Cusano T, Yuan Y. Identification of the immediate-early transcripts of Kaposi's Sarcoma-associated herpesvirus. J Virol. 1999;73:5556–5567. doi: 10.1128/jvi.73.7.5556-5567.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zimring J C, Goodbourn S, Offermann M K. Human herpesvirus 8 encodes an interferon regulatory factor (IRF) homolog that represses IRF-1-mediated transcription. J Virol. 1998;72:701–707. doi: 10.1128/jvi.72.1.701-707.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]