Abstract
Purpose
It is necessary to establish a mouse model of keratoconus (KC) for research and therapy. We aimed to determine corneal phenotypes in 3 Ppip5k2 mouse models.
Methods
Central corneal thickness (CCT) was determined using spectral domain optical coherence tomography (SD-OCT) in Ppip5k2+/K^ (n = 41 eyes), Ppip5k2K^/K^ (n = 17 eyes) and 2 knock-in mice, Ppip5k2S419A/+ (n = 54 eyes) and Ppip5k2S419A/S419A (n = 18 eyes), and Ppip5k2D843S/+ (n = 42 eyes) and Ppip5k2D843S/D843S (n = 44 eyes) at 3 and 6 months. Pachymetry maps were generated using the Mouse Corneal Analysis Program (MCAP) to process OCT images. Slit lamp biomicroscopy was used to determine any corneal abnormalities, and, last, hematoxylin and eosin (H&E) staining using corneal sections from these animals was used to examine morphological changes.
Results
CCT significantly decreased from 3 to 6 months in the Ppip5k2+/K^ and Ppip5k2K^/K^ mice compared to their littermate controls. OCT-based pachymetry maps revealed abnormally localized thinning in all three models compared to their wild-type (WT) controls. Slit lamp examinations revealed corneal abnormalities in the form of bullous keratopathy, stromal edema, stromal scarring, deep corneal neovascularization, and opacities in the heterozygous/homozygous mice of the three models in comparison with their controls. Corneal histological abnormalities, such as epithelial thickening and stromal layer damage, were observed in the heterozygous/homozygous mice of the three models in comparison with the WT controls.
Conclusions
We have identified phenotypic and histological changes in the corneas of three mouse lines that could be relevant in the development of animal models of KC.
Keywords: keratoconus (KC), mouse models, Ppip5k2, cornea, pachymetry mapping
Keratoconus (KC) is a bilateral, asymmetric corneal ectasia characterized by progressive thinning and steepening of the cornea into a conical shape.1 Despite occurring in both eyes, one eye is often more severely affected with symptoms such as corneal scarring, myopia, and irregular astigmatism.2,3 Within the corneal tissue, KC is identified by localized central thinning, compaction of stromal collagen fibers, Bowman's layer breaks, and epithelial degeneration.4–8 Symptoms often arise in adolescence, progressing into adulthood and stabilizing in the third or fourth decades of life.2 KC affects both sexes, and has progressively risen in frequency globally.1,9–13 KC is a multifactorial disease in which genetics, environmental, hormonal factors, inflammatory factors, and several comorbidities contribute to its susceptibility and pathogenesis.3,14–19 However, the precise molecular and cellular mechanisms of action and etiology remain unclear. Therefore, there is a critical need to understand KC pathogenesis for the development of therapeutic targets and diagnostic markers.
Genome wide association studies (GWAS) and linkage analysis have identified several loci of interest with various functional implications related to KC.20 In addition, studies using next generation sequencing and proteomics have identified many KC-related genes including thrombospondin 2 (THBS2), growth arrest specific 1 (GAS1), otopetrin 2 (OTOP2), and altered pathways such as reduced NRF2 signaling, activated integrated stress response, dysregulated cytokine production, altered TGFβ signaling, and increased oxidative stress implicated in the pathogenesis of KC.20–23
Recently, Khaled et al.18 identified two pathogenic variants (rs35671301, c. 1255 T > G, p. Ser419Ala, S419A), and (rs781831998, c.2528 A > G, p. Asn843Ser, and N843S) in the phosphatase domain of PPIP5K2 in 2 families with familial KC. These mutations impacted enzymatic activity by increasing kinase activity and decreasing phosphatase activity, respectively.18 The mechanistic implications of these changes in enzymatic activity are limited, although they likely impact cellular energy homeostasis that contribute to normal corneal function and thickness.18 A Ppip5k2 gene trap mouse model with elevated kinase activity and very little phosphatase activity showed abnormal corneal curvature and opacity via spectral domain-optical coherence tomography (SD-OCT) scanning and slit lamp examinations.18 However, these alterations were observed in only 40% of heterozygous Ppip5k2+/K^ (8 out of 20 mice) and homozygous Ppip5k2K^/K^ (4 out of 10 mice). With K^ referring to elevated kinase activity.18
Mice are essential laboratory animals and are widely used in biomedical research to model human disease due to their 99% genomic similarity to humans.24–26 However, the availability of appropriate animal models for KC is extremely limited.27 Tachibana et al.28 observed a conical morphology in the corneas of SKC mice. SKC female mice injected with testosterone exhibited a cone-like phenotype, whereas castrated male mice did not.28,29 This signified that the conical phenotype in SKC mice is androgen-dependent.28,29 A case report by Peiffer et al.30 observed a KC-like phenotype in a rhesus macaque, however, this study reported the phenotype in only one animal. Other studies have also attempted to induce KC experimentally in mice and rabbits with intrastromal collagenase injections.31,32 However, collagenase treatment studies were only reported in male mice and the long term-effects of collagenase in both models were not reported. Additionally, several mouse models have been reported to have decreased central corneal thickness (CCT), mimicking a disease condition called keratoglobus with a thin cornea, but not KC.33–37
Due to the scarcity of established mouse models of KC, this study sought to assess CCT and structural alterations in Ppip5k2 gene trap mice by increasing sample size and in 2 knock-in mouse lines carrying the point mutations identified in PPIP5K2. Observations were carried out using SD-OCT, slit lamp examinations, and histology analysis. These observations may help to identify KC-related phenotypes in these mouse models.
Methods
Mice Husbandry
The Institutional Animal Care and Use Committee (IACUC) at Augusta University approved the protocol, and all animal work was carried out in accordance with the Association for Research in Vision and Ophthalmology (ARVO) Statement for the Use of Animals in Vision and Ophthalmic Research. Breeding pairs of heterozygous B6N(Cg)-Ppip5k2tm1b (EUCOMM)Wtsi/J mice were obtained from the Jackson Laboratory (Bar Harbor, ME, USA), maintained and genotyped as previously described.18
CRISPR-Cas9 genome editing method was used to generate the two knock-in mouse lines at the Transgenic and Genome Editing Core Facility at Augusta University. Two different single guide RNAs (sgRNAs), which targeted sequences 5′-AGTGTGATGGATATAAATCAGGG-3′ (protospacer adjacent motif [PAM] site XGG) and 5′-CGCCTTATGCGATGTAAGTT CGG-3′ PAM site XGG were generated by (Synthego Corp., Redwood City, CA, USA) for S419A and D843S models, respectively. Two single-stranded DNA repair templates, 5′-GACTAACTGTCTTCTATCCTTTTATAAGGTTTTTTGACCTTTTTGAAAAGTGTGATGGATAcAAggCcGGcAAGTTAAAGCTCAAGAAGCCAAAACAATTACAGGCAAGTTTGTTGGGGTTTTTGTCGTTG-3′ and 5′-CCAGTGAGAGTCATGTGCATTCTTTACTATCCATTCTTCGGTATGGaGCacTgTGttcTGTAAGTTCGGGTTACTCCATGTAACTGACAGCAGTGGGCATGG-3′ were synthesized by (Integrated DNA Technologies Inc., Coralville, IA, USA) for both S419A and D843S models, respectively. The sgRNAs, ssDNA repair templates, and Cas9 protein (Alt-R S. p. Cas9 Nuclease V3 (Integrated DNA Technologies Inc., Coralville, IA, USA) were co-injected into C57BL/6J zygotes (Stock#009086; Jackson Laboratory, Bar Harbor, ME, USA). Founder mice for both S419A and D843S lines were obtained and confirmed by external PCR genotyping and Sanger sequencing. The primer sets used to verify the targeted allele for S419A mice were as follows: WT Forward- 5′-GCCAGGGAGACAAGGTGGAGGA-3′, WT Reverse- 5′-GAAGAGGTCCAAAGAGGTACTCACATCTCT-3′, Mutant Forward- 5′-GTGTGATGGATAcAAggCcGGcA-3′, Mutant Reverse- 5′-GCTTCTTGAGCTTTAACTTgCCgGccTTgT-3′ and for D843S were WT Forward- 5′-CAGGTATTCTAGAGGTGTTCTGTCTCCTGA-3′, WT Reverse- 5′-CTGAGCTTGGGACAGAGTTCAGGTGCA-3′, Mutant Forward- 5′-CCATTCTTCGGTATGGaGCacTgTGttc-3′, Mutant Reverse- 5′-CCCGAACTTACAgaaCAcAgtGCt-3′. The correctly targeted founders for both models were bred with C57BL/6J mice, followed by PCR genotyping and Sanger sequencing to confirm the germline transmission of the inserted sequences.
Droplet Digital PCR Mutation Detection Assays for Genotyping S419A and D843S Mice Models
Approximately 50 ng of mouse genomic DNA was used for the droplet digital PCR (ddPCR) assays in a total volume of 20 µL containing: 10 µL 2x Supermix for Probes without dUTP (Bio-Rad, Hercules, CA, USA), 1 µL DNA, 8 µL nuclease-free water (Fisher Scientific, Chicago, IL, USA), and 1 µL of single tube primer-probe mix ddPCR assays for S419A mutation detection (wild-type allele: HEX-TAAATCAGGG and mutant allele: FAM-CAAGGCCGGC) or for D843S mutation detection (wild-type allele: HEX-CGCCTTATGCGA and mutant allele: FAM-AGCACTGTGTTC) (Bio-Rad, Hercules, CA, USA). All the other steps followed the manufacturer's protocol. Two-dimensional amplitude plots displaying measured HEX and FAM amplitudes for each droplet were used to determine the genotype calls. Droplets with high FAM amplitude contain mutant amplified DNA (Ppip5k2S419A/S419A or Ppip5k2D843S/D843S). Droplets with high HEX amplitude contain wild-type amplified DNA. Droplets with both high HEX/FAM amplitudes indicate the presence of wild-type and mutant variants (Ppip5k2S419A/+ or Ppip5k2D843S/+).
Spectral Domain Optical Coherence Tomography
SD-OCT was used to visualize the anterior segment in mice using a Bioptigen Envisu-R2200 Spectral Domain Ophthalmic Imaging System (Leica Microsystems, Wetzlar, Germany) at 3 and 6 months of age.18,38 Briefly, mice were anesthetized with 100 mg/cc ketamine (NDC code 11695-0703-1; Covetrus, Portland, ME, USA) and 30 mg/cc xylazine (NDC code 59399-110-20) at a dosage of 10 µL/g of mouse weight. GenTeal Lubricant Eye Gel (Alcon, Fort Worth, TX, USA) was applied on the whiskers to prevent it from interfering with imaging. Mouse eyes were lubricated with Systane or GenTeal Tears eye drops (Alcon, Fort Worth, TX, USA) prior to and during assessment for clarity. A 12 mm telecentric probe was used with the reference arm set at position 245. Three scans were taken for each eye, producing one rectangular and two radial scans. A digital caliper tool available on the Bioptigen software was used to measure the CCT per eye at 3 and 6 months of age for each mouse. The central cornea was set on image 50 of each 100 image scans of the mouse cornea and the caliper was placed at the thickest portion of the scan, which typically correlated with zero on the x-axis image measuring scale. The caliper was then placed on the topmost surface of the cornea and then extended out to the bottom most surface of the cornea for the CCT measurement. Changes in CCT were derived between the two time points (3- and 6-months) per eye for each genotype of each mouse line.
Corneal Thickness Mapping
OCT-based corneal pachymetry of the mice was determined using the mouse cornea analysis program (MCAP) software, which was adapted from human corneal OCT images, as previously described.18,39 Each radial corneal OCT B scan was first segmented using a semi-automated, gradient-based algorithm to determine the corneal epithelial and endothelial surfaces by using a user-drawn seed segmentation for the first frame. All remaining frames are automatically segmented using this seed. Axial motion correction was performed on the radial corneal data which was then resampled uniformly to create a uniform Cartesian distribution. A sampled 3D representation of the imaged corneal surfaces was produced by fitting the sampled points using Zernike polynomial expansion. The vector form of Snell's Law was then used to adjust for refraction and optical path length distortions due to the optical nature of OCT. Last, pachymetry (corneal thickness) maps were created by direct z-axis subtraction of the epithelial and endothelial layers.
Slit Lamp Biomicroscopy Examinations
Mice were anesthetized with 100 mg/cc ketamine (NDC code 11695-0703-1; Covetrus, Portland, ME, USA) and 30 mg/cc xylazine (NDC code 59399-110-20) at a dosage of 10 µL/g of mouse body weight. The cornea and anterior chamber were examined with a slit lamp (SL-D7; Topcon, Tokyo, Japan), and images were documented with a digital camera (D100; Nikon, Tokyo, Japan).
Corneal Histology
Mice at 3 to 6 months old were euthanized for histology. Eyeballs were collected, fixed in Davidson's fixative for 24 hours, and transferred to 70% ethanol. Eyeballs were paraffin-embedded and cut on a sagittal plane through the optic nerve head (ONH) and the center of the cornea. Three sections (every other section) per eyeball with a thickness of 5 µm were processed for hematoxylin and eosin (H&E) staining at the AU Electron Microscopy and Histology Core. Corneal images were taken and analyzed with an ECHO revolve microscope (Discover Echo, a BICO company, San Diego, CA, USA).
Statistical Analyses
Statistical analysis was performed using GraphPad Prism (GraphPad Software, San Diego, CA, USA). Two-way ANOVA was used to test significance between genotypes at 3 and 6 months of age. A Student's t-test was used to test for differences between genotypes. The significance threshold was set at P < 0.05.
Results
CCT and Corneal Pathological Phenotypes in Ppip5k2 Animal Models Using SD-OCT
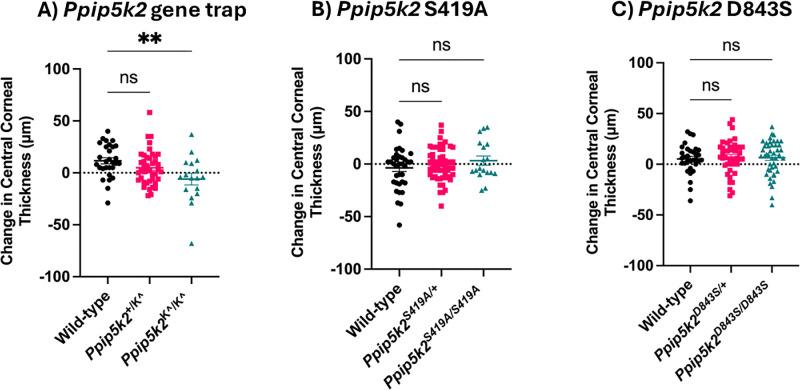
We sought to assess corneal phenotypes, curvature changes, and CCT at 3 and 6 months in Ppip5k2+/K^, Ppip5k2K^/K^, Ppip5k2S419A/+, Ppip5k2S419A/S419A, Ppip5k2D843S/+, and Ppip5k2D843S/D843S mice. Three-month-old mice were chosen because mouse corneal epithelial and stromal thickness stabilizes after 2 months of age, closely corresponding with adolescence in humans.40,41 Given that KC can develop into the mid-30s and 40s in humans before stabilizing, we chose to examine these alterations in 6-month-old mice, which is equivalent to 25 to 30 years of age in humans.42 Ppip5k2+/+ was referred to as wild-type (WT). Because KC typically affects one eye more than the other, we sought to derive changes in CCT between the two time points per eye. We observed a significant decrease in CCT between 3 and 6 months per eye in the homozygous Ppip5k2K^/K^ in comparison to WT (Fig. 1A). No significant changes in CCT were observed between 3 and 6 months per eye in the Ppip5k2S419/+ and Ppip5k2S419A/S419A or Ppip5k2D843/+ and Ppip5k2D843S/D843S mice in comparison with their WT (Figs. 1B, 1C).
Figure 1.
Change in CCT between the 2 time points per eye in 3 Ppip5k2 strains. Change in CCT in (A) Ppip5k2-gene trap mice, **P ≤ 0.006, wild-type (n = 24), Ppip5k2+/K^ heterozygous (n = 41), and Ppip5k2K^/K^ homozygous (n = 17). (B) Ppip5k2 S419A mice, ns: P > 0.05, wild-type (n = 36), Ppip5k2S419A/+ heterozygous (n = 54), and homozygous Ppip5k2S419A/S419A (n = 18). (C) Ppip5k2 D843S mice, ns: P > 0.05, wild-type (n = 32), Ppip5k2D843S/+ heterozygous (n = 42), and Ppip5k2D843S/D843S homozygous (n = 44). All comparisons were done with Student's t-test, data are mean ± SEM.
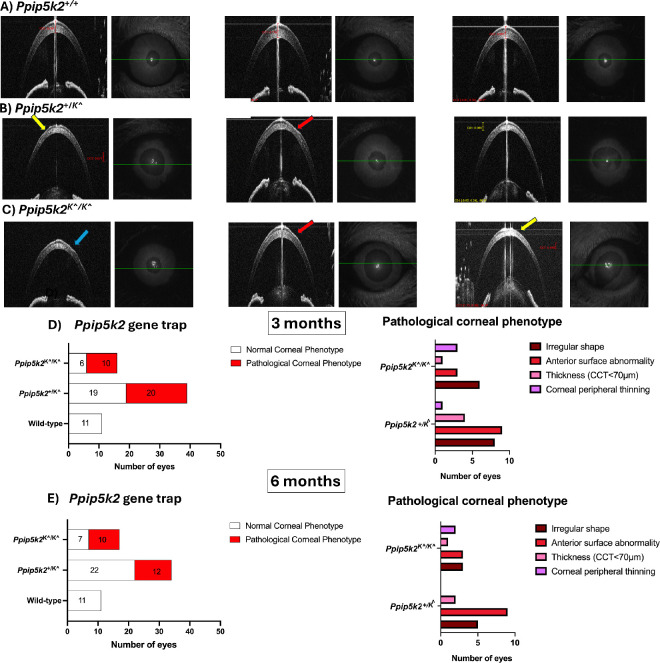
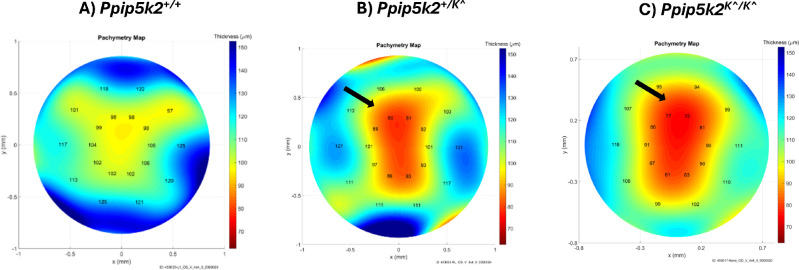
Next, we determined the different corneal phenotypes observed in the 3 strains at 3 and 6 months. Pathological corneal phenotypes, including irregular corneal shapes (blue arrow), anterior surface abnormalities (yellow arrow), CCT < 70 µm, and thinning at the corneal periphery (red arrow) were determined (Figs. 2A–C). In the Ppip5k2 gene trap mice, we observed about 51% and 62% of the corneas in the Ppip5k2+/K^ heterozygous and Ppip5k2K^/K^ homozygous had pathological phenotypes, respectively (Fig. 2D). At 6 months, 35% and 58% of the corneas in the Ppip5k2+/K^ heterozygous and Ppip5k2K^/K^ homozygous were diseased (Fig. 2E). We also observed that at both 3 and 6 months, some of the corneas in the Ppip5k2+/K^ heterozygous and Ppip5k2K^/K^ homozygous mice were normal (see Figs. 2D, 2E). Last, OCT-based pachymetry mapping also confirmed localized thinned areas in the Ppip5k2+/K^ heterozygous (Fig. 3B) and Ppip5k2K^/K^ homozygous mice (Fig. 3C) as compared to WT control (Fig. 3A).
Figure 2.
SD-OCT characterization of the Ppip5k2-gene trap mouse model at 3 and 6 months of age. (A) Representative images of normal corneas. (B, C) Corneal pathological phenotypes are evidenced by irregular shape (blue arrow), anterior surface abnormality (yellow arrow), and corneal peripheral thinning (red arrow). (D) Number of corneas with diseased and normal phenotype at 3 months (left) and the breakdown of the different phenotypes (right); (E) number of corneas with diseased and normal phenotypes at 6 months (left) and the breakdown of the different phenotypes (right).
Figure 3.
Corneal thickness mapping of the Ppip5k2-gene trap mice between 3 and 6 months of age. Thickness mapping of (A) Ppip5k2+/+ wild-type mice corneas. Corneal pathological phenotypes are evidenced in (B) Ppip5k2+/K^ heterozygous and (C) Ppip5k2K^/K^ homozygous by localized thinned areas with the black arrows and color gradient with red being the thinnest.
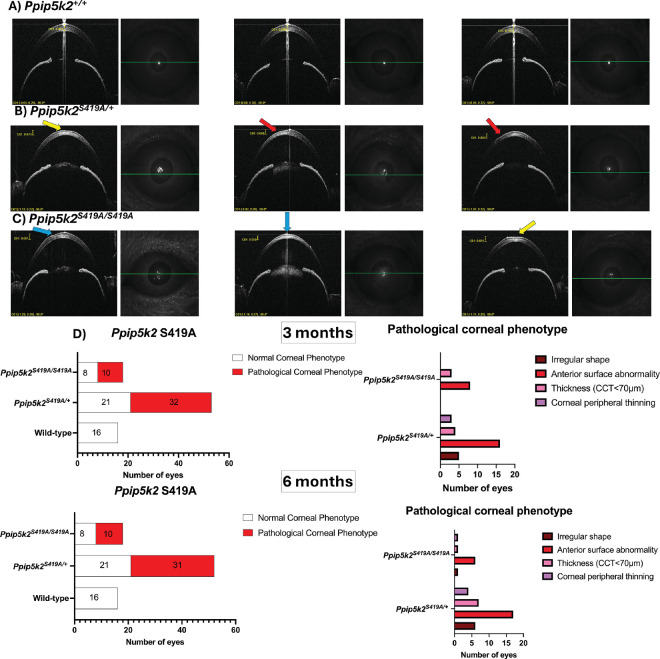
For the Ppip5k2 S419A cohort (Figs. 4A–E), in comparison with the WT, about 60% and 55% of the corneas Ppip5k2S419A/+ heterozygous and Ppip5k2S419A/S419A homozygous (see Figs. 4D, 4E), respectively, had diseased phenotypes at both 3 and 6 months. We also observed that at both 3 and 6 months some of the corneas in the Ppip5k2S419A/+ heterozygous and Ppip5k2S419A/S419A homozygous mice were normal (see Figs. 4D, 4E).
Figure 4.
SD-OCT characterization of the Ppip5k2 S419A knock-in mouse model at 3 and 6 months of age. (A) Representative images of normal corneas. (B, C) Corneal pathological phenotypes are evidenced by irregular shape (blue arrow), anterior surface abnormality (yellow arrow), and corneal peripheral thinning (red arrow). (D) Number of corneas with diseased and normal phenotype at 3 months (left) and the breakdown of the different phenotypes (right); (I) number of corneas with diseased and normal phenotypes at 6 months (left) and the breakdown of the different phenotypes (right).
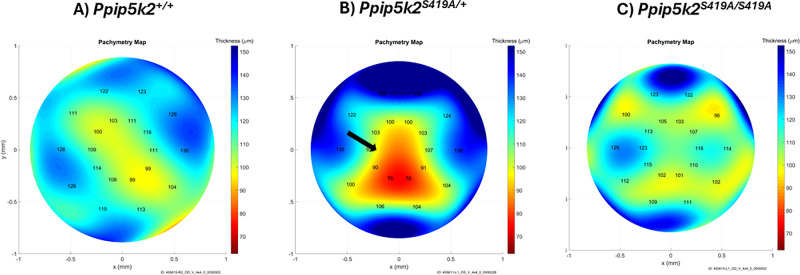
OCT-based pachymetry mapping also revealed more localized thinned areas in the Ppip5k2S419A/+ heterozygous (Fig. 5B) than the Ppip5k2S419A/S419A homozygous mice (Fig. 5C) as compared to the WT control mice (Fig. 5A).
Figure 5.
Corneal thickness mapping of the Ppip5k2 S419A knock-in mouse model between 3 and 6 months. Thickness mapping of (A) Ppip5k2+/+ wild-type mice corneas. Corneal pathological phenotypes are evidenced in (B) Ppip5k2S419A/+ heterozygous and (C) Ppip5k2S419A/S419A homozygous by localized thinned areas with the black arrows and color gradient with red being the thinnest.
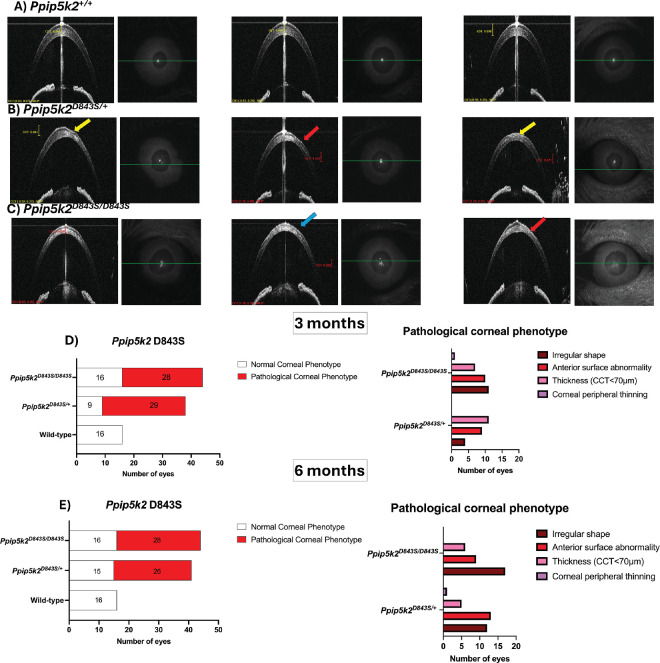
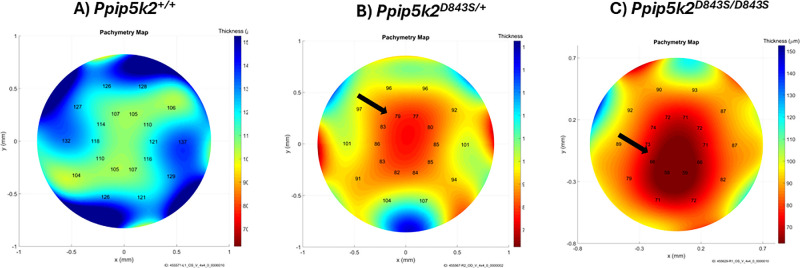
Last, for the Ppip5k2 D843S mice (Figs. 6A–E), about 76% and 64% of the corneas of Ppip5k2D843S/+ heterozygous and Ppip5k2D843S/D843S homozygous had pathological phenotypes out of a total of 38 and 44 corneas, respectively at 3 months (see Fig. 6D). At 6 months, 63% and 64% of the corneas in the Ppip5k2D843S/+ heterozygous and Ppip5k2D843S/D843S homozygous were diseased (Fig. 6F). We also observed that some of the corneas in the Ppip5k2D843S/+ heterozygous and Ppip5k2D843S/D843S homozygous mice were normal at both 3 and 6 months (see Figs. 6D, 6E). In addition, localized thinning was observed in the corneas of Ppip5k2D843S/+ heterozygous and Ppip5k2D843S/D843S homozygous mice, respectively (Figs. 7B, 7C) as compared to their WTs (n = 3; Fig. 7A) with pachymetry mapping.
Figure 6.
SD-OCT characterization of the Ppip5k2 D843S knock-in mouse model at 3 and 6 months. (A) Representative images of normal corneas. (B, C) Corneal pathological phenotypes are evidenced by irregular shape (blue arrow), anterior surface abnormality (yellow arrow), and corneal peripheral thinning (red arrow). (D) Number of corneas with diseased and normal phenotype at 3 months (left) and the breakdown of the different phenotypes (right); (E) number of corneas with diseased and normal phenotypes at 6 months (left) and the breakdown of the different phenotypes (right).
Figure 7.
Corneal thickness mapping of the Ppip5k2 D843S knock-in mouse model between 3 and 6 months. Thickness mapping of (A) Ppip5k2+/+ wild-type mice corneas. Corneal pathological phenotypes are evidenced in (B) Ppip5k2D843S/+ heterozygous and (C) Ppip5k2D843S/D843S homozygous by localized thinned areas with the black arrows by localized thinned areas with the black arrows and color gradient with red being the thinnest.
Slit Lamp Examinations of Corneas of Ppip5k2 Animals
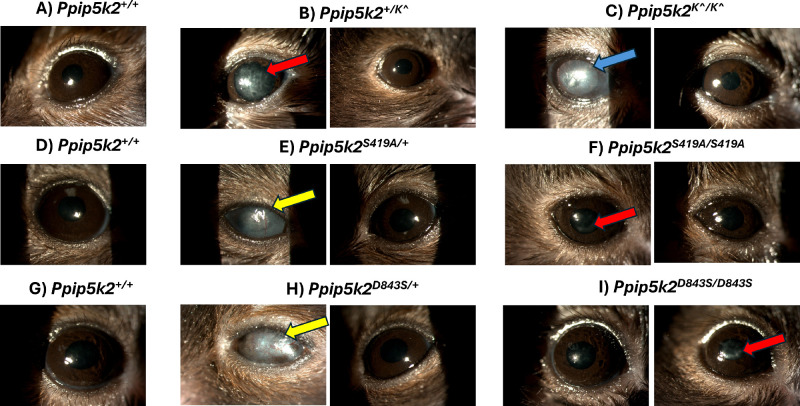
We sought to identify any changes in the corneas using slit lamp examinations in the heterozygous and homozygous knockout/knock-in mice compared with their WT littermates. As compared to Ppip5k2+/+ (Figs. 8A, 8D, 8G), about 15% out of 32 eyes and 40% out of 10 eyes of Ppip5k2+/K^ heterozygous (Fig. 8B) and Ppip5k2+/K^ homozygous (Fig. 8C) mice had some corneal abnormalities, such as bullous keratopathy/stromal edema/stromal scarring (blue arrow), deep corneal neovascularization (yellow arrow), and opacity (red arrows). Some similar observations were made in a smaller proportion of eyes in the Ppip5k2S419A/+ heterozygous (8% out of 24 eyes; Fig. 8E), Ppip5k2S419A/S419A homozygous mice (25% out of 4 eyes; Fig. 8F), Ppip5k2D843S/+ heterozygous (14% out of 14 eyes; Fig. 8H), and Ppip5k2D843S/D843S homozygous mice (25% out of 4 eyes; Fig. 8I).
Figure 8.
Slit lamp examination of Ppip5k2 animal models. Images of mouse corneas for (A) Ppip5k2+/+ wild-type (B) Ppip5k2+/K^ heterozygous, (C) Ppip5k2 K^/K^ homozygous, (D) Ppip5k2+/+ wild-type, (E) Ppip5k2S419A/+ heterozygous, (F) Ppip5k2S419A/S419A homozygous, and (G) Ppip5k2+/+ wild-type (H) Ppip5k2D843S/+ heterozygous and (I) Ppip5k2D843S/D843S homozygous mice. Arrows are representative images of corneal abnormalities, such as bullous keratopathy/stromal edema/stromal scarring (blue arrow), deep corneal neovascularization (yellow arrow), and opacity (red arrows). All other mice without arrows have normal corneal phenotype.
Histological Examinations of Corneas of Ppip5k2 Animal Models
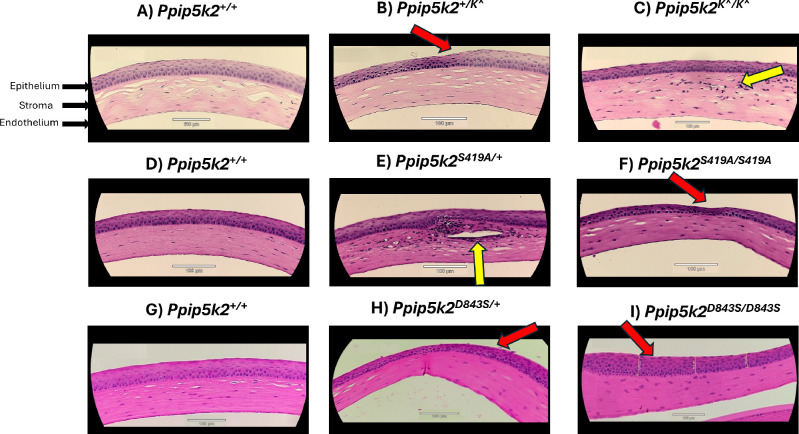
We then sought to determine any histopathological changes in the mouse corneas, such as interruptions in the basement membrane, epithelial thickness, structural changes in the stroma, and fibrosis in the stromal layer, as reported by Grieve et al.43 in histological changes in KC cornea in humans. As compared with Ppip5k2+/+ (Figs. 9A, 9D, 9G), about 20% of 20 eyes from Ppip5k2+/K^ heterozygous (Fig. 9B) and 12% out of 16 eyes from Ppip5k2 K^/K^ mice (Fig. 9C) had some corneal abnormalities in the form of epithelial thickening (red arrow), possible damage of keratocytes, and/or stromal structural changes (yellow arrows). Similar changes were seen in the Ppip5k2S419A/+ heterozygous (30% out of 10 eyes; Fig. 9E) and Ppip5k2S419A/S419A mice (25% out of 16 eyes; Fig. 9F) and the Ppip5k2D843S/+ heterozygous (15% eyes out of 20 eyes; Fig. 9H) and Ppip5k2D843S/D843S homozygous mice (21% eyes out of 24 eyes; Fig. 9I).
Figure 9.
Histopathological changes in the 3 Ppip5k2 animal models. Hematoxylin and eosin staining of mouse cornea sections for (A) Ppip5k2+/+ wild-type, (B) Ppip5k2+/K^ heterozygous, (C) Ppip5k2K^/K^ homozygous, (D) Ppip5k2+/+ wild-type, (E) Ppip5k2S419A/+ heterozygous, (F) Ppip5k2S419A/S419A homozygous and (G) Ppip5k2+/+ wild-type, (H) Ppip5k2D843S/+ heterozygous, and (I) Ppip5k2D843S/D843S homozygous mice. Black arrows indicate the different layers of the cornea. Red arrows represent corneal abnormalities in the form of epithelial thickening and yellow arrows represent possible damage of keratocytes and/or stromal structural changes. Magnification 20x; Scale bars indicate 100 µm.
Discussion
Genetic risk factors are significant contributors to KC pathogenesis. Many KC-associated genomic loci and altered pathways have been identified and are involved in various aspects of corneal health, structure, and development.19–21,44–51 Although several genes and pathways have been implicated in the etiology of KC, fundamental advancements in understanding the disease mechanism have been constrained due to the lack of animal models for KC.27 Therefore, we sought to characterize the corneal phenotypes in three different mouse models with varying degrees of Ppip5k2 kinase activity changes: Ppip5k2+/K^ and Ppip5k2K^/K^, Ppip5k2S419A/+ and Ppip5k2S419A/S419A, and Ppip5k2D843S/+ and Ppip5k2D843S/D843S to determine any KC-related phenotype.18 We observed a significant decrease in CCT from 3 to 6 months per eye in the Ppip5k2K^/K^ mice in comparison with their WT littermates.
CCT is an important measure of corneal health which has been related to an increased risk of KC.51–55 In mice, there are conflicting reports of CCT measurements.56–58 This is possibly due to the strains of mice, age, and the measuring techniques used.56,57,59 For example, using confocal microscopy, Zhang et al.56 reported a CCT of 170 µm in the BALB/c mice whereas Schulz et al.57 reported a CCT of 106 ± 3.45 µm in the same strain using an optical low coherence reflectometer. Moreover, using a histological approach, Henriksson et al.58 reported a CCT of 122.68 ± 4.8 µm, 137.02 ± 14.0 µm, and 134.16 ± 12.9 µm in 129/SVJ, C57BL/6, and BALB/c mice, respectively. Using ultrasound pachymetry, Lively et al.59 reported wide variations in CCT among 17 strains of mice with different genetic backgrounds. Of note, C57BL/6J mice, the most widely used inbred strain, had a CCT of 100.8 ± 5.7 µm.59 In line with these findings, the significance of CCT measurements for our gene trap mice using SD-OCT in comparison with the transgenic models could be attributed to the differences in the genetic makeup of the three models.
We also identified corneal structural changes in the three models. Irregularly shaped corneas were selected based on the corneal curvature changes in KC.60 Anterior surface abnormalities were a phenotypic criterion as they were observed in a previous study on KC-related mouse model.18 CCT < 70 µm was reported because it is significantly less than the CCT measurements in WT mice,56–59 and most of the mice with a CCT < 70 µm in our study showed anterior surface abnormalities. Peripheral corneal thinning was recorded due to the reported thinner peripheral corneal thickness than CCT in mice.58
In our study, at 3 months, different degrees of pathological corneal phenotypes were observed in the corneas of the Ppip5k2 gene trap, and the S419A and D843S lines suggesting incomplete penetrance, as reported in a previous study and in patients with KC.18 The varying differences in expression of these ocular parameters in these three models could be associated with mouse genetic backgrounds and sample sizes. However, lack of animal models of KC limits the interpretation of this data. Interestingly, OCT-based pachymetry mapping in mice, as reported first in previous findings, confirmed abnormal localized corneal thinning in all three diseased models as compared with their WT controls.18
Slit lamp biomicroscopy has been used to identify ocular defects in mice.18,61,62 Although we did not identify the typical cone-shaped protrusion of keratoconus in our three models, we observed corneal abnormalities in the form of bullous keratopathy, stromal edema, stromal scarring, deep corneal neovascularization, and opacities in comparison to WT littermates. In fact, one of the major drawbacks in animal models is the incomplete or partial representation of the clinical characteristics in human patients.63 In addition, considering the multifactorial nature of KC, our models may not show the expected phenotypes relevant to KC because they have not been exposed to the numerous environmental factors as humans.64 It is necessary to combine the genetic and environmental factors in modeling KC phenotypes in mice.
With the histological assessment in our mouse models, we identified corneal abnormalities in the form of epithelial thickening and stromal layer damage in the heterozygous/homozygous knockout and knock-in mice in comparison to WT littermates as reported in humans.43 Taking into consideration the anatomic corneal differences in humans and mice, where C57BL/6J lacks Bowman's layer, some of these indicators, such as absence or interruption of Bowman's layer in humans with KC may be very difficult to replicate in mice.58
Our study has some limitations. First, direct quantification of the products of PPIP5K2 enzymatic activity, 5-InsP7 and 1,5-InsP8, has been challenging due to limited sensitivity of the current technology and the very low levels in cells and tissues.65–68 Thus, indirect measurement of the levels of 5-InsP7 and 1,5-InsP8 is ascertained by measuring the in vitro kinase and phosphatase activities.69 Any alterations in the sensitivity of enzyme-based assays could affect the enzymatic activity readouts, which in turn may affect the levels of 5-InsP7 and 1, 5-InsP8.18,69,70 It could contribute to the non-significance in our findings from our models. It would be beneficial to determine the actual 5-InsP7 and 1,5-InsP8 levels in the corneal tissues. Second, the anatomic differences between the cornea of humans and mice may limit the ability to obtain a perfect mouse model of KC. Third, it would be interesting to track the CCT changes in the three mouse models from 2-month-old mice instead of 3-month-old mice. However, repeating the mouse study for individual CCT tracking with age would take at least 1 year with all 3 models. Last, the multifactorial nature of KC limits the possibility of KC development with only genetic factors. Thus, it may be beneficial to combine other factors to determine if and how these factors work together and lead to the development of KC in mice.
In summary, we have identified the consequences of Ppip5k2 mutations in the corneas of 3 mouse models. We have identified phenotypic and histological changes in mouse corneas that could be relevant in the development of animal models of KC. Because there are no well-established models for KC, some of these features could be determined in future studies. In addition, our mouse models suggest that genetics alone may not be enough to cause KC-relevant phenotypes and that combining with additional environmental and biomechanical factors could recapitulate some KC findings in humans.
Acknowledgments
Supported by NIH R01EY023242, R21EY033961, R01EY032960, R21EY028671, and P30EY031631.
Disclosure: T. Akoto, None; R. Hadvina, None; S. Jones, None; J. Cai, None; H. Yu, None; H. McCord, None; C.X.J. Jin, None; A.J. Estes, None; L. Gan, None; A. Kuo, None; S.B. Smith, None; Y. Liu, None
References
- 1. Karamichos D, Escandon P, Vasini B, et al.. Anterior pituitary, sex hormones, and keratoconus: beyond traditional targets. Prog Retin Eye Res. 2021; 88: 101016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Romero-Jiménez M, Santodomingo-Rubido J, Wolffsohn JS. Keratoconus: a review. Cont Lens Anterior Eye. 2010; 33(4): 157–166; quiz 205. [DOI] [PubMed] [Google Scholar]
- 3. Santodomingo-Rubido J, Carracedo G, Suzaki A, Villa-Collar C, Vincent SJ, Wolffsohn JS. Keratoconus: an updated review. Cont Lens Anterior Eye. 2022; 45(3): 101559. [DOI] [PubMed] [Google Scholar]
- 4. Rabinowitz YS. Keratoconus. Surv Ophthalmol. 1998; 42(4): 297–319. [DOI] [PubMed] [Google Scholar]
- 5. Scroggs MW, Proia AD. Histopathological variation in keratoconus. Cornea. 1992; 11(6): 553–9. [DOI] [PubMed] [Google Scholar]
- 6. Naderan M, Jahanrad A, Balali S. Histopathologic findings of keratoconus corneas underwent penetrating keratoplasty according to topographic measurements and keratoconus severity. Int J Ophthalmol. 2017; 10(11): 1640–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lozano V, Martín C, Blanco N, et al.. Exosomes released by corneal stromal cells show molecular alterations in keratoconus patients and induce different cellular behavior. Biomedicines. 2022; 10(10): 2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sykakis E, Carley F, Irion L, Denton J, Hillarby MC. An in depth analysis of histopathological characteristics found in keratoconus. Pathology. 2012; 44(3): 234–239. [DOI] [PubMed] [Google Scholar]
- 9. Torres Netto EA, Al-Otaibi WM, Hafezi NL, et al.. Prevalence of keratoconus in paediatric patients in Riyadh, Saudi Arabia. Br J Ophthalmol. 2018; 102(10): 1436–1441. [DOI] [PubMed] [Google Scholar]
- 10. Jonas JB, Nangia V, Matin A, Kulkarni M, Bhojwani K.. Prevalence and associations of keratoconus in rural maharashtra in central India: the central India eye and medical study. Am J Ophthalmol. 2009; 148(5): 760–5. [DOI] [PubMed] [Google Scholar]
- 11. Millodot M, Shneor E, Albou S, Atlani E, Gordon-Shaag A. Prevalence and associated factors of keratoconus in Jerusalem: a cross-sectional study. Ophthalmic Epidemiol. 2011; 18(2): 91–97. [DOI] [PubMed] [Google Scholar]
- 12. Waked N, Fayad AM, Fadlallah A, El Rami H. Keratoconus screening in a Lebanese students' population [Article in French]. J Fr Ophtalmol. 2012; 35(1): 23–29. [DOI] [PubMed] [Google Scholar]
- 13. Gomes JAP, Rodrigues PF, Lamazales LL. Keratoconus epidemiology: a review. Saudi J Ophthalmol. 2022; 36(1): 3–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rabinowitz YS. The genetics of keratoconus. Ophthalmol Clin North Am. 2003; 16(4): 607–620, vii. [DOI] [PubMed] [Google Scholar]
- 15. Sahebjada S, Al-Mahrouqi HH, Moshegov S, et al.. Eye rubbing in the aetiology of keratoconus: a systematic review and meta-analysis. Graefe's Arch Clin Exp Ophthalmol. 2021; 259(8): 2057–2067. [DOI] [PubMed] [Google Scholar]
- 16. Moussa S, Grabner G, Ruckhofer J, Dietrich M, Reitsamer H. Genetics in keratoconus - what is new? Open Ophthalmol J. 2017; 11: 201–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mas Tur V, MacGregor C, Jayaswal R, O'Brart D, Maycock N. A review of keratoconus: diagnosis, pathophysiology, and genetics. Surv Ophthalmol. 2017; 62(6): 770–783. [DOI] [PubMed] [Google Scholar]
- 18. Khaled ML, Bykhovskaya Y, Gu C, et al.. PPIP5K2 and PCSK1 are candidate genetic contributors to familial keratoconus. Sci Rep. 2019; 9(1): 19406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Abu-Amero KK, Al-Muammar AM, Kondkar AA. Genetics of keratoconus: where do we stand? J Ophthalmol. 2014; 2014: 641708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bykhovskaya Y, Rabinowitz YS. Update on the genetics of keratoconus. Exp Eye Res. 2021; 202: 108398. [DOI] [PubMed] [Google Scholar]
- 21. Shinde V, Hu N, Mahale A, et al.. RNA sequencing of corneas from two keratoconus patient groups identifies potential biomarkers and decreased NRF2-antioxidant responses. Sci Rep. 2020; 10(1): 9907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Soiberman U, Foster JW, Jun AS, Chakravarti S. Pathophysiology of keratoconus: what do we know today. Open Ophthalmol J. 2017; 11: 252–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Soiberman US, Shehata AEM, Lu MX, et al.. Small molecule modulation of the integrated stress response governs the keratoconic phenotype in vitro. Invest Ophthalmol Vis Sci. 2019; 60(10): 3422–3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhu F, Nair RR, Fisher EMC, Cunningham TJ. Humanising the mouse genome piece by piece. Nat Commun. 2019; 10(1): 1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Macdonald LE, Karow M, Stevens S, et al.. Precise and in situ genetic humanization of 6 Mb of mouse immunoglobulin genes. Proc Natl Acad Sci USA. 2014; 111(14): 5147–5152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kitsios GD, Tangri N, Castaldi PJ, Ioannidis JPA. Laboratory mouse models for the human genome-wide associations. PLoS One. 2010; 5(11): e13782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hadvina R, Estes A, Liu Y. Animal models for the study of keratoconus. Cells. 2023; 12(23): 2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tachibana M, Okamoto M, Sakamoto M, Matsushima Y. Hereditary keratoconus-like keratopathy in Japanese wild mice mapped to mouse chromosome 13. Mamm Genome. 2002; 13(12): 692–695. [DOI] [PubMed] [Google Scholar]
- 29. Tachibana M, Adachi W, Kinoshita S, et al.. Androgen-dependent hereditary mouse keratoconus: linkage to an MHC region. Invest Ophthalmol Vis Sci. 2002; 43(1): 51–57. [PubMed] [Google Scholar]
- 30. Peiffer RL Jr., Werblin TP, Patel AS. Keratoconus in a rhesus monkey. J Med Primatol. 1987; 16(6): 403–6. [PubMed] [Google Scholar]
- 31. Adhami-Moghadam F, Hadipour-Jahromy M, Fazelipour S, Khakpour S, Yonesian MS. Keratoconus experimentally produced in mice using collagenase. Physiol Pharmacol. 2009; 13(2): 209–215. [Google Scholar]
- 32. Qiao J, Li H, Tang Y, et al.. A rabbit model of corneal Ectasia generated by treatment with collagenase type II. BMC Ophthalmol. 2018; 18(1): 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Puk O, Dalke C, Calzada-Wack J, et al.. Reduced corneal thickness and enlarged anterior chamber in a novel ColVIIIa2G257D mutant mouse. Invest Ophthalmol Vis Sci. 2009; 50(12): 5653–5661. [DOI] [PubMed] [Google Scholar]
- 34. Akoto T, Li JJ, Estes AJ, Karamichos D, Lliu Y. The underlying relationship between keratoconus and Down syndrome. Int J Mol Sci. 2022; 23(18): 10796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wallang BS, Das S. Keratoglobus. Eye (Lond). 2013; 27(9): 1004–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hopfer U, Fukai N, Hopfer H, et al.. Targeted disruption of Col8a1 and Col8a2 genes in mice leads to anterior segment abnormalities in the eye. FASEB J. 2005; 19(10): 1232–1244. [DOI] [PubMed] [Google Scholar]
- 37. Segev F, Héon E, Cole WG, et al.. Structural abnormalities of the cornea and lid resulting from collagen V mutations. Invest Ophthalmol Vis Sci. 2006; 47(2): 565–573. [DOI] [PubMed] [Google Scholar]
- 38. Markand S, Saul A, Roon P, et al.. Retinal ganglion cell loss and mild vasculopathy in methylene tetrahydrofolate reductase (Mthfr)-deficient mice: a model of mild hyperhomocysteinemia. Invest Ophthalmol Vis Sci. 2015; 56(4): 2684–2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Liu AS, Brown DM, Conn RE, McNabb RP, Pardue MT, Kuo AN. Topography and pachymetry maps for mouse corneas using optical coherence tomography. Exp Eye Res. 2020; 190: 107868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sharif R, Bak-Nielsen S, Hjortdal J, Karamichos D. Pathogenesis of keratoconus: the intriguing therapeutic potential of Prolactin-inducible protein. Prog Retin Eye Res. 2018; 67: 150–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hanlon SD, Patel NB, Burns AR. Assessment of postnatal corneal development in the C57BL/6 mouse using spectral domain optical coherence tomography and microwave-assisted histology. Exp Eye Res. 2011; 93(4): 363–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jackson SJ, Andrews N, Ball D, et al.. Does age matter? The impact of rodent age on study outcomes. Lab Anim. 2017; 51(2): 160–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Grieve K, Georgeon C, Andreiuolo F, et al.. Imaging microscopic features of keratoconic corneal morphology. Cornea. 2016; 35(12): 1621–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. He W, Han X, Ong JS, et al.. Association of novel loci with keratoconus susceptibility in a multitrait genome-wide association study of the UK Biobank Database and Canadian Longitudinal Study on Aging. JAMA Ophthalmol. 2022; 140(6): 568–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bykhovskaya Y, Margines B, Rabinowitz YS. Genetics in Keratoconus: where are we? Eye Vis (Lond). 2016; 3: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hardcastle AJ, Liskova P, Bykhovskaya Y, et al.. A multi-ethnic genome-wide association study implicates collagen matrix integrity and cell differentiation pathways in keratoconus. Commun Biol. 2021; 4(1): 266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hosoda Y, Miyake M, Meguro A, et al.. Keratoconus-susceptibility gene identification by corneal thickness genome-wide association study and artificial intelligence IBM Watson. Commun Biol. 2020; 3(1): 410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Choquet H, Melles RB, Yin J, et al.. A multiethnic genome-wide analysis of 44,039 individuals identifies 41 new loci associated with central corneal thickness. Commun Biol. 2020; 3(1): 301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. McComish BJ, Sahebjada S, Bykhovskaya Y, et al.. Association of genetic variation with keratoconus. JAMA Ophthalmol. 2020; 138(2): 174–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Khawaja AP, Rojas Lopez KE, Hardcastle AJ, et al.. Genetic variants associated with corneal biomechanical properties and potentially conferring susceptibility to keratoconus in a genome-wide association study. JAMA Ophthalmol. 2019; 137(9): 1005–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Iglesias AI, Mishra A, Vitart V, et al.. Cross-ancestry genome-wide association analysis of corneal thickness strengthens link between complex and Mendelian eye diseases. Nat Commun. 2018; 9(1): 1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Landers JA, Hewitt AW, Dimasi DP, et al.. Heritability of central corneal thickness in nuclear families. Invest Ophthalmol Vis Sci. 2009; 50(9): 4087–4090. [DOI] [PubMed] [Google Scholar]
- 53. Zheng Y, Ge J, Huang G, et al.. Heritability of central corneal thickness in Chinese: the Guangzhou Twin Eye Study. Invest Ophthalmol Vis Sci. 2008; 49(10): 4303–4307. [DOI] [PubMed] [Google Scholar]
- 54. Ali W, Shakir M, Wasim S, Afza R. Central corneal thickness: ultrasound pachymetry verus anterior segment optical coherence tomography. Pakistan Journal of Ophthalmology. 2021; 37(4): 1260. [Google Scholar]
- 55. Williams R, Fink BA, King-Smith PE, Mitchell GL. Central corneal thickness measurements: using an ultrasonic instrument and 4 optical instruments. Cornea. 2011; 30(11): 1238–1243. [DOI] [PubMed] [Google Scholar]
- 56. Zhang EP, Schründer S, Hoffmann F. Orthotopic corneal transplantation in the mouse–a new surgical technique with minimal endothelial cell loss. Graefes Arch Clin Exp Ophthalmol. 1996; 234(11): 714–719. [DOI] [PubMed] [Google Scholar]
- 57. Schulz D, Iliev ME, Frueh BE, Goldblum D. In vivo pachymetry in normal eyes of rats, mice and rabbits with the optical low coherence reflectometer. Vision Res. 2003; 43(6): 723–728. [DOI] [PubMed] [Google Scholar]
- 58. Henriksson JT, McDermott AM, Bergmanson JP. Dimensions and morphology of the cornea in three strains of mice. Invest Ophthalmol Vis Sci. 2009; 50(8): 3648–3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lively GD, Jiang B, Hedberg-Buenz A, et al.. Genetic dependence of central corneal thickness among inbred strains of mice. Invest Ophthalmol Vis Sci. 2010; 51(1): 160–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Liduma S, Luguzis A, Krumina G. The impact of irregular corneal shape parameters on visual acuity and contrast sensitivity. BMC Ophthalmol. 2020; 20(1): 466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Chaurasia SS, Perera PR, Poh R, Lim RR, Wong TT, Mehta JS. Hevin plays a pivotal role in corneal wound healing. PLoS One. 2013; 8(11): e81544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Moore BA, Roux MJ, Sebbag L, et al.. A population study of common ocular abnormalities in C57BL/6N rd8 mice. Invest Ophthalmol Vis Sci. 2018; 59(6): 2252–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Li H, Auwerx J. Mouse systems genetics as a prelude to precision medicine. Trends Genet. 2020; 36(4): 259–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Seok J, Warren HS, Cuenca AG, et al.. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc Natl Acad Sci USA. 2013; 110(9): 3507–3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Lee S, Kim MG, Ahn H, Kim S. Inositol pyrophosphates: signaling molecules with pleiotropic actions in mammals. Molecules. 2020; 25(9): 2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Chakraborty A. The inositol pyrophosphate pathway in health and diseases. Biol Rev Camb Philos Soc. 2018; 93(2): 1203–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Shah A, Ganguli S, Sen J, Bhandari R. Inositol pyrophosphates: energetic, omnipresent and versatile signalling molecules. J Indian Inst Sci. 2017; 97(1): 23–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Chabert V, Kim GD, Qiu D, et al.. Inositol pyrophosphate dynamics reveals control of the yeast phosphate starvation program through 1,5-IP(8) and the SPX domain of Pho81. Elife. 2023; 12: RP87956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Gu C, Nguyen HN, Ganini D, et al.. KO of 5-InsP7 kinase activity transforms the HCT116 colon cancer cell line into a hypermetabolic, growth-inhibited phenotype. Proc Natl Acad Sci USA. 2017; 114(45): 11968–11973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Yousaf R, Gu C, Ahmed ZM, et al.. Mutations in Diphosphoinositol-Pentakisphosphate Kinase PPIP5K2 are associated with hearing loss in human and mouse. PLoS Genet. 2018; 14(3): e1007297. [DOI] [PMC free article] [PubMed] [Google Scholar]