Abstract
In addition to the usual retroviral promoter, the mouse mammary tumor virus (MMTV) long terminal repeat carries a second promoter located in the U3 region. Here we show that both of these promoters are independently able to give rise to superantigen activity in transgenic mice. The ability of multiple MMTV promoters to drive superantigen expression underscores its importance in the virus life cycle.
The long terminal repeat (LTR) of mouse mammary tumor virus (MMTV) encodes a superantigen (Sag). After infection of mice with exogenous virus, expression of Sag in antigen-presenting cells, such as B lymphocytes, specifically stimulates the proliferation of whole classes of T cells bearing the cognate Vβ chain as part of their T-cell receptor (1). This T-cell stimulation results in local cytokine production and, in turn, proliferation of B cells in the vicinity, including those that were infected with MMTV. The virus thus uses the host immune system to establish and amplify a reservoir of infected cells. Later, the virus is passed to mammary epithelial cells by an as-yet-unknown mechanism and causes mammary tumors in susceptible mice (12).
Expression of Sag from endogenous, germ line-transmitted MMTVs leads to deletion and/or anergy of specific reactive Vβ-bearing classes of T cells (1). The deletion thereby protects mice from subsequent challenge with an MMTV encoding a Sag with the same Vβ specificity (4).
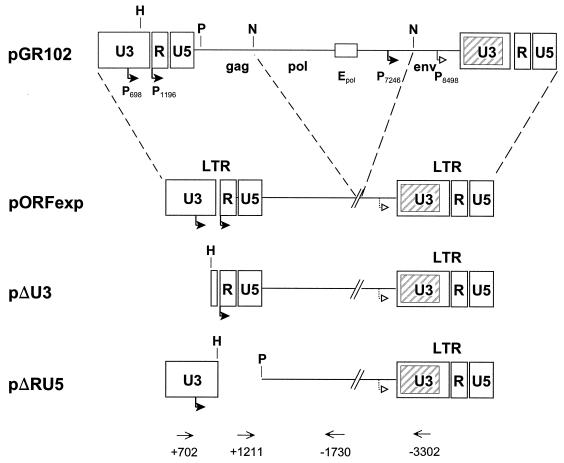
MMTV is unusual among retroviruses in that it carries, in addition to the classic retroviral promoter (P1196), a second promoter (P698) (Fig. 1) (8, 16, 17). Intriguingly, both of these promoters independently give rise to Sag activity in mixed-lymphocyte reactions when coupled 5′ to a Sag encoding the 3′ LTR (16, 17). To determine which of the two LTR-associated promoters is used in vivo for Sag expression, a number of transgenic mouse lines carrying the coding region for the Sag of the Mtv-2 provirus linked to either P1196, P698, or both promoters were established. Construction of the plasmids pORFexp, pΔU3, and pΔRU5 has been previously described (17). The inserts containing the miniproviruses were excised from each plasmid and used for microinjection into freshly isolated C57BL/B6 mouse oocytes. These were reimplanted after 12 h of cultivation into recipient pseudopregnant NMRI mice as previously described (3). DNA was prepared from tail clips of 6-week-old offspring and analyzed by PCR using the previously described primers located at +702 or +1211 together with primer −1730 (8) or a new primer, −3302 (5′-GCAACTTCCCCCAATAGCC-3′) (Fig. 1). Since all mice carry endogenous MMTV copies, signals specific for the deleted miniproviruses were indicative of transgenicity. Mice transgenic for pΔRU5 gave an indicative 0.5-kb fragment with the primers +702 and −1730 as the result of the deletion of the R and U5 sequences. Both the pORFexp and the pΔU3 mice gave an indicative 2.1-kb fragment with the primers +1211 and −3302 as the result of the deletion of the pol and env sequences (data not shown).
FIG. 1.
Schematic representation of the MMTV constructs used. The plasmid pGR102 carries a hybrid provirus in which the 5′ half is from Mtv-8 and the 3′ half is from Mtv-2 (13). The Sag, encoded by the open reading frame located in the 3′ LTR (shaded box), is thus derived from the Mtv-2 provirus and specifically interacts with Vβ14-bearing T cells (2). The plasmid pORFexp was derived from pGR102 by deletion of the internal NcoI fragment. Also shown are the locations of the classic promoter (P1196) and other promoters present in the LTR (P698) or in the env gene (P7246 and P8498) (nomenclature as given in reference 11). The enhancer (Epol) shown to be important for Sag activity from P7246 is indicated as an open box, as are the HpaI (H), PvuII (P), and NcoI (N) restriction sites used in the construction of the plasmids pORFexp, pΔU3, and pΔRU5. At the bottom of the figure, the positions of the primers used for the PCR analysis are shown.
Expression of the Mtv-2 Sag results in the deletion or anergy of Vβ14-bearing T cells (8). After 100 μl of blood from the retro bulbar plexus of anesthetized transgenic mice was sampled using a glass capillary, the T cells were stained with R-phycoerythrin-labeled anti-CD3 monoclonal antibody and a fluorescein-conjugated anti-Vβ14 monoclonal antibody and analyzed by flow cytofluorometry (fluorescence-activated cell sorting) (Elite Coulter Inc.) to determine the percentage of Vβ14+ T cells. Table 1 shows that these T cells were deleted (between <1 and 1.4%) in mice regardless of the promoter carried by the construct but that this deletion did not occur in nontransgenic mice (8.3%). Thus, both LTR-associated promoters have the potential to give rise to Sag activity in vivo in the temporal window during ontogeny, allowing deletion of Vβ14-bearing T cells. Additionally, these results show that both promoters are active in the antigen-presenting cells involved in the establishment of Sag-mediated tolerance. These data are consistent with previous data obtained with a mixed-lymphocyte reaction in vitro (17) in that the pΔRU5 construct, which carries only the new P698 promoter, shows the best Sag activity. Although we cannot rule out the possibility that the sequences deleted in our constructs normally silence one promoter or the other, this seems unlikely, since transcripts from both promoters can be detected in B cells (and mammary tumor GR cells) carrying the Mtv-2 locus (8).
TABLE 1.
Levels of Vβ14+ T cells in normal and transgenic mice
| Construct | LTR promoter(s) present | Mean % Vβ14+ T-cellsa |
|---|---|---|
| Nontransgenic | None | 8.3 |
| pORFexp | P698 and P1196 | 1.3 |
| pΔU3 | P1196 | 1.4 |
| pΔRU5 | P698 | <1.0 |
Data are the percentage measured in nontransgenic mice and the averages of the measurements for at least five transgenic mice from each line carrying a construct.
Evidence has been presented that a promoter located within the env coding sequences (P7246) (10, 15, 18), in association with an enhancer element located in the pol region (Epol), directs between 98 and 99.7% of Sag gene expression in B cells (11). However, these elements are not included in the constructs described here (Fig. 1). There is an additional promoter (P8498) in our construct; however, only low-level expression (0.3% maximum) has been attributed to this promoter (11). Thus, taken together, our data suggest that, in the absence of the P7246 promoter, either of the LTR promoters can also drive expression of a functional Sag in vivo, during ontogeny. We have previously shown the presence of spliced messages from these promoters to the Sag open reading frame (8, 9).
The presence of multiple promoters in MMTV may act as a fail-safe feature, ensuring Sag expression. Such Sag expression from endogenous MMTVs protects mice against infection with exogenous virus since the cognate T-cell classes are deleted (4), whereas the use of redundant promoters in exogenous viruses may ensure establishment of infection. Further, transcripts from the P698 promoter in particular may be important for the generation of new MMTV variants, in order to overcome infection restrictions imposed by the Sag-mediated T-cell deletion (14). Such MMTV variants arising from recombinations between pre-existing endogenous and exogenous viruses have been recently detected (5–7).
REFERENCES
- 1.Acha-Orbea H, MacDonald H R. Superantigens of mouse mammary tumor virus. Annu Rev Immunol. 1995;13:459–486. doi: 10.1146/annurev.iy.13.040195.002331. [DOI] [PubMed] [Google Scholar]
- 2.Acha-Orbea H, Shakov A N, Scarpellino L, Kolb E, Muller V, Vessaz-Shaw A, Fuchs R, Blochlinger K, Rollini P, Billotte J, Sarafidou M, MacDonald H R, Diggelmann H. Clonal deletion of Vβ14-bearing T cells in mice transgenic for mammary tumour virus. Nature. 1991;350:207–211. doi: 10.1038/350207a0. [DOI] [PubMed] [Google Scholar]
- 3.Brem G. Transgene nutztiere. Zuchtungskunde. 1998;60:248–262. [Google Scholar]
- 4.Golovkina T V, Chervonsky A, Dudley J P, Ross S R. Transgenic mouse mammary tumor virus superantigen expression prevents viral infection. Cell. 1992;69:637–645. doi: 10.1016/0092-8674(92)90227-4. [DOI] [PubMed] [Google Scholar]
- 5.Golovkina T V, Jaffe A B, Ross S R. Co-expression of exogenous and endogenous mouse mammary tumor virus RNA in vivo results in viral recombination and broadens the virus host range. J Virol. 1994;68:5019–5026. doi: 10.1128/jvi.68.8.5019-5026.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Golovkina T V, Prakash O, Ross S R. Endogenous mouse mammary tumor virus Mtv-17 is involved in Mtv-2-induced tumorigenesis in GR mice. Virology. 1996;218:14–22. doi: 10.1006/viro.1996.0161. [DOI] [PubMed] [Google Scholar]
- 7.Golovkina T V, Piazzon I, Nepomnaschy I, Buggiano V, de Olano Vela M, Ross S R. Generation of a tumorigenic milk-borne mouse mammary tumor virus by recombination between endogenous and exogenous viruses. J Virol. 1997;71:3895–3903. doi: 10.1128/jvi.71.5.3895-3903.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Günzburg W H, Heinemann F, Wintersperger S, Miethke T, Wagner H, Erfle V, Salmons B. Endogenous superantigen expression controlled by a novel promoter located in the MMTV long terminal repeat. Nature. 1993;354:154–158. doi: 10.1038/364154a0. [DOI] [PubMed] [Google Scholar]
- 9.Günzburg W H, Saller R M, Salmons B. Retroviral vectors directed to predefined cell types for gene therapy. Biologicals. 1995;23:5–12. doi: 10.1016/1045-1056(95)90003-9. [DOI] [PubMed] [Google Scholar]
- 10.Miller C L, Garner R, Paetkau V. An activation-dependent, T-lymphocyte-specific transcriptional activator in the mouse mammary tumor virus env gene. Mol Cell Biol. 1992;12:3262–3272. doi: 10.1128/mcb.12.7.3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reuss F, Coffin J M. Mouse mammary tumor virus superantigen expression in B cells is regulated by a central enhancer within the pol gene. J Virol. 1998;72:6073–6082. doi: 10.1128/jvi.72.7.6073-6082.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salmons B, Günzburg W H. Current perspectives in the biology of mouse mammary tumour virus. Virus Res. 1987;8:81–102. doi: 10.1016/0168-1702(87)90022-0. [DOI] [PubMed] [Google Scholar]
- 13.Salmons B, Groner B, Calberg-Bacq C-M, Ponta H. Production of mouse mammary tumour virus upon transfection of a recombinant proviral DNA into cultured cells. Virology. 1985;144:101–114. doi: 10.1016/0042-6822(85)90309-5. [DOI] [PubMed] [Google Scholar]
- 14.Salmons B, Wintersperger S, Günzburg W H. On the generation of endogenous superantigen diversity. Immunol Today. 1994;15:191–192. doi: 10.1016/0167-5699(94)90318-2. [DOI] [PubMed] [Google Scholar]
- 15.Sambasivarao D, Paetkau V. Interactions of a transcriptional activator in the env gene of the mouse mammary tumor virus with activation-dependent, T cell-specific transacting factors. J Biol Chem. 1996;271:8942–8950. doi: 10.1074/jbc.271.15.8942. [DOI] [PubMed] [Google Scholar]
- 16.Wintersperger S, Indraccolo S, Miethke T, Günzburg W H, Salmons B. A transient assay for gene expression studies in B-lymphocytes and its use for superantigen assays. BioTechniques. 1994;16:882–886. [PubMed] [Google Scholar]
- 17.Wintersperger S, Salmons B, Miethke T, Erfle V, Wagner H, Günzburg W H. Negative acting factor and superantigen are separable activities encoded by the mouse mammary tumor virus long terminal repeat. Proc Natl Acad Sci USA. 1995;92:2745–2749. doi: 10.1073/pnas.92.7.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang D-J, Tsiagbe V K, Huang C, Thorbecke G J. Control of endogenous mouse mammary tumor virus superantigen expression in SJL lymphomas by a promoter within the env region. J Immunol. 1996;157:3510–3517. [PubMed] [Google Scholar]