TO THE EDITOR:
The transcription factor signal transducer and activator of transcription 5B (STAT5B) plays a critical role as a downstream mediator of various growth factors and oncogenes1,2. The STAT5B N642H mutation is recognized as a gain of function mutation3-5 and is predominantly observed in lymphoid neoplasms initially, such as T cell large granular lymphocytic leukemia (T-LGLL) and T acute lymphoblastic leukemia1,6-10. Recent emerging evidence has indicated occurrences of STAT5B N642H in myeloid neoplasms, both with or without eosinophilia6,11,12. In lymphoid neoplasms, concomitant mutations are infrequent. In contrast, myeloid neoplasms with STAT5B mutations show more intricate genetic profiles, marked by a higher diversity and number of concurrent mutations11,13. However, discerning whether the STAT5B mutation originates in the lymphoid or myeloid lineage presents challenges in some cases. This report presents a distinctive scenario that began with an initial diagnosis of T-LGLL with eosinophilia, where the STAT5B mutated myeloid population remained obscured. Subsequently, disease progression led to the development of acute myeloid leukemia (AML) originating specifically from the STAT5B mutated myeloid cells.
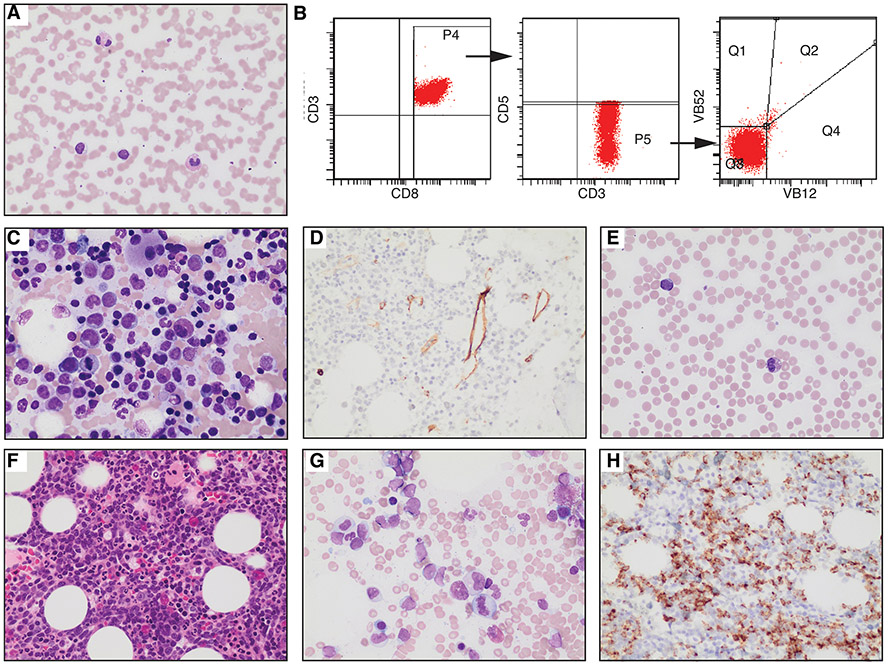
A 67-year-old woman presented with anemia (hemoglobin 10.9g/dL) and thrombocytopenia (platelet 68 x109/L). Her white cell count was 6.5x109/L and showed neutropenia, eosinophilia, and lymphocytosis. The differential was: neutrophils 0.82 x109/L, 12.5%; lymphocytes 3.66 x109/L, 56.2%; monocyte 0.7 x109/L, 10.8%; eosinophils 1.25 x109/L, 19.2%; basophils 0.03 x109/L, 0.5%; immature granulocyte 0.05 x109/L, 0.7%). Peripheral blood smear analysis depicted increased eosinophils and small lymphocytes displaying irregular nuclear contours, condensed chromatin, and cytoplasmic granules (Figure 1A). Peripheral blood flow cytometry identified aberrant T cells (27% of total cells, CD3+, CD4−, CD8+, CD7dim/−, CD5dim/−) without Vbeta expression, consistent with T-LGLL (Figure 1B). T cell clonality was confirmed by gene rearrangement analysis. The in-house 30-gene lymphoid NGS analysis of peripheral blood detected a STAT5B p.N642H mutation with a variant allele frequency (VAF) of 26%. A bone marrow biopsy showed 80% cellularity, displaying increased eosinophils, mild megakaryocytic dysplasia (Figure 1C), and 10% abnormal T cells with similar immunophenotype as peripheral blood. Blasts were not increased by manual count or CD34 immunohistochemical staining (Figure 1D). Due to the hypercellularity and atypical megakaryocytes, a myeloid NGS (STAT5B gene was not covered) analysis was conducted on the bone marrow aspirate, revealing several somatic mutations: PHF6 p. I314T (VAF 27%), TET2 p.M1333Nfs*6 (VAF 27%), TET2 p.E1413* (VAF 14%), and TET2 splice site (VAF 30%). The karyotype was normal: 46, XX [20], and FISH for PDGFRA, PDGRB, FGFR1, ETV6, and ABL1 rearrangements yielded negative results. The final diagnosis was T-LGLL with eosinophilia and clonal cytopenia of undetermined significance. The patient received methotrexate treatment.
Figure 1. Morphological and immunophenotypic characterization of T-LGLL and Subsequent AML.
A-D, samples from 2018 specimens. E-H, samples from 2022 specimens. (A) Peripheral blood smear of T-LGLL (Giemsa stain, 500X). Increased eosinophils and small, atypical lymphocytes with cytoplasmic granules are observed. (B) Flow cytometry of the patient’s peripheral blood showed atypical T cells [CD3+, CD8+, CD5dim/−, CD7dim/− (not shown), and Vbeta-]. (C) Bone marrow aspirate (C: 500X) shows scattered small lymphocytes, increased eosinophils, and dysplastic megakaryocytes. (D) Immunohistochemical staining for CD34 (400X). (E) Peripheral blood smear (Giemsa stain, 500X) showing increased eosinophils and occasional circulating myeloblasts. (F) Bone marrow core biopsy (400X) exhibiting hypercellularity with a left shift. (G) Bone marrow aspirate (500X) indicating elevated myeloblasts. (H) CD34 immunostaining of the bone marrow core (400X).
Four years later, she presented with anemia, thrombocytopenia, and significantly increased eosinophilia (49%, absolute count 1960/μl), accompanied by occasional circulating myeloblasts (Figure 1E). A bone marrow biopsy revealed a hypercellular marrow (Figure 1F) with 15% myeloblasts on a suboptimal aspirate (Figure 1G), confirmed by CD34 immunohistochemical staining (Figure 1H). By flow cytometry, myeloblasts accounted for 20-25% of total cells and were CD34+, CD117+, CD38+, HLA−DR+. Additionally, a small aberrant B cell population (negative for CD5, CD10, and surface light chains, <1% of total cells), and a tiny clonal T cell population (positive for CD4 and CD8dim, <0.5% of total cells) were detected. Molecular NGS studies revealed the previously detected STAT5B p.N642H (VAF 26%), TET2 p.M1333Nfs*6 (VAF 41%), PHF6 p.I314T (VAF 41%), and TET2 splice site (VAF 42%) mutations, with several additional mutations, including RUNX1 p.S388Ffs*167 (VAF 11%), NRAS p.A59delA (VAF 4%), and U2AF1 p.34F (VAF 3%). TET2 p.E1413*, which was detected in the previous bone marrow, was not identified. Cytogenetic analysis revealed an abnormal karyotype: 47, XX, +8 [3]/46, XX [17]. The patient was diagnosed with AML with eosinophilia and passed away shortly after the diagnosis.
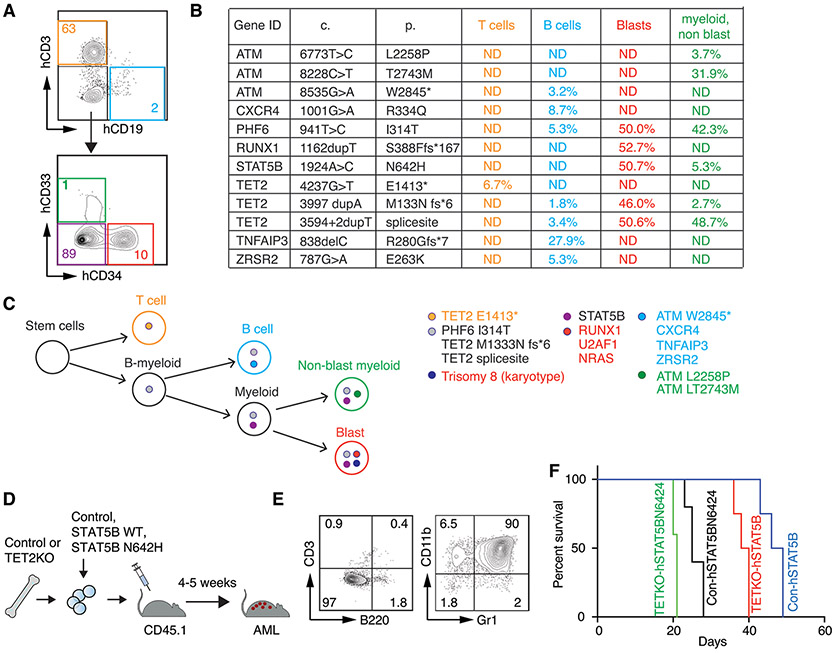
To elucidate the clonal relationships and evolution among distinct cell populations, we performed comprehensive lymphoid and myeloid NGS analyses using genomic DNAs isolated from flow cytometry sort-purified bone marrow cells collected in 2022: CD34+ AML blasts, CD33+CD34− non-AML myeloid cells, CD3+ T cells, and CD19+ B cells (Figure 2A). The examination of shared and distinct somatic mutations among these populations (Figure 2B) along with newly detected trisomy 8 unveiled the clonal relationship between these populations (Figure 2C). Notably, mutations including TET2 p.M1333Nfs*6, PHF6 p.I314T, and TET2 splice site were found shared among AML blasts, non-AML myeloid cells, and B cells, indicating these cells derived from a common precursor – likely a B-myeloid precursor. The presence of STAT5B mutation in both AML myeloblasts and non-AML myeloid cells suggests its emergence after myeloid commitment from B-myeloid precursors, further driven by newly acquired RUNX1 mutation and trisomy 8, contributing to AML development. Conversely, the absence of NRAS and U2AF1 mutations in any of these four populations retrospectively suggests their potential presence in the CD34−CD33− cells (purple fraction in Figure 2A), which unfortunately were not collected. Intriguingly, STAT5B mutation was not detected in the sorted T cells, which exclusively contained the TET2 p.E1413* mutation, implying persistence of the original T cell clone. The absence of this mutation in unsorted cells is probably attributable to the low frequency of T cells in this marrow sample. The predominance of the T cell clone in the initial specimen likely masked the presence of the myeloid clone carrying the STAT5B mutation, thereby leading to the interpretation associating the STAT5B mutation with T cell-related eosinophilia in T-LGLL. Furthermore, the cell sorting uncovered TNFAIP3 and ZRSR mutations solely in B cells suggesting a clone emerged post B cell commitment from the B-myeloid precursors.
Figure 2. Clonal evolution of STAT5B mutated myeloid neoplasm and its role in leukemogenesis.
(A) Flow cytometry sorting strategy highlighting different cell populations labeled by various colors. (B) Summary of mutations detected in each cell population using an in-house myeloid and lymphoid NGS panel. (C) Hypothesized clonal evolution steps based on NGS mutational testing results from (B). (D) Schematic representation of the investigation into the role of STAT5B overexpression in leukemogenesis in a mouse model. Mouse hematopoietic stem/progenitor cells (HSPCs) were isolated from the bone marrows of control or TET2 knockout mice, then transduced with MSCV-IRES-GFP based retroviruses carrying an empty vector (Con), wild type (WT) or STAT5B N642H, and subsequently injected into lethally irradiated recipient mice. (E) Representative flow cytometry profile illustrating the development of myeloid leukemia induced by both WT and N642H STAT5B overexpression. (F) Survival curve of mice receiving STAT5B WT or mutant transduced HSPCs, showing accelerated myeloid leukemia development in the STAT5B mutant group compared to the STAT5B wild-type group (p<0.05), with further expedited leukemia development upon TET2 deletion (p<0.01).
Research has elucidated that STAT5B but not STAT5A can drive the self-renewal of hematopoietic and leukemia stem cells. This distinction clarifies why STAT5B is preferentially activated downstream of several oncogenes, leading to the frequent occurrence of gain of function mutations in STAT5B2. To explore whether a STAT5B mutation alone could transform hematopoietic stem and progenitor cells (HSPCs) into AML or necessitates cooperation with other mutations, such as TET2 mutation, an animal experiment was conducted using control and TET2 knockout (KO) mice, along with STAT5B retroviral transduction (Figure 2D). Remarkably, recipient mice injected with either wild type (WT) or N642H STAT5B transduced HSPCs developed AML within 3-7 weeks, irrespective of TET2 deletion (Figure 2E). Notably, all recipient mice, whether transplanted with Control or TET2KO HSPCs, and expressing either WT or N642H STAT5B, develop myeloid leukemia with a consistent immunophenotype co-expressing myeloid markers Gr1 and CD11b. However, the combined deletion of TET2 and presence of STAT5B N642 demonstrated more rapid induction of AML (Figure 2F). These findings substantiate the principle that STAT5B alone can induce myeloid leukemia, and the simultaneous occurrence of somatic mutations enhances this process.
In this case, the pivotal question revolved around whether T-LGLL and the subsequent AML shared the STAT5B mutation and were clonally related. Typically, most diagnosticians may assume that the two diseases are related. However, our study, conducted through flow sort-purified cell populations, demonstrated the absence of a clonal relationship between the T and myeloid cells. These findings align with recent research indicating the rarity of the STAT5B mutation in CD8+ T-LGLLs compared to myeloid neoplasms11,13 and CD4+ T-LGLL9,14. However, the absence of the STAT5B N642H mutation on sorted T-cells may be attributed to the lack of the prior T-LGL, and thus, the possibility of the STAT5B N642H mutation in T-LGL of the initial specimen cannot be completely ruled out. Nonetheless, transgenic mice expressing STAT5B N642H under the Vav1 promoter, expected to express STAT5B mutant in all hematopoietic cells, develop CD8+ T lymphocytic leukemia/lymphoma5. This, combined with instances where STAT5B mutation occurs in T, B, myeloid, and eosinophils4, suggests that the same mutation in different cell lineage at distinct developmental stage with varying co-mutations may induce diverse types of neoplasms. It is important to note that the expression levels of STAT5B are also a key factor, as even the overexpression of WT STAT5B alone can induce myeloid leukemia. Further research endeavors are warranted to unravel the pathogenesis of STAT5B-mutated neoplasms and justify the development of specific STAT5B inhibitors.
Supplementary Material
FUNDING SOURCES
This work was supported by the National Institutes of Health/National Cancer Institute (R01CA237006) and the U.S. Department of Veterans Affairs (I01 BX004255), and the Department of Pathology startup funds (Case Western Reserve University and University Hospitals) to C.Z.
Footnotes
CONFLICT OF INTEREST DISCLOSURE
All authors declare no competing financial interests.
ETHICS APPROVAL STATEMENT
This case report was conducted ethically in accordance with the guideline of the University Hospitals (IRB# 04-95-91).
PATIENT CONSENT STATEMENT
This study was conducted with patient’s consent and adhered to the principles outlined in the Declaration of Helsinki.
DATA AVAILABILITY STATEMENT
The data of this study are available on request from the corresponding author, C.Z.
REFERENCE
- 1.Smith MR, Satter LRF, Vargas-Hernandez A. STAT5b: A master regulator of key biological pathways. Front Immunol. 2022;13:1025373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kollmann S, Grausenburger R, Klampfl T, et al. A STAT5B-CD9 axis determines self-renewal in hematopoietic and leukemic stem cells. Blood. 2021;138(23):2347–2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ariyoshi K, Nosaka T, Yamada K, et al. Constitutive activation of STAT5 by a point mutation in the SH2 domain. J Biol Chem. 2000;275(32):24407–24413. [DOI] [PubMed] [Google Scholar]
- 4.Ma CA, Xi L, Cauff B, et al. Somatic STAT5b gain-of-function mutations in early onset nonclonal eosinophilia, urticaria, dermatitis, and diarrhea. Blood. 2017;129(5):650–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pham HTT, Maurer B, Prchal-Murphy M, et al. STAT5BN642H is a driver mutation for T cell neoplasia. J Clin Invest. 2018;128(1):387–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cross NCP, Hoade Y, Tapper WJ, et al. Recurrent activating STAT5B N642H mutation in myeloid neoplasms with eosinophilia. Leukemia. 2019;33(2):415–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Araujo ED, Erdogan F, Neubauer HA, et al. Structural and functional consequences of the STAT5B(N642H) driver mutation. Nat Commun. 2019;10(1):2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rajala HL, Eldfors S, Kuusanmaki H, et al. Discovery of somatic STAT5b mutations in large granular lymphocytic leukemia. Blood. 2013;121(22):4541–4550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andersson EI, Tanahashi T, Sekiguchi N, et al. High incidence of activating STAT5B mutations in CD4-positive T-cell large granular lymphocyte leukemia. Blood. 2016;128(20):2465–2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kucuk C, Jiang B, Hu X, et al. Activating mutations of STAT5B and STAT3 in lymphomas derived from gammadelta-T or NK cells. Nat Commun. 2015;6:6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kavesh M, Mohebnasab M, Angel MR, et al. Distinguishing STAT3/STAT5B-mutated large granular lymphocyte leukemia from myeloid neoplasms by genetic profiling. Blood Adv. 2023;7(1):40–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sreedharanunni S, Jamwal M, Balakrishnan A, et al. Chronic eosinophilic leukemia with recurrent STAT5B N642H mutation-An entity with features of myelodysplastic syndrome/ myeloproliferative neoplasm overlap. Leuk Res. 2022;112:106753. [DOI] [PubMed] [Google Scholar]
- 13.Qu S, Jia Y, Wang H, et al. STAT3 and STAT5B mutations have unique distribution in T-cell large granular lymphocyte proliferations and advanced myeloid neoplasms. Leuk Lymphoma. 2021;62(6):1506–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lamy T, Moignet A, Loughran TP Jr. LGL leukemia: from pathogenesis to treatment. Blood. 2017;129(9):1082–1094. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data of this study are available on request from the corresponding author, C.Z.