Abstract
Feline leukemia virus subgroup B (FeLV-B) and gibbon ape leukemia virus (GALV) utilize the human protein Pit1 but not the related protein, Pit2, as receptor. A stretch of 9 amino acids, named region A, was identified in the putative fourth extracellular loop of Pit1 (residues 550 through 558) as critical for FeLV-B and GALV receptor function. However, the presence of Pit1 region A did not confer receptor function for FeLV-B upon Pit2, while it did so for GALV. We have here shown that the presence of two Pit1-specific loop 4 residues (tyrosine 546 and valine 548) in addition to Pit1 region A is sufficient to make Pit2 an efficient FeLV-B receptor; that is, a stretch of 13 amino acids encompassing all loop 4 amino acid differences between Pit1 and Pit2 comprises a C-terminal determinant for FeLV-B receptor function. Thus, the same limited receptor region is sufficient to confer receptor function for both viruses upon Pit2.
Feline leukemia virus subgroup B (FeLV-B), Gibbon ape leukemia virus (GALV), Amphotropic murine leukemia virus (A-MuLV), and 10A1 MuLV all belong to the genus of mammalian type C retroviruses. Their cellular receptors have been cloned from various species including humans, hamsters, cats, rats, and mice (7, 8, 16, 17, 19, 24, 29, 30, 32, 33). The human receptor for FeLV-B and GALV is Pit1 (formerly GLVR1) (19, 28), and the human receptor for A-MuLV is the related protein (62% amino acid identity) Pit2 (formerly GLVR2) (30). Pit1 is not an efficient A-MuLV receptor, and similarly, Pit2 does not support FeLV-B or GALV entry, while 10A1 can efficiently use both Pit1 and Pit2 as receptors (17, 22).
Based on hydropathy plots and the observation that the Pit genes seem to have arisen by a gene duplication, Pit1 and Pit2 have been predicted to be multipass transmembrane proteins with 10 transmembrane regions, 5 extracellular loops, and a large intracellular hydrophilic domain (3, 7, 30). Their cellular function is sodium-dependent phosphate transport (9, 20, 31).
Within the putative fourth extracellular loop, a stretch of 9 amino acids (Pit1 positions 550 through 558), named region A, shows considerable amino acid variation among Pit1, Pit2, and their homologs from other species. Several studies have shown that amino acid residues in region A are critical for both FeLV-B and GALV receptor function (5, 8, 17, 22, 23, 25–27). For GALV, the presence of human Pit1 region A conferred receptor function upon MusPit1, while it was abolished in the reciprocal chimera (7, 8). These chimeras exhibited the same receptor pattern for FeLV-B as observed for GALV, demonstrating a similar critical role of region A in FeLV-B entry (27). Moreover, substitution of Pit1 region A for that of Pit2 abolished receptor function for both viruses (22). FeLV-B and GALV do, however, not entirely share the same receptor requirements, in that they exhibit different requirements for region A sequences (27) and for sequences outside region A, e.g., Pit1 region A conferred GALV but not FeLV-B receptor function upon Pit2 (22). Interestingly, the presence of the N-terminal two-thirds (encompassing loops 1, 2, and 3 and the large intracellular domain) of Pit1 in addition to Pit1 region A resulted in a functional FeLV-B receptor, and a chimera in which the entire C-terminal third (encompassing loops 4 and 5) was Pit1 derived also supported FeLV-B entry (22). Similar results were obtained by Tailor and Kabat upon testing comparable Pit1-RatPit2 chimeras for FeLV-B receptor function (26). Thus, N- or C-terminal Pit1 sequences, in addition to region A, are necessary to confer FeLV-B receptor function upon Pit2 (22); however, the exact nature of these sequences is not known.
For A-MuLV, an N-terminal determinant was recently mapped to loop 2 (11, 12); moreover, loop 4 has also been shown to play a role in A-MuLV entry (12, 13, 17, 22). Indeed, results obtained with chimeras between Pit1 and a related protein from Neurospora crassa, Pho-4, suggest that loop 2 and loop 4 sequences interdependently specify A-MuLV entry (12). For 10A1, a role of both loop 2 and region A sequences in entry has also been indicated (13). Recently, using Pit1-RatPit2 chimeras, Tailor and Kabat investigated the receptor function of vector pseudotypes carrying FeLV-B–A-MuLV SU chimeric proteins (26). Based on the results obtained, the authors suggested that wild-type FeLV-B SU proteins interact specifically with Pit1 loop 2 and Pit1 loops 4 and 5 (26).
By making chimeras between Pit1 and Pit2 and testing these for FeLV-B receptor function, we investigated whether the C-terminal FeLV-B receptor determinant(s) in loops 4 and 5 could be narrowed down and whether sequences in Pit1 loop 2 constitute the N-terminal FeLV-B receptor determinant.
Receptor function of the chimeras shown in Tables 1 and 2 was tested by a transient-transfection-infection assay essentially as described elsewhere (22). The constructs pOJ74, pOJ75, pOJ80, and pOJ102 have been described previously (22); the remaining constructs were made by site-directed mutagenesis using the Altered Sites II kit (Promega) (10); all constructs are based on the expression vector pcDNA1ArtkpA (30). To verify the presence of chimeric protein on the cell surface, receptor functions for GALV and—for selected constructs—for 10A1 were tested. GALV (SEATO) pseudotypes (5 × 105 CFU/ml) were produced by PG13GBN cells (14, 15). 10A1 pseudotypes (106 CFU/ml) were obtained by infection of an NIH 3T3 cell clone harboring the G1BgSvN vector (reference 14 and unpublished data) with viruses derived from the plasmid pRR151, which encodes Moloney Gag-Pol and 10A1 Env (21). All pseudotypes were titrated on D17 cells (22). FeLV-B receptor function was determined on Mus dunni tail fibroblasts (MDTF) (ATCC CRL-2017) using two rounds of exposure to FeLV-B pseudotypes with a titer of 6 × 102 CFU/ml. The FeLV-B pseudotypes used carried the LacZ-encoding vector LNPOZ (1) and were obtained by infecting a D17 cell clone harboring LNPOZ (22) with viruses derived from feline 355-511 cells transfected with a pBR322 clone of the λHF60 insert of FeLV-B (Gardner-Arnstein [G/A]) (4, 6, 18); FeLV-B(G/A) was a kind gift from Joyce Dunn.
TABLE 1.
Permissivity for infection in cells transiently expressing Pit1, Pit2, or chimerasa
| Plasmidb | Representationc | Permissivity (%)d for infection with:
|
||
|---|---|---|---|---|
 |
FeLV-B | GALV | 10A1e | |
| pOJ75 | 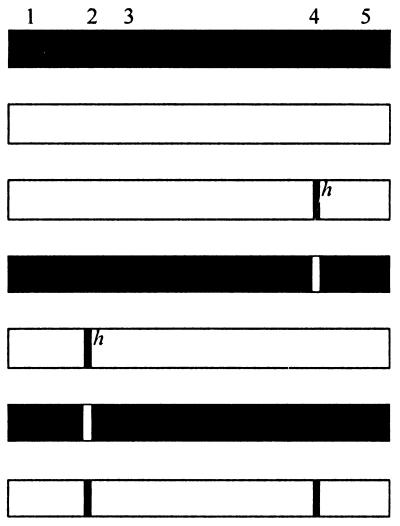 |
100 ± 11 | 100 ± 49 | NDf |
| pOJ74 | <0.05g | <0.002 | 100 ± 10 | |
| pOJ80 | <0.05 | 83 ± 30 | ND | |
| pOJ102 | <0.05 | <0.002 | 24 ± 1 | |
| pA1 | <0.05 | <0.002 | 61 ± 7 | |
| pA2 | 44 ± 25 | 25 ± 2 | ND | |
| pA3 | <0.05 | 33 ± 5 | ND | |
| Empty vector | <0.05 | <0.002 | <0.02 | |
Cells were transfected with DNA of the indicated plasmids and were exposed to FeLV-B (MDTF), GALV (MDTF), or 10A1 (CHO K1 cells) pseudotypes of LacZ-encoding vectors.
pOJ75 encodes Pit1, and pOJ74 encodes Pit2.
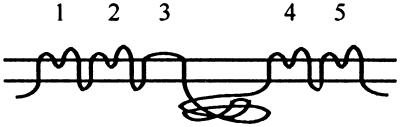
Putative topology of Pit proteins; numbers indicate extracellular loops.
The data are averages of three independent transfections ± standard deviations. The average number of blue cells per 60-mm-diameter dish transfected with the Pit1- or the Pit2-encoding plasmid was assigned a value of 100 corresponding to about 740 (FeLV-B), 30,000 (GALV), and 1,700 (10A1) blue cells. The experiment was repeated with an independent preparation of plasmid DNA, and similar results were obtained.
Tested in a separate experiment.
ND, not determined.
Based on one blue cell per three 60-mm-diameter dishes.
TABLE 2.
Permissivity for infection in MDTF transiently expressing Pit1, Pit2, or chimerasa
| Plasmidb | Representationc | Permissivity (%)d for infection with:
|
|
|---|---|---|---|
 |
FeLV-B | GALV | |
| pOJ75 | 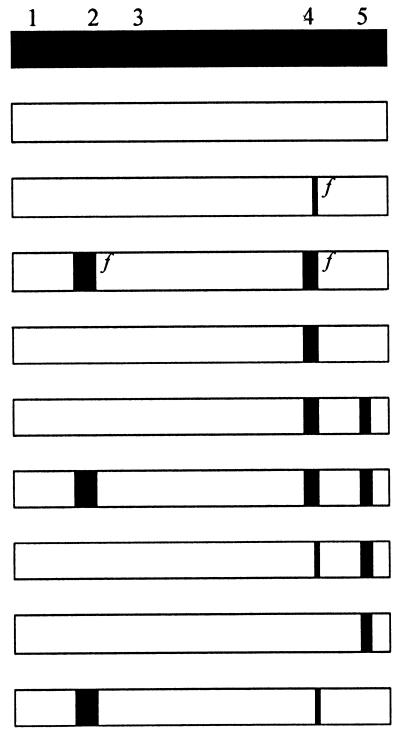 |
100 ± 23 | 100 ± 23 |
| pOJ74 | <0.02e | <0.002 | |
| pOJ80 | 0.4 ± 0.4 | 279 ± 12 | |
| pA6 | 10 ± 2 | 33 ± 9 | |
| pA7 | 57 ± 18 | 142 ± 6 | |
| pA8 | 39 ± 19 | 87 ± 38 | |
| pA9 | 4.0 ± 0.3 | 25 ± 2 | |
| pA10 | <0.02 | 101 ± 14 | |
| pA11g | <0.05 | <0.002 | |
| pA14 | <0.02 | 82 ± 9 | |
| Empty vector | <0.02 | <0.002 | |
MDTF were transfected with DNA of the indicated plasmids and were exposed to FeLV-B or GALV pseudotypes of LacZ-encoding vectors.
pOJ75 encodes Pit1, and pOJ74 encodes Pit2.
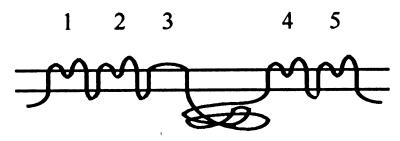
Putative topology of Pit proteins; numbers indicate extracellular loops.
The data are averages of three independent transfections ± standard deviations. The average number of blue cells per 60-mm-diameter dish transfected with the Pit1-encoding plasmid was assigned a value of 100 corresponding to about 1,400 and 26,000 blue cells for FeLV-B and GALV, respectively. The experiment was repeated with an independent preparation of plasmid DNA, and similar results were obtained.
Based on one blue cell per three 60-mm-diameter dishes.
Slim vertical bars in loop 4 represent region A (Fig. 1). Broad bars in loop 4 represent the entire loop 4 (Fig. 1). Bars in loop 2 represent the entire loop 2 (Fig. 1).
Tested in a separate experiment.
A stretch of 13 Pit1-specific amino acids in loop 4 confers FeLV-B receptor function upon Pit2.
Previous results have shown that the presence of Pit1 region A in Pit2 was not sufficient to confer FeLV-B receptor function upon Pit2, while the presence of the entire C-terminal third of Pit1 comprising loops 4 and 5 resulted in FeLV-B receptor function (22, 26). Region A (Pit1 residues 550 through 558) contains seven of the nine amino acid differences between Pit1 and Pit2 in loop 4, with the remaining two residues positioned immediately upstream of region A (Pit1 positions 546 and 548 [Fig. 1]). In accordance with previous results, we found that a Pit2-derived chimera harboring Pit1 region A did not support FeLV-B entry (pOJ80 [Tables 1 and 2]). However, the chimera A7, which differs from pOJ80 by the presence of the Pit1-specific residues, Y and V, in positions W518 and I520 (Pit2 numbering), respectively, supported FeLV-B entry at a level of approximately 60% of that obtained on cells expressing Pit1 (Table 2; Fig. 1). In line with this observation, all constructs harboring an entire Pit1 loop 4 sequence (A6, A7, A8, and A9 [Table 2]) supported FeLV-B entry, while no chimera harboring only a Pit1-specific region A sequence did so (A3, A10, A14, and pOJ80 [Tables 1 and 2]). Thus, we have mapped a C-terminal determinant of FeLV-B receptor function to a 13-amino-acid stretch in loop 4 comprising Pit1 residues 546 through 558. This observation is in agreement with Pit1 region A being sufficient to confer FeLV-B receptor function upon MusPit1 and the C-terminal third of Pit1 being sufficient to confer FeLV-B receptor function upon Pit2 and RatPit2, in that these three chimeras all harbor the Pit1-specific 13-amino-acid stretch identified here (Fig. 1) (22, 26, 27).
FIG. 1.
Alignment of putative extracellular loop sequences for Pit1, Pit2, RatPit2, and MusPit1. Numbers at the left of the sequences correspond to the positions of the first and last amino acids shown. Dots indicate amino acid identity to Pit1. Gaps introduced for alignment are indicated by dashes. The sequences underlined are region A and the loop 2 sequence mutated in the chimeras A1, A2, and A3. All residues mutated in this study are in boldface.
In agreement with the mapping of the C-terminal FeLV-B receptor determinant to loop 4, no role in receptor function could be assigned to Pit1 loop 5 residues 632, 636, and 639 (Fig. 1). Pit2 carrying Pit1-specific residues in these positions either alone (A11) or together with Pit1 region A (A10), respectively, did not support FeLV-B entry; A11 supported 10A1 entry (data not shown).
In previous experiments, the C-terminal determinant of FeLV-B receptor function was mapped to the C-terminal third of the protein comprising extracellular loops 4 and 5 (22, 26); we have here narrowed it down to 13 amino acids in loop 4 comprising Pit1 residues 546 to 558. While region A previously has been shown to be critical for FeLV-B and GALV receptor function and involved in A-MuLV—and possibly 10A1—receptor function (5, 8, 12, 13, 17, 22, 23, 25–27), the Pit1-specific residues in positions 546 and 548 have not previously been shown to be involved in receptor function for viruses utilizing Pit proteins as receptors. Whether all of the 13 amino acids in loop 4 here identified are necessary for conferring FeLV-B receptor function upon Pit2 is not known. Also, infection studies as employed here do not allow for determining whether this sequence provides a binding site for FeLV-B SU or, e.g., allows the viral SU to interact with other regions of the receptor.
Pit1-specific loop 2 and region A sequences do not confer FeLV-B receptor function upon Pit2.
Although Pit1 region A is not sufficient to confer FeLV-B receptor function upon Pit2, a chimera harboring the N-terminal two-thirds of Pit1 comprising loops 1, 2, and 3 and the large intracellular domain together with Pit1 region A was an efficient FeLV-B receptor (chimera pOJ103 in reference 22). For A-MuLV and 10A1, results have indicated that an N-terminal determinant of receptor function can be mapped to loop 2 (11–13); moreover, a model has been proposed in which FeLV-B variable region B (VRB) interacts with Pit1 loop 2 (26). In an attempt to identify the N-terminal determinant of FeLV-B receptor function in the chimera encoded by pOJ103, we have investigated the ability of Pit1 loop 2 to confer receptor function upon pOJ80 (Tables 1 and 2). The presence of the most divergent C-terminal part of Pit1 loop 2 (pA3 [Table 1]) or of the entire Pit1 loop 2 sequence (pA14 [Table 2]) did not confer receptor function upon pOJ80. Thus, since pOJ103 is an efficient FeLV-B receptor (22), we conclude that Pit1 loop 2 does not constitute the N-terminal determinant of FeLV-B receptor function; however, whether it is part of a larger N-terminal determinant cannot be determined.
A two-step model for the interaction between FeLV-B SU and Pit1 was recently put forward by Tailor and Kabat, the authors suggesting a specific and discrete interaction of FeLV-B VRA with Pit1 loops 4 and 5 and of FeLV-B VRB with Pit1 loop 2 (26). The proposed two-step model cannot explain the observation that FeLV-B can utilize chimeras harboring Pit1 loops 4 and 5 together with Pit2 or RatPit2 loops 1, 2, and 3 and the large intracellular domain (22, 26) or, as shown here, a Pit2-derived chimera harboring only Pit1 loop 4 residues in positions 518 through 530 (pA7 [Table 2]). All these chimeras are efficient FeLV-B receptors; thus, a Pit1-specific loop 2 sequence is not essential for FeLV-B receptor function in chimeras harboring the 13 amino acids in loop 4 identified here. Moreover, in line with this, a Pit1-specific loop 2 sequence was not sufficient to confer FeLV-B receptor function upon a Pit2-derived chimera already harboring Pit1 region A (chimera A14 [Table 2]).
The presence of Pit1 region A was sufficient to confer receptor function for GALV upon Pit2 (pOJ80 in Tables 1 and 2) (22), while FeLV-B receptor function required the presence of other N- or C-terminal Pit1-specific sequences (22). These observations led to the impression that FeLV-B might have somewhat different receptor requirements than those of GALV (22). However, we here found that for both FeLV-B and GALV the same limited protein region confined within a stretch of 13 amino acids in loop 4 is sufficient to confer receptor function upon Pit2. From previous results of our work and others' work and from the results presented here, it is clear that FeLV-B receptor function is only dependent on Pit1-specific N-terminal sequences when the four amino acids upstream of region A are Pit2 specific (22, 26). Interestingly, the ability of loop 4 sequences to confer GALV receptor function was also shown to be context dependent: e.g., while certain region A mutants were compatible with receptor function in a Pit1 background, they only very inefficiently supported GALV entry when placed in a Pit2 background, although the presence of a wild-type Pit1 region A sequence in Pit2 results in an efficient GALV receptor (2). Thus, FeLV-B and GALV have the common trait that sequences in loop 4 are critical for their receptor functions and that certain loop 4 sequences have specific backbone requirements in order to allow receptor function.
Acknowledgments
We thank Maribeth V. Eiden for the PG13GBN cell line and Alan Rein for the pRR151 plasmid. We furthermore thank Bente Andersen for excellent technical assistance.
This work was supported by the Karen Elise Jensen Foundation, the Danish Medical Research Council (9802349), NASTRA (540011047), the Novo Nordisk Foundation, and the Danish Biotechnology Programme.
REFERENCES
- 1.Adam M A, Ramesh N, Miller A D, Osborne W R. Internal initiation of translation in retroviral vectors carrying picornavirus 5′ nontranslated regions. J Virol. 1991;65:4985–4990. doi: 10.1128/jvi.65.9.4985-4990.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chaudry G J, Eiden M V. Mutational analysis of the proposed gibbon ape leukemia virus binding site in Pit1 suggests that other regions are important for infection. J Virol. 1997;71:8078–8081. doi: 10.1128/jvi.71.10.8078-8081.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chien M L, Foster J L, Douglas J L, Garcia J V. The amphotropic murine leukemia virus receptor gene encodes a 71-kilodalton protein that is induced by phosphate depletion. J Virol. 1997;71:4564–4570. doi: 10.1128/jvi.71.6.4564-4570.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dunn K J, Yuan C C, Blair D G. A phenotypic host range alteration determines RD114 virus restriction in feline embryonic cells. J Virol. 1993;67:4704–4711. doi: 10.1128/jvi.67.8.4704-4711.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eiden M V, Farrell K B, Wilson C A. Substitution of a single amino acid residue is sufficient to allow the human amphotropic murine leukemia virus receptor to also function as a gibbon ape leukemia virus receptor. J Virol. 1996;70:1080–1085. doi: 10.1128/jvi.70.2.1080-1085.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elder J H, Mullins J I. Nucleotide sequence of the envelope gene of Gardner-Arnstein feline leukemia virus B reveals unique sequence homologies with a murine mink cell focus-forming virus. J Virol. 1983;46:871–880. doi: 10.1128/jvi.46.3.871-880.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johann S V, Gibbons J J, O'Hara B. GLVR1, a receptor for gibbon ape leukemia virus, is homologous to a phosphate permease of Neurospora crassa and is expressed at high levels in the brain and thymus. J Virol. 1992;66:1635–1640. doi: 10.1128/jvi.66.3.1635-1640.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johann S V, van Zeijl M, Cekleniak J, O'Hara B. Definition of a domain of GLVR1 which is necessary for infection by gibbon ape leukemia virus and which is highly polymorphic between species. J Virol. 1993;67:6733–6736. doi: 10.1128/jvi.67.11.6733-6736.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kavanaugh M P, Miller D G, Zhang W, Law W, Kozak S L, Kabat D, Miller A D. Cell-surface receptors for gibbon ape leukemia virus and amphotropic murine retrovirus are inducible sodium-dependent phosphate symporters. Proc Natl Acad Sci USA. 1994;91:7071–7075. doi: 10.1073/pnas.91.15.7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kunkel T A, Roberts J D, Zakour R A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 11.Leverett B D, Farrell K B, Eiden M V, Wilson C A. Entry of amphotropic murine leukemia virus is influenced by residues in the putative second extracellular domain of its receptor, Pit2. J Virol. 1998;72:4956–4961. doi: 10.1128/jvi.72.6.4956-4961.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lundorf M D, Pedersen F S, O'Hara B, Pedersen L. Amphotropic murine leukemia virus entry is determined by specific combinations of residues from receptor loops 2 and 4. J Virol. 1999;73:3169–3175. doi: 10.1128/jvi.73.4.3169-3175.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lundorf M D, Pedersen F S, O'Hara B, Pedersen L. Single amino acid insertion in loop 4 confers amphotropic murine leukemia virus receptor function upon murine Pit1. J Virol. 1998;72:4524–4527. doi: 10.1128/jvi.72.5.4524-4527.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McLachlin J R, Mittereder N, Daucher M B, Kadan M, Eglitis M A. Factors affecting retroviral vector function and structural integrity. Virology. 1993;195:1–5. doi: 10.1006/viro.1993.1340. [DOI] [PubMed] [Google Scholar]
- 15.Miller A D, Garcia J V, von Suhr N, Lynch C M, Wilson C, Eiden M V. Construction and properties of retrovirus packaging cells based on gibbon ape leukemia virus. J Virol. 1991;65:2220–2224. doi: 10.1128/jvi.65.5.2220-2224.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller D G, Edwards R H, Miller A D. Cloning of the cellular receptor for amphotropic murine retroviruses reveals homology to that for gibbon ape leukemia virus. Proc Natl Acad Sci USA. 1994;91:78–82. doi: 10.1073/pnas.91.1.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller D G, Miller A D. A family of retroviruses that utilize related phosphate transporters for cell entry. J Virol. 1994;68:8270–8276. doi: 10.1128/jvi.68.12.8270-8276.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mullins J I, Casey J W, Nicolson M O, Burck K B, Davidson N. Sequence arrangement and biological activity of cloned feline leukemia virus proviruses from a virus-productive human cell line. J Virol. 1981;38:688–703. doi: 10.1128/jvi.38.2.688-703.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O'Hara B, Johann S V, Klinger H P, Blair D G, Rubinson H, Dunn K J, Sass P, Vitek S M, Robins T. Characterization of a human gene conferring sensitivity to infection by gibbon ape leukemia virus. Cell Growth Differ. 1990;1:119–127. [PubMed] [Google Scholar]
- 20.Olah Z, Lehel C, Anderson W B, Eiden M V, Wilson C A. The cellular receptor for gibbon ape leukemia virus is a novel high-affinity sodium-dependent phosphate transporter. J Biol Chem. 1994;269:25426–25431. [PubMed] [Google Scholar]
- 21.Ott D, Rein A. Basis for receptor specificity of nonecotropic murine leukemia virus surface glycoprotein gp70SU. J Virol. 1992;66:4632–4638. doi: 10.1128/jvi.66.8.4632-4638.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pedersen L, Johann S V, van Zeijl M, Pedersen F S, O'Hara B. Chimeras of receptors for gibbon ape leukemia virus/feline leukemia virus B and amphotropic murine leukemia virus reveal different modes of receptor recognition by retrovirus. J Virol. 1995;69:2401–2405. doi: 10.1128/jvi.69.4.2401-2405.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pedersen L, van Zeijl M, Johann S V, O'Hara B. Fungal phosphate transporter serves as a receptor backbone for gibbon ape leukemia virus. J Virol. 1997;71:7619–7622. doi: 10.1128/jvi.71.10.7619-7622.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rudra-Ganguly N, Ghosh A K, Roy-Burman P. Retrovirus receptor PiT-1 of the Felis catus. Biochim Biophys Acta. 1998;1443:407–413. doi: 10.1016/s0167-4781(98)00241-3. [DOI] [PubMed] [Google Scholar]
- 25.Schneiderman R D, Farrell K B, Wilson C A, Eiden M V. The Japanese feral mouse Pit1 and Pit2 homologs lack an acidic residue at position 550 but still function as gibbon ape leukemia virus receptors: implications for virus binding motif. J Virol. 1996;70:6982–6986. doi: 10.1128/jvi.70.10.6982-6986.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tailor C S, Kabat D. Variable regions A and B in the envelope glycoproteins of feline leukemia virus subgroup B and amphotropic murine leukemia virus interact with discrete receptor domains. J Virol. 1997;71:9383–9391. doi: 10.1128/jvi.71.12.9383-9391.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tailor C S, Takeuchi Y, O'Hara B, Johann S V, Weiss R A, Collins M K. Mutation of amino acids within the gibbon ape leukemia virus (GALV) receptor differentially affects feline leukemia virus subgroup B, simian sarcoma-associated virus, and GALV infections. J Virol. 1993;67:6737–6741. doi: 10.1128/jvi.67.11.6737-6741.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takeuchi Y, Vile R G, Simpson G, O'Hara B, Collins M K, Weiss R A. Feline leukemia virus subgroup B uses the same cell surface receptor as gibbon ape leukemia virus. J Virol. 1992;66:1219–1222. doi: 10.1128/jvi.66.2.1219-1222.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tatsumi S, Segawa H, Morita K, Haga H, Kouda T, Yamamoto H, Inoue Y, Nii T, Katai K, Taketani Y, Miyamoto K I, Takeda E. Molecular cloning and hormonal regulation of PiT-1, a sodium-dependent phosphate cotransporter from rat parathyroid glands. Endocrinology. 1998;139:1692–1699. doi: 10.1210/endo.139.4.5925. [DOI] [PubMed] [Google Scholar]
- 30.van Zeijl M, Johann S V, Closs E, Cunningham J, Eddy R, Shows T B, O'Hara B. A human amphotropic retrovirus receptor is a second member of the gibbon ape leukemia virus receptor family. Proc Natl Acad Sci USA. 1994;91:1168–1172. doi: 10.1073/pnas.91.3.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilson C A, Eiden M V, Anderson W B, Lehel C, Olah Z. The dual-function hamster receptor for amphotropic murine leukemia virus (MuLV), 10A1 MuLV, and gibbon ape leukemia virus is a phosphate symporter. J Virol. 1995;69:534–537. doi: 10.1128/jvi.69.1.534-537.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilson C A, Farrell K B, Eiden M V. Comparison of cDNAs encoding the gibbon ape leukaemia virus receptor from susceptible and non-susceptible murine cells. J Gen Virol. 1994;75:1901–1908. doi: 10.1099/0022-1317-75-8-1901. [DOI] [PubMed] [Google Scholar]
- 33.Wilson C A, Farrell K B, Eiden M V. Properties of a unique form of the murine amphotropic leukemia virus receptor expressed on hamster cells. J Virol. 1994;68:7697–7703. doi: 10.1128/jvi.68.12.7697-7703.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]



