Abstract
After the polyprotein precursor of retroviral envelope proteins is proteolytically cleaved, the surface (SU) and transmembrane (TM) subunits remain associated with each other by noncovalent interactions or by disulfide bonds. Disulfide linkages confer a relatively stable association between the SU and TM envelope protein subunits of Rous sarcoma virus and murine leukemia virus. In contrast, the noncovalent association between SU and TM of human immunodeficiency virus leads to significant shedding of SU from the surface of infected cells. The SU and TM proteins of bovine leukemia virus (BLV) initially were reported to be disulfide linked but later were concluded not to be, since TM is often lost during purification of SU protein. Here, we show that SU and TM of BLV do, indeed, associate through disulfide bonds, whether the envelope proteins are overexpressed in transfected cells, are produced in virus-infected cells, or are present in newly produced virions.
Retroviral envelope proteins confer infectivity on the virus. These proteins are first synthesized as a polyprotein precursor whose amino terminus is inserted through the membrane of the endoplasmic reticulum. In the lumen of the endoplasmic reticulum, the precursor protein becomes glycosylated. Protein folding and disulfide bond formation are aided by protein disulfide isomerase and other chaperone proteins (reviewed in references 8 and 10). Oligomers of precursor proteins formed within this compartment are transported to the Golgi apparatus, where carbohydrates are further processed. Cleavage of an envelope precursor protein by a cellular dibasic endoprotease yields a surface glycoprotein (SU) that is anchored to the lipid bilayer of cellular membranes by covalent or noncovalent association with a transmembrane protein (TM). Transport to the plasma membrane of oligomers made up of cleaved envelope subunits places SU outside the cell and makes the envelope protein complex available for incorporation into the viral envelope during the budding of particles from the cell. Infection of the host cell is initiated when SU mediates binding of virions to cell surface receptors and TM induces fusion of viral and cellular membranes.
To function properly, SU and TM envelope protein subunits must remain associated with one another either through disulfide bonds linking two cysteine residues or through noncovalent interactions. The gp85-SU and gp37-TM envelope subunits of Rous sarcoma virus are covalently linked by disulfide bonds (18). After purified viral particles are lysed in sodium dodecyl sulfate (SDS), Rous sarcoma virus envelope subunits migrate together as a large complex on nonreducing SDS-polyacrylamide gels but migrate separately as discrete polypeptides on reducing gels. In contrast, the gp120-SU and gp41-TM envelope subunits of human immunodeficiency virus (HIV) associate noncovalently; the two separate on sucrose density gradients whether or not reducing agents have been used to break disulfide bonds (21). Lack of covalent association with TM means that gp120-SU is easily shed into culture medium after cleavage of the precursor protein and transport of the envelope proteins to the cell surface (9, 17, 30, 34). Substitution of amino acids other than cysteine within the N termini of SU and TM can release even more gp120-SU (13, 17), indicating that amino acids other than those directly forming disulfide bonds affect the ability of HIV SU and TM to associate.
Whether the SU and TM proteins of bovine leukemia virus (BLV) are disulfide bonded has been unclear. A 1978 review (2) stated that the two envelope subunits are linked by disulfide bonds in virions, but more recent reviews (3, 16) have said that they are not. Dietzschold et al. (7) and Rohde et al. (31) showed in 1978 that glycosylated proteins of 60 and 32 kDa were disulfide bonded when BLV virions were disrupted either with nonionic detergent or with SDS in the presence of the alkylating agent iodoacetamide. However, the two proteins shared a number of tryptic peptides (7), calling into question their identification as distinct envelope subunits. Bex et al. (1) reported in 1979 that under nonreducing conditions, a 94-kDa complex of 60- and 30-kDa glycoproteins was purified by gel filtration from virions solubilized with nonionic detergent. Uckert et al. (39) later demonstrated by two-dimensional polyacrylamide gel electrophoresis that glycoproteins of 60 and 30 kDa were linked if no reducing agent was present during isolation of viral particles. However, using virion lysates prepared in the absence of reducing agents, Schultz et al. (35) purified 60- and 30-kDa proteins as separate entities and showed that their respective amino-terminal sequences were distinct and identical to those predicted for SU and TM by the nucleotide sequences of a BLV provirus (29). Gatot et al. (12) recently stated that TM is lost during purification of SU, citing results of earlier experiments (27) in which virions were lysed with 1% Triton X-100 and the viral proteins were purified by ion-exchange chromatography. Under those conditions, SU was immunoprecipitated from the eluate without coprecipitation of TM.
Disulfide-linked SU and TM subunits were shown some years ago to be present in both murine leukemia virus (MuLV)-infected cells and virions, although only a small fraction of the mature envelope proteins was involved (19, 22, 25, 33, 40). That fraction could be increased upon addition of the thiol-reactive alkylating agent N-ethylmaleimide (NEM). The disulfide linkage between subunits has recently been shown to involve virtually all cell-associated SU and TM (20). Two cysteine motifs, CXXC in SU and CX6CC in TM, are highly conserved in avian and murine C- and D-type retroviruses, as well as in BLV and its relative human T-cell leukemia virus (24; R. Patarca and W. A. Haseltine, Letter, Nature 312:496, 1984). The cysteine motif CWLC in gp70-SU of MuLV is involved in the linkage to p15E-TM (24). Based on the disulfide bonding pattern of the bacterially produced extracellular domain of p15E-TM (11), the third cysteine of the conserved CX6CC motif of MuLV TM has been proposed to be involved in the intersubunit disulfide bond (24). Since BLV envelope proteins contain these cysteine motifs, we asked whether SU and TM of BLV are similarly linked by disulfide bonds and if so, whether their association is stabilized by blocking free sulfhydryl groups during preparation of lysates.
Linkage of SU and TM in transfected cells overexpressing BLV envelope proteins.
We initially investigated the nature of SU and TM association using protein expressed from a cloned BLV env gene. The construct pcDNA3-ENV was derived from env mRNA present in cultured peripheral blood mononuclear cells obtained from a BLV-infected sheep, and its sequence was shown to be wild type (14). An expression plasmid (pcDNA3-CAT) encoding chloramphenicol acetyltransferase (CAT) served as a negative control.
Antibodies recognizing epitopes within the BLV Env precursor protein, mature SU, and mature TM were identified by their recognition on immunoblots of glycoproteins expressed in COS-1 cells transfected with Env or CAT expression vectors. Forty-eight hours after transfection, cells were disrupted in lysis buffer (25 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1% [wt/vol] Nonidet P-40, 1% [wt/vol] sodium deoxycholate, 2 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 1 mg of ovalbumin per ml, 0.5 μg of leupeptin per ml, 2 μg of aprotinin per ml, 10 μg of pepstatin A per ml). Lysates were incubated on ice for 20 min, and then insoluble material was removed by centrifugation at 16,000 × g for 30 min at 4°C. To enrich glycosylated proteins, cleared lysates (1 ml) were gently rocked for 4 h at 4°C with 60 μl of 50% [wt/vol] Sepharose 4B conjugated to lentil lectin (Sigma). The beads were washed once with lysis buffer, the glycoproteins were eluted with an equal volume of double-strength gel sample buffer (125 mM Tris-HCl [pH 6.8], 4% [wt/vol] SDS, 20% [vol/vol] glycerol, 0.01% [wt/vol] bromophenol blue, 100 mM dithiothreitol [DTT]), and the eluates were boiled for 4 min. Following electrophoresis on an SDS–12% polyacrylamide gel, glycoproteins were transferred to polyvinylidene difluoride membranes by electrophoresis for 16 h at 15 V. Sections of the blot were probed with serum from BLV-infected sheep 410 (28), with hyperimmune rabbit serum raised against the 16 C-terminal amino acids of TM (6), or with monoclonal antibody specific for the linear D epitope (4) of SU (BLV2; Veterinary Medical Research and Development, Inc., Pullman, Wash.). Washed strips were incubated with biotinylated, species-specific anti-immunoglobulins and then washed again before being incubated with biotinylated alkaline phosphatase linked to avidin. To reveal sites of antibody binding, strips were incubated with Lumi-Phos Plus substrate (GIBCO-Bethesda Research Laboratories) for 1 min and then realigned and exposed to X-ray film.
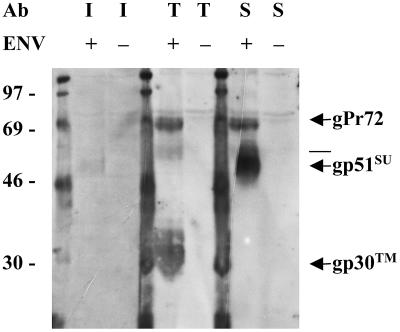
Serum from the BLV-infected sheep reacted weakly with gp51-SU (Fig. 1); the D-specific monoclonal anti-SU antibody reacted strongly with gp51-SU, as well as with the glycosylated, 72-kDa envelope precursor protein (gPr72-Env) that contains SU determinants. Anti-TM serum strongly recognized gp30-TM and gPr72-Env, both of which contain the C-terminal amino acids of TM. In addition, the anti-TM serum recognized a faint band slightly larger than gp51-SU, which could represent a dimer of TM. Others have demonstrated that multimers of HIV TM can persist in SDS-polyacrylamide gels (23).
FIG. 1.
Antigenic specificity of antibodies (Ab) recognizing BLV envelope proteins. Strips of a membrane containing glycoproteins enriched from lysates of COS-1 cells transfected with a BLV Env expression vector (+) or a CAT expression vector (−) were probed with serum from a BLV-infected sheep (I) diluted 1:250, with rabbit anti-TM antibody (T) diluted 1:4,000, or with monoclonal antibody specific for the D epitope of SU (S) diluted 1:2,000. Binding sites were revealed with luminescence created by reaction of alkaline phosphatase with substrate. An X-ray film exposed for 4 h was developed and scanned at 300 pixels/in., and the image was printed using Adobe Photoshop. The molecular masses of marker proteins are indicated to the left, and BLV proteins are identified on the right. The line between gPr72 and gp51-SU marks the mobility of a band that may represent a dimer of TM, as indicated in the text. Note that when the membrane was cut into sections, the segment to be probed with anti-TM extended partway into the Env-containing lane that was probed with anti-SU. Careful examination of the blot shows that the intermediate band recognized by anti-TM migrated more slowly than the large, heavy band representing gp51-SU.
To determine whether SU and TM could be retrieved together by antibodies specific for one subunit, we immunoprecipitated radiolabeled Env proteins from lysates of transfected cells. Cells that had been transfected with plasmid 24 h beforehand were washed and incubated for 16 h at 37°C in cysteine-free medium containing [35S]cysteine (80 μCi/ml; >1,000 Ci/mmol; Amersham). Washed cells were disrupted with cold lysis buffer. Glycoproteins were enriched by adsorption to lentil lectin and eluted by two 4-h incubations with 400 μl of 0.2 M methyl-α-d-mannopyranoside (Sigma) (26). Eluates were precleared with normal sheep serum and protein G-Sepharose or normal rabbit serum and protein A-Sepharose.
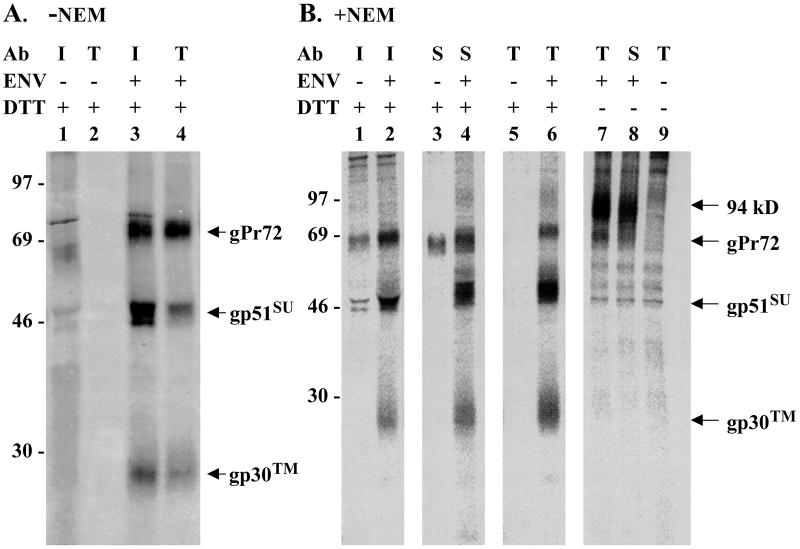
Immunoprecipitates formed using immune sheep serum contained the two BLV Env proteins having SU determinants, gp51-SU and gPr72, but also contained a protein with the mobility expected for gp30-TM (Fig. 2A, lane 3). Antibodies present in the immune sheep serum were unlikely, at the concentration used, to have cleared all envelope glycoproteins from the eluate. The remaining supernatant was therefore exposed to rabbit anti-TM serum to learn which proteins would be retrieved. The resulting immunoprecipitates included the 30-kDa TM glycoprotein and gPr72, both of which contain the C-terminal TM peptide (Fig. 2A, lane 4). Significantly, gp51-SU was also present, indicating that SU could be retrieved in association with TM. Thus, antibodies recognizing determinants in either SU or TM appeared to coprecipitate the other subunit from lysates of cells overexpressing Env protein, suggesting that a fairly stable association exists between the two subunits. However, such an association could be created by disulfide bond formation in lysates.
FIG. 2.
Disulfide linkage of SU and TM proteins in transfected cells overexpressing BLV envelope protein. COS-1 cells transfected with a BLV Env expression vector (+) or a CAT expression vector (−) were radiolabeled with [35S]cysteine, and glycoproteins concentrated from cell lysates were subjected to immunoprecipitation. (A) Cells were lysed in the absence of NEM. Glycoproteins recovered from 2 × 105 transfected cells were precipitated with serum (diluted 1:266) from a BLV-infected sheep (I), and the supernatants were then precipitated with rabbit anti-TM (T) antibody (Ab) diluted 1:1,000. Bound proteins were eluted from protein G- or A-Sepharose with double-strength SDS gel sample buffer containing 100 mM DTT. The eluates were boiled for 4 min, and proteins were separated by electrophoresis on SDS–12% polyacrylamide gels. The gel was fixed with 30% methanol–10% acetic acid, impregnated with En3Hance (DuPont), dried, and then exposed to X-ray film for 14 days. The developed film was scanned at 300 pixels/in., and the image was printed using Adobe Photoshop. (B) Before being lysed in buffer containing 10 mM NEM, cells were incubated in cold phosphate-buffered saline containing 10 mM NEM. Each lane represents glycoproteins from 105 transfected cells that were present in immunoprecipitates obtained using infected sheep serum (I) diluted 1:133, monoclonal antibody specific for the D epitope of SU (S) diluted 1:200, or anti-TM antibody (T) diluted 1:1,000. Prior to electrophoresis, precipitated proteins were heated with or without DTT, as indicated. A PhosphorImager screen was exposed to the fluor-impregnated gel for 4 days before being scanned on a Storm 860 (Molecular Dynamics). Images digitized using Image-Quant software were printed using Adobe Photoshop. The values to the left are molecular sizes in kilodaltons.
To learn whether SU and TM are disulfide linked in cells, formation of disulfide bonds has to be prevented during cell lysis, glycoprotein enrichment, and immunoprecipitation. Prior to lysis, cells were treated for 15 min with cold phosphate-buffered saline containing 10 mM NEM, which is membrane permeant. Any cysteines that are part of a disulfide linkage will not react with NEM, and once all free cysteines have reacted with NEM, they are not available to form disulfide bonds. NEM was also included at 10 mM in the lysis buffer. Again, immune sheep serum precipitated the 72-kDa glycosylated envelope precursor protein, as well as the SU and TM proteins (Fig. 2B, lane 2). Rabbit anti-TM serum immunoprecipitated all three proteins as well (lane 6), again indicating that SU was retrieved by its association with TM or gPr72. To determine whether the converse was true, the monoclonal antibody specific for the D epitope of SU was used to precipitate envelope proteins. It, too, retrieved TM protein in addition to the two proteins containing SU antigenic determinants (Fig. 2B, lane 4). The heavy chain of the monoclonal antibody did not compress SU (lane 4) as did the abundant heavy chain in sheep serum (lane 2). Proteins precipitated by both the immune sheep serum and the SU-specific monoclonal antibody from control lysates of CAT-expressing cells included a cellular protein (lanes 1 and 3) that migrated in the same area as the uncleaved envelope precursor; this background protein augmented the apparent abundance of gPr72 in the adjoining positive samples (lanes 2 and 4). However, this cellular background protein was not present in precipitates of the peptide-specific TM antibody (lane 5). In summary, antibodies specific for determinants found in either SU or TM could retrieve the other envelope subunit under conditions preventing new disulfide bond formation during manipulation of cellular lysates, indicating that these proteins are disulfide bonded in Env-expressing cells.
If SU and TM are disulfide linked but the bonds are not reduced prior to electrophoresis, the subunits should migrate on gels as a complex with greater mass. Without DTT in the gel sample buffer, a high-molecular-weight complex of approximately 94 kDa was present when either anti-TM antibody (lane 7) or anti-SU monoclonal antibody (lane 8) was used. The gPr72 precursor band migrated just ahead of the 94-kDa complex; bands representing gp51-SU and gp30-TM were no longer present in the precipitates (lanes 7 and 8). Hence, all gp51-SU and gp30-TM molecules retrieved in the absence of a reducing agent were engaged in high-molecular-weight complexes, reinforcing the conclusion that SU and TM of BLV are linked by disulfide bonds when overexpressed in cells.
Linkage of SU and TM in BLV-producing cells and in virions.
Since these experiments were done with envelope protein that was overexpressed in the absence of other viral proteins, we asked whether SU and TM would similarly exhibit disulfide bonding in BLV-infected cells and in viral particles. BLV-negative B cells of the BL3 tumor cell line (37) and BLV-infected B cells of the BL3* line (32) were radiolabeled with [35S]cysteine for 7 h. NEM (10 mM) was then added to half of the cultures, and all were incubated at room temperature for 15 min. After cells were harvested by centrifugation at 110 × g for 4 min, the radioactive medium was saved for isolation of virions. Cell pellets were washed twice with cold phosphate-buffered saline and disrupted in 2 ml of cold lysis buffer, and the lysates were cleared. To remove residual cells, the radioactive medium was centrifuged at 3,000 × g for 5 min and then to pellet virus, the resulting supernatant was centrifuged at 47,000 × g for 20 min in the cold. Viral pellets were resuspended in 1 ml of lysis buffer (with or without NEM) and cleared.
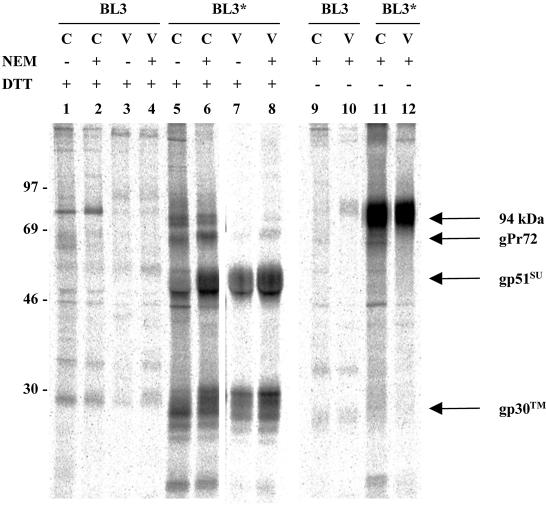
Anti-TM antibody immunoprecipitated envelope precursor protein and TM from lysates of NEM-treated BL3* cells; SU was also present (Fig. 3, lane 6). Only SU and TM were retrieved from lysates of viral pellets (lane 8). Without reduction of disulfide bonds by DTT (lanes 11 and 12), SU and TM migrated together as a high-molecular-weight 94-kDa complex and no free SU or TM was detected. To determine whether initial treatment with NEM was necessary to preserve disulfide bonds, lysates were prepared in its absence. Even without NEM, both SU and TM were present in precipitates prepared with the anti-TM antibody (lanes 5 and 7). Interestingly, the amounts of TM and SU present in precipitates of cells and viral pellets were less than those present when cells had first been treated with NEM (lanes 6 and 8), suggesting that stabilization of disulfide bonds enhances the binding of this antibody to TM. Callebaut et al. (5) have similarly reported that binding of antipeptide antibody to BLV SU is enhanced when free sulfhydryl groups are alkylated by treatment with iodoacetamide.
FIG. 3.
Recovery of SU in immunoprecipitates of TM from lysates of BLV-infected cells and viral pellets. Radiolabeled BL3 and BL3* cells (C) and virions (V) were incubated with or without 10 mM NEM for 15 min at room temperature before being lysed with or without NEM. Glycoproteins recovered from lysates of 5 × 107 cells or from virions produced by that number of cells were precipitated with anti-TM serum, and then the immune complexes were bound to protein A-Sepharose beads and eluted with double-strength sample buffer with or without DTT. The image is of a 4-day exposure of a dried, fluor-impregnated gel to a PhosphorImager screen. The values on the left are molecular sizes in kilodaltons.
These results indicate that gp51-SU and gp30-TM of BLV are linked covalently through bonds that are reduced by treatment with DTT. Furthermore, all detectable BLV envelope protein subunits that are retrieved by binding to lentil lectin appear to be disulfide bonded since no free SU or TM was detected under nonreducing conditions. Recently obtained evidence indicates that when MuLV Env is expressed in the absence of other viral proteins, all proteolytically processed SU and TM is disulfide bonded (20). When proteins are solubilized in nonionic detergent, the linkage is disrupted in a time- and temperature-dependent fashion unless disulfide exchange reactions are blocked by adding NEM or acidifying the solution. The envelope subunits of BLV are likely to dissociate under similar circumstances, which may explain why some previously obtained results indicated that these proteins are not linked by disulfide bonds. Under the conditions of the experiments reported here, treatment with NEM was not required to preserve disulfide bonds. However, in our own previous work, we did not retrieve TM when SU was immunoprecipitated (15); one difference is that those immunoprecipitations were done from cell extracts rather than from enriched glycoproteins. In the absence of NEM, some isomerization of the cysteine residues around the disulfide bond may occur. Treatment with NEM or iodoacetamide may stabilize envelope protein conformation and allow better recognition by peptide-specific antibodies, as shown here and by others for BLV envelope protein (5) and for HIV envelope protein (17).
Attachment of the maleimide group of NEM through a thioether linkage to free cysteine residues prevents them from participating in a thiol exchange reaction with cysteines in existing disulfide bonds. Pinter et al. (24) have proposed that such a reaction can occur during solubilization of MuLV envelope protein, affecting the two cysteine motifs proposed to be involved in the disulfide bond between SU and TM. They point out that the CXXC motif within SU is similar to the active-site sequence of thiol exchange enzymes and have proposed that an isomerization catalyzed by CXXC is necessary for the conformational changes required for viral entry into cells (24). All five of the cysteine residues within the two motifs are important for correct folding of MuLV envelope protein; mutating any one diminishes proteolytic processing of envelope precursor (38). HIV envelope protein has two conserved cysteine residues in the external portion of TM in a position analogous to that of the CX6CC motif of oncogenic retroviruses. Changing either of these cysteines in HIV TM also impairs proteolytic processing (36). We have found that the third cysteine of the CX6CC motif in BLV TM is essential for proteolytic processing, as substitution of arginine at this position results in almost completely unprocessed protein that does not support syncytium formation (14). Knowing that SU and TM are linked by a disulfide bond is important for understanding how these proteins interact in vivo. This linkage may play a key role in conformation changes during receptor binding and the initiation of fusion.
Acknowledgments
We thank Glen Cantor for the kind gift of antipeptide antibodies specific for BLV TM and David Sanders for discussions about intermolecular disulfide bonds in retroviruses. Debbie Grossman provided expert technical assistance.
This work was supported by Public Health Service grant CA-46374 from the National Cancer Institute and by the UC Davis Cancer Center. E.R.J. was a predoctoral trainee supported by Public Health Service grant GM-07377 from the National Institute of Medicine. Instrumentation at the NSF-funded Plant Genetics Facility, UC Davis, was used to acquire images.
REFERENCES
- 1.Bex F, Bruck C, Mammerickx M, Portetelle D, Ghysdael J, Cleuter Y, Leclercq M, Dekegel D, Burny A. Humoral antibody response to bovine leukemia virus infection in cattle and sheep. Cancer Res. 1979;39:1118–1123. [PubMed] [Google Scholar]
- 2.Burny A, Bex F, Chantrenne H, Cleuter Y, Dekegel D, Ghysdael J, Kettmann R, Leclercq M, Leunen J, Mammerickx M, Portetelle D. Bovine leukemia virus involvement in enzootic bovine leukosis. Adv Cancer Res. 1978;28:251–311. doi: 10.1016/s0065-230x(08)60649-1. [DOI] [PubMed] [Google Scholar]
- 3.Burny A, Cleuter Y, Kettmann R, Mammerickx M, Marbaix G, Portetelle D, van den Broeke A, Willems L, Thomas R. Bovine leukaemia: facts and hypotheses derived from the study of an infectious cancer. Cancer Surv. 1987;6:139–159. [PubMed] [Google Scholar]
- 4.Callebaut I, Burny A, Krchnak V, Gras-Masse H, Wathelet B, Portetelle D. Use of synthetic peptides to map sequential epitopes recognized by monoclonal antibodies on the bovine leukemia virus external glycoprotein. Virology. 1991;185:48–55. doi: 10.1016/0042-6822(91)90752-w. [DOI] [PubMed] [Google Scholar]
- 5.Callebaut I, Burny A, Portetelle D. Iodoacetamide treatment of bovine leukemia virus glycoprotein gp51 enhances the Western blotting reactivity of anti-peptide antibodies. FEBS Lett. 1991;292:148–150. doi: 10.1016/0014-5793(91)80854-v. [DOI] [PubMed] [Google Scholar]
- 6.Cantor G H, Pritchard S M, Orlik O, Splitter G A, Davis W C, Reeves R. Bovine leukemia virus transmembrane protein gp30 physically associates with the down-regulatory phosphatase SHP-1. Cell Immunol. 1999;193:117–124. doi: 10.1006/cimm.1999.1475. [DOI] [PubMed] [Google Scholar]
- 7.Dietzschold B, Kaaden O, Frenzel B, Schneider L C. Subunit and fine structure of the glycoprotein of bovine leukemia virus. Ann Rech Vet. 1978;9:613–617. [PubMed] [Google Scholar]
- 8.Doms R W, Lamb R A, Rose J K, Helenius A. Folding and assembly of viral membrane proteins. Virology. 1993;193:545–562. doi: 10.1006/viro.1993.1164. [DOI] [PubMed] [Google Scholar]
- 9.Earl P L, Moss B, Doms R W. Folding, interaction with GRP78-BiP, assembly, and transport of the human immunodeficiency virus type 1 envelope protein. J Virol. 1991;65:2047–2055. doi: 10.1128/jvi.65.4.2047-2055.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Einfeld D. Maturation and assembly of retroviral glycoproteins. Curr Top Microbiol Immunol. 1996;214:131–176. doi: 10.1007/978-3-642-80145-7_5. [DOI] [PubMed] [Google Scholar]
- 11.Fass D, Kim P S. Dissection of a retrovirus envelope protein reveals structural similarity to influenza hemagglutinin. Curr Biol. 1995;5:1377–1383. doi: 10.1016/s0960-9822(95)00275-2. [DOI] [PubMed] [Google Scholar]
- 12.Gatot J S, Callebaut I, Mornon J P, Portetelle D, Burny A, Kerkhofs P, Kettmann R, Willems L. Conservative mutations in the immunosuppressive region of the bovine leukemia virus transmembrane protein affect fusion but not infectivity in vivo. J Biol Chem. 1998;273:12870–12880. doi: 10.1074/jbc.273.21.12870. [DOI] [PubMed] [Google Scholar]
- 13.Helseth E, Olshevsky U, Furman C, Sodroski J. Human immunodeficiency virus type 1 gp120 envelope glycoprotein regions important for association with the gp41 transmembrane glycoprotein. J Virol. 1991;65:2119–2123. doi: 10.1128/jvi.65.4.2119-2123.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnston E R. Ph.D. thesis. University of California, Davis; 1999. [Google Scholar]
- 15.Johnston E R, Powers M A, Kidd L C, Radke K. Peripheral blood mononuclear cells from sheep infected with a variant of bovine leukemia virus synthesize envelope glycoproteins but fail to induce syncytia in culture. J Virol. 1996;70:6296–6303. doi: 10.1128/jvi.70.9.6296-6303.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kettmann R, Burny A, Callebaut I, Droogmans L, Mammerickx M, Willems L, Portetelle D. Bovine leukemia virus. In: Levy J A, editor. The retroviridae. Vol. 3. New York, N.Y: Plenum Press; 1994. pp. 39–81. [Google Scholar]
- 17.Kowalski M, Potz J, Basiripour L, Dorfman T, Goh W C, Terwilliger E, Dayton A, Rosen C, Haseltine W, Sodroski J. Functional regions of the envelope glycoprotein of human immunodeficiency virus type 1. Science. 1987;237:1351–1355. doi: 10.1126/science.3629244. [DOI] [PubMed] [Google Scholar]
- 18.Leamnson R N, Halpern M S. Subunit structure of the glycoprotein complex of avian tumor virus. J Virol. 1976;18:956–968. doi: 10.1128/jvi.18.3.956-968.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leamnson R N, Shander M H M, Halpern M S. A structural protein complex in Moloney leukemia virus. Virology. 1977;76:437–439. doi: 10.1016/0042-6822(77)90318-x. [DOI] [PubMed] [Google Scholar]
- 20.Opstelten D E, Wallin M, Garoff H. Moloney murine leukemia virus envelope protein subunits, gp70 and Pr15E, form a stable disulfide-linked complex. J Virol. 1998;72:6537–6545. doi: 10.1128/jvi.72.8.6537-6545.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Owens R J, Compans R W. The human immunodeficiency virus type 1 envelope glycoprotein precursor acquires aberrant intermolecular disulfide bonds that may prevent normal proteolytic processing. Virology. 1990;179:827–833. doi: 10.1016/0042-6822(90)90151-g. [DOI] [PubMed] [Google Scholar]
- 22.Pinter A, Fleissner E. The presence of disulfide-linked gp70-p15(E) complexes in AKR murine leukemia virus. Virology. 1977;83:417–422. doi: 10.1016/0042-6822(77)90187-8. [DOI] [PubMed] [Google Scholar]
- 23.Pinter A, Honnen W J, Tilley S A, Bona C, Zaghouani H, Gorny M K, Zolla-Pazner S. Oligomeric structure of gp41, the transmembrane protein of human immunodeficiency virus type 1. J Virol. 1989;63:2674–2679. doi: 10.1128/jvi.63.6.2674-2679.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pinter A, Kopelman R, Li Z, Kayman S C, Sanders D A. Localization of the labile disulfide bond between SU and TM of the murine leukemia virus envelope protein complex to a highly conserved CWLC motif in SU that resembles the active-site sequence of thiol-disulfide exchange enzymes. J Virol. 1997;71:8073–8077. doi: 10.1128/jvi.71.10.8073-8077.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pinter A, Lieman-Hurwitz J, Fleissner E. The nature of the association between the murine leukemia virus envelope proteins. Virology. 1978;91:345–351. doi: 10.1016/0042-6822(78)90382-3. [DOI] [PubMed] [Google Scholar]
- 26.Pique C, Tursz T, Dokhelar M C. Mutations introduced along the HTLV-1 envelope gene result in a non-functional protein: a basis for envelope conservation? EMBO J. 1990;9:4243–4248. doi: 10.1002/j.1460-2075.1990.tb07872.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Portetelle D, Bruck C, Mammerickx M, Burny A. In animals infected by bovine leukemia virus (BLV) antibodies to envelope glycoprotein gp51 are directed against the carbohydrate moiety. Virology. 1980;105:223–233. doi: 10.1016/0042-6822(80)90169-5. [DOI] [PubMed] [Google Scholar]
- 28.Radke K, Sigala T J, Grossman D. Transcription of bovine leukemia virus in peripheral blood cells obtained during early infection in vivo. Microb Pathog. 1992;12:319–331. doi: 10.1016/0882-4010(92)90095-6. [DOI] [PubMed] [Google Scholar]
- 29.Rice N R, Stephens R M, Couez D, Deschamps J, Kettmann R, Burny A, Gilden R V. The nucleotide sequence of the env gene and post-env region of bovine leukemia virus. Virology. 1984;138:82–93. doi: 10.1016/0042-6822(84)90149-1. [DOI] [PubMed] [Google Scholar]
- 30.Robey W G, Arthur L O, Matthews T J, Langlois A, Copeland T D, Lerche N W, Oroszlan S, Bolognesi D P, Gilden R V, Fischinger P J. Prospect for prevention of human immunodeficiency virus infection: purified 120-kDa envelope glycoprotein induces neutralizing antibody. Proc Natl Acad Sci USA. 1986;83:7023–7027. doi: 10.1073/pnas.83.18.7023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rohde W, Pauli G, Paulsen J, Harms E, Bauer H. Bovine and ovine leukemia viruses. I. Characterization of viral antigens. J Virol. 1978;26:159–164. doi: 10.1128/jvi.26.1.159-164.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Romano M J, Stewart J A, Lewin H A. Phenotypic characterization of bovine lymphoblastoid cell lines. Vet Immunol Immunopathol. 1989;23:293–307. doi: 10.1016/0165-2427(89)90142-6. [DOI] [PubMed] [Google Scholar]
- 33.Schneider J, Hunsmann G. Surface expression of murine leukemia virus structural polypeptides on host cells and the virion. Int J Cancer. 1978;22:204–213. doi: 10.1002/ijc.2910220215. [DOI] [PubMed] [Google Scholar]
- 34.Schneider J, Kaaden O, Copeland T D, Oroszlan S, Hunsmann G. Shedding and interspecies type sero-reactivity of the envelope glycopolypeptide gp120 of the human immunodeficiency virus. J Gen Virol. 1986;67:2533–2538. doi: 10.1099/0022-1317-67-11-2533. [DOI] [PubMed] [Google Scholar]
- 35.Schultz A, Copeland T, Oroszlan S. The envelope proteins of bovine leukemia virus: purification and sequence analysis. Virology. 1984;135:417–427. doi: 10.1016/0042-6822(84)90197-1. [DOI] [PubMed] [Google Scholar]
- 36.Syu W, Lee W R, Du B, Yu Q C, Essex M, Lee T H. Role of conserved gp41 cysteine residues in the processing of human immunodeficiency virus envelope precursor and viral infectivity. J Virol. 1991;65:6349–6352. doi: 10.1128/jvi.65.11.6349-6352.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Theilen G H, Miller J M, Higgins J, Ruppanner R N, Garrett W. Vaccination against bovine leukaemia virus infection. In: Straub O C, editor. Fourth International Symposium on Bovine Leukosis. The Hague, The Netherlands: Martinus Nijhoff; 1982. pp. 547–560. [Google Scholar]
- 38.Thomas A, Roth M J. Analysis of cysteine mutations on the transmembrane protein of Moloney murine leukemia virus. Virology. 1995;211:285–289. doi: 10.1006/viro.1995.1402. [DOI] [PubMed] [Google Scholar]
- 39.Uckert W, Westermann P, Wunderlich V. Nearest neighbor relationships of major structural proteins within bovine leukemia virus particles. Virology. 1982;121:240–250. doi: 10.1016/0042-6822(82)90164-7. [DOI] [PubMed] [Google Scholar]
- 40.Witte O N, Tsukamoto-Adey A, Weissman I L. Cellular maturation of oncornavirus glycoproteins: topological arrangement of precursor and product forms in cellular membranes. Virology. 1977;76:539–553. doi: 10.1016/0042-6822(77)90236-7. [DOI] [PubMed] [Google Scholar]