Summary:
Embryonic development is remarkably robust, but temperature stress can degrade its ability to generate animals with invariant anatomy. Phenotypes associated with environmental stress suggest that some cell types are more sensitive to stress than others, but the basis of this sensitivity is unknown. Here, we characterize hundreds of individual zebrafish embryos under temperature stress using whole-animal single cell RNA-seq to identify cell types and molecular programs driving phenotypic variability. We find that temperature perturbs the normal proportions and gene expression programs of numerous cell types and also introduces asynchrony in developmental timing. The notochord is particularly sensitive to temperature, which we map to a specialized cell type, sheath cells. These cells accumulate misfolded protein at elevated temperature, leading to a cascading structural failure of the notochord and anatomic defects. Our study demonstrates that whole-animal scRNA-seq can identify mechanisms for developmental robustness and pinpoint cell types that constitute key failure points.
Introduction
Temperature and developmental rate are commonly correlated across animal development 1–3. The acceleration of development at higher temperatures has been attributed to increased metabolic rate and protein synthesis 4. Within a species-specific tolerated range, embryos raised in elevated temperatures are phenotypically normal, suggesting that the developmental program is synchronously accelerated. While the mechanisms underlying this synchrony are not fully understood, recent studies have emphasized the role of proteostasis—the maintenance of proper protein folding, synthesis, and degradation—in determining developmental rate differences across species 5–7. Mechanisms regulating cellular proteostasis are also key for understanding the relationship between environmental stress and phenotype 8. Regulation of protein folding and synthesis is sensitive to diverse environmental signals 9 and misregulation of these processes has phenotypic consequences 10–13. Because the burden of proteostasis varies considerably across cell types 14,15, we sought to use transcriptional signatures of proteostasis to investigate how each lineage is able to buffer the effects of temperature stress. Perturbation of proteostasis during early development introduces phenotypic variability 13,16,17, but these phenotypes are non-random, suggesting that some cell types or developmental processes may be more sensitive to stress than others.
Zebrafish embryos are naturally exposed to temperature fluctuation and display temperature-induced phenotypes and alterations in developmental rate. Furthermore, temperature influences body size scaling, presumably through coordinated effects on the program of development,18 and also has consequences for aging. While the extended lifespan of zebrafish makes aging studies challenging, fast-growing annual killifish show reduced lifespan when raised at elevated temperature 19. Wild populations of Danio rerio have a natural range spanning from 26°C to as high as 38°C 20. In lab populations of zebrafish, embryos cannot survive chronic exposure to 36°C and beyond, while they display an increasing number of phenotypic abnormalities from 32°C and beyond 1. This phenomenon is not limited to ectotherms; thermal limits are evident in mammals and other vertebrates, with species-specific differences defining the critical temperature thresholds beyond which developmental phenotypes arise 21,22.
We sought to understand the cellular basis for stress-induced developmental phenotypes and, specifically, whether cell types respond differently to temperature. Single cell genomic techniques can resolve heterogeneity in developmental timing and in levels of molecular processes promoting proteostasis across cell types, but these techniques are limited in their ability to capture variability in the form of biological replicates due to the standard practice of pooling embryos. To overcome this, we use single-cell combinatorial indexing combined with DNA oligo hashing to capture transcriptional states and cell type abundances for hundreds of individual zebrafish embryos at multiple temperatures and time points. We leverage this large number of replicates to make statistical inferences about the source of environmentally-induced phenotypes, as well as to identify molecular sources and cell type-specific contributions to the loss of developmental robustness. From these analyses, we find that temperature affects both developmental rate and synchrony among cell types, and that the unfolded protein response is required for temperature-induced acceleration. Furthermore, temperature stress alters cell type composition, leaving a lasting imprint on the embryo that cannot be explained by differences in developmental stage. Finally, we show that perturbed proteostasis reveals cell type-specific temperature sensitivity in the notochord. Taken together, we identify cell type-specific mechanisms of developmental robustness by integrating individual-to-individual variation and accounting for differential responses to temperature across cell types.
Results
Multiplexed single-cell RNA-seq profiles individual embryos developing under temperature stress
To measure effects of temperature on developmental robustness, we raised zebrafish embryos over a range of increasingly stressful temperatures below the threshold for heat shock 23. In addition to the standard temperature of 28°C, we raised embryos in the standard condition as well as at two elevated temperatures, 32°C and 34°C. At elevated temperatures, embryos develop at a faster rate 24, with a substantial fraction of individuals showing axial defects of varying severity, along with other previously documented phenotypes (Fig. 1A) 1,25–27. Because we can profile individuals, we included embryos raised at elevated temperatures that were either phenotypically normal or had severe, temperature-induced phenotypes. To identify individual embryos, we used nuclear “hashing” wherein polyadenylated DNA oligos with unique barcodes for each embryo are added and fixed along with liberated nuclei after whole-embryo dissociation (Fig. 1B) 28. To account for increased variability in elevated temperature samples, additional embryos were sampled in these conditions (16 control, 40 32°C and 40 34°C embryos from each timepoint). Treated and control embryos from three time points (24, 30, 36 hours post-fertilization [hpf]) were dissociated and hashed individually and subsequently processed with a single-cell combinatorial indexing protocol to isolate single nuclei with quality comparable to previous sci-RNA-seq experiments (Fig. S1A, S1B) for transcriptome profiling 29. We recovered 5–10% of the cells (Fig. S1A–C) from each individual embryo profiled (assuming 20,000–40,000 cells per embryo), which is sufficient to capture all major cell types for each individual. Cell types were represented more evenly (Gini index = 0.67) than in previous whole-embryo studies (Gini index = 0.80 and 0.71 for atlases from 30 and 31, respectively), possibly owing to our use of single-nucleus rather than whole-cell RNA-seq. (Fig. S1D). By combining the data from all 288 profiled individuals (403,992 total cells) we built a comprehensive atlas of cell type-specific responses to temperature stress (Fig. 1C, Fig. 1C). We projected our temperature perturbation data onto our annotated zebrafish development reference dataset produced from analyzing 1.2 million cells from 18 hpf to 96 hpf (see related manuscript, 32) and identified 85 distinct cell types at these stages. Consistent with the robust progression of development despite elevated temperature, all expected cell types were represented in embryos raised at 32°C and 34°C, and we observed no specific activation of heat shock markers hsp90aa1.1 or hsp70l (Fig. S1E–F), nor global activation of the heat shock response (Fig. S1G), which can diminish cell type-specific expression patterns 33.
Figure 1. Effects of stress on phenotypic variability is captured via individual animal hashing of single-cell transcriptomes.
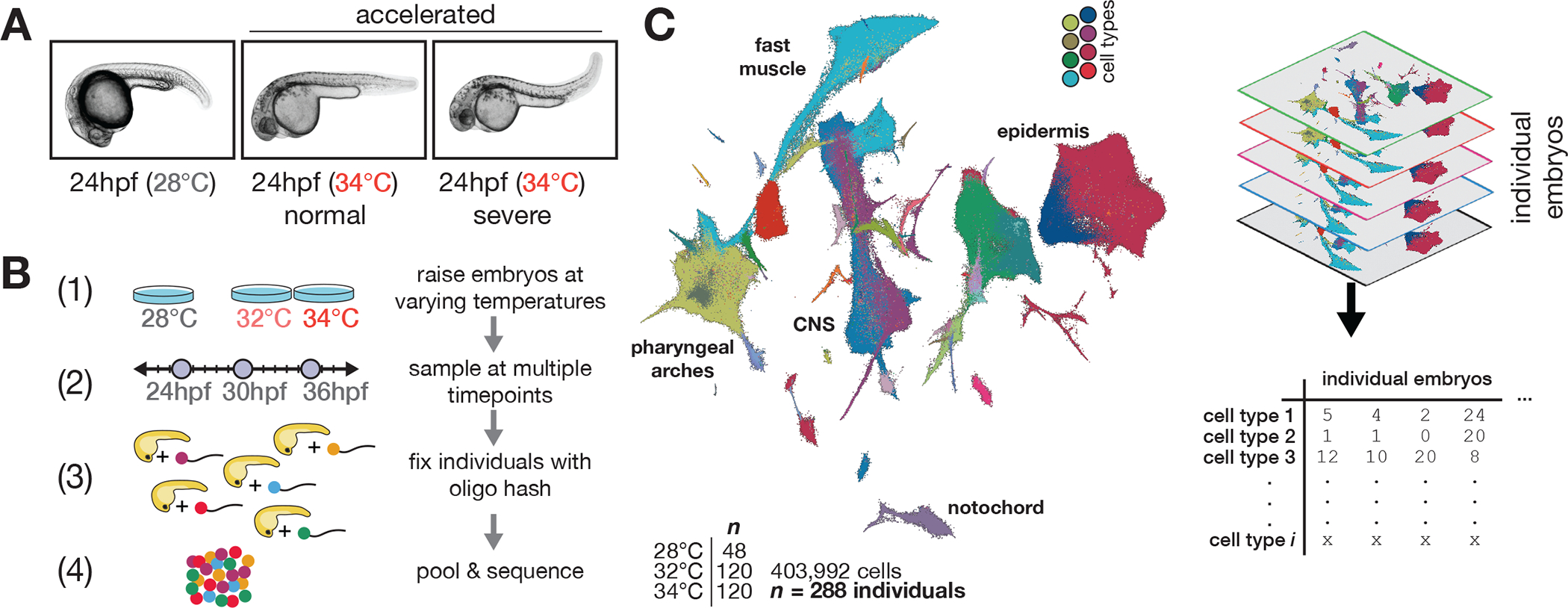
(A), Representative images of 24 hpf embryos raised at standard and elevated temperature; individual embryos with normal-looking and bent-tail phenotypes were included in the dataset. (B), Experimental workflow for temperature perturbation experiment and individual embryo hashing. (C), UMAP of temperature-perturbation dataset, projected into coordinate space of reference atlas (see related manuscript, 32). Right side shows how single cell data are transformed to generate cell composition matrices.
Variation in developmental stage can be determined from cell type composition
Because of variation in developmental progression over time and temperature, zebrafish studies categorize embryos based on a “staging series” of visible landmarks evident during development at standard temperature. For example, the proper stage-matched control for an embryo raised to 24 hpf at 34°C would be a 31 hpf embryo at 28°C 24. We therefore sought to develop a method to quantify the degree to which an embryo is accelerated directly from our single-cell data, as staging each embryo is essential for isolating the effects of temperature on cell type composition.
Using our reference dataset (see related manuscript, 32), we analyzed individual embryos in two dimensions; embryos were grouped according to similarity in cell type composition, and this grouping produced a trajectory defined primarily by sample time point (Fig. 2A, middle, landmarks labeled on left). Individual temperature-treated embryos were projected into this low dimensional space and assigned a “pseudostage” value based on their relative position in the trajectory. Because temperature treatment distorts the relationship between clock time and embryo stage, we next tested whether pseudostaging captured the expected increases in developmental rate at high temperature. We projected high temperature embryos onto the reference embryo UMAP and found that treated embryos progressed further along the pseudostage trajectory than contemporaneous controls (Fig. 2A, right). Pseudostage values were strongly correlated (R^2 = 0.82) to acceleration predicted by a proposed linear model of temperature and developmental stage (Fig. 2B, see Methods) 24. We further analyzed individual cell types whose abundance was predictive of whole-embryo pseudostage, and found that fin fold cells could be used to precisely stage the embryos (Fig S1H, S1I). Pseudostage values were also strongly correlated with whole-embryo stage values computed from nearest-neighbor average time points in individuals, rather than whole-embryo cell type composition (Fig. 2C), with the greatest differences in these metrics appearing at earlier developmental stages (Fig. S2A). Thus, whole-embryo cell type composition data captures variation in developmental stage, including that found in untreated embryos sampled at the same clock time (Fig. 2B). Staging embryos based on cell type composition is therefore sufficient to control for both biological and technical variation in developmental progression.
Figure 2. Staging embryos by cell type composition captures temperature-induced acceleration of development and allows isolation of temperature-dependent effects on cell abundance.
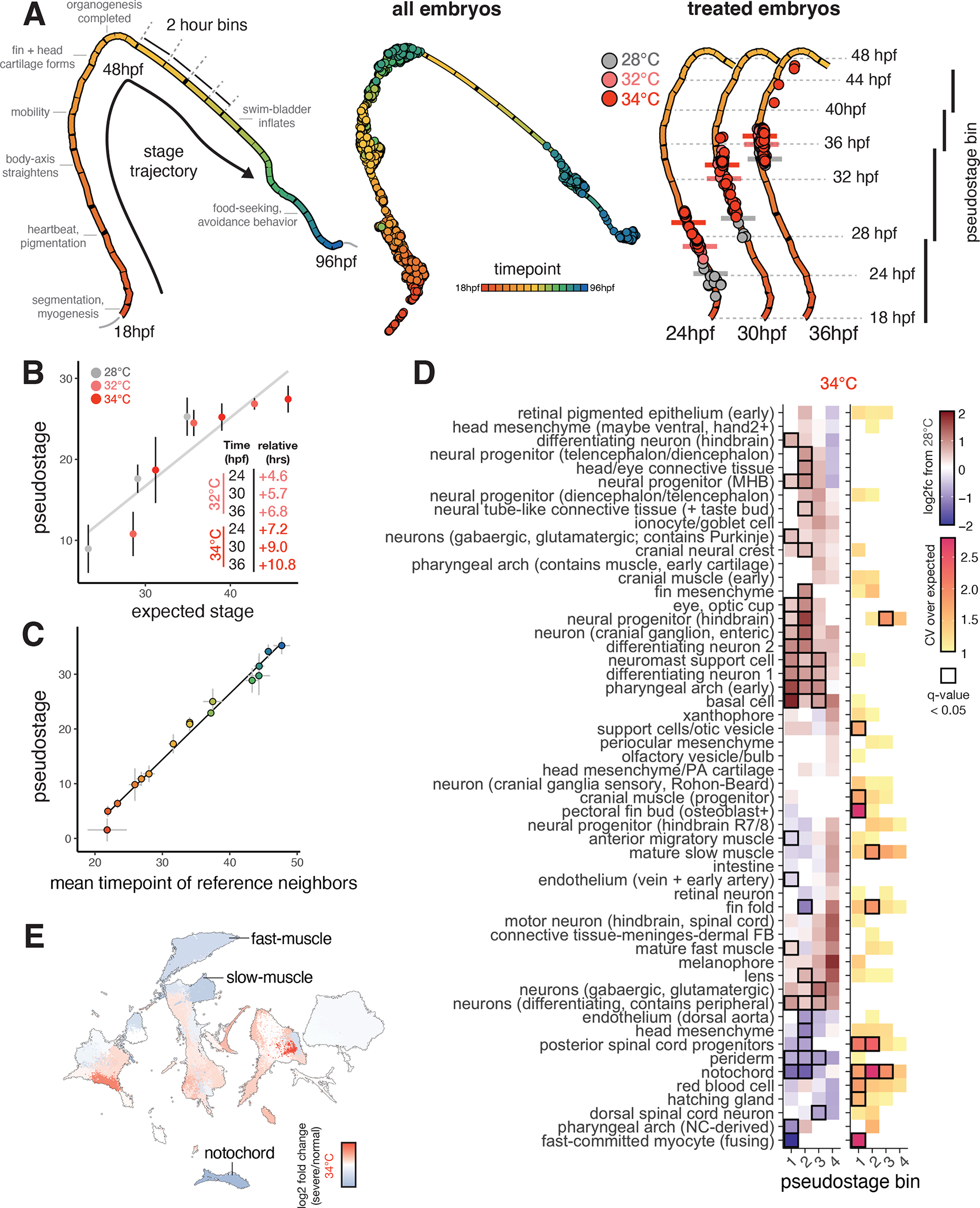
(A), UMAP projection of embryo trajectory produced using all individual embryos from the developmental reference; each segment of the principal graph represents a 2-hour window of development, with key events noted. Right panel shows how, at each time point, embryos from elevated temperature are “ahead” of their 28°C counterparts on the embryo stage trajectory (median pseduostage for each temperature indicated with horizontal bars). (B), Scatterplot showing mean pseudostage values are correlated with expectations from a linear model of temperature-induced acceleration of developmental rate; error bars represent standard deviation. Result of linear regression is shown in black. (C), Scatterplot showing mean pseudostage values for all embryos in the reference dataset compared to a nearest-neighbor label transfer in transcriptome space; error bars on both axes represent standard deviation; both cell composition and transcriptomes contain ample information on developmental stage. (D), Heatmap showing the effects of temperature on mean cell type abundance relative to untreated, stage-matched controls (left) and on variability (CV relative to controls) of cell counts (right). Significant tests (q < 0.05) from beta binomial regression are indicated with a black box. Each column represents a pseudostage bin, wherein embryos from untreated and treated samples are stage-matched. (E), UMAP projection of all cell types, colored by relative abundance change in severe (bent) individuals raised at 34°C compared to normal-looking embryos also raised at 34°C.
Temperature stress alters cell type proportions and injects variability into embryogenesis
Next, we sought to explicitly evaluate the assumption that cell types develop synchronously and in proper proportions in embryos raised at different temperatures. Specifically, we wondered whether we could detect cell type-specific sensitivity to temperature in cell abundance data, as this may contribute to the stereotyped phenotypes that arise in embryos raised at high temperature. We grouped control and treated embryos of similar developmental stage into four pseudostage bins (Fig. 2A, right). Using both absolute cell counts, and relative proportions of each cell type per embryo, we performed a regression test using the beta binomial distribution, which is well-suited for compositional data 34,35. After accounting for differences in embryo stage in the model due to the different temperatures, we identified 20–30 cell types that showed significant (q-value < 0.05) increases or decreases in abundance with temperature (Fig. 2D, left, table S1A), with a greater number of affected cell types at 34°C than 32°C. (Fig. S2B). Of these, the most strongly affected cell types were notochord, dorsal aorta, and head mesenchyme, showing large reductions in cell number. Among the decreased cell types, there were two groups with physical proximity in the anterior (pharyngeal arch and surrounding head mesenchyme) and posterior body (dorsal aorta, posterior spinal cord progenitors, notochord, and fin fold mesenchyme). Surprisingly, neural progenitor cell types showed significant increases in abundance, but this was not due to increased signatures of cell cycle activation at the sampled timepoints. Other cell types such as periocular mesenchyme, xanthophore pigment cells, and red blood cells did not show significant changes in abundance as a function of temperature, suggesting that these cell types are not as sensitive to stress (Fig. 2D). The effects of 32°C and 34°C treatments on cell abundances were correlated (R^2 = 0.69), suggesting that these sub-heat shock temperatures affected developmental processes similarly (Fig. S2C). Overall, many cell types showed consistent changes in proportion at higher temperatures, even after accounting for developmental stage, reinforcing the notion that stress fundamentally alters cell composition in embryos, rather than perturbing development of just a few lineages.
To test whether elevated temperature increased variability in cell composition globally, we compared multinomial models informed by pseudostage alone, or by pseudostage and temperature. We found 42% of the variation in whole-embryo cell composition between 24 and 36 hpf was explained by sample time point, with temperature explaining an additional 12% (Fig. S2D), confirming a global increase in anatomical variability. To isolate specific sources of variability during embryogenesis, we identified cell types whose counts showed increased variance over a control expectation (shown as relative coefficient of variation, CV). After correcting for the dependence of variance on the mean abundance of each cell type, we identified eleven cell types showing significant increases in variability at one of the elevated temperatures (Fig. 2D, examples in Fig. S2E, table S1B). However, within cell types, the largest dilations in CV occurred in a pseudostage-specific manner (Fig. 2D, right). For example, counts of posterior spinal cord progenitors showed increased variability at high temperature in earlier stages, but stabilized at later stages (Fig. 2D, inset). Across all cell types on average, relative CV values increased at higher temperatures (Fig. S2F) and peaked between pseudostages 20–25, consistent with the previous observation that some developmental stages may be more sensitive to stress (Fig. S2F, upper panel) 36.
We observed phenotypes of varying severity in embryos raised at high temperatures (severe phenotypes, Fig. 1A). We next sought to test whether altered cell composition contributed to differences in phenotypic severity. For embryos raised at 34°C, we classified the phenotype severity for each embryo (table S2), and, within a single temperature, compared cell type compositions of severe embryos to normal embryos. Embryos with more severe phenotypes showed reduced abundance of several cell types, including notochord, muscle, fin mesenchyme, pharyngeal arch (Fig. 2E, Fig. S2G), and posterior spinal cord progenitors, suggesting that variation in cell type composition contributes to overall phenotypic variation, even within a temperature. Of these, reduced levels of muscle recruitment in adult zebrafish that experience high temperature during embryonic development37 has been previously observed, and the body axis defects observed at high temperatures might be expected to affect posterior spinal cord neurons. Profiling cell type-specific sources of phenotype severity highlights the value of profiling large numbers of replicate embryos. Sensitive detection of temperature-dependent effects on cell abundances would not be possible with just a few replicate embryos, as resultant phenotypes can be obscured by individual-level phenotypic variability.
In summary, we find that some, but not all, cell types show altered abundance as a function of temperature, suggesting the response to temperature is integrated non-uniformly across cell types. This unexpected result raises the question: what is the molecular basis for these cell type-specific effects?
Temperature introduces asynchrony in developmental rate across cell types
We next explored whether the temperature-dependent changes in cell type composition were attributable to changes in cell type-specific developmental rates. While the embryo as a whole develops 4–6 hours more quickly when raised at 32°C/34°C than at the standard 28°C (Fig. 2B, inset), variation in this acceleration across cell types has not previously been measured. To determine whether cell type-specific developmental rates vary, we examined relative differences in each cell type’s transcriptional “age,” a measure derived from comparison to our developmental reference, in temperature treatments compared to controls (Fig. S3A, see Methods). Taking the mean age of all cells for each embryo captures whole-embryo acceleration similar to the pseudostage metric (Fig. S3B). For example, notochord cells in embryos raised at 32°C and 34°C showed +1.6hr and +2.2hr acceleration of transcriptional age relative to the whole-embryo expectation (Fig. S3C). Several additional cell types, including the dorsal aorta, melanophore and hypochord cells (Fig. 3A, middle), showed a similar relative advancement, and we classified these cells as highly accelerated (Fig. 3B). In contrast, basal cells, pigment progenitors, and fin fold cells were unresponsive or decelerated at high temperature (Fig. S3C). To ensure that this signal of developmental acceleration was indeed driven by core genes related to progression of each cell type, rather than a subset of temperature-responsive genes, we analyzed the number of differentially expressed genes associated with temperature after accounting for developmental stage. For most cell types (85%), the number of significantly differentially expressed genes associated with developmental stage is greater than the number associated with temperature, highlighting the importance of including developmental stage information to properly isolate the effects of an environmental perturbation from effects on developmental delay or acceleration (Fig. S3D). Furthermore, developmental acceleration is associated with changes across the transcriptome, rather than being concentrated in a small number of sensitive genes. In the notochord, which showed strong temperature-dependent acceleration, we found no apparent relationship between genes differentially expressed at high temperature and those changing significantly with developmental age, but temperature-induced genes tended to be more highly expressed than those associated with developmental age (Fig. S5E–F).
Figure 3. Temperature introduces asynchrony in developmental rate across cell types.
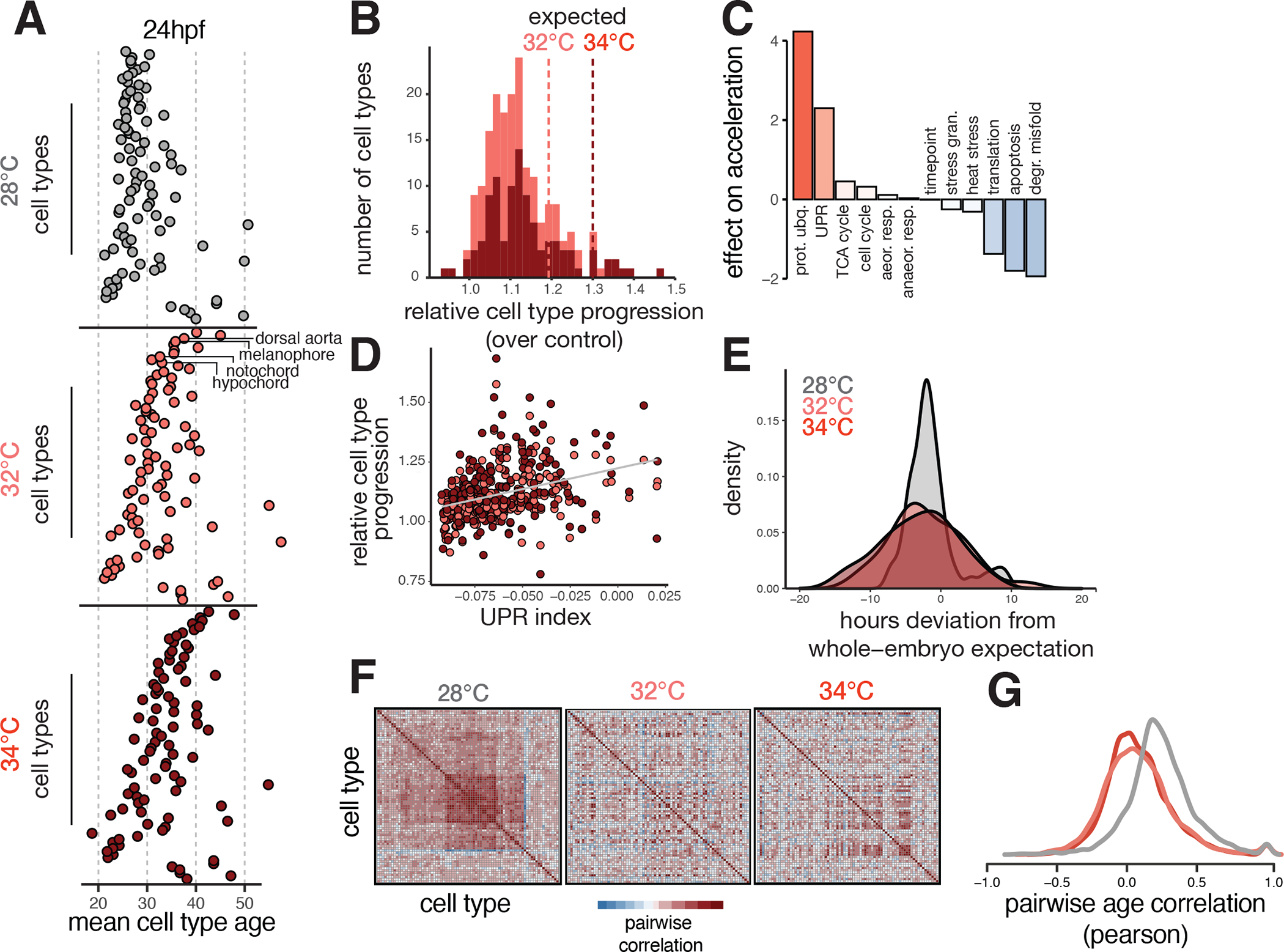
(A), Dotplots showing the transcriptional ages of all cell types at 24 hpf, faceted by temperature. Cell types on the y-axis are ordered by their relative acceleration at 34°C, highlighting the most sensitive cell types near the top, and insensitive types near the bottom. Cell type ordering is the same for all temperatures; specific examples are indicated with labels. (B), Histogram showing distributions of relative cell type progression at each temperature, with vertical lines showing the relative progression expected for the whole embryo. (C), Barplot showing the effect sizes for expression levels of several cellular processes related to metabolism, protein folding, proliferation, and stress response in an additive model predicting relative progression of the cell type at high temperature. (D), Scatterplot showing basal levels of the unfolded protein response in each cell type against its relative progression, revealing a positive trend for both temperatures. (E), Density histogram showing increased variance in developmental stage for embryos raised at elevated temperature. (F), Heatmaps showing pairwise correlation coefficients of transcriptional age for all cell types in the embryo; at 28°C, most cell type pairs are positively correlated across individual embryos, whereas this correlation structure is diminished at elevated temperatures. (G), Density histogram of pairwise correlation values at each temperature, summarizing the loss in correlation structure seen in panel (F).
Given that some cell types were more developmentally accelerated than others, we next sought to directly quantify the synchrony of development across cell types. We first assessed variation in developmental stage at the embryo level by examining pseudostage deviations at each temperature; both temperatures increased variability in developmental stage (Fig. 3E). We also found that coordinated developmental timing between cell types could be observed in the covariance structure of cell type ages across all embryos (Fig. 3F). As expected, there was a positive correlation in age between all cell type pairs in control embryos (average r = 0.24) (Fig. 3G). However, we found that the covariance structure between cell types was disrupted in both of our elevated temperature conditions; the correlation of transcriptional age between cell type pairs was reduced by three-fold on average (Fig. 3G). Furthermore, many cell type pairs that showed coordinated activation of cell cycle-related genes also showed reduced correlation in elevated temperature conditions, suggesting loss of coordinated growth (Fig. S3G). Together, these results highlight how temperature introduces a non-uniform increase in developmental rate across different cell types and disrupts the developmental synchrony between cell types. Together, these results highlight an additional source of temperature-induced variability during development: asynchrony in developmental rate emerging from cell type-specific sensitivity to temperature.
Without an obvious anatomical or lineal relationship uniting highly accelerated cell types, we wondered whether any underlying molecular processes could explain differential developmental acceleration. Differences in developmental rate under temperature stress and between species have been variously associated with metabolic rate, cell division, protein degradation, and protein synthesis. Accordingly, we generated signature scores for these processes, as well as two stress pathways, the unfolded protein response and the heat shock response, to identify those with the strongest effect in predicting relative acceleration of cell type-specific developmental rate. To test this, we modeled the developmental rate increases across cell types as a function of baseline (during normal development at standard temperature) expression of genes important for each process (Fig. 3C, inset). We verified that neither the basal or induced heat shock response was strongly correlated with temperature-dependent acceleration, consistent with a temperature treatment that challenges proteostasis but does not activate a canonical heat-shock response (Fig. S3H–I, Fig. S1E–G). Cell types expressing higher levels of genes involved in protein synthesis, apoptosis, and degradation of misfolded protein showed less temperature-dependent acceleration of development (Fig. 3C), slowing down relative to whole-embryo expectation. We validated the effect of protein synthesis on slowing developmental rate by treating embryos with low-dose cyclohexamide (CHX) and 20°C cold-treatment; delayed progression was evident in both treatments (Fig. S4A–B). Furthermore, the effects of protein synthesis inhibition on gene expression were strongly correlated with effects of cold-treatment (Fig. S4C–D). Lastly, we identified genes with opposing transcriptional responses to both 20°C and 34°C treatment, including translation elongation factor eef2b in muscle cells (Fig. S4E–F), suggesting that control of protein levels may contribute to cell type-specific temperature response. Cell types with high expression of genes involved in protein ubiquitination and the unfolded protein response (UPR) showed more temperature-dependent acceleration of development than those with lower expression of these processes. That temperature-dependent acceleration across all cell types could be partially explained by baseline levels of UPR suggests a role for ER stress in modulating developmental rate under normal conditions (Fig. 3D). Activation of UPR during normal development has been observed in both the notochord and the hatching gland 38–41 in fish, as well as during cell differentiation and growth in other models 42,43, but has not been previously linked to developmental rate.
The unfolded protein response buffers notochord-specific temperature sensitivity
To explore whether transcriptional responses to temperature, similar to changes in cell abundances and developmental age, were non-uniform across cell types, we analyzed genes with differential expression at higher temperatures. We identified several modules of genes that tended to be upregulated in response to elevated temperature (Fig. S5A, table S3A). These modules were enriched for genes related to cell cycle, chromatin organization, transcription, ion transport, and protein processing in the ER, but not the canonical heat-shock response (Fig. S5A, table S3B). In contrast to global activation of the heat-shock response 33, these temperature-responsive modules tended to be upregulated in a cell type-specific manner (Fig. S5B). For example, only blood cells showed activation of the cell cycle module, while the notochord showed concomitant activation of translation and ER stress modules (Fig. S5B, boxed). Of all the temperature-response modules, the ER stress gene cluster showed the broadest response across tissues.
The unexpected appearance of the UPR (Fig. 3C) and a gene module for translation and ER stress (Fig. S5A) among cell type-specific responses to temperature led us to explore this phenomenon more deeply across cell types. We generated signature scores for translation and the UPR in all cell types in individual embryos and found that cells tended to express high levels of either one signature or the other, consistent with the UPR’s role in attenuating translation (Fig. 4A, table S3C) 44. In contrast, we observe in the notochord a simultaneous upregulation of UPR and translation that is specific to elevated temperature conditions (Fig. 4B, Fig. S5C). This exceptional upregulation of translation in the presence of UPR is pronounced to a lesser extent in the hypochord and pectoral fin bud; nearly all other cell types showed reductions in the number of cells expressing both signatures (Fig. 4B). Notably, the notochord showed temperature-dependent effects on cell abundance, as well as a marked acceleration of developmental age (Fig. 2E, Fig. 3A, Fig. S5D). We wondered whether this exceptional upregulation of UPR and translation might be related to the notochord’s apparent sensitivity to temperature.
Figure 4. Exceptional regulation of the unfolded protein response underlies temperature sensitivity in the notochord.
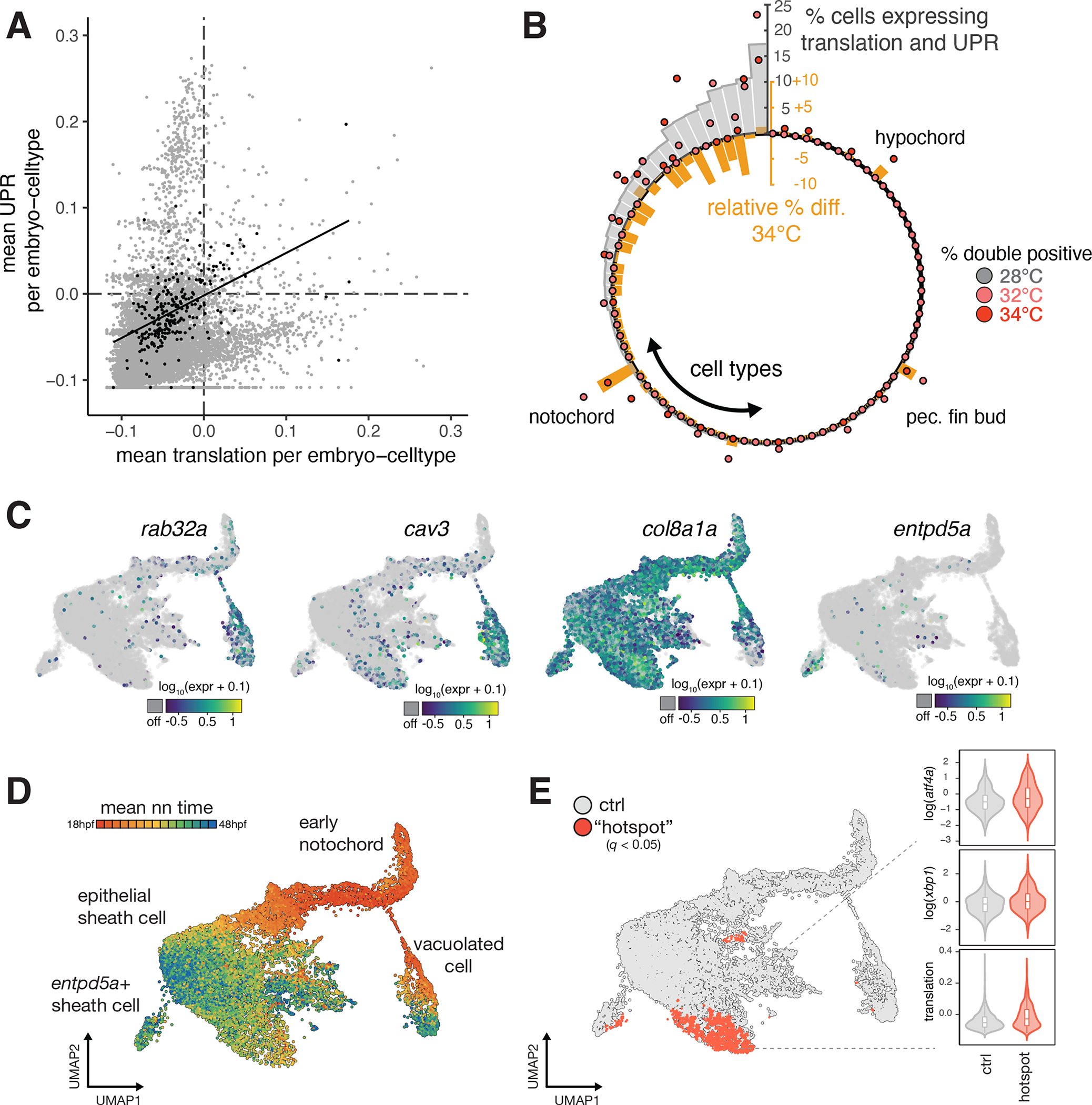
(A), Scatterplot showing levels of translation signature and UPR signature across all embryo-cell types. Consistent with known translational attenuation by UPR, these processes are generally uncorrelated across cell types. The notochord (plotted as black dots) is an exception, with a positive correlation (Pearson’s r = 0.51) between these processes, and appreciable levels of both UPR and translation in a subset of embryos. (B), Circular barplot showing occurrence of translation + UPR co-expression, displayed as % double positive in each cell type (grey bars). Overlaid dots show raw % double positive cells for embryos raised at elevated temperature, colored pink and dark red for 32°C and 34°C, respectively. Orange bars show the relative change in the fraction of double positive cells in each cell type at elevated temperatures. For the large majority of cell types, this difference is negative. The notochord, hypochord, and pectoral fin bud are the sole exceptions, where double positive cells increase in response to temperature increase. (C), Marker gene plots showing expression of genes defining notochord sub-types (rab32a and cav3 in vacuolated cells; col8a1a in sheath cells; entpd5a in pre-mineralization sheath cells). (D), UMAP showing co-embedded notochord cells from reference and temperature perturbation experiment, colored according to the mean time point label of nearest neighbors in the reference. Annotations for each cell type are indicated. (E), UMAP showing sheath cells from temperature perturbation experiment with significant spatial bias using hotspot test (q < 0.05) in UMAP space (see Methods). Cells with a significant spatial base are shown in red. Inset shows levels of UPR markers and translation signature in these cells increasing with temperature.
The notochord has unique mechanical and signaling functions, with specific cell sub-types carrying out these roles. These notochord sub-types are apparent in the reference dataset, including the vacuolated inner cells (rab32a+, cav3+), epithelial-like sheath cells (col8a1+), and pre-mineralization sheath cells (entpd5a+) (Fig. 4C), with these sub-types diverging earlier in development (Fig 4D). To identify which of these types might be responsible for its temperature-sensitivity, we compared notochord cells from embryos raised in elevated temperatures with notochord cells from our wild-type reference dataset (Fig. 4E). We detected significant (q < 0.05) enrichment for cells from the elevated temperature condition in focal regions of the notochord UMAP, including a “hotspot” among the epithelial-like sheath cells, which expressed genes for translation and UPR at higher levels than other notochord subtypes (Fig. 5A). The primary function of the epithelial-like sheath cells is to produce, process, and package large amounts of collagen and ECM proteins that will ultimately be secreted to produce the extracellular notochord sheath, which is essential for the rigidity of the notochord and the proper formation of the embryonic body 45.
Figure 5. Loss of Atf6 limits temperature-induced acceleration of developmental rate and increases temperature sensitivity of the notochord.
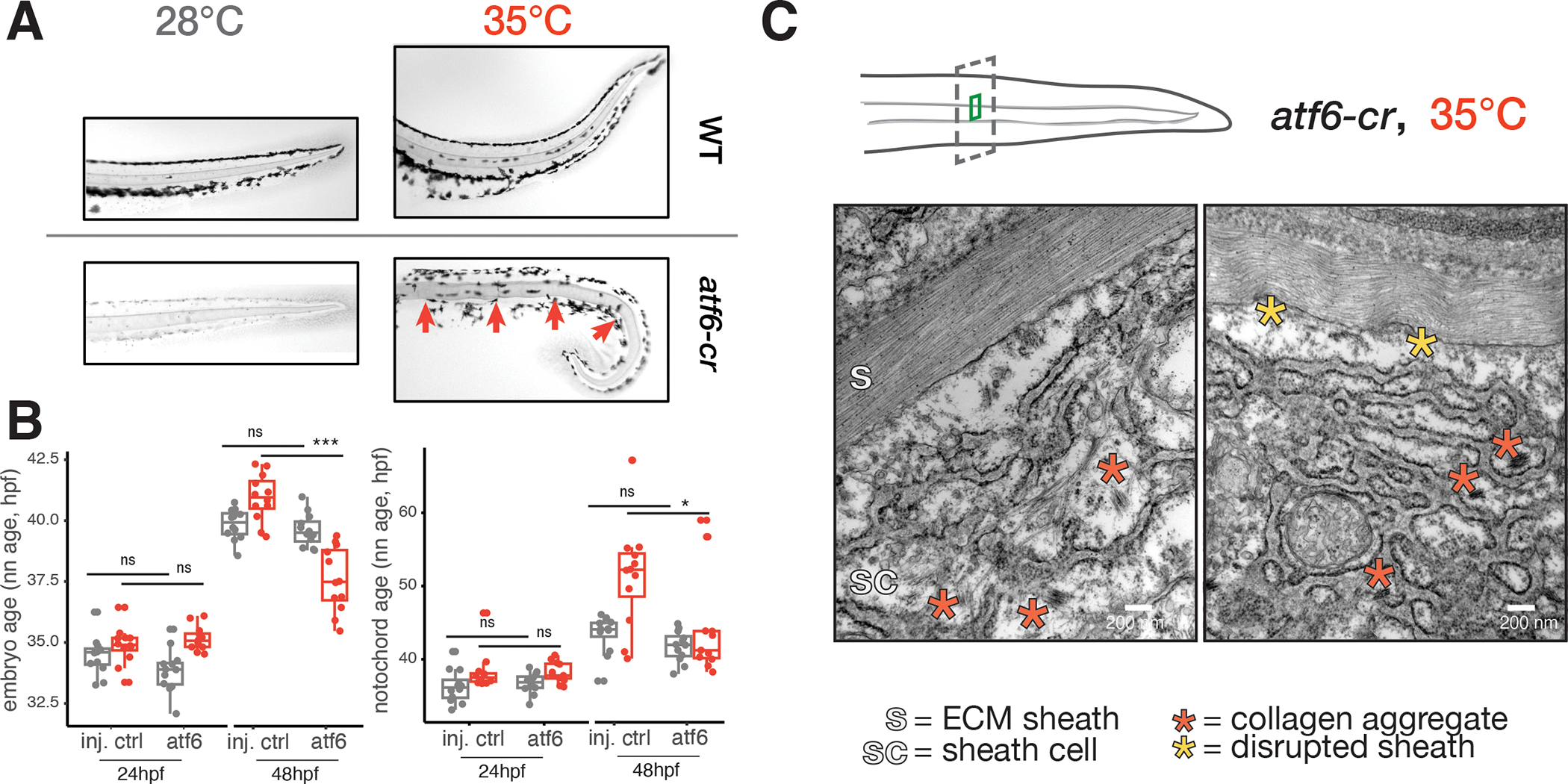
(A), Images of Alcian-blue stained notochords (camera image in black-and-white) in the tails of wild-type and atf6 crispant embryos raised at 28°C and 35°C. Examples of notochord defects in crispants raised at elevated temperature, kinks indicated with red arrows (phenylthiourea was used for imaging 28°C crispants). (B), Boxplots showing reduced capacity for acceleration of development at 34°C for the entire embryo (left) and notochord (right), while no developmental delay is apparent at 28°C. (C), Results of TEM of notochord sheath cells in atf6 crispants raised at elevated temperature, showing disrupted ER structure and aggregated collagen fibrils (red asterisks). Scale bar = 200nm; sc = sheath cell; s = sheath.
To test whether UPR buffers temperature-mediated ER stress in the developing notochord, we used CRISPR in F0 embryos to knock out the ER stress-sensing transcription factor Atf6 and examined notochord morphology using Alcian blue stain. In accordance with previous descriptions in medaka fish 40, we observed slight notochord defects at both 24 hpf and 48 hpf when fish were raised at standard temperature (Fig. 5A) and nearly all atf6 crispants were viable past 24 hpf. We next tested if defects in atf6 crispants were exacerbated when these embryos were raised at elevated temperature, especially given the proteostatic stress expected from temperature-dependent upregulation of translation in the notochord. Notochords from control fish raised at elevated temperature looked normal except for the usual bends, while atf6 crispant fish showed kinks, bends, or deformations of the notochord sheath (Fig. 5A). These phenotypes evoke notochord mutants that affect the structural integrity of the sheath, such as col8a1, rather than the mutants affecting vacuolation, such as rab32a 46,47.
To further test how the role of Atf6 in activating UPR in the notochord affects its ability to withstand temperature stress, we performed an additional sci-Plex experiment on 96 individual embryos of atf6 CRISPANTS and injection controls raised at normal and high temperature. We found that both whole-embryo and notochord-specific developmental age was lower than expected when atf6 CRISPANT embryos were raised at high temperature (Fig. 5B). Additionally, pharmacological inhibition of UPR with ISRIB in embryos raised at high temperature prevented notochord sheath cells from entering the stress-induced “hotspot” transcriptional state (Fig. 4D), though temperature-induced developmental acceleration was still apparent at 48hpf with this treatment, suggesting some functional separation between components of the UPR in control of acceleration (Fig. S5E, S5F). While atf6 CRISPANTS show reduced temperature-dependent acceleration of development, both notochord-specific and embryo-wide developmental ages are unaffected when mutants are raised at the normal temperature (Fig. 5B). Therefore, atf6 CRISPANTS do not show generalized developmental delay, but rather a deficiency in accelerating development in response to increased temperature. This result is consistent with our finding that basal UPR levels in each cell type are correlated with temperature-dependent acceleration of their development (Fig. 3C), highlighting the role of the UPR in coordinating cell type-specific changes of developmental timing in the embryo. Based on the requirement of UPR for temperature-accelerated development in individual cell types, we next sought to understand cell type-specific consequences of impaired control of timing and phenotypic severity of atf6 CRISPANTS.
Because the sheath cells showed a potent upregulation of genes required for translation in response to increased temperature (Fig. 5A), we wondered whether this cell type was functioning properly in deposition of the notochord sheath. We examined the ultrastructure of sheath cells and the sheath using transmission electron microscopy. We confirmed that the notochord sheath showed increased disorder at 35°C in both wild-type and atf6 crispant fish compared to controls raised at standard temperature (Fig. 5C, see comparison in Fig. S5G), suggesting that proper modification and organization of collagen is challenged at elevated temperature. Furthermore, we note a dense, altered ER structure in this condition (Fig. S5G). The most striking difference in atf6 mutants raised at high temperature was an accumulation of misfolded, non-secreted collagen fibers in the sheath cells themselves (Fig. 5B). Both protein aggregates and the severe disruption of ER structure in these fish suggest a failure of export, likely owing to the increased burden of protein synthesis at high temperatures combined with a lack of a properly functioning ER stress response. Taken together, these experiments show that in order for the notochord to structurally support the developing embryo, one of its cell types must operate near its proteostatic limits, requiring management by the UPR even at normal temperatures. Elevated temperature pushes the notochord sheath cells past their proteostatic limits, leading to protein aggregation, cellular defects, and a compromised notochord sheath that ultimately leads to a bent tail.
Discussion
Here, we used large-scale single-cell profiling on individual zebrafish embryos to explore how water temperature impacts their development. To do so, we introduce novel computational approaches to detect and account for effects on the abundance and transcriptomes of individual cell types.
In characterizing cell type-specific responses to temperature in embryos, we identify previously overlooked sources of compromised developmental robustness. First, we find variability in the temperature-induced developmental acceleration across cell types, introducing asynchrony in the coordinated and timely traversal of developmental trajectories. This phenomenon is not unlike the consequence of genetic mutants causing stalling or mis-timed events of single cell types in the coordinated program of development 48. Unlike in these genetic mutants, elevated temperature does not induce stalling in any single cell type but rather introduces asynchrony across cell types; the degree of accelerations among cell types ranges from negligible to over 40% and correlated progression between cell types is lost. This loss of synchrony between cell types that signal to one another during embryonic development may have broad consequences for tissue morphogenesis and may be related to documented non cell-autonomous effects of cell type-specific temperature-sensing.49,50 Further efforts to examine pairs of cell types with known signaling interactions may explain the observed patterns of synchrony loss. In sum, elevated temperature challenges the robustness of the developmental program by introducing non-uniform acceleration of developmental timing across cell types, breaking synchrony across the embryo.
What mechanisms could allow for this critical vulnerability in development? All cells have the capacity to sense and respond to temperature. The classic example is heat shock, a threshold phenomenon wherein cells experiencing a upward shift in temperature rapidly activate a suite of heat shock proteins to manage the acute stress. The temperatures we used in this study do not meet this threshold, and accordingly, we observe no activation of the canonical heat shock response (Fig. S1D–F). Even so, developmental rate of the embryos shows a continuous, linear relationship with temperature in the 25°C-35°C range (Kimmel 1994), revealing the existence of a temperature-sensitive gene program outside of the heat shock response linked to control of timing. Surprisingly, we found that the relationship between temperature and developmental rate was non-uniform at the level of cell type, with some cell types accelerating more than others in response to the same elevated temperature. Therefore, distinct cell types appear to be differentially sensitive to temperature.
We used two additional treatments to push developmental rate in the opposite direction, cooler temperatures and a non-lethal dose of the protein synthesis inhibitor cycloheximide. In comparing these treatments, we detected effects of each perturbation on global gene expression even within embryos at the same developmental stage. We found that the majority of gene expression differences found in both of these delay-inducing treatments are the same genes affected during warm temperature treatment, but with the opposite sign (see Fig. S8B-C), suggesting a common sensing mechanism to adjust developmental rate.
Cell type-specific sensitivity to temperature is also evident in the altered cell composition of embryos raised under stress. Whether the cellular phenotypes displayed by embryos raised at high temperature are of any adaptive value is unclear. Further study of natural populations and related species will be required to further explore this question, since lab populations of zebrafish show reduced plasticity and thermal tolerance relative to their wild counterparts 51. The notochord was especially sensitive, owing to the burden of protein synthesis in epithelial sheath cells which, under high temperature conditions, accumulate misfolded protein. The developing notochord depends on high levels of protein synthesis 52, ER chaperone function 53, and specialized secretion 41 to construct a well-ordered, collagen-rich sheath. This results in considerable stress on the ER and physiological activation of the UPR. The surprising upregulation of translation in these cells despite presumed translational attenuation by the UPR suggests that sheath cells might bypass translation regulation to achieve their exceptional protein synthesis capacity 54. Several other cell types with high demands of protein expression also depend on stress responses during normal development; the UPR is activated in the hatching gland, periderm, and vascular smooth muscle and via the heat shock response (HSR), cytoplasmic chaperones are activated in red blood cells, fast muscle, and cardiomyocytes. While the consequences of excessive burden on protein homeostasis in cells are evident in the aggregation associated with neurodegenerative diseases55, we note the impact of this burden during embryogenesis, when even a transient stress may irreversibly perturb normal development. Furthermore, while basal activation of homeostatic mechanisms such as the UPR bestow some cell types (like the notochord) with additional capacity for pushing the limits of proteostasis during normal development, they necessarily bring along additional risk: the evolution of developmental programs that overdraw on the essential stress response mechanisms required to deal with unexpected environmental changes. As with the heat shock response, the UPR shows deep evolutionary conservation in managing proteostatic stress, but the additional challenges of multicellularity may have driven these stress-associated pathways toward new functions. Indeed, the UPR pathway shows increasingly elaborate control through evolutionary time, such as the capacity for protein synthesis control in the transition from unicellular to multicellular eukaryotes 56.
Proteostasis is maintained throughout different cell types and stages of development, but can be broken under environmental stress. The consequences of this breakdown are borne unevenly across cell types, each according to their respective demands on the pathways required to maintain homeostasis. Despite these heterogeneous demands on proteostasis control, how is a uniform acceleration of all cell types coordinated to achieve more rapid development at increased temperature? First, we have shown that this acceleration is not uniform; significant variability in acceleration is apparent at higher temperatures, and synchrony among cell types is reduced (Fig. 3E–G). Even so, temperature has a predictable mean effect on acceleration (Kimmel et al), and also pushes most cell types forward along their differentiation trajectories, suggesting some level of global control. We propose that this global control arises at least in part through management of proteostasis by the UPR; cell types with higher basal activation of UPR show greater temperature-induced acceleration (Fig. 3B) and UPR is required for temperature-induced acceleration (Fig. 5B). While the precise mechanism is not yet clear, UPR-dependent control of developmental timing may arise from a cell autonomous phenomenon in all cell types (such as control of protein synthesis levels) or from a non-cell-autonomous coordination via a subset of sensitive cell types (such as the notochord) with the ability to influence developmental rate of surrounding cells. These two mechanisms are not mutually exclusive, but further experiments will be required to directly test the requirement for non-autonomous control of synchrony in temperature-induced acceleration. Furthermore, the UPR may converge on other aspects of the broader environmental stress response (ESR) to help synchronize cells during development; further genetic experiments may clarify which of the many sensors and signal transduction components of the ESR contribute to its control, as well as the relative importance of chromatin organization, transcriptional and translational control, and other cellular processes. Our study provides a methodological path forward for such studies.
We propose that cell types with high demands on proteostasis during normal development may be more susceptible to further perturbations of homeostasis by temperature or other environmental stressors. Further exploration into the differential demands placed by each cell type on ancient and general molecular processes may shed new light on how stress on a molecular pathway in a specific cell type generates phenotypes at the tissue, organ, or organismal scale.
Limitations of the study
The technology presented in this study allows for theoretically limitless sample multiplexing, opening up analysis of biological replicate embryos. However, this approach depends on nuclear rather than whole-cell sc-RNA-seq data and, while this kind of data produces less biased cellular sampling of the embryo, it brings limitations in the total number of RNA molecules captured per cell. Further improvements to mRNA capture efficiency in this protocol would add statistical power in identifying more subtle gene expression differences induced by temperature stress.
Our study shows that zebrafish embryos raised at elevated temperature have altered cell type composition and differential gene expression at important junctures of organogenesis. While the high time resolution and cellular coverage of many biological replicate embryos from the commonly studied AB strain indeed supports this finding, the adaptive relevance in wild zebrafish remains to be ascertained. Warming oceans create an urgent need for further studies in natural populations and related species to explore this question. Lab populations of zebrafish do show reduced plasticity and thermal tolerance relative to their wild counterparts 51 and may therefore be less resilient when developing in warm water, but both the cellular mechanisms and cell types central to temperature-dependent developmental acceleration are highly conserved across vertebrates. The changes we observe are therefore likely to be broadly relevant. Supported by imaging data, we find that the reduced abundance of notochord cells is associated with failure of sheath cell function at high temperature, but we have not quantitatively assessed the nature of other cell abundance changes we observed. Why do some cell types increase in abundance relative to untreated embryos? Follow-up studies will be required to link these cell-type composition changes to whole-embryo or persistent adult phenotypes. Further, the groups of cell types with affected abundance may reflect underlying cell-cell signaling interactions. Individual embryo data opens up many opportunities to infer how cell types interact with each other, but identifying these interactions is a nontrivial statistical problem.
Finally, our model of developmental robustness implies that building an embryo of correct proportions depends on precise timing and coordination among developmental trajectories of different cell types. We have shown that this timing and coordination is achieved, in part, through control of cellular proteostasis through via the UPR. However, beyond direct visualization of large proteins in the notochord sheath, we have not been able to directly connect a protein-level measurement with our single-cell analysis of developmental trajectories throughout the embryo. To fully realize this goal would require technical advances in single-cell proteomic tools. The ability to track protein levels, or of protein synthesis activity, in many cell types through developmental time, and in response to temperature, would permit a general test of our model.
STAR Methods
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Cole Trapnell (coletrap@uw.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
Single-cell RNA-seq data and raw sequencing are publicly available as of the date of publication via the NCBI Gene Expression Omnibus (GEO) accession GSE202294.
All original code, including notebooks for performing data processing, statistical analysis and generating plots, has been deposited at Github and is publicly available as of the date of publication: https://github.com/cole-trapnell-lab/sdg-zfish. Pipelines for generating count matrices from sci-RNA-seq3 sequencing data are available at https://github.com/bbi-lab/bbi-dmux and https://github.com/bbi-lab/bbi-sci. Analyses of the single cell transcriptome data were performed using Monocle3, which was updated to include methods from this study; a general tutorial can be found at http://cole-trapnell-lab.github.io/monocle-release/monocle3.
All materials used in this study are available by request from the corresponding author.
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
Animals
Fish stocks used in this study were: wild-type AB. Adult fish were maintained at 28.5°C under 14hr:10hr light:dark cycles and fed daily. Embryo clutches were collected from several pair-wise adult crosses. Embryos were shifted to elevated (32°C, 34°C, or 35°C) or reduced temperatures (20°C) at 6 hpf, and remained at these temperatures until sampling; staging followed Kimmel24. Embryos for this study were sampled between 24hpf and 72hpf. The sex of the embryos is not yet determined at stages profiled in this study. All procedures involving live animals followed federal, state and local guidelines for humane treatment and protocols approved by the Institutional Animal Care and Use Committee (protocol #2997–01) of the University of Washington.
METHOD DETAILS
Preparation of barcoded nuclei from individual embryos
Individual zebrafish embryos (24 to 36 hpf) were manually dechorionated and transferred to separate wells of a 96-well V-bottom plate containing 75 uL of 1X TrypLE + 2mg/mL Collagenase P (Millipore Sigma, cat. 11213865001). Embryos were then dissociated manually at 30°C by pipetting every 5 minutes for about 20 minutes until no visible chunks were present under a dissecting scope. Stop solution (dPBS, 5% FBS) was then added to each well and cells were spun down at 600×g for 5 min. Cells were then rinsed 1X in cold, 1X dPBS and spun down again. After rinses, supernatant was fully removed and cells were resuspended in 50 uL of CLB (Nuclei buffer, 0.1% IGEPAL, 1% SuperaseIn RNase Inhibitor (20 U/μL, Ambion), 1% BSA (20 mg/ml, NEB)) + hash oligos (1 uM, IDT) and incubated for 3 min on ice to liberate nuclei and integrate hash barcodes. To each well, 200 ul of ice cold, 5% Paraformaldehyde was added to each well. After an additional round of mixing, nuclei were fixed on ice for 15 minutes. All wells were then pooled together in a 15 mL conical tube and spun down for 15 min at 750×g. Supernatant was decanted and cells rinsed in 2mL of cold NBB (Nuclei Buffer, 1% BSA, 1% SuperaseIn) at 750×g for 6 min. Supernatant was then removed and cells were resuspended in 1mL of NBB and flash frozen in LN2 and stored at −80.
sci-RNA-seq3 library construction
The fixed nuclei were processed similarly to the published sci-RNA-seq3 protocol29. For paraformaldehyde fixed cells, frozen fixed cells were thawed on ice, spun down at 750×g for 6 min, and incubated with 500ul NBB (Nuclei buffer, 1% BSA, 1% SuperaseIn) including 0.2% Triton X-100 for 3 min on ice. Cells were pelleted and resuspended in 400ul NBB. The cell suspension was sonicated on low speed for 12s. Cells were then pelleted at 750×g for 5 min prior to resuspension in NB + dNTPs. The subsequent steps were similar with the sci-RNA-seq3 protocol (with paraformaldehyde fixed nuclei) with slight modifications: (1) We distributed 25,000 fixed cells (instead of 80,000 nuclei) per well for reverse transcription. (2) Centrifugation speeds for all spins were increased to 750×g.
Sequencing, read processing and cell filtering
Libraries were sequenced on an Illumina Novaseq 6000 (S4 200 cycle kit) with sequencing chemistries compatible with library construction and kit specifications. Standard chemistry: I1 – 10bp, I2 – 10bp, R1 – 28bp, R2 - remaining cycles (> 45). Read alignment and gene count matrix generation was performed using the Brotman Baty Institute (BBI) pipeline for sci-RNA-seq3 (https://github.com/bbi-lab/bbi-sci). After the single cell gene count matrix was generated, cells with fewer than 200 UMIs were filtered out. For mitochondrial signatures, we aggregated all reads from the mitochondrial chromosome. Each cell was assigned to a specific zebrafish embryo based on the total hash count (hash_umi >= 5) and the enrichment of a particular hash oligo (hash enrichment cutoff >= 3).
Count matrix pre-processing
Transcript by cell count matrix was pre-processed using a standard Monocle3 workflow: estimate_size_factors() -> detect_genes(min_expr = 0.1) -> preprocess_cds() with 100 principal components for whole-embryo and 50 principle components for subsets, align_cds(residual_model_formula_str = “~log10(n.umi)”), reduce_dimension(max_components = 3, preprocess_method = ‘Aligned’), and finally cluster_cells(resolution = 1e-4).
Cell annotation by projection of query data to reference
Cell type annotations were assigned via a label transfer procedure from the reference dataset (see related manuscript,32) to the query temperature-treated dataset. Briefly, the PCA rotation matrix, log10(n.umi) batch correction linear model, and non-linear UMAP transformation from the reference data were computed and saved for subsequent transformation of query data. The query temperature-treated dataset was then projected into the reference space using the following procedure: coefficients to transformed gene expression values into PCA loadings were applied, the linear batch correction was then applied to remove effects of total UMIs per cell, and, lastly, the UMAP transformation was applied to the batch-adjusted PCA loadings to project query data into the reference coordinate space. A similar overall approach is used in Andreatta et al.57
After an initial transfer of the ‘major group’ label (four possibilities: mesoderm, mesenchyme-fin, periderm + other, CNS) in the global space of all cells, cells that gained major group labels were projected in a sub-space corresponding to that major group. In this sub-space, finer resolution annotations (germ layer, tissue, broad cell type, sub cell type) were transferred using the majority vote of reference neighbors (k=10).
Estimation of transcriptional age for each cell and cell type
Alongside the process for transferring cell type annotation labels as above, each cell from the query temperature-treated dataset was assigned an estimated timepoint label (mean_nn_time) by taking the average sample timepoint of its 10 nearest neighbors in the developmental reference dataset. For each embryo, we bulked and averaged all cells of the same type to compute a cell type-specific “transcriptional age.”
Comparison of embryo pseudostage with conventional staging
A linear model for the effects of temperature on developmental stage from Kimel et al was used to generate stage expectations: h = H * (0.055T − 0.57), where h = hours of development to reach the stage at 28.5°C and H = hours of development at temperature T.24
Additive model for cell type-specific acceleration
Signatures scores were computed using the aggregate_gene_expression() function in Monocle3, using the log-normalized average counts across gene sets representing several biological processes (table S3C). To assess the baseline contributions of each process to acceleration by temperature, signature scores were computed for embryos raised at the standard 28°C. A linear model was used (vglm from VGAM package58) to examine the relationship of each signature with relative developmental acceleration, and coefficients for each process were compared.
Perturbation of proteostasis via CRISPR/Cas9 mutagenesis and inhibitors
gRNAs were designed using either the IDT or CRISPOR59 online tool. gRNAs were synthesized by IDT as crRNAs, annealed to tracrRNAs and assembled into RNPs with spCas9 protein (IDT). Preparation and subsequent injection of RNPs was performed as previously described;60 two gRNAs were simultaneously injected for atf6. To inhibit protein synthesis globally, embryos were treated with a non-lethal dose of 5μM cyclohexamide (CHX, Sigma, C4859) starting at 6hpf. To inhibit the unfolded protein response, embryos were treated with 100μM ISRIB (Sigma, SML0843) starting at 6hpf.
Transmission Electron Microscopy
Fish were euthanized then fixed in sodium cacodylate buffered 4% glutaraldehyde overnight at 4°C. Embryos were stained in 2% osmium tetroxide for 30 min, washed, and then stained in 1% uranyl acetate overnight at 4°C. Samples were dehydrated with a graded ethanol series then infiltrated with a 1:1 propylene oxide:Durcupan resin for 2 hours followed by fresh Durcupan resin overnight and flat embedded prior to polymerization. Blocks of posterior tissue, from yolk extension to tail, were thin sectioned on a Leica EM UC7 and sections imaged on a JEOL 1230 transmission electron microscope.
Notochord staining
Alcian blue staining followed an online procedure (SDB Online Short Course, Zebrafish Alcian Blue), except that embryos were raised in 1-phenyl-2-thiourea to suppress pigment formation instead of bleaching.
Differential gene expression analysis
To capture biological variation in gene expression between stage-matched embryos, cells of each type were first pseudobulked (sums of size-factor-normalized gene counts) by embryo. Using this embryo-level pseudobulk data as input, differentially expressed genes were determined in each cell type using the fit_models() function in Monocle3. The model formula included additive terms for temperature (as a factor), a spline of embryo pseudostage (degrees of freedom = 3), and the total number of cells. Global analysis of temperature-dependent gene expression was performed by extracting temperature coefficients from the model output, filtering for genes with significant q-values (q < 0.05). Temperature coefficients in each cell type were combined into a single matrix, and processed analogously to the find_gene_modules() function in Monocle3 29, which uses a transposition of the typical gene x cell matrix to group genes with similar patterns across cells, rather than grouping cells. Temperature coefficients (no normalization) were pre-processed with PCA and dimensionality reduction was performed with UMAP (n_neighbors = 25L, min_dist = 0.2). Clusters of genes with similar temperature responses across cell types were identified using the cluster_cells() function.
Hotspot analysis
The local spatial statistic Getis-Ord index (Gi) was used to identify statistically significant regions of the UMAP embedding that were enriched or depleted of perturbed cells. A high-value Gi indicates that a perturbed cell is surrounded by other cells with the same perturbation, whereas a Gi close to zero indicates that a perturbed cell is surrounded by cells with other perturbation labels. A Gi was calculated for each cell’s local neighborhood (k=15) using the “localG()” function in the spdep package. This returns a z-score that indicates whether the observed spatial clustering is more pronounced than expected by random. Multiple testing correction was done using a Bonferroni correction. Areas of the UMAP where a given perturbation is enriched are called “hot spots.”
QUANTIFICATION AND STATISTICAL ANALYSIS
Cell abundance and variance statistics
Changes in the proportions of each cell type were assessed by first counting the number of each annotated cell type in each embryo. To control for technical differences in cell recovery across embryos, “size factors” by dividing the total number of cells recovered from an embryo by the geometric mean of total cell counts across all embryos. The number of cells of each type recovered from each embryo were then divided by that embryo’s size factor. No explicit sample-size calculation was performed for number of embryos, but individual-embryo protocols were optimized to sample enough cells to capture all major cell types in the embryo between 24 – 96hpf. No individual samples were excluded. Within samples, we excluded cells that did not pass quality filters from subsequent single-cell analysis; these include minimum UMI counts, mitochondrial read fraction, and doublets. No explicit randomized design was deployed, but individual replicate embryos were pooled and processed together, reducing technical batch effects. No blinding was used in this study.
Normalized counts for each cell type at different developmental stages were then compared across treatments using a generalized linear model defined by the equations:
Where the normalized counts of cell type at time is modeled as a beta-binomially distributed random variable with mean and overdispersion with respect to the binomial distribution. We modeled both parameters of the beta binomial response as a function of temperature, reasoning that treated embryos might exhibit greater variability than wild-type embryos. The binary indicator variable denotes treatment in each embryo. Separate models for each temperature in each cell type and at each pseudostage bin were fit using the VGAM package58. Significance of temperature effects in each model were assessed by a Wald test. In all figures, relevant comparisons and total values of n is denoted in Figure legends.
Supplementary Material
Fig. S1. Individual-level capture of high-quality single cell transcriptomes, Related to Figure 1. (A), Barplot showing the number of individual cells that pass (Y = yes, N = no) a Hash Enrichment Ratio (count of most abundant hash relative to second-most abundant) threshold of 3. Below is a histogram showing the full distribution of Hash Enrichment Ratios; cells where all reads were derived from a single hash are labeled as ‘Inf.’ (B), Boxplots showing the number of cells in each embryo (left) and number of UMIs per cell in each embryo (right), faceted by sample time point and colored by temperature treatment. (C), Knee plot showing the threshold for UMI count (red line) in called cells. (D), Barplots showing tissue compositions as per-embryo cell counts in two additional scRNA-seq studies of zebrafish whole-embryo samples. A measure of composition bias (Gini index) is listed above each barchart; higher values indicate higher bias in tissue recovery, or less even coverage across tissues. (E), Marker plot showing no induction of hsp90aa1.1 at 32°C or 34°C, including cells from all timepoints. (F), Marker plot for hsp70l, a gene used for transgenic reporters of heat shock in zebrafish, shown as in (E). (G), Histogram showing no systemic activation of the heat shock response (HSR) across cell types at any of the temperatures profiled. While the total number of cell types with negligible HSR levels increases with temperature, nearly all cell types should have HSR signature values greater than 0 in the event of global activation of the HSR. (H), Dotplot of each cell type, ordered by the Pearson correlation between that cell type’s abundance and the whole-embryo pseudostage. Bellwether cell types with the highest correlations are labeled, with colors indicating tissue of origin. (I), Two examples from the highly correlated types shown in (A), with raw cell type abundance data plotted against embryo stage for a strong negatively correlated type (fin mesenchyme) and strong positively correlated type (differentiated neurons); inset at right shows additional examples.
Fig. S2. Temperature increases variability in specific cell types and stages of development, Related to Figure 2. (A), Scatterplot comparing whole-embryo staging determined by either cell composition (x-axis) or transcriptional data (y-axis); inset barplots show coefficient of variation for each metric in groups of embryos sampled at various timepoints (timepoints in hpf indicated in grey text above each bar). Loess fit lines are shown for embryos raised at each temperature, 28°C (grey), 32°C (pink), 34°C (red). (B), Stacked barchart showing how the number of cell types with significant abundance changes increases with temperature; this phenomenon held for all pseudostage bins (in color) except bin 4, which lacked sufficient control embryos to analyze statistically. (C), Scatterplot showing positive relationship of log2 fold change in abundances in both elevated temperatures, colored as in (A). (D), Histogram showing changes in cell composition resulting from 34°C treatment, as predicted by multinomial model. (E), Scatterplots showing raw data (black points) and model outputs of mean (solid line) and variability (ribbons with dotted line) for three temperature-sensitive cell types. (F), Heatmap of each cell type showing relative increases in observed variability compared to variability expected during wildtype development through developmental time (pseudostage bins); shades of red indicate increased variability while white to blue shades show neutral or decreased variability. The upper panel shows the average CVs for each temperature across all stages, highlighting pseudostages with increased variability at higher temperatures. (G), Barplot showing the log2 fold change in abundance for all cell types in 34°C embryos with severe phenotypes relative to those that were phenotypically normal.
Fig. S3. Temperature accelerates development non-uniformly across cell types, Related to Figure 3. (A), Schematic showing method for computing transcriptional age of each cell through nearest-neighbor averaging of time point labels in the reference. (B), Scatterplot as in Fig. 2B, but showing how well whole-embryo staging by transcriptional data (nn-transcriptional age), rather than cell composition, predicts temperature-induced developmental acceleration. (C), Boxplots showing individual examples of cell types with greater-than-expected acceleration (left) and less-than-expected acceleration (righta), with whole embryo expectation from Kimmel et al for each temperature shown as dotted lines.24 (D) Barplots showing the number of significantly (q < 0.05) differentially expressed genes in each cell type, separated by whether the gene has a significant association for developmental age (green) or temperature (red). Cell types along the x-axis are ordered by the difference between the number of significant temperature genes vs significant developmental age genes; the large majority show more genes changing with developmental age rather than temperature. (E) Scatterplot showing significant genes separated by developmental age and temperature effects for a specific cell type, the notochord. Temperature and age coefficients are shown for each gene, and are colored based on significant associations with each factor, neither, or both. (F), Density histogram showing that genes with significant associations with temperature tended to show higher expression levels than developmental age genes. (G) Density histogram of pairwise Pearson correlations for cell cycle signature values across all cell type pairs, with separate densities for each temperature. Inset shows boxplots of pairwise cell cycle correlations grouped into cell type pairs that showed lower or higher levels of developmental timing synchrony. (H), Scatterplot as in Fig. 3C, showing that the basal level of heat shock response signature in each cell type is not a strong predictor of that cell type’s developmental acceleration at 32°C or 34°C. (I), Barplots showing the level of heat shock response signature induction for all cell types at 34°C; cell types on the y-axis are ordered by relative developmental acceleration, with the most strongly accelerated types at the top, and columns for each timepoint. No induction of heat shock response signature is apparent in the most accelerated cell types.
Fig. S4. Cold treatment slows development and reveals cell type-specific temperature-responsive genes in the normal range of zebrafish development, Related to Figure 3. (A), Dotplot showing the sampling timing for cold-treated embryos (x-axis is clock time), as well as their predicted whole-embryo stage based on integration with the 28°C reference data; the predicted stage roughly matched the expectation of the Kimmel et al. (B), Boxplot showing the effect of 5μM cyclohexamide (CHX) on temperature-induced acceleration of whole-embryo stage; developmental delay is evident in CHX-treated embryos raised at 28°C, and this effect is not overridden by 34°C temperature treatment. (C), Scatterplot showing temperature-dependent activation of genes of the fast muscle, expressed as model coefficients for each temperature after accounting for developmental stage. Most genes showed either heat-induced increases and cold-induced decreases in expression (red box in upper-left), or heat-induced decreases and cold-induced increases in expression (blue box in lower left). (D), Scatterplot as in (C), but for slow muscle. (E), Scatterplot for fast muscle similar to (B), but showing cold vs cyclohexamide treatment, which shows an overall strong positive correlation with gene expression similarly affected by both sources of developmental delay. (F), Scatterplot as in (E), but for slow muscle.
Fig. S5. Transcriptional responses to elevated temperature tend to be cell type-specific, revealing burdens on proteostasis in the notochord, Related to Figure 4. (A), UMAP of all genes with significant temperature-dependent transcriptional changes (see Methods). Each point represents an individual gene, and genes with similar responses (coefficient in differential expression model after accounting for stage) across cell types are grouped together. Each cluster is labeled with a different color, and a representative GO term for the group of genes in each cluster is indicated. Clusters in which gene groups returned no significant GO terms are labeled ‘NA’. (B), Heatmap of expression levels for each gene module in (a) as an average for each cell type at elevated temperatures. Consistent with the heterogeneous grouping of temperature responses in (a), elevated module expression tends to be cell type-specific (examples boxed). Module 5, which contains genes related to translation and ER processing, has the most broad response across cell types, but the magnitude of the response is greatest within the notochord. (C), Scatterplot showing levels of UPR signature in the notochord cells of all individual embryos, colored by temperature. Embryos raised at higher temperature show higher levels of UPR signature at nearly all profiled stages. Lines and ribbons are derived from loess fits across pseudostage. (D), Boxplot showing marked reduction of notochord cells in embryos raised at high temperature, with raw counts in control and temperature-treated embryos shown as points. (E), Boxplot showing the effect of the UPR inhibitor ISRIB on whole-embryo stage at 28°C and 34°C; embryos treated with ISRIB showed significant increases over expected temperature-induced acceleration at 24hpf, but this effect was not evident in 48hpf embryos. (F), Hotspot analysis, as in Fig. 4D, showing enriched transcriptional state of notochord sheath cells in embryos treated with ISRIB; note that ISRIB-treated cells do not occupy the high-temperature state identified in wild-type embryos. (G), TEM of sheath cells in wild-type and atf6 crispants, showing effects in both untreated and temperature-treated embryos. ER structure is perturbed in atf6 crispants, and severe sheath defects are visible in the temperature-treated atf6 crispants. Scale bar = 200nm.
Table S2. Phenotypic severity classification for each embryo, related to Figure 2.
Methods S1. Protocol for individual-embryo sci-RNA-seq library preparation, Related to Figure 1.
Table S1. Cell abundance model outputs, Related to Figure 2. A) Model outputs from mean abundance analysis. B) Model outputs from variance analysis.
Table S3. Global analysis of temperature-induced gene expression changes across cell types, Related to Figure 4. A) Model coefficients for temperature-induced differential expression in each cell type. B) GO enrichments from differentially expressed genes. C) Genes used to compute signature scores.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| 20% Paraformaldehyde Aqueous Solution, EM Grade | EMS | 50-980-492 |
| Collagenase P | Millipore-Sigma | 11213865001 |
| 1X TrypLE Express | Thermo Fisher | 12604013 |
| DPBS, no calcium, no magnesium | Thermo Fisher | 14190144 |
| SUPERase In RNase Inhibitor | Thermo Fisher | AM2696 |
| BSA | NEB | B9000S |
| IGEPAL CA-630 | Millipore Sigma | I8896 |
| Triton X-100 for molecular biology | Millipore Sigma | T8787 |
| Superscript IV reverse transcriptase | Thermo Fisher | 18090200 |
| RNaseOUT Recombinant Ribonuclease Inhibitor | Thermo Fisher | 10777019 |
| NEBNext High Fidelity 2x PCR master mix | NEB | M0541L |
| NEBNext mRNA Second Strand Synthesis Module | NEB | E6111L |
| Agencourt AMPure XP Beads | Beckman Coulter | A63882 |
| N,N-Dimethylformamide | Millipore Sigma | D4551 |
| Experimental models: Organisms/strains | ||
| Zebrafish/Wild-type AB strain | U Washington | N/A |
| Deposited data | ||
| Raw FASTQ files | GEO | GSE202294 |
| Processed count matrix | GEO | GSE202294 |
| Cell metadata | GEO | GSE202294 |
| Oligonucleotides | ||
| Indexed reverse transcription oligos (5′- /5Phos/CAGAGCNNNNNNNN[10bpRTindex]TTTTTTTTTTTTTTTTTTTTTTTTTTTTTT-3′) | IDT | https://cole-trapnell-lab.github.io/zscape/ |
| Indexed ligation oligos (5’- GCTCTG[10bp barcode A]/ideoxyU/ACGACGCTCTTCCGATCT[reverse complement of barcode A]-3') | IDT | https://cole-trapnell-lab.github.io/zscape/ |
| Indexed PCR oligos (P5) (5′- AATGATACGGCGACCACCGAGATCTACAC[i5]ACACTCTTTCCCTACACGACGCTCTTCCG ATCT-3′) | IDT | https://cole-trapnell-lab.github.io/zscape/ |
| Indexed PCR oligos (P7) (5′- CAAGCAGAAGACGGCATACGAGAT[i7]GTCTCGTGGGCTCGG-3′ | IDT | https://cole-trapnell-lab.github.io/zscape/ |
| Tn5-N7 oligo (5′-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAG-3′) | Eurofins | N/A |
| Tn5 Mosaic End (ME) oligo (5′-/5Phos/CTGTCTCTTATACACATCT-3′) | Eurofins | N/A |
| Software and Algorithms | ||
| Monocle 3 v1.3.1 | This paper, Saunders et al, 2022 | https://cole-trapnell-lab.github.io/monocle3/ |
| R 4.1.3 | CRAN | https://cran.r-project.org/ |
| bbi-demux v1.0 | Github | https://github.com/bbi-lab/bbi-dmux |
| bbi-sci v1.0 | Github | https://github.com/bbi-lab/bbi-sci |
| Nextflow 20.07.1 | Nextflow | https://www.nextflow.io/ |
ACKNOWLEDGEMENTS
We thank Nola Klemfuss and the Brotman Baty Institute Advanced Technology Lab for support with sequencing and data processing. We also thank Frank Steemers and Fan Zhang for additional sequencing support. We thank Ed Parker for assistance with TEM.
Funding
This work was supported by a grant from the Paul G. Allen Frontiers Group (Allen Discovery Center for Cell Lineage Tracing to C.T. and J.S.) and the National Institutes of Health (UM1HG011586 to C.T. and J.S.; 1R01HG010632 to C.T. and J.S.). J.S. is an Investigator of the Howard Hughes Medical Institute. M.W.D. was supported by UM1HG011586 to C.T. and NHGRI 1RM1HG010461 to C.Q.
Footnotes
Declaration of interests
C.T. is a SAB member, consultant and/or co-founder of Algen Biotechnologies, Altius Therapeutics, and Scale Biosciences. J.S. is a SAB member, consultant and/or co-founder of Cajal Neuroscience, Guardant Health, Maze Therapeutics, Camp4 Therapeutics, Phase Genomics, Adaptive Biotechnologies and Scale Biosciences.
ADDITIONAL RESOURCES
A website containing 3-level, individual-embryo indexing sci-RNA-seq protocol, as well as additional resources for visualizing wild-type and temperature-treated atlases can be found at: https://cole-trapnell-lab.github.io/zscape/.
References and Notes
- 1.Schirone RC, and Gross L (1968). Effect of temperature on early embryological development of the zebra fish,Brachydanio rerio. J. Exp. Zool. 169, 43–52. [Google Scholar]
- 2.Sharpe PJ, and DeMichele DW (1977). Reaction kinetics of poikilotherm development. J. Theor. Biol. 64, 649–670. [DOI] [PubMed] [Google Scholar]
- 3.Kuntz SG, and Eisen MB (2014). Drosophila embryogenesis scales uniformly across temperature in developmentally diverse species. PLoS Genet. 10, e1004293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gillooly JF, Charnov EL, West GB, Savage VM, and Brown JH (2002). Effects of size and temperature on developmental time. Nature 417, 70–73. [DOI] [PubMed] [Google Scholar]
- 5.Rayon T, Stamataki D, Perez-Carrasco R, Garcia-Perez L, Barrington C, Melchionda M, Exelby K, Lazaro J, Tybulewicz VLJ, Fisher EMC, et al. (2020). Species-specific pace of development is associated with differences in protein stability. Science 369. 10.1126/science.aba7667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matsuda M, Hayashi H, Garcia-Ojalvo J, Yoshioka-Kobayashi K, Kageyama R, Yamanaka Y, Ikeya M, Toguchida J, Alev C, and Ebisuya M (2020). Species-specific segmentation clock periods are due to differential biochemical reaction speeds. Science 369, 1450–1455. [DOI] [PubMed] [Google Scholar]
- 7.Min M, Rong Y, Tian C, and Spencer SL (2020). Temporal integration of mitogen history in mother cells controls proliferation of daughter cells. Science 368, 1261–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jarosz DF, Taipale M, and Lindquist S (2010). Protein homeostasis and the phenotypic manifestation of genetic diversity: principles and mechanisms. Annu. Rev. Genet. 44, 189–216. [DOI] [PubMed] [Google Scholar]
- 9.Balchin D, Hayer-Hartl M, and Hartl FU (2016). In vivo aspects of protein folding and quality control. Science 353, aac4354. [DOI] [PubMed] [Google Scholar]
- 10.Queitsch C, Sangster TA, and Lindquist S (2002). Hsp90 as a capacitor of phenotypic variation. Nature 417, 618–624. [DOI] [PubMed] [Google Scholar]
- 11.Jarosz DF, and Lindquist S (2010). Hsp90 and environmental stress transform the adaptive value of natural genetic variation. Science 330, 1820–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rohner N, Jarosz DF, Kowalko JE, Yoshizawa M, Jeffery WR, Borowsky RL, Lindquist S, and Tabin CJ (2013). Cryptic variation in morphological evolution: HSP90 as a capacitor for loss of eyes in cavefish. Science 342, 1372–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rutherford SL, and Lindquist S (1998). Hsp90 as a capacitor for morphological evolution. Nature 396, 336–342. [DOI] [PubMed] [Google Scholar]
- 14.Isaacs WB, and Fulton AB (1987). Cotranslational assembly of myosin heavy chain in developing cultured skeletal muscle. Proc. Natl. Acad. Sci. U. S. A. 84, 6174–6178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hetz C (2021). Adapting the proteostasis capacity to sustain brain healthspan. Cell. 10.1016/j.cell.2021.02.007. [DOI] [PubMed] [Google Scholar]
- 16.Rutherford SL (2003). Between genotype and phenotype: protein chaperones and evolvability. Nat. Rev. Genet. 4, 263–274. [DOI] [PubMed] [Google Scholar]
- 17.Goldschmidt R (1929). „Experimentelle Mutation und das Problem der sogenannten Parallelinduktion.“Biol. Zentralbl., Bd 49. [Google Scholar]
- 18.Audzijonyte A, Richards SA, Stuart-Smith RD, Pecl G, Edgar GJ, Barrett NS, Payne N, and Blanchard JL (2020). Fish body sizes change with temperature but not all species shrink with warming. Nat Ecol Evol 4, 809–814. [DOI] [PubMed] [Google Scholar]
- 19.Hsu C-Y, and Chiu Y-C (2009). Ambient temperature influences aging in an annual fish (Nothobranchius rachovii). Aging Cell 8, 726–737. [DOI] [PubMed] [Google Scholar]
- 20.Engeszer RE, Patterson LB, Rao AA, and Parichy DM (2007). Zebrafish in the wild: a review of natural history and new notes from the field. Zebrafish 4, 21–40. [DOI] [PubMed] [Google Scholar]
- 21.Cockroft DL, and New DA (1978). Abnormalities induced in cultured rat embryos by hyperthermia. Teratology 17, 277–283. [DOI] [PubMed] [Google Scholar]
- 22.Germain MA, Webster WS, and Edwards MJ (1985). Hyperthermia as a teratogen: parameters determining hyperthermia-induced head defects in the rat. Teratology 31, 265–272. [DOI] [PubMed] [Google Scholar]
- 23.Shoji W, and Sato-Maeda M (2008). Application of heat shock promoter in transgenic zebrafish. Dev. Growth Differ. 50, 401–406. [DOI] [PubMed] [Google Scholar]
- 24.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, and Schilling TF (1995). Stages of embryonic development of the zebrafish. Dev. Dyn. 203, 253–310. [DOI] [PubMed] [Google Scholar]
- 25.Schmidt K, and Starck JM (2010). Developmental plasticity, modularity, and heterochrony during the phylotypic stage of the zebra fish, Danio rerio. J. Exp. Zool. B Mol. Dev. Evol. 314, 166–178. [DOI] [PubMed] [Google Scholar]
- 26.Sfakianakis DG, Leris I, and Kentouri M (2012). Exercise–related muscle lactate metabolism in zebrafish juveniles: The effect of early life temperature. Ital. J. Zool. 79, 568–573. [Google Scholar]
- 27.Ackerly KL, and Ward AB (2016). How temperature-induced variation in musculoskeletal anatomy affects escape performance and survival of zebrafish (Danio rerio). J. Exp. Zool. A Ecol. Genet. Physiol. 325, 25–40. [DOI] [PubMed] [Google Scholar]
- 28.Srivatsan SR, McFaline-Figueroa JL, Ramani V, Saunders L, Cao J, Packer J, Pliner HA, Jackson DL, Daza RM, Christiansen L, et al. (2020). Massively multiplex chemical transcriptomics at single-cell resolution. Science 367, 45–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cao J, Spielmann M, Qiu X, Huang X, Ibrahim DM, Hill AJ, Zhang F, Mundlos S, Christiansen L, Steemers FJ, et al. (2019). The single-cell transcriptional landscape of mammalian organogenesis. Nature 566, 496–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wagner DE, Weinreb C, Collins ZM, Briggs JA, Megason SG, and Klein AM (2018). Single-cell mapping of gene expression landscapes and lineage in the zebrafish embryo. Science 360, 981–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Farnsworth DR, Posner M, and Miller AC (2021). Single cell transcriptomics of the developing zebrafish lens and identification of putative controllers of lens development. Exp. Eye Res. 206, 108535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saunders LM, Srivatsan SR, Duran M, Dorrity MW, Ewing B, Linbo T, Shendure J, Raible DW, Moens CB, Kimelman D, et al. (2022). Deep molecular, cellular and temporal phenotyping of developmental perturbations at whole organism scale. bioRxiv, 2022.08.04.502764. 10.1101/2022.08.04.502764. [DOI] [Google Scholar]
- 33.Jean-Baptiste K, McFaline-Figueroa JL, Alexandre CM, Dorrity MW, Saunders L, Bubb KL, Trapnell C, Fields S, Queitsch C, and Cuperus JT (2019). Dynamics of Gene Expression in Single Root Cells of Arabidopsis thaliana. Plant Cell 31, 993–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martin BD, Witten D, and Willis AD (2020). MODELING MICROBIAL ABUNDANCES AND DYSBIOSIS WITH BETA-BINOMIAL REGRESSION. Ann. Appl. Stat. 14, 94–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Phipson B, Sim CB, Porrello ER, Hewitt AW, Powell J, & Oshlack A (2022). propeller: testing for differences in cell type proportions in single cell data. Bioinformatics, 38(20), 4720–4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eberlein S (1985). Stage specific embryonic defects following heat shock inDrosophila. Dev. Genet. 6, 179–197. [Google Scholar]
- 37.Johnston IA, Lee H-T, Macqueen DJ, Paranthaman K, Kawashima C, Anwar A, Kinghorn JR, and Dalmay T (2009). Embryonic temperature affects muscle fibre recruitment in adult zebrafish: genome-wide changes in gene and microRNA expression associated with the transition from hyperplastic to hypertrophic growth phenotypes. J. Exp. Biol. 212, 1781–1793. [DOI] [PubMed] [Google Scholar]
- 38.Bennett JT, Joubin K, Cheng S, Aanstad P, Herwig R, Clark M, Lehrach H, and Schier AF (2007). Nodal signaling activates differentiation genes during zebrafish gastrulation. Dev. Biol. 304, 525–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ishikawa T, Kashima M, Nagano AJ, Ishikawa-Fujiwara T, Kamei Y, Todo T, and Mori K (2017). Unfolded protein response transducer IRE1-mediated signaling independent of XBP1 mRNA splicing is not required for growth and development of medaka fish. Elife 6. 10.7554/eLife.26845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ishikawa T, Okada T, Ishikawa-Fujiwara T, Todo T, Kamei Y, Shigenobu S, Tanaka M, Saito TL, Yoshimura J, Morishita S, et al. (2013). ATF6α/β-mediated adjustment of ER chaperone levels is essential for development of the notochord in medaka fish. Mol. Biol. Cell 24, 1387–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ishikawa T, Toyama T, Nakamura Y, Tamada K, Shimizu H, Ninagawa S, Okada T, Kamei Y, Ishikawa-Fujiwara T, Todo T, et al. (2017). UPR transducer BBF2H7 allows export of type II collagen in a cargo- and developmental stage-specific manner. J. Cell Biol. 216, 1761–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Iwakoshi NN, Lee A-H, Vallabhajosyula P, Otipoby KL, Rajewsky K, and Glimcher LH (2003). Plasma cell differentiation and the unfolded protein response intersect at the transcription factor XBP-1. Nat. Immunol. 4, 321–329. [DOI] [PubMed] [Google Scholar]
- 43.Angelos E, and Brandizzi F (2022). The UPR regulator IRE1 promotes balanced organ development by restricting TOR-dependent control of cellular differentiation in Arabidopsis. Plant J. 109, 1229–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pavitt GD, and Ron D (2012). New insights into translational regulation in the endoplasmic reticulum unfolded protein response. Cold Spring Harb. Perspect. Biol. 4. 10.1101/cshperspect.a012278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yasuoka Y (2020). Morphogenetic mechanisms forming the notochord rod: The turgor pressure-sheath strength model. Dev. Growth Differ. 62, 379–390. [DOI] [PubMed] [Google Scholar]
- 46.Stemple DL, Solnica-Krezel L, Zwartkruis F, Neuhauss SC, Schier AF, Malicki J, Stainier DY, Abdelilah S, Rangini Z, Mountcastle-Shah E, et al. (1996). Mutations affecting development of the notochord in zebrafish. Development 123, 117–128. [DOI] [PubMed] [Google Scholar]
- 47.Gansner JM, and Gitlin JD (2008). Essential role for the alpha 1 chain of type VIII collagen in zebrafish notochord formation. Dev. Dyn. 237, 3715–3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Griffin KJ, Amacher SL, Kimmel CB, and Kimelman D (1998). Molecular identification of spadetail: regulation of zebrafish trunk and tail mesoderm formation by T-box genes. Development 125, 3379–3388. [DOI] [PubMed] [Google Scholar]
- 49.van Oosten-Hawle P, Porter RS, and Morimoto RI (2013). Regulation of organismal proteostasis by transcellular chaperone signaling. Cell 153, 1366–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Prahlad V, Cornelius T, and Morimoto RI (2008). Regulation of the cellular heat shock response in Caenorhabditis elegans by thermosensory neurons. Science 320, 811–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Morgan R, Finnøen MH, Jensen H, Pélabon C, and Jutfelt F (2020). Low potential for evolutionary rescue from climate change in a tropical fish. Proc. Natl. Acad. Sci. U. S. A. 117, 33365–33372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Trapani V, Bonaldo P, and Corallo D (2017). Role of the ECM in notochord formation, function and disease. J. Cell Sci. 130, 3203–3211. [DOI] [PubMed] [Google Scholar]
- 53.Köhler A, Mörgelin M, Gebauer JM, Öcal S, Imhof T, Koch M, Nagata K, Paulsson M, Aumailley M, Baumann U, et al. (2020). New specific HSP47 functions in collagen subfamily chaperoning. FASEB J. 34, 12040–12052. [DOI] [PubMed] [Google Scholar]
- 54.Cagnetta R, Wong HH-W, Frese CK, Mallucci GR, Krijgsveld J, and Holt CE (2019). Noncanonical Modulation of the eIF2 Pathway Controls an Increase in Local Translation during Neural Wiring. Mol. Cell 73, 474–489.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hetz C, and Mollereau B (2014). Disturbance of endoplasmic reticulum proteostasis in neurodegenerative diseases. Nat. Rev. Neurosci. 15, 233–249. [DOI] [PubMed] [Google Scholar]
- 56.Hollien J (2013). Evolution of the unfolded protein response. Biochim. Biophys. Acta 1833, 2458–2463. [DOI] [PubMed] [Google Scholar]
- 57.Andreatta M, Corria-Osorio J, Müller S, Cubas R, Coukos G, and Carmona SJ (2021). Interpretation of T cell states from single-cell transcriptomics data using reference atlases. Nat. Commun. 12, 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yee TW (2010). The VGAM Package for Categorical Data Analysis. J. Stat. Softw. 32, 1–34. [Google Scholar]
- 59.Concordet J-P, and Haeussler M (2018). CRISPOR: intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res. 46, W242–W245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hoshijima K, Jurynec MJ, Klatt Shaw D, Jacobi AM, Behlke MA, and Grunwald DJ (2019). Highly Efficient CRISPR-Cas9-Based Methods for Generating Deletion Mutations and F0 Embryos that Lack Gene Function in Zebrafish. Dev. Cell 51, 645–657.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Individual-level capture of high-quality single cell transcriptomes, Related to Figure 1. (A), Barplot showing the number of individual cells that pass (Y = yes, N = no) a Hash Enrichment Ratio (count of most abundant hash relative to second-most abundant) threshold of 3. Below is a histogram showing the full distribution of Hash Enrichment Ratios; cells where all reads were derived from a single hash are labeled as ‘Inf.’ (B), Boxplots showing the number of cells in each embryo (left) and number of UMIs per cell in each embryo (right), faceted by sample time point and colored by temperature treatment. (C), Knee plot showing the threshold for UMI count (red line) in called cells. (D), Barplots showing tissue compositions as per-embryo cell counts in two additional scRNA-seq studies of zebrafish whole-embryo samples. A measure of composition bias (Gini index) is listed above each barchart; higher values indicate higher bias in tissue recovery, or less even coverage across tissues. (E), Marker plot showing no induction of hsp90aa1.1 at 32°C or 34°C, including cells from all timepoints. (F), Marker plot for hsp70l, a gene used for transgenic reporters of heat shock in zebrafish, shown as in (E). (G), Histogram showing no systemic activation of the heat shock response (HSR) across cell types at any of the temperatures profiled. While the total number of cell types with negligible HSR levels increases with temperature, nearly all cell types should have HSR signature values greater than 0 in the event of global activation of the HSR. (H), Dotplot of each cell type, ordered by the Pearson correlation between that cell type’s abundance and the whole-embryo pseudostage. Bellwether cell types with the highest correlations are labeled, with colors indicating tissue of origin. (I), Two examples from the highly correlated types shown in (A), with raw cell type abundance data plotted against embryo stage for a strong negatively correlated type (fin mesenchyme) and strong positively correlated type (differentiated neurons); inset at right shows additional examples.
Fig. S2. Temperature increases variability in specific cell types and stages of development, Related to Figure 2. (A), Scatterplot comparing whole-embryo staging determined by either cell composition (x-axis) or transcriptional data (y-axis); inset barplots show coefficient of variation for each metric in groups of embryos sampled at various timepoints (timepoints in hpf indicated in grey text above each bar). Loess fit lines are shown for embryos raised at each temperature, 28°C (grey), 32°C (pink), 34°C (red). (B), Stacked barchart showing how the number of cell types with significant abundance changes increases with temperature; this phenomenon held for all pseudostage bins (in color) except bin 4, which lacked sufficient control embryos to analyze statistically. (C), Scatterplot showing positive relationship of log2 fold change in abundances in both elevated temperatures, colored as in (A). (D), Histogram showing changes in cell composition resulting from 34°C treatment, as predicted by multinomial model. (E), Scatterplots showing raw data (black points) and model outputs of mean (solid line) and variability (ribbons with dotted line) for three temperature-sensitive cell types. (F), Heatmap of each cell type showing relative increases in observed variability compared to variability expected during wildtype development through developmental time (pseudostage bins); shades of red indicate increased variability while white to blue shades show neutral or decreased variability. The upper panel shows the average CVs for each temperature across all stages, highlighting pseudostages with increased variability at higher temperatures. (G), Barplot showing the log2 fold change in abundance for all cell types in 34°C embryos with severe phenotypes relative to those that were phenotypically normal.
Fig. S3. Temperature accelerates development non-uniformly across cell types, Related to Figure 3. (A), Schematic showing method for computing transcriptional age of each cell through nearest-neighbor averaging of time point labels in the reference. (B), Scatterplot as in Fig. 2B, but showing how well whole-embryo staging by transcriptional data (nn-transcriptional age), rather than cell composition, predicts temperature-induced developmental acceleration. (C), Boxplots showing individual examples of cell types with greater-than-expected acceleration (left) and less-than-expected acceleration (righta), with whole embryo expectation from Kimmel et al for each temperature shown as dotted lines.24 (D) Barplots showing the number of significantly (q < 0.05) differentially expressed genes in each cell type, separated by whether the gene has a significant association for developmental age (green) or temperature (red). Cell types along the x-axis are ordered by the difference between the number of significant temperature genes vs significant developmental age genes; the large majority show more genes changing with developmental age rather than temperature. (E) Scatterplot showing significant genes separated by developmental age and temperature effects for a specific cell type, the notochord. Temperature and age coefficients are shown for each gene, and are colored based on significant associations with each factor, neither, or both. (F), Density histogram showing that genes with significant associations with temperature tended to show higher expression levels than developmental age genes. (G) Density histogram of pairwise Pearson correlations for cell cycle signature values across all cell type pairs, with separate densities for each temperature. Inset shows boxplots of pairwise cell cycle correlations grouped into cell type pairs that showed lower or higher levels of developmental timing synchrony. (H), Scatterplot as in Fig. 3C, showing that the basal level of heat shock response signature in each cell type is not a strong predictor of that cell type’s developmental acceleration at 32°C or 34°C. (I), Barplots showing the level of heat shock response signature induction for all cell types at 34°C; cell types on the y-axis are ordered by relative developmental acceleration, with the most strongly accelerated types at the top, and columns for each timepoint. No induction of heat shock response signature is apparent in the most accelerated cell types.
Fig. S4. Cold treatment slows development and reveals cell type-specific temperature-responsive genes in the normal range of zebrafish development, Related to Figure 3. (A), Dotplot showing the sampling timing for cold-treated embryos (x-axis is clock time), as well as their predicted whole-embryo stage based on integration with the 28°C reference data; the predicted stage roughly matched the expectation of the Kimmel et al. (B), Boxplot showing the effect of 5μM cyclohexamide (CHX) on temperature-induced acceleration of whole-embryo stage; developmental delay is evident in CHX-treated embryos raised at 28°C, and this effect is not overridden by 34°C temperature treatment. (C), Scatterplot showing temperature-dependent activation of genes of the fast muscle, expressed as model coefficients for each temperature after accounting for developmental stage. Most genes showed either heat-induced increases and cold-induced decreases in expression (red box in upper-left), or heat-induced decreases and cold-induced increases in expression (blue box in lower left). (D), Scatterplot as in (C), but for slow muscle. (E), Scatterplot for fast muscle similar to (B), but showing cold vs cyclohexamide treatment, which shows an overall strong positive correlation with gene expression similarly affected by both sources of developmental delay. (F), Scatterplot as in (E), but for slow muscle.
Fig. S5. Transcriptional responses to elevated temperature tend to be cell type-specific, revealing burdens on proteostasis in the notochord, Related to Figure 4. (A), UMAP of all genes with significant temperature-dependent transcriptional changes (see Methods). Each point represents an individual gene, and genes with similar responses (coefficient in differential expression model after accounting for stage) across cell types are grouped together. Each cluster is labeled with a different color, and a representative GO term for the group of genes in each cluster is indicated. Clusters in which gene groups returned no significant GO terms are labeled ‘NA’. (B), Heatmap of expression levels for each gene module in (a) as an average for each cell type at elevated temperatures. Consistent with the heterogeneous grouping of temperature responses in (a), elevated module expression tends to be cell type-specific (examples boxed). Module 5, which contains genes related to translation and ER processing, has the most broad response across cell types, but the magnitude of the response is greatest within the notochord. (C), Scatterplot showing levels of UPR signature in the notochord cells of all individual embryos, colored by temperature. Embryos raised at higher temperature show higher levels of UPR signature at nearly all profiled stages. Lines and ribbons are derived from loess fits across pseudostage. (D), Boxplot showing marked reduction of notochord cells in embryos raised at high temperature, with raw counts in control and temperature-treated embryos shown as points. (E), Boxplot showing the effect of the UPR inhibitor ISRIB on whole-embryo stage at 28°C and 34°C; embryos treated with ISRIB showed significant increases over expected temperature-induced acceleration at 24hpf, but this effect was not evident in 48hpf embryos. (F), Hotspot analysis, as in Fig. 4D, showing enriched transcriptional state of notochord sheath cells in embryos treated with ISRIB; note that ISRIB-treated cells do not occupy the high-temperature state identified in wild-type embryos. (G), TEM of sheath cells in wild-type and atf6 crispants, showing effects in both untreated and temperature-treated embryos. ER structure is perturbed in atf6 crispants, and severe sheath defects are visible in the temperature-treated atf6 crispants. Scale bar = 200nm.
Table S2. Phenotypic severity classification for each embryo, related to Figure 2.
Methods S1. Protocol for individual-embryo sci-RNA-seq library preparation, Related to Figure 1.
Table S1. Cell abundance model outputs, Related to Figure 2. A) Model outputs from mean abundance analysis. B) Model outputs from variance analysis.
Table S3. Global analysis of temperature-induced gene expression changes across cell types, Related to Figure 4. A) Model coefficients for temperature-induced differential expression in each cell type. B) GO enrichments from differentially expressed genes. C) Genes used to compute signature scores.
Data Availability Statement
Single-cell RNA-seq data and raw sequencing are publicly available as of the date of publication via the NCBI Gene Expression Omnibus (GEO) accession GSE202294.
All original code, including notebooks for performing data processing, statistical analysis and generating plots, has been deposited at Github and is publicly available as of the date of publication: https://github.com/cole-trapnell-lab/sdg-zfish. Pipelines for generating count matrices from sci-RNA-seq3 sequencing data are available at https://github.com/bbi-lab/bbi-dmux and https://github.com/bbi-lab/bbi-sci. Analyses of the single cell transcriptome data were performed using Monocle3, which was updated to include methods from this study; a general tutorial can be found at http://cole-trapnell-lab.github.io/monocle-release/monocle3.
All materials used in this study are available by request from the corresponding author.


