Version Changes
Revised. Amendments from Version 1
The Background section has been edited to be more concise and focused.
Abstract
We present a genome assembly from an individual Zeus faber (the John Dory; Chordata; Actinopteri; Zeiformes; Zeidae). The genome sequence is 804.7 megabases in span. Most of the assembly is scaffolded into 22 chromosomal pseudomolecules. The mitochondrial genome has also been assembled and is 16.72 kilobases in length.
Keywords: Zeus faber, John Dory, genome sequence, chromosomal, Zeiformes
Species taxonomy
Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Deuterostomia; Chordata; Craniata; Vertebrata; Gnathostomata; Teleostomi; Euteleostomi; Actinopterygii; Actinopteri; Neopterygii; Teleostei; Osteoglossocephalai; Clupeocephala; Euteleosteomorpha; Neoteleostei; Eurypterygia; Ctenosquamata; Acanthomorphata; Paracanthopterygii; Zeiogadaria; Zeariae; Zeiformes; Zeidae; Zeus; Zeus faber Linnaeus, 1758 (NCBI:txid64108).
Background
Zeus faber Linnaeus, 1758, known as John Dory or St Peter’s fish, is a solitary, demersal marine fish with a laterally compressed, golden-brown body marked by a black spot on either side and long dorsal spines ( Wheeler, 1978). It is widely distributed in the eastern Atlantic, Mediterranean, Pacific and Indian Oceans, and along the entire West African coast, occurring at depths of 0–200 m ( Iwamoto, 2015; Maravelias et al., 2007). It has recently been recorded for the first time in the Black Sea ( Aydın & Karadurmuş, 2023). Its large, protrusible mouth and well-developed eyes enable it to prey on relatively large fish ( Kim et al., 2020; Stergiou & Fourtouni, 1991).
In the eastern Mediterranean, juveniles initially feed on zooplankton like mysids, then shift to small benthopelagic fishes as they grow, eventually preying on larger schooling pelagic species ( Kim et al., 2020; Stergiou & Fourtouni, 1991). In Korean coastal waters there is also varying diet composition with size and age ( Kim et al., 2020). Off the Portuguese coast, however, there is no prey switching from juvenile to adult life stages ( Silva, 1999). Z. faber is considered an opportunistic feeder, switching prey depending on food availability and abundance which can vary seasonally and with life stage ( Kim et al., 2020).
Z. faber is known to make ‘croaking’ or ‘barking’ noises upon capture onboard ( Radford et al., 2018). These vocalisations have since been documented in situ in Australia and were found to induce an escape response in conspecifics and heterospecifics such as the Australian Snapper ( Pagurus auratus), suggesting they make sounds as a territorial display against competitors ( Radford et al., 2018).
John Dory is commercially significant, valued for consumption, fish meal, and oil, and has a presence in the gamefish and aquarium trades ( Iwamoto, 2015). It is a key species in mixed trawl fisheries in the British Isles and a common by-catch globally ( Dunn, 2001; Iwamoto, 2015). The most recent stock assessment in the British Isles occurred between 1994 and 1996, focusing on landings and biological data from the English Channel ( Dunn, 2001). The English Channel appears to be a nursery ground, with seasonal peaks in landings correlating with recruitment during the third and fourth quarters, at approximately 23 cm TL ( Dunn, 2001). Most landed individuals range from 23–29 cm TL, with a maximum observed TL of 59 cm.
The IUCN assessed the global conservation status of Z. faber as ‘Data Deficient’ in 2013 ( Iwamoto, 2015), citing limited biological and historical data ( Dunn, 2001), which contributes to uncertainties in stock status and fishing pressure.
Molecular investigation of this species has shown significant genetic differentiation (7.44%) between clades in the North Atlantic/Mediterranean region and Australasia, indicating potential speciation ( Ward et al., 2008). The first genome of Z. faber was generated in 2016 for a study suggesting immune-related genes play an important role in teleost evolution and speciation ( Malmstrøm et al., 2016). This data note presents the second published genome of John Dory, collected and sequenced as part of the Darwin Tree of Life project ( Blaxter et al., 2022). This dataset will be important for furthering our understanding of teleost pathology, immunology, evolution and phylogenetics ( Malmstrøm et al., 2016; Ward et al., 2008).
Genome sequence report
The genome was sequenced from an individual Zeus faber ( Figure 1) collected from Bigbury Bay, UK (50.27, –3.97). A total of 43-fold coverage in Pacific Biosciences single-molecule HiFi long reads was generated. Primary assembly contigs were scaffolded with chromosome conformation Hi-C data. Manual assembly curation corrected 9 missing joins or mis-joins, reducing the scaffold number by 1.04%.
Figure 1. Photograph of the Zeus faber (fZeuFab8) specimen used for genome sequencing.
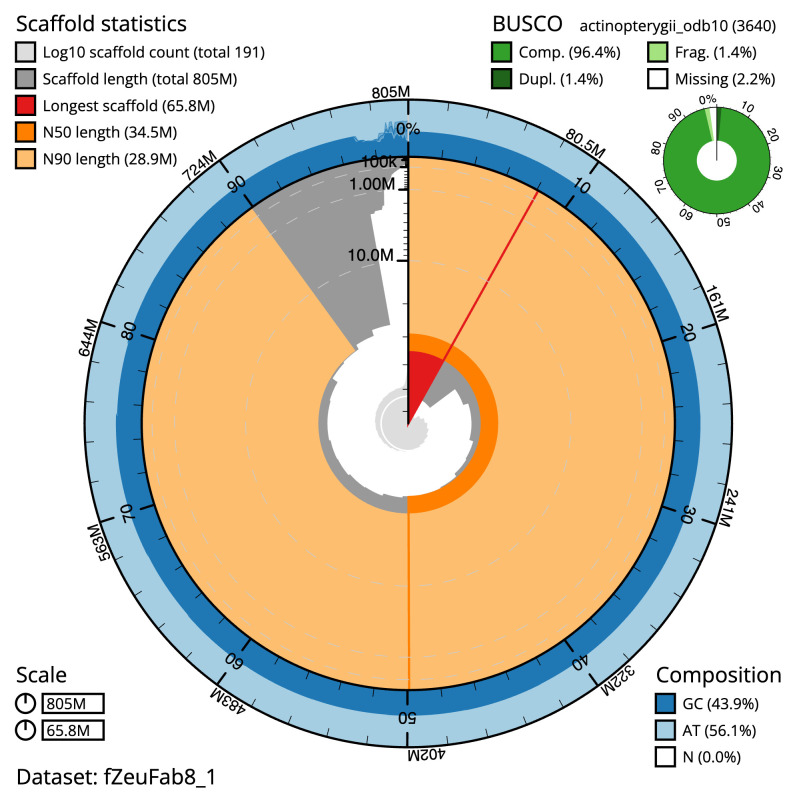
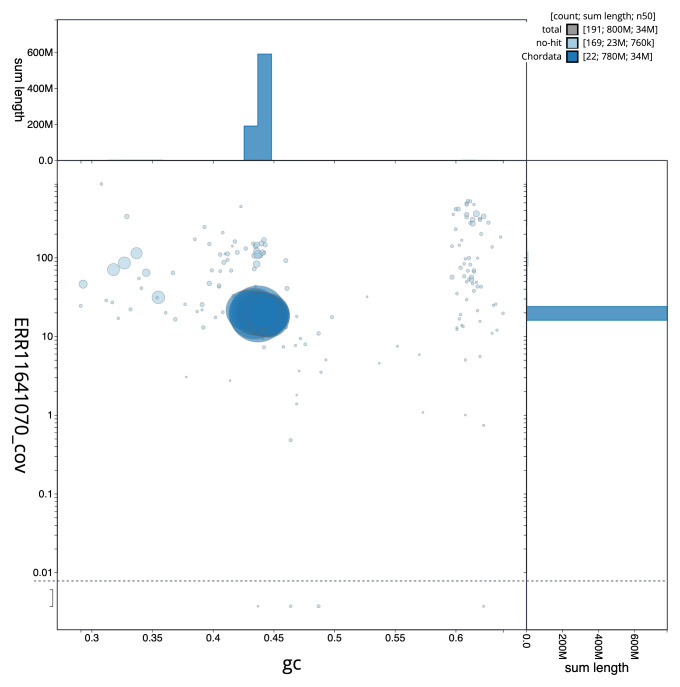
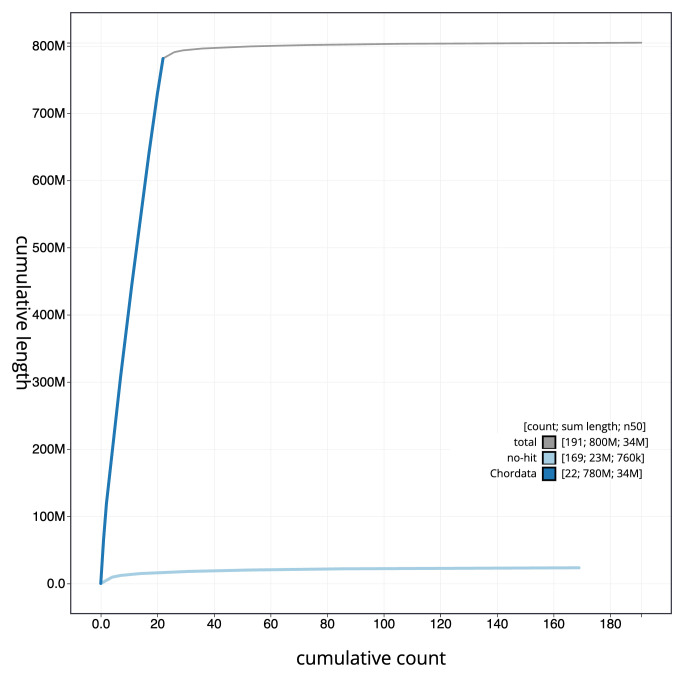
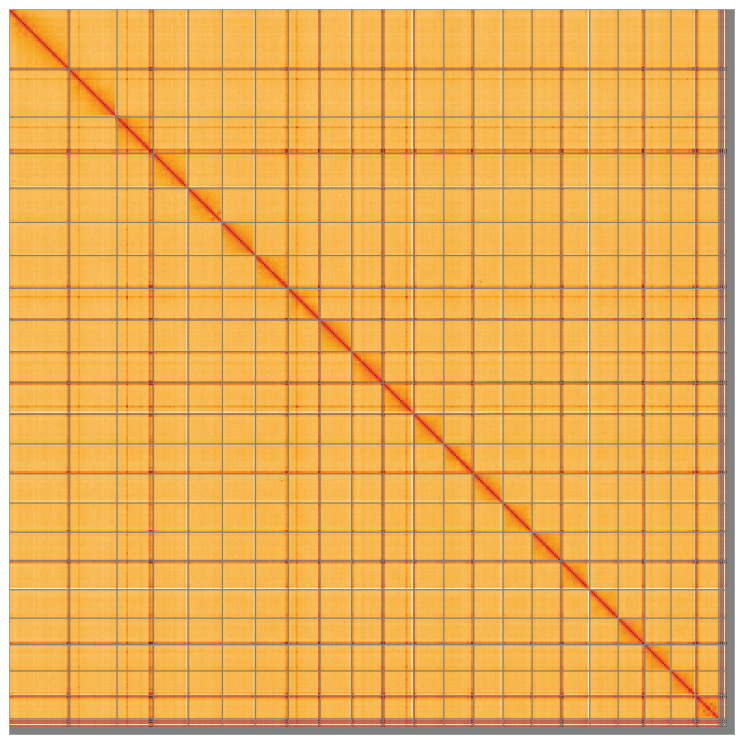
The final assembly has a total length of 804.7 Mb in 190 sequence scaffolds with a scaffold N50 of 34.5 Mb ( Table 1). The snail plot in Figure 2 provides a summary of the assembly statistics, while the distribution of assembly scaffolds on GC proportion and coverage is shown in Figure 3. The cumulative assembly plot in Figure 4 shows curves for subsets of scaffolds assigned to different phyla. Most (97.08%) of the assembly sequence was assigned to 22 chromosomal-level scaffolds. Chromosome-scale scaffolds confirmed by the Hi-C data are named in order of size ( Figure 5; Table 2). While not fully phased, the assembly deposited is of one haplotype. Contigs corresponding to the second haplotype have also been deposited. The mitochondrial genome was also assembled and can be found as a contig within the multifasta file of the genome submission.
Figure 2. Genome assembly of Zeus faber, fZeuFab8.1: metrics.
The BlobToolKit snail plot shows N50 metrics and BUSCO gene completeness. The main plot is divided into 1,000 size-ordered bins around the circumference with each bin representing 0.1% of the 804,731,948 bp assembly. The distribution of scaffold lengths is shown in dark grey with the plot radius scaled to the longest scaffold present in the assembly (65,762,550 bp, shown in red). Orange and pale-orange arcs show the N50 and N90 scaffold lengths (34,476,449 and 28,869,016 bp), respectively. The pale grey spiral shows the cumulative scaffold count on a log scale with white scale lines showing successive orders of magnitude. The blue and pale-blue area around the outside of the plot shows the distribution of GC, AT and N percentages in the same bins as the inner plot. A summary of complete, fragmented, duplicated and missing BUSCO genes in the actinopterygii_odb10 set is shown in the top right. An interactive version of this figure is available at https://blobtoolkit.genomehubs.org/view/fZeuFab8_1/dataset/fZeuFab8_1/snail.
Figure 3. Genome assembly of Zeus faber, fZeuFab8.1: BlobToolKit GC-coverage plot.
Sequences are coloured by phylum. Circles are sized in proportion to sequence length. Histograms show the distribution of sequence length sum along each axis. An interactive version of this figure is available at https://blobtoolkit.genomehubs.org/view/fZeuFab8_1/dataset/fZeuFab8_1/blob.
Figure 4. Genome assembly of Zeus faber, fZeuFab8.1: BlobToolKit cumulative sequence plot.
The grey line shows cumulative length for all sequences. Coloured lines show cumulative lengths of sequences assigned to each phylum using the buscogenes taxrule. An interactive version of this figure is available at https://blobtoolkit.genomehubs.org/view/fZeuFab8_1/dataset/fZeuFab8_1/cumulative.
Figure 5. Genome assembly of Zeus faber, fZeuFab8.1: Hi-C contact map of the fZeuFab8.1 assembly, visualised using HiGlass.
Chromosomes are shown in order of size from left to right and top to bottom. An interactive version of this figure may be viewed at https://genome-note-higlass.tol.sanger.ac.uk/l/?d=ODHdK-fnRfy3tLmo69JGwQ.
Table 1. Genome data for Zeus faber, fZeuFab8.1.
| Project accession data | ||
|---|---|---|
| Assembly identifier | fZeuFab8.1 | |
| Species | Zeus faber | |
| Specimen | fZeuFab8 | |
| NCBI taxonomy ID | 64108 | |
| BioProject | PRJEB63619 | |
| BioSample ID | SAMEA111562156 | |
| Isolate information | fZeuFab8 (DNA, Hi-C and RNA sequencing) | |
| Assembly metrics * | Benchmark | |
| Consensus quality (QV) | 52.6 | ≥ 50 |
| k-mer completeness | 99.98% | ≥ 95% |
| BUSCO ** | C:96.4%[S:94.9%,D:1.4%],
F:1.4%,M:2.2%,n:3,640 |
C ≥ 95% |
| Percentage of
assembly mapped to chromosomes |
97.08% | ≥ 95% |
| Sex chromosomes | None |
localised
homologous pairs |
| Organelles | Mitochondrial genome: 16.72 kb |
complete single
alleles |
| Raw data accessions | ||
| PacificBiosciences
SEQUEL II |
ERR11641070, ERR11641069 | |
| Hi-C Illumina | ERR11641144, ERR11641145 | |
| PolyA RNA-Seq Illumina | ERR11641143 | |
| Genome assembly | ||
| Assembly accession | GCA_960531495.1 | |
|
Accession of alternate
haplotype |
GCA_960530785.1 | |
| Span (Mb) | 804.7 | |
| Number of contigs | 1,078 | |
| Contig N50 length (Mb) | 1.4 | |
| Number of scaffolds | 190 | |
| Scaffold N50 length (Mb) | 34.5 | |
| Longest scaffold (Mb) | 65.76 | |
* Assembly metric benchmarks are adapted from column VGP-2020 of “Table 1: Proposed standards and metrics for defining genome assembly quality” from Rhie et al. (2021).
** BUSCO scores based on the actinopterygii_odb10 BUSCO set using version 5.3.2. C = complete [S = single copy, D = duplicated], F = fragmented, M = missing, n = number of orthologues in comparison. A full set of BUSCO scores is available at https://blobtoolkit.genomehubs.org/view/fZeuFab8_1/dataset/fZeuFab8_1/busco.
Table 2. Chromosomal pseudomolecules in the genome assembly of Zeus faber, fZeuFab8.
| INSDC
accession |
Chromosome | Length
(Mb) |
GC% |
|---|---|---|---|
| OY482845.1 | 1 | 65.76 | 43.5 |
| OY482846.1 | 2 | 52.95 | 43.5 |
| OY482847.1 | 3 | 40.01 | 43.5 |
| OY482848.1 | 4 | 38.03 | 43.5 |
| OY482849.1 | 5 | 37.98 | 44.0 |
| OY482850.1 | 6 | 36.47 | 44.5 |
| OY482851.1 | 7 | 36.1 | 44.0 |
| OY482852.1 | 8 | 35.01 | 44.0 |
| OY482853.1 | 9 | 34.68 | 44.5 |
| OY482854.1 | 10 | 34.48 | 44.0 |
| OY482855.1 | 11 | 33.96 | 43.5 |
| OY482856.1 | 12 | 33.1 | 44.0 |
| OY482857.1 | 13 | 32.89 | 44.5 |
| OY482858.1 | 14 | 32.3 | 43.5 |
| OY482859.1 | 15 | 32.12 | 44.5 |
| OY482860.1 | 16 | 31.67 | 44.5 |
| OY482861.1 | 17 | 31.65 | 42.5 |
| OY482862.1 | 18 | 31.31 | 44.5 |
| OY482863.1 | 19 | 29.26 | 44.0 |
| OY482864.1 | 20 | 28.87 | 44.5 |
| OY482865.1 | 21 | 27.29 | 45.0 |
| OY482866.1 | 22 | 25.41 | 44.0 |
| OY482867.1 | MT | 0.02 | 42.5 |
The estimated Quality Value (QV) of the final assembly is 52.6 with k-mer completeness of 99.98%, and the assembly has a BUSCO v5.3.2 completeness of 96.4% (single = 94.9%, duplicated = 1.4%), using the actinopterygii_odb10 reference set ( n = 3,640).
Metadata for specimens, barcode results, spectra estimates, sequencing runs, contaminants and pre-curation assembly statistics are given at https://links.tol.sanger.ac.uk/species/64108.
Methods
Sample acquisition and nucleic acid extraction
A Zeus faber specimen (specimen ID MBA-211116-004A, ToLID fZeuFab8) was collected from Bigbury Bay, UK (latitude 50.27, longitude –3.97) on 2021-11-16 using an otter trawl deployed from the RV Sepia. The collectors were Patrick Adkins, Joanna Harley, Rachel Brittain, Kesella Scott-Somme (all Marine Biological Association) and identified by Rachel Brittain, and then preserved in liquid nitrogen. The fish died as part of a trawl attached to another project and was opportunistically taken and dissected by the DToL team who were also on board the Sepia that day.
The workflow for high molecular weight (HMW) DNA extraction at the Wellcome Sanger Institute (WSI) includes a sequence of core procedures: sample preparation; sample homogenisation, DNA extraction, fragmentation, and clean-up. In sample preparation, the fZeuFab8 sample was weighed and dissected on dry ice ( Jay et al., 2023). Tissue was homogenised using a PowerMasher II tissue disruptor ( Denton et al., 2023a). HMW DNA was extracted in the WSI Scientific Operations core using the Automated MagAttract v2 protocol ( Oatley et al., 2023). The DNA was sheared into an average fragment size of 12–20 kb in a Megaruptor 3 system with speed setting 31 ( Bates et al., 2023). Sheared DNA was purified by solid-phase reversible immobilisation ( Strickland et al., 2023): in brief, the method employs a 1.8X ratio of AMPure PB beads to sample to eliminate shorter fragments and concentrate the DNA. The concentration of the sheared and purified DNA was assessed using a Nanodrop spectrophotometer and Qubit Fluorometer and Qubit dsDNA High Sensitivity Assay kit. Fragment size distribution was evaluated by running the sample on the FemtoPulse system.
RNA was extracted from tissue of fZeuFab8 in the Tree of Life Laboratory at the WSI using the RNA Extraction: Automated MagMax™ mirVana protocol ( do Amaral et al., 2023). The RNA concentration was assessed using a Nanodrop spectrophotometer and a Qubit Fluorometer using the Qubit RNA Broad-Range Assay kit. Analysis of the integrity of the RNA was done using the Agilent RNA 6000 Pico Kit and Eukaryotic Total RNA assay.
Protocols developed by the WSI Tree of Life laboratory are publicly available on protocols.io ( Denton et al., 2023b).
Sequencing
Pacific Biosciences HiFi circular consensus DNA sequencing libraries were constructed according to the manufacturers’ instructions. Poly(A) RNA-Seq libraries were constructed using the NEB Ultra II RNA Library Prep kit. DNA and RNA sequencing was performed by the Scientific Operations core at the WSI on Pacific Biosciences SEQUEL II (HiFi) and Illumina NovaSeq 6000 (RNA-Seq) instruments. Hi-C data were also generated from tissue of fZeuFab8 using the Arima2 kit and sequenced on the Illumina NovaSeq 6000, Illumina NovaSeq 6000 instrument.
Genome assembly, curation and evaluation
Assembly was carried out with Hifiasm ( Cheng et al., 2021) and haplotypic duplication was identified and removed with purge_dups ( Guan et al., 2020). The assembly was then scaffolded with Hi-C data ( Rao et al., 2014) using YaHS ( Zhou et al., 2023). The assembly was checked for contamination and corrected as described previously ( Howe et al., 2021). Manual curation was performed using HiGlass ( Kerpedjiev et al., 2018) and PretextView ( Harry, 2022). The mitochondrial genome was assembled using MitoHiFi ( Uliano-Silva et al., 2023), which runs MitoFinder ( Allio et al., 2020) or MITOS ( Bernt et al., 2013) and uses these annotations to select the final mitochondrial contig and to ensure the general quality of the sequence.
A Hi-C map for the final assembly was produced using bwa-mem2 ( Vasimuddin et al., 2019) in the Cooler file format ( Abdennur & Mirny, 2020). To assess the assembly metrics, the k-mer completeness and QV consensus quality values were calculated in Merqury ( Rhie et al., 2020). This work was done using Nextflow ( Di Tommaso et al., 2017) DSL2 pipelines “sanger-tol/readmapping” ( Surana et al., 2023a) and “sanger-tol/genomenote” ( Surana et al., 2023b). The genome was analysed within the BlobToolKit environment ( Challis et al., 2020) and BUSCO scores ( Manni et al., 2021; Simão et al., 2015) were calculated.
Table 3 contains a list of relevant software tool versions and sources.
Table 3. Software tools: versions and sources.
| Software
tool |
Version | Source |
|---|---|---|
| BlobToolKit | 4.1.7 | https://github.com/blobtoolkit/blobtoolkit |
| BUSCO | 5.3.2 | https://gitlab.com/ezlab/busco |
| Hifiasm | 0.19.5-r587 | https://github.com/chhylp123/hifiasm |
| HiGlass | 1.11.6 | https://github.com/higlass/higlass |
| Merqury | MerquryFK | https://github.com/thegenemyers/MERQURY.FK |
| MitoHiFi | 3 | https://github.com/marcelauliano/MitoHiFi |
| PretextView | 0.2 | https://github.com/wtsi-hpag/PretextView |
| purge_dups | 1.2.5 | https://github.com/dfguan/purge_dups |
| sanger-tol/
genomenote |
v1.0 | https://github.com/sanger-tol/genomenote |
| sanger-tol/
readmapping |
1.1.0 | https://github.com/sanger-tol/readmapping/tree/1.1.0 |
| YaHS | 1.2a.2 | https://github.com/c-zhou/yahs |
Wellcome Sanger Institute – Legal and Governance
The materials that have contributed to this genome note have been supplied by a Darwin Tree of Life Partner. The submission of materials by a Darwin Tree of Life Partner is subject to the ‘Darwin Tree of Life Project Sampling Code of Practice’, which can be found in full on the Darwin Tree of Life website here. By agreeing with and signing up to the Sampling Code of Practice, the Darwin Tree of Life Partner agrees they will meet the legal and ethical requirements and standards set out within this document in respect of all samples acquired for, and supplied to, the Darwin Tree of Life Project.
Further, the Wellcome Sanger Institute employs a process whereby due diligence is carried out proportionate to the nature of the materials themselves, and the circumstances under which they have been/are to be collected and provided for use. The purpose of this is to address and mitigate any potential legal and/or ethical implications of receipt and use of the materials as part of the research project, and to ensure that in doing so we align with best practice wherever possible. The overarching areas of consideration are:
• Ethical review of provenance and sourcing of the material
• Legality of collection, transfer and use (national and international)
Each transfer of samples is further undertaken according to a Research Collaboration Agreement or Material Transfer Agreement entered into by the Darwin Tree of Life Partner, Genome Research Limited (operating as the Wellcome Sanger Institute), and in some circumstances other Darwin Tree of Life collaborators.
Funding Statement
This work was supported by Wellcome through core funding to the Wellcome Sanger Institute [206194, <a href=https://doi.org/10.35802/206194>https://doi.org/10.35802/206194</a>] and the Darwin Tree of Life Discretionary Award [218328, <a href=https://doi.org/10.35802/218328>https://doi.org/10.35802/218328 </a>].
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 2; peer review: 2 approved, 1 not approved]
Data availability
European Nucleotide Archive: Zeus faber (John dory). Accession number PRJEB63619; https://identifiers.org/ena.embl/PRJEB63619 ( Wellcome Sanger Institute, 2023). The genome sequence is released openly for reuse. The Zeus faber genome sequencing initiative is part of the Darwin Tree of Life (DToL) project. All raw sequence data and the assembly have been deposited in INSDC databases. The genome will be annotated using available RNA-Seq data and presented through the Ensembl pipeline at the European Bioinformatics Institute. Raw data and assembly accession identifiers are reported in Table 1.
Author information
Members of the Marine Biological Association Genome Acquisition Lab are listed here: https://doi.org/10.5281/zenodo.8382513.
Members of the Darwin Tree of Life Barcoding collective are listed here: https://doi.org/10.5281/zenodo.4893703.
Members of the Wellcome Sanger Institute Tree of Life Management, Samples and Laboratory team are listed here: https://doi.org/10.5281/zenodo.10066175.
Members of Wellcome Sanger Institute Scientific Operations: Sequencing Operations are listed here: https://doi.org/10.5281/zenodo.10043364.
Members of the Wellcome Sanger Institute Tree of Life Core Informatics team are listed here: https://doi.org/10.5281/zenodo.10066637.
Members of the Tree of Life Core Informatics collective are listed here: https://doi.org/10.5281/zenodo.5013541.
Members of the Darwin Tree of Life Consortium are listed here: https://doi.org/10.5281/zenodo.4783558.
References
- Abdennur N, Mirny LA: Cooler: scalable storage for Hi-C data and other genomically labeled arrays. Bioinformatics. 2020;36(1):311–316. 10.1093/bioinformatics/btz540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allio R, Schomaker-Bastos A, Romiguier J, et al. : MitoFinder: efficient automated large-scale extraction of mitogenomic data in target enrichment phylogenomics. Mol Ecol Resour. 2020;20(4):892–905. 10.1111/1755-0998.13160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aydin M, Karadurmus U: First record of the benthopelagic fish John dory Zeus faber (Linnaeus, 1758) in the Black Sea coasts of Türkiye. Aquat Res. 2023;6(2):159–165. 10.3153/AR23016 [DOI] [Google Scholar]
- Bates A, Clayton-Lucey I, Howard C: Sanger Tree of Life HMW DNA fragmentation: diagenode Megaruptor ®3 for LI PacBio. protocols.io. 2023. 10.17504/protocols.io.81wgbxzq3lpk/v1 [DOI] [Google Scholar]
- Bernt M, Donath A, Jühling F, et al. : MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 2013;69(2):313–319. 10.1016/j.ympev.2012.08.023 [DOI] [PubMed] [Google Scholar]
- Blaxter M, Mieszkowska N, Di Palma F, et al. : Sequence locally, think globally: the Darwin Tree of Life project. Proc Natl Acad Sci U S A. 2022;119(4): e2115642118. 10.1073/pnas.2115642118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challis R, Richards E, Rajan J, et al. : BlobToolKit - interactive quality assessment of genome assemblies. G3 (Bethesda). 2020;10(4):1361–1374. 10.1534/g3.119.400908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Concepcion GT, Feng X, et al. : Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat Methods. 2021;18(2):170–175. 10.1038/s41592-020-01056-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denton A, Oatley G, Cornwell C, et al. : Sanger Tree of Life sample homogenisation: PowerMash. protocols.io. 2023a. 10.17504/protocols.io.5qpvo3r19v4o/v1 [DOI] [Google Scholar]
- Denton A, Yatsenko H, Jay J, et al. : Sanger Tree of Life wet laboratory protocol collection V.1. protocols.io. 2023b. 10.17504/protocols.io.8epv5xxy6g1b/v1 [DOI] [Google Scholar]
- Di Tommaso P, Chatzou M, Floden EW, et al. : Nextflow enables reproducible computational workflows. Nat Biotechnol. 2017;35(4):316–319. 10.1038/nbt.3820 [DOI] [PubMed] [Google Scholar]
- do Amaral RJV, Bates A, Denton A, et al. : Sanger Tree of Life RNA Extraction: Automated MagMax TM mirVana. protocols.io. 2023. 10.17504/protocols.io.6qpvr36n3vmk/v1 [DOI] [Google Scholar]
- Dunn M: The biology and exploitation of John dory, Zeus faber (Linnaeus, 1758) in the waters of England and Wales. ICES J Mar Sci. 2001;58(1):96–105. 10.1006/jmsc.2000.0993 [DOI] [Google Scholar]
- Guan D, McCarthy SA, Wood J, et al. : Identifying and removing haplotypic duplication in primary genome assemblies. Bioinformatics. 2020;36(9):2896–2898. 10.1093/bioinformatics/btaa025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harry E: PretextView (Paired REad TEXTure Viewer): a desktop application for viewing pretext contact maps.2022; [Accessed 19 October 2022]. Reference Source
- Howe K, Chow W, Collins J, et al. : Significantly improving the quality of genome assemblies through curation. GigaScience. Oxford University Press,2021;10(1): giaa153. 10.1093/gigascience/giaa153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto T: Zeus faber, The IUCN Red List of Threatened Species.2015; [Accessed 2 February 2024]. 10.2305/IUCN.UK.2015-4.RLTS.T198769A42390771.en [DOI] [Google Scholar]
- Jay J, Yatsenko H, Narváez-Gómez JP, et al. : Sanger Tree of Life sample preparation: triage and dissection. protocols.io. 2023. 10.17504/protocols.io.x54v9prmqg3e/v1 [DOI] [Google Scholar]
- Kerpedjiev P, Abdennur N, Lekschas F, et al. : HiGlass: web-based visual exploration and analysis of genome interaction maps. Genome Biol. 2018;19(1): 125. 10.1186/s13059-018-1486-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Kim HG, Oh CW: Diet composition and feeding strategy of John Dory, Zeus faber, in the coastal waters of Korea. J Ecol Environ. 2020;44(1): 8. 10.1186/s41610-020-00153-y [DOI] [Google Scholar]
- Malmstrøm M, Matschiner M, Tørresen OK, et al. : Evolution of the immune system influences speciation rates in teleost fishes. Nat Genet. 2016;48(10):1204–1210. 10.1038/ng.3645 [DOI] [PubMed] [Google Scholar]
- Manni M, Berkeley MR, Seppey M, et al. : BUSCO update: novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes. Mol Biol Evol. 2021;38(10):4647–4654. 10.1093/molbev/msab199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maravelias CD, Tsitsika EV, Papaconstantinou C: Seasonal dynamics, environmental preferences and habitat selection of John Dory ( Zeus faber). Estuar Coast Shelf S. 2007;72(4):703–710. 10.1016/j.ecss.2006.12.002 [DOI] [Google Scholar]
- Oatley G, Denton A, Howard C: Sanger Tree of Life HMW DNA extraction: Automated MagAttract v.2. protocols.io. 2023. 10.17504/protocols.io.kxygx3y4dg8j/v1 [DOI] [Google Scholar]
- Radford CA, Putland RL, Mensinger AF: Barking mad: the vocalisation of the John Dory, Zeus faber. PLoS One. 2018;13(10): e0204647. 10.1371/journal.pone.0204647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SSP, Huntley MH, Durand NC, et al. : A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159(7):1665–1680. 10.1016/j.cell.2014.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhie A, McCarthy SA, Fedrigo O, et al. : Towards complete and error-free genome assemblies of all vertebrate species. Nature. 2021;592(7856):737–746. 10.1038/s41586-021-03451-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhie A, Walenz BP, Koren S, et al. : Merqury: Reference-free quality, completeness, and phasing assessment for genome assemblies. Genome Biol. 2020;21(1): 245. 10.1186/s13059-020-02134-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva A: Feeding habits of John Dory, Zeus faber, off the Portuguese continental coast. J Mar Biol Assoc U K. 1999;79(2):333–340. 10.1017/S002531549800037X [DOI] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, et al. : BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31(19):3210–3212. 10.1093/bioinformatics/btv351 [DOI] [PubMed] [Google Scholar]
- Stergiou KI, Fourtouni H: Food habits, ontogenetic diet shift and selectivity in Zeus faber Linnaeus, 1758. J Fish Biol. 1991;39(4):589–603. 10.1111/j.1095-8649.1991.tb04389.x [DOI] [Google Scholar]
- Strickland M, Cornwell C, Howard C: Sanger Tree of Life fragmented DNA clean up: manual SPRI. protocols.io. 2023. 10.17504/protocols.io.kxygx3y1dg8j/v1 [DOI] [Google Scholar]
- Surana P, Muffato M, Qi G: sanger-tol/readmapping: sanger-tol/readmapping v1.1.0 - Hebridean Black (1.1.0). Zenodo. 2023a. 10.5281/zenodo.7755669 [DOI] [Google Scholar]
- Surana P, Muffato M, Sadasivan Baby C: sanger-tol/genomenote (v1.0.dev). Zenodo. 2023b. 10.5281/zenodo.6785935 [DOI] [Google Scholar]
- Uliano-Silva M, Ferreira JGRN, Krasheninnikova K, et al. : MitoHiFi: a python pipeline for mitochondrial genome assembly from PacBio high fidelity reads. BMC Bioinformatics. 2023;24(1): 288. 10.1186/s12859-023-05385-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasimuddin M, Misra S, Li H, et al. : Efficient architecture-aware acceleration of BWA-MEM for multicore systems. In: 2019 IEEE International Parallel and Distributed Processing Symposium (IPDPS).IEEE,2019;314–324. 10.1109/IPDPS.2019.00041 [DOI] [Google Scholar]
- Ward R, Costa F, Holmes B, et al. : DNA barcoding of shared fish species from the North Atlantic and Australasia: minimal divergence for most taxa, but Zeus faber and Lepidopus caudatus each probably constitute two species. Aquat Biol. 2008;3(1):71–78. 10.3354/ab00068 [DOI] [Google Scholar]
- Wellcome Sanger Institute: The genome sequence of the John Dory, Zeus faber Linnaeus, 1758. European Nucleotide Archive, [dataset], accession number PRJEB63619,2023.
- Wheeler A: Key to the Fishes of Northern Europe: a guide to the identification of more than 350 species. London: Frederick Warne (Publishers) Ltd,1978. Reference Source [Google Scholar]
- Zhou C, McCarthy SA, Durbin R: YaHS: yet another Hi-C scaffolding tool. Bioinformatics. 2023;39(1): btac808. 10.1093/bioinformatics/btac808 [DOI] [PMC free article] [PubMed] [Google Scholar]