Abstract
Clink, a 20-kDa protein of faba bean necrotic yellows virus, a single-stranded DNA plant virus, interacts with pRB family members and a SKP1 homologue from Medicago sativa. An LxCxE motif and an F-box of Clink mediate the interactions with the respective proteins. The capacity of Clink to bind pRB correlates with its ability to stimulate viral replication. Interaction of a single protein with the cell cycle regulator pRB and SKP1, a constituent of the ubiquitin-protein turnover pathway, appears to be a novel feature. Hence, Clink may represent a new class of viral cell cycle modulators.
A common strategy of DNA viruses is the creation of an environment favorable for efficient replication of their genome by subverting the cell cycle control of the host and forcing cells into DNA synthesis or S phase. In mammalian cells, a key cell cycle regulator is the retinoblastoma tumor suppressor protein pRB, which represses onset and progression into S phase by interacting with a wide range of cell cycle-related proteins. Among those are transcription factors of the E2F family that form complexes with hypophosphorylated pRB. During the G1/S transition, pRB is progressively phosphorylated by the action of cyclin-dependent kinases, and as a result, E2F is released from the complex and becomes available to activate the expression of S-phase-specific genes (14, 30).
Oncoproteins of certain mammalian DNA tumor viruses, such as E1A of adenovirus type 6, E7 of human papillomavirus type 16, or the large T antigen protein of simian virus 40, stimulate the entry of cells into S phase by interaction with pRB through a short protein sequence comprising essentially the sequence motif LxCxE (10, 45). This interaction abrogates the pRB-mediated block of cell cycle progression and may contribute to tumor formation (24). In addition, these or other viral proteins are involved in the neutralization of additional cell cycle regulators, in particular of the growth suppressor p53 (24, 39). The interaction between the papillomavirus E7 and E6 proteins with pRB and p53, respectively, mediates the degradation of the latter by the ubiquitin-proteasome pathway (8, 25, 44). Hence, in addition to the interaction with growth suppressors, papillomaviruses make use of the protein degradation machinery to target these proteins to the 26S proteasome.
Ubiquitination of proteins destined for degradation by the 26S proteasome is mediated by the action of the enzymatic complexes E1, E2, and E3 (21). E1 and E2 activate ubiquitin and catalyze the polyubiquitination of the substrate, which is thus marked for degradation by the 26S proteasome. A diverse class of complexes, the E3 ubiquitin-ligases, contains the elements of specificity for the substrates to be ubiquitinated. The core components of one particularly versatile class of E3s, designated SCFs, are SKP1, Cdc53/Cullin, and RBX/ROC1, which assemble with different F-box proteins (31, 33). The F-box, which mediates binding to SKP1, is a conserved domain found in a large number of proteins (4). F-box proteins serve as substrate-specific adapter subunits to recruit various substrates to a core ubiquitination complex. Characterization of evolutionarily conserved SCF complexes has revealed the importance of the ubiquitin 26S proteasome pathway to the G1/S transition of the cell cycle (2, 4, 33, 40).
Geminiviruses and nanoviruses are single-stranded DNA (ssDNA) viruses of plants with exclusively nuclear replication cycles (6, 11). Their genome comprises one or two circular DNAs (2.7 to 3.0 kb) for geminiviruses (35) and until now an undetermined number (at least six) of circular ssDNAs (all of about 1 kb) for nanoviruses (7, 9, 28, 42). Both groups of viruses encode replication initiator proteins (Rep proteins) that act as sequence-specific origin recognition endonucleases and trigger replication initiation of the virus genome (18, 34). These viruses exploit the DNA synthesis of the host, and geminivirus infection induces the accumulation of proliferating-cell nuclear antigen in differentiated plant cells (38). Moreover, it was shown that a geminivirus Rep protein interacts via an LxCxE motif with a pRB family member (51). This suggests that mammalian tumor viruses and plant geminiviruses have common cellular targets and employ similar strategies to modulate the host cell cycle control. Indications for the conservation of cell cycle-regulating proteins in animal and plant cells stem from the cloning of several plant pRB homologues, cyclins, and cyclin-dependent kinases (16, 20), as well as of components of the ubiquitin-dependent proteolytic pathway (13, 48).
The nanovirus faba bean necrotic yellows virus (FBNYV) encodes five different replication initiator proteins, each of them on an individual circular DNA (27, 28). None of these Rep proteins possesses a pRB-binding motif. However, a 20-kDa protein encoded by FBNYV genome component 10 (C10) contains an LxCxE motif (28). Here we show that in addition to a pRB-binding motif, the FBNYV C10 protein contains an F-box and binds to MsSKP1, a plant SKP1 homologue which we identified in alfalfa (Medicago sativa). The C10 protein is capable of binding to members of the pRB family, and this interaction correlates with a stimulation of viral DNA replication. To the best of our knowledge, the combination of a pRB interaction motif and an F-box on a single protein is without precedent. For its potential to link viral DNA replication with key regulatory pathways of the cell cycle, we named the FBNYV C10 protein Clink, for “cell cycle link.”
MATERIALS AND METHODS
Plasmid constructions.
Purification of FBNYV DNA from infected Vicia faba and cloning of full-length Rep- and Clink-encoding genome components have been described elsewhere (27, 28, 46). The clink gene of FBNYV C10 was amplified by PCR with Pfu polymerase (Stratagene) using primer pair C10BamHI (ACAGGATCCATGGGTCTGAAATATTTC) and C10SalI (TACGTCGACTCAACTAATAACAATATC). The amplified DNA was digested by BamHI and SalI (sites included in the respective primers) and inserted into the corresponding sites of plasmids pQE30 (Qiagen) and pK18 (43) to generate plasmids pQE30-C10 and pK18-C10, respectively. An EcoRI-SalI fragment from pK18-C10 containing the clink gene was transferred into the corresponding sites of plasmids pGBT9 (Clontech) (GAL4 DNA-BD vector) and pBD-GAL4 Cam (Stratagene), giving rise to plasmids pGBT9-Clink and pBD-Clink, respectively.
Plasmids pGEX-RB and pGEX-RBC706F contain the coding sequence of human pRB (amino acids 379 to 928) (26), and plasmid pGAD424-ZmRb1 consists of the activation domain of Gal4 fused to ZmRb1 (50).
The entire coding sequence of MsSKP1 was excised as an EcoRI-XhoI fragment from pAD-MsSKP1 and inserted between the EcoRI and XhoI sites of pGEX-4T-1 (Pharmacia), generating plasmid pGEX-SKP1.
Site-directed mutagenesis.
Mutagenesis was performed using the Quik Change site-directed mutagenesis kit (Stratagene) on plasmid pBSKII:C10 carrying the full-length FBNYV component 10 (46) or on plasmid pK18-C10 carrying the clink gene. To change the Clink LxCxE motif (from amino acids LSCRE to LSRRA), we used primer pair C10C112R, E114A(+) (GACGACTTATCTAGACGTGCATTACTGCCG) and C10C112R, E114A(−) (CGGCAGTAATGCACGTCTAGATAAGTCGTC). Mutagenesis of the F-box was carried out using primer pair C10P10L(+) (CTCTCATCTTCTCGAGGAGCTTAG) and C10P10L(−) (CTAAGCTCCTCGAGAAGATGAGAG) to change the proline codon into leucine and primer pair C10I17A(+) (CTTAGAGAGAAGGCTGTGCACGATCATC) and C10I17A(−) (GATGATCGTGCACAGCCTTCTCTCTAAG) to change the isoleucine codon into alanine.
All PCRs performed to amplify wild-type clink for subsequent cloning or to introduce point mutations have been verified by complete sequencing of the gene.
Dimers of mutated full-length C10 DNA were inserted into pBin19 in the same way as described for wild-type C10 (46). The mutated clink genes from plasmids pK18-C10C112R, E114A, pK18-C10P10L, and pK18-C10I17A were transferred into plasmids pQE30 and pGBT9 as described for wild-type clink.
Protein-protein interactions in vitro.
Expression of the glutathione S-transferase (GST) and His6 fusion proteins encoded by pGEX and pQE30 plasmid derivatives respectively, took place in a derivative of Escherichia coli strain BL21 essentially as described previously (46) with slight modifications. Bacteria were harvested, resuspended in NETN buffer (100 mM NaCl, 1 mM EDTA, 20 mM Tris-HCl [pH 8.0], 0.5% Nonidet P-40, 1 mM phenylmethylsulfonyl fluoride, 0.1 KI unit of aprotinin, 1 mM dithiothreitol). After sonication and clarification by centrifugation, the protein supernatant from bacteria expressing GST fusion proteins was incubated for 2 h at 4°C with glutathione-Sepharose beads (Sigma). The beads were washed and further incubated for 2 h with a protein supernatant from bacteria expressing a His6 fusion protein. After being washed, the beads were resuspended in Laemmli sample buffer and boiled. Proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to Hybond-C Extra membranes (Amersham). Immunoblotting and detection of the proteins were performed as described previously (3), using anti-pRB (PharMingen), anti-GST (Pharmacia), and anti-His6 (Qiagen) as primary antibodies.
Yeast two-hybrid assays.
The yeast strain Y166 (MATa leu2-3,112 ura3-52 trp1-901 his3-200 ade2-101 gal4-542 gal80-538 GAL-lacZ GAL-URA3) (a gift of S. J. Elledge, Baylor College of Medicine, Houston, Tex.) contains the reporter genes lacZ and URA3 inducible by the Gal4 transcription factor. Yeast cotransformation with bait and prey plasmids was performed as described in the Clontech MATCHMAKER two-hybrid system protocol. The transformation mixture was plated on selection medium devoid of or supplemented with uracil. To confirm the interaction between the two fusion proteins, β-galactosidase activity was tested by a colorimetric assay in extracts of yeast with o-nitrophenyl-β-d-galactopyranoside (ONPG) as the substrate. One unit of β-galactosidase is defined as the amount of activity hydrolyzing 1 nmol of ONPG per min per mg of total protein.
The M. sativa cDNA expression library was constructed using the HybriZAP Two Hybrid cloning kit (Stratagene) and used as prey in the yeast two-hybrid assay. cDNA synthesis of RNA from a M. sativa cv. RegenS cell suspension (RA3) was primed by a 3′ oligo-(dT)–XhoI adapter primer. After second-strand synthesis, an EcoRI adapter was ligated to the cDNA, which was subsequently digested by XhoI, size fractionated to eliminate molecules of <400 bp, and directionally cloned in the EcoRI-XhoI-predigested HybriZAP phagemid. About 23.4 × 106 primary phages were excised in vivo and harvested in 30 independent lots to generate the pAD-GAL4 expression library. The average insert size was 800 to 1,000 bp, and 120 μg of the library DNA was used to transform the yeast strain Y166 carrying plasmid pBD-Clink as bait.
Replication of FBNYV DNA in N. benthamiana.
Viral DNA replication was assayed in Nicotiana benthamiana leaf discs following agroinoculation as described previously (46). Leaf discs were kept on Murashige and Skoog (Sigma) agar plates supplemented (or not) with 0.1 mg of 1-naphthaleneacetic acid per liter and 1 mg of 6-benzylaminopurine per liter. Several days postinoculation, total DNA from leaf tissue was isolated and fractionated on agarose gels. Replicative forms of viral DNA were identified by Southern hybridization (3) using 32P-labeled rep component-specific probes.
Nucleotide sequence accession number.
MsSKP1 cDNA sequence has been assigned GenBank accession no. AF135596.
RESULTS
FBNYV Clink interacts with pRB family proteins through the LxCxE motif.
FBNYV Clink protein and its homologues in other nanoviruses (Fig. 1) contain an LxCxE motif, a sequence found to be essential for the interaction of animal virus oncoproteins with pRB (10). Therefore, we investigated whether Clink could interact with members of the pRB family.
FIG. 1.
Amino acid sequences of FBNYV Clink and its homologues in other nanoviruses. The GenBank accession numbers of the corresponding genomic DNAs are as follows: banana bunchy top virus (BBTV), L41518; FBNYV, AJ132187; milk vetch dwarf virus (MDV), AB000923; subterranean clover stunt virus (SCSV), U16732. Comparison was done using the PileUp program of the University of Wisconsin Genetics Computer Group. Conserved amino acids of the pRB-binding and F-box motifs are marked in black. Amino acids of FBNYV Clink altered by mutation are indicated by asterisks.
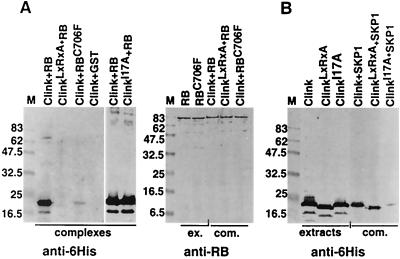
The interaction between Clink and human pRB was tested in vitro using a protein-binding assay involving a GST fusion protein of pRB (GST-pRB) and Clink tagged by an amino-terminal hexahistidine extension (His6-Clink). GST-pRB was bound to glutathione-Sepharose beads, which were then incubated with protein extracts of bacteria expressing His6-Clink. Proteins from the complexes retained by the beads were identified by Western blotting using antibodies to pRB or to the hexahistidine tag of Clink. Results of a typical interaction assay (Fig. 2A) revealed that Clink was bound via a complex with pRB to glutathione-Sepharose. The interaction between these two proteins was strongly reduced when either Clink in its LxCxE motif (ClinkC112R, E114A) or pRB in its pocket domain (pRBC706F) (26) was mutated. These results showed that the interaction was specific and depended on a functional LxCxE sequence of Clink and on an intact pocket domain of pRB.
FIG. 2.
In vitro interaction of Clink and its mutants with human pRB and MsSKP1 proteins. Western blots of protein complexes retained by glutathione-Sepharose beads incubated with different combinations of bacterial extracts expressing wild-type and mutant His6-Clink and GST-pRB fusion proteins (A) or His6-Clink and GST-MsSKP1 fusion proteins (B) are shown. The antibodies used are indicated at the bottom, and the respective proteins or protein combinations are indicated at the top. Lanes loaded with crude extracts from bacteria are labeled “extracts” or “ex.,” and those loaded with complexes retained on the beads are labeled “complexes” or “com.” The term “ClinkLxRxA” stands for ClinkC112R, E114A. Molecular masses of marker proteins (M) are indicated on the left in kilodaltons. The expected sizes of the fusion proteins are 44.3 kDa for GST-MsSKP1, 91 kDa for GST-pRB, and 21.1 kDa for His6-Clink. Even though wild-type and two mutated Clink proteins have nearly identical molecular masses, faster migration of the ClinkLxRxA protein is observed and may be due to a higher isoelectric point resulting from the amino acid substitutions (6.28 for wild-type Clink and 6.58 for ClinkLxRxA).
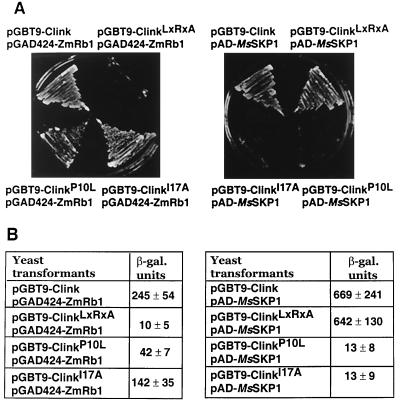
To corroborate the results of the in vitro assays, the yeast two-hybrid system was used to show the interaction between Clink and the plant pRB homologue ZmRb1 from maize. Activation of the two reporter genes URA3 and lacZ was observed only in yeast transformants expressing wild-type Clink- and ZmRb1-GAL4 fusion proteins. No activation of the reporter genes was observed in yeast transformed by a plasmid encoding ZmRb1-Gal4 along with a plasmid encoding ClinkC112R, E114A-Gal4 fusion protein (Fig. 3). Thus, mutations in the LxCxE motif of Clink also abolished the interaction with ZmRb1 in the yeast two-hybrid assay, providing additional and independent evidence that these amino acids are essential for complex formation.
FIG. 3.
Interaction of Clink and its mutants with ZmRb1 and MsSKP1 proteins in yeast. (A) Growth of double transformants on selective medium lacking uracil. The respective combinations of plasmids encoding fusion proteins with the GAL4 DNA-binding domain (pGBT9 derivatives) and fusion proteins with the GAL4 activation domain (pAD and pGAD424 derivatives) are indicated. (B) β-Galactosidase (β-gal.) activity of yeast extracts containing the indicated plasmid combinations. Each value is the mean of at least three independent transformants.
Clink interacts with a plant SKP1 homologue via an F-box motif.
To identify plant proteins that interact with FBNYV Clink, we used the yeast two-hybrid system to screen an M. sativa cDNA expression library fused to the Gal4 activation domain-encoding sequence, using pBD-Clink as bait. A total of 1.2 × 107 transformants were assayed for uracil-independent growth. Of these transformants, 41 were able to activate both URA3 and lacZ reporter genes. Plasmids from five transformants were isolated, and the DNA sequence of the inserts was determined. The deduced amino acid sequences derived from the five M. sativa cDNAs were almost identical and showed similarity to members of the SKP1 protein family. They were named MsSKP1, and the sequence of the cDNA used for further studies has been deposited in the EMBL and GenBank databases. Of the remaining 36 transformants capable of uracil-independent growth, 31 contained SKP1 cDNA sequences as identified by PCR using vector-specific (T7) and SKP1-specific primers. After further analysis, five clones were identified as false positives.
Various independent studies have shown that SKP1 is capable of binding to numerous proteins whose common features are the presence of an F-box and other motifs involved in protein-protein interaction, e.g., WD 40- or leucine-rich repeats (4, 40). We identified a putative F-box sequence in the amino-terminal part of Clink (Fig. 1). To assess the significance of that motif for the interaction of Clink with SKP1, two independent F-box mutants, ClinkP10L and ClinkI17A (Fig. 1), were constructed and tested for interaction with MsSKP1 in the yeast two-hybrid assay. Double transformants containing pAD-MsSKP1 and pGBT9-ClinkP10L or pGBT9-ClinkI17A did not activate either the URA3 or the lacZ reporter gene, indicating that the altered amino acids of Clink are crucial for its binding to SKP1 (Fig. 3).
These results were confirmed by an in vitro protein-protein binding assay performed with the fusion proteins GST-MsSKP1 and His6-Clink or His6-ClinkI17A. Interaction was detected between recombinant MsSKP1 and Clink, whereas the F-box mutant, ClinkI17A, showed strongly reduced binding to GST-MsSKP1 (Fig. 2B).
Clink enhances the replication of FBNYV DNA.
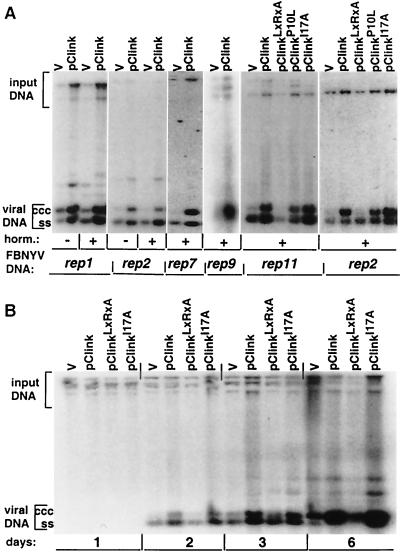
The results of the yeast two-hybrid assays and the in vitro interaction of FBNYV Clink with pRB and SKP1 suggested that Clink may influence the cell cycle, allowing efficient viral DNA synthesis. We therefore tested the effect of Clink on the replication of FBNYV DNAs that encode replication initiator proteins (rep components). Five such rep components had been identified in two FBNYV isolates (27, 28). Each of these DNAs replicates autonomously in cells of N. benthamiana leaves when introduced by agrobacteria on a T-DNA transfection vector (46). When agrobacteria carrying the Clink-encoding genome component along with agrobacteria carrying a rep component were used to coinoculate leaf discs, a three- to sevenfold stimulation of replication was observed for all FBNYV rep components tested (Fig. 4A). The replication was independent of growth-promoting plant hormones. To test the influence of the pRB-binding motif and the F-box of Clink on the stimulation of replication, mutations causing the following alterations of the protein were introduced into the clink gene: ClinkC112R, E114A, ClinkP10L, and ClinkI17A (Fig. 1). The mutation of the pRB-binding motif (C112R, E114A) abolished the stimulation of FBNYV DNA replication, whereas Clink-enhanced replication remained unaffected when the F-box mutants were used in the replication assay (Fig. 4A); the same tendencies on replication of rep2 (Fig. 4B) and rep1 (data not shown) components were observed at different times after agroinoculation of leaf discs. The slight reduction of stimulation observed with ClinkP10L was in line with the finding that its ability to bind pRB was also reduced by this mutation (Fig. 3B). In the absence of structural data, we can only speculate that replacing this particular proline with leucine may affect the global conformation of the protein, also altering its affinity for pRB.
FIG. 4.
Replication enhancement of FBNYV rep DNAs by Clink. Southern hybridization assays of total DNA extracted from leaf discs of N. benthamiana after agroinoculation with pairwise combinations of agrobacteria carrying pBin19 with dimers of a respective rep component (indicated at the bottom [FBNYV DNA]), along with agrobacteria carrying the pBin19 empty vector (v) or wild-type or mutated Clink-encoding FBNYV DNA (indicated at the top) are shown. Leaf disc medium was supplemented (+) or not (−) with plant hormones (horm.). rep-specific probes used for hybridization are shown at the bottom. Replicative forms of viral DNA are marked by ccc (covalently closed circular DNA) and ss (ssDNA). “Input DNA” denotes hybridization with residual pBin19-rep plasmids from agrobacteria used as the inoculum. In each series of experiments, equal amounts of total DNA were loaded, as judged by ethidium bromide staining of the gels (data not shown). The intensities of all DNA bands were quantified upon analysis of the blots with a PhosphorImager (Molecular Dynamics), and stimulation of replication was normalized to the signal produced by the input DNA. (A) DNA was extracted at 3 days after agroinoculation. (B) Leaf discs were agroinoculated with rep2 component and wild-type or mutated Clink-encoding DNA (indicated at the top), and DNA was extracted 1, 2, 3, and 6 days after agroinoculation, as indicated at the bottom.
In summary, the results suggest that the interaction of Clink with a pRB-like protein of N. benthamiana stimulates FBNYV DNA replication whereas its F-box-dependent interaction with SKP1 is apparently not required for enhancement of replication in this assay.
DISCUSSION
We have shown that Clink, the protein encoded by FBNYV C10, contains distinct functional domains required for binding two cellular proteins, pRB and SKP1. The property of Clink to stimulate viral DNA replication correlated to its capacity to bind pRB. Whereas the LxCxE sequence of Clink was required to enhance replication, the presence of a functional Clink protein by itself was not an absolute requirement for viral replication in N. benthamiana. Hence, if S-phase entry is necessary for FBNYV replication, Clink may not be the only viral trigger for that step. Some additional cell cycle-modulating activity might be brought about by the nanovirus Rep proteins. For instance, interaction of a Rep protein with a pRB-homologue has been shown for a geminivirus (51). Lack of an LxCxE motif in Rep proteins does not exclude the possibility of pRB binding, as shown for several geminivirus Rep proteins without such a sequence (1; A. D. Meyer, A. Bezier, and B. Gronenborn, unpublished data). Alternatively, cell division factors induced in leaf tissue of N. benthamiana following agroinoculation might suffice to warrant a basal level of FBNYV DNA replication, which is then increased by the action of Clink. Nevertheless, Clink may still be essential for FBNYV DNA replication upon infection of leguminous hosts. The lack of an experimental infection assay for FBNYV using cloned copies of all its DNA components has so far precluded a definite answer to that question.
In the context of a nanovirus infection, Clink would be required early. This assumption is supported by the finding that virions of banana bunchy top virus, another nanovirus, predominantly contain DNA primers derived from DNA encoding the Clink homologue (17). Upon infection, these primers would initiate efficient conversion of this ssDNA into transcriptionally active double-stranded DNA (dsDNA) and hence would lead to synthesis of the encoded gene product, Clink.
The fact that we isolated a number of independent cDNA clones of SKP1 homologues from Medicago (MsSKP1) in the two-hybrid screen indicated a relative abundance of MsSKP1 RNA in rapidly dividing suspension cells and a high affinity of Clink for MsSKP1. This interaction was specific, since we did not isolate MsSKP1 cDNAs in a screen using the F-box-mutated ClinkI17A. Our screens failed to identify a pRB homologue, which may reflect a low concentration of the respective transcripts in a population of rapidly dividing cells. For instance, accumulation of mRNA of the maize pRB homologue ZmRb1 was observed only in differentiated nondividing tissues (22). Alternatively, if present in suspension cells, pRB-specific transcripts may have been missed during generation of the cDNA library due to size limitations or frameshifts.
The capacity of Clink to bind both pRB and MsSKP1 offers the possibility that a plant pRB is the target for ubiquitin-mediated degradation. However, F-box mutations did not affect the enhancing effect of Clink on virus DNA replication in N. benthamiana. Hence, in the assay system used, the available data do not allow a conclusion about whether targeted degradation of pRB contributes to cell cycle progression in plants. In the context of a natural infection, Clink may not suffice to disrupt by simple interaction all of the pRB-E2F complexes in the cell and degradation may represent an additional and more efficient mechanism of inactivating pRB. Thus, nanovirus Clink protein may also inactivate pRB by targeting its degradation, as was described for the papillomavirus E7 protein (8, 25). In this case, however, the SCF complex is probably not involved, since a direct interaction of E7 with the 26S proteasome was found (5).
Participation of proteolysis has been described for different steps of cell cycle regulation (29, 47). In yeast, SKP1 is required for the G1/S transition but also for the G2/M transition (4). Therefore, the interaction of Clink with an SKP1 homologue may mediate the degradation of proteins in a later phase of the cell cycle and/or of virus infection. ssDNA plant viruses probably encode functions to restore normal cell cycle control in the host cells, once a critical amount of viral genome products has accumulated, since neither geminiviruses nor nanoviruses cause uncontrolled cell proliferation. Binding to Clink may render SKP1 inaccessible for other proteins involved in the control of mitosis. If this were the case, cell divisions would be inhibited and viral DNA replication might be achieved during a process of endoreduplication. In yeast, for instance, F-box proteins and the ubiquitin-proteasome pathway are involved in the control of the ploidy level (32). Finally, Clink itself might be targeted for degradation, thereby restoring (part of) the host cell cycle control. A comparable degradation via ubiquitination of F-box proteins was recently described for yeast Cdc4p and Grr1p (52) and MEKKα of Dictyostelium (12).
Alternatively, Clink may mediate the degradation of substrates with no cell cycle link. These could include viral proteins that need to be degraded at a particular stage of infection and/or cellular proteins. A remarkable example of virus-induced degradation of a cellular target protein is represented by CD4, the receptor of human immunodeficiency virus type 1 (37).
In plants, F-box proteins such as TIR1 and COI1 are involved in plant hormone action (13, 15, 41, 49) or in flower development (FIM and UFO) (23, 36). Whereas F-box proteins and their targets destined for degradation have been quite well studied in many organisms (2, 19, 33, 52), nothing is known about the ubiquitination targets of plant F-box proteins.
Clink is the first plant virus protein with a functional F-box and most probably modulates the plant's cell cycle via binding to pRB. The capacity of Clink to interact with pRB and also with SKP1 involved in the ubiquitin-mediated protein turnover pathway is a hitherto unique feature of a single protein. Hence, this adds to the complexity of virus-host interaction.
ACKNOWLEDGMENTS
We thank Crisanto Gutiérrez and Didier Trouche for providing ZmRb1 and human pRB encoding plasmids, respectively, and Stephen J. Elledge for providing yeast strain Y166. We are indebted to Françoise de Kouchkovsky for excellent technical assistance and Liliane Troussard for DNA sequencing. We thank Dominique Thomas for critical comments on the manuscript and Jeff Leung for fruitful discussions.
M.N.A. was supported by the French Ministry of Research, A.D.M. received a Swiss National Fonds TMR fellowship (83EU050209), and J.G. was supported by an EMBO long-term fellowship. This work was supported in part by the European Commission under the INCO-DC Programme (ERBIC18-CT96-0121).
REFERENCES
- 1.Ach R A, Durfee T, Miller A B, Taranto P, Hanley-Bowdoin L, Zambryski P C, Gruissem W. RRB1 and RRB2 encode maize retinoblastoma-related proteins that interact with a plant D-type cyclin and geminivirus replication protein. Mol Cell Biol. 1997;17:5077–5086. doi: 10.1128/mcb.17.9.5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amati B, Vlach J. Kip1 meets SKP2: new links in cell-cycle control. Nat Cell Biol. 1999;1:E91–E93. doi: 10.1038/12087. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Brooklyn, N.Y: Greene Publishing Associates; 1987. [Google Scholar]
- 4.Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper J W, Elledge S J. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86:263–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- 5.Berezutskaya E, Bagchi S. The human papillomavirus E7 oncoprotein functionally interacts with the S4 subunit of the 26 S proteasome. J Biol Chem. 1997;272:30135–30140. doi: 10.1074/jbc.272.48.30135. [DOI] [PubMed] [Google Scholar]
- 6.Bisaro D M. Geminivirus DNA replication. In: DePamphilis M L, editor. DNA replication in eukaryotic cells. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 833–854. [Google Scholar]
- 7.Boevink P, Chu P W, Keese P. Sequence of subterranean clover stunt virus DNA: affinities with the geminiviruses. Virology. 1995;207:354–361. doi: 10.1006/viro.1995.1094. [DOI] [PubMed] [Google Scholar]
- 8.Boyer S N, Wazer D E, Band V. E7 protein of human papilloma virus-16 induces degradation of retinoblastoma protein through the ubiquitin-proteasome pathway. Cancer Res. 1996;56:4620–4624. [PubMed] [Google Scholar]
- 9.Burns T M, Harding R M, Dale J L. The genome organization of banana bunchy top virus: analysis of six ssDNA components. J Gen Virol. 1995;76:1471–1482. doi: 10.1099/0022-1317-76-6-1471. [DOI] [PubMed] [Google Scholar]
- 10.Chellappan S, Kraus V B, Kroger B, Munger K, Howley P M, Phelps W C, Nevins J R. Adenovirus E1A, simian virus 40 tumor antigen, and human papillomavirus E7 protein share the capacity to disrupt the interaction between transcription factor E2F and the retinoblastoma gene product. Proc Natl Acad Sci USA. 1992;89:4549–4553. doi: 10.1073/pnas.89.10.4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chu P, Boevink P, Surin B, Larkin P, Keese P, Waterhouse P. Non-geminated single-stranded DNA plant viruses. In: Singh R P, Singh U S, Kohmoto K, editors. Pathogenesis and host specificity in plant diseases. III. Oxford, United Kingdom: Pergamon Press; 1995. pp. 311–341. [Google Scholar]
- 12.Chung C Y, Reddy T B, Zhou K, Firtel R A. A novel, putative MEK kinase controls developmental timing and spatial patterning in Dictyostelium and is regulated by ubiquitin-mediated protein degradation. Genes Dev. 1998;12:3564–3578. doi: 10.1101/gad.12.22.3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.del Pozo J C, Estelle M. Function of the ubiquitin-proteasome pathway in auxin response. Trends Plant Sci. 1999;4:107–112. doi: 10.1016/s1360-1385(99)01382-5. [DOI] [PubMed] [Google Scholar]
- 14.Dyson N. The regulation of E2F by pRB-family proteins. Genes Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 15.Gray W M, del Pozo J C, Walker L, Hobbie L, Risseeuw E, Banks T, Crosby W L, Yang M, Ma H, Estelle M. Identification of an SCF ubiquitin-ligase complex required for auxin response in Arabidopsis thaliana. Genes Dev. 1999;13:1678–1691. doi: 10.1101/gad.13.13.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gutiérrez C. The retinoblastoma pathway in plant cell cycle and development. Curr Opin Plant Biol. 1998;1:492–497. doi: 10.1016/s1369-5266(98)80041-1. [DOI] [PubMed] [Google Scholar]
- 17.Hafner G J, Harding R M, Dale J L. A DNA primer associated with banana bunchy top virus. J Gen Virol. 1997;78:479–486. doi: 10.1099/0022-1317-78-2-479. [DOI] [PubMed] [Google Scholar]
- 18.Hafner G J, Stafford M R, Wolter L C, Harding R M, Dale J L. Nicking and joining activity of banana bunchy top virus replication protein in vitro. J Gen Virol. 1997;78:1795–1799. doi: 10.1099/0022-1317-78-7-1795. [DOI] [PubMed] [Google Scholar]
- 19.Harper J W, Elledge S J. Skipping into the E2F1-destruction pathway. Nat Cell Biol. 1999;1:E5–E7. doi: 10.1038/8952. [DOI] [PubMed] [Google Scholar]
- 20.Hemerly, A. S., P. C. G. Ferreira, M. Van Montagu, and D. Inzé. 1999. Cell cycle control and plant morphogenesis: is there an essential link? Bioessays:29–37.
- 21.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 22.Huntley R, Healy S, Freeman D, Lavender P, de Jager S, Greenwood J, Makker J, Walker E, Jackman M, Xie Q, Bannister A J, Kouzarides T, Gutiérrez C, Doonan J H, Murray J A. The maize retinoblastoma protein homologue ZmRb-1 is regulated during leaf development and displays conserved interactions with G1/S regulators and plant cyclin D (CycD) proteins. Plant Mol Biol. 1998;37:155–169. doi: 10.1023/a:1005902226256. [DOI] [PubMed] [Google Scholar]
- 23.Ingram G C, Doyle S, Carpenter R, Schultz E A, Simon R, Coen E S. Dual role for fimbriata in regulating floral homeotic genes and cell division in Antirrhinum. EMBO J. 1997;16:6521–6534. doi: 10.1093/emboj/16.21.6521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jansen-Dürr P. How viral oncogenes make the cell cycle. Trends Genet. 1996;12:270–275. doi: 10.1016/0168-9525(96)81455-7. [DOI] [PubMed] [Google Scholar]
- 25.Jones D L, Thompson D A, Munger K. Destabilization of the RB tumor suppressor protein and stabilization of p53 contribute to HPV type 16 E7-induced apoptosis. Virology. 1997;239:97–107. doi: 10.1006/viro.1997.8851. [DOI] [PubMed] [Google Scholar]
- 26.Kaelin W G, Jr, Pallas D C, DeCaprio J A, Kaye F J, Livingston D M. Identification of cellular proteins that can interact specifically with the T/E1A-binding region of the retinoblastoma gene product. Cell. 1991;64:521–532. doi: 10.1016/0092-8674(91)90236-r. [DOI] [PubMed] [Google Scholar]
- 27.Katul L, Maiss E, Vetten H J. Sequence analysis of a faba bean necrotic yellows virus DNA component containing a putative replicase gene. J Gen Virol. 1995;76:475–479. doi: 10.1099/0022-1317-76-2-475. [DOI] [PubMed] [Google Scholar]
- 28.Katul L, Timchenko T, Gronenborn B, Vetten H J. Ten distinct circular ssDNA components, four of which encode putative replication-associated proteins, are associated with the faba bean necrotic yellows virus genome. J Gen Virol. 1998;79:3101–3109. doi: 10.1099/0022-1317-79-12-3101. [DOI] [PubMed] [Google Scholar]
- 29.King R W, Deshaies R J, Peters J M, Kirschner M W. How proteolysis drives the cell cycle. Science. 1996;274:1652–1659. doi: 10.1126/science.274.5293.1652. [DOI] [PubMed] [Google Scholar]
- 30.Knudsen E S, Buckmaster C, Chen T T, Feramisco J R, Wang J Y. Inhibition of DNA synthesis by RB: effects on G1/S transition and S-phase progression. Genes Dev. 1998;12:2278–2292. doi: 10.1101/gad.12.15.2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koepp D M, Harper J W, Elledge S J. How the cyclin became a cyclin: regulated proteolysis in the cell cycle. Cell. 1999;97:431–434. doi: 10.1016/s0092-8674(00)80753-9. [DOI] [PubMed] [Google Scholar]
- 32.Kominami K, Toda T. Fission yeast WD-repeat protein pop1 regulates genome ploidy through ubiquitin-proteasome-mediated degradation of the CDK inhibitor Rum1 and the S-phase initiator Cdc18. Genes Dev. 1997;11:1548–1560. doi: 10.1101/gad.11.12.1548. [DOI] [PubMed] [Google Scholar]
- 33.Krek W. Proteolysis and the G1-S transition: the SCF connection. Curr Opin Genet Dev. 1998;8:36–42. doi: 10.1016/s0959-437x(98)80059-2. [DOI] [PubMed] [Google Scholar]
- 34.Laufs J, Jupin I, David C, Schumacher S, Heyraud-Nitschke F, Gronenborn B. Geminivirus replication: genetic and biochemical characterization of Rep protein function, a review. Biochimie. 1995;77:765–773. doi: 10.1016/0300-9084(96)88194-6. [DOI] [PubMed] [Google Scholar]
- 35.Lazarowitz S G. Geminiviruses: genome structure and function. Crit Rev Plant Sci. 1992;11:327–349. [Google Scholar]
- 36.Lee I, Wolfe D S, Nilsson O, Weigel D. A LEAFY co-regulator encoded by UNUSUAL FLORAL ORGANS. Curr Biol. 1997;7:95–104. doi: 10.1016/s0960-9822(06)00053-4. [DOI] [PubMed] [Google Scholar]
- 37.Margottin F, Bour S P, Durand H, Selig L, Benichou S, Richard V, Thomas D, Strebel K, Benarous R. A novel human WD protein, h-beta TrCp, that interacts with HIV-1 Vpu connects CD4 to the ER degradation pathway through an F-box motif. Mol Cell. 1998;1:565–574. doi: 10.1016/s1097-2765(00)80056-8. [DOI] [PubMed] [Google Scholar]
- 38.Nagar S, Pedersen T J, Carrick K M, Hanley-Bowdoin L, Robertson D. A geminivirus induces expression of a host DNA synthesis protein in terminally differentiated plant cells. Plant Cell. 1995;7:705–719. doi: 10.1105/tpc.7.6.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neil J C, Cameron E R, Baxter E W. p53 and tumor viruses: catching the guardian off-guard. Trends Microbiol. 1997;5:115–120. doi: 10.1016/S0966-842X(96)10083-4. [DOI] [PubMed] [Google Scholar]
- 40.Patton E E, Willems A R, Tyers M. Combinatorial control in ubiquitin-dependent proteolysis: don't Skp the F-box hypothesis. Trends Genet. 1998;14:236–243. doi: 10.1016/s0168-9525(98)01473-5. [DOI] [PubMed] [Google Scholar]
- 41.Ruegger M, Dewey E, Gray W M, Hobbie L, Turner J, Estelle M. The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast Grr1p. Genes Dev. 1998;12:198–207. doi: 10.1101/gad.12.2.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sano Y, Wada M, Hashimoto Y, Matsumoto T, Kojima M. Sequences of ten circular ssDNA components associated with the milk vetch dwarf virus genome. J Gen Virol. 1998;79:3111–3118. doi: 10.1099/0022-1317-79-12-3111. [DOI] [PubMed] [Google Scholar]
- 43.Schafer A, Tauch A, Jager W, Kalinowski J, Thierbach G, Puhler A. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene. 1994;145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- 44.Scheffner M, Huibregtse J M, Vierstra R D, Howley P M. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell. 1993;75:495–505. doi: 10.1016/0092-8674(93)90384-3. [DOI] [PubMed] [Google Scholar]
- 45.Taya Y. RB kinases and RB-binding proteins: new points of view. Trends Biochem Sci. 1997;22:14–17. doi: 10.1016/s0968-0004(96)10070-0. [DOI] [PubMed] [Google Scholar]
- 46.Timchenko T, de Kouchkovsky F, Katul L, David C, Vetten H J, Gronenborn B. A single Rep protein initiates replication of multiple genome components of faba bean necrotic yellows virus, a single-stranded DNA virus of plants. J Virol. 1999;73:10173–10182. doi: 10.1128/jvi.73.12.10173-10182.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Townsley F M, Ruderman J V. Proteolytic ratchets that control progression through mitosis. Trends Cell Biol. 1998;8:238–244. doi: 10.1016/s0962-8924(98)01268-9. [DOI] [PubMed] [Google Scholar]
- 48.Vierstra R D. Proteolysis in plants: mechanisms and functions. Plant Mol Biol. 1996;32:275–302. doi: 10.1007/BF00039386. [DOI] [PubMed] [Google Scholar]
- 49.Xie D X, Feys B F, James S, Nieto-Rostro M, Turner J G. COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science. 1998;280:1091–1094. doi: 10.1126/science.280.5366.1091. [DOI] [PubMed] [Google Scholar]
- 50.Xie Q, Sanz-Burgos A P, Hannon G J, Gutiérrez C. Plant cells contain a novel member of the retinoblastoma family of growth regulatory proteins. EMBO J. 1996;15:4900–4908. [PMC free article] [PubMed] [Google Scholar]
- 51.Xie Q, Suarez-Lopez P, Gutiérrez C. Identification and analysis of a retinoblastoma binding motif in the replication protein of a plant DNA virus: requirement for efficient viral DNA replication. EMBO J. 1995;14:4073–4082. doi: 10.1002/j.1460-2075.1995.tb00079.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou P, Howley P M. Ubiquitination and degradation of the substrate recognition subunits of SCF ubiquitin-protein ligases. Mol Cell. 1998;2:571–580. doi: 10.1016/s1097-2765(00)80156-2. [DOI] [PubMed] [Google Scholar]