Abstract
TT virus (TTV) is a recently discovered infectious agent originally obtained from transfusion-related hepatitis. However, the causative link between the TTV infection and liver disease remains uncertain. Recent studies demonstrated that genome sequences of different TTV strains are significantly divergent. To assess genetic heterogeneity of the TTV genome in more detail, a sequence analysis of PCR fragments (271 bp) amplified from open reading frame 1 (ORF1) was performed. PCR fragments were amplified from 5 to 40% of serum specimens obtained from patients with different forms of hepatitis who reside in different countries (e.g., China, Egypt, Vietnam, and the United States) and from normal human specimens obtained from U.S. residents. A total of 170 PCR fragments were sequenced and compared to sequences derived from the corresponding TTV genome region deposited in GenBank. Genotypes 2 and 3 were found to be significantly more genetically related than any other TTV genotype. Moreover, three sequences were shown to be almost equally related to both genotypes 2 and 3. These observations suggest a merger of genotypes 2 and 3 into one genotype, 2/3. Additionally, five new groups of TTV sequences were identified. One group represents a new genotype, whereas the other four groups were shown to be more evolutionary distant from all known TTV sequences. The evolutionary distances between these four groups were also shown to be greater than between TTV genotypes. The phylogenetic analysis suggested that these four new genetic groups represent closely related yet different viral species. Thus, TTV exists as a “swarm” of at least five closely related but different viruses. These observations suggest a high degree of genetic complexity within the TTV population. The finding of the additional TTV-related species should be taken into consideration when the association between TTV infections and human diseases of unknown etiology is studied.
A new TT virus (TTV) was recently discovered in a serum specimen obtained from a Japanese patient with posttransfusion hepatitis of unknown etiology (16, 20). In the same study, TTV DNA was detected by PCR in serum specimens from three of five patients with posttransfusion non-A through non-G hepatitis. It was shown that TTV DNA titers closely correlated with the level of aminotransferase in these three patients (16). Additionally, TTV DNA was found in liver tissues in titers that are equal to or 10 to 100 times greater than those in the corresponding serum specimens (20). In combination with these findings, the identification of TTV DNA in 47% of patients with fulminant hepatitis and 46% of patients with chronic liver disease of unknown etiology (20) was used to hypothesize that TTV may be responsible for at least a part of acute and chronic liver disease of unknown etiology (16, 20). However, the detection of TTV DNA in 12% of normal blood donors in Japan (20) indicates that this hypothesis should be thoroughly tested before a final conclusion is drawn regarding an association between TTV and viral hepatitis. Subsequent studies have demonstrated that TTV can be frequently found in blood donors and that TTV transmission through transfusion of blood products may have occurred extensively (25). In addition, fecal excretion indicated that TTV may be transmitted nonparenterally by a fecal-oral route (19). The widespread occurrence of TTV infection around the world is well documented by several groups of researchers from different countries (2, 3, 5–7, 9, 10, 14, 15, 31–34), yet none of these studies have been able to establish a firm correlation between the TTV infection and liver disease.
The TTV genome is a circular negative-stranded DNA of 3,852 nucleotides (nt) (11, 13). The positive DNA strand contains two large (202 and 770 amino acids) open reading frames (ORF1 and ORF2) (13, 20). Similar to chicken anemia virus, a member of the Circoviridae, the TTV ORF1-encoded protein contains an N-terminal arginine-rich hydrophilic region and two of the four conserved Rep protein motifs, suggesting that TTV replicates by a rolling circle mechanism (13). Although TTV shares some features such as a negative-stranded circular DNA genome with members of the Circoviridae (11), it is distinctly different from viruses of this family. For example, the TTV genome is almost twice as large as the genome of Circoviridae members. Additionally, the circular nature of the TTV genome and lack of notable sequence homology rule out classification of this virus into the Parvoviridae. Therefore, TTV has been tentatively classified as the sole member of a new virus family Circinoviridae (13).
The TTV genome is heterogeneous. The first publication presenting TTV sequences reported that the TTV genome can be classified into two different genotypes (20). This observation was later confirmed by other researchers (2, 9, 15, 35) and expanded to identify a third TTV genotype (25). Subsequently, the existence of three TTV genotypes was substantiated by several other groups (3, 7, 10, 13, 31). Recently, a phylogenetic analysis performed by using TTV isolates from Asia, Africa, and South America revealed three additional TTV genotypes (30). The existence of six TTV genotypes was confirmed in a subsequent publication, which also reported a seventh TTV genotype (29). Thus, within a short period of time seven TTV genotypes were identified, verifying significant sequence heterogeneity of the TTV genome.
In this study, numerous serum specimens obtained from different parts of the world were tested for TTV by PCR. The sequences obtained in this study and those retrieved from GenBank were subjected to a phylogenetic analysis, which resulted in the identification of five additional genetic groups. One of these groups represents a new TTV genotype; the other four were found to be significantly more evolutionarily distant from TTV and from each other than TTV genotypes are from each other. This observation implies that these four new genetic groups represent closely related yet different TTV-like viruses. We hypothesize that a whole “swarm” of numerous TTV-related species circulates in the human population worldwide. The data obtained in this study suggest that in addition to the prototype TTV variants, any association of these new TTV-related species with human diseases of unknown etiology should be further investigated.
MATERIALS AND METHODS
Serum samples.
A total of 1,122 serum specimens obtained from the United States (n = 994), China (n = 41), Vietnam (n = 41), and Egypt (n = 48) were tested by PCR. All specimens from China, Vietnam, and Egypt were obtained from hepatitis C virus (HCV)-positive patients. The TTV PCR product was identified in 12, 16, and 14 specimens from China, Vietnam, and Egypt, respectively. The U.S. specimens were obtained from normal blood donors (n = 80), paid HCV-positive plasma donors (n = 25; Boston Biomedica Inc., West Bridgewater, Mass.), patients identified through the Sentinel Counties Study of community-acquired viral hepatitis (n = 584; Centers for Disease Control and Prevention, Atlanta, Ga.) (1), and patients with acute hepatitis of various viral etiologies (n = 173). A total of 128 U.S. specimens were found to be TTV PCR positive: 11 specimens among normal blood donors (13.8%), 7 among paid plasma donors (28.0%), 32 among patients from the Sentinel Counties Study (5.1%), and 34 among acute hepatitis patients (19.8%). Additionally, among 130 specimens obtained from 77 liver transplant recipients, 44 specimens from 29 recipients were found positive for TTV.
Primers.
The set of primers used in this study was almost completely based on the set of primers NG059, NG061, and NG063, originally designed by Okamoto et al. (20). However, every primer was slightly modified by removal of a few nucleotides from the 3′ end. Thus, the set contained the external primer 059 (nt 1898 to 1920) with sequence 5′-CACAGACAGAGGAGAAGGCAAC, the internal primer 061 (nt 1914-1934) with sequence 5′-GGCAACATGTTATGGATAGA, and the external/internal primer 063 (nt 2163 to 2185) with sequence 5′-CTGGCATTTTACCATTTCCAAA. The internal primers were used for sequencing as well. The amplified PCR product was 271 bp.
Additionally, three sets of primers were designed to amplify different regions of the TTV ORF1 at nt 587 to 1194 (fragment 130, 607 bp), 1770 to 2463 (fragment 138, 587 bp), and 2323 to 2919 (fragment 142, 596 bp). Fragment 130 was amplified by using external primers MC129 (5′-CCCGGAGATAGAGCACCATGGCC) and MC132 (5′-ATCTAAAATTCGATCCATAGG) and internal primers MC130 (5′-CCATGGCCTATGGCTGGTGG) and MC131 (ATAGGTAGAACGAGAAAACAG). Fragment 138 was amplified by using external primers MC137 (5′-CAACAAAAACCTTGCTAGGAAA) and MC140 (5′-AGAGGCCACGTCTGAAGTCCCA) and internal primers MC138 (5′-AAACACCTTCACAAATGAGGA) and MC139 (5′-CGGTGAGTTGTATTTCGGGTC). Fragment 142 was amplified by using external primers MC141 (5′-ATACCACTCAGACATTAAAAA) and MC144 (5′-ATTACTGGAAGGGTTGTAAGG) and internal primers MC142 (5′-ATGAAATACCGTTTTAAGTGG) and MC143 (5′-TGGGGTCACCAAACATGTTTA).
Extraction of TTV DNA and PCR.
Serum DNA was extracted from 50 μl of serum by the standard method, which included pretreatment of samples with proteinase K-sodium dodecyl sulfate and extraction with phenol and chloroform. DNA samples were dissolved in 20 μl of Tris-HCl buffer (pH 8.0), denatured at 95°C for 5 min, and then quickly placed on ice. Two microliters of the denatured DNA was used for the first-round PCR with external primers, and 2 μl of the first-round PCR product was used to initiate the second-round PCR with internal primers. The first-round PCR program included 30 cycles of amplification, and the second-round PCR included 25 cycles. The first-round amplification cycle consisted of 94°C for 1 min, 50°C for 40 s, and 72°C for 1 min; the second-round amplification cycle was 94°C for 40 s, 55°C for 40 s, and 72°C for 45 s.
Cloning.
PCR fragments from two specimens, hm05 and hm26, were cloned with plasmid pPCR-Script Amp SK(+) (Stratagene, La Jolla, Calif.). Fragments 130, 138, and 142 (see above) from serum specimen hm217 were cloned with plasmid pGEX-4T-2 (Pharmacia Biotech Inc., Piscataway, N.J.).
Sequencing.
All TTV PCR products were purified for sequencing by using the Wizard PCR Preps DNA purification system (Promega, Madison, Wis.). The primary structure of purified PCR fragments was determined by using an automated sequencer (ABI model 373 DNA sequencer; Applied Biosystems, Foster City, Calif.) according to the manufacturer's protocol. In all cases, internal PCR primers were used as sequencing primers.
Computer-assisted sequence analysis.
The nucleotide sequences were aligned by using the PileUp program (Wisconsin Package, version 9.0). The phylogenetic analysis was performed by using the PHYLIP package, version 3.57. Evolutionary distances between nucleotide sequences were determined by using the DNADIST program. Frequency distribution analysis was performed with the Microsoft Excel program. The programs Neighbor and Drawtree were used to calculate and produce unrooted phylogenetic trees. To confirm the reliability of the phylogenetic tree, a bootstrap test was performed with 100 resamplings on randomly selected sequences representing every major branch of the unrooted tree. The reconstruction of topology of phylogenetic trees was also performed by using the maximum likelihood method, based on the quartet-puzzling algorithm (26, 27). The program PUZZLE, version 4.0, was used to perform this analysis. In addition to the tree topology, this program provides reliability values for each internal branch. In the present study, the number of unresolved quartets never exceeded 10.0%.
Nucleotide sequence accession numbers.
All sequences obtained in this study were deposited in GenBank with accession no. af173089 to af173153 and af173424 to af173468 for PCR fragments used in the phylogenetic analysis, af173475, af173476, and af174579 for PCR fragment 130, af173477 and af0174580 for fragment 138, af173478, af173479, and af174581 for fragment 142, and af173469 to af173473 and af173474 for PCR fragments obtained from follow-up specimens of patients hm05 and hm26, respectively. The accession numbers of published TTV sequences used for the phylogenetic analysis are as follows: af084106 to af084137 (35) and ab008394, ab011486 to ab011491, ab011493, ab011494, ab013688, ab013692, ab013694, ab013696, ab013700, ab013705, ab013707, ab013712, ab016935, ab017767, ab017769 to ab017771, af073794, ab017886 to ab017889, ab018885, ab018858, ab018960, ab018961, ab020174, ab020181, ab021797, ab021804, ab021805, ab021816, ab021820, af060545 to af060550, af072746, af072749, af079537, af079541, and af079543.
RESULTS
TTV sequence homology.
All PCR fragments (see Materials and Methods) obtained in this study were directly sequenced. These sequences (n = 108) were compared to the corresponding sequences deposited in GenBank (n = 89). A significant degree of heterogeneity was observed between the compared sequences. The average homology between all TTV nucleotide sequences was 72.2%. The minimal homology detected was as low as 44.7% between sequences janbnc10 (30) and lt2699 (accession no. af173427). All nucleotide sequences were translated into amino acid sequences. A comparison of the derived protein sequences showed that the average homology was 71.8%. The minimal homology between protein sequences was found to be 44.1% between janbnc10 (30) on one side and sequences hm97317, lt1493, egt216 (accession no. af173131, af173141, and af173109, respectively) and af073794 on the other.
Premature stop codon within the ORF1.
One sequence, egt209 (accession no. af173108), could not be translated to generate a protein product encoded by the entire PCR fragment. This sequence contained a premature stop codon (data not shown). The presence of this stop codon was confirmed by repetitive sequencing of this fragment. Because sequencing was performed directly on the PCR fragment without preliminary cloning, the TTV species containing this premature stop codon within ORF1 must represent the dominant species in this serum specimen. Alternatively, the identified mutation can be explained by the PCR error that occurred during early cycles of amplification. However, because this mutation could be originally introduced into only one strand of only one double-stranded DNA molecule, with a second strand being intact, and because this is a point mutation, which hardly could provide amplification advantages to DNA molecules carrying it, this alternative explanation seems less probable.
TTV genetic groups.
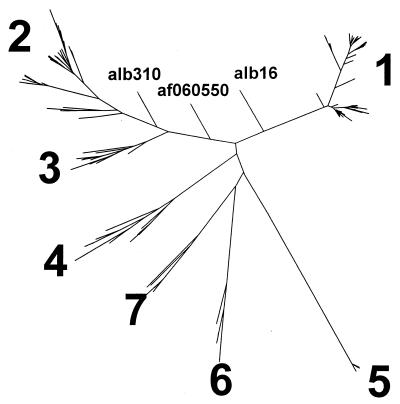
Phylogenetic analysis performed on all sequences determined in this study (n = 108) and sequences retrieved from GenBank (n = 89) revealed the existence of seven major TTV genetic groups or genotypes. Figure 1 shows a phylogenetic tree reconstructed by the neighbor-joining method using evolutionary distances calculated by the DNADIST program. Additionally, the phylogenetic tree of a very similar topological configuration was constructed using a maximum likelihood approach based on the quartet-puzzling algorithm (see Materials and Methods). The number of unresolved quartets was less than 10.0%.
FIG. 1.
TTV phylogenetic tree.
Genotypes 1 and 2 were originally identified by Okamoto et al. (20), while genotype 3 was found by Simmonds et al. (25). The existence of these three genotypes was subsequently confirmed by several groups of researchers (3, 7, 10, 13, 31). TTV genotypes 4, 5, and 6 were recently identified in serum specimens obtained from Asia, Africa, and South America (30). Subsequently, the existence of these six TTV genotypes was confirmed in the study of TTV heterogeneity in Japanese hemophiliacs (29). TTV genotype 7 represents a new genotype found in the present study.
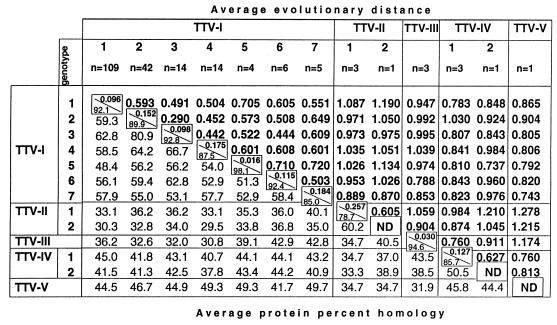
The average evolutionary distances between TTV genotypes, in general, vary between 0.442 (between genotypes 3 and 4) and 0.720 (between genotypes 5 and 7). However, the average evolutionary distance between genotypes 2 and 3 is only 0.290 (Fig. 2, TTV-1). The average protein percent homology between TTV genotypes, in general, varies from 48.4% (between genotypes 1 and 5) to 66.7% (between genotypes 3 and 4). However, protein sequences derived from TTV genotypes 2 and 3 are 80.9% homologous (Fig. 2, TTV-I). These observations strongly suggest that the previously described genotypes 2 and 3 are much more genetically related than are any other genotypes identified in this study.
FIG. 2.
Average evolutionary distances between TTV genotypes. ND, not determined.
Genetic relatedness of genotype 2 and 3 sequences.
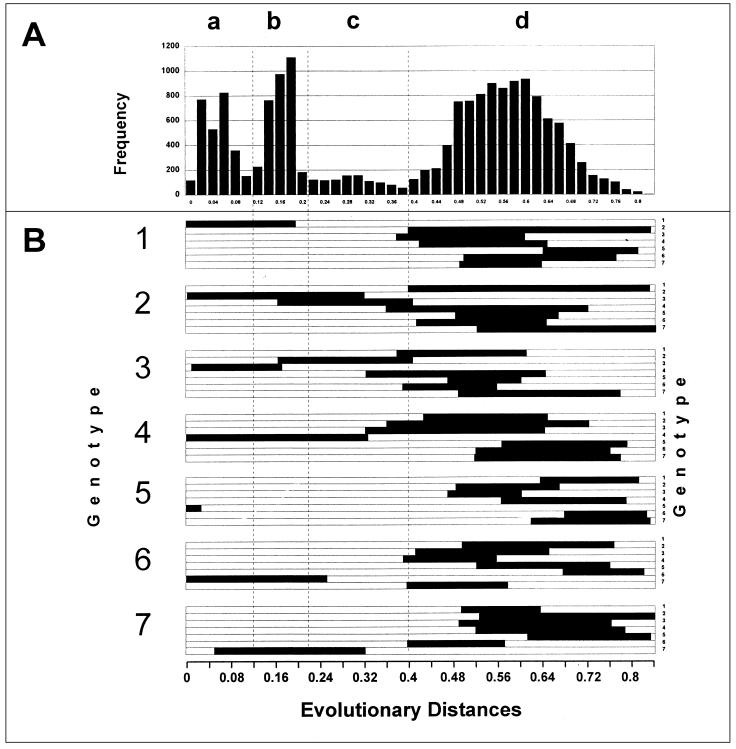
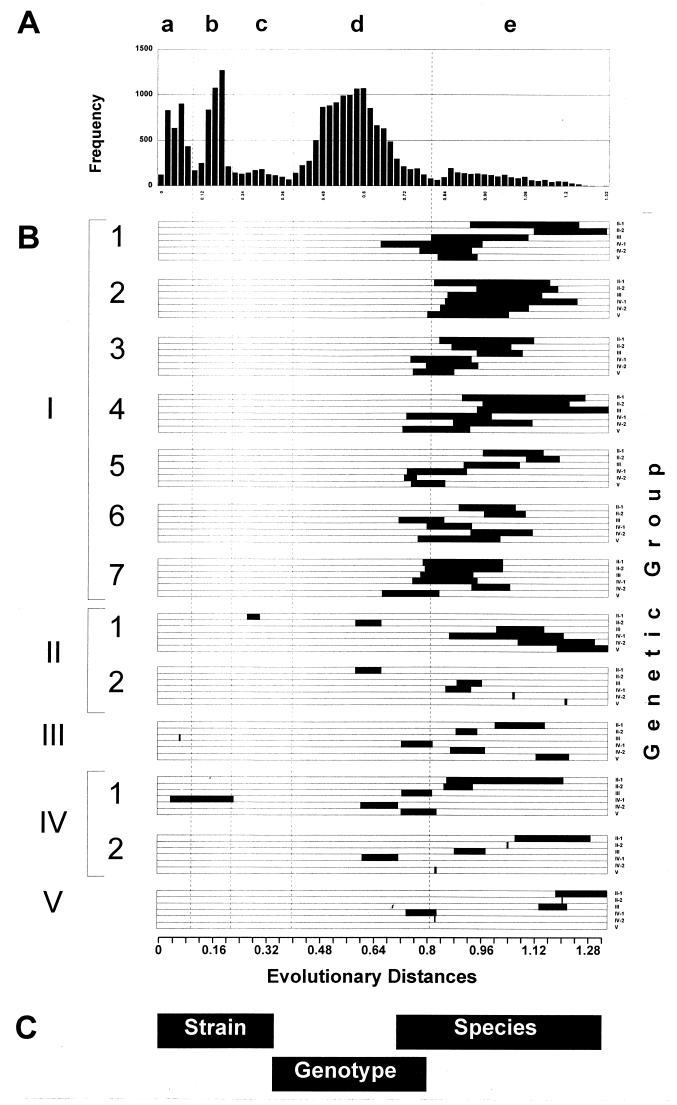
Because genotypes 2 and 3 were found to be more closely related than other genotypes, an additional analysis of genetic heterogeneity was performed. To elucidate the phylogenetic relationship between sequences of these two genotypes, we performed a frequency distribution analysis of evolutionary distances using evolutionary distances calculated by the program DNADIST and maximum likelihood (see Materials and Methods). The patterns of the frequency distribution of evolutionary distances obtained by these two programs were very similar, although the range of evolutionary distances calculated by the maximum likelihood program was more narrow than ones calculated by the DNADIST program. Figure 3A shows this frequency distribution, and Fig. 3B shows ranges of evolutionary distances calculated by the DNADIST program. These DNADIST evolutionary distances are used throughout the paper.
FIG. 3.
Frequency distribution (A) and ranges (B) of evolutionary distances between different groups of TTV sequences.
An analysis of the frequency distribution revealed noticeable discrete heterogeneity of the TTV sequences, as indicated by four major peaks (Fig. 3A). Peaks a and b represent evolutionary distances between sequences that belong to the same genotype (Fig. 3B). Peak a comprises evolutionary distances between sequences of different TTV strains within the same genotype. Peak b represents more distant phylogenetic relationships between sequences than those found in peak a and may represent subtype relationships between sequences. However, a careful analysis of evolutionary distances within the range of peak b does not allow for the consistent identification of subtype groupings. This observation may be explained by the lack of sufficient genealogical information in the region used in this study. It is conceivable that better separation between peaks in this graph might be discerned by using different regions of the genome or different methods of analysis, either of which may result in subtype identification. It was shown, for example, for the hepatitis G virus that applying different regions of the genome of different sizes significantly improved the detection of different genetic groups (4, 12). Alternatively, however, as may be the case with TTV, the absence of subtypes may be associated with a very complex genealogical relationship between sequences, which may be the result of a long evolutionary process.
Peak d represents evolutionary distances between different genotypes except for the evolutionary distances between genotypes 2 and 3, which fall within peak c (Fig. 3B). As can be seen in Fig. 3B, evolutionary distances between some sequences from genotypes 4, 6, and 7 may be found within the range covered by peak c. However, this peak is composed mainly of distances between sequences of genotypes 2 and 3. At the same time, evolutionary distances between genotype 2 or 3 sequences and sequences from other TTV genotypes are all within the genotype range of peak d (Fig. 3B). Thus, sequence relatedness between genotypes 2 and 3 is of a particular nature. These sequences are much more related to each other than to any other genotype. A similar observation has been made for the HCV genotype 6 sequences (24), suggesting that genotypes 7, 9, 10, and 11, previously identified in the southeastern part of the world, are unusually distant subtypes of genotype 6 (24). Analysis of a frequency distribution of evolutionary distances between various HCV sequences demonstrated the existence of three evenly separated peaks, corresponding to evolutionary distances between strains, subtypes, and genotypes (24). The evolutionary distances between subtypes of HCV genotype 6 occupy space between peaks, corresponding to regular subtypes and genotypes, and generate a shoulder of these peaks rather than an individual peak. However, the evolutionary distances between sequences of TTV genotypes 2 and 3 do not constitute just a shoulder of peak b (possible subtypical relationships between sequences) or of peak d (genotypical relationships between sequences) but rather occupy their own discrete space along the x axis between b and d (Fig. 3A). Based on this observation, we suggest combining genotypes 2 and 3 into one genotype, 2/3, and tentatively defining genotypes 2 and 3 as quasitypes rather than very distant subtypes of this combined genotype.
Phylogenetic analysis of sequences alb16, alb310, and af060550.
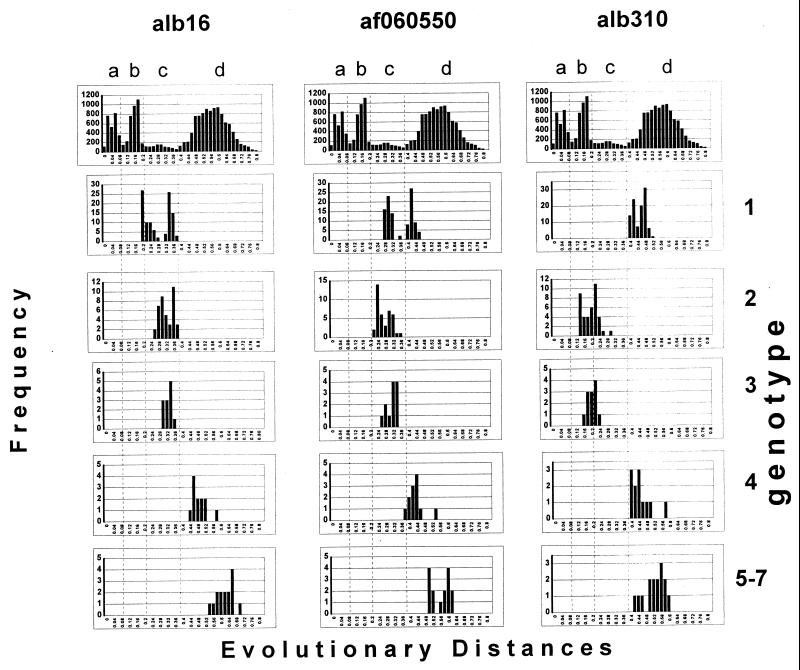
These three nucleotide sequences could not be assigned unambiguously to any TTV genotype with the methods used in this study. However, as can be seen in Fig. 1, these sequences are most closely related to genotypes 1 and 2/3. Sequence alb310 was identified in the study, whereas sequences alb16 and af060550 were previously reported (8, 35). The evolutionary distances between these sequences fall within the range of peak b in Fig. 3. As calculated by the DNADIST program (see Materials and Methods), the distances between alb310 on one side and alb16 and af060550 on the other are 0.2 and 0.1993, respectively. The sequences alb16 and af060550 are a little more closely related, the evolutionary distance between them being 0.1571. Very similar evolutionary distances were calculated by the maximum likelihood program (see Materials and Methods). This observation strongly suggests that based on the frequency distribution of evolutionary distances (Fig. 3), these three sequences (alb16, alb310, and af060550) represent different TTV strains which may belong to the same genotype.
Analysis of frequency distribution of evolutionary distances revealed that the distances between alb310 and sequences of TTV genotypes 1 and 4 to 7 fell within peak d (Fig. 4) and therefore demonstrated genotypical relationships between these sequences. However, the evolutionary distances between alb310 and sequences of the quasitypes 2 and 3 fell mainly within the range of peak b (Fig. 4), which is a strong indication that alb310 belongs to genotype 2/3. This observation provides additional support for the suggestion of combining the quasitypes 2 and 3 into one genotype. Since alb310 appears to be closely related to alb16 and af060550, the last two sequences should also belong to genotype 2/3. Additional analysis of the alb16 sequence, however, demonstrated that alb16 fell within the genotype range from sequences of genotype 4 to 7 and within the quasitype range from sequences of two different genotypes, namely, genotypes 1 and 2/3 (Fig. 4). This observation implies that while alb310 clearly belongs to genotype 2/3, alb16, which is most closely related to alb310, is almost equally distant from two different genotypes and therefore cannot be clearly assigned to a single genotype. It is interesting that evolutionary distances between genotype 1 sequences and alb16 or af060550 have a bimodal pattern of frequency distribution (Fig. 4). However, if, as in the case of the alb16 sequence, these distances fall within the peak c range, evolutionary distances for af060550 are distributed almost equally between the c and d areas of the graph. This observation suggests that af060550 is in a genotype-specific relationship with some sequences of genotype 1 and in a quasitype relationship with the other sequences of this genotype.
FIG. 4.
Frequency distribution of evolutionary distances between individual sequences alb16, alb310, and af060550 and sequences of genotypes 1 to 4 and a combined group composed of TTV genotypes 5 to 7. The top panel in every column is frequency distribution of evolutionary distances for all TTV sequences reproduced from Fig. 1.
Thus, sequences alb16, alb310, and af060550 are in uncertain genealogical relationships with sequences of genotype 1 and genotype 2/3. Without any additional data, this observation might be considered a mere artifact of the phylogenetic methods used in this study. Alternatively, however, it may reflect an actual complex TTV evolutionary process. The phylogenetic relationships between sequences alb16, alb310, and af060550, and to some extent between sequences of genotypes 1 and 2/3, may be indicative of an active ongoing evolutionary process that generates and maintains TTV variants with different degrees of genetic fitness. The existence of such variants may be responsible for the lack of a strict discrete heterogeneity of these TTV sequences.
As follows from the phylogenetic analysis described above, the three sequences alb16, alb310, and af060550 may be considered a link between genotypes 1 and 2/3. Such a link combines these two genotypes into a higher-hierarchy supertype. The presence of short branches, such as those seen for alb16, alb310, and af060550, close to the bifurcation point of this supertype in the phylogenetic tree (Fig. 1) represents a visual reflection of the existence of some transient intermediate phylogenetic forms of TTV. The detection of these forms may indicate an active but slow evolutionary process, which allows for the coexistence of TTV variants of different genetic fitness for a long time when none of these variants can rapidly outcompete the others.
TTV sequences from serial serum specimens.
Serial specimens were obtained from four liver transplant recipients infected with TTV. In one patient (three serum specimens), no changes in the TTV sequence were found over 6 months; only one nucleotide change was observed over a 7-month period in the other patient. This nucleotide substitution caused an amino acid change from Gly to Ala (accession no. af173436, af173142, and af173143). Two serum specimens separated by 8 months were available from the third liver transplant recipient. When the TTV PCR fragment was sequenced from these specimens, a significant difference was found. These two sequences (accession no. af173149 and af173140) were only 82.5% homologous. This finding is a strong indication that these two sequences belong to two different TTV strains. A total of six serum specimens were available from the fourth recipient over a 9-month period. Five sequences obtained from five follow-up specimens collected within 6 months were identical. The sixth sequence, however, obtained from the specimen collected 9 months later, was significantly different from the others. This sequence (GenBank accession no. af173437) shared only 55.3% similarity with the other sequences obtained from this patient. The phylogenetic analysis of these sequences showed that the sixth sequence was of genotype 2/3, whereas the other sequences from this patient belonged to genotype 1.
TTV sequence heterogeneity in one serum specimen.
Some PCR fragments could not be directly sequenced without detecting a number of ambiguous nucleotide positions. Two such fragments obtained from two different serum specimens, hm26 and hm05, were cloned with the pPCR-Script vector (see Materials and Methods). For each fragment, 10 clones were randomly used to determine the nucleotide sequence. It was found that five clones obtained from specimen hm26 and 1 clone obtained from specimen hm05 contained fragments of sizes that were different from the expected size. The primary structure of these cloned fragments was drastically different from that of TTV. No homology was found between these fragments and any sequence in GenBank (data not shown). Thus, these sequences were not considered to be of TTV origin. The other five clones obtained from specimen hm26 contained TTV sequences 98.2 to 100% homologous to each other. All of these sequences belong to the quasitype 3 of genotype 2/3. However, the other nine clones obtained from specimen hm05 contained sequences that belong to three different TTV genotypes, namely, 1 (five sequences), 2/3 (quasitype 2, one sequence), and 4 (three sequences) (accession no. af173469 to af173473). Homology between these sequences from different genotypes varied from 52.8 to 62.4%, and homology between sequences that belong to the same genotype varied from 93.4 to 100%. Interestingly, two clones comprising the TTV genotype 1 sequence contained a single nucleotide deletion within the 5′-end region of the PCR fragment (accession no. af173469 and af173470). Translation of sequences derived from specimen hm26 and from genotype 4 sequences derived from specimen hm05 yielded protein sequences that were 98.7 to 100% homologous in each specimen. The finding of such a high similarity between these sequences suggests that all five TTV sequences obtained from specimen hm26 and all genotype 4 sequences found in specimen hm05 represent quasispecies rather than different TTV strains. On the other hand, some TTV genotype 1 fragments derived from specimen hm510 shared only 93.4% similarity of nucleotide sequences or, when translated, demonstrated only 95.6% similarity of amino acid sequences (data not shown). This observation suggests that these sequences may belong to different TTV strains. Thus, specimen hm26 contains several quasispecies of the same TTV strain, whereas specimen hm05 contains three different TTV genotypes (1, 2/3, and 4). Genotype 1 was represented in this specimen with two different TTV strains, one of which contained three different quasispecies. Genotype 4 sequences in this specimen were represented with only one TTV strain containing three quasispecies.
Additionally, three PCR fragments derived from different regions of the TTV ORF1 at nt 587 to 1194 (fragment 130), 1770 to 2463 (fragment 138), and 2323 to 2919 (fragment 142) (see Materials and Methods) were amplified from specimen BBI217. All three fragments were sequenced and cloned with the vector pGEX-4T-2 (Materials and Methods). The primary structure was determined for four clones of fragment 130 and for three clones of fragments 138 and 142 (accession no. af173475 to af173479) and compared to each other and to the sequence of the original PCR fragments (accession no. af174579 to af174581). One of the cloned fragment 130 sequences contained a premature stop codon in phase with ORF1. Similarity between different clones of fragment 130 varied from 93.8 to 98.6%. Similarly, when cloned sequences were compared to the PCR fragment 130 itself, similarity was within the range of 92.3 to 98.8%. The derived amino acid sequences of the cloned fragments were 96.9 to 97.5% homologous (data not shown). In comparison to fragment 130, the nucleotide sequences of fragments 138 and 142 were less heterogeneous. Similarity between clones of fragment 138 varied from 99.5 to 99.8%, and that between clones of fragment 142 varied from 98.5 to 100%. It is interesting that none of cloned fragment sequences was identical to the sequence obtained by direct sequencing of these PCR fragments.
TTV-related viruses.
Recently, new TTV sequences were discovered in Japanese hemophiliacs (29). A phylogenetic analysis performed on these sequences revealed that they may belong to a new, seventh TTV genotype (29). In the course of our study, we found the sequence den4624 (accession no. af173104), which was most homologous to these sequences recovered from Japanese hemophiliacs. However, more careful analysis of all these sequences showed that the average evolutionary distance between den4624 and the putative TTV genotype 7 from the Japanese hemophiliacs was 0.605, which is a strong indication that these sequences are two different genotypes rather than a single new TTV genotype. Comparison of these new sequences to the TTV sequences used in this study showed average evolutionary distances of 0.870 to 1.190. This observation indicates that the den4624 sequence and the genotype 7 sequences from Japanese hemophiliacs (29) are significantly more distant from all other TTV sequences than are any two TTV genotypes identified in this study. Furthermore, an additional analysis of sequences obtained from different patients in this study revealed eight more sequences of low similarity with TTV sequences. The average evolutionary distances between these sequences and those from TTV genotypes 1 to 7 varied from 0.743 to 1.030. When a frequency distribution analysis compared the evolutionary distances between all TTV sequences, genotype 7 sequences from Japanese hemophiliacs (29) and these additional nine sequences identified in the present study comprised a new peak e, as shown in Fig. 5A.
FIG. 5.
Frequency distribution (A) and ranges (B) of evolutionary distances between all TTV related viruses. Panel B shows ranges of evolutionary distances specific for strains, genotypes, and TTV species. Numerals I through V denote species TTV-I through TTV-V, respectively; numerals 1 through 7 identify genotypes within TTV species.
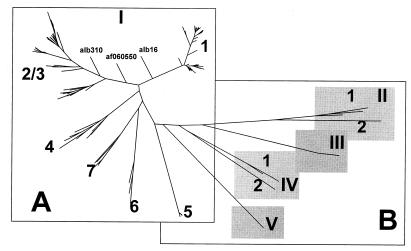
The phylogenetic tree built using all of these sequences contained four additional major branches, II, III, IV, and V (Fig. 6B). When sequences constituting these new branches were compared to each other and to the TTV sequences, collectively identified as branch I in Fig. 6A, the evolutionary distances were mainly distributed within area e in Fig. 5B. This observation suggests that the major branches of the phylogenetic tree shown in Fig. 6 represent sequences that are more distant from each other than any previously identified TTV genotypes. We hypothesize that these branches represent very closely related but different viruses that can be considered TTV-related species (Fig. 5C). Therefore, all TTV variants shown in Fig. 1 and 6A can be tentatively designated TTV-I. The viruses identified by branches II, III, IV, and V in Fig. 6B can be designated TTV-II, TTV-III (af173429), TTV-IV (af173118, af173431, af173090, and af173120), and TTV-V (af173103), respectively. As was shown in this study, TTV-I can be classified into at least six different genotypes (Fig. 1 and 6A). TTV-II and TTV-IV can be classified into two genotypes each (Fig. 2, 5B, and 6B). A single genotype was identified for TTV-III and TTV-V (Fig. 6B). The TTV genotype 7 sequences found in the Japanese hemophiliacs (29) constitute genotype 1 and den4624 constitutes genotype 2 of TTV-II (Fig. 2 and 6B).
FIG. 6.
TTV phylogenetic tree for TTV-I (A) and TTV-II through TTV-V (B). Numerals 1 through 7 identify genotypes within TTV species.
The minimal homology of 44.7% was observed between the TTV-I nucleotide sequences lt2699 (accession no. af173427) and janbc10 (30). When all TTV sequences were compared to each other, the minimal homology of nucleotide sequences of only 31.3% was observed between den4624 of TTV-II genotype 2 and rus1 (accession no. af173104) of TTV-I genotype 1. A comparison of derived amino acid sequences revealed that the lowest similarity between amino acid sequences derived from different TTV species was 26.4% between af079537 of TTV-I genotype 1 and den4624 of TTV-II genotype 2, or between ab021805 of TTV-II genotype 1 and lt866 of TTV-I genotype 4, whereas the lowest similarity between TTV-I protein sequences was found to be 44.1%.
DISCUSSION
TTV heterogeneity.
As was observed by others (2, 3, 7–10, 13, 15, 20, 22, 23, 25, 29–31, 35) and shown in this paper, the TTV genome can be classified into several genotypes. This observation is supported by phylogenetic analysis. The identification of distinct peaks in the pattern of the frequency distribution of evolutionary distances (Fig. 3) strongly identifies discrete heterogeneity of TTV sequences. However, several factors have a distorting effect on the discreteness of the TTV genome heterogeneity, suggesting a departure from the simple strain-subtype-genotype classification. One of these factors is a significant difference in relatedness of different genotypes. For example, genotypes 2 and 3, which have been previously identified (2, 9, 15, 20), are very closely related, with an average evolutionary distance between them of 0.290 (Fig. 2), whereas the average evolutionary distances between all other genotypes vary from 0.442 to 0.720. A tight genetic relatedness between genotypes 2 and 3 resulted in an additional peak c in the graph of frequency distribution of evolutionary distances (Fig. 3). A similar frequency distribution pattern of evolutionary distances was observed by other investigators (13). As discussed above, this observation was used as the basis for tentatively designating genotypes 2 and 3 as quasitypes and combining them into a single genotype, 2/3. Another factor distorting the discreteness of the TTV sequence heterogeneity is the phylogenetic relationships between TTV variants alb16 (35), alb330, and af060550 (9) on one side and genotypes 1 and 2/3 on the other. The phylogenetic analysis performed in this study failed to unambiguously assign these three sequences to any TTV genotype or to generate a separate genotype or genotypes (see above). Other researchers also noticed a unique particular nature of the af060550 sequence and also experienced difficulties in classifying this sequence into one of the known TTV genotypes (3, 9). The phylogenetic relationships of these three sequences with different TTV genotypes observed in the present study are very complex (Fig. 1 and 4). To some extent, these sequences can be considered a genetic link between genotypes 1 and 2/3. The identification of such sequences as alb16, alb330, and af060550 confounds the distinction between these two genotypes. This observation led us to suggest combining all of these sequences into a supertype (see above). It is conceivable that the identification of more TTV variants may further strengthen the genetic link between genotypes 1 and 2/3 and may help to find similar relationships between the other TTV genotypes. Another example of an unclear distinction between genetic groups was observed with more closely related sequences, mainly within genotypes 1 and 2/3, where the largest number of sequences was identified. Despite the fact that the frequency distribution pattern of evolutionary distances contains a separate and distinct peak b (Fig. 3), the presence of which suggests the existence of some subgroups within the TTV genotypes, the intricate and uncertain phylogenetic relationships between some sequences within these genotypes did not allow for the identification of distinct subtypes. In accordance with this observation, other studies, where a large number of sequences was used for the phylogenetic analysis, also failed to clearly identify subtypes within these genotypes (3, 13).
When analyzing the complex nature of TTV sequence heterogeneity observed in this study, one should take into consideration that all of these observations are based on analysis of only one small fragment of the TTV genome. Selection of sequences for phylogenetic analysis is very important. The identification of some genetic groups of HCV, for example, may be confusing if a highly variable region such as the E2 hypervariable region 1 or a highly conserved region such as the 5′ noncoding region or core region is used for the phylogenetic analysis (17, 19, 24, 28, 36). Another example that highlights the importance of sequence selection is the hepatitis G/GB-C virus (HGV/GBV-C). Some relatively small regions of the HGV/GBV-C genome are entirely unsuitable for the identification of consistent genetic groups, whereas larger fragments were successfully used to classify the HGV/BGV-C genome in a few genetic types (4, 12). Both examples emphasize that caution should be taken when interpreting results obtained from phylogenetic analysis based on a single region of the viral genome. However, a recent publication provided strong support to validate the phylogenetic analysis performed in the present study. It was shown that the TTV genetic grouping identified with a short sequence used in our study was entirely consistent with genetic grouping revealed by using a longer sequence of more than 2,000 bp (23). Nevertheless, more extensive studies using different regions of the TTV genome are warranted to support conclusions drawn in the present paper on the genetic classification of this virus.
TTV evolution.
The titer of the TTV DNA detected in plasma by PCR is usually very low, varying from 1 to 103 (21). The titration experiments performed on some serum specimens used in this study revealed a TTV DNA titer not exceeding 101. A low abundance of TTV DNA in plasma suggests a low rate of TTV replication in infected individuals. In general, a low rate of replication should correlate with a low rate of new mutations, generated mainly during genome replication by polymerase. In accordance with this concept, the TTV genome primary structure was found to be highly stable. As shown in this study and observed by others (3), the number of nucleotide changes within the TTV genome over time is very limited. For example, one study demonstrated that the TTV genome remained unchanged over a 31-month observation period (3). These data on the genetic stability of the TTV genome, however, contradict the enormous TTV sequence heterogeneity described in some studies (29, 30) and further elaborated in our study. The TTV genome exists in quasispecies and in every higher-level genetic group up to genotypes and even in supertypes and species (see above). To resolve this apparent contradiction, it was hypothesized that TTV may have had a very long history of evolution in humans (30).
The other important factor regarding TTV evolution is the capability of multiple genotypes and genetic variants (see above) to persist for a long period in one infected person (3, 21, 29, 31). One study, for example, showed that the nucleotide sequence of TTV genotypes 1 and 2 could be detected over a 15-month follow-up period without any alterations (3). This factor combined with the observation of very low TTV DNA titers in plasma (see above) suggests that all or the majority of TTV variants have a relatively low genetic fitness, which allows for a stable coexistence of several TTV variants for a very long time without significant changes in the relative abundance of any of them. As was hypothesized above, features of TTV evolution such as low rates of replication and generation of mutations, long history of human infections, low genetic fitness, and lack of strong competition between different TTV variants may explain the semidiscrete heterogeneity of the TTV genome. The observed heterogeneity pattern may be a reflection of a long, ongoing TTV evolutionary process which may permit the temporary coexistence of transient intermediate TTV variants that only slightly differ by their genetic fitness.
Another interesting feature of the TTV genome heterogeneity is the frequent detection of mutations that cause premature stop codons within ORF1. For example, two clones among nine obtained by cloning the TTV PCR fragment from specimen hm510 contained a 1-nt deletion (see above). It is interesting that the same 1-nt deletion was found in 1 of 10 cloned fragments obtained from plasma and in all 6 cloned fragments obtained from peripheral blood mononuclear cells (21). The presence of such a deletion changes the phase of translation and allows for the expression of alternative ORFs as C-terminal extensions of the abortive translation products of the incomplete ORF1. A relatively high frequency of such deletions in the TTV population suggests a potential functional significance of these products of alternative translation.
Mutations causing premature stop codons can be also detected by direct sequencing of PCR fragments, as was, for example, detected for specimen egt209 (see above). This finding indicates that the TTV genome containing such a mutation is predominant in this serum specimen. The presence of premature stop codons demonstrates that at least in some cases, a significant proportion of the TTV population does not contain the full-size ORF1. Taking into consideration that the analyzed TTV sequences are only 217 bp and speculating that the rate of occurrence of premature stop codons is approximately uniform within different parts of the TTV genome, only a small fraction, if any, of the TTV genome molecules should be capable of expressing the full-length ORF1 protein. The evidence that some other regions of ORF1 also may contain in-phase stop codons was found in the present study when fragments derived from different regions within the ORF1 were cloned and sequenced. One cloned sequence of fragment 130 at nt 586 to 1194 contained a nucleotide change from G in the tryptophan codon to A, which generated a stop codon TAG in the center of this fragment. This observation of the frequent occurrence of mutations preventing expression of the full-size ORF1-encoded protein suggests that the full-size ORF1 product is expressed at a very low rate. It is very tempting to speculate that low titer and low rate of replication of the TTV genome may be part of the mechanism of TTV persistence, and a significant fraction, if not all, of the TTV genome molecules may be unable to express all functions required for productive replication. Thus, the TTV population may be considered a population composed of a major proportion of incapacitated TTV genomes, and replication depends either on a collective expression of different proteins by all members of the population (when every member contributes at least one specific product of expression which cannot be efficiently expressed by the other members) and/or on a minor proportion of a TTV population able to express the full-size protein products.
TTV swarm.
One of the most interesting findings of this study is the detection of sequences amplified by PCR using TTV-specific primers (see Materials and Methods) that are distinctly less homologous to the sequences of any of the six identified TTV genotypes (see above). The comparison of these sequences to the other TTV sequences generated a separate peak e in the frequency distribution of evolutionary distances (Fig. 5). We interpreted the presence of this additional peak as a direct indication for the existence of a whole swarm of TTV-related viruses or TTV species, tentatively designated TTV-I (the prototype TTV), TTV-II, TTV-III, TTV-IV, and TTV-V (Fig. 6). This observation of the existence of a great degree of diversity among different TTV variants beyond genotypes was recently supported by the identification of the SANBAN TTV isolate, which shared a very low percent homology of nucleotide and amino acid sequences with the prototype TTV strain. This TTV isolate was hypothesized to belong to a new viral species (8).
The finding of these closely related viruses, which are more distant from each other than are other genotypes, points to a very interesting problem: what molecular criteria should be used for discrimination of different genotypes of one virus from different viruses? Undoubtedly, the frequency distribution of evolutionary distances or even mere percent homology may be used as one of several possible criteria for distinguishing one virus from another. The development of these molecular criteria is critical for molecular evolutionary studies and taxonomy and can be instrumental in solving many confusing issues in phylogenetic analysis. For example, 16 different TTV genotypes that are separated by a sequence divergence of more than 30% or evolutionary distances of more than 0.30 were recently identified (22). However, phylogenetic analysis performed accordingly to criteria developed in the present study showed that sequences of genotype 7 and 8 (22, 23) can be classified as genotype 1 and a new genotype 3 of TTV-II, whereas sequences of genotypes 10 to 16 (22, 23) can be classified into three different TTV-related species (data not shown). The development of new criteria for a better distinction between genotypes and viruses is urgently needed.
By using evolutionary distances calculated by the DNADIST program (see Materials and Methods), we found that the distances falling within the range from 0 to 0.34 were specific for different TTV strains. The sequences with evolutionary distances between 0.34 to 0.80 may be classified as genotypes, and those between 0.71 to 1.32 may be interpreted as different viral species. However, because of the overlap between ranges specific for different classification groups, a complete phylogenetic analysis should be performed to identify the genealogical relationships between sequences.
Finally, the identification of a swarm of closely related TTV-like viruses significantly substantiates the hypothesis of a long and slow evolutionary process of these viruses in humans. This swarm should have many more members than those found in this study, where only one set of PCR primers designed for the amplification of the prototype TTV-I sequences was used. The application of only one primer set can significantly limit the number of identified sequence variants. However, the bias introduced with these primers cannot affect genetic relatedness between already identified different genetic groups. It is conceivable that the use of primers derived from other TTV-like viruses may help to discover new TTV species from humans and possibly from animals. Moreover, the application of these new primers will allow a more accurate estimate of the prevalence of TTV-like infections in humans and may bring new light on the significance of these infections in causing diseases of unknown etiology.
REFERENCES
- 1.Alter M, Margolis H S, Krawczynski K, Judson F N, Mares A, Alexander W J, Hu P Y, Miller J K, Gerber M A, Samplner R E, Meeks E L, Beach M J. The natural history of community-acquired hepatitis C in the United States. N Engl J Med. 1992;327:1899–1905. doi: 10.1056/NEJM199212313272702. [DOI] [PubMed] [Google Scholar]
- 2.Biagini P, Gallian P, Cantaloube J-F, de Micco P, de Lamballerie X. Presence of TT virus in French blood donors and intravenous drug users. J Hepatol. 1998;29:684–685. doi: 10.1016/s0168-8278(98)80167-0. [DOI] [PubMed] [Google Scholar]
- 3.Biagini P, Gallian P, Houssam A, Cantaloube J-F, de Micco P, de Lamballerie X. Determination and phylogenetic analysis of partial sequences from TT virus isolates. J Gen Virol. 1999;80:419–424. doi: 10.1099/0022-1317-80-2-419. [DOI] [PubMed] [Google Scholar]
- 4.Cong M, Fried M W, Lambert S, Lopareva E N, Zhan M, Pujol F H, Thyagarajan S P, Byun K S, Fields H A, Khudyakov Y E. Sequence heterogeneity within three different regions of the hepatitis G virus genome. Virology. 1999;255:250–259. doi: 10.1006/viro.1998.9592. [DOI] [PubMed] [Google Scholar]
- 5.Davidson F, MacDonald D, Mokili J L, Prescott L E, Graham S, Simmonds P. Early acquisition of TT virus (TTV) in an area endemic for TTV infection. J Infect Dis. 1999;179:1070–1076. doi: 10.1086/314730. [DOI] [PubMed] [Google Scholar]
- 6.Desai S M, Muerhoff A S, Leary T P, Erker J C, Simons J N, Chalmers M L, Birkenmeyer L G, Pilot-Matias T J, Mushahwar I K. Prevalence of TT virus infection in US blood donors and populations at risk for acquiring parenterally transmitted viruses. J Infect Dis. 1999;179:1242–1244. doi: 10.1086/314735. [DOI] [PubMed] [Google Scholar]
- 7.Goto K, Sugiyama K, Terabe K, Mizutani F, Wada Y. Detection rates of TT virus among children who visited a general hospital in Japan. J Med Virol. 1999;57:405–407. doi: 10.1002/(sici)1096-9071(199904)57:4<405::aid-jmv13>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 8.Hijikata M, Takahashi K, Mishiro S. Complete circular DNA genome of a TT virus variant (isolate name SANBAN) and 44 partial ORF2 sequences implicating a great degree of diversity beyond genotypes. Virology. 1999;260:17–22. doi: 10.1006/viro.1999.9797. [DOI] [PubMed] [Google Scholar]
- 9.Hohne M, Berg T, Muller A R, Schreier E. Detection of sequences of TT virus, a novel DNA virus, in German patients. J Gen Virol. 1998;79:2761–2764. doi: 10.1099/0022-1317-79-11-2761. [DOI] [PubMed] [Google Scholar]
- 10.Ikeda H, Takasu M, Inoue K, Okamoto H, Miyakawa Y, Mayumi M. Infection with an unenveloped DNA virus (TTV) in patients with acute or chronic liver disease of unknown etiology and in those positive for hepatitis C virus RNA. J Hepatol. 1999;30:205–212. doi: 10.1016/s0168-8278(99)80063-4. [DOI] [PubMed] [Google Scholar]
- 11.Miyata H, Tsunoda H, Kazi A, Yamada A, Khan M A, Murakami J, Kamahora T, Shiraki K, Hino S. Identification of a novel GC-rich 113-nucleotide region to complete the circular, single-stranded DNA genome of TT virus, the first human circovirus. J Virol. 1999;73:3582–3586. doi: 10.1128/jvi.73.5.3582-3586.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muerhoff A S, Smith D B, Leary T P, Erker J C, Desai S M, Mushahwar I K. Identification of GB virus C variants by phylogenetic analysis of 5′-untranslated and coding region sequences. J Virol. 1997;71:6501–6508. doi: 10.1128/jvi.71.9.6501-6508.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mushahwar I K, Erker J C, Muerhoff A S, Leary T P, Simons J N, Birkenmeyer L G, Chalmers M L, Pilot-Matias T J, Desai S M. Molecular and biophysical characterization of TT virus: evidence for a new virus family infecting humans. Proc Natl Acad Sci USA. 1999;96:3177–3182. doi: 10.1073/pnas.96.6.3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakano T, Park Y M, Mizokami M, Choi J Y, Orito E, Ohno T, Kato T, Kondo Y, Tanaka Y, Kato H, Kato T, Kim B S. TT virus infection among blood donors and patients with non-B, non-C liver diseases in Korea. J Hepatol. 1999;30:389–393. doi: 10.1016/s0168-8278(99)80095-6. [DOI] [PubMed] [Google Scholar]
- 15.Niel C, de Oliveira J M, Ross R S, Gomes S A, Roggendorf M, Viazov S. High prevalence of TT virus infection in Brazilian blood donors. J Med Virol. 1999;57:259–263. [PubMed] [Google Scholar]
- 16.Nishizawa T, Okamoto H, Konishi K, Yoshizawa H, Miyakawa Y, Mayumi M. A novel DNA virus (TTV) associated with elevated transaminase levels in posttransfusion hepatitis of unknown etiology. Biochem Biophys Res Commun. 1997;241:92–97. doi: 10.1006/bbrc.1997.7765. [DOI] [PubMed] [Google Scholar]
- 17.Ohba K, Mizokami M, Ohno T, Suzuki K, Orito E, Ina Y, Lau J Y N, Gojobori T. Classification of hepatitis C virus into major types and subtypes based on molecular evolutionary analysis. Virus Res. 1995;36:201–214. doi: 10.1016/0168-1702(95)00007-d. [DOI] [PubMed] [Google Scholar]
- 18.Okamoto H, Mishiro S. Genetic heterogeneity of hepatitis C virus. Intervirology. 1994;37:68–76. doi: 10.1159/000150360. [DOI] [PubMed] [Google Scholar]
- 19.Okamoto H, Akahane Y, Ukita M, Fukuda M, Tsuda F, Miyakawa Y, Mayumi M. Fecal excretion of a nonenveloped DNA virus (TTV) associated with posttransfusion non-A-G hepatitis. J Med Virol. 1998;56:128–132. [PubMed] [Google Scholar]
- 20.Okamoto H, Nishizawa T, Kato N, Ukita M, Ikeda H, Iizuka H, Miyakawa Y, Mayumi M. Molecular cloning and characterization of a novel DNA virus (TTV) associated with posttransfusion hepatitis of unknown etiology. Hepatol Res. 1998;10:1–16. [Google Scholar]
- 21.Okamoto H, Kato N, Iizuka H, Tsuda F, Miyakawa Y, Mayumi M. Distinct genotypes of a nonenveloped DNA virus associated with posttransfusion non-A to G hepatitis (TT virus) in plasma and peripheral blood mononuclear cells. J Med Virol. 1999;57:252–258. doi: 10.1002/(sici)1096-9071(199903)57:3<252::aid-jmv7>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 22.Okamoto H, Takahashi M, Nishizawa T, Ukita M, Fukuda M, Tsuda F, Miyakawa Y, Mayumi M. Marked genomic heterogeneity and frequent mixed infection of TT virus demonstrated by PCR with primers from coding and noncoding regions. Virology. 1999;259:428–436. doi: 10.1006/viro.1999.9770. [DOI] [PubMed] [Google Scholar]
- 23.Okamoto H, Nizhizawa T, Ukita M, Takahashi M, Fukuda M, Iizuka H, Miyakawa Y, Mayumi M. The entire nucleotide sequence of a TT virus isolate from the United States (TUS01): comparison with reported isolates and phylogenetic analysis. Virology. 1999;259:437–448. doi: 10.1006/viro.1999.9769. [DOI] [PubMed] [Google Scholar]
- 24.Simmonds P, Mellor J, Sakuldamrongpanich T, Nuchaprayoon C, Tanprasert S, Holmes E C, Smith D B. Evolutionary analysis of variants of hepatitis C virus found in South-East Asia: comparison with classifications based upon sequence similarity. J Gen Virol. 1996;77:3013–3024. doi: 10.1099/0022-1317-77-12-3013. [DOI] [PubMed] [Google Scholar]
- 25.Simmonds P, Davidson F, Lycett C, Prescott L E, MacDonald D M, Ellender J, Yap P L, Ludlam C A, Haydon G H, Gillon J, Jarvis L M. Detection of a novel virus (TTV) in blood donors and blood products. Lancet. 1998;352:191–195. doi: 10.1016/s0140-6736(98)03056-6. [DOI] [PubMed] [Google Scholar]
- 26.Strimmer K, von Haeseler A. Quartet puzzling: a quartet maximum-likelihood method for reconstructing tree topologies. Mol Biol Evol. 1996;13:964–969. [Google Scholar]
- 27.Strimmer K, von Haeseler A. Likelihood-mapping: a simple method to visualize phylogenetic content of a sequence alignment. Proc Natl Acad Sci USA. 1997;94:6815–6819. doi: 10.1073/pnas.94.13.6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stuyver L, Wyseur A, van Arnhem W, Lunel F, Laurent-Puig P, Pawlotsky J M, Kleter B, Bassit L, Nkengasong J, van Doorn L J. Hepatitis C virus genotyping by means of 5′-UR/core line probe assays and molecular analysis of untypeable samples. Virus Res. 1995;38:137–157. doi: 10.1016/0168-1702(95)00052-r. [DOI] [PubMed] [Google Scholar]
- 29.Takayama S, Yamazaki S, Matsuo S, Sugii S. Multiple infection of TT virus (TTV) with different genotypes in Japanese hemophiliacs. Biochem Biophys Res Commun. 1999;256:208–211. doi: 10.1006/bbrc.1999.0270. [DOI] [PubMed] [Google Scholar]
- 30.Tanaka Y, Mizokami M, Orito E, Ohno T, Nakano T, Kato T, Kato H, Mukaide M, Park Y-M, Kim B-S, Ueda R. New genotypes of TT virus (TTV) and a genotyping assay based on restriction fragment length polymorphism. FEBS Lett. 1998;437:201–206. doi: 10.1016/s0014-5793(98)01231-9. [DOI] [PubMed] [Google Scholar]
- 31.Tanaka H, Okamoto H, Luengrojanakul P, Chainuvati T, Tsuda F, Tanaka T, Miyakawa Y, Mayumi M. Infection with an unenveloped DNA virus (TTV) associated with posttransfusion non-A to G hepatitis in hepatitis patients and healthy blood donors in Thailand. J Med Virol. 1998;56:234–238. doi: 10.1002/(sici)1096-9071(199811)56:3<234::aid-jmv10>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 32.Tanaka Y, Mizokami M, Orito E, Nakano T, Kato T, Ding X, Ohno T, Ueda R, Sonoda S, Tajima K, Miura T, Hayami M. A new genotype of TT virus (TTV) infection among Colombian native indians. J Med Virol. 1999;57:264–268. doi: 10.1002/(sici)1096-9071(199903)57:3<264::aid-jmv9>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 33.Tangkijvanich P, Hirsch P, Theamboonlers A, Nuchprayoon I, Poovorawan Y. Association of hepatitis viruses with hepatocellular carcinoma in Thailand. J Gastroenterol. 1999;34:227–233. doi: 10.1007/s005350050248. [DOI] [PubMed] [Google Scholar]
- 34.Tsuda F, Okamoto H, Ukita M, Tanaka T, Akahane Y, Konishi K, Yoshizawa H, Miyakawa Y, Mayumi M. Determination of antibodies to TT virus (TTV) and application to blood donors and patients with post-transfusion non-A to G hepatitis in Japan. J Virol Methods. 1999;77:199–206. doi: 10.1016/s0166-0934(98)00154-2. [DOI] [PubMed] [Google Scholar]
- 35.Viazov, S., R. S. Ross, C. Niel, J. M. de Oliveira, C. Varenholz, G. Da Villa, and M. Roggendorf. Sequence variability in the putative coding region of TT virus: evidence for two rather than several major types. J. Gen. Virol. 79:3085–3089. [DOI] [PubMed]
- 36.Yun Z, Lara C, Johansson B, de Rivera I L, Sonnerborg A. Discrepancy of hepatitis C virus genotypes as determined by phylogenetic analysis of partial NS5 and core sequences. J Med Virol. 1996;49:155–160. doi: 10.1002/(SICI)1096-9071(199607)49:3<155::AID-JMV1>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]