Abstract
A 20-year-old female hypogammaglobulinemic patient received monotypic Sabin 3 vaccine in 1962. The patient excreted type 3 poliovirus for a period of 637 days without developing any symptoms of poliomyelitis, after which excretion appeared to have ceased spontaneously. The evolution of Sabin 3 throughout the entire period of virus excretion was studied by characterization of seven sequential isolates from the patient. The isolates were analyzed in terms of their antigenic properties, virulence, sensitivity for growth at high temperatures, and differences in nucleotide sequence from the Sabin type 3 vaccine. The isolates followed a main lineage of evolution with a rate of nucleotide substitution that was very similar to that estimated for wild-type poliovirus during person-to-person transmission. There was a delay in the appearance of antigenic variants compared to sequential type 3 isolates from healthy vaccines, which could be one of the possible explanations for the long-term excretion of virus from the patient. The distribution of mutations in the isolates identified regions of the virus possibly involved in adaptation for growth in the human gut and virus persistence. None of the isolates showed a full reversion of the attenuated and temperature-sensitive phenotypes of Sabin 3. Information of this sort will help in the assessment of the risk of spread of virulent polioviruses from long-term excretors and in the design of therapies to stop long-term excretion. This will make an important contribution to the decision-making process on when to stop vaccination once wild poliovirus has been eradicated.
Poliovirus, the agent responsible for paralytic poliomyelitis, is a member of the Picornaviridae family, a group of nonenveloped positive-strand RNA viruses. The coding region of the genome is translated as a single polyprotein and is then processed to generate the viral capsid and nonstructural proteins (Fig. 1). The coding region is preceded by a long 5′ noncoding region (NCR) of approximately 740 nucleotides that contains important determinants of virulence (32, 46).
FIG. 1.
Organization of the genome of poliovirus. The protease (P3C) and polymerase (P3D) can undergo an alternative cleavage within 3D mediated by P2A to give P3C′ and P3C′. Reprinted from the Journal of General Microbiology (32) with permission of the publisher.
The Sabin oral poliovirus vaccine, composed of live-attenuated strains of the three poliovirus serotypes (38), has been used to control and reduce greatly the incidence of poliomyelitis around the world during the last 30 years. Sabin strains replicate in the guts of immunocompetent persons for a limited period after vaccination (2). During this period, which ranges between several days and 3 months, vaccinees excrete viruses in which mutations and genetic rearrangements are selected very rapidly (32). These changes occur in a sequential manner, probably as a response to different selective pressures in the gut (29).
In the case of Sabin 3-derived viruses, the changes always involve a reversion at nucleotide 472 of the 5′ NCR (U to C) (9) and one or several coding mutations in the capsid proteins that restore a defect in virus assembly caused by a mutation at capsid residue VP3-91 (22). These mutations result in partial or total reversion of the attenuation phenotype of Sabin 3 (22). More strikingly, virtually all type 3 isolates excreted by healthy vaccinees (given Sabin trivalent vaccine) more than 11 days after vaccination display major genetic rearrangements as a consequence of genetic recombination events (3; A. J. Macadam, personal communication). These recombination events generate type 3 viruses containing the 5′ region of the genome, including the capsid coding region, from type 3 vaccine virus and large portions of the 3′ half of the genome, which involves the nonstructural coding region, from either type 1, type 2, or both viral genomes. Molecular analysis of these viruses has revealed that type 3 recombinant isolates excreted from healthy vaccinees share common genetic features (3, 18; A. J. Macadam, personal communication). The results indicate that viruses containing the carboxyl-terminal region of the nonstructural protein 2C and the amino-terminal region of polymerase 3D from Sabin 3 are selected against in the gut. One of the possible explanations for this is that Sabin 3 lacks a proteolytic cleavage site located in the amino-terminal region of 3D which is present in most poliovirus strains and therefore might confer a selective advantage for virus replication in vivo (32). This site is incorporated into the genome of type 3 recombinant viruses from type 1 or type 2 sequences. Cleavage at this site results in an alternative processing of the protease-polymerase 3CD polypeptide. The biological significance of this alternative processing of 3CD has not yet been determined (21).
Although poliovirus strains excreted by vaccines often show an increase in virulence with respect to the vaccine strains, the rate of vaccine-associated poliomyelitis (VAP) among healthy vaccinees is very low and the vaccine is considered to be very safe (34). However, the situation is different in persons with defects in their immune systems. Immunodeficient people, particularly those with antibody deficiencies, have a much higher risk of VAP (iVAP) than do immunocompetent individuals (an estimated 3,000-fold excess) (41). Sabin viruses can replicate for very long periods in these patients, and in some cases, this leads to the fatal disease (13, 17, 26, 39; G. Dunn, unpublished results). Recent genetic analysis of virus isolates from an asymptomatic immunodeficient person (Dunn, unpublished) or from an immunodeficient person with iVAP (17) have allowed us and others to estimate that excretion of poliovirus can continue for as long as 10 years after vaccination of immunocompromised persons.
Early studies proposed that reversion of some vaccine phenotypes might be slower in chronically infected antibody-deficient patients (27). However, there is very little information about the molecular properties of polioviruses excreted by immunodeficient patients from the time of vaccination. Patients with diagnosed immunological anomalies are generally not vaccinated with the live-attenuated strains, and only virus isolates recovered after the onset of iVAP are regularly available.
In this paper we describe the genetic and phenotypic properties of isolates from a hypogammaglobulinemic patient, who was fed monotypic Sabin 3 vaccine as part of a Medical Research Council study in the late 1950s and early 1960s (26). Seven sequential isolates were studied to monitor the evolution of the virus during the entire period of excretion. The isolates extend from day 36 until day 637 after vaccination, when excretion appeared to have ceased spontaneously.
The results are discussed in terms of evolution of the Sabin 3 poliovirus strain in the human gut and the possible implications of chronic virus excretion for the strategy of global eradication of polio.
MATERIALS AND METHODS
Clinical history of the patient.
The patient (26) was a 20-year-old woman (222f), who had begun intermittent replacement therapy in April 1957 at the age of 15 (at which time her immunoglobulin G [IgG] level was 400 mg/ml) because she was suffering from recurrent infections. Her IgG level fell during the next year to 200 mg/ml in November 1958 and 120 mg/ml in March 1959. When her serum was examined before any injections of immunoglobulin, the titer of neutralizing antibodies for all three polioviruses was 1:16. In November 1958, the titers were about 1:4. At the time when she was given monovalent type 1 virus orally in December 1961, her IgG level was about 500 mg/ml and the titers of poliovirus neutralizing antibodies were 1:8, 1:8, and 1:32 for types 1, 2, and 3, respectively. No type 1 virus was recovered, and monovalent type 3 was given on 20 January 1962. Type 3 virus was recovered at regular intervals from then until December 1962, when virus excretion became intermittent; it continued so until 30 October 1963. A stool sample collected on 27 November 1963 was negative, as were six consecutive stool samples collected at weekly intervals until the end of January 1964. Thus excretion occurred for about 21 months. The number of excreted virus was never greater than 102 tissue culture infective doses per g of stool. An attempt to interfere with the excretion of type 3 virus by giving type 2 virus after excretion had been occurring for 3 months was unsuccessful, and no type 2 virus was recovered.
Viruses.
Virus samples taken from days 36, 136, 307, 391, 442, 480, and 637 (isolates H36 to H637) after patient 222f was fed with monotypic Sabin 3 vaccine were analyzed in this study. Isolation of viruses was performed as previously described (27). The rest of the type 3 polioviruses examined were obtained from laboratory stocks. Working virus preparations were collected by growth of the viruses in HEp-2C cells in minimal essential medium without fetal calf serum at 35°C. The virus stocks were stored at −70°C.
Cells.
HEp-2C cells were used in the different assays described in this paper and were grown in culture as described previously (22).
Nucleotide sequencing.
Viral cDNA was synthesized by reverse transcription of purified viral RNA followed by amplification by PCR, and the products were sequenced by the use of an ABI Prism 310 Genetic Analyzer as specified by the manufacturer.
Neutralization of virus isolates with monoclonal antibodies.
Neutralization of virus infectivity with monoclonal antibodies specific for the four different antigenic sites identified in Sabin 3 was performed as described elsewhere (29).
Temperature sensitivity.
Temperature sensitivity was assayed by observation of plaque formation on HEp-2C cells at 39.5 and 40.0°C as described previously (28).
Neurovirulence.
Viruses were assayed by the standard World Health Organization-approved test for vaccine safety (44), except that fewer animals were used per virus.
In vivo labelling of viral proteins.
Viral polypeptides were labelled with 35S essentially as described previously (23), except that Tran35S-label and methionine- and cysteine-free medium (ICN) were used. Infected HEp-2C cell lysates were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
RESULTS
Genomic analysis of the type 3 strains isolated from the immunodeficient patient.
To study the sequence evolution undergone by Sabin 3 during long-term replication in the immunodeficient patient, the genomes of seven sequential strains (H36 to H637) isolated from the patient were analyzed by nucleotide sequencing (nucleotide 55 to the 3′ end). The isolates corresponded to days 36, 136, 307, 391, 442, 480, and 637 after patient 222f was fed with monotypic Sabin 3 vaccine, so that intertypic recombination was unlikely.
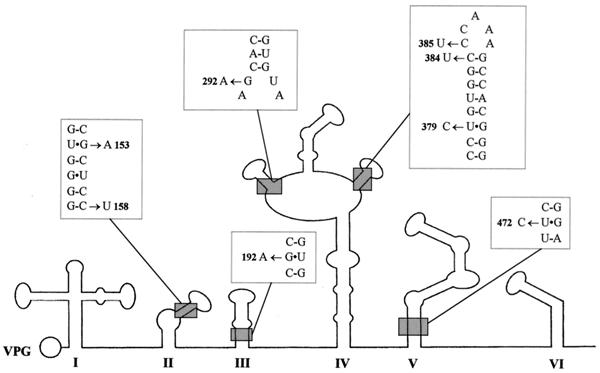
Table 1 shows the differences in the sequences of the strains in the 5′-end NCR with respect to Sabin 3. All isolates had a reversion (U to C) at position 472 of the 5′ NCR. Nucleotide 472 is located in domain V of the internal ribosome entry site (IRES), where mutations play a very important role in the attenuation phenotype of all three Sabin strains (15, 25, 36, 43). This change restores a G · C base pair in domain V, and it is known to occur very rapidly upon replication of the virus in the human gut (8). Viral isolates contained other changes in the 5′ NCR including mutations in other domains of the IRES from day 391 (Table 1 and Fig. 2). Some of the changes had a potential effect on the secondary-structure stability of the IRES by strengthening or weakening the predicted base-paired composition of the stems in domains II, III, and IV (Fig. 2) (40). All isolates except H480 showed a reversion (G to A) at position 7432 of the 3′ NCR just prior to the poly(A) tract. This change has been observed in some isolates from healthy vaccinees and patients with VAP but does not seem to play a significant role in attenuation (43).
TABLE 1.
Nucleotide differences between isolates from the immunodeficient patient and Sabin 3 in the different domains of the 5′ NCR of the genome
| Strain | Nucleotide change(s) in:
|
|||||
|---|---|---|---|---|---|---|
| Domain II | Domain III | Domain IV | Domain V | Hypervariable region | Other regions | |
| H36 | U472C | C680U | ||||
| H136 | U472C | U676A | ||||
| H307 | U472C | U676A | 101Ca | |||
| H391 | G147A, G153A | G192A | U261C, G292A, C384U | U472C | U676A | U92C |
| H442 | G147A, G153A | G192A | U261C, G292A, C384U | U472C | U676A | U92C |
| H480 | G147A, G153A | G192A | U261C, G292A, C384U, C385U | U472C | C649U, U676A | U92C, A115U |
| H637 | G147A, G153A, C158U | G192A | G292A, U379C, C384U, C385U | U472C | C649U, U676A, A722G | U92C, A115U, G445A |
Nucleotide insertion.
FIG. 2.
Predicted secondary structure of the first 620 bases of the 5′ NCR of poliovirus type 3. The locations of mutations with a potential effect on the secondary-structure stability of the region found in isolates from patient 222f are indicated.
Amino acid substitutions.
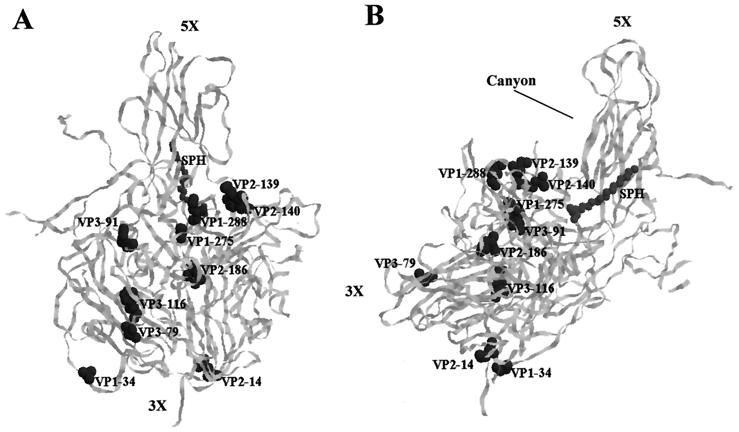
Amino acid changes in the region of the genome coding for the structural (capsid) proteins of the different strains with respect to Sabin 3 are shown in Table 2. Several coding mutations arose and persisted in the capsid proteins of the sequential isolates. As represented in Fig. 3, these included mutations at different locations in the virion. All strains showed a reversion to wild-type sequence at amino acid VP3-91, which is located at the interface between protomers and is involved in capsid assembly and the attenuation phenotype of the vaccine strain (10, 23). Reversion of this phenotype occurs in isolates from vaccinees and patients with VAP either by direct back mutation at position VP3-91 or by secondary changes in other capsid residues that suppress the effect of VP3-91 Phe (22). Residues VP2-139 and VP2-140, which had changed on days 391 and 136, respectively, are located in the south rim of the surface depression (“canyon”) that surrounds the fivefold axis of symmetry and form part of the putative receptor footprint (4, 35). Amino acid VP1-34, mutated from day 136, maps to the amino-terminal loop of VP1, which makes contact with VP4 and forms part of the complex internal network of the virion essential for capsid stability (5). VP2-14 also changed from day 136 and is situated on the border of a seven-stranded β-sheet structure that is responsible for the stable interactions between pentameric subunits (5, 10). Mutations at VP1-275 and VP2-186 were acquired early in the infection (before day 36); they are located relatively close to each other in the proximity of antigenic site 3a (30), where a change was also detected from day 136 (residue VP1-288). A mutation in antigenic site 4, at amino acid 79 of VP3, and another at internal residue 116 of VP3 were detected on day 391 and persisted until the end of the infection.
TABLE 2.
Amino acid changes between isolates from the immunodeficient patient and Sabin 3 in the capsid region
| Protein | Amino acid | Amino acid in strain:
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Sabin 3 | H36 | H136 | H307 | H391 | H442 | H480 | H637 | ||
| VP1 | 12 | Gly | —a | — | — | — | Ser | — | — |
| 34 | Ala | — | Val | Val | Val | Val | Val | Val | |
| 54 | Ala | — | — | Thr | — | — | — | — | |
| 105 | Met | — | — | Thr | — | — | — | — | |
| 166 | Lys | — | — | Arg | — | — | — | — | |
| 256 | Val | — | — | — | — | — | — | Ile | |
| 275 | Ala | Thr | Ser | Thr | — | Thr | Thr | Thr | |
| 288 | Asn | — | Asp | Asp | Asp | Asp | Asp | Asp | |
| VP2 | 14 | Leu | — | Met | Met | Met | Met | Met | Met |
| 76 | Lys | — | Ile | — | — | — | — | — | |
| 139 | Gln | — | — | Tyr | His | His | His | His | |
| 140 | Arg | — | Met | Met | Met | Met | Met | Met | |
| 155 | Lys | — | — | — | — | Ile | — | — | |
| 168 | Ser | — | Pro | — | — | — | — | — | |
| 170 | Lys | — | — | — | — | — | — | Arg | |
| 186 | Leu | Met | Met | Met | Met | Met | Met | Met | |
| VP3 | 59 | Ser | — | — | Asn | — | — | — | — |
| 79 | Ser | — | — | — | Thr | Thr | Thr | Thr | |
| 91 | Phe | Ser | Ser | Ser | Ser | Ser | Ser | Ser | |
| 116 | Phe | — | — | — | Leu | Leu | Leu | Leu | |
| 209 | Ser | — | — | — | — | — | — | Cys | |
—, same as in Sabin 3.
FIG. 3.
Ribbon diagram of the α-carbon trace of the Sabin 3 promoter (10), viewed from the front (A) and side (B) of the virion (in panel B, the outside is toward the top left of the image and the inside is toward the bottom right). The virus particle consists of 60 protomers, each containing a single copy of VP1, VP2, VP3, and VP4, arranged in icosahedral symmetry. The fivefold and threefold axes of symmetry and the canyon are labelled. Amino acid substitutions selected and maintained in sequential isolates from patient 222f are highlighted. The location of sphingosine (SPH) in the hydrocarbon-binding pocket is also indicated.
Other mutations were located in regions of possible biological importance but were not maintained during the infection course (Table 2). Amino acids VP1-105, VP1-166, and VP1-256, for example, are situated in or close to the north rim of the canyon on the roof of the conserved hydrophobic pocket, which is normally occupied by an endogenous lipid and where drugs that prevent virus uncoating are inserted (10, 33).
Coding mutations were also present in nonstructural proteins and involved protease 2A; proteins 2B, 2C, and 3A; VPg; protease 3C; and the viral polymerase 3D (Table 3). Less is known of the structural and functional significance of these changes, but some mutations appeared at different stages of the infection and were maintained from then on, presumably because they conferred some biological advantage. Remarkably, several mutations were found in the carboxyl-terminal region of protein 2C from isolate H307 onwards (Table 3). The changes map in a cysteine-rich domain (2C-269–302) that is conserved in enteroviruses and rhinoviruses and in the proximity of an arginine-rich region that constitutes a putative RNA binding motif (2C-312–317) (37, 42). Mutation at residue 3D-147 (Ala to Thr), also present from day 307, generates a recognition site (TY/G) for cleavage by the viral protease 2A.
TABLE 3.
Amino acid changes between isolates from the immunodeficient patient and Sabin 3 in the nonstructural region
| Protein | Amino acid | Amino acid in strain:
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Sabin 3 | H36 | H136 | H307 | H391 | H442 | H480 | H637 | ||
| 2A | 41 | Val | —a | — | — | Ile | Ile | Ile | Ile |
| 101 | His | — | — | — | Tyr | — | Tyr | Tyr | |
| 2B | 88 | Thr | — | — | Ile | — | — | — | — |
| 2C | 33 | Arg | — | — | Lys | Lys | Lys | Lys | Lys |
| 35 | Arg | — | — | Lys | — | — | — | — | |
| 103 | Lys | — | Arg | Arg | Arg | Arg | Arg | Arg | |
| 263 | Ala | — | — | Ser | — | — | — | — | |
| 268 | Thr | — | — | Met | Met | Met | Met | Met | |
| 270 | Lys | — | — | — | Glu | Glu | Glu | Glu | |
| 295 | Lys | — | — | — | Arg | Arg | Arg | Arg | |
| 311 | Ile | — | — | — | Val | Val | Val | Val | |
| 318 | Ser | — | — | — | — | Phe | — | — | |
| 3A | 12 | Ile | — | — | — | — | — | Val | Val |
| 86 | His | — | — | — | — | — | Gln | Gln | |
| VPg | 18 | Ala | — | Thr | Thr | Thr | Thr | Thr | Thr |
| 3C | 16 | Val | Ile | — | — | — | — | — | — |
| 51 | Asp | — | — | Glu | Glu | Glu | Glu | Glu | |
| 60 | Lys | — | — | — | Arg | Arg | Arg | Arg | |
| 74 | Ile | Val | — | — | — | — | — | — | |
| 131 | Ile | Thr | Thr | Thr | Thr | Thr | Thr | Thr | |
| 3D | 4 | Gln | — | His | His | His | His | His | His |
| 52 | Thr | — | — | — | — | — | Ala | Ala | |
| 147 | Ala | — | — | Thr | Thr | Thr | Thr | Thr | |
| 158 | Val | — | — | — | — | — | Ile | Ile | |
| 166 | Thr | — | — | — | — | — | — | Ala | |
| 170 | Gln | Leu | — | — | — | — | — | — | |
| 176 | Arg | — | Lys | — | — | — | — | — | |
| 200 | Arg | Lys | — | Lys | — | — | — | Lys | |
| 421 | Leu | — | — | — | — | — | — | Phe | |
| 501 | Ile | — | Leu | Leu | Leu | Leu | Leu | Leu | |
—, same as in Sabin 3.
Significance of the mutations in the carboxyl-terminal region of protein 2C and at amino acid 3D-147 (alternative cleavage site).
Both the carboxyl-terminal region of protein 2C and amino acid 3D-147 are found within the region of the genome of type 3 recombinant viruses, isolated from healthy vaccinees or patients with VAP, that is exchanged by recombination with either Sabin 1 or Sabin 2 genomes (3, 22; Macadam, unpublished). Interestingly, Sabin 1 and Sabin 2 strains also contain differences in sequence in those regions with respect to Sabin 3. To evaluate the relevance of these mutations, the sequences in the carboxyl-terminal region of protein 2C and at amino acid 3D-147 of the isolates from patient 222f were compared to those of four distantly related wild type 3 viruses, two Sabin 3-derived VAP strains, and Sabin 1 and 2 strains. As shown in Table 4, all the viruses except the two first isolates from the immunodeficient patient showed changes with respect to Sabin 3. Amino acid 2C-268 changed in patient 222f from day 307, from a threonine to a methionine, and was found to be a methionine in all the rest of the viruses except for Sabin 3 and one VAP isolate. Sabin 1 and 2 strains, the four wild type 3 poliovirus strains, and the two Sabin 3-derived VAP isolates contain a change at residue 2C-270 with respect to Sabin 3, from an aspartic acid to an asparagine, whereas amino acid 2C-271 changed from day 391, from a lysine to a glutamic acid, in isolates from patient 222f. Isolates from the immunodeficient patient also incorporated changes from day 391 at residue 2C-295, from a lysine to an arginine, and at amino acid 2C-310, from an isoleucine to a valine, which is also a valine in Sabin 2, two of the wild type 3 polioviruses, and one VAP isolate. Sabin 1 and Sabin 2 strains and the four wild type 3 viruses have a threonine at position 3D-147, which changed in the immunodeficient patient from day 307 from an alanine to a threonine, generating the alternative proteolytic cleavage site in the 3CD polypeptide.
TABLE 4.
Sequence differences between isolates from the immunodeficient patient, wild type 3 polioviruses, Sabin 3-derived viruses from VAP patients, and Sabin strains in the carboxyl-terminal region of protein 2C and at amino acid 3D-147
| Origin | Strain | Amino acid at protein 2C
|
Amino acid at 3C′/3D′ (3D-147) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 268 | 270 | 271 | 295 | 310 | 313 | 318 | |||
| Vaccine | Sabin 3 | Thr | Lys | Asp | Lys | Ile | Arg | Ser | Ala |
| Sabin 1 | Met | —d | Asn | — | — | — | — | Thr | |
| Sabin 2 | Met | — | Asn | — | Val | — | — | Thr | |
| Wilda | SK (1950) | Met | — | Asn | — | — | — | — | Thr |
| 22 (1959) | Met | — | Asn | — | — | — | — | Thr | |
| 655 (1971) | Met | — | Asn | — | Val | — | — | Thr | |
| 23127 (1984)b | Met | — | Asn | — | Val | Lys | — | Thr | |
| Sabin 3 derivedc | 4968 (1983) | Met | — | Asn | — | Val | — | — | Ala |
| 2073 (1990) | — | — | Asn | — | — | — | — | Ala | |
| Patient 222f | H36 | — | — | — | — | — | — | — | Ala |
| H136 | — | — | — | — | — | — | — | Ala | |
| H307 | Met | — | — | — | — | — | — | Thr | |
| H391 | Met | Glu | — | Arg | Val | — | — | Thr | |
| H442 | Met | Glu | — | Arg | Val | — | Phe | Thr | |
| H480 | Met | Glu | — | Arg | Val | — | — | Thr | |
| H637 | Met | Glu | — | Arg | Val | — | — | Thr | |
Wild viruses: SK (P3/Saukett/USA), 22 (P3/III-22/USA), 655 (P3/655/Sweden), 23127 (P3/23127/Finland). The year of isolation is indicated in parentheses.
Data from reference 14.
Sabin 3-derived isolates from suspected VAP patients 4968 (P3/4968/Saudi Arabia), 2073 (VI/2073/Singapore). The year of isolation is indicated in parentheses.
—, same as in Sabin 3.
Genetic evolution of the strains.
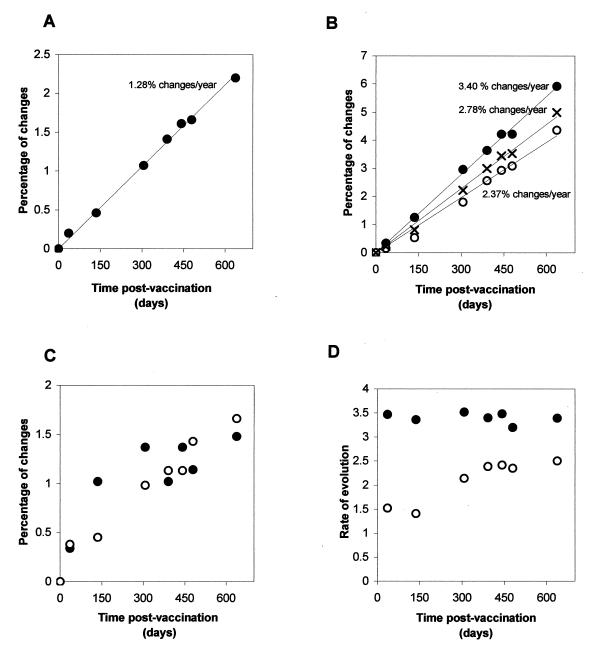
Figure 4 shows the accumulation of changes in the genome of the type 3 strains excreted by the immunodeficient patient during the 637 days that virus excretion lasted. Incorporation of nucleotide changes over the whole genome, expressed as a percentage of changes with respect to the Sabin 3 genome at the different excretion points, is shown in Fig. 4A. The data could be adjusted to a linear function (y = 0.0035x; R2 = 0.9971) which gave a rate of evolution of 1.28% of nucleotide changes over the entire genome. This rate corresponds to 1.83 nucleotide changes per week over the entire genome, which is in the range of the rate of between 1 and 2 changes per week that has been estimated for wild-type poliovirus evolution during person-to-person transmission (16).
FIG. 4.
Accumulation of changes in the genome of type 3 isolates from the immunodeficient patient with respect to the Sabin 3 genome during the period of excretion. (A) Percentage of nucleotide changes with respect to Sabin 3 over the whole genome. (B) Percentage of changes in synonymous third-base codon positions. (C) Percentage of amino acid substitutions. (D) Rate of sequence evolution at synonymous third-base codon positions calculated at each excretion point independently and extrapolated as percentage of changes per year. Symbols: ●, capsid region; ○, nonstructural region. ×, whole coding region.
To obtain a better estimate of the rate of sequence evolution, synonymous changes at third-base codon positions in the coding regions of the genome were analyzed alone. Changes at those positions are assumed to have a neutral effect on virus fitness and have been shown to more accurately reflect the genetic relationships between poliovirus strains (17, 19). The values were represented together or separately for the capsid and nonstructural regions and are displayed in Fig. 4B. The data adjusted to linear functions in the entire coding region (y = 0.0076x; R2 = 0.9954), the capsid region (y = 0.0093x; R2 = 0.9973), and the nonstructural region (y = 0.0065x; R2 = 0.9873). These data produced estimated rates of evolution of 2.78, 3.40, and 2.37% of nucleotide substitutions at synonymous third-base codon positions/year over the entire coding region, the capsid region, and the nonstructural regions, respectively.
To evaluate if the rate of sequence evolution had varied during the excretion period, the rate of change in synonymous third-base codon positions was calculated by considering each excretion point independently. As shown in Fig. 4D, the rate of evolution was rather constant during the excretion period. However, isolates from days 36 and 136 showed values in the nonstructural region that were significantly lower than those for the rest of the isolates.
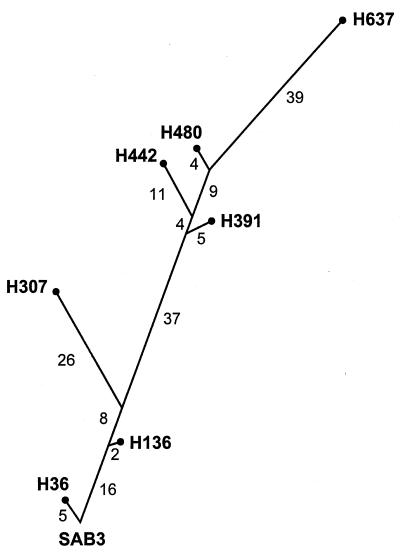
The data from these analyses were used to elaborate a phylogenetic tree representing the genetic relationships between the strains. As shown in Fig. 5, the phylogenetic tree revealed a branched structure where a main genotypic lineage and various minor branches could be distinguished. The branches extended for different lengths of time from the main lineage, presumably reflecting times during the infection when viruses coreplicated independently in different sites of the intestinal tract. Viruses from genotypic branches that led to isolates H307 and H442, for example, seem to have coexisted separately with strains from the main lineage for approximately 150 and 70 days, respectively (Fig. 5). Analysis of the accumulation of amino acid changes within each branch strongly supports this idea (Tables 2 and 3). Viruses from minor lineages seemed to have subsequently disappeared or been outcompeted by strains from the main genotypic lineage.
FIG. 5.
Phylogenetic tree showing the genetic relationships between the isolates excreted by the immunodeficient patient. The tree is a mere representation of the differences at synonymous third-base codon positions between sequential isolates over the entire coding region. The number in each isolate represents the date of isolation (days after vaccination). The number of changes accumulated in each segment is shown.
The selection of changes in the IRES in the 5′ NCR seemed to have varied during the infection. Isolate H307 contained only two changes in this region, while strain H391 and the strains thereafter had eight or more (Table 1).
Changes in amino acid in the capsid and nonstructural regions followed different patterns. Viruses acquired changes in capsid residues more rapidly during the initial stages of infection, whereas mutations in nonstructural proteins seemed to have accumulated in a more uniform manner (Fig. 4C).
Antigenic structure.
The antigenic structure of the strains excreted by the immunodeficient patient was determined by studying the reactivity of the viruses with a panel of monoclonal antibodies of known specificity (30). The results are presented in Table 5. Antibodies specific for the immunodominant site 1 neutralized all strains. Isolates H136 and H637 failed to react with three or two antigenic site 2 antibodies, respectively. All isolates except H36 were resistant to neutralization by antibodies specific for site 3. Finally, strains H391, H442, H480, and H637 lost reactivity with monoclonal antibody 1281, which is specific for antigenic site 4. The results were consistent with the observed amino acid changes at the predefined antigenic sites (Table 2).
TABLE 5.
Reactivity of isolates from the immunodeficient patient with monoclonal antibodies specific for different antigenic sites of Sabin 3
| Strain | Reactivity with antibody toa:
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Site 1
|
Site 2
|
Site 3
|
Site 4
|
||||||||
| 134 | 194 | 204 | 472 | 838 | 877 | 880 | 882 | 138 | 885 | 1281 | |
| Sabin 3 | + | + | + | + | + | + | + | + | + | + | + |
| H36 | + | + | + | + | + | + | + | + | + | + | + |
| H136 | + | + | + | + | − | − | − | − | − | − | + |
| H307 | + | + | + | + | + | + | + | + | − | − | + |
| H391 | + | + | + | + | + | + | + | + | − | − | − |
| H442 | + | + | + | + | + | + | + | + | − | − | − |
| H480 | + | + | + | + | + | + | + | + | − | − | − |
| H637 | + | + | + | + | − | − | − | + | − | − | − |
+ and −, neutralization or not, respectively, of infection of HEp-2C cells with 100 PFU of virus using a 1:50 dilution of antibody, as described in reference 29.
Temperature sensitivity and neurovirulence.
The results for the temperature sensitivity and neurovirulence of the strains are shown in Table 6. All strains exhibited some degree of temperature sensitivity for growth in HEp-2C cells compared to the wild strain Leon. Isolates H136, H307, H442, and H637 showed a similar or greater reduction in virus titer with respect to Sabin 3 at 39.5 and 40.0°C. Only strains H36 and H391 exhibited a partial loss of the temperature-sensitive phenotype of Sabin 3. Comparison of the sequences of the strains suggests that several mutations could be responsible for the differences in the temperature-sensitive phenotype between the strains. As suggested previously, the observed phenotypes could be the result of the combination of the effect of different mutations that affect different steps of virus replication (32).
TABLE 6.
Temperature sensitivity and neurovirulence of isolates from the immunodeficient patient
| Virus | Log titer reduction
|
No. of clinically affected animals/total no. | Mean histological lesion score | |
|---|---|---|---|---|
| 35/39.5°C | 35/40.0°C | |||
| Sabin 3 | 3.3 | >6.0 | 0/12 | 0.44 |
| H36 | 0.4 | 5.0 | 2/4 | 1.03 |
| H136 | 4.5 | >6.0 | ||
| H307 | 3.3 | >6.0 | ||
| H391 | 0.3 | 2.7 | 3/4 | 1.62 |
| H442 | 3.1 | >6.0 | ||
| H480 | 1.4 | >6.0 | ||
| H637 | 3.8 | >6.0 | 1/2 | 0.93 |
| Leon | 0.2 | 0.5 | 12/12 | 3.13 |
Strains H36, H391, and H637 were examined for neurovirulence. As shown in Table 6, all three strains exhibited an increase in neurovirulence compared to Sabin 3. However, all three isolates showed a significantly lower mean lesion score with respect to the Sabin 3 wild parent virus, the Leon strain.
Alternative cleavage of 3CD polyprotein.
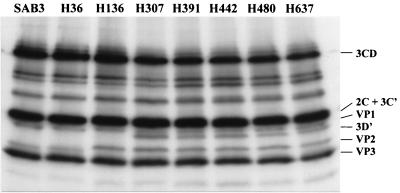
To confirm that mutation at 3D-147 (Ala→Thr) introduced a new proteolytic site in the 3CD polypeptide, the protein synthesis of these viruses in tissue culture was studied. Figure 6 shows the results of the sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis of 35S-labelled HEp-2C cell extracts infected with the different viruses. As predicted from the nucleotide sequence, strains from day 307 showed a protein band with an electrophoretic mobility compatible with that of protein 3D′; a product of the alternative cleavage of 3CD by the viral protease 2A. A difference in the electrophoretic mobility of VP2 in isolates from day 136 could clearly be observed in the gel. The difference is very likely to be the consequence of changes in the amino acid sequence of VP2 in which several mutations were incorporated from day 136 (Table 2).
FIG. 6.
Autorradiography of sodium dodecyl sulfate-polyacrylamide protein gel of 35S-labelled HEp-2C cell extracts infected with the isolates excreted from the immunodeficient patient or Sabin 3. The arrow indicates the position of 3D′. 3C′ comigrated with other viral proteins and could not be distinguished from them.
DISCUSSION
The study of the evolution of vaccine-derived polioviruses in humans has been limited to the few weeks that virus excretion after vaccination normally lasts (2, 32). Here we report a detailed study of the genetic and phenotypic characteristics of seven sequential type 3 poliovirus isolates from an immunodeficient patient (patient 222f) who excreted type 3 poliovirus for 637 days after being fed with monovalent Sabin 3 vaccine (26).
Analysis of nucleotide changes at synonymous third-base codon positions in the coding region indicated that sequence evolution was fairly constant throughout the period of virus excretion. The estimated rate of sequence evolution over the entire coding region (2.78% of nucleotide changes at synonymous third-base codon positions/year) is similar to the rate (3.07% of nucleotide substitutions at synonymous third-base codon positions/year) calculated for wild-type poliovirus during person-to-person transmission (16). The rate of sequence evolution in the capsid region (3.40% of nucleotide substitutions at synonymous third-base codon positions/year) was remarkably similar to that in the capsid VP1 recently estimated from a series of type 1 poliovirus isolates over a period of 200 days from an iVAP case (3.30% of nucleotide changes at synonymous third-base codon positions/year) (17). These observations indicate that poliovirus follows similar patterns of molecular evolution under different conditions of infection. Differences in selection pressures, particularly in humoral immune responses, do not seem to affect the evolution of these presumably neutral positions. Sequence evolution in isolates from patient 222f appeared to be slower in the region of the genome coding for nonstructural proteins (2.37% of nucleotide substitutions at synonymous third-base positions/year), particularly during the first 136 days of virus excretion. One of the possible explanations is the presence of RNA secondary-structure motifs that may function as cis-acting signals for virus replication and might have limited the incorporation of viable nucleotide mutations in this region.
The viral isolates displayed a branched phylogenetic tree in which a major genotypic lineage could be distinguished (Fig. 5). Identification of minor branches that diverted from the main lineage and seemed to have survived for considerable lengths of time suggests that viruses from different lineages coreplicated in the gut as the virus was presumably colonizing different sites in the gastrointestinal tract. Generation of virus genetic lineages within individual patients was observed during the Finnish outbreak of poliomyelitis caused by a wild type 3 virus (19).
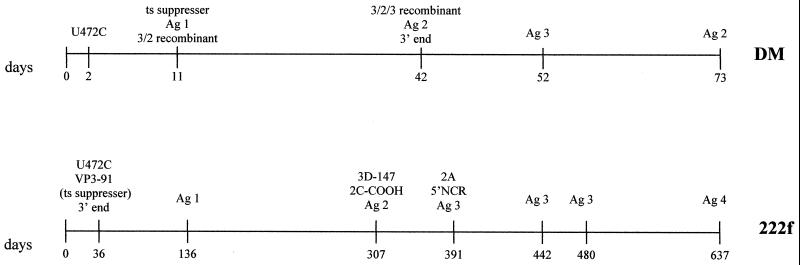
Characteristic mutations were selected in viruses replicating in the immunodeficient patient at different times during the infection, suggesting that particular selection pressures may have operated in the gut at different times. Figure 7 shows some of the mutations observed in the immunodeficient patient, displayed in a chronological order and compared to those found in a healthy vaccinee (29) during their respective entire period of virus excretion. By day 36, both of the major determinants of attenuation of Sabin 3 (43), nucleotide 472 in the 5′ NCR at domain V of the IRES and capsid residue VP3-91 (which is also responsible for temperature sensitivity in Sabin 3), had reverted, confirming the importance of these two positions for virus fitness in the gut. Both changes were maintained during the entire period of infection. However, none of the isolates tested showed a full reversion to the neurovirulent phenotype of the P3/Leon strain, the Sabin 3 wild parent virus. Moreover, all the strains exhibited some degree of inhibition of growth at high temperatures compared to the P3/Leon virus. As described for type 3 isolates from healthy vaccinees or patients with VAP (22), the isolates from the immunodeficient patient appeared to show a correlation between loss of temperature sensitivity and neurovirulence, suggesting that at least some mutations were responsible for both phenomena. Contrary to the common perception, long-term replication of vaccine-derived type 3 poliovirus in the human gut does not necessarily lead to strains insensitive for growth at high temperatures and highly neurovirulent. From day 391, viruses incorporated further changes in the 5′ NCR at domains II, III, and IV of the IRES. Domains II and IV, together with domain V, are thought to contain the most important cis-acting signals in the 5′ NCR, which are essential for the unusual cap-independent initiation of translation of picornaviruses (25). Notably, none of the changes resulted in the disruption of the predicted base-paired structure (40), although some resulted in the weakening of a predicted base pair (Fig. 2). These observations emphasize the biological significance of maintaining a particular RNA secondary structure in this region.
FIG. 7.
Antigenic and molecular evolution of type 3 poliovirus in healthy baby DM (29) and in the immunodeficient patient 222f. Ag, antigenic variant; 2A, protease 2A; 2C-COOH, carboxyl-terminal region of protein 2C; ts, temperature sensitive.
Capsid mutations involved amino acids in the canyon, the drug-lipid binding pocket, antigenic sites, internal sequences, monomeric and pentameric interfaces, and other surface residues of unknown phenotype (Fig. 3). Several changes were located at positions known to affect receptor binding or the subsequent receptor-mediated conformational changes that lead to virus cell entry, including mutations in putative receptor contact residues. Interestingly, type 3 mutant viruses generated from the Leon strain that exhibited persistence in HEp-2C cells possess altered receptor binding properties (7). The authors proposed that in this manner the mutant strains reduced their lytic capacity that allowed persistence in tissue culture.
A progressive antigenic drift is normally observed in sequential isolates from the same individual in healthy vaccines (29; A. J. Macadam, personal communication). Changes in antigenic sites can be detected only a few days after vaccination and, for type 3 viruses, usually involve antigenic sites 2 and 4 and occasionally site 3 (29; A. J. Macadam, personal communication). Changes in antigenic site 2 were detected in isolates from the immunodeficient patient only on days 136 and 637 (the last isolate), a mutation in antigenic site 3 was found on day 136 and thereafter, and antigenic site 4 changed from day 391. The delay in the appearance of antigenic variants could be associated with the deficient immune pressure exerted against the virus in the patient and, consequently, with the long-term excretion of virus. Human antipoliovirus antibodies provided by immunotherapy were insufficient to eliminate the virus but were probably responsible for the observed antigenic variation. As reported for type 3 isolates from healthy vaccinees, no changes were detected in antigenic site 1, which reinforces the hypothesis of a lack of immune pressure against site 1 in humans (31).
Isolates obtained from day 307 onward incorporated several changes in the carboxyl-terminal region of protein 2C, important for RNA replication (37, 42), and a mutation at residue 3D-147 (Ala→Thr), which generated a recognition site (TY/G) for cleavage by the viral protease 2A that is conserved among most poliovirus strains. Remarkably, four distantly related wild type 3 polioviruses and two different Sabin 3-derived VAP isolates that were also analyzed in this study contained similar amino acid changes with respect to the Sabin 3 strain (Table 4). These results indicate that the changes may provide the viruses with some selective advantage for growth in the gut. This suggestion is in agreement with the common observation that type 3 isolates from healthy vaccinees given Sabin trivalent vaccine show a recombinant genome in which both the carboxyl-terminal region of 2C and the amino-terminal region of 3D are derived from Sabin 1 or Sabin 2 genomes, which also contain similar changes in these regions with respect to Sabin 3 (Table 4). These results support the idea that genetic recombination is a common and efficient mechanism of poliovirus evolution, in that several changes could be acquired at once while stepwise mutation might be less easy although clearly not impossible.
Other changes in nonstructural proteins included two mutations in protease 2A in strains collected from day 391. Curiously, the appearance of mutations in protease 2A coincides in time with changes in the IRES region in the 5′ NCR. Sequence changes in both 5′ NCR and protease 2A are known to influence protein synthesis, and in some cases, mutations in 2A compensate for defects caused by mutations in the 5′ NCR (24).
The reason why patient 222f stopped excreting poliovirus is unclear. Only 2 of 30 patients who formed part of the same study excreted virus for prolonged periods after being fed with Sabin vaccine (26). The second patient excreted type 1 poliovirus for 32 months. At this point, a rapid disappearance of virus was observed, associated with the shedding of the intestinal mucosa as a result of infection with Shigella sonnei (26). In another case, an immunodeficient patient who excreted poliovirus type 2 for 3.5 years stopped excreting the virus 2 months after he started receiving treatment with secretory IgA (13).
Long-term excretion of poliovirus by immunodeficient patients has important implications for the program for global eradication of polio (6). The fact that some immunodeficiencies are often acquired late in life and/or not diagnosed before polio immunization is carried out makes the prevention and control of poliomyelitis in the immunodeficient population a difficult task. Patients with primary immunodeficiencies are therefore a potential reservoir of persistent poliovirus excretion. The World Health Organization considers this circumstance to be one of the priorities for the establishment of strategies for stopping immunization once wild poliomyelitis has been eradicated (45). It is therefore crucial to evaluate and control the prevalence of persistent poliovirus infection among immunodeficient individuals. Identification of molecular markers associated with long-term replication of vaccine-derived viruses in humans from studies such as the one presented here could be essential for assessing these issues. Development of efficient treatments that complement immunoglobulin therapy to eliminate poliovirus from these patients is under active investigation (11). The use of more genetically stable live-attenuated poliovirus vaccines (1, 12, 20; A. J. Macadam, personal communication) could help to reduce the incidence of VAP in nondiagnosed immunodeficient patients and may contribute to reduce the risk of virulent virus spreading into the population in the postvaccination era.
ACKNOWLEDGMENTS
We thank Andrew Macadam and David Wood for fruitful discussions and Andrew Davies for the artwork. Claire Shorrock was extremely helpful with the automatic sequencing. We are also indebted to Sue Marsden, Sophie Reid, and Gary Beaven for important contributions.
REFERENCES
- 1.Agol V I, Pilipenko E V, Slobodskaya O R. Modification of translational control elements as a new approach to design of attenuated picornavirus strains. J Biotechnol. 1996;44:119–128. doi: 10.1016/0168-1656(95)00088-7. [DOI] [PubMed] [Google Scholar]
- 2.Alexander J P, Jr, Gary H E, Jr, Pallansch M A. Duration of poliovirus excretion and its implications for acute flaccid paralysis surveillance: a review of the literature. J Infect Dis. 1997;175(Suppl. 1):S176–S182. doi: 10.1093/infdis/175.supplement_1.s176. [DOI] [PubMed] [Google Scholar]
- 3.Cammack N, Phillips A, Dunn G, Patel V, Minor P D. Intertypic genomic rearrangements of poliovirus strains in vaccinees. Virology. 1988;167:507–514. [PubMed] [Google Scholar]
- 4.Chapman M S, Rossmann M G. Comparison of surface properties of picornaviruses: strategies for hiding the receptor site from immune surveillance. Virology. 1993;195:745–756. doi: 10.1006/viro.1993.1425. [DOI] [PubMed] [Google Scholar]
- 5.Chow M, Basavappa R, Hogle J M. The role of conformational transitions in poliovirus pathogenesis. In: Chiu W, Burnett R M, Garcea R L, editors. Structural biology of viruses. Oxford, United Kingdom: Oxford University Press; 1997. pp. 157–186. [Google Scholar]
- 6.Dowdle W R, Birmingham M E. The biologic principles of poliovirus eradication. J Infect Dis. 1997;175(Suppl. 1):S286–S292. doi: 10.1093/infdis/175.Supplement_1.S286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duncan G, Pelletier I, Colbere-Garapin F. Two amino acid substitutions in the type 3 poliovirus capsid contribute to the establishment of persistent infection in HEp-2c cells by modifying virus-receptor interactions. Virology. 1998;241:14–29. doi: 10.1006/viro.1997.8955. [DOI] [PubMed] [Google Scholar]
- 8.Dunn G, Begg N T, Cammack N, Minor P D. Virus excretion and mutation by infants following primary vaccination with live oral poliovaccine from two sources. J Med Virol. 1990;32:92–95. doi: 10.1002/jmv.1890320205. [DOI] [PubMed] [Google Scholar]
- 9.Evans D M, Dunn G, Minor P D, Schild G C, Cann A J, Stanway G, Almond J W, Currey K, Maizel J V., Jr Increased neurovirulence associated with a single nucleotide change in a noncoding region of the Sabin type 3 poliovaccine genome. Nature. 1985;314:548–550. doi: 10.1038/314548a0. [DOI] [PubMed] [Google Scholar]
- 10.Filman D J, Syed R, Chow M, Macadam A J, Minor P D, Hogle J M. Structural factors that control conformational transitions and serotype specificity in type 3 poliovirus. EMBO J. 1989;8:1567–7159. doi: 10.1002/j.1460-2075.1989.tb03541.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fromtling J, Castañer J. Pleconaril (Win-63843) Drugs Future. 1997;22:40–44. [Google Scholar]
- 12.Gromeier M, Alexander L, Wimmer E. Internal ribosomal entry site substitution eliminates neurovirulence in intergeneric poliovirus recombinants. Proc Natl Acad Sci USA. 1996;93:2370–2375. doi: 10.1073/pnas.93.6.2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hara M, Saito Y, Komatsu t, Kodama H, Abo W, Chiba S, Nakao T. Antigenic analysis of poliovirus isolated from a child with agammaglobulinemia and paralytic poliomyelitis after Sabin vaccine administration. Microbiol Immunol. 1981;25:905–913. doi: 10.1111/j.1348-0421.1981.tb00095.x. [DOI] [PubMed] [Google Scholar]
- 14.Hughes P J, Evans D M A, Minor P D, Schild G C, Almond J W, Stanway G. The nucleotide sequence of a type 3 poliovirus isolated during a recent outbreak of poliomyelitis in Finland. J Gen Virol. 1986;67:2093–2102. doi: 10.1099/0022-1317-67-10-2093. [DOI] [PubMed] [Google Scholar]
- 15.Kawamura N, Kohara M, Abe S, Komatsu T, Tago K, Arita M, Nomoto A. Determinants in the 5′ noncoding region of poliovirus Sabin 1 RNA that influence the attenuation phenotype. J Virol. 1989;63:1302–1309. doi: 10.1128/jvi.63.3.1302-1309.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kew O M, Mulders M N, Lipskaya G Y, da Silva E E, Pallansch M A. Molecular epidemiology of polioviruses. Semin Virol. 1995;6:401–414. [Google Scholar]
- 17.Kew O M, Sutter R W, Nottay B K, McDonough M J, Prevots D R, Quick L, Pallansch M A. Prolonged replication of a type 1 vaccine-derived poliovirus in an immunodeficient patient. J Clin Microbiol. 1998;36:2893–2899. doi: 10.1128/jcm.36.10.2893-2899.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.King A M. Preferred sites of recombination in poliovirus RNA: an analysis of 40 intertypic cross-over sequences. Nucleic Acids Res. 1988;16:11705–11723. doi: 10.1093/nar/16.24.11705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kinnunen L, Poyry T, Hovi T. Generation of virus genetic lineages during an outbreak of poliomyelitis. J Gen Virol. 1991;72:2483–2489. doi: 10.1099/0022-1317-72-10-2483. [DOI] [PubMed] [Google Scholar]
- 20.Kohara M, Abe S, Komatsu T, Tago K, Arita M, Nomoto A. A recombinant virus between the Sabin 1 and Sabin 3 vaccine strains of poliovirus as a possible candidate for a new type 3 poliovirus live vaccine strain. J Virol. 1988;62:2828–2835. doi: 10.1128/jvi.62.8.2828-2835.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee C K, Wimmer E. Proteolytic processing of poliovirus polyprotein: elimination of 2Apro-mediated, alternative cleavage of polypeptide 3CD by in vitro mutagenesis. Virology. 1988;166:405–414. doi: 10.1016/0042-6822(88)90511-9. [DOI] [PubMed] [Google Scholar]
- 22.Macadam A J, Arnold C, Howlett J, John A, Marsden S, Taffs F, Reeve P, Hamada N, Wareham K, Almond J, Cammack N, Minor P D. Reversion of the attenuated and temperature-sensitive phenotypes of the Sabin type 3 strain of poliovirus in vaccinees. Virology. 1989;172:408–414. doi: 10.1016/0042-6822(89)90183-9. [DOI] [PubMed] [Google Scholar]
- 23.Macadam A J, Ferguson G, Arnold C, Minor P D. An assembly defect as a result of an attenuating mutation in the capsid proteins of the poliovirus type 3 vaccine strain. J Virol. 1991;65:5225–5235. doi: 10.1128/jvi.65.10.5225-5231.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Macadam A J, Ferguson G, Fleming T, Stone D M, Almond J W, Minor P D. Role for poliovirus protease 2A in cap independent translation. EMBO J. 1994;13:924–927. doi: 10.1002/j.1460-2075.1994.tb06336.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Macadam A J, Stone D M, Almond J W, Minor P D. The 5′ noncoding region and virulence of poliovirus vaccine strains. Trends Microbiol. 1994;2:449–454. doi: 10.1016/0966-842x(94)90803-6. [DOI] [PubMed] [Google Scholar]
- 26.MacCallum F O. Hypogammaglobulinaemia in the United Kingdom. VII. The role of humoral antibodies in protection against and recovery from bacterial and virus infections in hypogammaglobulinaemia. Spec Rep Ser Med Res Counc G B. 1971;310:72–85. [PubMed] [Google Scholar]
- 27.Magrath D I, Boulger L R, Hartley E G. 10th Symposium of the European Association against Poliomyelitis. Brussels, Belgium: European Association against Poliomyelitis; 1966. Strains of poliovirus from individuals fed Sabin vaccine studied by in vitro markers and the monkey neurovirulence test; pp. 334–345. [Google Scholar]
- 28.Minor P D. Comparative biochemical studies of type 3 poliovirus. J Virol. 1980;34:73–84. doi: 10.1128/jvi.34.1.73-84.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Minor P D, John A, Ferguson M, Icenogle J P. Antigenic and molecular evolution of the vaccine strain of type 3 poliovirus during the period of excretion by a primary vaccinee. J Gen Virol. 1986;67:693–706. doi: 10.1099/0022-1317-67-4-693. [DOI] [PubMed] [Google Scholar]
- 30.Minor P D. Antigenic structure of picornaviruses. Curr Top Microbiol Immunol. 1990;161:121–154. doi: 10.1007/978-3-642-75602-3_5. [DOI] [PubMed] [Google Scholar]
- 31.Minor P D, Ferguson M, Phillips A, Magrath D I, Huovilainen A, Hovi T. Conservation in vivo of protease cleavage sites in antigenic sites of poliovirus. J Gen Virol. 1987;68:1857–1865. doi: 10.1099/0022-1317-68-7-1857. [DOI] [PubMed] [Google Scholar]
- 32.Minor P D. The molecular biology of poliovaccines. J Gen Virol. 1992;73:3065–3077. doi: 10.1099/0022-1317-73-12-3065. [DOI] [PubMed] [Google Scholar]
- 33.Mosser A G, Sgro J Y, Rueckert R R. Distribution of drug resistance mutations in type 3 poliovirus identifies three regions involved in uncoating functions. J Virol. 1994;68:8193–8201. doi: 10.1128/jvi.68.12.8193-8201.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nkowane B M, Wassilak S G, Orenstein W A, Bart K J, Schonberger L B, Hinman A R, Kew O M. Vaccine-associated paralytic poliomyelitis. United States: 1973 through 1984. JAMA. 1987;257:1335–1340. [PubMed] [Google Scholar]
- 35.Racaniello V R. Early events in poliovirus infection: virus-receptor interactions. Proc Natl Acad Sci USA. 1996;93:11378–11381. doi: 10.1073/pnas.93.21.11378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ren R, Moss E G, Racaniello V R. Identification of two determinants that attenuate vaccine-related type 2 poliovirus. J Virol. 1991;65:1377–1382. doi: 10.1128/jvi.65.3.1377-1382.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodríguez P L, Carrasco L. Poliovirus protein 2C contains two regions involved in RNA binding activity. J Biol Chem. 1995;270:10105–10112. doi: 10.1074/jbc.270.17.10105. [DOI] [PubMed] [Google Scholar]
- 38.Sabin A B, Boulger L R. History of Sabin attenuated poliovirus oral live vaccines. J Biol Stand. 1973;1:115–118. [Google Scholar]
- 39.Savilahti E, Klemola T, Carlsson B, Mellander L, Stenvik M, Hovi T. Inadequacy of mucosal IgM antibodies in selective IgA deficiency: excretion of attenuated polioviruses is prolonged. J Clin Immunol. 1988;8:89–94. doi: 10.1007/BF00917895. [DOI] [PubMed] [Google Scholar]
- 40.Skinner M A, Racaniello V R, Dunn G, Cooper J, Minor P D, Almond J W. New model for the secondary structure of the 5′ non-coding RNA of poliovirus is supported by biochemical and genetic data that also show that RNA secondary structure is important in neurovirulence. J Mol Biol. 1989;207:379–392. doi: 10.1016/0022-2836(89)90261-1. [DOI] [PubMed] [Google Scholar]
- 41.Sutter R W, Prevots R. Vaccine-associated paralytic poliomyelitis among immunodeficient persons. Infect Med. 1994;11:426–438. doi: 10.1086/379791. [DOI] [PubMed] [Google Scholar]
- 42.Teterina N L, Gorbalenya A E, Egger D, Bienz K, Ehrenfeld E. Poliovirus 2C protein determinants of membrane binding and rearrangements in mammalian cells. J Virol. 1997;71:8962–8972. doi: 10.1128/jvi.71.12.8962-8972.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Westrop G D, Wareham K A, Evans D M, Dunn G, Minor P D, Magrath D I, Taffs F, Marsden S, Skinner M A, Schild G C, Almond J W. Genetic basis of attenuation of the Sabin type 3 oral poliovirus vaccine. J Virol. 1989;63:1338–1344. doi: 10.1128/jvi.63.3.1338-1344.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.World Health Organization. Requirements for poliomyelitis vaccine (oral) WHO Tech Rep Ser. 1983;687:107–175. [Google Scholar]
- 45.World Health Organization. Draft report on the scientific basis for stopping immunisation against poliomyelitis meeting, March 23–25. Geneva, Switzerland: World Health Organization; 1998. [Google Scholar]
- 46.Wimmer E, Hellen C U T, Cao X. Genetics of Poliovirus. Annu Rev Genet. 1993;27:353–435. doi: 10.1146/annurev.ge.27.120193.002033. [DOI] [PubMed] [Google Scholar]