Abstract
Background
Although several studies have addressed plasma proteomics in heart failure with preserved ejection fraction, limited data are available on the prognostic value of urinary proteomics. The objective of our study was to identify urinary proteins/peptides associated with death and heart failure admission in patients with heart failure with preserved ejection fraction.
Methods and Results
The study population included participants enrolled in TOPCAT (Treatment of Preserved Cardiac Function Heart Failure With an Aldosterone Antagonist Trial). The relationship between urine protein levels and the risk of death or heart failure admission was assessed using Cox regression, in both nonadjusted analyses and adjusting for urine creatinine levels, and the MAGGIC (Meta‐Analysis Global Group in Chronic Heart Failure) score. A total of 426 (12.4%) TOPCAT participants had urinary protein data and were included. There were 40 urinary proteins/peptides significantly associated with death or heart failure admission in nonadjusted analyses, 21 of which were also significant adjusted analyses. Top proteins in the adjusted analysis included ANGPTL2 (angiopoietin‐like protein 2) (hazard ratio [HR], 0.5731 [95% CI, 0.47–0.7]; P=3.13E‐05), AMY2A (α amylase 2A) (HR, 0.5496 [95% CI, 0.44–0.69]; P=0.0001), and DNASE1 (deoxyribonuclease‐1) (HR, 0.5704 [95% CI, 0.46–0.71]; P=0.0002). Higher urinary levels of proteins involved in fibrosis (collagen VI α‐1, collagen XV α‐1), metabolism (pancreatic α‐amylase 2A/B, mannosidase α class 1A member 1), and inflammation (heat shock protein family D member 1, inducible T cell costimulatory ligand) were associated with a lower risk of death or heart failure admission.
Conclusions
Our study identifies several novel associations between urinary proteins/peptides and outcomes in heart failure with preserved ejection fraction. Many of these associations are independent of clinical risk scores and may aid in risk stratification in this patient population.
Keywords: biomarkers, heart failure with preserved ejection fraction, prognosis, proteomics, urine proteomics
Subject Categories: Heart Failure
Nonstandard Abbreviations and Acronyms
- HFpEF
heart failure with preserved ejection fraction
- MAGGIC
Meta‐Analysis Global Group in Chronic Heart Failure
- TOPCAT
Treatment of Preserved Cardiac Function HF with an Aldosterone Antagonist Trial
- UPP
urinary protein/peptide
Clinical Perspective.
What Is New?
Our study quantified urine protein/peptide levels from participants in TOPCAT (Treatment of Preserved Cardiac Function HF With an Aldosterone Antagonist Trial) and evaluated the relationship between urine biomarker levels and the risk of death or heart failure admission.
We found multiple urine proteins/peptides to be associated with the risk of death or heart failure admission, including proteins involved in fibrosis, metabolism, and inflammation.
What Are the Clinical Implications?
Our study suggests a potential use for several novel urine proteins/peptides as prognostic markers in heart failure with preserved ejection fraction.
Further studies are needed to assess whether urine protein/peptide level measurements can improve clinical decision making in patients with heart failure with preserved ejection fraction.
Heart failure (HF) with preserved ejection fraction (HFpEF) represents approximately half of all HF diagnoses, and the rate of HFpEF is increasing relative to HF with reduced ejection fraction. 1 , 2 Unlike heart failure with reduced ejection fraction, there are no medical therapies that have been established to reduce all‐cause mortality in HFpEF.
Broad proteomics discovery approaches can reveal novel biomarkers of risk in patients with HFpEF, which could lead to the identification of novel therapeutic targets, or prognostic biomarkers that could aid in risk stratification in clinical research and practice. Although previous studies have focused on plasma proteomics approaches in HFpEF, 3 , 4 , 5 , 6 , 7 , 8 , 9 , 10 , 11 , 12 , 13 , 14 , 15 data on urinary proteomics are scarce, with only 1 study assessing differences in urinary proteins between participants with HF and controls. 16 Data on the prognostic value of UPPs (urinary proteins/peptides) in HFpEF are not available.
UPPs have the advantage of being noninvasive, inexpensive to collect, and usually without side effects or complications. 17 Previous urinary biomarker studies have identified urine peptides associated with coronary artery disease, kidney disease, and arterial stiffness. 18 , 19 Of note, the urinary proteome contains information not only from the kidney and the urinary tract but also from other organs via glomerular filtration of some plasma proteins. 20
In this study, we performed de novo urinary protein/peptide measurements in frozen urine samples available from 426 participants enrolled in TOPCAT (Treatment of Preserved Cardiac Function HF with an Aldosterone Antagonist Trial). We evaluated the relationship between urine biomarker levels and the risk of the death or HF‐related hospital admission (DHFA) in this cohort.
METHODS
The raw data and analytical methods of this article are not publicly available for purposes of reproducing the results or replicating the procedures. These data might be available subject to the establishment of appropriate data‐sharing agreements and regulatory approvals. The parent TOPCAT data are available through the US National Institutes of Health BioLINCC website.
Study Population
Individuals included in this analysis were participants of TOPCAT. TOPCAT was a multicenter, double‐blinded, placebo‐controlled randomized control trial of spironolactone that enrolled 3445 adults with HFpEF (left ventricular ejection fraction ≥45%) from 6 countries from 2006 to 2012. The design, characteristics, inclusion/exclusion criteria, and results of the trial have been previously published. 21 , 22 All study participants provided written informed consent. The study received approval from our institution's review board.
Urine Biomarker Samples
Urine samples for biomarker analyses were obtained from a subset of TOPCAT study participants who had available samples for de novo proteomic analyses. A total of 2308 protein groups were assessed in unbiased, data‐dependent, label‐free proteomic profiling, including imputation of values with minimum label‐free quantification intensity. Approximately 270 μL of urine from participants were aliquoted into 96 well plates. Each plate included 4 wells with pooled normal healthy volunteer urine samples that were designated as quality control samples. Urine sample plates were stored at −80 °C until commencement of the experiment. Each plate was prepared for proteomics analysis on a separate day. Urinary proteins were subjected to reduction and alkylation by addition of 30 μL 0.1 mol/L dithiothreitol and 0.2 mol/L iodoacetamide, followed by incubation at 60 °C at 1000 rpm for 1 hour in a thermo shaker. Samples were cooled to room temperature, then combined with 900 μL of cold acetonitrile and incubated overnight at −20 °C. The plate was centrifuged at room temperature for 20 minutes at 2500g in a plate centrifuge. Supernatants were aspirated using a 1.2 mL pipette using a multichannel pipette. Protein pellets were washed by adding 1 mL of 100% acetonitrile at room temperature. The plate was shaken on a plate shaker for 5 minutes, and subsequently centrifuged for 10 minutes at 2500g at room temperature. Finally, the supernatants were aspirated leaving behind the clean pellet, which was air dried for 5 minutes. Protein pellets were dissolved in 100 μL of freshly prepared 8 mol/L urea containing 100 mmol/L tris HCL and digested with 0.5 μg LysC (Wako Chemicals, Richmond, VA) at 37 °C for 4 hours at 1000 rpm. The partially digested samples were diluted with 400 μL LCMS grade water, and 1 μg of trypsin/LysC mix (Promega, Madison, WI) was added. The plate was incubated at 37 °C overnight at 1000 rpm to allow complete digestion of proteins. Peptide concentrations were measured using a tryptophan fluorescence method.
Liquid Chromatography With Tandem Mass Spectrometry Analysis
A total of 500 ng of each sample was loaded onto individual Evotips (Evosep, Odense, Denmark) and washed with 50 μL 0.1% formic acid followed by the addition of 100 μL storage solvent (0.1% formic acid) to keep the Evotips wet until analysis. The Evosep One system was coupled online to a QExactive HF mass spectrometer (Thermo Fisher Scientific, Waltham, MA) with a nanoelectrospray ion source (Thermo Fisher Scientific). Peptides were eluted from Evotips onto a Pepsep C‐18 reversed phase column (ReproSil 3 μm, 120 Å, 8 cm in length and 75 μm inner diameter), and separated with a preset 30 samples per day gradient provided by the Evosep One system. Mass spectrometry data were acquired using Xcalibur software. A data‐dependent method was used to dynamically choose the top 10 most abundant precursor ions from the survey scan using high‐energy collision dissociation fragmentation. Survey scans were acquired with a mass range of 400 to 1000 thompson units at a resolution of 60 000 at 200 m/z. The maximum ion injection times for the survey scan and the tandem mass spectrometry scans were 50 and 100 milliseconds, respectively, and the automatic gain control target values were set to 3E6 and 1E5, respectively. The isolation window was set to 1.5 thompson units, and ions were fragmented with a normalized collision energy of 27. Unassigned precursor ion charge states, singly charged ions, as well as ions of charge states above 8 were rejected. Peptide match was preferred, and dynamic exclusion was set to 40 seconds.
Sample Bioinformatics Analysis
Mass spectra were analyzed using MaxQuant software version 1.6.6.0. The maximum allowed mass deviation was set to 4.5 ppm for monoisotopic precursor ions and 0.5 Da for tandem mass spectrometry peaks. Enzyme specificity was set to Trypsin/P, and a maximum of 2 missed cleavages were allowed. Carbamidomethyl cysteine was set as a fixed modification, and N‐terminal acetylation and methionine oxidation as variable modifications. The spectra were searched against the human Uniprot sequence database combined with common contaminants and concatenated with the reversed versions of all sequences. Protein identification required at least 1 unique or razor peptide per protein group. Quantification in MaxQuant was performed using the label free quantification algorithm with fast label free quantification and a minimum ratio count of 1. The false‐positive rate was set to 1% at both peptide and protein level. Match between runs was selected with an alignment time window of 20 minutes and match time window of 0.7 minutes. Contaminants, reversed sequence identification, and proteins only identified by site were excluded from further data analysis. Missing values were imputed with sample minimum label free quantification intensities, which were performed for 57% of values across the cohort.
Ingenuity Pathway Analysis
Pathway enrichment analysis was performed using ingenuity pathway analysis software (Qiagen, Hilden, Germany; www.qiagen.com/ingenuity). 23 UPPs were identified according to UniProt identification and included in overrepresentation analyses if associated at a nominal P value threshold of 0.01. The analysis calculates a P value (Fisher exact test) quantifying the overlap, and a Z score quantifying the likelihood and direction (upregulated or downregulated), between the proteomics pattern and known canonical pathways.
Statistical Analysis
Characteristics of participants were assessed with mean and SD for normally distributed variables and median and interquartile range for nonnormally distributed variables. We compared clinical characteristics between subjects with and without proteomics data available. We used the nonpaired Student t test for continuous normally distributed variables, Kruskal–Wallis test for nonnormally distributed continuous variables, and χ 2 or Fisher exact test for categorical variables.
The primary outcome was the composite of DHFA, as defined. 12 , 13 , 14 We evaluated the relationship between urine biomarker levels and the risk of DHFA using Cox regression. We ran 3 different analyses to assess the impact of correcting for different clinical factors: (1) models without adjustment, (2) models that adjusted for urine creatinine levels, and (3) models that adjusted for urine creatinine levels and the MAGGIC (Meta‐Analysis Global Group in Chronic HF) risk score, which incorporates multiple demographic, clinical, and laboratory parameters. 24
Statistical significance was defined as a 2‐tailed P value <0.05. We corrected the α level for multiple comparisons based on the principal components underlying the variability of all measured UPPs, as previously described. 9 , 11 , 12 , 25 , 26 , 27 , 28 All probability values presented are 2‐tailed. We then performed interaction tests by: (1) location (Americas versus Eastern Europe) and (2) arm of trial (spironolactone versus placebo) to assess potential effect modification by these characteristics between the baseline urinary proteome and the outcome. Analyses were performed using the MATLAB statistics and machine learning toolbox R2022a. 29
RESULTS
TOPCAT Population
A comparison of subjects with and without available urinary protein data is shown in Table 1. Those with urine proteomic data also tended to exhibit slightly higher prevalence of atrial fibrillation and a history of myocardial infarction and were somewhat more likely to use statins. Renal function (estimated glomerular filtration rate) was similar between groups (65.4 in participants without urine samples versus 65.7 in participants with urine samples). Systolic and diastolic blood pressures differed between the groups, but notably the difference between groups is on average <5 mm Hg.
Table 1.
Characteristics of TOPCAT Participants With and Without Urine Proteomics Data
| Characteristic | Participants without urine proteomic samples (n=3016) | Participants with urine proteomic samples (n=426) |
|---|---|---|
| Demographic characteristics | ||
| Age, y | 69 (61–76) | 69 (61–77) |
| Female sex | 1587 (52.3) | 196 (46) |
| Race | ||
| White | 2668 (88.46) | 391 (91.78) |
| Black | 274 (9.08) | 28 (6.57) |
| Asian | 16 (0.53) | 3 (0.70) |
| Other | 67 (2.22) | 3 (0.70) |
| Location | ||
| Americas | 1535 (50.90%) | 230 (53.99%) |
| Eastern Europe | 1481 (49.10%) | 196 (46.01%) |
| Systolic BP, mm Hg | 130 (120–140) | 128 (120–134) |
| Diastolic BP, mm Hg | 80 (70–81) | 75 (68–80) |
| eGFR, mL/min per 1.73 m2 | 65.4 (53.7–79.3) | 65.7 (53.7–77.9) |
| BMI, kg/m2 | 30.8 (27.1–35.6) | 32 (27.8–36.4) |
| Randomized to spironolactone arm | 1524 (50.53%) | 198 (46.48%) |
| Medical history | ||
| Myocardial infarction | 757 (25.11) | 136 (31.92) |
| Stroke | 234 (7.76) | 31 (7.28) |
| COPD | 349 (11.58) | 54 (12.68) |
| Hypertension | 2742 (90.95) | 404 (94.84) |
| Atrial fibrillation | 1033 (34.26) | 180 (42.25) |
| Diabetes | 980 (32.50) | 138 (32.39) |
| Medication use | ||
| β‐Blockers | 2334 (77.41) | 342 (80.28) |
| Calcium channel blocker | 1141 (37.84) | 152 (35.68) |
| ACE/ARB use | 2558 (84.84) | 341 (80.05) |
| Aspirin use | 1972 (65.41) | 278 (65.26) |
| Statin use | 1528 (50.68) | 277 (65.02) |
Values represent median (interquartile range) or n (percent). ACE indicates angiotensin‐converting enzyme; ARB, angiotensin receptor blocker; BMI, body mass index; BP, blood pressure; COPD, chronic obstructive pulmonary disease; eGFR, estimated glomerular filtration rate; and TOPCAT, Treatment of Preserved Cardiac Function HF With an Aldosterone Antagonist Trial.
Association of UPP Levels and the Incidence of DHFA: Nonadjusted Analyses
Among the individuals included in this analysis (n=426), there were 64 deaths, 54 participants who had experienced an HF‐related admission, and 100 participants who reached the composite outcome of DHFA. In nonadjusted analyses, we found 40 UPPs to be significantly associated with DHFA after α error correction. Figure [A] shows a volcano plot representing the relationship between UPPs and the risk of DHFA. Table 2 lists UPPs that were significantly associated with DHFA, along with standardized HRs and 95% CIs. Table S1 list the full list of name, function, and category of these proteins.
Figure . Volcano plot showing standardized hazard ratios for urinary proteins associated with the composite outcome of death or heart failure‐related admissions in nonadjusted analyses (A) and after adjustment for urinary creatinine and the MAGGIC score (B).
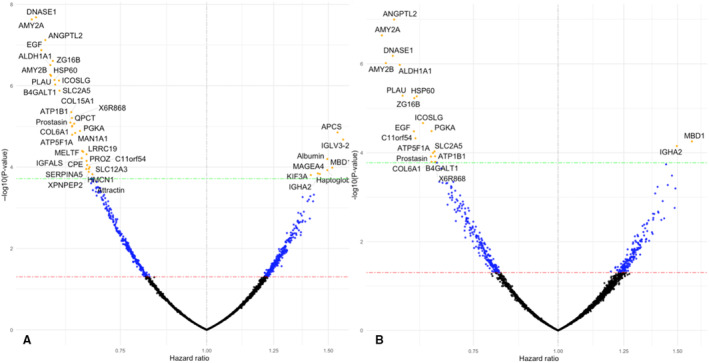
The dashed lines represent the uncorrected (red) and corrected (green) significance level. MAGGIC indicates Meta‐Analysis Global Group in Chronic Heart Failure. The dots represent urinary proteins either below statistical significance (black), uncorrected significance (blue), and corrected significance (yellow).
Table 2.
Urinary Biomarkers Associated With DHFA With and Without Adjustment
| Uniprot ID | Short name | Nonadjusted | Adjusted | ||||||
|---|---|---|---|---|---|---|---|---|---|
| β | HR | 95% CI | P value | β | HR | 95% CI | P value | ||
| Q9UKU9 | ANGPTL2 | −0.54 | 0.57 | 0.48–0.71 | 7.54E‐08 | −0.56 | 0.57 | 0.47–0.7 | 3.13E0‐5 |
| P04746 | AMY2A | −0.58 | 0.56 | 0.46–0.69 | 2.34E‐08 | −0.6 | 0.55 | 0.44–0.69 | 0.0001 |
| P24855 | DNASE1 | −0.57 | 0.58 | 0.46–0.69 | 2.09E‐08 | −0.56 | 0.57 | 0.46–0.71 | 0.0002 |
| P00352 | ALDH1A1 | −0.52 | 0.60 | 0.49–0.73 | 0.0001 | −0.54 | 0.56 | 0.47–0.72 | 0.0003 |
| P19961 | AMY2B | −0.52 | 0.59 | 0.48–0.73 | 0.0002 | −0.58 | 0.58 | 0.44–0.7 | 0.0003 |
| P00749 | PLAU | −0.52 | 0.60 | 0.49–0.73 | 0.0002 | −0.53 | 0.59 | 0.47–0.74 | 0.0016 |
| P10809 | HSP60 | −0.51 | 0.60 | 0.49–0.74 | 0.0002 | −0.48 | 0.62 | 0.5–0.76 | 0.0017 |
| A0A0C4DGN4 | ZG16B | −0.51 | 0.59 | 0.49–0.73 | 0.0001 | −0.49 | 0.61 | 0.5–0.76 | 0.0018 |
| K4DIA0 | ICOSLG | −0.49 | 0.61 | 0.5–0.74 | 0.0002 | −0.46 | 0.63 | 0.51–0.78 | 0.0067 |
| P00558 | PGKA | −0.42 | 0.66 | 0.54–0.79 | 0.004 | −0.43 | 0.65 | 0.53–0.8 | 0.0101 |
| P01133 | EGF | −0.55 | 0.58 | 0.47–0.71 | 1.33E‐07 | −0.49 | 0.61 | 0.49–0.77 | 0.0102 |
| Q9H0W9 | C11orf54 | −0.4 | 0.67 | 0.55–0.82 | 0.0274 | −0.48 | 0.62 | 0.49–0.78 | 0.0146 |
| K7EPZ6 | MBD1 | 0.42 | 1.52 | 1.23–1.88 | 0.0317 | 0.45 | 1.58 | 1.26–1.96 | 0.0172 |
| A0A075B6N7 | IGHA2 | 0.38 | 1.46 | 1.2–1.77 | 0.045 | 0.4 | 1.50 | 1.23–1.83 | 0.0218 |
| P22732 | SLC2A5 | −0.49 | 0.61 | 0.5–0.75 | 0.0004 | −0.42 | 0.66 | 0.53–0.81 | 0.0286 |
| P25705 | ATP5F1A | −0.45 | 0.64 | 0.52–0.78 | 0.005 | −0.42 | 0.65 | 0.53–0.81 | 0.0309 |
| P05026 | ATP1B1 | −0.45 | 0.64 | 0.52–0.78 | 0.0025 | −0.42 | 0.66 | 0.53–0.81 | 0.0367 |
| Q16651 | Prostasin | −0.45 | 0.64 | 0.52–0.78 | 0.003 | −0.43 | 0.65 | 0.52–0.81 | 0.0372 |
| A0A087X0S5 | COL6A1 | −0.45 | 0.64 | 0.52–0.78 | 0.0031 | −0.43 | 0.65 | 0.52–0.81 | 0.0486 |
| X6R868 | X6R868 | −0.45 | 0.64 | 0.53–0.78 | 0.0019 | −0.42 | 0.66 | 0.53–0.82 | 0.0488 |
| P15291 | B4GALT1 | −0.5 | 0.60 | 0.49–0.74 | 0.0003 | −0.41 | 0.66 | 0.53–0.82 | 0.0499 |
| P02743 | APCS | 0.44 | 1.55 | 1.27–1.88 | 0.0044 | 0.37 | 1.44 | 1.19–1.75 | 0.0552 |
| A0A087X0K0 | COL15A1 | −0.45 | 0.64 | 0.53–0.77 | 0.0014 | −0.4 | 0.67 | 0.55–0.83 | 0.0677 |
| P02768 | Albumin | 0.4 | 1.50 | 1.23–1.82 | 0.0196 | 0.39 | 1.47 | 1.19–1.82 | 0.0967 |
| P33908 | MAN1A1 | −0.44 | 0.65 | 0.53–0.79 | 0.0045 | −0.4 | 0.67 | 0.53–0.84 | 0.1303 |
| Q16769 | QPCT | −0.44 | 0.64 | 0.53–0.78 | 0.0027 | −0.38 | 0.68 | 0.55–0.85 | 0.1414 |
| P01717 | IGLV3‐25 | 0.45 | 1.58 | 1.28–1.94 | 0.0065 | 0.37 | 1.45 | 1.18–1.79 | 0.1528 |
| P43358 | MAGEA4 | 0.37 | 1.45 | 1.2–1.75 | 0.0436 | 0.34 | 1.40 | 1.16–1.7 | 0.1668 |
| Q96RW7 | HMCN1 | −0.39 | 0.68 | 0.55–0.82 | 0.0338 | −0.36 | 0.70 | 0.56–0.86 | 0.2185 |
| J3KPF9 | KIF3A | 0.35 | 1.42 | 1.18–1.69 | 0.0476 | 0.33 | 1.39 | 1.15–1.69 | 0.2265 |
| O75882 | Attractin | −0.38 | 0.68 | 0.56–0.83 | 0.0455 | −0.37 | 0.69 | 0.55–0.86 | 0.2313 |
| P35858 | IGFALS | −0.42 | 0.66 | 0.54–0.81 | 0.0186 | −0.36 | 0.70 | 0.56–0.87 | 0.2734 |
| P16870 | CPE | −0.4 | 0.67 | 0.55–0.82 | 0.0217 | −0.36 | 0.70 | 0.56–0.87 | 0.3026 |
| Q9H756 | LRRC19 | −0.41 | 0.66 | 0.54–0.81 | 0.0131 | −0.35 | 0.70 | 0.56–0.88 | 0.4487 |
| P08582 | MELTF | −0.41 | 0.66 | 0.54–0.81 | 0.0125 | −0.33 | 0.72 | 0.58–0.89 | 0.4703 |
| P00738 | Haptoglobin | 0.4 | 1.50 | 1.22–1.83 | 0.0363 | 0.33 | 1.39 | 1.12–1.71 | 0.5238 |
| P55017 | SLC12A3 | −0.39 | 0.68 | 0.56–0.82 | 0.0307 | −0.32 | 0.73 | 0.6–0.89 | 0.5271 |
| P22891 | PROZ | −0.4 | 0.67 | 0.55–0.81 | 0.0149 | −0.31 | 0.74 | 0.6–0.9 | 0.6072 |
| P05154 | SERPINA5 | −0.4 | 0.67 | 0.55–0.82 | 0.0341 | −0.3 | 0.74 | 0.6–0.92 | 0.8695 |
| O43895 | XPNPEP2 | −0.39 | 0.68 | 0.55–0.83 | 0.04 | −0.28 | 0.76 | 0.61–0.94 | 0.983 |
Urinary proteins significantly associated with the outcome either in unadjusted analyses (left) or analyses that adjusted for urine creatinine levels and the MAGGIC risk score (right). HR indicates hazard ratio; and ID, identification.
ALDH1A1 indicates Aldehyde dehydrogenase 1A1; AMY2A, Pancreatic alpha‐amylase; AMY2B, Pancreatic alpha‐amylase 2B; ANGPTL2, Angiopoietin‐related protein 2; APCS, Serum amyloid P‐component; ATP1B1, Sodium/potassium‐transporting ATPase subunit beta‐1; ATP5F1A, ATP synthase subunit alpha, mitochondrial; B4GALT1, Beta‐1,4‐galactosyltransferase 1; C11orf54, Ester hydrolase C11orf54; COL15A1, Collagen alpha‐1(XV) chain; COL6A1, Collagen alpha‐1(VI) chain; CPE, Carboxypeptidase E; DNASE1, Deoxyribonuclease‐1; EGF, Pro‐epidermal growth factor; HMCN1, Hemicentin‐1; HSPD1, 60 kDa heat shock protein, mitochondrial; ICOSLG, ICOS ligand; IGFALS, Insulin‐like growth factor‐binding protein complex acid labile subunit; IGHA2, Immunoglobulin heavy constant alpha 2 (IGHA2); IGLV3‐25, Immunoglobulin lambda variable 3‐25; KIF3A, Kinesin‐like protein; LRRC19, Leucine‐rich repeat‐containing protein 19; MAGEA4, Melanoma‐associated antigen 4; MAN1A1, Mannosyl‐oligosaccharide 1,2‐alpha‐mannosidase IA; MBD1, Methyl‐CpG‐binding domain protein 1; MELTF, Melanotransferrin; PGKA, Phosphoglycerate kinase 1; PLAU, Urokinase‐type plasminogen activator; PROZ, Vitamin K‐dependent protein Z; QPCT, Glutaminyl‐peptide cyclotransferase; SERPINA5, Plasma serine protease inhibitor; SLC12A3, Solute carrier family 12 member 3; SLC2A5, Solute carrier family 2, facilitated glucose transporter member 5; X6R868, Bile salt activated lipase; XPNPEP2, Xaa‐Pro aminopeptidase 2; and ZG16B, Zymogen granule protein 16 homolog B.
Of the 40 UPPs associated with DHFA, 8 were positively associated, whereas 32 were inversely associated with the risk of DHFA. The top 3 UPPs positively associated with the risk of DFHA were IGLV3‐25 (immunoglobulin lambda variable 3–25) (hazard ratio [HR], 1.576 [95% CI, 1.28–1.94]; P=0.0065), APCS (serum amyloid P‐component) (HR, 1.547 [95% CI, 1.27–1.88]; P=0.0044), and MBD1 (methyl‐CpG‐binding domain protein 1) (HR, 1.52 [95% CI, 1.23–1.88]; P=0.0317). All other UPPs were negatively associated with DHFA. The top 3 negatively associated UPPs included: DNASE1 (deoxyribonuclease‐1) (HR, 0.584 [95% CI, 0.46–0.69]; P=2.09E‐08), AMY2A (α amylase 2A) (HR, 0.588 [95% CI, 0.46–0.69]; P=2.34E‐08), and ANGPTL2 (angiopoietin‐like protein 2) (HR, 0.566 [95% CI, 0.48–0.71]; P=7.54E‐08).
Association of Urinary Biomarker Levels and the Incidence of DHFA: Adjusted Analyses
In models that adjusted for urine creatinine, we found 39 proteins significantly associated with the incidence of DHFA. All except for 1 (glutathione S‐transferase Mu 3) was also significantly associated with the outcome in the nonunadjusted analyses.
In the subsample with urinary protein data, the MAGGIC risk score was significantly associated with death (standardized HR, 1.90 [95% CI, 1.45–2.47]; P<0.0001), heart failure‐related hospitalizations (HR, 1.77 [95% CI, 1.33–2.34]; P<0.0001), and the composite of DHFA (HR, 1.79 [95% CI, 1.45–2.20]; P<0.0001). In models that adjusted for urinary creatinine and the MAGGIC score, we found 21 UPPs to be significantly associated with the risk of DHFA, all of which were significantly associated with this outcome in the nonadjusted analyses (Figure [B]). The top UPPs associated with the risk of DHFA in these adjusted models included: ANGPTL2 (HR, 0.57 [95% CI, 0.47–0.70]; P<0.0001), AMY2A (HR, 0.55 [95% CI, 0.44–0.69]; P=0.0001), and DNASE1 (HR, 0.57 [95% CI, 0.46–0.71]; P=0.0002).
Additionally, we conducted an analysis adjusting for (1) urinary creatinine and estimated glomerular filtration rate, (2) total urine protein, and (3) the inverse of urine creatinine (1/urine creatinine) to assess the sensitivity of our results to kidney function specifically. In each analysis, all proteins were present in the primary analysis except for 4 proteins (Tables S2 through S4). Finally, we conducted an analysis adjusting for the trial arm (placebo versus spironolactone administration). We found 38 significant UPPs, all of which were present in the primary analyses (Table S5). The 2 UPPs that were not significant after adjusting for randomization arm were KIF3A (kinesin family member 3A) and attractin.
Interaction Testing
We investigated potential interactions between location (Americas versus Russia/Georgia) and all urinary protein levels as predictors of DHFA. At corrected significance, we found a significant interaction with complement C1q B chain (P for interaction=0.00458). This protein was negatively associated with DHFA in Russia/Georgia (standardized HR, 0.63 [95% CI, 0.45–0.90]; P=0.01) and positively associated with DHFA in the Americas (HR, 1.49 [95% CI, 1.19–1.86]; P=0.0004). Notably, none of the urinary proteins that were associated with DHFA in the overall cohort exhibited significant interactions with enrollment continent, even at nominal significance. Similarly, there were no significant interactions between urinary protein levels at baseline and randomization arm as predictors of DHFA, either in the entire urinary proteome or in the subset of proteins associated with DHFA in the main effects analysis.
Ingenuity Pathway Analysis
Ingenuity pathway analysis pathway overrepresentation analysis based on the UPPs associated with the risk of DHFA identified 5 canonical signaling pathways associated with the outcome. Four of these were fibrosis‐related pathways (hepatic fibrosis/hepatic stellate cell activation, wound healing, idiopathic pulmonary fibrosis, and GP6 signaling), whereas the remainder pathway (nicotinamide adenine dinucleotide [NAD] signaling) is related to cell metabolism.
DISCUSSION
We conducted a proteomic analysis of urinary biomarkers associated with the risk of DHFA in HFpEF. We identified 40 UPPs associated with this outcome, which are related to fibrosis, metabolism, and inflammation. We identified 21 proteins that were associated with the risk of DHFA after adjustment for the MAGGIC risk score and urinary creatinine levels.
Multiple previous studies have examined plasma proteomics approaches in HFpEF. 3 , 4 , 5 , 6 , 7 , 8 , 9 , 10 , 11 , 12 , 13 , 14 , 15 In contrast to the extensive previous work with plasma proteomics, data on urinary proteomics in HFpEF are scarce. Only 1 previous study assessed differences in urinary proteins between participants with HF and controls. 16 Studies assessing the relationship between the urinary proteome and outcomes in HFpEF are not available.
We found several UPPs to be inversely associated with the risk of DHFA in HFpEF. Some of our results may be counterintuitive considering prior research in plasma proteomics. For example, whereas increased urinary levels of ANGPTL2 were associated with a decreased risk of DHFA in our study, previous literature has implicated ANGPTL2 as playing a causal role in cardiovascular disease and HF via its proinflammatory properties. 30 , 31 However, it is important to note that several potential mechanisms of dissociation between plasma and urinary protein associations with outcomes may exist. It is known that plasma and urine protein levels are in general poorly correlated. 32 , 33 Whereas urine is produced predominantly as a result of plasma filtration in the kidney, the origin of larger proteins and peptides present in the urine is less clear. On one hand, low molecular weight proteins and peptides can be filtered and variably reabsorbed by the renal tubules. On the other hand, it has also been shown that renal tubules can secrete extracellular vesicles that contain multiple molecules, including a variety of proteins. 34 Finally, the remainder of the urinary tract itself may be a source of proteins. Prior research has suggested that the kidney may be a source of production of ANGPLT2. 35 , 36 , 37 , 38 A urinary source of ANGPTL2 or another protein could be a reason why elevated urinary levels of a protein would have different associations than the circulating protein, and suggest other mechanisms for pathophysiology. It is possible that the urinary system may use compensatory mechanisms to increase or decrease clearance of proteins, which at present are poorly understood. Finally, some filtered proteins or peptides may undergo proteolytic or other chemical modifications in the urinary tract, which could significantly impact their function and quantification, leading to clinical associations unrelated to its degree of filtration or secretion. Our findings should therefore be interpreted with caution, because we only studied associations with incident outcomes, rather than underlying mechanisms. Further research should be performed to better understand the determinants of differences between plasma and urinary protein levels.
Increased Levels of Fibrosis Peptides in Urine Are Associated With a Decreased Risk of DHFA
We found increased urinary levels of 2 collagen‐derived proteins COL15A1 (collagen α‐1[XV] chain) and COL6A1 (collagen α‐1[VI] chain) to be associated with a lower risk of DHFA. COL6A1 is a chain of type VI collagen, which is a major structural component of microfibrils. 39 , 40 Interestingly, plasma levels of endotrophin, a peptide derived from the collagen VI α‐3 chain, has been reported to be strongly and positively associated with the risk of adverse outcomes in HFpEF. 15 COL15A1 is part of type XV collagen, which is widely expressed but frequently localized to the basement membrane. 41 Derangements in collagen turnover and organization in cardiomyopathies are well established. 42 They have also been specifically implicated as part of the pathophysiology of HFpEF. 43 , 44 One study of urinary proteomics found elevated levels of urinary biomarkers associated with collagen metabolism, including COL6A1 and COL15A1, to be generally enriched in patients with both HFpEF and heart failure with reduced ejection fraction. However, this study did not assess their association with outcomes. 16
Our findings on inverse associations between COL6A1 and COL15A1 and the risk of DHFA is counterintuitive, given the well‐known association between plasma biomarkers of tissue fibrosis and adverse outcomes in HFpEF. 45 , 46 It should be noted that UPPs have multiple determinants and are not a direct representation of plasma levels. In addition to glomerular filtration, proteins can be secreted in the urinary tract by tubular cells and epithelial cells. For instance, it has been reported that the secretion of collagen by renal tubular and epithelial cells can be affected by exposure to albumin. 47 Also, there is variable reabsorption of filtered proteins by tubular cells, and there may be interindividual differences in the degradation of filtered or secreted proteins that may ultimately affect their measured levels in urine. Finally, it is possible that increased levels in urine could represent increased clearance of plasma collagen‐derived peptides. Further research is required to assess the mechanistic determinants of the urinary proteome/peptidome in HFpEF.
Metabolic Pathways Implicated in Outcomes
We identified several UPPs that are related to metabolic processes, including carbohydrate metabolism (AMY2A [pancreatic α‐amylase], AMY2B [α amylase 2B], MAN1A1 [mannosyl‐oligosaccharide 1,2‐α‐mannosidase IA]), and lipid metabolism (ALDH1A1 [aldehyde dehydrogenase 1A1], B4GALT1 [β‐1,4‐galactosyltransferase 1]).
We found that increased urinary pancreatic α‐amylase (AMY2A and AMY2B) protein is associated with a lower risk of DHFA. There is limited literature on the association of AMY2A and HF, but 1 study found that levels of plasma AMY2A were lower in patients with HF compared with healthy controls. 48 However, another study reported that plasma amylase levels are elevated in patients with severe but not mild HF. 49
Other Novel Associations
We found prostasin to be inversely associated with the risk of DHFA. Prostasin is an epithelial sodium channel stimulator. Decreased prostasin expression is associated with poor outcomes in colorectal cancer and oral squamous cell carcinoma. 50 , 51 However, a recent study found that plasma prostasin levels are positively associated with diabetes risk and cancer mortality, 52 whereas another study found that serum prostasin has an inverse association with physical activity in a population‐based cohort. 53 Whether the association between lower urinary prostasin levels and the increased risk of DHFA seen in our study is related to physical activity in these patients remains to be assessed in future research. To our knowledge, prostasin has not been associated with HFpEF.
Study Limitations
Our study should be interpreted in the context of its strengths and limitations. Strengths of our study include its well‐characterized cohort, relatively long period of follow‐up, prospectively adjudicated outcomes with a stringent criteria and methodology, and the unbiased nature of UPP measurements. Our study also has limitations. Urinary samples were not available from all TOPCAT participants. In addition, the inclusion of some participants with undiagnosed cardiac or renal amyloidosis cannot be excluded, which may confound the urinary proteome. Our study did not include an external validation cohort, given the limited availability of HFpEF cohorts with prospective follow‐up and available urine samples. Our study identifies multiple novel urinary proteins that are prognostic in HFpEF, but further research in larger samples should be performed to assess the optimal analytical techniques, prognostic cut points, and prognostic performance. Finally, it is worth noting that no significant interactions were found between urinary proteins and either continent of origin or randomized spironolactone therapy, except for 1 protein that interacted with continent of origin, which has unclear biologic and clinical significance. However, given the overall limited sample size, interaction analyses may have failed to detect some effect modifications, and stratified analyses would be underpowered and challenging to interpret. Finally, the origin of the UPPs measured in our study (contribution from circulating blood, secretion by the urinary tract, differential catabolism) could not be assessed and should be the focus of future research.
CONCLUSIONS
Our study reports the relationship between UPPs and the risk of DHFA in HFpEF. We identify several novel associations between UPPs and adverse outcomes in this patient population. This work forms a basis for both validation in larger and different data sets and for mechanistic experimentation. Further research should assess the clinical value of urinary proteomic biomarkers in HFpEF, including its potential role in prognosis and clinical management, as well as HFpEF pathophysiology.
Sources of Funding
This work was funded by a grant from Bristol‐Myers Squibb to the University of Pennsylvania (J.A.C.).
Disclosures
J.B.C. is supported by National Institutes of Health grants K23‐HL133843 and R01‐HL153646. P.H.S. is an employee and shareholder of Bristol Myers Squibb. E.R. has received unrestricted educational grants from Amgen, Merck Sharp & Dohme, AstraZeneca, Sanofi, and Unilever; speakers' or consultancy fees from Daiichi Sankyo, Novonordisk, Boehringer Ingelheim, Servier, Amgen, Sanofi, Novartis, and Teva; all paid directly to Ghent University. P.Z. is supported by grants R01 HL155599, R01 HL157264, R01 HL149722, U01‐HL160277, UH3DK128298. He also receives research support from Amgen. He has consulted for Pfizer and Vyaire. J.A.C., L.Z., and A.D. are named inventors in a patent application related to urine protein biomarkers in HFpEF.
J.A.C has recently consulted for Bayer, Sanifit, Fukuda‐Denshi, Bristol Myers Squibb, Johnson & Johnson, Edwards Life Sciences, Merck, and the Galway‐Mayo Institute of Technology. He received University of Pennsylvania research grants from the National Institutes of Health, Fukuda‐Denshi, Bristol Myers Squibb, and Microsoft. He is named as an inventor in a University of Pennsylvania patent for the use of inorganic nitrates/nitrites in Heart Failure with Preserved Ejection Fraction. He is named as inventor on patent applications for the use of protein biomarkers in heart failure. He has received research device loans from Atcor Medical, Fukuda‐Denshi, Uscom, NDD Medical Technologies, Microsoft, and MicroVision Medical.
J.B.C is supported by R01‐HL153646, R01‐HL157108, R01‐HL155599, R01‐HL157264, U01‐HL160277, U24‐DK060990, and R01‐AG074989, and an American Heart Association Bugher Award. A.M.R is a holder of NZ Heart Foundation Chair of Cardiovascular Studies and recipient of grant support in cash or kind from Roche Diagnostics. He is an advisory board member for Roche Dx, Ad Board member Novartis. He has recieved grants from Novo Nordisk, grants in kind from Abbott Laboratories and Sphingotec. He is a shareholder in BMS and Upstream Medical Technologies. D.G., L.N.C are employees and shareholder at BMS. S.H. is an independent board member, Sampled Personalized Medicine Coalition Board Member Board Observer, Precede Biosciences Shareholder of BMS, JNJ and Novartis stock. R.N.D holds the New Zealand Heart Foundation Chair of Heart Health.
Supporting information
Acknowledgments
This article was prepared using data and biosamples from TOPCAT obtained from the National Heart, Lung, and Blood Institute Biologic Specimen and Data Repository Information Coordinating Center and does not necessarily reflect the opinions or views of the TOPCAT investigators or the National Heart, Lung, and Blood Institute.
This article was sent to Sula Mazimba, MD, MPH, Associate Editor, for review by expert referees, editorial decision, and final disposition.
Supplemental Material is available at https://www.ahajournals.org/doi/suppl/10.1161/JAHA.123.033410
For Sources of Funding and Disclosures, see page 9.
References
- 1. Pfeffer MA, Shah AM, Borlaug BA. Heart failure with preserved ejection fraction in perspective. Circ Res. 2019;124:1598–1617. doi: 10.1161/CIRCRESAHA.119.313572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Oktay AA, Rich JD, Shah SJ. The emerging epidemic of heart failure with preserved ejection fraction. Curr Heart Fail Rep. 2013;10:401–410. doi: 10.1007/s11897-013-0155-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Regan JA, Truby LK, Tahir UA, Katz DH, Nguyen M, Kwee LC, Deng S, Wilson JG, Mentz RJ, Kraus WE, et al. Protein biomarkers of cardiac remodeling and inflammation associated with HFpEF and incident events. Sci Rep. 2022;12:20072. doi: 10.1038/s41598-022-24226-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tonry C, McDonald K, Ledwidge M, Hernandez B, Glezeva N, Rooney C, Morrissey B, Pennington SR, Baugh JA, Watson CJ. Multiplexed measurement of candidate blood protein biomarkers of heart failure. ESC Heart Fail. 2021;8:2248–2258. doi: 10.1002/ehf2.13320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hanff TC, Cohen JB, Zhao L, Javaheri A, Zamani P, Prenner SB, Rietzschel E, Jia Y, Walsh A, Maranville J, et al. Quantitative proteomic analysis of diabetes mellitus in heart failure with preserved ejection fraction. JACC Basic Transl Sci. 2021;6:89–99. doi: 10.1016/j.jacbts.2020.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Faxen UL, Venkateshvaran A, Shah SJ, Lam CSP, Svedlund S, Saraste A, Beussink‐Nelson L, Fermer ML, Gan LM, Hage C, et al. Generalizability of HFA‐PEFF and H(2)FPEF diagnostic algorithms and associations with heart failure indices and proteomic biomarkers: insights from PROMIS‐HFpEF. J Card Fail. 2021;27:756–765. doi: 10.1016/j.cardfail.2021.02.005 [DOI] [PubMed] [Google Scholar]
- 7. Adamo L, Yu J, Rocha‐Resende C, Javaheri A, Head RD, Mann DL. Proteomic signatures of heart failure in relation to left ventricular ejection fraction. J Am Coll Cardiol. 2020;76:1982–1994. doi: 10.1016/j.jacc.2020.08.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sanders‐van Wijk S, Tromp J, Beussink‐Nelson L, Hage C, Svedlund S, Saraste A, Swat SA, Sanchez C, Njoroge J, Tan RS, et al. Proteomic evaluation of the comorbidity‐inflammation paradigm in heart failure with preserved ejection fraction: results from the PROMIS‐HFpEF study. Circulation. 2020;142:2029–2044. doi: 10.1161/CIRCULATIONAHA.120.045810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tromp J, Khan MA, Klip IT, Meyer S, de Boer RA, Jaarsma T, Hillege H, van Veldhuisen DJ, van der Meer P, Voors AA. Biomarker profiles in heart failure patients with preserved and reduced ejection fraction. J Am Heart Assoc. 2017;6:6. doi: 10.1161/JAHA.116.003989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kresoja KP, Rommel KP, Wachter R, Henger S, Besler C, Klöting N, Schnelle M, Hoffmann A, Büttner P, Ceglarek U, et al. Proteomics to improve phenotyping in obese patients with heart failure with preserved ejection fraction. Eur J Heart Fail. 2021;23:1633–1644. doi: 10.1002/ejhf.2291 [DOI] [PubMed] [Google Scholar]
- 11. Javaheri A, Diab A, Zhao L, Qian C, Cohen JB, Zamani P, Kumar A, Wang Z, Ebert C, Maranville J, et al. Proteomic analysis of effects of spironolactone in heart failure with preserved ejection fraction. Circ Heart Fail. 2022;15:e009693. doi: 10.1161/CIRCHEARTFAILURE.121.009693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chirinos JA, Zhao L, Jia Y, Frej C, Adamo L, Mann D, Shewale SV, Millar JS, Rader DJ, French B, et al. Reduced apolipoprotein M and adverse outcomes across the spectrum of human heart failure. Circulation. 2020;141:1463–1476. doi: 10.1161/CIRCULATIONAHA.119.045323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chirinos JA, Orlenko A, Zhao L, Basso MD, Cvijic ME, Li Z, Spires TE, Yarde M, Wang Z, Seiffert DA, et al. Multiple plasma biomarkers for risk stratification in patients with heart failure and preserved ejection fraction. J Am Coll Cardiol. 2020;75:1281–1295. doi: 10.1016/j.jacc.2019.12.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cohen JB, Schrauben SJ, Zhao L, Basso MD, Cvijic ME, Li Z, Yarde M, Wang Z, Bhattacharya PT, Chirinos DA, et al. Clinical phenogroups in heart failure with preserved ejection fraction: detailed phenotypes, prognosis, and response to spironolactone. JACC Heart Fail. 2020;8:172–184. doi: 10.1016/j.jchf.2019.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chirinos JA, Zhao L, Reese‐Petersen AL, Cohen JB, Genovese F, Richards AM, Doughty RN, Díez J, González A, Querejeta R, et al. Endotrophin, a collagen VI formation–derived peptide, in heart failure. NEJM Evid. 2022;1:EVIDoa2200091. doi: 10.1056/EVIDoa2200091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. He T, Mischak M, Clark AL, Campbell RT, Delles C, Díez J, Filippatos G, Mebazaa A, McMurray JJV, González A, et al. Urinary peptides in heart failure: a link to molecular pathophysiology. Eur J Heart Fail. 2021;23:1875–1887. doi: 10.1002/ejhf.2195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Njoku K, Chiasserini D, Jones ER, Barr CE, O'Flynn H, Whetton AD, Crosbie EJ. Urinary biomarkers and their potential for the non‐invasive detection of endometrial cancer. Front Oncol. 2020;10:559016. doi: 10.3389/fonc.2020.559016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wei D, Melgarejo JD, Thijs L, Temmerman X, Vanassche T, Van Aelst L, Janssens S, Staessen JA, Verhamme P, Zhang ZY. Urinary proteomic profile of arterial stiffness is associated with mortality and cardiovascular outcomes. J Am Heart Assoc. 2022;11:e024769. doi: 10.1161/JAHA.121.024769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Latosinska A, Siwy J, Faguer S, Beige J, Mischak H, Schanstra JP. Value of urine peptides in assessing kidney and cardiovascular disease. PROTEOMICS–Clin Appl. 2021;15:2000027. doi: 10.1002/prca.202000027 [DOI] [PubMed] [Google Scholar]
- 20. Decramer S, de Peredo AG, Breuil B, Mischak H, Monsarrat B, Bascands J‐L, Schanstra JP. Urine in clinical proteomics. Mol Cell Proteomics. 2008;7:1850–1862. doi: 10.1074/mcp.R800001-MCP200 [DOI] [PubMed] [Google Scholar]
- 21. Pitt B, Pfeffer MA, Assmann SF, Boineau R, Anand IS, Claggett B, Clausell N, Desai AS, Diaz R, Fleg JL, et al. Spironolactone for heart failure with preserved ejection fraction. N Engl J Med. 2014;370:1383–1392. doi: 10.1056/NEJMoa1313731 [DOI] [PubMed] [Google Scholar]
- 22. Desai AS, Lewis EF, Li R, Solomon SD, Assmann SF, Boineau R, Clausell N, Diaz R, Fleg JL, Gordeev I, et al. Rationale and design of the treatment of preserved cardiac function heart failure with an aldosterone antagonist trial: a randomized, controlled study of spironolactone in patients with symptomatic heart failure and preserved ejection fraction. Am Heart J. 2011;162:966–972. doi: 10.1016/j.ahj.2011.09.007 [DOI] [PubMed] [Google Scholar]
- 23. Krämer A, Green J, Pollard J Jr, Tugendreich S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics. 2014;30:523–530. doi: 10.1093/bioinformatics/btt703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pocock SJ, Ariti CA, McMurray JJ, Maggioni A, Kober L, Squire IB, Swedberg K, Dobson J, Poppe KK, Whalley GA, et al. Predicting survival in heart failure: a risk score based on 39 372 patients from 30 studies. Eur Heart J. 2013;34:1404–1413. doi: 10.1093/eurheartj/ehs337 [DOI] [PubMed] [Google Scholar]
- 25. Gao X, Starmer J, Martin ER. A multiple testing correction method for genetic association studies using correlated single nucleotide polymorphisms. Genet Epidemiol. 2008;32:361–369. doi: 10.1002/gepi.20310 [DOI] [PubMed] [Google Scholar]
- 26. Auro K, Joensuu A, Fischer K, Kettunen J, Salo P, Mattsson H, Niironen M, Kaprio J, Eriksson JG, Lehtimäki T, et al. A metabolic view on menopause and ageing. Nat Commun. 2014;5:4708. doi: 10.1038/ncomms5708 [DOI] [PubMed] [Google Scholar]
- 27. Chirinos JA, Cohen JB, Zhao L, Hanff T, Sweitzer N, Fang J, Corrales‐Medina V, Anmar R, Morley M, Zamani P, et al. Clinical and proteomic correlates of plasma ACE2 (angiotensin‐converting enzyme 2) in human heart failure. Hypertension. 2020;76:1526–1536. doi: 10.1161/HYPERTENSIONAHA.120.15829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vidula MK, Orlenko A, Zhao L, Salvador L, Small AM, Horton E, Cohen JB, Adusumalli S, Denduluri S, Kobayashi T, et al. Plasma biomarkers associated with adverse outcomes in patients with calcific aortic stenosis. Eur J Heart Fail. 2021;23:2021–2032. doi: 10.1002/ejhf.2361 [DOI] [PubMed] [Google Scholar]
- 29. MATLAB. version 7.10.0 (R2010a) ed. The MathWorks Inc; 2012. [Google Scholar]
- 30. Tian Z, Miyata K, Kadomatsu T, Horiguchi H, Fukushima H, Tohyama S, Ujihara Y, Okumura T, Yamaguchi S, Zhao J, et al. ANGPTL2 activity in cardiac pathologies accelerates heart failure by perturbing cardiac function and energy metabolism. Nat Commun. 2016;7:1–19. doi: 10.1038/ncomms13016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Oike Y, Tian Z, Miyata K, Morinaga J, Endo M, Kadomatsu T. ANGPTL2―a new causal player in accelerating heart disease development in the aging. Circ J. 2017;81:1379–1385. doi: 10.1253/circj.CJ-17-0854 [DOI] [PubMed] [Google Scholar]
- 32. Magalhaes P, Pontillo C, Pejchinovski M, Siwy J, Krochmal M, Makridakis M, Carrick E, Klein J, Mullen W, Jankowski J, et al. Comparison of urine and plasma peptidome indicates selectivity in renal peptide handling. PROTEOMICS–Clin Appl. 2018;12:1700163. doi: 10.1002/prca.201700163 [DOI] [PubMed] [Google Scholar]
- 33. Jia L, Zhang L, Shao C, Song E, Sun W, Li M, Gao Y. An attempt to understand kidney's protein handling function by comparing plasma and urine proteomes. PLoS One. 2009;4:e5146. doi: 10.1371/journal.pone.0005146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Giulia C, Bellucci L, Montini G, Collino F. Urinary extracellular vesicles: uncovering the basis of the pathological processes in kidney‐related diseases. Int J Sci. 2021;22:22. doi: 10.3390/ijms22126507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schmidt T, Samaras P, Frejno M, Gessulat S, Barnert M, Kienegger H, Krcmar H, Schlegl J, Ehrlich HC, Aiche S, et al. ProteomicsDB. Nucleic Acids Res. 2018;46:D1271–D1281. doi: 10.1093/nar/gkx1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nishi H. Angiopoietin‐like protein 2 and kidney fibrosis: lessons from knockout mice. Kidney Int. 2016;89:272–274. doi: 10.1016/j.kint.2015.12.022 [DOI] [PubMed] [Google Scholar]
- 37. Desjardins MP, Thorin‐Trescases N, Sidibe A, Fortier C, De Serres SA, Lariviere R, Thorin E, Agharazii M. Levels of angiopoietin‐Like‐2 are positively associated with aortic stiffness and mortality after kidney transplantation. Am J Hypertens. 2017;30:409–416. doi: 10.1093/ajh/hpw208 [DOI] [PubMed] [Google Scholar]
- 38. Thorin‐Trescases N, Thorin E. High circulating levels of ANGPTL2: beyond a clinical marker of systemic inflammation. Oxidative Med Cell Longev. 2017;2017:1096385–1096312. doi: 10.1155/2017/1096385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. GeneCards . COL6A1. Accessed October 1, 2022. https://www.genecards.org/cgi‐bin/carddisp.pl?gene=COL6A1#summaries.
- 40. Safran M, Rosen N, Twik M, BarShir R, Stein TI, Dahary D, Fishilevich S, Lancet D. The Genecards Suite. Practical Guide to Life Science Databases. Springer; 2021:27–56. [Google Scholar]
- 41. GeneCards . COL15A1. Accessed October 1, 2022. https://www.genecards.org/cgi‐bin/carddisp.pl?gene=COL15A1#summaries.
- 42. López B, González A, Ravassa S, Beaumont J, Moreno MU, San José G, Querejeta R, Díez J. Circulating biomarkers of myocardial fibrosis: the need for a reappraisal. J Am Coll Cardiol. 2015;65:2449–2456. doi: 10.1016/j.jacc.2015.04.026 [DOI] [PubMed] [Google Scholar]
- 43. Mohammed SF, Hussain S, Mirzoyev SA, Edwards WD, Maleszewski JJ, Redfield MM. Coronary microvascular rarefaction and myocardial fibrosis in heart failure with preserved ejection fraction. Circulation. 2015;131:550–559. doi: 10.1161/CIRCULATIONAHA.114.009625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Borlaug BA. The pathophysiology of heart failure with preserved ejection fraction. Nat Rev Cardiol. 2014;11:507–515. doi: 10.1038/nrcardio.2014.83 [DOI] [PubMed] [Google Scholar]
- 45. Paulus WJ, Zile MR. From systemic inflammation to myocardial fibrosis: the heart failure with preserved ejection fraction paradigm revisited. Circ Res. 2021;128:1451–1467. doi: 10.1161/CIRCRESAHA.121.318159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Prenner SB, Pillutla R, Yenigalla S, Gaddam S, Lee J, Obeid MJ, Ans AH, Jehangir Q, Kim J, Zamani P, et al. Serum albumin is a marker of myocardial fibrosis, adverse pulsatile aortic hemodynamics, and prognosis in heart failure with preserved ejection fraction. J Am Heart Assoc. 2020;9:e014716. doi: 10.1161/JAHA.119.014716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wohlfarth V, Drumm K, Mildenberger S, Freudinger R, Gekle M. Protein uptake disturbs collagen homeostasis in proximal tubule‐derived cells. Kidney Int Suppl. 2003;63:S103–S109. doi: 10.1046/j.1523-1755.63.s84.13.x [DOI] [PubMed] [Google Scholar]
- 48. Hou LN, Li F, Zeng QC, Su L, Chen PA, Xu ZH, Zhu DJ, Liu CH, Xu DL. Excretion of urinary orosomucoid 1 protein is elevated in patients with chronic heart failure. PLoS One. 2014;9:e107550. doi: 10.1371/journal.pone.0107550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Parissis JT, Adamopoulos SN, Venetsanou KF, Karas SM, Kremastinos DT. Elevated plasma amylase levels in advanced chronic heart failure secondary to ischemic or idiopathic dilated cardiomyopathy: correlation with circulating interleukin‐6 activity. J Interf Cytokine Res. 2003;23:329–333. doi: 10.1089/107999003766628179 [DOI] [PubMed] [Google Scholar]
- 50. Yamamoto K, Yamashita F, Kawaguchi M, Izumi A, Kiwaki T, Kataoka H, Kaneuji T, Yamashita Y, Fukushima T. Decreased prostasin expression is associated with aggressiveness of oral squamous cell carcinoma. Hum Cell. 2021;34:1434–1445. doi: 10.1007/s13577-021-00575-3 [DOI] [PubMed] [Google Scholar]
- 51. Bao Y, Li K, Guo Y, Wang Q, Li Z, Yang Y, Chen Z, Wang J, Zhao W, Zhang H, et al. Tumor suppressor PRSS8 targets Sphk1/S1P/Stat3/Akt signaling in colorectal cancer. Oncotarget. 2016;7:26780–26792. doi: 10.18632/oncotarget.8511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bao X, Xu B, Muhammad IF, Nilsson PM, Nilsson J, Engström G. Plasma prostasin: a novel risk marker for incidence of diabetes and cancer mortality. Diabetologia. 2022;65:1642–1651. doi: 10.1007/s00125-022-05771-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Stattin K, Lind L, Elmståhl S, Wolk A, Lemming EW, Melhus H, Michaëlsson K, Byberg L. Physical activity is associated with a large number of cardiovascular‐specific proteins: cross‐sectional analyses in two independent cohorts. Eur J Prev Cardiol. 2019;26:1865–1873. doi: 10.1177/2047487319868033 [DOI] [PubMed] [Google Scholar]
- 54. UniProt: the universal protein knowledgebase in 2023. Nucleic Acids Res. 2023;51:D523–D531. doi: 10.1093/nar/gkac1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Stelzer G, Rosen N, Plaschkes I, Zimmerman S, Twik M, Fishilevich S, Stein TI, Nudel R, Lieder I, Mazor Y, et al. The GeneCards suite: from gene data mining to disease genome sequence analyses. Curr Protoc Bioinformatics. 2016;54:1.30.1–1.30.33. doi: 10.1002/cpbi.5 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


