Abstract
Rubella virus particles, consisting of a nucleocapsid surrounded by a lipid envelope in which two virus-encoded glycoproteins E1 and E2 are embedded, assemble on intracellular membranes and are secreted from cells, possibly via the cellular secretory pathway. We have recently demonstrated that the cytoplasmic domain of E1 (residues 469 to 481, KCLYYLRGAIAPR) is required for virus release. Alteration of cysteine 470 to alanine did not affect virus release, whereas mutation of leucine 471 to alanine reduced virus production by 90%. In the present study, substitutions of remaining amino acids in the E1 cytoplasmic domain were made in order to investigate the role of each amino acid in regulating rubella virus release. Generated mutants were analyzed in the context of infectious full-length cDNA clone and virus-like particles using combined genetic, biochemical, and electron microscopic approaches. Substitution of a single residue of tyrosine 472 to alanine or tyrosine 473 to serine resulted in a block in virus release without affecting protein transport and virus budding into the lumen of the Golgi complexes. Infectious RNA transcripts bearing these mutations were incapable of forming plaques. Mutants with substitutions at the amino-terminal region (leucine 474, arginine 475, and glycine 476) in the E1 cytoplasmic domain had reduced virus release and small-plaque phenotype, while mutants with substitutions at the carboxy-terminal region (alanine 477, isoleucine 478, alanine 479, proline 480, and arginine 481) had only marginal defects in virus release. Plaque-forming revertants could be isolated from mutants Y472A and Y473S. Sequencing analysis revealed that the substituted serine residue in mutant Y473S reverted to the original tyrosine residue, whereas the substituted alanine residue in mutant Y472A was retained. These results indicate that the E1 cytoplasmic domain modulates virus release in a sequence-dependent manner and that the tyrosine residues are critical for this function. We postulate that residues YYLRG constitute a domain in the E1 tail that may interact with other proteins and this interaction is involved in regulating virus release.
Rubella virus (RV), the etiological agent of German measles, is the only member of the Rubivirus genus in the Togaviridae family (7, 8, 17). The RV virion consists of three structural proteins: a capsid protein which forms nucleocapsid inside the virion; and two membrane glycoproteins, E1 and E2, embedded in the viral envelope (26). The structural proteins are synthesized as a polyprotein precursor in the order capsid-E2-E1 (5, 6, 20, 21). This polyprotein is translocated into the endoplasmic reticulum (ER) by two independently functioning signal peptides within the carboxy (C) terminus of capsid and E2 (10, 13). In the ER, the polyprotein precursor is cleaved by cellular signal peptidases into the three structural proteins, capsid, E2, and E1 (11). The capsid protein is associated with membranes, presumably mediated by the E2 signal sequence attached at its C terminus (10, 18). It is unknown how nucleocapsid is formed. The glycoproteins E2 and E1 form a specific heterodimer in the ER shortly after synthesis (2). The E2-E1 heterodimers are transported out of the ER and to the Golgi complexes, where virus buds through cellular membranes (12, 22). The budded viral particles are then released from the cells, possibly via the cellular secretory pathway. It has been proposed that the E1 cytoplasmic domain may drive virus budding by interaction with nucleocapsids (12). By using an infectious cDNA clone derived from RV M33 strain (27), we have recently demonstrated that the E1 cytoplasmic domain is required for virus release. Mutation of leucine 471 at the amino (N) terminus of the E1 cytoplasmic domain reduced virus production by 90% (27). Garbutt et al. (9) also reported that replacement of E1 cytoplasmic domain with the analogous region from other type I membrane glycoproteins results in arrest in release of virus-like particles but does not affect virus budding into the Golgi complexes. In this study, we continued the investigation of the role of the remaining amino acid residues in the E1 cytoplasmic domain in regulating virus release. Our results show that substitutions of most of the N-terminal amino acids in the E1 tail affected virus release and plaque phenotype, while substitutions in the C-terminal amino acids had only marginal effects on virus release. Of the substitutions, a single-amino-acid substitution of alanine for tyrosine 472 or serine for tyrosine 473 in the E1 tail blocked virus release without affecting virus budding into the lumen of the Golgi complexes, indicating that the E1 tail regulates virus release in a sequence-dependent manner and that the tyrosines are the key residues for virus release.
MATERIALS AND METHODS
Virus and cells.
Vero cells were grown in Eagle's minimum essential medium (MEM) supplemented with 5% fetal bovine serum, penicillin (100 U/ml), and streptomycin (50 μg/ml). BHK-21 cells were grown in MEM containing 10% fetal bovine serum, and 10% tryptose phosphate broth.
Construction of mutants.
A series of mutations was introduced into the E1 coding region by PCR-mediated mutagenesis with appropriate primers containing the desired nucleotide changes using the full-length cDNA clone pBRM34 template. This contains a new SphI site created by changing T to A at nucleotide (nt) 9647 (27).
To construct mutants Y472A and Y473S, PCR amplifications were performed in reactions with sense primers 5′-TTACTCGCATGCTGTGCCAAATGCTTGGCCTACTTG-3′ for mutant Y472A and 5′-TTACTCGCATGCTGTGCCAAATGCTTGTACAGCTTGCGC-3′ for mutant Y473S (nucleotide changes are underlined) and with antisense primer 5′-GAATTCAAGCT17-3′ (the latter contains a HindIII site). The amplified DNA fragments were cut with SphI and HindIII and reintroduced into the full-length cDNA clone pBRM34. To construct mutants L474S, R475S, G476C, A477S, I478V, A479S, P480S, and R481S, PCR amplifications were done with sense primer 5′-AATGCCCGAGTGGATCCA-3′ and antisense primers 5′-TACTCCGCGGCGCTATAGCACCGCGCTAGCGGGCC-3′ for L474S, 5′-TTGAGCGGCGCTATAGCACCGCGCTAGCGGGCC-3′ for R475S, 5′-CGCAGCGCTATAGCACCGCGCTAGCGGGCC-3′ for R476C, 5′-GGCTCTATAGCACCGCGCTAGCGGGCC-3′ for A477S, 5′-GCTGTAGCACCGCGCTAGCGGGCC-3′ for I478V, 5′-ATATCACCGCGCTAGCGGGCC-3′ for A479S, 5′-ATAGCATCGCGCTAGCGGGCC-3′ for P480S, and 5′-ATAGCACCGAGCTAGCGGGCC-3′ for R481S (nucleotide changes are underlined). The amplified DNA fragments were cut with BamHI and NheI and reinserted into the full-length cDNA clone pBRM34. All of the mutations were verified by sequencing.
To construct wild-type 24S (24S/WT), the DNA fragment containing full-length RV subgenomic cDNA was isolated from plasmid 24S/pSPT19 (11) and cloned into pSFV-1 vector (15). To construct mutants 24S/Y472A, 24S/Y473S, 24S/L474S, 24S/R475S, 24S/G476C, 24S/A477S, and 24S/R481S, BamHI-HindIII fragments containing these mutations were isolated from mutants Y472A, Y473S, L474S, R475S, G476C, A477S, and R481S and inserted to replace the corresponding fragment in 24S/WT.
RNA transcription and transfection.
RNA transcripts were synthesized using SP6 RNA polymerase in the presence of m7Gpp(5′)G cap analog as described previously (27). Vero cells were transfected by a Lipofectin-mediated transfection method. Briefly, 10 μl of RNA transcription reaction was mixed with 10 μl of Lipofectin (Gibco/BRL) at room temperature. The mixtures were added to Vero cells that had been washed twice with MEM. After incubation at 37°C for 2 h, the solutions were removed and replaced with growth medium. At day 5 posttransfection, the liquid medium was harvested as viral stock for virus titration. BHK cells were transfected by electroporation as described previously (27, 28). Briefly, BHK cells were harvested by trypsin treatment and washed twice with ice-cold phosphate-buffered saline without Ca2+ and Mg2+ and resuspended at a concentration of 107 cells/ml; 20 μl of RNA transcript was mixed with 0.5 ml of cells, and the mixture was transferred to a 2-mm cuvette. Electroporation was at room temperature with two consecutive 1.5-kV, 25-μF pulses with a Gene-Pulser (Bio-Rad). After electroporation, the cells were diluted into 10 ml of culture medium and distributed into six-well culture plates.
Metabolic labeling, immunoprecipitation, and endoglycosidase H (endo H) digestion.
Analysis of viral structural protein synthesis and release of viral particles was as described previously (27). Briefly, BHK cells transfected with mutant RNAs by electroporation were incubated at 37°C for 40 h, washed with MEM, and starved in methionine-free medium for 30 min at 37°C. This medium was replaced with one containing [35S]methionine (200 μCi/ml; NEN), and the cells were pulse-labeled for 80 min. After labeling, the cells were chased for 10 min or 4 h in chase medium containing unlabeled methionine at 10 times the usual concentration. At either chase point, the medium was harvested for assay of released virus particles. The cells were washed with ice-cold phosphate-buffered saline and lysed in 100 μl of Triton-TNE lysis buffer (1% Triton X-100, 10 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1 mM EDTA, 100 μg of phenymethylsulfonyl fluoride per ml). The lysates were centrifuged to remove nuclei and immunoprecipitated with human anti-RV serum. After incubation at 4°C for 1 h, 40 μl of a 50% suspension of protein A-Sepharose beads (Pharmacia Biotech) were added and incubated for a further 1 h at room temperature with shaking. The beads were washed three times with lysis buffer, resuspended in sample buffer (0.1 M citrate [pH 5.5], 0.15% sodium dodecyl sulfate [SDS]), and boiled for 5 min. After centrifugation, the immunoprecipitates were collected and mixed with SDS-gel loading buffer (62.5 mM Tris-HCl [pH 6.0], 2% SDS, 5% 2-mercaptoethanol, 500 mM sucrose) and then analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) on 10% gels under reducing conditions.
For assay of released virus particles from transfected cells, the viral particles in the chase medium were precipitated with 40% polyethylene glycol (PEG) containing 2.5M NaCl and resuspended in Triton-TNE buffer. The suspension was immunoprecipitated with human anti-RV serum, and SDS-PAGE analysis of the immunoprecipitates was done as described above.
For endo H digestion, the immunoprecipitates were digested with endo H (1 mU per 10 μl of immunoprecipitate) in the presence of phenylmethylsulfonyl fluoride for 14 h at 37°C. After digestion, the immunoprecipitates were mixed with SDS-gel loading buffer and analyzed on SDS-PAGE.
Electron microscopy.
Transfected BHK cells were treated with 2% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.2), immediately scraped from the plate, and kept at 4°C for 15 min. After pelleting, the cells were postfixed, dehydrated, and infiltrated with Epon. Sections were cut, stained with 2% uranyl acetate and lead citrate, and examined by electron microscopy.
RESULTS
Construction and growth characteristics of E1 cytoplasmic domain mutants.
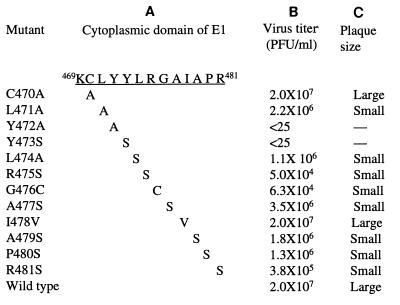
The E1 cytoplasmic domain of RV is predicted to contain 13 amino acids in sequence KCLYYLRGAIAPR (residues 469 to 481). In our previous studies, we found that mutation of the cysteine residue at position 470 to alanine did not affect virus release but mutation of the leucine residue at position 471 to alanine reduced virus production by 90% (27). To further determine the role of each amino acid in the E1 cytoplasmic domain in virus release and to identify the residues within this domain that are essential for the process, we mutated each of the remaining amino acid residues in the E1 cytoplasmic domain as shown in Fig. 1. The nucleotide sequences (nt 9673 to 9700) encoding amino acids from tyrosine 473 to arginine 481 form a prominent stem-loop (SL) structure (4, 8). The loop contains a sequence UAUA that exhibits eukaryotic promoter activity in the negative polarity (3), and the GC-rich stem is implicated to play an important role in viral RNA replication (4). Thus, amino acid substitutions were chosen to maintain this SL structure (checked by computer modeling of RNA folding). The small polar serine residue was chosen to substitute for tyrosine 473, leucine 474, arginine 475, alanine 477, alanine 479, and proline 480. Glycine 476 was mutated to cysteine, of similar polarity to serine, by changing G to U at nucleotide 9680 and isoleucine 478 to valine by changing A to G at nucleotide 9686 (resulting in UAUA to UACA in the negative-polarity strand). The exception was mutant R481S, in which replacement of arginine 481 (CGC) with serine (AGC) led to abolition of this SL structure. To compare the role of tyrosine 472 in virus release with that of leucine 471 (27), tyrosine 472 was mutated to alanine. These mutations were introduced into a full-length infectious cDNA clone, pBRM34, derived from the wild-type RV M33 strain (27).
FIG. 1.
Mutations in E1 cytoplasmic domain. (A) Amino acid sequence of the E1 cytoplasmic domain and the substitutions produced by mutagenesis. Numbering is based on the sequence published by Clarke et al. (5). (B) Vero cells were transfected with RNAs using Lipofectin and overlaid with growth medium. After 5 days of transfection, the medium was removed for virus titration. Mean titers of two independent plaque assays are shown. (C) Plaque sizes of some of the mutants shown in Fig. 2.
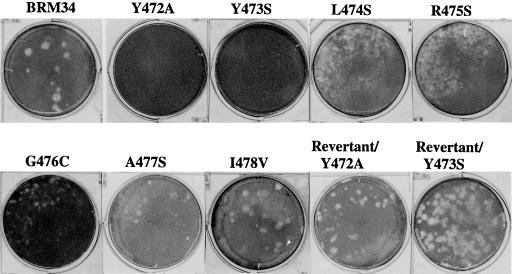
To characterize the E1 cytoplasmic domain mutants, RNAs transcribed in vitro from each mutant and parental pBRM34 clone were transfected into Vero cells by using Lipofectin. After transfection, culture medium was harvested at 5 days posttransfection, and the virus titers in the medium were determined by plaque assay on Vero cells. As shown in Fig. 1, all of the mutants produced lower amounts of viruses than did parental BRM34 except for mutant I478V. Substitutions of alanine for tyrosine 472 and serine for tyrosine 473 dramatically impaired virus production. Virus in the medium from these two mutants was not detectable by plaque assay on Vero cells (Fig. 2). Mutations of leucine 474, alanine 477, alanine 479, and proline 480 resulted in 10- to 20-fold reduction in virus production compared to parental BRM34 virus. Mutation of arginine 481 to serine had a moderate effect on virus production, 50-fold lower than that of parental BRM34 virus. Mutations of arginine 475 to serine and glycine 476 to cysteine reduced virus production by 400-fold compared to the parental virus. The plaques formed by these mutants were smaller than those formed by the parental virus (Fig. 2). Change of isoleucine 478 to valine showed no effect on virus production; the virus titer and plaque size were similar to that of the parental virus (Fig. 1 and 2).
FIG. 2.
Morphology of plaques produced by parental BRM34 virus and mutant viruses. Vero cells were infected with parental BRM34 virus or mutant viruses, and the infected cells were overlaid with agarose medium. After incubation for 6 days at 35°C, the cells were stained with neutral red. Only representative plates are shown.
Substitutions of alanine for tyrosine 472 and serine for tyrosine 473 in the E1 cytoplasmic domain block virus release.
We next examined and compared the effects of these substitutions on viral structural protein synthesis and release as virus particles by pulse-chase analysis of BHK cells transfected with mutant RNAs by electroporation. After 40 h of transfection, the cells were pulse-labeled for 80 min and chased for 10 min or 4 h, after which the cell lysates were analyzed by immunoprecipitation using human anti-RV serum. For analysis of virus release, the virus particles in the corresponding chase medium were precipitated with PEG and then immunoprecipitated with human anti-RV serum. The results are shown in Fig. 3. Immunoprecipitation of the cell lysates after 10 min of chase revealed that E1, E2, and capsid proteins were correctly produced in all of the mutants, indicating that the cleavage of mutant polyprotein precursors proceeded normally. Two forms of E2 were observed: the 39-kDa ER form and the 45-kDa Golgi form. After 4 h of chase, the ER form of E2 in all of the mutants was converted to the Golgi form, indicating that transport of the structural proteins is not affected. The level of protein synthesis, however, differed between the mutants. After quantitation by densitometry of autoradiographs of the radiolabel in intracellular E1 after 10 min of chase, it was found that mutants L474S and I478V synthesized levels of proteins comparable to that of the parental BRM34 virus. Lower levels of protein synthesis were observed in mutants R475S, G476C, A477S, A479S, and P480S, which produced about 47, 53, 73, 43, and 80%, respectively, of E1 produced by the parental BRM34 virus. Protein synthesis in mutants Y472A, Y473S, and R481S was, however, significantly reduced. Observed differences in levels of mutant protein synthesis is not due to different transfection efficiency for each mutant, since a similar proportion of cells (about 80%) was transfected in mutants, as determined by immunofluorescence staining at 40 h posttransfection. A possible explanation for the reduced protein synthesis may be that the nucleotide changes introduced in these mutants significantly affect viral RNA replication or protein translation. It is known that the nucleotide sequences (nt 9673 to 9700) encoding amino acids from tyrosine 473 to arginine 481 form an SL structure. This SL structure has been shown to bind to calreticulin in vitro and may have a functional role in virus replication (4). Indeed, the change of C to A (arginine CGC→ serine AGC) at nucleotide 9695 in mutant R481S completely abolishes the SL structure (data not shown), suggesting that its maintenance may be required for viral replication.
FIG. 3.
Synthesis, transport, and release of structural proteins following transfection of mutant RNAs. BHK cells were transfected with in vitro-transcribed mutant or parental BRM34 RNAs by electroporation. At 40 h postinfection, the infected cells were pulse-labeled for 80 min with [35S]methionine and chased with medium containing unlabeled methionine for 10 min or 4 h. The chase medium was harvested, and the labeled cells were lysed with lysis buffer. The lysates were immunoprecipitated with human anti-RV serum and analyzed by SDS-PAGE on 10% gels under reducing conditions. Virus particles in the chase medium were precipitated with PEG, and pelleted virus particles were resuspended in Triton-TNE buffer. The suspension was immunoprecipitated with human anti-RV serum and analyzed by SDS-PAGE (10% gel) and subsequent autoradiography. Positions of migration of RV structural proteins E1, E2, and C are shown.
Immunoprecipitation of 4-h-chase medium revealed that the amounts of virus released from the infected cells between the mutants were very different. The most significant reduction in released virus was observed in mutants Y472A, Y473S, and R481S, which released barely detectable level of virus (Fig. 3). To compare the amounts of virus release among the mutants, the quantity of extracellular E1 after 4 h of chase was quantitated by densitometry. The relative amount of virus release was calculated after normalizing the mutant/BRM34 ratio of the radiolabel in the extracellular E1 to the mutant/BRM34 ratio in the intracellular E1. It was found that virus release in mutants L474S, R475S, and G476C was only 20 to 40% of the amount of parental virus. Mutants A477S and P480S produced half of the amount of parental virus. In contrast, only a slight reduction in virus release was observed in mutant A479S, and no reduction was observed in mutant I478V. These results indicate that individual substitutions of most of the N-terminal amino acids in the E1 tail significantly affected virus release. The reduction in virus release seen in the mutants was not sufficient to account for the reduction in virus infectivity, particularly in mutants R475S and G476C, which showed a 400-fold reduction in virus titer. Thus, the released mutant viruses may have reduced infectivity. Lack of plaque formation by mutants Y472A and Y473S also indicates that the released mutant viruses are defective in infectivity. In contrast, the infectivity of mutant R481S appeared not to be significantly affected since it showed only a 50-fold reduction in virus titer despite its protein synthesis and virus release being similar to those of mutants Y472A and Y473S.
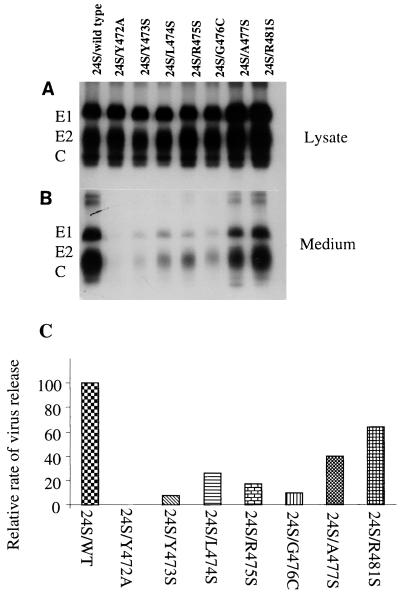
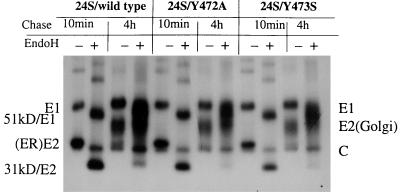
Since protein synthesis was greatly reduced in mutants Y472A, Y473S, and R481S, it is possible that the reduced virus release observed in these mutants could be due to the reduced level of viral protein synthesis. To exclude the effects of mutations on viral RNA replication or protein synthesis, we next examined virus release and protein synthesis in mutants Y472A, Y473S, and R481S, using a system in which RV structural protein synthesis and virus release were not dependent on viral RNA replication. It has been shown that expression of three RV structural proteins in transfected BHK (22) or CHO cells (9, 12) results in assembly and release of virus-like particles in the absence of genomic RNA. The subgenomic cDNA (24S) encoding the three structural proteins of wild-type or mutants was inserted into the pSFV-1 vector (15), a novel expression system based on Semliki Forest virus replicon. Protein synthesis and release as virus-like particles were examined by pulse-chase analysis of the cells transfected with RNAs from 24S/WT or 24S mutants. Levels of protein synthesis observed between 24S mutants and 24S/WT using pSFV-1 vector were similar to those seen in the immunoprecipitates of cell lysates after pulse-labeling of the transfected cells for 30 min and chasing for 10 min (Fig. 4A). However, after 4 h of chase, lower amounts of secreted virus-like particles in the chase medium were clearly observed in mutants 24S/Y472A, 24S/Y473S, 24S/L474S, 24S/R475S, 24S/G476C, and 24S/A477S compared to 24S/WT (Fig. 4B). To compare the release of virus-like particles between the mutants, the amounts of extracellular E1 in the medium and intracellular E1 in the lysate were quantitated by densitometry of the autoradiographs. The relative rate of virus release for each mutant was calculated after normalizing the mutant/wild type ratio of radiolabel in the extracellular E1 to the ratio in the intracellular E1 and is presented in Fig. 4C. Again, the most dramatic reduction in secretion of virus-like particles was found in mutants 24S/Y472A and 24S/Y473S; mutant 24S/Y472A released a barely detectable level of virus-like particles; mutant 24S/Y473S released about 8% of the wild-type level. The released virus-like particles in mutants 24S/L474S, 24S/R475S, and 24S/G476C ranged from 10 to 26% of the wild-type level; 40% of the amount of wild-type particles was secreted by mutant 24S/A477S. However, 64% of the level of wild-type particles was released in mutant 24S/R481S, suggesting that the change of arginine 481 to serine had only a minor effect on virus release, and the reduction in virus titer and the defect in virus release observed in mutant R481S (Fig. 3) were mostly due to altered protein synthesis or viral RNA replication. Taken together, these results clearly show that tyrosine residues at positions 472 and 473 in the E1 cytoplasmic domain are essential for virus release. Interestingly, the mutations introduced in mutants Y472A and Y473S may also significantly affect viral protein synthesis or viral RNA replication.
FIG. 4.
Expression and secretion of virus-like particles in 24S mutants. BHK cells were transfected with in vitro-transcribed RNA derived from pSFV-1 vector containing wild-type or mutant 24S. At 12 h posttransfection, the transfected cells were pulse-labeled for 30 min with [35S]methionine and chased with medium containing unlabeled methionine for 10 min or 4 h. The chase medium was harvested, and the labeled cells were lysed with Triton-TNE buffer. Immunoprecipitation of the lysates and chase medium were carried out as described for Fig. 3 and analyzed by 10% SDS-PAGE (10% gel). (A) Immunoprecipitation of cell lysates after 10 min of chase; (B) immunoprecipitation of virus particles in 4-h-chase medium. Positions of migration of RV structural proteins E1, E2, and C are shown. (C) Relative rate of virus release. The amounts of radiolabel in intracellular E1 in the lysate and in extracellular E1 in the medium after chase were quantitated by densitometry of the autoradiographs. The mutant/wild type ratios of radiolabel in the extracellular and intracellular E1 were calculated. The relative rate of virus release for each mutant is shown after normalization of the ratio of the radiolabel in the extracellular E1 to the ratio in the intracellular E1.
The inhibition in virus release in mutants Y472A and Y473S is not due to either a defect in protein transport or a block in virus budding.
To elucidate the mechanisms underlying the dramatic inhibition in virus release observed in mutants Y472A and Y473S, we next investigated the effects of these mutations on protein transport and viral budding. To examine protein transport to the Golgi complexes, immunoprecipitates of cell lysates were treated with endo H to monitor the maturation of glycoproteins. RV structural proteins undergo extensive glycosylation during transport from the ER to the Golgi, and the acquisition of resistance to endo H digestion by RV structural proteins signifies that they have reached the Golgi compartment (11). As shown in Fig. 5, after 10 min of chase, the ER forms of E2 and E1 in the mutants were sensitive to endo H digestion, resulting in a reduction in molecular mass from 39 to 31 kDa in E2 and from 58 to 51 kDa in E1, respectively. After 4 h of chase, the E2 in the mutants became completely endo H resistant. Part of E1 also became resistant. No change in molecular weight was found in capsid protein after endo H digestion. A similar digestion pattern was observed in the wild-type E2 and E1. Thus, these results indicate that the structural proteins in the mutants were transported normally to the Golgi.
FIG. 5.
Endo H treatment of structural proteins. Immunoprecipitates were isolated as described for Fig. 4, digested with endo H at 37°C for 14 h (+) or not treated with endo H (−), and then analyzed by SDS-PAGE (10% gel) and autoradiography.
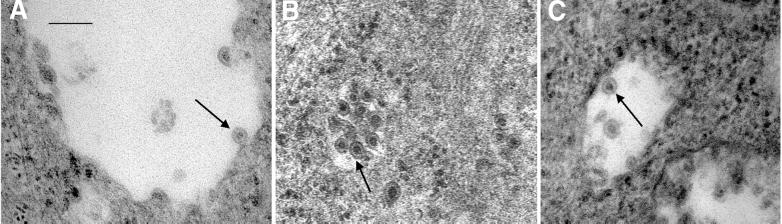
Virus budding was examined by electron microscopy of BHK cells transfected with mutants 24S/Y472A and 24S/Y473S or 24S/WT. As shown in Fig. 6, budding of virus-like particles into the lumen of Golgi complexes was observed in cells transfected with either 24S mutants or 24S/WT. Thus, substitutions of tyrosine residues at positions 472 and 473 did not affect the virus budding process. From these results, we conclude that the block of virus release seen in mutants Y472A and Y473S involves the pathway of virus secretion from the infected cells after virus budding.
FIG. 6.
Electron microscopic analysis of intracellular virus budding. Transfected BHK cells were treated with 2% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.2), immediately scraped from the plate, and kept at 4°C for 15 min. After being pelleted, the cells were postfixed, dehydrated, and infiltrated with Epon. Sections were cut, stained, and observed by electron microscopy. (A) 24S/WT; (B) 24S/Y472A; (C) 24S/Y473S. Images were scanned with a UMAX Astra 1220U scanner with Adobe Photoshop 5.0 software.
Analysis of Y472A and Y473S revertants.
As shown in Fig. 2, mutants Y472A and Y473S were found to be incapable of producing viral plaques. Vero cells infected with mutants Y472A and Y473S also showed no signs of cytopathic effect (CPE). However, a small amount of released virus or virus-like particles could be detected in the culture medium (Fig. 3 and 4). Thus it is possible to passage these mutant viruses in cells to isolate revertants that grow better and form viral plaques. Molecular analysis of these revertants would provide some insights toward the structure-function relationship of the E1 cytoplasmic domain in virus secretion. The culture medium harvested from Vero cells transfected with mutants Y472A and Y473S RNAs after 6 days of incubation was inoculated into a new monolayer of Vero cells and incubated for another 6 days. The emergence of revertants was monitored by appearance of CPE in infected cells. After five passages, Vero cells inoculated with passaged culture medium showed strong CPE. Plaque assays revealed that the viruses in the passage 5 culture medium were similar in plaque size and virus titer to parental BRM34 virus (Fig. 2). To determine the nature of the reversion, viral RNA was extracted at passage 5 and used for cDNA synthesis and subsequent PCR amplification. The sequences encoding the E1 cytoplasmic domain were examined. It was found that the tyrosine-to-serine substitution introduced in mutant Y473S had reverted to tyrosine in the passage 5 virus although the substitution was based on two nucleotides changes (Table 1), indicating that this tyrosine residue at position 473 is essential for the life cycle of RV. Sequencing analysis of the E1 cytoplasmic domain in the passage 5 virus from mutant Y472A showed that the tyrosine-to-alanine substitution is conserved in the revertants. No other mutations were found in the E1 cytoplasmic domain (Table 1), indicating that second-site suppressor mutations arose in other regions of the viral genome. Preliminary characterization of Y472A revertants revealed that the second-site suppressor mutations in the E1 gene were not sufficient to reverse the defective phenotype imposed by the tyrosine change (data not shown), suggesting some additional suppressor mutation(s) in C or E2 genes.
TABLE 1.
Y472A and Y473S revertantsa
| Construct | Plaqueb | Revertant plaqueb | Reversionc |
|---|---|---|---|
| WT(Y472Y473) | Large | ||
| Y473S TAC → AGC | — | Large | AGC→ TAC |
| (Tyr → Ser) | (Ser → Tyr) (3) | ||
| Y472A TAC → GCC | — | Large/ | GCC—GCC |
| (Tyr → Ala) | Small | (Ala—Ala) (4) |
Revertants were isolated by passaging of the culture medium (1/4 volume) from transfected Vero cells with mutant RNAs to a new monolayer of Vero cells. After incubation for 6 days at 37°C, the culture medium was harvested and passaged in the same manner. After five passages, plaque assay of the culture medium was performed to determine the formation of plaques and virus titers. Viral RNA was extracted from the viruses in passage 5 and was used for cDNA synthesis and PCR amplification and cloning.
Plaque assay was performed and the plaques are shown in Fig. 2.
Only the E1 cytoplasmic domain coding region was sequenced; numbers indicate numbers of sequenced DNA clones.
DISCUSSION
We have combined genetic, biochemical, and microscopic approaches to investigate the role of each amino acid residue in the E1 cytoplasmic domain in virus assembly and release in the context of infectious viruses and virus-like particles. Substitutions of most of the N-terminal amino acids in the E1 cytoplasmic domain affected virus growth, virus release, and plaque phenotype, indicating the functional importance of the E1 cytoplasmic domain in RV replication. In particular, a single-amino-acid substitution of either of the two tyrosine residues in this domain resulted in a block in virus release. Infectious RNA transcripts bearing these mutations were incapable of forming plaques. Furthermore, analysis of revertants from mutant Y473S showed that the serine substitution in the mutant quickly reverted to the original tyrosine residue. Altogether, these results indicate that the E1 cytoplasmic domain modulates virus release in a sequence-dependent manner and these two tyrosine residues are critical for this function.
It is not clear at present how a single substitution of tyrosine residue in the E1 cytoplasmic domain blocks virus release. The block is probably not at the steps of intracellular protein transport and viral budding at the Golgi complexes, since the budding of tyrosine mutants at the Golgi complexes proceeded normally, consistent with the results from E1 cytoplasmic domain deletion mutant reported by Garbutt et al. (9). It is also unlikely that the rate of budding of the mutants is much reduced, thereby leading to less virus in the culture fluid, since the viral budding of mutants at the Golgi complexes was readily seen and the rate was similar to that for the wild type. We postulate that the block in virus release may be due to a defect in transport of the budded viral particles from the Golgi complexes to the plasma membrane for release. In this case, it is still unclear how a single alteration of a tyrosine residue in the E1 cytoplasmic domain could modulate viral particle transport since the tyrosine residue is located within the viral particles. The simplest explanation is that substitutions of the tyrosine residues in the E1 tail dramatically affect the overall conformation of the ectodomains of E2-E1 heterodimer such that the mutant viruses have altered structure and are incompetent for incorporation into transport vesicles for release. Although this explanation is straightforward, we think it is unlikely since all mutations near tyrosine 472 and 473 (mutations of leucine 471 to alanine [24], leucine 474 to serine, arginine 475 to serine and glycine 476 to cysteine) would not be expected to give such clear negative effects on virus release. Instead, we speculate that these residues YYLRG may constitute a domain in the E1 tail that may interact with other proteins and that the two tyrosines are the essential residues. Another explanation is that RV, after budding at the Golgi complexes, undergoes a maturation process accompanied by structural changes; i.e., structural maturation is required for viral particle transport and release. This maturation step cannot proceed in the mutant viruses because of alterations to the tyrosine residues, leading to arrest in virus secretion. As the tyrosine residues are located inside viral particles, we propose that the structural changes during virus maturation are brought by a process which involves interaction of the E1 tail with other viral structural proteins, possibly nucleocapsids. We favor this explanation, since the single substitution of tyrosine 472 or tyrosine 473 blocked virus release and the serine substitution in mutant Y473S quickly reverted to the original tyrosine in its revertants. The emergence of Y472A revertants containing second-site mutations which reverse the defect imposed by the tyrosine change also provides support for the notion that the intermolecular interactions between E1 tail with other viral structural proteins occur during the process of virus release. In related alphaviruses, a similar tyrosine residue in the cytoplasmic domain of E2 glycoprotein has been identified as being essential for the interaction between glycoproteins and a hydrophobic pocket of the nucleocapsids that is required for virus budding and maturation (14, 24). Lee et al. (14) further suggested that upon binding of the tyrosine to the capsid packet, the nucleocapsid undergoes a conformational change such that the core is matured and readied for disassembly. Without the optimal nucleocapsid-E2 tail interaction as evidenced in nucleocapsid mutants, the core does not undergo conformational changes required for subsequent disassembly.
Although evidence supporting structural maturation required for RV release remains to be established, it is conceivable that the conformational structure of extracellular viruses is different from that of intracellularly budded viruses. Structural maturation during virus release has recently been reported for transmissible gastroenteritis coronavirus (TGEV), which has two types of virus-related particles in infected cells: large annular viral particles and small dense viral particles (23). The large annular viral particles are the immature precursors of small dense viral particles. The latter are the infectious TGEV virions in the culture supernatants. Monensin treatment of TGEV-infected cells caused an accumulation of large annular particles in perinuclear elements of the ER-Golgi intermediate compartment. Removal of monensin led to the release of small dense viral particles into secretory vesicles and culture supernatants.
It is also possible that the tyrosine residues at positions 472 and 473 may be required to recruit targeting signals to a specific vesicle containing budded viruses by interacting with cellular proteins. There is a body of evidence showing that many transmembrane proteins harbor a tyrosine-based signal in their cytoplasmic domains for transport. Examples are the membrane envelope glycoproteins of the retroviruses, vesicular stomatitis virus, and varicella-zoster virus (1, 16, 25). Sorting of the membrane glycoprotein of retrovirus is reported to require interaction of the tyrosine-based motif in the cytoplasmic domain with the clathrin-associated adapter complexes (19). Although substitutions of tyrosine residues in the E1 cytoplasmic domain did not affect RV structural protein transport, it is still possible that RV has evolved to use a tyrosine based signal in its E1 tail for sorting and release of assembled virus. These possible mechanisms are currently under investigation.
In this study, we observed that nucleotide changes introduced into mutants Y472A, Y473S, and R481S greatly reduced the level of viral structural protein synthesis. RNA secondary structure analysis of the 3′-terminal 305 nt of RV genomic RNA reveals four SL structures; SL1 is located in the E1 transmembrane and ectodomain coding regions, SL2 is located in the exact E1 tail coding region, while SL3 and SL4 are within the 59-nt 3′-terminal nontranslated region preceding the poly(A) tract (4). The change of C (arginine CGC) to A (serine AGC) in mutant R481S completely abolishes the SL2 structure, whereas the nucleotide changes introduced in mutants L474S, R475S, G476C, A477S, I478V, A479S, and P480S do not and the overall SL2 structure is maintained in these mutants. However, some minor alterations in the SL2 are shown in mutants R475S, G476C, A477S, A479S, and P480S by RNA folding. Chen and Frey (4) analyzed this structure in the context of a full-length infectious cDNA clone by mutagenesis and found that the overall maintenance of the SL structure is important in viral replication, presumably by binding to calreticulin in vivo and regulating negative-sense RNA replication. Thus, the reduced protein synthesis in mutant R481S could be explained by a similar mechanism. The lower levels of protein synthesis observed in mutants R475S, G476C, and A479S indicate that some alterations in the SL2 structure due to the nucleotide changes in these mutants probably affect the calreticulin binding activity of SL2. However, the changes of UA to GC (tyrosine UAC→ alanine GCC) in mutant Y472A and UA to AG (tyrosine UAC→ serine AGC) in mutant Y473S do not affect overall SL2 structure as examined by RNA folding. Thus, it is not clear how these nucleotide changes reduced viral protein synthesis. It will be of interest in the future to elucidate the mechanisms underlying the effects on protein synthesis by the nucleotide changes introduced in mutants Y472A and Y473S.
ACKNOWLEDGMENTS
This work was supported by a grant from the Medical Research Council of Canada. Shirley Gillam is an investigator of the British Columbia's Children's Hospital Foundation.
REFERENCES
- 1.Alconda A, Bauer U, Hoffack B. A tyrosine-based motif and a casein kinase II phosphorylation site regulate the intracellular trafficking of the varicella-zoster virus glycoprotein I, a protein localized in the trans-Golgi network. EMBO J. 1996;15:6096–6110. [PMC free article] [PubMed] [Google Scholar]
- 2.Baron M, Forsell K. Oligomerization of the structural proteins of rubella virus. Virology. 1991;185:811–819. doi: 10.1016/0042-6822(91)90552-m. [DOI] [PubMed] [Google Scholar]
- 3.Cao X Q, Liu T Y, Nakhasi H L. The cis-acting 3′-element of rubella virus RNA has DNA promoter activity. Gene. 1992;114:251–256. doi: 10.1016/0378-1119(92)90583-b. [DOI] [PubMed] [Google Scholar]
- 4.Chen M H, Frey T K. Mutagenic analysis of the 3′ cis-acting elements of the rubella virus genome. J Virol. 1999;73:3386–3403. doi: 10.1128/jvi.73.4.3386-3403.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clarke D M, Loo T W, Hui I, Chong P, Gillam S. Nucleotide sequence and in vitro expression of rubella virus 24S subgenomic mRNA encoding the structural proteins E1, E2 and C. Nucleic Acids Res. 1987;15:3041–3057. doi: 10.1093/nar/15.7.3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clarke D M, Loo T W, McDonald H, Gillam S. Expression of rubella virus cDNA coding for the structural proteins. Gene. 1988;65:23–30. doi: 10.1016/0378-1119(88)90413-1. [DOI] [PubMed] [Google Scholar]
- 7.Dominguez G, Wang C, Frey T K. Sequence of the genome RNA of rubella virus: evidence for genetic rearrangement during Togavirus evolution. Virology. 1990;177:225–238. doi: 10.1016/0042-6822(90)90476-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frey T K. Molecular biology of rubella virus. Adv Virus Res. 1994;44:69–160. doi: 10.1016/S0065-3527(08)60328-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garbutt M, Law L M, Chan H, Hobman T C. Role of rubella virus glycoprotein domains in assembly of virus-like particles. J Virol. 1999;73:3524–3533. doi: 10.1128/jvi.73.5.3524-3533.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hobman T C, Gillam S. In vitro and in vivo expression of rubella virus glycoprotein E2: the signal peptide is contained in the C-terminal region of capsid protein. Virology. 1989;173:241–250. doi: 10.1016/0042-6822(89)90240-7. [DOI] [PubMed] [Google Scholar]
- 11.Hobman T C, Lundstrom M L, Gillam S. Processing and intracellular transport of rubella virus structural proteins in COS cells. Virology. 1990;178:122–133. doi: 10.1016/0042-6822(90)90385-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hobman T C, Lundstrom M L, Mauracher C A, Woodward L, Gillam S, Farquhar M G. Assembly of rubella virus structural proteins into virus-like particles in transfected cells. Virology. 1994;202:574–585. doi: 10.1006/viro.1994.1379. [DOI] [PubMed] [Google Scholar]
- 13.Hobman T C, Shukin R, Gillam S. Translocation of rubella virus glycoprotein E1 into the endoplasmic reticulum. J Virol. 1988;62:4259–4264. doi: 10.1128/jvi.62.11.4259-4264.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee S, Owen K E, Choi H-K, Lee H, Lu G, Wangle G, Brown D T, Rossmann M G, Kuhn R J. Identification of a protein binding site on the surface of the alphavirus nucleocapsid and its implication in virus assembly. Structure. 1996;4:531–541. doi: 10.1016/s0969-2126(96)00059-7. [DOI] [PubMed] [Google Scholar]
- 15.Lilijestrom P, Garoff H. A new generation of animal cell expression vectors based on the Semliki Forest virus replicon. Bio/Technology. 1991;9:1356–1361. doi: 10.1038/nbt1291-1356. [DOI] [PubMed] [Google Scholar]
- 16.Lodge R, Lalonde J P, Lemay G, Cohen E. The membrane-proximal intracytoplasmic tyrosine residue of HIV-1 envelope glycoprotein is critical for basolateral targeting of viral budding in MDCK cell. EMBO J. 1997;16:695–705. doi: 10.1093/emboj/16.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matthews R E F. Classification and nomenclature of viruses. Intervirology. 1982;17:1–99. doi: 10.1159/000149278. [DOI] [PubMed] [Google Scholar]
- 18.McDonald H, Hobman T C, Gillam S. The influence of capsid protein cleavage on the processing of E2 and E1 glycoproteins of rubella virus. Virology. 1991;183:52–56. doi: 10.1016/0042-6822(91)90117-t. [DOI] [PubMed] [Google Scholar]
- 19.Ohno H, Aguilar R C, Fournier M C, Henneck S, Cosson P, Bonifacino J S. Interaction of envelope of endocytic signals from the HIV-1 envelope glycoprotein complex with members of adaptor medium chain family. Virology. 1997;238:305–315. doi: 10.1006/viro.1997.8839. [DOI] [PubMed] [Google Scholar]
- 20.Oker-Blom C. The gene order for rubella virus structural proteins is NH2-C-E2-E1-COOH. J Virol. 1984;51:964–973. doi: 10.1128/jvi.51.2.354-358.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oker-Blom C, Ulmanen I, Kääriäinen L, Pettersson R F. Rubella virus 40S genome RNA specifies a 24S subgenomic mRNA that codes for a precursor to structural proteins. J Virol. 1984;49:403–408. doi: 10.1128/jvi.49.2.403-408.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qiu Z, Ou D, Wu H, Hobman T C, Gillam S. Expression and characterization of virus-like particles containing rubella virus structural proteins. J Virol. 1994;68:4086–4091. doi: 10.1128/jvi.68.6.4086-4091.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salanueva I J, Carrascosa J L, Risco C. Structure maturation of the transmissible gastroenteritis coronavirus. J Virol. 1999;73:7952–7964. doi: 10.1128/jvi.73.10.7952-7964.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Skoging U, Vihinen M, Nilsson L, Liljestrom P. Aromatic interactions define the binding of the alphavirus spike to its nucleocapsid. Structure. 1996;4:519–529. doi: 10.1016/s0969-2126(96)00058-5. [DOI] [PubMed] [Google Scholar]
- 25.Thomas D C, Brewer C B, Roth M G. Vesicular stomatitis virus glycoprotein contains a dominant cytoplasmic basolateral sorting signal critically dependent upon a tyrosine. J Biol Chem. 1993;288:3313–3320. [PubMed] [Google Scholar]
- 26.Waxham M N, Wolinsky J S. A model of the structural organization of rubella virions. Rev Infect Dis. 1985;7(Suppl. 1):S133–S139. doi: 10.1093/clinids/7.supplement_1.s133. [DOI] [PubMed] [Google Scholar]
- 27.Yao J, Gillam S. Mutational analysis, using a full-length rubella virus cDNA clone, of rubella virus E1 transmembrane and cytoplasmic domains required for virus release. J Virol. 1999;73:4622–4630. doi: 10.1128/jvi.73.6.4622-4630.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yao J, Strauss E G, Strauss J H. Interaction between PE2, E1, and 6K required for assembly of alphavirus studied with chimeric viruses. J Virol. 1996;70:7910–7920. doi: 10.1128/jvi.70.11.7910-7920.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]