Abstract
Tamoxifen has been the mainstay therapy to treat early, locally advanced, and metastatic estrogen receptor-positive (ER + ) breast cancer, constituting around 75% of all cases. However, the emergence of resistance is common, necessitating the identification of novel therapeutic targets. Here, we demonstrated that long-noncoding RNA LINC00152 confers tamoxifen resistance by blocking tamoxifen-induced ferroptosis, an iron-mediated cell death. Mechanistically, inhibiting LINC00152 reduces the mRNA stability of phosphodiesterase 4D (PDE4D), leading to activation of the cAMP/PKA/CREB axis and increased expression of the TRPC1 Ca2+ channel. This causes cytosolic Ca2+ overload and generation of reactive oxygen species (ROS) that is, on the one hand, accompanied by downregulation of FTH1, a member of the iron sequestration unit, thus increasing intracellular Fe2+ levels; and on the other hand, inhibition of the peroxidase activity upon reduced GPX4 and xCT levels, in part by cAMP/CREB. These ultimately restore tamoxifen-dependent lipid peroxidation and ferroptotic cell death which are reversed upon chelating Ca2+ or overexpressing GPX4 or xCT. Overexpressing PDE4D reverses LINC00152 inhibition-mediated tamoxifen sensitization by de-activating the cAMP/Ca2+/ferroptosis axis. Importantly, high LINC00152 expression is significantly correlated with high PDE4D/low ferroptosis and worse survival in multiple cohorts of tamoxifen- or tamoxifen-containing endocrine therapy-treated ER+ breast cancer patients. Overall, we identified LINC00152 inhibition as a novel mechanism of tamoxifen sensitization via restoring tamoxifen-dependent ferroptosis upon destabilizing PDE4D, increasing cAMP and Ca2+ levels, thus leading to ROS generation and lipid peroxidation. Our findings reveal LINC00152 and its effectors as actionable therapeutic targets to improve clinical outcome in refractory ER+ breast cancer.
Subject terms: Breast cancer, Cancer
Introduction
Estrogen receptor-positive (ER + ) breast cancer accounts for almost 75% of all breast cancers. Endocrine therapies, modulating ER level and/or activity are the standard of care in ER+ breast cancer. The selective ER modulator (SERM) tamoxifen has been the backbone of endocrine therapy for over 40 years, especially for premenopausal women [1, 2]. Tamoxifen is recommended to treat early, locally advanced and metastatic ER+ breast cancers [3] and significantly improves overall survival [3, 4]. Despite its initial clinical success, patients inevitably develop resistance and experience poor clinical outcomes [5]. The molecular mechanisms of tamoxifen resistance are partially understood and involve ER downregulation, ESR1 mutations, modulation of cell cycle regulators, activation of receptor tyrosine kinases and downstream effectors, and modulation of non-coding RNAs [6–10]. The clinical management of tamoxifen resistance involves the use of other endocrine therapies or CDK4/6 inhibitors [11]. However, resistance to second-line therapies is also inevitable [12]. Therefore, there is still an unmet clinical need for the identification of key processes and novel targets to overcome tamoxifen resistance and improve patient outcome.
Ferroptosis is a relatively newly discovered iron-dependent oxidative form of cell death that is characterized by lipid peroxidation, cell volume shrinkage and increased mitochondrial membrane density [13–15]. It is under tight control by the interplay between pro-oxidant and antioxidant systems [16]. Major indicators of ferroptosis are the increase in lipid peroxidation and decreased expression or activity of the glutathione peroxidase 4 (GPX4), a major component of the antioxidant system [17]. Certain anti-cancer therapies have been shown to activate ferroptosis, such as sorafenib [18], lapatinib [19], and radiation therapy [20], and vulnerability to ferroptotic cell death has been demonstrated to be a feature of certain therapy-resistant cancer cells [21]. Along these lines, ferroptosis induction is emerging as an effective strategy to eradicate therapy-resistant cancer cells. However, the potential roles of ferroptosis in tamoxifen-mediated cell death or activating ferroptosis in overcoming resistance to endocrine therapies used in ER+ breast cancer, including tamoxifen, are mostly unknown.
Long non-coding RNAs (lncRNAs) are non-coding RNAs with >200 nucleotides in length that carry out diverse functions, including transcriptional regulation, regulation of proteins or RNA molecules by direct binding and stabilization, and sponging miRNAs [22]. LncRNAs may also drive resistance to anti-cancer therapies, including endocrine therapy via protecting ER against downregulation [23], enhancing ER signaling [24] or activating downstream PI3K/AKT or mTOR signaling [25]. LINC00152 is an oncogenic lncRNA that promotes survival, proliferation, epithelial-mesenchymal transition (EMT) and invasiveness in cancer cells [26–28]. It has been associated with tumor aggressiveness [28, 29] and demonstrated to be a biomarker in various cancers, such as breast cancer, colorectal cancer, and glioma [30–32]. Despite being a driver in several key oncogenic processes, the potential functions of LINC00152 in regulating distinct forms of cell death, such as ferroptosis to drive resistance to endocrine therapy, including tamoxifen in ER+ breast cancer remain to be elucidated.
Here we showed that LINC00152 is a driver of tamoxifen resistance, and its inhibition causes tamoxifen sensitization via reducing the mRNA stability of its novel interactor PDE4D, a cAMP-degrading phosphodiesterase. This leads to the activation of cAMP/PKA/CREB axis, which increases the expression of the TRPC1 Ca2+ channel, causing cytosolic Ca+2 overload, generation of reactive oxygen species (ROS) and increase in lipid peroxidation, leading to restoration of tamoxifen-induced ferroptosis via deactivation of the xCT/GPX4 axis. Notably, overexpressing PDE4D rescues LINC00152-mediated tamoxifen resistance by blocking tamoxifen-induced ferroptosis.
Results
LINC00152 is upregulated in tamoxifen resistance, and high LINC00152 expression correlates with worse prognosis
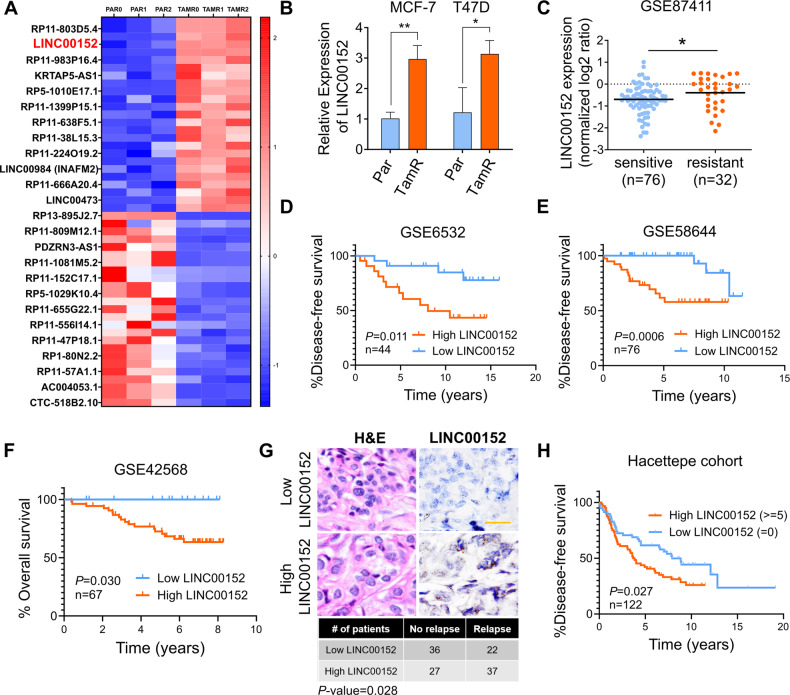
To identify differentially expressed lncRNAs involved in tamoxifen resistance, we performed RNA-sequencing (RNA-seq) (Supplementary Fig. S1A, B) using our previously generated acquired tamoxifen-resistant (TamR) MCF-7 cells along with the parental (Par) counterparts, which were cultured alongside resistant cells without drug treatment, and therefore remained sensitive to tamoxifen [33]. Comprehensive GSEA analysis showed enrichment of tamoxifen resistance and loss of estrogen response-related genes within TamR cells (Supplementary Data 1, Supplementary Fig. S1C), validating resistance. We identified 330 differentially expressed lncRNAs (q-value = 0.05) between resistant and parental isogenic cells (Supplementary Data 2). LINC00152 was among the most significantly upregulated (FDR < 0.001, log2FC = 1.39), highly abundant (average FPKM ~ 25, top 5th highest among all lncRNAs), validated and oncogenic [34] lncRNAs in tamoxifen-resistant cells compared to parental counterparts (Fig. 1A, Supplementary Fig. S1B, Supplementary Data 2). We further validated its upregulation via qRT-PCR analysis in MCF-7.TamR and T47D.TamR cells, a second tamoxifen-resistant ER+ breast cancer cell line that we developed [33] compared to parental cells (Fig. 1B).
Fig. 1. LINC00152 is upregulated in tamoxifen resistance and its higher expression correlates with poor prognosis.
A Heatmap of the z-scores of the FPKM values of the top 25 most abundant, upregulated, and top 25, least abundant, downregulated lncRNAs in MCF-7 tamoxifen-resistant (TAMR) cells compared to parental (Par) cells in three biological replicates (0, 1, 2). The names of 25 genes are depicted on the heatmap. Red indicates upregulation and blue indicates downregulation in TamR cells. B qRT-PCR analysis of LINC00152 in both MCF-7 and T47D.Par and TamR cells. C LINC00152 levels in tamoxifen sensitive vs. resistant tumors at baseline from GSE87411. D, E The disease-free survival in tamoxifen-treated ER+ breast cancer patients with tumors expressing low vs. high LINC00152 expression from GSE6532 (D) and GSE58644 (E). F The overall survival in patients with tumors expressing low vs. high LINC00152 expression from GSE42568. G Representative in situ hybridization (ISH) staining of LINC00152 in low vs. high expressers among endocrine therapy-treated ER+ breast carcinoma specimens from the Hacettepe cohort. Scale bar = 20 µm. The numbers of Hacettepe patients having low vs high LINC00152 and with recurrence vs. no recurrence are provided at the bottom of the panel. H The disease-free survival in Hacettepe patients with tumors expressing low vs. high LINC00152 expression as quantified by ISH staining. Data are presented as mean values ± standard deviation (SD). P-values for the bar graphs and boxplots were calculated with the unpaired, two-tailed Student’s t-test. The significance for the Kaplan–Meier survival graphs was calculated with the Log-rank test. The chi-square test was used for (G). *P < 0.05, **P < 0.01.
To test the clinical relevance of LINC00152 upregulation in tamoxifen resistance, we first analyzed available gene expression profiling data of tamoxifen-treated ER+ breast cancer patients. We demonstrated a significantly higher expression of LINC00152 in patients with no response to tamoxifen therapy compared to patients exhibiting tamoxifen response in a dataset with available response information, GSE87411 (Fig. 1C). A list of genes previously reported to be upregulated in tamoxifen or endocrine resistance were enriched in ER+ breast cancer patients expressing higher LINC00152 (Supplementary Fig. S1D, E). Furthermore, we demonstrated that higher LINC00152 expression is significantly associated with worse disease-free and overall survival (Fig. 1D–F). We then performed in-situ hybridization (ISH) of LINC00152 in our own cohort of endocrine therapy-treated ER+ breast cancer patients and demonstrated that higher LINC00152 levels are associated with significantly worse disease-free survival (Fig. 1G, H). The LINC00152 ISH score was also higher in tumors with high Ki67 score, which is a marker of cell proliferation [35] and high grade (Supplementary Fig. S1F, G). Moreover, we showed that LINC00152 levels are higher in tumors compared to normal tissues (Supplementary Fig. S2A, B) and that its higher expression is associated with worse overall survival (Supplementary Fig. S2C, D) also in patients with other tumors, e.g., hepatocellular carcinoma and lung adenocarcinoma. These data suggest that in addition to its roles in driving tamoxifen resistance, LINC00152 may also play key roles in tumorigenesis in general. Overall, we identified LINC00152 as a clinically relevant lncRNA upregulated in tamoxifen resistance.
Inhibiting LINC00152 overcomes tamoxifen resistance by inducing tamoxifen-dependent ferroptosis
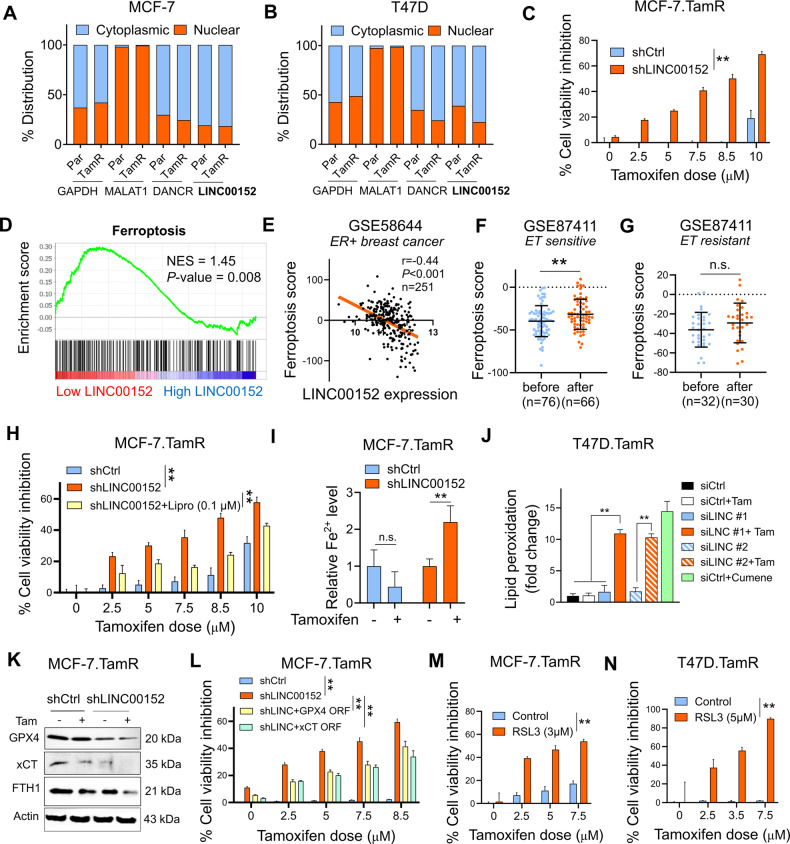
To determine the roles of LINC00152 in tamoxifen resistance, we first determined its cellular localization. We showed that LINC00152 is primarily a cytoplasmic lncRNA in both Par and TamR MCF-7 and T47D cells (Fig. 2A, B). Here, lncRNAs, DANCR [36] and MALAT1 [37] are shown as positive controls as they are known to be cytoplasmic and nuclear, respectively. Then, we showed that stable LINC00152 knockdown (MCF-7.TamR.shLINC00152) (Supplementary Fig. S3A) led to a significant increase in cell viability inhibition in combination with tamoxifen (Fig. 2C). These results were also recapitulated by using two different siRNAs against LINC00152 (Supplementary Fig. S3B, C). On the contrary, overexpressing LINC00152 reduced the viability inhibition induced by tamoxifen (Supplementary Fig. S3D–G).
Fig. 2. Inhibiting LINC00152 overcomes tamoxifen resistance by inducing tamoxifen-dependent ferroptosis.
A, B The nuclear vs. cytoplasmic localization of LINC00152 in MCF-7 (A) and T47D (B) Par and TamR cells. MALAT1 and DANCR were used as controls for nuclear and cytoplasmic RNAs, respectively. C Percent cell viability inhibition in MCF-7.TamR cells expressing shLINC00152 upon treatment with tamoxifen. D GSEA analysis showing the ferroptosis-related genes enriched in low LINC00152-expressing ER+ breast cancer patients treated with endocrine therapy from GSE87411. E Pearson correlation analysis of LINC00152 expression with ferroptosis score in ER+ patients from GSE58644. F, G The levels of ferroptosis score before and after endocrine therapy in sensitive (F) and resistant (G) ER+ patients from GSE87411. H Percentage cell viability inhibition in shLINC00152-expressing MCF-7.TamR cells treated with tamoxifen with or without 0.1 µM of liproxstatin-1 (Lipro). I Relative intracellular Fe2+ levels in shCtrl or shLINC00152-expressing MCF-7.TamR cells that were treated with 7.5 µM tamoxifen for 1.5 h. J Lipid peroxidation assay in T47D.TamR cells transfected with siLINC00152 (siLINC) in combination with tamoxifen, showing fold change of lipid peroxidation. Cumene was used as a positive control. K Western blot analysis of the negative regulators of ferroptosis, xCT, GPX4, and FTH1 in MCF-7.TamR cells expressing shLINC00152 and treated with tamoxifen. Actin is used as a loading control in all Western blots unless stated otherwise. L Percentage cell viability inhibition in shLINC00152-expressing MCF-7.TamR cells transfected with GPX4 or xCT open reading frame (ORF). M, N Percentage cell viability inhibition in MCF-7.TamR (M) and T47D.TamR (N) cells treated with tamoxifen alone or in combination with 3 or 5 µM RSL3. Data are presented as mean values ± standard deviation (SD). P-values for (D) were calculated using the GSEA software (Broad Institute) while others were calculated with the paired (cell viability) or unpaired (others), two-tailed Student’s t-test. *P < 0.05, **P < 0.01, n.s. not significant.
Next, to identify the molecular mechanisms of LINC00152-mediated tamoxifen resistance, we performed RNA-seq upon siRNA-mediated knockdown of LINC00152 in the MCF-7.TamR cells. We generated a LINC00152 knockdown (KD) score comprising of genes down or upregulated upon LINC00152 knockdown (0.67 > FC > 1.5, P < 0.1). Accordingly, patients who express high LINC00152 KD score are expected to express low levels of LINC00152. Tamoxifen sensitivity-related genes were enriched among patients who express high LINC00152 KD score, i.e., showing low LINC00152 expression, supporting the role of LINC00152 in tamoxifen resistance (Supplementary Fig. S4A). Then, we performed pathway enrichment analysis and observed a significant enrichment of genes involved in ferroptosis among LINC00152-modulated mRNAs (Supplementary Fig. S4B). Gene set enrichment analysis (GSEA) among endocrine therapy-treated ER+ breast cancer patients separated based on LINC00152 expression revealed significant enrichment of a validated ferroptosis gene signature, generated using FerrDb V2 database [38] in patients with low LINC00152 expression (Fig. 2D). Furthermore, several ferroptosis-related pathways e.g., glutathione metabolism, sphingolipid pathway and selenoamino acid metabolism were enriched (Supplementary Fig. S4C). We observed a significant negative correlation between LINC00152 expression and ferroptosis signature score among ER+ breast cancer patients in an independent dataset (Fig. 2E). Furthermore, there was a significant increase in the ferroptosis score in sensitive tumors, but not in resistant ones, after treatment with endocrine therapy in the GSE87411 dataset in which samples were collected before and after endocrine therapy (Fig. 2F, G), suggesting that ferroptosis induction is associated with clinical endocrine therapy sensitivity.
To experimentally test the role of ferroptosis induction in LINC00152 inhibition-mediated tamoxifen sensitization, we inhibited ferroptosis using the ferroptosis inhibitor liproxstatin-1 [39] and showed partial rescue of cell viability (Fig. 2H). Furthermore, knockdown of LINC00152 in combination with tamoxifen significantly increased intracellular Fe2+ levels (Fig. 2I) that were followed by lipid peroxidation (Fig. 2J), and deactivation of xCT, GPX4, and FTH1 (Fig. 2K), major antioxidant proteins [17, 40] or iron chelators [41, 42]. Importantly, overexpressing xCT or GPX4 reduced LINC00152 inhibition-mediated tamoxifen sensitization (Fig. 2L, Supplementary Fig. S5A, B). Supporting these results, knocking down xCT or GPX4 resulted in the reversal of LINC00152-mediated tamoxifen resistance that was accompanied by reduction of LINC00152-induced xCT/GPX4 levels (Supplementary Fig. S5C–F). Likewise, inhibiting GPX4 pharmacologically using RSL3 phenocopied the effect of LINC00152 knockdown on tamoxifen sensitization (Fig. 2M, N). To test if ferroptosis is a mechanism of tamoxifen-induced cell death in parental cells, we inhibited ferroptosis using liproxstatin-1 which partially rescued cell viability in parental cells under tamoxifen treatment in a dose-dependent manner (Supplementary Fig. S6A, B). Tamoxifen-induced cell viability inhibition was also reversed by vitamin E, a lipid soluble antioxidant with anti-ferroptosis activity [43] (Supplementary Fig. S6C). Supporting these results, tamoxifen reduced the expression of xCT and FTH1, and increased PTGS2, a marker of ferroptosis in parental cells (Supplementary Fig. S6D). Of note, while inhibitors of necrosis (Necrox-5) or necroptosis (GSK-872) did not alter tamoxifen sensitivity, inhibiting apoptosis, using Q-Vd-OPh partially rescued cell viability under treatment with tamoxifen (Supplementary Fig. S7A-C). Markers of apoptosis such as p21, c. PARP and c. caspase 7 were also induced by tamoxifen in parental cells (Supplementary Fig. S7D). This is in line with previous studies showing apoptotic cell death as one of the mechanisms of action of tamoxifen [44]. Overall, our data suggest that LINC00152 confers tamoxifen resistance in part via counteracting tamoxifen-induced ferroptosis, thus rendering TamR cells dependent on LINC00152, whose inhibition restores tamoxifen-induced ferroptosis and inhibition of cell viability.
LINC00152 inhibition increases TRPC1 expression and cytosolic Ca2+ downstream of cAMP signaling
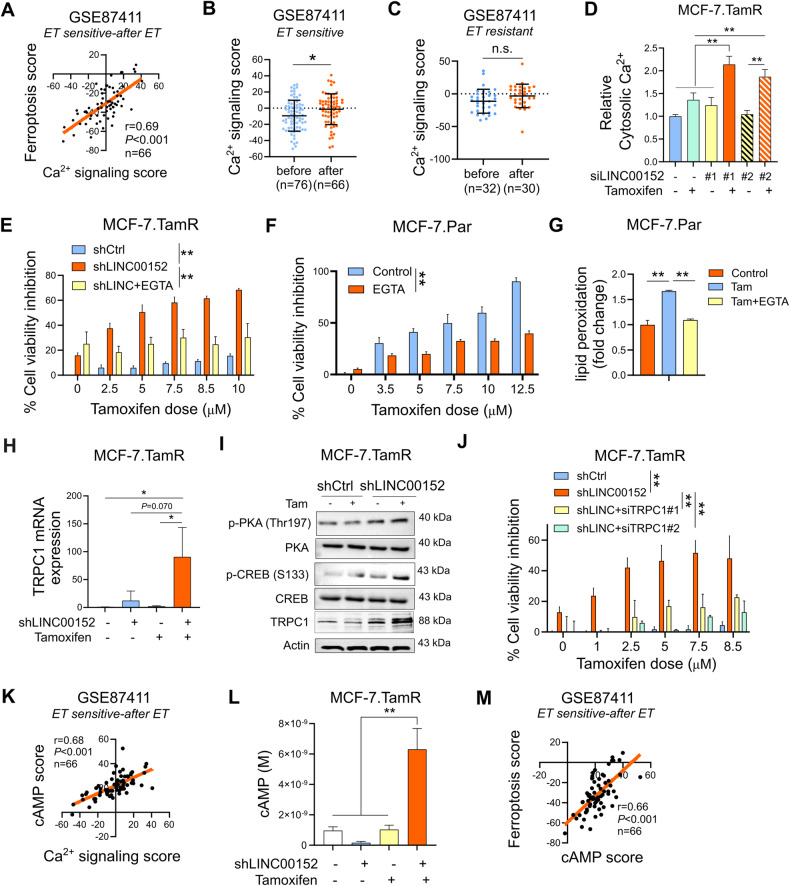
Increase in cytosolic Ca2+ has been shown to be a major regulator of ferroptosis [45]. In line with this, we found a significant positive correlation between the scores of validated Ca2+ signaling [46] and ferroptosis [38] signatures in post endocrine therapy-treated sensitive ER+ breast cancer patients (Fig. 3A). Ca2+ signaling score was also significantly upregulated after endocrine therapy in sensitive but not in resistant patients (Fig. 3B, C), like the ferroptosis score in the same patient population (Fig. 2F, G). Supporting these results, we observed a significant increase in Ca2+ levels in siLINC00152-transfected MCF-7.TamR cells treated with tamoxifen (Fig. 3D). Importantly, chelating Ca2+ using EGTA reversed LINC00152 inhibition-induced tamoxifen sensitization (Fig. 3E). Notably, EGTA treatment also reduced tamoxifen-induced viability inhibition via inhibiting tamoxifen-induced ferroptosis (Fig. 3F, G), suggesting that Ca2+ induction is one of the key mechanisms of tamoxifen-induced ferroptosis and sensitivity, which is restored in TamR cells upon LINC00152 inhibition.
Fig. 3. LINC00152 inhibition increases TRPC1 expression and cytosolic Ca2+ downstream of cAMP signaling.
A The Pearson correlation analysis of Ca2+ signaling with ferroptosis score in endocrine therapy sensitive ER+ patients after therapy from GE87411. B, C The levels of Ca2+ signaling score before and after endocrine therapy in sensitive (B) and resistant (C) ER+ patients from GSE87411. D The levels of cytosolic Ca2+ in MCF-7.TamR cells that were transfected with two different siRNAs against LINC00152 and treated with tamoxifen. E Percentage cell viability in shLINC00152-expressing MCF-7.TamR cells treated with tamoxifen with or without EGTA. F Percentage cell viability inhibition in MCF-7.Par cells treated with tamoxifen with or without EGTA. G Relative lipid peroxidation in cells from (F). H qRT-PCR analysis of TRPC1 mRNA in shCtrl or shLINC00152-expressing MCF-7.TamR cells treated with tamoxifen. I Western Blot analysis of PDE4D, p-CREB (S133), p-PKA (Thr197) and TRPC1 in shCtrl or shLINC00152-expressing MCF-7.TamR cells treated with tamoxifen. J Percentage cell viability in shLINC00152-expressing MCF-7.TamR cells transfected with 2 different siRNAs against TRPC1. K The Pearson correlation analysis of Ca2+ signaling score with cAMP score in endocrine therapy sensitive ER+ patients after therapy from GSE124647. L cAMP levels in MCF-7.TamR shCtrl or shLINC00152 cells treated with tamoxifen. M The Pearson correlation analysis of cAMP score and ferroptosis score in endocrine therapy sensitive ER+ patients after therapy from GSE87411. Data are presented as mean values ±standard deviation (SD). P-values were calculated with the paired (cell viability) or unpaired (others), two-tailed Student’s t-test. *P < 0.05, **P < 0.01, n.s. not significant.
To identify the regulator of Ca2+ signaling under the control of LINC00152, we analyzed the RNA-seq data of siLINC00152-transfected cells and found that TRPC1, a Ca2+ channel located on cell or endoplasmic reticulum (EnR) membrane and regulating the Ca2+ levels within the cytosol [47], was upregulated upon LINC00152 knockdown (Supplementary Fig. S8A). This was validated at mRNA and protein levels by qRT-PCR and Western blot, respectively, and its level was further increased in combination with tamoxifen in TamR cells (Fig. 3H, I). Importantly, knocking down TRPC1 using two different siRNAs rescued cell viability under tamoxifen treatment (Fig. 3J, Supplementary Fig. S8B), while overexpressing TRPC1 in TamR cells increased tamoxifen sensitivity (Supplementary Fig. S8C, D), showing the functional role of TRPC1 in LINC00152 inhibition-mediated tamoxifen sensitization.
Increase in cAMP is known to stimulate Ca2+ flux by activating the calcium channels [48]. We identified a significant positive correlation between calcium signaling score and cAMP signaling score [49] after treatment with endocrine therapy (Fig. 3K) in the same endocrine-sensitive patients from GSE87411 showing positive correlation between Ca2+ signaling and ferroptosis scores (Fig. 3A). Supporting these results, we detected a significant increase in cAMP upon knocking down LINC00152 in combination with tamoxifen (Fig. 3L) that was followed by phosphorylation of the downstream effectors, PKA and CREB (Fig. 3I). Notably, inhibiting CREB rescued the expression of xCT in shLINC00152-expressing MCF-7.TamR cells (Supplementary Fig. S8E) and tamoxifen-treated parental cells (Supplementary Fig. S8F), suggesting that cAMP/CREB is not only important for Ca2+/ROS induction but also for suppressing xCT/GPX4 to promote LINC00152 inhibition-induced and tamoxifen-dependent ferroptosis. Furthermore, the levels of cAMP score were significantly positively correlated with the ferroptosis score in post-endocrine therapy-treated sensitive ER+ breast cancer patients (Fig. 3M), similar to calcium signaling score (Fig. 3A). Altogether, these data demonstrate that LINC00152 inhibition in combination with tamoxifen activates cAMP signaling and increases TRPC1 levels, thus triggering cytoplasmic Ca2+ overload that likely initiates ferroptosis in TamR cells.
LINC00152 inhibits tamoxifen-dependent ferroptosis by regulating PDE4D and downstream cAMP/Ca2+ signaling in tamoxifen-resistant cells
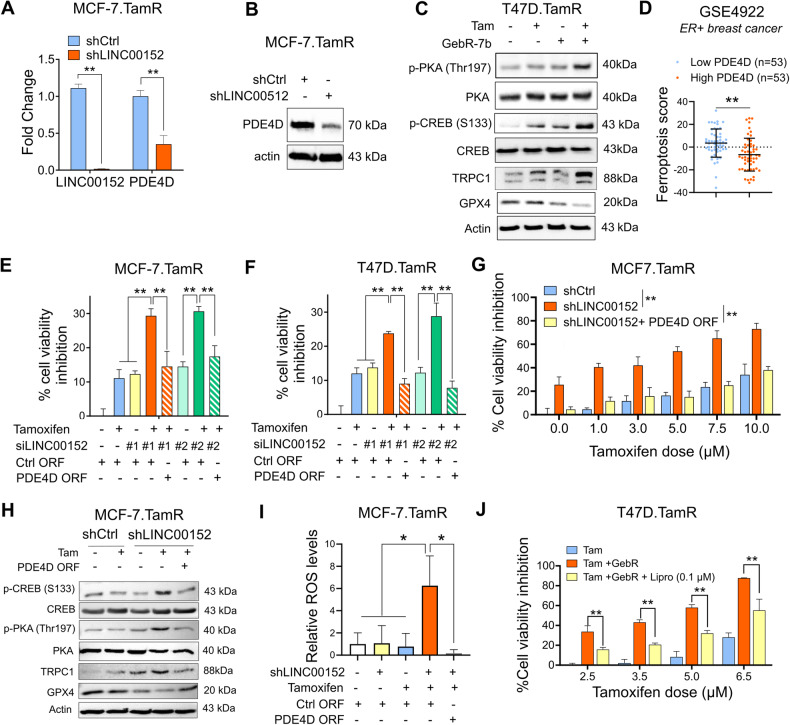
Phosphodiesterase 4D (PDE4D) is one of the cAMP-specific phosphodiesterases that degrades cAMP [50]. We found that PDE4D mRNA is downregulated in the RNA-seq data of siLINC00152-transfected MCF-7.TamR cells (Supplementary Fig. S9A). The reduction was further validated at both mRNA and protein levels by qRT-PCR and Western blotting, respectively, upon siRNA or shRNA-mediated LINC00152 knockdown in TamR cells (Fig. 4A, B, Supplementary Fig. S9B, C). Conversely, overexpressing LINC00152 in parental MCF-7 and T47D cells significantly increased the mRNA and protein levels of PDE4D (Supplementary Fig. S9D, E). Notably, combining tamoxifen with the PDE4D inhibitor GebR-7b [51] increased TRPC1 levels along with activation of PKA/CREB and reduction of GPX4 (Fig. 4C), phenocopying the effects of LINC00152 knockdown (Fig. 3I). Next, we demonstrated a significantly lower ferroptosis score in patients expressing high levels of PDE4D (Fig. 4D), showing the clinical relevance.
Fig. 4. LINC00152 inhibits tamoxifen-dependent ferroptosis by regulating PDE4D and downstream cAMP/Ca2+ signaling in tamoxifen-resistant cells.
A qRT-PCR analysis of LINC00152 and PDE4D in the shLINC00152-expressing MCF-7.TamR cells. B Western blot analysis of PDE4D in the shLINC00152-expressing MCF-7.TamR cells. C Western blot analysis of p-CREB (S133), p-PKA (Thr197), TRPC1 and ferroptosis inhibitor GPX4 in T47D.TamR cells that were treated with tamoxifen in combination with GebR-7b. D The levels of ferroptosis score in low vs. high PDE4D-expressing ER+ patients from GSE4922. E, F Percentage cell viability inhibition in MCF-7 and T47D.TamR cells co-transfected with siLINC00152 and PDE4D ORF and treated with tamoxifen. G Percentage of viability inhibition in shLINC00152-expressing MCF-7.TamR cells stably overexpressing PDE4D and that were treated with tamoxifen. H Western blot analysis of p-CREB (S133), p-PKA (Thr197), TRPC1 and GPX4 in shLINC00152-expressing MCF-7.TamR cells stably overexpressing PDE4D and that were treated with tamoxifen. I Relative ROS levels in MCF-7.TamR cells expressing shLINC00152 and treated with tamoxifen. J Percentage viability inhibition in T47D.TamR cells treated with tamoxifen and GebR-7b with or without 0.1 µM of liproxstatin-1 (Lipro). Data are presented as mean values ± standard deviation (SD). P-values were calculated with the paired (cell viability) or unpaired (others), two-tailed Student’s t-test. *P < 0.05, **P < 0.01.
We showed that PDE4D overexpression completely rescued cell viability in the presence of siRNA or shRNA-mediated LINC00152 knockdown in combination with tamoxifen (Fig. 4E–G) via reducing TRPC1 expression along with de-activation of PKA and CREB and increased expression of GPX4 (Fig. 4H). These data demonstrate the key contribution of PDE4D in LINC00152 mediated tamoxifen resistance. Importantly, we detected a strong induction of ROS, a crucial initiator of ferroptosis [42], upon LINC00152 knockdown in combination with tamoxifen, which was completed reversed upon PDE4D overexpression (Fig. 4I). Along these lines, inhibiting ferroptosis using liproxstatin-1 partially rescued cell viability under treatment with tamoxifen and GebR-7b (Fig. 4J). Overall, these data suggest that LINC00152 blocks tamoxifen-induced ferroptosis and mediates tamoxifen resistance via regulating PDE4D/cAMP/Ca2+ signaling axis.
LINC00152 stabilizes PDE4D mRNA by interacting with its 3’UTR
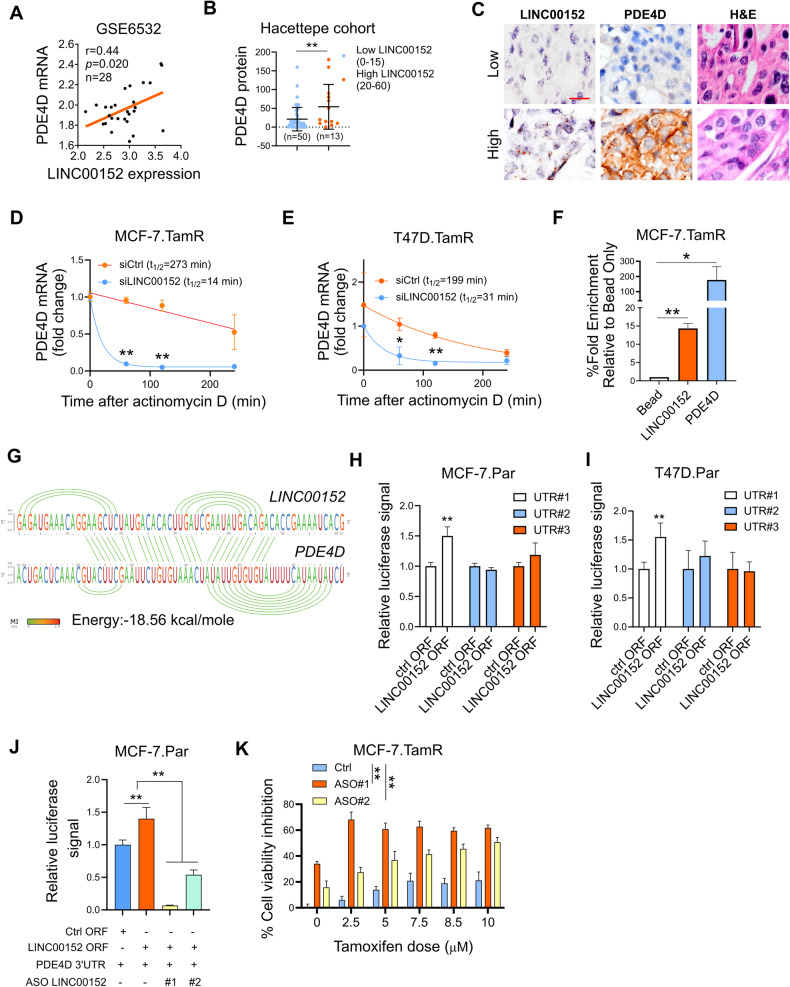
lncRNAs can modulate gene expression in multiple ways, one of which is to regulate the stability of the target mRNA [52]. We observed a positive correlation between LINC00152 and PDE4D mRNA in ER+ breast cancer patients progressed upon tamoxifen therapy in GSE6532 dataset, suggesting mRNA level regulation of PDE4D by LINC00152 (Fig. 5A). PDE4D also showed a positive correlation with LINC00152 at the protein level in our own cohort of ER+ breast cancer patients (Fig. 5B, C). mRNA stability assay demonstrated that LINC00152 knockdown reduces the half-life of PDE4D mRNA in both MCF-7 and T47D.TamR cells (Fig. 5D, E). Furthermore, RNA pull-down demonstrated the direct binding between LINC00152 and PDE4D mRNA in TamR cells (Fig. 5F). The 3’UTR region of mRNAs is usually responsible for regulating the stability of mRNAs [53]. Therefore, we reasoned that LINC00152 is potentially interacting with the 3’UTR region of the PDE4D mRNA to modulate its stability. To test this hypothesis, we first examined the predicted binding sites between PDE4D 3’UTR and LINC00152 using the IntaRNA tool 2.0 [54] and identified three potential interaction sites on the PDE4D 3’UTR (Fig. 5G, Supplementary Fig. S10). We then cloned the 3’UTR of PDE4D in 3 different pieces, each containing one predicted binding site for LINC00152 (UTR#1, 2, 3) downstream of the luciferase gene. Both MCF-7 and T47D cell lines were co-transfected with PDE4D 3’UTR vectors together with LINC00152 overexpression vector. This resulted in an increase in luciferase signal from UTR#1 (Fig. 5H, I), suggesting that LINC00152 potentially binds to 3’UTR region#1 (binding energy: −18.56 kcal/mol, Fig. 5G) and increases PDE4D mRNA stability. Supporting these results, two different ASO sequences that block the interaction site between LINC00152 and PDE4D 3’UTR reduced the binding between LINC00152 and PDE4D 3’UTR (Fig. 5J) and mediated tamoxifen sensitization in a dose-dependent manner (Fig. 5K). Overall, these data demonstrate that LINC00152 interacts with PDE4D 3’ UTR in TamR cells, increasing its mRNA stability and thereby mediating tamoxifen resistance.
Fig. 5. LINC00152 stabilizes PDE4D mRNA by interacting with its 3’UTR.
A The Pearson correlation analysis of LINC00152 and PDE4D mRNA in patients from GSE6532. B PDE4D protein expression determined by IHC in low vs. high LINC00152-expressing patients from the Hacettepe cohort. C Representative images of LINC00152 ISH, PDE4D IHC, and H&E staining in patients from the Hacettepe cohort. Scale bar = 20 µm. D, E qRT-PCR analysis of PDE4D in MCF-7.TamR (D) and T47D.TamR (E) cells transfected with siLINC00152 for 48 h, followed by treatment with 5 µg/ml of actinomycin (D). F Percentage fold enrichment of PDE4D in LINC00152 pull down in MCF-7.TamR cells. G Interaction between LINC00152 and PDE4D 3’UTR predicted by the IntaRNA tool. The visualization of the interaction was obtained using RILogo. H, I Luciferase assay in parental MCF7 (H) and T47D (I) cells co-transfected with LINC00152 overexpression vector and PDE4D 3’UTR, demonstrating binding to PDE4D 3’UTR. J Luciferase assay in MCF-7.Par cells co-transfected with LINC00152 overexpression vector and PDE4D 3’UTR with or without ASOs targeting the interaction site between LINC00152 and PDE4D 3’UTR. K Percent cell viability inhibition in ASO-transfected MCF-7.TamR cells under tamoxifen treatment. Data are presented as mean values ± standard deviation (SD). P-values were calculated with the paired (cell viability) or unpaired (others), two-tailed Student’s t-test. *P < 0.05, **P < 0.01.
LINC00152 inhibition overcomes tamoxifen resistance in vivo
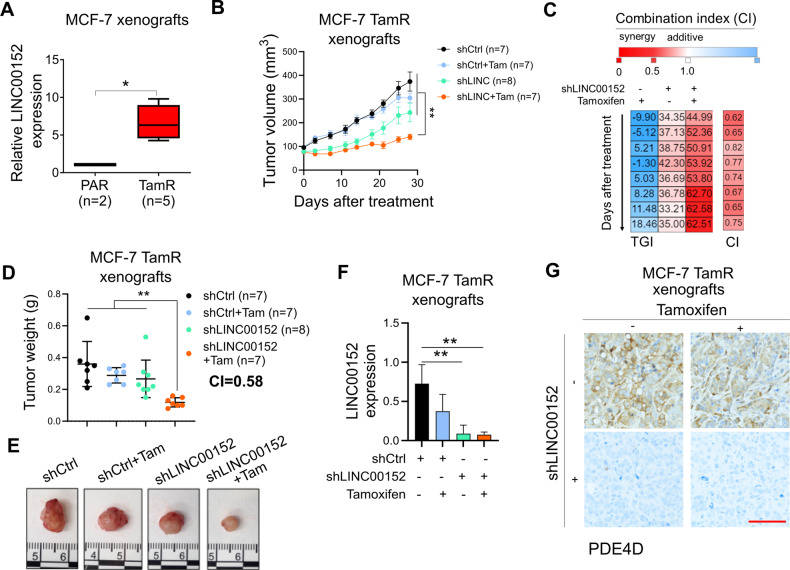
To test the effects of LINC00152 knockdown on in vivo tamoxifen sensitization, we first analyzed the expression of LINC00152 in MCF-7.TamR vs. parental tumors grown in mice and observed a significant upregulation of LINC00152 also within in vivo-grown MCF-7.TamR xenografts (Fig. 6A) like cultured cells (Fig. 1B). We then generated xenografts of MCF-7.TamR.shCtrl and shLINC00152 cells and treated the mice with vehicle (corn oil) or tamoxifen (3 mg/kg, daily) for a month. As shown in Fig. 6B–E, there was a significant synergistic reduction of tumor growth and tumor weight upon LINC00152 knockdown in combination with tamoxifen. qRT-PCR analysis of LINC00152 and immunohistochemistry (IHC) staining of PDE4D in tumors collected at the end of the experiment demonstrated that LINC00152 knockdown was sustained in vivo (Fig. 6F) and that PDE4D expression was strongly downregulated (Fig. 6G), validating our in vitro findings. Overall, we showed that LINC00152 inhibition overcomes tamoxifen resistance in vivo, at least in part, via downregulating PDE4D expression.
Fig. 6. LINC00152 inhibition overcomes tamoxifen resistance in vivo.
A PDE4D mRNA expression in MCF-7.Par (PAR) vs. TamR xenograft tumors. B Tumor volumes of MCF-7.TamR xenografts expressing shLINC00152 and treated with tamoxifen (3 mg/kg, daily). C Combination indices showing synergy between LINC00152 knockdown and tamoxifen in terms of tumor growth inhibition. D Tumor weights from mice in B. Combination index showing synergy between LINC00152 knockdown and tamoxifen in terms of tumor weight reduction is provided. E Tumor images from mice in (B). F qRT-PCR analysis of LINC00152 in tumors from (B). G IHC staining of PDE4D in tumors from (B). Scale bar = 100 µm. Data for the bar graphs and box plots are represented as mean values ± SD, while data for the tumor volume graph are represented as mean values ± standard error of the mean (SEM). P-values for the bar graphs and box plots were calculated with the unpaired, two-tailed Student’s t-test. The significance of the tumor volume graph was calculated with two-way ANOVA. *P < 0.05, **P < 0.01.
Discussion
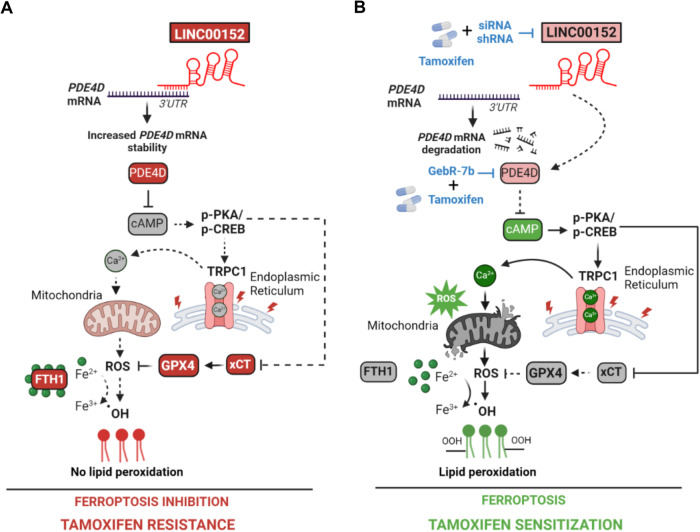
Tamoxifen is the first ER modulator approved by the FDA in the 1970s, and it has been the backbone of adjuvant hormone therapy [2]. However, resistance develops in ~20–30% of high-risk, advanced ER-positive breast cancer patients, decreasing survival [55]. Therefore, there is a dire need to identify novel therapeutic strategies to improve the clinical outcome for patients with resistance to tamoxifen. In this study, we demonstrated that ferroptosis is a mechanism of tamoxifen-induced cell death which is abrogated in resistance. We found that LINC00152 is overexpressed in tamoxifen resistant cells and confers tamoxifen resistance by inhibiting tamoxifen-induced ferroptosis while its inhibition confers sensitivity to tamoxifen-induced ferroptosis. We showed that LINC00152 enhances the mRNA stability of its novel target PDE4D, thus de-activating cAMP signaling and Ca2+ release and inducing xCT/GPX4, leading to inhibition of tamoxifen-dependent ferroptosis (Fig. 7A). On the other hand, LINC00152 inhibition destabilizes PDE4D and activates cAMP/PKA/CREB, like targeting PDE4D with GebR-7b. This increases TRPC1, cytosolic Ca2+, ROS and Fe2+ levels, accompanied by reduced levels of the antioxidant proteins FTH1, GPX4, and xCT, ultimately leading to lipid peroxidation, ferroptotic cell death, and tamoxifen sensitization (Fig. 7B). Notably, we showed that high LINC00152 expression is significantly correlated with high PDE4D/low ferroptosis and worse survival in multiple cohorts of tamoxifen- or tamoxifen-containing endocrine therapy-treated ER+ breast cancer patients, thereby demonstrating the clinical relevance of the axis we identified.
Fig. 7. Schematic summary of LINC00152/PDE4D-driven tamoxifen resistance.
A LINC00152 is overexpressed in tamoxifen resistance, binds to PDE4D 3’UTR, and increases its mRNA stability. This results in reduced cAMP levels and de-activation of PKA/CREB, leading to decreased expression of TRPC1, reduction of cytosolic Ca2+, and blockage of reactive oxygen species (ROS) generation. Furthermore, the negative regulators of ferroptosis, xCT, GPX4, and FTH1, are upregulated, leading to increased intracellular Fe2+ and inhibition of lipid peroxidation that ultimately blocks tamoxifen-dependent ferroptosis and mediates tamoxifen resistance. B Targeting LINC00152 in combination with tamoxifen leads to reduced mRNA stability of PDE4D due to loss of binding to its 3’UTR, thereby activating cAMP/PKA/CREB, similar to targeting PDE4D with GebR-7b in combination with tamoxifen. This further increases TRPC1 levels, elevates cytosolic Ca2+, generates reactive oxygen species (ROS), and increases Fe2+ upon downregulation of xCT, FTH1, and GPX4, leading to lipid peroxidation and tamoxifen-dependent ferroptosis. Dashed lines indicate that the connection is not active.
Activation of ferroptosis is initiated by an imbalance between prooxidant and antioxidant systems [16] and is emerging as an attractive strategy to overcome resistance to certain drugs, such as cisplatin [56, 57]. The ROS-mediated lipid peroxidation is one of the key steps driving ferroptosis upon oxidative degradation of lipids and the subsequent membrane damage [42]. On the other hand, xCT and GPX4 inhibit ferroptosis by promoting the synthesis of reduced glutathione (GSH), the primary antioxidant molecule [40], and reducing phospholipid hydroperoxide production [17], respectively. The current ferroptosis-inducing strategies used to eliminate therapy-resistant cancer cells usually target GPX4 by downregulating its expression [58] or by inhibiting its activity via the reduction of its cofactor, GSH [59], thus disturbing the balance between prooxidant and antioxidant systems. Notably, we showed that our novel combination of tamoxifen and LINC00152 knockdown induces tamoxifen-dependent ferroptosis by not only deactivating the antioxidant defense via reducing xCT and GPX4, but also by increasing Ca2+ overload through modulating the PDE4D/cAMP/PKA/CREB axis, thus causing ROS generation, accompanied by loss of the iron chelator, FTH1 that ultimately increases Fe2+ levels, and results in lipid peroxidation. Furthermore, we recently showed that endocrine therapy and CDK4/6 inhibitors commonly increase ROS levels in cancer cells and lead to cAMP-induced DNA damage, leading to PARP trapping and transcriptional blockage [60]. Future studies may investigate whether sensitivity to ferroptosis-mediated cell death is also altered in resistance to other endocrine therapies or CDK4/6 inhibitors, and whether LINC00152 targeting could also be used as an effective strategy to overcome resistance to these agents or in other resistant models vulnerable to ferroptosis induction.
Elevated levels of second messengers, such as cAMP and Ca2+, are known to be involved in stress response, controlling cell survival and/or death [61, 62]. The EnR is the center for managing cellular stress [63] and is the main storage site for Ca+2 [64]. A few anti-cancer agents are known to mediate cell death via activating EnR stress, such as bortezomib [65] and cisplatin [66]. However, the mechanisms of stress response induction in endocrine resistance are mostly undiscovered. We previously reported a novel mechanism of tamoxifen sensitization via inducing EnR stress, leading to activation of p38/JNK signaling and apoptosis in tamoxifen-resistant ER+ breast cancer cells [33]. Here, we characterized yet another novel type of stress response leading to tamoxifen sensitization, involving Ca2+ overload upon LINC00152/PDE4D inhibition. Recent evidence suggests that manipulating Ca2+ homeostasis stands as an attractive strategy to restore ferroptosis sensitivity. For instance, a novel tetraspanin, MS4A15, located on EnR membrane, blocks ferroptosis via depleting luminal Ca2+ stores and reprogramming lipids [67]. We demonstrated that tamoxifen-induced ferroptosis is dependent on elevated intracellular Ca2+ levels, and that chelating Ca2+ reverses tamoxifen sensitivity. In TamR cells, we identified TRPC1, a Ca2+ channel that can be located on a cell or EnR membrane [47], as a potentially novel regulator of ferroptosis. We showed that TRPC1 is upregulated upon activation of cAMP/PKA/CREB under LINC00152 knockdown, increases cytosolic Ca2+ and ROS levels. While inhibition of TRPC1 using siRNAs reverses LINC00152 inhibition-mediated tamoxifen sensitization, its overexpression confers tamoxifen sensitivity. Importantly, EnR is known to be in crosstalk with mitochondria [68], and an excessive flux of Ca2+ from EnR to mitochondria can lead to increased respiratory chain activity and production of excessive ROS [69], supporting our findings. Of note, since excessive Ca2+ may also trigger apoptotic cell death [70], elevated Ca2+ levels may also partially be responsible for tamoxifen-induced apoptosis. Although TRPC1 has not been reported as a CREB-induced gene, knockdown of CREB was shown to downregulate one of the G-protein coupled dopamine receptors responsible for Ca2+ influx, thereby reducing calcium signaling [69]. This raises the possibility that TRPC1 could also be transcribed in a CREB-dependent manner upon downregulation of LINC00152/PDE4D. Furthermore, the complete rescue of cell viability upon overexpressing PDE4D, in the presence of LINC00152 knockdown and tamoxifen, by reduction of TRPC1 and blockage of ROS, suggest that PDE4D is a novel and crucial modulator of tamoxifen-dependent ferroptotic cell death.
We demonstrated the clinical relevance of the LINC00152/PDE4D/cAMP/Ca2+/ferroptosis axis and showed that higher LINC00152 levels are associated with worse survival in multiple cohorts of endocrine therapy-treated ER+ breast cancer patients as well as other cancers. These results suggest that LINC00152 could be a potential biomarker for prognosis. However, future studies are warranted to test its sensitivity and specificity as well as its clinical association with the processes in our axis with larger cohorts of patients. In addition to being a potential biomarker, LINC00152 could also be targeted in clinics. LncRNAs are highly attractive targets for cancer therapy given their tissue-specific expression, thus enabling specific targeting of tumors [71]. Importantly, in the last decade, RNA-based therapies, such as ASOs or siRNAs, have undergone substantial improvements and are now being tested in clinical trials for various diseases, including cancer [72]. Overall, our data not only identify a novel mechanism of tamoxifen sensitization involving the regulation of ferroptosis by LINC00152 but also demonstrate the potential of LINC00152 as a clinically relevant resistance mediator, thus encouraging the clinical testing of LINC00152 targeting to overcome tamoxifen resistance.
Materials and methods
Cell lines, drugs, and culture conditions
Human breast cancer cell lines, MCF-7 and T47D, were purchased from ATCC and were cultured in phenol red–free DMEM (Gibco) with 10% FBS, 0.1% insulin, 50 U/mL penicillin/streptomycin, 1% nonessential amino acids (Gibco). Tamoxifen-resistant (TamR) MCF-7 and T47D cells were generated previously [33]. The parental counterparts (Par) were cultured alongside resistant cells without drug treatment, and therefore remained sensitive to tamoxifen. Tamoxifen and GebR-7b were purchased from Sigma. Liproxstatin-1, CREB inhibitor 666-15, Vitamin E, GSK-872, Q-Vd-OPh, and RSL3 were purchased from MedChem Express, and Necrox-5 was purchased from Cayman Chemicals. Cells were routinely tested for mycoplasma contamination using MycoAlert detection kit (Lonza) and were authenticated by STR sequencing.
ER+ human tumor samples
To analyze the effects of LINC00152 expression on the survival of endocrine therapy-treated female ER+ breast cancer patients and its correlation with PDE4D protein, we performed in situ hybridization (ISH) of LINC00152 and immunohistochemistry (IHC) staining of PDE4D in primary tumor samples from 124 ER+ breast cancer patients that were diagnosed between 2001 and 2018 at Hacettepe University School of Medicine, Ankara, Turkey and treated with endocrine therapy with or without radiotherapy and chemotherapy. The study was approved by the Non-Interventional Clinical Research Ethics Committee of Hacettepe University (approval no: 2020/02-40).
Whole-transcriptome sequencing (RNA-seq) and data analysis
RNA-seq of MCF-7.Par vs. TamR cells were performed in triplicates using the Illumina HiSeq 2000 platform at McGill University and Genome Quebec Innovation Centre as described previously [33]. The 330 differentially expressed lncRNAs between parental vs. TamR cells (q-value = 0.05) were retrieved (Supplementary Data 2). RNA-seq of MCF-7.TamR cells transfected with siControl, siLINC00152#1, or siLINC00152#2 were performed for each condition in duplicates at the University of South Carolina Functional Genomics Core. The genes differentially expressed upon LINC00152 knockdown (siControl vs. siLINC00152#1 and/or siControl vs. siLINC00152#2) with the cut-off of 0.67 > FC > 1.5 and P < 0.1 were determined. The details are provided in Supplementary Methods.
Lipid peroxidation assay
For measuring the levels of lipid peroxidation as an indicator of ferroptosis, Image-iT™ Lipid Peroxidation Kit (ThermoFisher) was utilized following manufacturer’s protocol. Cells that were stained with the BODIPY™ 581/591 C11 dye provided in the kit for 30 min were analyzed by flow cytometry. The shift from red to green signal that occurs upon oxidation by lipid hydroperoxides were quantified by taking the ratio of the mean fluorescence emission at ∼590 nm to ∼510 nm and plotted as the relative ferroptosis levels.
Cytosolic Ca2+ measurement
To detect cytosolic Ca2+ levels, the MCF-7.TamR cells were co-transfected with two different siLINC00152 and the GFP-based Ca2+ indicator, GCaMP3 (Addgene #64853) in 96-well format. One day later, cells were treated with 10 µM tamoxifen for 10 min, and the fluorescence signal was measured with SpectraMax microplate reader.
RNA pulldown
T47D.TamR cells were lysed in freshly prepared lysis buffer with the addition of RNase inhibitor and Protease inhibitors (20 mM Tris-HCl, pH 7.4, 200 mM KCl, 150 mM NaCl, 1% Triton-X-100, 0.1% SDS, 1 mM DTT, protease inhibitor cocktail and 200 U/mL of Riboblock). 1 mg of total lysate was mixed with 100 pmol of biotinylated probe against LINC00152 (Supplementary Table S1) and incubated for 1 h at room temperature. Then, the lysates were incubated with blocked streptavidin beads and incubated overnight at 4 °C. The next day, beads were incubated with Proteinase K buffer (10 mM Tris-HCl pH 7.0, 100 mM NaCl, 1 mM EDTA, 0.5% SDS) and 5 μL of proteinase-K (20 mg/mL) at 56 °C for 1 h and boiled at 95 °C for 5 min. RNA was extracted via an RNA purification kit (Zymo, R1013), converted to cDNA, and analyzed by RT-qPCR using the primers listed in Supplementary Table S2. %Input method was used to calculate enrichment relative to input samples and normalized to only bead control.
Intracellular cAMP measurements
To measure intracellular cAMP levels, MCF-7.TamR.shLINC cells that were treated with tamoxifen for 30 min were washed once with PBS and detached using Tryple. Three thousand cells/well were collected in Eppendorf tubes and lysed with ice-cold 2.5 mol/L perchloric acid, followed by neutralization with 2 M KOH. Cells were incubated on ice for 30 min and then centrifuged at 10,000 rpm for 10 min to collect the clear lysate. Twenty microliters of the clear lysate were transferred per well of a white plate, and cAMP measurement was done using the LANCE Ultra cAMP detection kit (PerkinElmer) according to the manufacturer’s protocol.
Intracellular ROS measurement
The intracellular ROS was measured using the DCFDA / H2DCFDA - Cellular ROS Assay Kit from Abcam according to the manufacturer’s protocol. Briefly, 8000 MCF-7.TamR.shCtrl and shLINC00152 cells were seeded on clear bottom, black wall plates and, the next day, transfected with PDE4D ORF. One day later, cells were treated with tamoxifen for 9 h, followed by staining with 20 µM DCFDA for 1 h. Then, the fluorescence was detected by a Multimode SpectraMax plate reader at 37 °C with excitation/emission at 485 nm/535 nm. The background GFP signal of the cells and the media were subtracted, and ROS levels were represented as relative change to shCtrl cells without any treatment.
Intracellular Fe2+ measurement
The intracellular Fe2+ levels were measured using the FerroOrange Assay Kit from Dojindo Molecular Technologies (JAPAN) (F374) according to the manufacturer’s instructions. Briefly, MCF-7.TamR shCtrl vs. shLINC00152 cells were seeded on clear bottom, black wall plates, and the next day, treated with 7.5 µM tamoxifen. The FerroOrange dye was added to the media at a 1:1000 dilution immediately after treatment. After 1.5 h, the fluorescence signal was quantified at Ex/Em = 561 nm/600 nm. The signal from only media was subtracted from all measurements, and the relative Fe2+ levels were calculated by taking the ratio of tamoxifen-treated shCtrl or shLINC00152 cells to their corresponding untreated counterparts.
In vivo mice experiments
All the in vivo studies were carried out in accordance with the Institutional Animal Care and Use Committee of the Medical University of South Carolina. Six-to-eight-week-old female Nu/J mice were housed with a temperature-controlled and 12-h light/12-h dark cycle environment. Tumors of MCF-7.Par, TamR, TamR.shCtrl or TamR.shLINC00152 cells were developed by injecting 1 × 107 cells subcutaneously near mammary fat pads. Slow-release estradiol pellets (0.36 mg, 60 days; Innovative Research of America) were implanted subcutaneously (s.c) at the nape of the neck 1 day before injection. Tumor growth was regularly monitored, and size measurements were performed two times per week. Tumor volume was calculated using the formula (length × width2)/2. After MCF-7.TamR.shCtrl tumor size reached 100 mm3, the mice with shCtrl and shLINC00152 tumors were randomly divided into 2 groups, with 7–8 mice per group. Animals were treated with vehicle (corn oil) or tamoxifen (3 mg/kg in corn oil, by oral gavage) daily. The tumor size of each mouse was recorded twice a week. Mice were sacrificed 28 days after initiation of the treatment. There was no blinding during in vivo experiments. Sample sizes were determined based on previous studies [33, 60].
Bioinformatics and statistical analysis
The normalized expression matrices of the microarray data sets, GSE6532 [73], GSE58644 [74], GSE42568 [75], GSE87411 [76] and GSE4922 [77] were downloaded from the GEO database, and the TCGA data were downloaded from XenaBrowser (UCSC Genomics Institute) [78] and KM-Plotter database [79]. Selection of datasets were based on availability of the probes for the genes of interest, e.g., LINC00152, the availability of normal tissue in addition to cancerous tissue, the availability of clinical data, such as treatment response and sample collection time, e.g., before and after treatment with endocrine therapy and the survival data. The LINC00152 knockdown (KD) signature was generated using the cumulative list of genes down or upregulated upon LINC00152 knockdown using siLINC00152#1 or siLINC00152#2 in MCF-7.TamR cells. The KD score was then calculated by subtracting the sum of the z-score of the genes downregulated from the sum of the z-scores of the genes upregulated using the SPSS Statistics software. The ferroptosis gene signature comprises genes driving ferroptosis and was obtained from FerrDb V2 database [38]. The Ca2+ signaling signature was obtained from the KEGG database [46], and the cAMP signature was generated using genes significantly upregulated (P-value < 0.001 and FC > 2) upon treatment of cancer cells with the cAMP analog, 8-CPT-cAMP for 24 h [49]. All ferroptosis, calcium, and cAMP scores were generated by summing up the z scores of the genes in each list separately. The GSEA analyses in low LINC00152 KD score-expressing or low LINC00152-expressing patients were performed using the GSEA tool (Broad Institute).
The results are represented as mean ± standard deviation (SD) or mean ± standard error of the mean (SEM), as indicated in the figure legends. All statistical analyses were performed in GraphPad Prism Software. Comparisons between the two groups were made using unpaired two-sided Student’s t-test. Comparison of the tumor volume graphs was done using two-way ANOVA with Dunnett’s multiple comparisons test. Survival curves were generated using the Kaplan–Meier method. Separation of patients was done based on median (GSE6532, GSE42568 and the Hacettepe cohort) or quartile (GSE58644) expression of LINC00152, which are two commonly used patient stratification methods [80]. For survival analysis with the TCGA data, the best cut-off option of KM-Plotter database was utilized. Significance between groups was calculated by the Log-rank test. For correlation analysis, Pearson correlation coefficients were calculated. For chi-square testing of Hacettepe samples, number of patients showing relapse or no relapse within 5-years were compared based on low vs. high LINC00152 expression separated from median. Outlier analysis was performed using Grubbs’ test. Experiments were repeated two to three times independently with similar results.
All other methods, including RNA-seq and data analysis, transient transfection with siRNAs, stable transfections using lentiviral vectors, quantitative RT-PCR analysis, mRNA stability assay using actinomycin D, 3’UTR assay, Western blotting, in situ hybridization and immunohistochemistry are provided in Supplementary Methods. The Supplementary Tables for the sequences of shRNAs (Supplementary Table S3), siRNAs and ASOs (Supplementary Table S4), overexpression vectors (Supplementary Table S5), and the list of antibodies (Supplementary Table S6) are also provided in the Supplementary Information file.
Supplementary information
Acknowledgements
We are thankful to the members of the Ozgur Sahin laboratory for invaluable discussion and advice. We also thank the Flow Cytometry & Cell Sorting Shared Resource and Translational Science Laboratory of the Medical University of South Carolina.
Author contributions
Ozge S. designed and performed experiments, acquired and analyzed data, interpreted data, and prepared the paper; R.A. performed rescue experiments with PDE4D overexpression, lipid peroxidation and cAMP/Ca2+ measurement experiments and helped with shRNA generation; K-T.H. performed rescue experiments with LINC00152 overexpression; A.I. performed ISH of LINC00152 in ER+ breast cancer samples and contributed to data interpretation; M.U. performed the staining of PDE4D in ER+ breast cancer samples and contributed to data interpretation; N.B. contributed to cell viability and qRT-PCR experiments; P.G.E. contributed to bioinformatics analysis and determining LINC00152 expression in tumors; U.M.T. contributed to bioinformatic and experimental validations of LINC00152-PDE4D binding and analysis of the RNA-seq data; B.U. performed rescue experiments with inhibitors of other cell death mechanisms and EGTA, and sensitization with RSL3; M.C. performed the RNA pulldown experiments and test the LINC00152-PDE4D interaction blocking ASOs; K.C. contributed to analyses of RNA-seq data and performed patient data analysis; M.E.G. contributed to lipid peroxidation assay and Ca2+ measurements; H.B. was contributed to analysis of the differentially expressed lncRNAs in TamR cells; O.S.S. contributed to the in vivo experiment; Y.R. performed the RNA-seq and contributed to data interpretation; D.T. contributed to the bioinformatics analyses and data interpretation; D.G. contributed to the analyses of RNA-seq and data interpretation; B.O. contributed to the design of experiments and the interpretation of data; S.A., provided the clinical information of ER+ patients; A.U. contributed to the staining of PDE4D and data interpretation and A.A. contributed to the ISH staining of LINC00152 in ER+ breast cancer samples and data interpretation; O.S. designed the study, oversaw experiments and data analyses, and prepared the paper. All authors reviewed and commented on the paper.
Funding
This work was supported by research funding from American Cancer Society Research Scholar Grant RSG-19-194-01-CSM (O.S.), and in part, from the National Institutes of Health (NIH; R01CA267101 and R01CA251374 to O.S.), SmartState Endowment in Lipidomics and Drug Discovery (O.S.) and previously by TUBITAK Grant 215Z357 (O.S.), TUBITAK-CNRS Bilateral Grant (TUBITAK, 214S364 (O.S.) and CNRS (D.T.)), and Susan G. Komen Interdisciplinary Graduate Training to Eliminate Cancer Disparities (IGniTE-CD) GTDR17500160 (OzgeS). The core facilities utilized are supported by NIH (C06 RR015455), Hollings Cancer Center Support Grant (P30 CA138313) or Center of Biomedical Research Excellence (COBRE) in Lipidomics and Pathobiology (P30 GM103339).
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request. Gene expression data were downloaded from the NCBI Gene Expression Omnibus database under GSE6532, GSE58644, GSE42568, GSE87411, and GSE4922. The RNA-seq data of MCF-7.TamR cells compared to parental were uploaded to the Gene Expression Omnibus database under the BioProject ID: PRJNA1040324. The RNA-seq data of MCF-7.TamR siLINC00152 cells compared to siCtrl were uploaded to the Gene Expression Omnibus database under the BioProject ID: PRJNA1036119. The raw data for western blotting are provided as a source data file.
Competing interests
O.S. is the co-founder of OncoCube Therapeutics LLC, the founder and president of LoxiGen, Inc., and the scientific advisory board member of A2A Pharmaceuticals. The other authors declare no potential competing interests.
Ethics
The study was approved by the Non‐Interventional Clinical Research Ethics Committee of Hacettepe University (approval no: 2020/02-40). Informed consent was obtained from all patients.
Footnotes
Edited by Rami Aqeilan
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41419-024-06814-3.
References
- 1.Early Breast Cancer Trialists’ Collaborative G. Davies C, Godwin J, Gray R, Clarke M, Cutter D, et al. Relevance of breast cancer hormone receptors and other factors to the efficacy of adjuvant tamoxifen: patient-level meta-analysis of randomised trials. Lancet. 2011;378:771–84. doi: 10.1016/S0140-6736(11)60993-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Howell A, Howell SJ. Tamoxifen evolution. Br J Cancer. 2023;128:421–5. doi: 10.1038/s41416-023-02158-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clemons M, Danson S, Howell A. Tamoxifen (“Nolvadex”): a review. Cancer Treat Rev. 2002;28:165–80. doi: 10.1016/S0305-7372(02)00036-1. [DOI] [PubMed] [Google Scholar]
- 4.Ekholm M, Bendahl PO, Fernö M, Nordenskjöld B, Stål O, Rydén L. Two Years of Adjuvant Tamoxifen Provides a Survival Benefit Compared With No Systemic Treatment in Premenopausal Patients With Primary Breast Cancer: Long-Term Follow-Up (> 25 years) of the Phase III SBII:2pre Trial. J Clin Oncol 34(19):2232-2238. 10.1200/JCO.2015.65.6272. [DOI] [PubMed]
- 5.Osborne CK, Fuqua SA. Mechanisms of tamoxifen resistance. Breast Cancer Res Treat. 1994;32:49–55. doi: 10.1007/BF00666205. [DOI] [PubMed] [Google Scholar]
- 6.Barone I, Brusco L, Fuqua SA. Estrogen receptor mutations and changes in downstream gene expression and signaling. Clin Cancer Res. 2010;16:2702–8. doi: 10.1158/1078-0432.CCR-09-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao A, Sun T, Ma G, Cao J, Hu Q, Chen L, et al. LEM4 confers tamoxifen resistance to breast cancer cells by activating cyclin D-CDK4/6-Rb and ERalpha pathway. Nat Commun. 2018;9:4180. doi: 10.1038/s41467-018-06309-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Massarweh S, Osborne CK, Creighton CJ, Qin L, Tsimelzon A, Huang S, et al. Tamoxifen resistance in breast tumors is driven by growth factor receptor signaling with repression of classic estrogen receptor genomic function. Cancer Res. 2008;68:826–33. doi: 10.1158/0008-5472.CAN-07-2707. [DOI] [PubMed] [Google Scholar]
- 9.Osborne CK, Shou J, Massarweh S, Schiff R. Crosstalk between estrogen receptor and growth factor receptor pathways as a cause for endocrine therapy resistance in breast cancer. Clin Cancer Res. 2005;11:865s–70s. doi: 10.1158/1078-0432.865s.11.2. [DOI] [PubMed] [Google Scholar]
- 10.Shi Q, Li Y, Li S, Jin L, Lai H, Wu Y, et al. LncRNA DILA1 inhibits Cyclin D1 degradation and contributes to tamoxifen resistance in breast cancer. Nat Commun. 2020;11:5513. doi: 10.1038/s41467-020-19349-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nabieva N, Fasching PA. CDK4/6 inhibitors-overcoming endocrine resistance is the standard in patients with hormone receptor-positive breast cancer. Cancers (Basel) 2023;15:1763. doi: 10.3390/cancers15061763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pandey K, An HJ, Kim SK, Lee SA, Kim S, Lim SM, et al. Molecular mechanisms of resistance to CDK4/6 inhibitors in breast cancer: a review. Int J Cancer. 2019;145:1179–88. doi: 10.1002/ijc.32020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dixon SJ, Lemberg KM, Lamprecht MR, Skouta R, Zaitsev EM, Gleason CE, et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149:1060–72. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yagoda N, von Rechenberg M, Zaganjor E, Bauer AJ, Yang WS, Fridman DJ, et al. RAS-RAF-MEK-dependent oxidative cell death involving voltage-dependent anion channels. Nature. 2007;447:864–8. doi: 10.1038/nature05859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang WS, Stockwell BR. Synthetic lethal screening identifies compounds activating iron-dependent, nonapoptotic cell death in oncogenic-RAS-harboring cancer cells. Chem Biol. 2008;15:234–45. doi: 10.1016/j.chembiol.2008.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen X, Li J, Kang R, Klionsky DJ, Tang D. Ferroptosis: machinery and regulation. Autophagy. 2021;17:2054–81. doi: 10.1080/15548627.2020.1810918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang WS, SriRamaratnam R, Welsch ME, Shimada K, Skouta R, Viswanathan VS, et al. Regulation of ferroptotic cancer cell death by GPX4. Cell. 2014;156:317–31. doi: 10.1016/j.cell.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao R, Kalathur RKR, Coto-Llerena M, Ercan C, Buechel D, Shuang S, et al. YAP/TAZ and ATF4 drive resistance to Sorafenib in hepatocellular carcinoma by preventing ferroptosis. EMBO Mol Med. 2021;13:e14351. doi: 10.15252/emmm.202114351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma S, Henson ES, Chen Y, Gibson SB. Ferroptosis is induced following siramesine and lapatinib treatment of breast cancer cells. Cell Death Dis. 2016;7:e2307. doi: 10.1038/cddis.2016.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lei G, Zhang Y, Koppula P, Liu X, Zhang J, Lin SH, et al. The role of ferroptosis in ionizing radiation-induced cell death and tumor suppression. Cell Res. 2020;30:146–62. doi: 10.1038/s41422-019-0263-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Viswanathan VS, Ryan MJ, Dhruv HD, Gill S, Eichhoff OM, Seashore-Ludlow B, et al. Dependency of a therapy-resistant state of cancer cells on a lipid peroxidase pathway. Nature. 2017;547:453–7. doi: 10.1038/nature23007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cairns J, Ingle JN, Kalari KR, Shepherd LE, Kubo M, Goetz MP, et al. The lncRNA MIR2052HG regulates ERalpha levels and aromatase inhibitor resistance through LMTK3 by recruiting EGR1. Breast Cancer Res. 2019;21:47. doi: 10.1186/s13058-019-1130-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xue X, Yang YA, Zhang A, Fong KW, Kim J, Song B, et al. LncRNA HOTAIR enhances ER signaling and confers tamoxifen resistance in breast cancer. Oncogene. 2016;35:2746–55. doi: 10.1038/onc.2015.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Z, Yu D, Li H, Lv Y, Li S. Long non‑coding RNA UCA1 confers tamoxifen resistance in breast cancer endocrinotherapy through regulation of the EZH2/p21 axis and the PI3K/AKT signaling pathway. Int J Oncol. 2019;54:1033–42. doi: 10.3892/ijo.2019.4679. [DOI] [PubMed] [Google Scholar]
- 26.Bian Z, Zhang J, Li M, Feng Y, Yao S, Song M, et al. Long non-coding RNA LINC00152 promotes cell proliferation, metastasis, and confers 5-FU resistance in colorectal cancer by inhibiting miR-139-5p. Oncogenesis. 2017;6:395. doi: 10.1038/s41389-017-0008-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Notzold L, Frank L, Gandhi M, Polycarpou-Schwarz M, Gross M, Gunkel M, et al. The long non-coding RNA LINC00152 is essential for cell cycle progression through mitosis in HeLa cells. Sci Rep. 2017;7:2265. doi: 10.1038/s41598-017-02357-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao J, Liu Y, Zhang W, Zhou Z, Wu J, Cui P, et al. Long non-coding RNA Linc00152 is involved in cell cycle arrest, apoptosis, epithelial to mesenchymal transition, cell migration and invasion in gastric cancer. Cell Cycle. 2015;14:3112–23. doi: 10.1080/15384101.2015.1078034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ji J, Tang J, Deng L, Xie Y, Jiang R, Li G, et al. LINC00152 promotes proliferation in hepatocellular carcinoma by targeting EpCAM via the mTOR signaling pathway. Oncotarget. 2015;6:42813–24. doi: 10.18632/oncotarget.5970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen Z, Cai X, Chang L, Xia Y, Wang L, Hou Y, et al. LINC00152 is a potential biomarker involved in the modulation of biological characteristics of residual colorectal cancer cells following chemoradiotherapy. Oncol Lett. 2018;15:4177–84. doi: 10.3892/ol.2018.7833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang W, Wu F, Zhao Z, Wang KY, Huang RY, Wang HY, et al. Long noncoding RNA LINC00152 is a potential prognostic biomarker in patients with high-grade glioma. CNS Neurosci Ther. 2018;24:957–66. doi: 10.1111/cns.12850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu J, Shuang Z, Zhao J, Tang H, Liu P, Zhang L, et al. Linc00152 promotes tumorigenesis by regulating DNMTs in triple-negative breast cancer. Biomed Pharmacother. 2018;97:1275–81. doi: 10.1016/j.biopha.2017.11.055. [DOI] [PubMed] [Google Scholar]
- 33.Mishra RR, Belder N, Ansari SA, Kayhan M, Bal H, Raza U, et al. Reactivation of cAMP Pathway by PDE4D Inhibition Represents a Novel Druggable Axis for Overcoming Tamoxifen Resistance in ER-positive Breast Cancer. Clin Cancer Res. 2018;24:1987–2001. doi: 10.1158/1078-0432.CCR-17-2776. [DOI] [PubMed] [Google Scholar]
- 34.Yu Y, Yang J, Li Q, Xu B, Lian Y, Miao L. LINC00152: A pivotal oncogenic long non-coding RNA in human cancers. Cell Prolif. 2017;50:e12349. doi: 10.1111/cpr.12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davey MG, Hynes SO, Kerin MJ, Miller N, Lowery AJ. Ki-67 as a prognostic biomarker in invasive breast cancer. Cancers (Basel) 2021;13:4455. doi: 10.3390/cancers13174455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhan Y, Chen Z, Li Y, He A, He S, Gong Y, et al. Long non-coding RNA DANCR promotes malignant phenotypes of bladder cancer cells by modulating the miR-149/MSI2 axis as a ceRNA. J Exp Clin Cancer Res. 2018;37:273. doi: 10.1186/s13046-018-0921-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39:925–38. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou N, Yuan X, Du Q, Zhang Z, Shi X, Bao J, et al. FerrDb V2: update of the manually curated database of ferroptosis regulators and ferroptosis-disease associations. Nucleic Acids Res. 2023;51:D571–D82. doi: 10.1093/nar/gkac935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Friedmann Angeli JP, Schneider M, Proneth B, Tyurina YY, Tyurin VA, Hammond VJ, et al. Inactivation of the ferroptosis regulator Gpx4 triggers acute renal failure in mice. Nat Cell Biol. 2014;16:1180–91. doi: 10.1038/ncb3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koppula P, Zhuang L, Gan B. Cystine transporter SLC7A11/xCT in cancer: ferroptosis, nutrient dependency, and cancer therapy. Protein Cell. 2021;12:599–620. doi: 10.1007/s13238-020-00789-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bogdan AR, Miyazawa M, Hashimoto K, Tsuji Y. Regulators of iron homeostasis: new players in metabolism, cell death, and disease. Trends Biochem Sci. 2016;41:274–86. doi: 10.1016/j.tibs.2015.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tang D, Chen X, Kang R, Kroemer G. Ferroptosis: molecular mechanisms and health implications. Cell Res. 2021;31:107–25. doi: 10.1038/s41422-020-00441-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hu Q, Zhang Y, Lou H, Ou Z, Liu J, Duan W, et al. GPX4 and vitamin E cooperatively protect hematopoietic stem and progenitor cells from lipid peroxidation and ferroptosis. Cell Death Dis. 2021;12:706. doi: 10.1038/s41419-021-04008-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang GJ, Kimijima I, Onda M, Kanno M, Sato H, Watanabe T, et al. Tamoxifen-induced apoptosis in breast cancer cells relates to down-regulation of bcl-2, but not bax and bcl-X(L), without alteration of p53 protein levels. Clin Cancer Res. 1999;5:2971–7. [PubMed] [Google Scholar]
- 45.Pedrera L, Espiritu RA, Ros U, Weber J, Schmitt A, Stroh J, et al. Ferroptotic pores induce Ca(2+) fluxes and ESCRT-III activation to modulate cell death kinetics. Cell Death Differ. 2021;28:1644–57. doi: 10.1038/s41418-020-00691-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cheng KT, Liu X, Ong HL, Swaim W, Ambudkar IS. Local Ca(2)+ entry via Orai1 regulates plasma membrane recruitment of TRPC1 and controls cytosolic Ca(2)+ signals required for specific cell functions. PLoS Biol. 2011;9:e1001025. doi: 10.1371/journal.pbio.1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gomez E, Pritchard C, Herbert TP. cAMP-dependent protein kinase and Ca2+ influx through L-type voltage-gated calcium channels mediate Raf-independent activation of extracellular regulated kinase in response to glucagon-like peptide-1 in pancreatic beta-cells. J Biol Chem. 2002;277:48146–51. doi: 10.1074/jbc.M209165200. [DOI] [PubMed] [Google Scholar]
- 49.Zhang L, Zambon AC, Vranizan K, Pothula K, Conklin BR, Insel PA. Gene expression signatures of cAMP/protein kinase A (PKA)-promoted, mitochondrial-dependent apoptosis. Comparative analysis of wild-type and cAMP-deathless S49 lymphoma cells. J Biol Chem. 2008;283:4304–13. doi: 10.1074/jbc.M708673200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Conti M, Richter W, Mehats C, Livera G, Park JY, Jin C. Cyclic AMP-specific PDE4 phosphodiesterases as critical components of cyclic AMP signaling. J Biol Chem. 2003;278:5493–6. doi: 10.1074/jbc.R200029200. [DOI] [PubMed] [Google Scholar]
- 51.Bruno O, Fedele E, Prickaerts J, Parker LA, Canepa E, Brullo C, et al. GEBR-7b, a novel PDE4D selective inhibitor that improves memory in rodents at non-emetic doses. Br J Pharm. 2011;164:2054–63. doi: 10.1111/j.1476-5381.2011.01524.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Statello L, Guo CJ, Chen LL, Huarte M. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol. 2021;22:96–118. doi: 10.1038/s41580-020-00315-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mayr C. What are 3’ UTRs doing? Cold Spring Harb Perspect Biol. 2019;11:a034728. doi: 10.1101/cshperspect.a034728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mann M, Wright PR, Backofen R. IntaRNA 2.0: enhanced and customizable prediction of RNA-RNA interactions. Nucleic Acids Res. 2017;45:W435–W9. doi: 10.1093/nar/gkx279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.International Breast Cancer Study G, Colleoni M, Gelber S, Goldhirsch A, Aebi S, Castiglione-Gertsch M, et al. Tamoxifen after adjuvant chemotherapy for premenopausal women with lymph node-positive breast cancer: International Breast Cancer Study Group Trial 13-93. J Clin Oncol. 2006;24:1332–41. doi: 10.1200/JCO.2005.03.0783. [DOI] [PubMed] [Google Scholar]
- 56.Du J, Wang X, Li Y, Ren X, Zhou Y, Hu W, et al. DHA exhibits synergistic therapeutic efficacy with cisplatin to induce ferroptosis in pancreatic ductal adenocarcinoma via modulation of iron metabolism. Cell Death Dis. 2021;12:705. doi: 10.1038/s41419-021-03996-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang Y, Wu X, Ren Z, Li Y, Zou W, Chen J, et al. Overcoming cancer chemotherapy resistance by the induction of ferroptosis. Drug Resist Updat. 2023;66:100916. doi: 10.1016/j.drup.2022.100916. [DOI] [PubMed] [Google Scholar]
- 58.Markowitsch SD, Schupp P, Lauckner J, Vakhrusheva O, Slade KS, Mager R, et al. Artesunate inhibits growth of sunitinib-resistant renal cell carcinoma cells through cell cycle arrest and induction of ferroptosis. Cancers (Basel) 2020;12:3150. doi: 10.3390/cancers12113150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hong T, Lei G, Chen X, Li H, Zhang X, Wu N, et al. PARP inhibition promotes ferroptosis via repressing SLC7A11 and synergizes with ferroptosis inducers in BRCA-proficient ovarian cancer. Redox Biol. 2021;42:101928. doi: 10.1016/j.redox.2021.101928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Saatci O, Cetin M, Uner M, Tokat UM, Chatzistamou I, Ersan PG, et al. Toxic PARP trapping upon cAMP-induced DNA damage reinstates the efficacy of endocrine therapy and CDK4/6 inhibitors in treatment-refractory ER+ breast cancer. Nat Commun. 2023;14:6997. doi: 10.1038/s41467-023-42736-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Galluzzi L, Vitale I, Aaronson SA, Abrams JM, Adam D, Agostinis P, et al. Molecular mechanisms of cell death: recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ. 2018;25:486–541. doi: 10.1038/s41418-017-0012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang Y, Tang Y, Hang H, Wang M, Pang Y, Yu Y, et al. cAMP induces cell apoptosis in multiple myeloma and overcomes bortezomib resistance. Am J Cancer Res. 2018;8:16–29. [PMC free article] [PubMed] [Google Scholar]
- 63.Sano R, Reed JC. ER stress-induced cell death mechanisms. Biochim Biophys Acta. 2013;1833:3460–70. doi: 10.1016/j.bbamcr.2013.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Meldolesi J, Pozzan T. The endoplasmic reticulum Ca2+ store: a view from the lumen. Trends Biochem Sci. 1998;23:10–4. doi: 10.1016/S0968-0004(97)01143-2. [DOI] [PubMed] [Google Scholar]
- 65.Nawrocki ST, Carew JS, Dunner K, Jr., Boise LH, Chiao PJ, Huang P, et al. Bortezomib inhibits PKR-like endoplasmic reticulum (ER) kinase and induces apoptosis via ER stress in human pancreatic cancer cells. Cancer Res. 2005;65:11510–9. doi: 10.1158/0008-5472.CAN-05-2394. [DOI] [PubMed] [Google Scholar]
- 66.Mandic A, Hansson J, Linder S, Shoshan MC. Cisplatin induces endoplasmic reticulum stress and nucleus-independent apoptotic signaling. J Biol Chem. 2003;278:9100–6. doi: 10.1074/jbc.M210284200. [DOI] [PubMed] [Google Scholar]
- 67.Xin S, Mueller C, Pfeiffer S, Kraft VAN, Merl-Pham J, Bao X, et al. MS4A15 drives ferroptosis resistance through calcium-restricted lipid remodeling. Cell Death Differ. 2022;29:670–86. doi: 10.1038/s41418-021-00883-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gordaliza-Alaguero I, Canto C, Zorzano A. Metabolic implications of organelle-mitochondria communication. EMBO Rep. 2019;20:e47928. doi: 10.15252/embr.201947928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang L, Liu L, Thompson R, Chan C. CREB modulates calcium signaling in cAMP-induced bone marrow stromal cells (BMSCs). Cell Calcium. 2014; 56(4):257-68. 10.1016/j.ceca.2014.07.013. [DOI] [PMC free article] [PubMed]
- 70.Moon DO. Calcium’s role in orchestrating cancer apoptosis: mitochondrial-centric perspective. Int J Mol Sci. 2023;24:8982. doi: 10.3390/ijms24108982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Balihodzic A, Prinz F, Dengler MA, Calin GA, Jost PJ, Pichler M. Non-coding RNAs and ferroptosis: potential implications for cancer therapy. Cell Death Differ. 2022;29:1094–106. doi: 10.1038/s41418-022-00998-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Reilley MJ, McCoon P, Cook C, Lyne P, Kurzrock R, Kim Y, et al. STAT3 antisense oligonucleotide AZD9150 in a subset of patients with heavily pretreated lymphoma: results of a phase 1b trial. J Immunother Cancer. 2018;6:119. doi: 10.1186/s40425-018-0436-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Loi S, Haibe-Kains B, Desmedt C, Lallemand F, Tutt AM, Gillet C, et al. Definition of clinically distinct molecular subtypes in estrogen receptor-positive breast carcinomas through genomic grade. J Clin Oncol. 2007;25:1239–46. doi: 10.1200/JCO.2006.07.1522. [DOI] [PubMed] [Google Scholar]
- 74.Tofigh A, Suderman M, Paquet ER, Livingstone J, Bertos N, Saleh SM, et al. The prognostic ease and difficulty of invasive breast carcinoma. Cell Rep. 2014;9:129–42. doi: 10.1016/j.celrep.2014.08.073. [DOI] [PubMed] [Google Scholar]
- 75.Clarke C, Madden SF, Doolan P, Aherne ST, Joyce H, O’Driscoll L, et al. Correlating transcriptional networks to breast cancer survival: a large-scale coexpression analysis. Carcinogenesis. 2013;34:2300–8. doi: 10.1093/carcin/bgt208. [DOI] [PubMed] [Google Scholar]
- 76.Ellis MJ, Suman VJ, Hoog J, Goncalves R, Sanati S, Creighton CJ, et al. Ki67 Proliferation index as a tool for chemotherapy decisions during and after neoadjuvant aromatase inhibitor treatment of breast cancer: results from the American College of Surgeons Oncology Group Z1031 Trial (Alliance) J Clin Oncol. 2017;35:1061–9. doi: 10.1200/JCO.2016.69.4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ivshina AV, George J, Senko O, Mow B, Putti TC, Smeds J, et al. Genetic reclassification of histologic grade delineates new clinical subtypes of breast cancer. Cancer Res. 2006;66:10292–301. doi: 10.1158/0008-5472.CAN-05-4414. [DOI] [PubMed] [Google Scholar]
- 78.Goldman MJ, Craft B, Hastie M, Repecka K, McDade F, Kamath A, et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat Biotechnol. 2020;38:675–8. doi: 10.1038/s41587-020-0546-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gyorffy B. Transcriptome-level discovery of survival-associated biomarkers and therapy targets in non-small-cell lung cancer. Br J Pharm. 2024;181:362–74. doi: 10.1111/bph.16257. [DOI] [PubMed] [Google Scholar]
- 80.Al-Ejeh F, Simpson PT, Saunus JM, Klein K, Kalimutho M, Shi W, et al. Meta-analysis of the global gene expression profile of triple-negative breast cancer identifies genes for the prognostication and treatment of aggressive breast cancer. Oncogenesis. 2014;3:e124. doi: 10.1038/oncsis.2014.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request. Gene expression data were downloaded from the NCBI Gene Expression Omnibus database under GSE6532, GSE58644, GSE42568, GSE87411, and GSE4922. The RNA-seq data of MCF-7.TamR cells compared to parental were uploaded to the Gene Expression Omnibus database under the BioProject ID: PRJNA1040324. The RNA-seq data of MCF-7.TamR siLINC00152 cells compared to siCtrl were uploaded to the Gene Expression Omnibus database under the BioProject ID: PRJNA1036119. The raw data for western blotting are provided as a source data file.