Abstract
Retroviral Gag polyproteins drive virion assembly by polymerizing to form a spherical shell that lines the inner membrane of nascent virions. Deletion of the nucleocapsid (NC) domain of the Gag polyprotein disrupts assembly, presumably because NC is required for polymerization. Human immunodeficiency virus type 1 NC possesses two zinc finger motifs that are required for specific recognition and packaging of viral genomic RNA. Though essential, zinc fingers and genomic RNA are not required for virion assembly. NC promiscuously associates with cellular RNAs, many of which are incorporated into virions. It has been hypothesized that Gag polymerization and virion assembly are promoted by nonspecific interaction of NC with RNA. Consistent with this model, we found an inverse relationship between the number of NC basic residues replaced with alanine and NC's nonspecific RNA-binding activity, Gag's ability to polymerize in vitro and in vivo, and Gag's capacity to assemble virions. In contrast, mutation of NC's zinc fingers had only minor effects on these properties.
Retroviral nucleocapsid proteins (NC) are expressed as part of a Gag polyprotein that is cleaved by the virally encoded protease during virion maturation (reviewed in references 20 and 28). For human immunodeficiency virus type 1 (HIV-1), the mature Gag proteins include matrix (MA, p17), capsid (CA, p24), nucleocapsid (NC, p7), and p6. With the exception of those of spumaviruses, all retroviral NCs contain either one or two Cys-His boxes (Cys-X2-Cys-X4-His-X4-Cys), reminiscent of zinc finger motifs found in many DNA binding proteins. In addition, and without exception, all retroviral NCs contain a large number of basic residues distributed throughout the protein.
NC serves multiple functions in the retroviral life cycle, many of which stem from its ability to bind nucleic acid. Incorporation of viral genomic RNA into virions results from a specific interaction between NC's Cys-His boxes and cis-acting packaging sequences (Ψ) in viral genomic RNA. However, in addition to its sequence-specific nucleic acid-binding activity, NC is also able to interact nonspecifically with nucleic acid (for reviews see references 3 and 14). NC's specific and nonspecific binding activities can act separately or synergistically on the same nucleic acid molecule, for example, in the selection of Ψ-RNA, as recently shown by the solution structure of HIV-1 NC bound to the 14-nucleotide SL3 stem-loop from the HIV-1 RNA packaging signal (16).
The nonspecific nucleic acid-binding activity of NC has been shown to be mediated by basic residues distributed throughout the protein (21, 29, 32, 42, 45) and is consistent with the observations that NC coats the entire viral genomic RNA in mature virions and that significant quantities of cellular RNAs, such as 7SL RNA, 5S, 18S, and 28S rRNA, and tRNA, are incorporated into retroviral virions (for a review see reference 3). Given that NC is the major and most abundant viral RNA-binding protein, incorporation of these RNAs into virions presumably occurs through nonspecific RNA binding mediated by NC.
In addition to its role in RNA packaging, the NC domain is the major domain promoting interactions among Gag polyproteins, and deletion of NC abrogates virion production. Cys-His boxes play no role in these processes since disruption of zinc fingers by mutation or by compounds that oxidize cysteine thiolates has minimal effect on virion production (for a review see reference 3). In addition, Ψ− virions assemble efficiently (1, 12, 33), indicating that viral genomic RNA incorporation is not necessary for virion assembly.
In contrast to what is found for zinc fingers, data suggest that HIV-1 basic residues mediate Gag-Gag interaction (2, 5) and are required for virion assembly (15). However, evidence for involvement of basic residues in Gag-Gag interaction is indirect, and their role in virion assembly remains unclear in that some have reported a minimal effect of basic residue mutations on assembly (39, 41).
If NC basic residues do play a role in virion assembly, it would suggest that the nonspecific RNA-binding activity of these residues might drive assembly. In support of this model, assembly defects associated with NC deletion mutants can be partially reversed if stretches of basic residues are fused to the Gag polyprotein (5) or if NC is replaced with a heterologous RNA-binding protein (53). Also, in vitro assembly of virion cores from recombinant protein depends on the presence of nonspecific nucleic acid (8, 23, 25).
To test the hypothesis that the nonspecific nucleic acid-binding activity of HIV-1 NC is necessary for Gag-Gag interaction and virion assembly, we studied a panel of HIV-1 NC mutants in which from 2 to 10 basic residues were replaced with alanine. We show that mutations that impair NC's nonspecific RNA-binding activity, negatively and proportionally, affect Gag-Gag interaction and virion assembly. Mutations in the zinc fingers do not affect NC's nonspecific RNA-binding activity and have minor effects on Gag-Gag interaction and virion assembly. These results support the hypothesis that NC's nonspecific RNA-binding activity is required for Gag-Gag interaction and provides a driving force for virion assembly.
MATERIALS AND METHODS
Plasmid DNA constructs.
HIV-1 proviral DNAs containing mutations in NC basic residues (41) were obtained from Anna Aldovini. Fragments encompassing these mutations, as well as the corresponding region from the wild type, were transferred from the HXB2 provirus into NL4-3 proviral DNA as described below. An SphI/EcoRI fragment from pNL4-3 (nucleotides 1443 to 5743 [38]) was inserted into the corresponding sites of pUC19 (pUC19NL4-3). An SphI/EcoRV fragment (nucleotides 1443 to 2977) encompassing the NC coding sequence was then transferred from HXB2 into pUC19NL4-3. The entire SphI/EcoRI fragment was then cloned back into pNL4-3. Proviral DNAs containing mutations in the Cys-His boxes of HIV-1 NC (18) were obtained from Heinrich Göttlinger.
The engineering of bacterial expression constructs encoding the glutathione S-transferase (GST)–NC fusion protein and the hemagglutinin (HA)-tagged Gag polyprotein has been described (11, 13).
DNA constructs for eukaryotic expression of myristylation-deficient Gag polyproteins were engineered as follows. A fragment encoding the entire Gag polyprotein (nucleotides 789 to 2292) with an HA epitope at its N terminus was excised from pBSHAGagX (13) using restriction enzymes NcoI and XhoI and cloned into the corresponding sites of pEF/myc/cyto (Invitrogen). The gag sequence in pBSHAGagX contains multiple conservative mutations that act at the RNA level to render gag expression Rev independent (46). Fusion of the HA epitope at the N terminus of the Gag polyprotein prevents Gag myristylation and therefore blocks release of Gag virions from cells. DNA fragments encompassing NC mutations were generated by PCR using the mutant proviral DNAs as templates and oligonucleotides 5′-GCGCCTGCAGAATGGGATAGATTGCATCCA-3′ and 5′-GCGCGCTCGAGTTATTGTGACGAGGGGTCG-3′. PCR products were cloned as PstI/XhoI fragments into the same sites of pEFHA-Gag, and their identities were confirmed by dideoxy sequencing.
Cell lines.
The human lymphocyte line Jurkat (52) was maintained in RPMI 1640 supplemented with 10% fetal bovine serum. Human 293T and HeLa fibroblasts were maintained in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum.
Replication assay.
Viral infections were initiated in 106 Jurkat cells by DEAE-dextran (250 μg/ml; Pharmacia Biotech Inc., Piscataway, N.J.) using 2 μg of proviral DNA in 1 ml of serum-free RPMI 1640 for 20 min at room temperature. Cells were then washed in serum-free medium and resuspended in 3 ml of conditioned medium. Every 2 days supernatant was harvested and frozen and cells were passaged. At the conclusion of the experiment the stored samples were analyzed for reverse transcriptase (RT) activity as described below.
Exogenous RT assay.
Ten microliters of cell culture supernatant was added to 50 μl of RT cocktail (60 mM Tris-HCl [pH 8.0], 180 mM KCl, 6 mM MgCl2, 6 mM dithiothreitol, 0.6 mM EGTA, 0.12% Triton X-100, 6 μg of oligo[dT]/ml, 12 μg of poly[rA]/ml, 0.05 mM [α-32P]dTTP [800 Ci/mmol]) for 1 h at 37°C. Two microliters was spotted onto DE-81 paper and washed three times with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) (43). A phosphorimager (Molecular Dynamics, Sunnyvale, Calif.) was used to quantitate the radioactivity incorporated.
Metabolic labeling and immunoprecipitation.
HeLa cells in 35-mm-diameter plates were transfected with proviral DNAs using calcium phosphate as previously described (11). Forty-eight hours posttransfection, cells were incubated for 1 h at 37°C with 2 ml of DMEM lacking methionine and cysteine prior to a 45-min pulse with 100 μCi of [35S]Met-[35S]Cys (Translabel; ICN). Cells were washed with phosphate-buffered saline (PBS), incubated with complete DMEM, and lysed 0, 1, 3, and 6 h later in radioimmunoprecipitation assay (RIPA) buffer (150 mM NaCl, 1% NP-40, 0.5% deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 50 mM Tris-Cl, pH 8.0). Virions were purified from the supernatant by ultracentrifugation for 2 h at 80,000 × g through a cushion of 25% (wt/vol) sucrose; the pellet was resuspended in RIPA buffer. Cell lysate- and virion-associated fractions were incubated with 100 μl of protein A-Sepharose beads (Sigma; 10% slurry in RIPA buffer) for 1 h at 4°C. Supernatant was removed from the beads and incubated with 25 μg of total immunoglobulin from an HIV-1-infected individual (sera were obtained through the AIDS Research and Reference Reagent Program; catalog no. 3957) for 2 h at 4°C. Protein A-Sepharose beads (100 μl) were then added for 1 h at 4°C. Beads were washed three times, and proteins bound to the beads were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) and phosphorimager quantification.
Virion purification.
293T cells or HeLa cells were transfected (see above) with Gag expression plasmids or with complete proviral DNAs. At 72 h posttransfection, culture supernatant was filtered with a syringe (0.45-μm pore size), layered on 2 ml of 25% (wt/vol) sucrose, and accelerated at 80,000 × g for 2 h. Pelleted virions were resuspended in the appropriate buffer and analyzed.
For linear sucrose density analysis, virions pelleted as described above were resuspended overnight in 200 μl of RPMI 1640 on ice. Virions were then loaded on a linear sucrose gradient (20 to 60% [wt/vol]) and accelerated at 80,000 × g for 24 h. Fractions were harvested, and exogenous RT activity and solution density were measured.
Analysis of HIV-1 virion morphology by electron microscopy.
293T cells were transfected with HIV-1 proviral DNAs. Cells were fixed 72 h posttransfection with freshly made 2.5% glutaraldehyde in phosphate buffer (pH 7.0). Cells were postfixed in 1% osmium tetroxide and then embedded in Epon. Poststaining was done with 1% uranyl acetate. Sections were cut approximately 60 nm thick to accommodate the volume of the core structure parallel to the section plane. Specimens were analyzed with a Zeiss CEM 902 electron microscope, equipped with a spectrometer to enhance image contrast, at an accelerating voltage of 80 kV. A liquid nitrogen cooling trap on the specimen holder was used throughout. For each mutant, a series of electron micrographs (200 to 500 virions) was used for the statistical evaluation of the different morphologies present in the sample. The only exception was M1-2/BR for which only 20 particles were identified.
Protein expression and in vitro protein-binding experiments.
Proteins were expressed in Escherichia coli strain BL21(DE3)LysS (Novagen), as previously described (34). Cells were lysed in a solution consisting of 20 mM HEPES (pH 6.8), 150 mM potassium acetate, 2 mM magnesium acetate, 2 mM dithiothreitol, 0.1% Casamino Acids, 1% Tween 20, and 1 mM phenylmethylsulfonyl fluoride. In vitro GST pull-downs were performed as previously described (34) with the above-mentioned buffer. The quantity of bound protein was determined by densitometric analysis of the signals obtained after Western blotting.
Antibodies and Western blot analysis.
Murine monoclonal anti-HA antibody was purchased from Berkeley Antibody Company (Berkeley, Calif.). Rabbit polyclonal anti-cyclophilin A antibody was purchased from Affinity BioReagents (Golden, Colo.). Western blot analysis was performed as previously described (34).
In vitro RNA-binding assay.
GST-NC fusion proteins were immobilized onto glutathione-agarose beads (Sigma) and washed in binding buffer (50 mM Tris-HCl [pH 8.0], 50 mM NaCl, 10 μM NaCl2). In vitro-transcribed 32P-labeled RNA (4 × 104 cpm; specific activity, 5 × 105 to 6 × 105 cpm/μg) was added to immobilized GST-NC proteins for 30 min at 4°C in a total volume of 200 μl of binding buffer. After the washing, the radioactivity present in the samples was determined by direct counting using a beta counter (Beckman; L56000SC). Proteins bound to the beads were solubilized in SDS gel loading buffer and analyzed by Coomassie staining.
In vitro RNA transcription.
RNA was made with a kit (mMESSAGE-mMACHINE; Ambion) according to the manufacturer's directions. Two DNA templates were used. For synthesis of “nonspecific RNA” pBSKSII− (Stratagene) was linearized with XhoI and transcribed using T7 polymerase. The product was a linear 110-nucleotide RNA, encompassing sequences from the plasmid polylinker. To produce an RNA that encompasses the HIV-1 packaging sequence (nucleotides 455 to 788), pGL-leader-luciferase (11) was linearized with XhoI and transcribed with T7 polymerase.
Dot blot analysis.
Virions produced by calcium phosphate transfection of 293T cells were purified by ultracentrifugation through 25% (wt/vol) sucrose as described above and resuspended in PBS. Virions were normalized by exogenous RT activity and transferred to a nylon membrane using a dot blot apparatus (Bio-Rad). The membrane was incubated overnight at 42°C in 10% polyethylene glycol–1.5× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7])–7% SDS–100 μg of salmon sperm DNA/ml, with a 32P-end-labeled DNA oligonucleotide (5′-CTGACGCTCTCGCACCC-3′; antisense nucleotides 808 to 792 from pNL4-3) that hybridizes with HIV-1 genomic RNA (36). The membrane was washed in 0.1% SDS–0.2× SSC and exposed for phosphorimager analysis.
In vivo complementation assay.
293T fibroblasts were cotransfected with DNAs encoding a myristylation-deficient HA-Gag polyprotein and a complete NL4-3 provirus. The NL4-3 construct bears the protease-inactivating mutation D25R (49) and is wild type with respect to its gag sequence. NC mutations were expressed in the context of HA-Gag. Virions in the supernatant were purified by ultracentrifugation 48 h posttransfection, as described above. Pellets were resuspended in PBS, normalized by exogenous RT activity, and analyzed by Western blotting.
Analysis of intracellular detergent-resistant complexes.
HeLa cells were transfected by calcium phosphate with proviral DNAs in 35-mm-diameter plates. Forty-eight hours posttransfection, cells were incubated for 1 h at 37°C with 2 ml of DMEM lacking methionine and cysteine. Cells were pulsed for 2 h with 100 μCi of [35S]Met-[35S]Cys (Translabel; ICN). Cells were washed once with PBS and lysed in PBS containing 1% Triton X-100 and 1 mM phenylmethylsulfonyl fluoride for 4 h on ice. The soluble fraction was harvested from the plate and spun on a tabletop centrifuge for 5 min at 3,000 rpm, and the supernatant was loaded on a linear sucrose gradient (20 to 60% [wt/vol]) for 24 h at 80,000 × g. Thirteen fractions were harvested from the top of the gradient. The density of each fraction was measured. Proteins in each fraction were immunoprecipitated with serum from an HIV-1-infected individual as described above. Samples were subjected to SDS-PAGE, and gels were dried and analyzed by phosphorimager analysis.
RESULTS
Mutations in HIV-1 NC basic residues impair viral replication.
To determine the role of HIV-1 NC basic residues in virion assembly, NC mutants were selected for a study in which 1, 2, 4, 6, or 10 basic amino acids were replaced with alanine (Fig. 1). The engineering of these mutants has been described previously (41).
FIG. 1.
(A) Schematic representation of the HIV-1 Gag polyprotein showing the major domains. Vertical lines, positions of viral protease cleavage sites. (B) Amino acid sequence of HIV-1 NC. The sequences of the mutants used in this study are shown. The names of the mutant NC proteins are on the left. Dashes indicate amino acid residues identical to those of the wild type (WT).
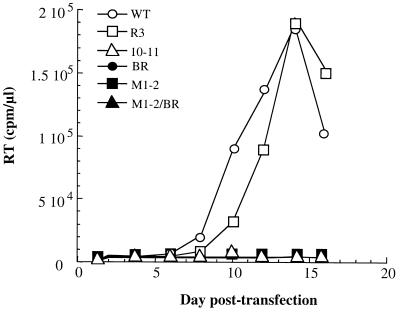
The effect of the NC mutations on viral replication was examined first (Fig. 2 and Table 1). Infections were initiated in T-cell lines by DEAE-dextran transfection of proviral DNAs bearing the NC mutations. Cells were passaged every 2 days, and supernatant was collected. Virus spread through the culture was detected by measuring exogenous RT activity in the supernatant of transfected cells.
FIG. 2.
HIV-1 replication kinetics following transfection of wild-type (WT) or NC mutant proviral DNAs into the Jurkat T-cell line. The accumulation of RT activity in the cell culture supernatant is shown for the indicated day posttransfection.
TABLE 1.
Summary of results obtained with HIV-1 NC mutants
| NC species | Infectivity | Level (% wild type) of:
|
|||
|---|---|---|---|---|---|
| Genomic RNA binding | Nucleic acid bindinga | Gag-Gag interactiona | Virion assembly | ||
| Wild type | + | 100 | 100 | 100 | 100 |
| R3 | + | 45 | 95 | 98 | 70 |
| 10-11 | − | 22 | 50 | 69 | 33 |
| BR | − | 19 | 26 | 56 | 12 |
| M1-2 | − | 4 | 17 | 31 | 5 |
| M1-2/BR | − | 2 | 8 | 5 | 4 |
As determined in vitro.
Substitution of one basic amino acid (mutant R3) had no significant effect on the kinetics of RT accumulation in the supernatant of transfected Jurkat T cells, compared to that of the wild type. No increase above background RT activity was detected with mutants in which 2, 4, 6, or 10 basic amino acids were replaced with alanine, indicating that each of these mutations abrogates HIV-1 replication. Identical results were obtained with each of the same viruses when replication assays were performed with H9 T cells (data not shown).
Mutations in HIV-1 NC basic residues impair virion assembly.
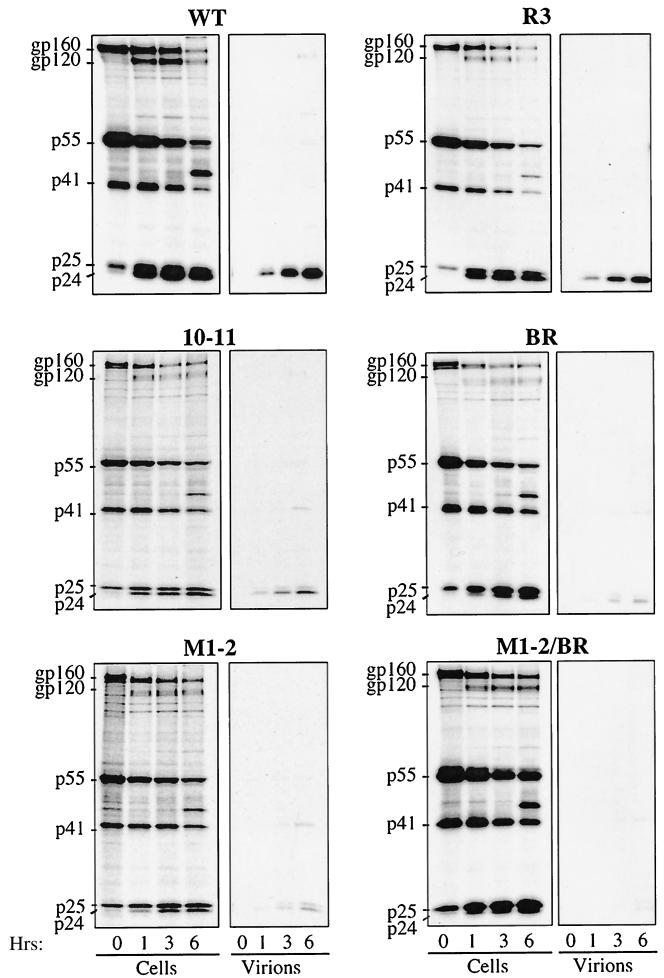
To identify the step in the retroviral replication cycle that was disrupted by the HIV-1 NC mutations, we began by examining virion assembly. Proviral DNAs were transfected into HeLa cells. Viral proteins were metabolically labeled with [35S]methionine and [35S]cysteine for 45 min and chased with cold medium for 0, 1, 3, and 6 h (Fig. 3). Cells were lysed, and viral proteins were immunoprecipitated using serum from an HIV-1-infected individual. Cell-free, virion-associated proteins were purified by ultracentrifugation through 25% sucrose prior to immunoprecipitation. Samples were processed by SDS-PAGE, and signal intensity was quantitated by phosphorimager analysis. For each mutant the amount of particulate protein released was normalized to the wild-type amount using the amount of cell-associated viral protein labeled at time zero.
FIG. 3.
Pulse-chase analysis of HIV-1 NC mutants. HeLa cells transfected with the indicated NL4-3 proviral DNAs were metabolically labeled with [35S]Met-[35S]Cys for 45 min and chased with unlabeled media for 0, 1, 3, and 6 h, as indicated. Virion-associated proteins were purified by ultracentrifugation through 25% sucrose. Virion- and cell-associated proteins were immunoprecipitated using sera from an HIV-1-infected individual and analyzed by SDS-PAGE. The mobilities of the envelope glycoprotein precursor (gp160), surface envelope protein (gp120), Pr55Gag precursor (p55), incompletely processed Gag precursors (p41 and p25), and completely processed CA (p24) are indicated on the left. WT, wild type.
Mutant R3 had assembly kinetics identical to that of the wild type, consistent with its normal replication in Jurkat cells. In contrast, mutants 10-11, BR, M1-2, and M1-2/BR showed defects in virion release that increased in severity as the number of basic residues replaced with alanine increased (Fig. 3 and Table 1). At the 6-h time point in the pulse-chase, mutant 10-11 had produced 33% as much particle-associated protein as the wild type. The corresponding value for mutant BR was 12%, and those for mutants M1-2 and M1-2/BR were 5 and 4%, respectively. Impairment of virion release was accompanied by a proportional increase in the level of viral protein that accumulated in the cell-associated fraction, arguing against an effect of NC mutations on protein stability. As additional support for this conclusion, pulse-chase analysis of mutants expressed in the context of a myristylation-deficient Gag polyprotein, in which the Gag molecules produced are retained inside the cell, revealed no difference between the stability of mutant proteins and that of the wild type (data not shown). Also, released mutant particles were as stable as the wild type, as judged by virion resistance to detergent or to heat (data not shown).
In addition to disrupting particle production, mutants 10-11, BR, M1-2, and M1-2/BR disrupted the efficiency of Gag polyprotein processing by the viral protease. This was evident from the abnormal accumulation of incompletely processed intermediates p41 (composed of MA-CA-SP1) and p25 (composed of CA-SP1) in both cell-associated and virion-associated material. As with the virion assembly defect, the magnitude of the protease processing defect was greatest with mutant M1-2/BR and was less severe with the remaining mutants in the following order: M1-2 > BR > 10-11 (Fig. 3). The defect in protease processivity was not due to decreased incorporation of the Gag-Pol polyprotein into virions; a normal ratio of virion-associated Gag-Pol polyprotein to Gag polyprotein was demonstrated by Western blotting and by determining the ratio of CA (by enzyme-linked immunosorbent assay) to exogenous RT activity (data not shown). The serum used to immunoprecipitate viral proteins does not recognize NC. The production of NC and the processing of its precursors were therefore monitored by Western blotting with a polyclonal anti-NC antibody; no significant differences were observed when the mutants were compared with the wild type (data not shown).
Effect of HIV-1 NC basic residue mutations on virion morphology.
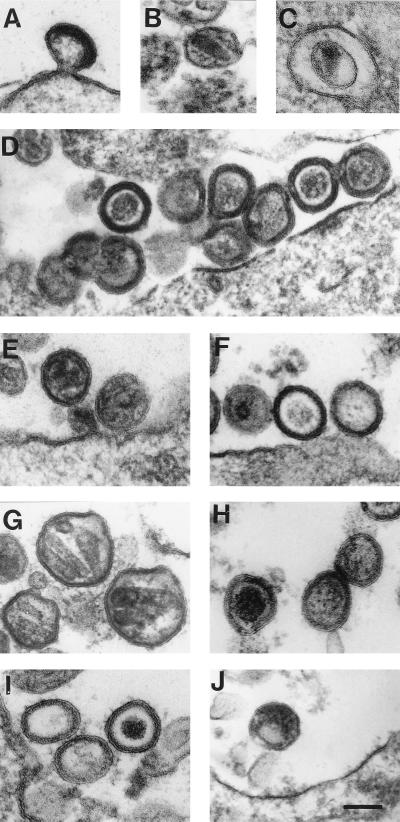
The effect of basic residue mutations on virion structure was examined by electron microscopy. Cells transfected with wild-type provirus produced mature, budding particles with typical cone-shaped, high-density core structures (Fig. 4A and B). Particles produced by mutant R3 had a morphology similar to that of the wild type (Fig. 4C and D), although 56% of the virions had an immature morphology with polyproteins tightly packed inside the envelope. Particles seen with mutant 10-11 were largely immature (79%) with a rim of high-density material inside the envelope (Fig. 4E and F).
FIG. 4.
Analysis of HIV-1 NC mutant virion morphology. 293T cells were transfected with proviral DNAs, either wild type (A and B), mutant R3 (C and D), mutant 10-11 (E and F), mutant BR (G and H), mutant M1-2 (I), or mutant M1-2/BR (J). Cells were fixed, stained, embedded, and visualized by electron microscopy. Bar, 100 nm.
The virion structures produced by mutant BR were more complex (Fig. 4G and H). Fifty-six percent of the particles were of immature morphology, 17% of the particles had a dense, globular core structure in the center, and 4% of the particles were of approximately twice the normal size and contained two or three elongated core structures within the envelope (Fig. 4G). The shape of these viral core structures was that of a cylinder or a cone of low density. Particles with a tubular core structure (2%) or with two circular core structures (6%), either with low or high density and probably sectioned perpendicular to the core structure, were also observed. Eighty-one percent of the particles produced by mutant M1-2 showed immature morphology, while 10% of the particles showed a dense, globular core structure in the center of the virion (Fig. 4I). Additionally, bigger particles, carrying one to three tubular core structures within the envelope, were seen in 3% of the particles examined (data not shown). Virion particles were detected at very low frequency with mutant M1-2/BR, but an irregular, dense core structure was occasionally observed (Fig. 4J). With none of the mutants did we detect accumulation of intracellular virions, indicating that the mutants disrupt virion assembly rather than virion release.
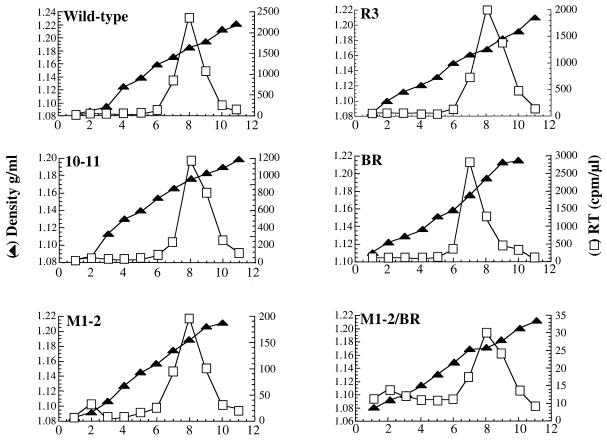
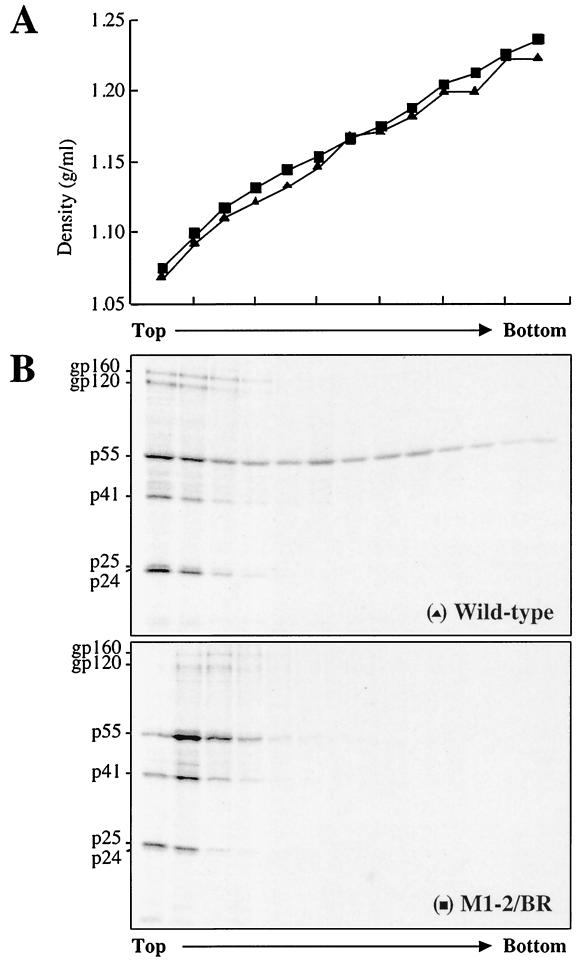
Virion density is not altered in HIV-1 NC mutants.
Since electron microscopy revealed alterations in the structure of the mutant virions, experiments were performed to determine if virion density was altered. Virions produced by transfection of 293T cells were loaded onto linear sucrose density gradients (20 to 60%) and accelerated at 80,000 × g for 24 h. Twelve fractions were collected, and solution density and exogenous RT activity were determined for each (Fig. 5). The densities of all mutant virions were determined to be 1.16 to 1.18 g/ml, within the normal limit for retroviral virions. Similar results were obtained with virions produced from HeLa cells and when virions were quantitated by p24 (data not shown).
FIG. 5.
Determination of HIV-1 NC mutant virion density. Virions produced by transfection of the indicated proviral DNAs into 293T cells were purified, concentrated, and layered onto a linear sucrose gradient (20 to 60%). Eleven fractions (abscissa) were collected from the top of the gradient. Left ordinate, fraction density; right ordinate, exogenous RT activity in each fraction.
Mutations in HIV-1 NC basic residues impair virion incorporation of viral genomic RNA.
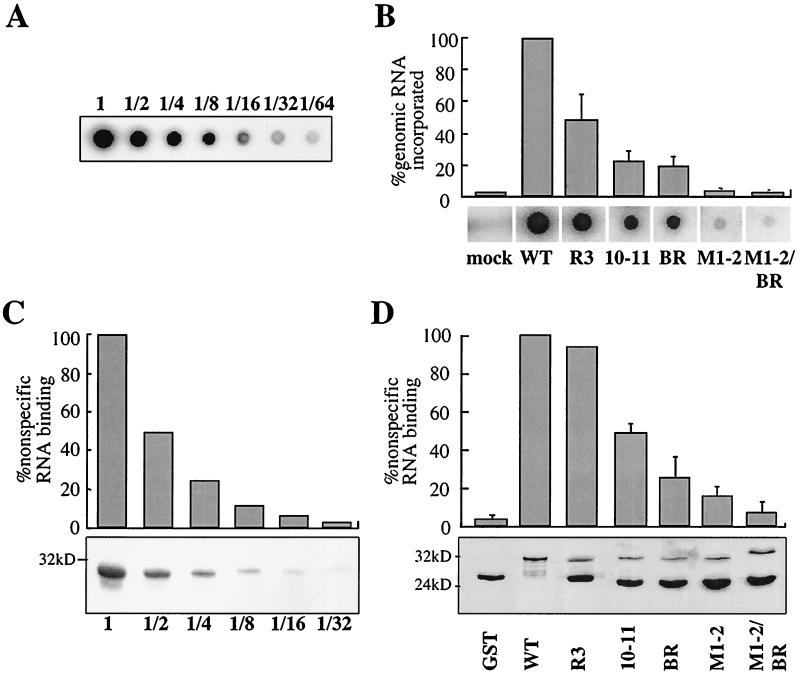
Two different assays were performed to test the ability of the NC mutants to interact with nucleic acid. In the first assay we determined the quantity of viral genomic RNA packaged into mutant virions as a measure of NC's ability to interact with RNA in vivo (Fig. 6A and B and Table 1). Virions produced by transfection of proviral DNAs into 293T cells were purified by ultracentrifugation through 25% sucrose. The quantities of particles produced by the wild type and by the different mutants were normalized to each other by measuring exogenous RT activity. Virions were blotted on a nylon membrane and probed with a 32P-labeled oligonucleotide specific for viral genomic RNA. A strong signal was obtained with wild-type virions (Fig. 6A). With twofold dilutions of the wild-type sample a linear decrease in signal was observed (Fig. 6A). The signal obtained after hybridization was not due to RNA from lysed cells or from transfected plasmid DNA since supernatant pelleted from cells transfected with a provirus encoding an assembly-defective virus gave no signal (Fig. 6B, mock lane). When NC mutant virions were tested, a proportional decrease in viral genomic RNA incorporation as more basic residues were replaced with alanine was observed. Compared to that for the wild type, the reduction in signal ranged from 45% for mutant R3 to 2% for mutant M1-2/BR (Fig. 6B).
FIG. 6.
RNA-binding activity of HIV-1 NC mutants. (A) Twofold dilutions of purified wild-type HIV-1 virions were loaded onto a nylon membrane and probed with a 32P-labeled DNA oligonucleotide specific for viral genomic RNA. (B) Wild-type (WT) and NC mutant virions were purified, normalized by exogenous RT activity, and subjected to dot blot analysis as for panel A. Results are presented as percentages of wild-type virus activity. The graph presents means from three independent experiments with standard errors of the means. Primary data from a representative experiment are shown underneath. Mock, “virion preparation” from cells transfected with a myristylation-deficient NL4-3. (C) GST protein fused to wild-type NC was expressed in bacteria. After serial twofold dilutions, the protein was purified using glutathione-agarose beads and bound in solution to 32P-labeled, nonspecific RNA. After a washing, RNA that remained bound was quantitated directly in a beta counter. Bound proteins were visualized by Coomassie staining after SDS-PAGE (bottom). (D) NC basic-residue mutants were expressed as GST fusion proteins and analyzed as described for panel C. RNA-binding activity is indicated as a percentage of the wild type. The graph presents means from three independent experiments with standard errors of the means. The bound fusion proteins were visualized by Coomassie staining after SDS-PAGE (bottom).
Mutations in HIV-1 NC basic residues impair NC's nonspecific RNA-binding activity.
An in vitro assay was established to determine if the NC mutations disrupted nonspecific binding to RNA. Each NC mutant was expressed as a GST fusion protein and immobilized on glutathione-agarose beads. Beads with bound protein were washed and resuspended in a solution containing 32P-labeled RNA that was transcribed in vitro using pBluescript as the template. After extensive washing, radioactivity that remained associated with the beads was counted as an indication of the quantity of RNA bound to NC.
A strong signal was obtained with wild-type GST-NC (Fig. 6C). The signal decreased in a linear fashion when decreasing amounts of GST-NC were bound to the beads. Compared to that of wild-type GST-NC, a decrease in RNA-binding ability was observed with the NC mutants as an increasing number of basic residues were replaced with alanine (Fig. 6D and Table 1). The disruptive effect of the NC mutations was confirmed in Northwestern and Southwestern assays using either 32P-labeled RNA or DNA probes, respectively (data not shown). The disruption of RNA binding by the NC mutants was not explained by differences in the loading of mutant proteins, as shown by Coomassie staining of SDS-PAGE gels (Fig. 6D, bottom).
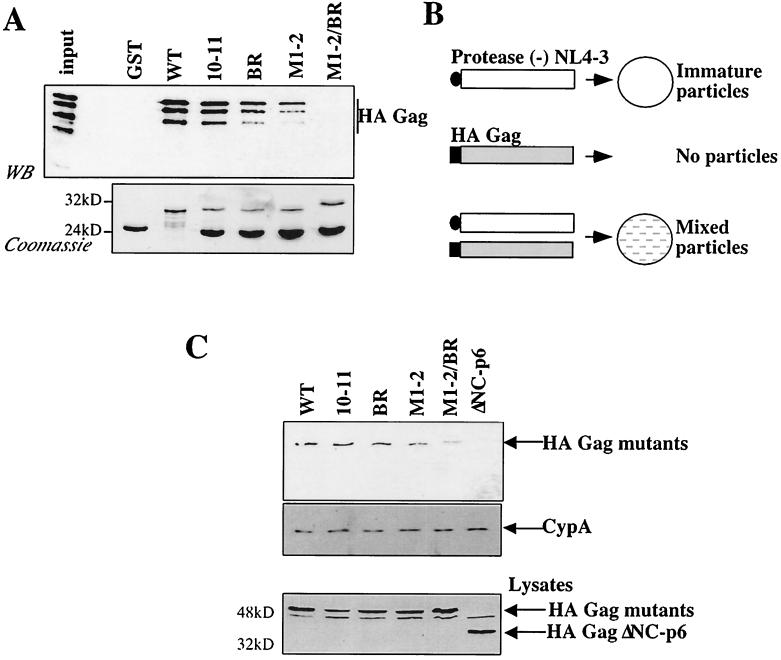
HIV-1 NC basic residues are required for Gag-Gag interaction.
The effect of the HIV-1 NC mutants on the Gag-Gag interaction was tested using two different assays. First, GST-NC mutant proteins were assayed for the ability to interact in vitro with full-length HA-tagged Gag (Fig. 7A). We have previously characterized the interaction between Gag and NC in vitro and have shown that the interaction is dependent on RNA (7). GST-NC fusion proteins were immobilized onto glutathione-agarose beads and incubated with a lysate of bacteria expressing full-length HA-Gag. Samples were washed, and, after SDS-PAGE, bound proteins were detected by Western blotting using an anti-HA antibody (Fig. 7A, top) and by Coomassie staining (Fig. 7A, bottom). The greater the number of basic residues that were mutated to alanine, the poorer was the ability of NC to bind HA-Gag. The R3 mutation, with only a single residue mutated, had no appreciable effect on the Gag-Gag interaction (data not shown and Table 1). The lysate of bacteria expressing HA-Gag shows four major bands resulting from C-terminal truncation. Previous analysis demonstrated that only the three lower-mobility bands retain the complete NC domain (35). Interestingly, these three bands and not the fourth, which lacks NC, bind to GST-NC.
FIG. 7.
NC basic-residue mutants disrupt Gag-Gag interaction. (A) NC mutants were expressed as GST fusion proteins in bacteria, immobilized onto glutathione-agarose beads and incubated with a lysate from bacteria expressing HA-Gag. After a washing, proteins bound to the beads were boiled, processed by SDS-PAGE, and analyzed by Western blotting (WB) with an anti-HA antibody (top) or by Coomassie staining (bottom). The input lane shows 10% of the total HA-Gag bacterial lysate used in the binding reactions. The positions of migration of molecular mass markers (in kilodaltons) are indicated on the left of the Coomassie gel. WT, wild type. (B) Schematic representation of in vivo complementation assay. Expression of protease-defective HIV-1 provirus (white rectangle) by transfection of 293T cells leads to the release of immature particles in the supernatant. HA-Gag (gray rectangle) produced by transfection does not release particles because this protein is not myristylated. Coexpression of protease-defective HIV-1 and HA-Gag in the same cell produces mixed particles if HA-Gag is rescued via interaction with the provirally encoded Gag. (C) HA-Gags, wild type or bearing the NC mutations indicated, were coexpressed with protease-defective HIV-1 provirus. Particles purified from the supernatant were normalized by exogenous RT and analyzed by Western blotting using anti-HA (top) or anti-cyclophilin A (middle) antibodies. Transfected cell lysates were analyzed by Western blotting using anti-HA (bottom). A Gag polyprotein with a deletion of the NC and p6 domains (ΔNC-p6) was included as a negative control. The positions of migration of the proteins are on the right.
The effect of the NC mutants on the Gag-Gag interaction was then tested with an in vivo complementation assay (Fig. 7B). This assay relies on the fact that Gag must be myristylated in order to associate with the plasma membrane and produce virions. A myristic acid-deficient Gag polyprotein was produced by exchanging Gag's amino-terminal glycine with an HA epitope tag. This protein cannot produce virions by itself but can be rescued into virions via association with wild-type Gag when the two proteins are expressed within the same cell. The amount of HA-Gag rescued into virions is therefore a measure of the ability of the two proteins to interact with each other. Similar complementation assays have been described previously (2, 5, 7). In our assay, the wild-type Gag polyprotein was expressed from a protease-deficient, though otherwise complete, provirus; the NC mutants were expressed in the context of an HA-tagged Gag expression construct. The use of a complete provirus as a source of wild-type Gag allowed us to use exogenous RT activity to quantify virion release. A protease-deficient provirus was chosen since the NC mutants disrupt protease processing to different degrees (Fig. 3) and this would have complicated analysis of the data.
Virions produced by 293T cells that had been cotransfected with the two plasmids were purified by ultracentrifugation through 25% sucrose. Samples were normalized by exogenous RT activity and analyzed by Western blotting with an anti-HA antibody (Fig. 7C, top). HA-Gag bearing a wild-type NC domain was rescued efficiently into virions. Rescue into virions of HA-Gag lacking the entire NC and p6 domains (ΔNC-p6) could not be detected under the conditions used here. As an increasing number of NC basic residues were replaced with alanine, the HA-Gags showed a progressive defect in their abilities to be rescued into virions by wild-type Gag. In contrast, incorporation of cyclophilin A, a cellular protein that binds the CA domain (6, 34), was unaffected by the presence of the NC mutations (Fig. 7C, middle). Differences in the efficiency of mutant HA-Gag incorporation into virions could not be explained by instability of the mutant proteins as judged by Western blotting (Fig. 7C, bottom) or by pulse-chase analysis (data not shown).
Intracellular, detergent-resistant Gag complexes are disrupted by basic-residue mutations.
Detergent-resistant, intracellular complexes of the HIV-1 Gag polyprotein have been observed in virus-producing cells (31). Deletion of NC abolishes their formation (30), suggesting that these complexes are dependent on the Gag-Gag interaction. To determine if mutation of basic residues disrupts the formation of these complexes, HeLa cells were transfected with proviral DNAs, those of either the wild type or mutant M1-2/BR. Cells were pulsed with [35S]methionine and [35S]cysteine for 2 h and incubated for 4 h in the presence of 1% Triton X-100. The soluble fraction was loaded onto a linear sucrose gradient (20 to 60%) and accelerated at 80,000 × g for 24 h. Thirteen fractions were collected from the top of the gradient. Density was measured for each fraction prior to immunoprecipitation of viral proteins using sera from an HIV-1-infected individual (Fig. 8A). For both wild-type and mutant viruses, Env protein localized at the top of the gradient. For the wild-type virus, unprocessed Gag polyprotein was found in low-density fractions but a significant portion also formed complexes of higher density that extended the length of the gradient. With the M1-2/BR mutant the Gag polyprotein complexes were found only in the lower-density fractions, indicating that, by disrupting the Gag-Gag interaction, this mutant impaired the formation of intracellular complexes (Fig. 8B).
FIG. 8.
NC basic residues are required for the formation of intracellular, detergent-resistant Gag complexes. HeLa cells were transfected with either wild-type or M1-2/BR-bearing proviruses as indicated. Proteins were metabolically labeled for 2 h, and cells were lysed in 1% Triton X-100. The cytoplasmic fraction was loaded onto a linear sucrose gradient (20 to 60%) and accelerated at 80,000 × g for 24 h. Thirteen fractions were harvested from top to bottom as indicated. The solution density of each fraction was measured (A). Viral proteins were immunoprecipitated, processed by SDS-PAGE, and detected with a phosphorimager (B).
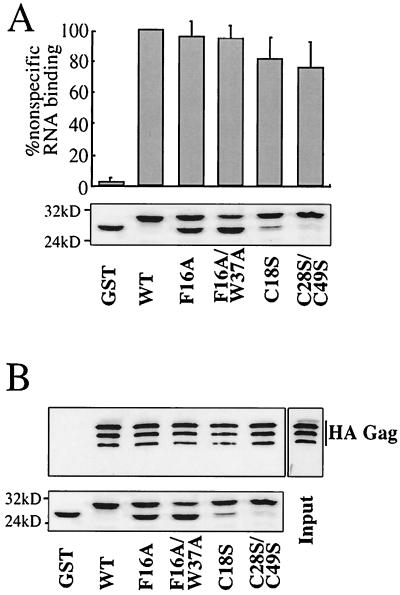
HIV-1 NC zinc fingers are dispensable for nonspecific RNA binding activity and Gag-Gag interaction.
NC zinc fingers are required for packaging viral genomic RNA and for virion infectivity, but, unlike the NC basic residues, they are dispensable for virion assembly (for a review see reference 3). We therefore examined the effect of zinc finger mutants on our in vitro assays for nonspecific RNA-binding activity and for Gag-Gag interaction. Previously characterized zinc finger mutants (18) F16A, C18S, F16A/W37A, and C28/C49S were selected for comparison with the basic-residue mutants. Each of these mutations has been shown to have drastic effects on the packaging of viral genomic RNA into virions, with relatively modest effects on virion assembly (18) (data not shown).
GST-NC fusion proteins, either wild type or bearing one of the zinc finger mutations, were analyzed for the ability to bind to nonspecific RNA in vitro (Fig. 9A). Disruption of one (F16A, C18S) or two (F16/W37A, C28/C49S) zinc fingers had only a minor effect on the ability of these mutants to associate with RNA. These results contrast with the dramatic effect of the basic-residue mutations in this same assay (Fig. 6D).
FIG. 9.
Analysis of HIV-1 NC Cys-His box mutants. (A) The indicated HIV-1 NC Cys-His box mutants were expressed as GST fusion proteins in bacteria and immobilized onto glutathione-agarose beads. Binding of 32P-labeled RNA was assessed as described for Fig. 6. The graph presents means from three independent experiments with standard errors of the means. (B) GST-NC proteins bearing Cys-His box mutants were assayed for the ability to bind full-length HA-tagged Gag in vitro, as described in the legend of Fig. 7A. Samples were analyzed by Western blotting with an anti-HA antibody (top) or by Coomassie staining (bottom). The input lane shows 10% of the total HA-Gag bacterial lysate used in the binding reaction. The positions of migration of molecular mass markers (in kilodaltons) are indicated on the left of the Coomassie gel.
We then tested GST-NC fusion proteins bearing the zinc finger mutations for the ability to associate with HA-Gag in vitro. The GST-NC proteins were bound to glutathione-agarose beads and incubated in lysate from bacteria expressing HA-Gag. Proteins that remained bound to the beads after extensive washing were processed by SDS-PAGE and detected by Western blotting using an anti-HA antibody (Fig. 9B, top) or by Coomassie staining (Fig. 9B, bottom). None of the zinc finger mutations significantly impaired NC's association with HA-Gag. Again, these results are in dramatic contrast to the results obtained with the basic-residue mutations.
DISCUSSION
By analyzing the phenotype of a panel of HIV-1 NC basic-residue mutants we have obtained data in support of a model in which HIV-1 Gag polymerization and virion assembly are promoted by interaction of the NC domain with RNA. We demonstrated that the number of NC basic residues that are mutated correlates with the magnitude of the defects in virion assembly. The assembly defect was not due to decreased translation efficiency or decreased protein half-life. Some have reported the viral protease dependence of assembly defects associated with Gag mutants (26). We examined the effect of mutational inactivation of the viral protease on our mutants and saw no correction of the impairment in virion release (data not shown).
Defects in virion assembly due to NC basic-residue mutations have been reported by some (15) but not by others (41). Our results are contrary to those described by the latter group. Differences in the techniques used to study virion assembly may account for this discrepancy. Poon et al. (41) used Western blot analysis, a method that assays steady-state protein accumulation. We used pulse-chase analysis, which provides a detailed kinetic analysis of virion assembly. By using this technique, we clearly showed a defect in the assembly of NC basic-residue mutants. This defect was not cell line dependent, since the same results were observed using HeLa, COS, and 293T cells (data not shown).
In addition to being associated with the assembly defect, the NC mutations were associated with decreased processing of the Gag polyprotein by the viral protease. Consistent with the biochemical evidence for a protease processing defect, visualization of virus-producing cells by electron microscopy showed that a higher proportion of mutant virions exhibit immature morphology. Similar effects of NC basic-residue mutations on virion morphology have been reported (4, 39). The viral protease defect is not explained by differential incorporation of Gag versus Gag-Pol into mutant virions since the Gag/Gag-Pol ratio is normal (data not shown). Although it is possible that the protease processing defect results from decreased numbers of Gag and Gag-Pol molecules per virion, we believe that it results from decreased RNA-binding activity by the NC mutants. HIV-1 viral protease processing has been shown to be RNA dependent in vitro (47, 48), and in our mutants, the protease processivity defect is proportional to the number of basic residues mutated. Although the RNA dependence of viral protease processing has only been demonstrated with a p15 substrate (NC-p6) in vitro, this requirement may be applicable to other protease cleavage sites in vivo. RNA dependence may be most obvious at sites with low affinity for the viral protease, thus explaining why we observe the accumulation of the CA/SP1 intermediate in our mutants.
Our data show that NC basic residues mediate nonspecific RNA recognition in vitro and contribute to viral genomic RNA incorporation into virions. These findings provide a link between previous studies on the importance of NC basic residues for RNA binding in vitro (17, 29, 45) and in vivo (15). The defect in nucleic acid binding was proportional to the number of basic residues of NC replaced, indicating that the number of basic charges determines NC's ability to bind nonspecifically to RNA. Our results are in agreement with nuclear magnetic resonance data showing that NC basic residues associate nonspecifically with phosphodiester groups on RNA (16). However, our data do not exclude the possibility that specific basic residues make larger contributions than others to NC's nonspecific RNA-binding activity or to virion assembly.
Many groups have demonstrated that NC is required for virion assembly (10, 15, 24, 27). Evidence that NC's role in assembly is to promote Gag-Gag interaction has been provided by trans-complementation assembly assays (2), two-hybrid experiments (19, 51), ligand affinity blotting (10), GST pull-down assays with recombinant protein in vitro (7), and cysteine cross-linking (37). Our data show that basic residues distributed throughout NC contribute to virion assembly by promoting interactions among Gag molecules. This was demonstrated by showing a correlation between the number of NC basic residues replaced with alanine and defects in the Gag-Gag interaction in vitro and in vivo. In addition, mutations in the basic residues of NC disrupt the formation of intracellular, detergent-resistant Gag complexes. These complexes are formed shortly after protein synthesis and have high density, indicating that they are multimeric structures of the Gag polyprotein. Though these high-density complexes have not been proven to be assembly intermediates, we believe that by disrupting Gag polymerization, NC basic-residue mutations might directly prevent the formation of these complexes and the subsequent nucleation of nascent virions at the cell surface. Alternatively, by impairing the Gag-Gag interaction the NC mutations might interfere with Gag's ability to bind the plasma membrane, as suggested by others (40, 44), and thus block assembly.
Virions produced by NC deletion mutants have decreased density (2, 15). It has been proposed that the decreased density is due to the formation of virions processing fewer Gag molecules per virion, secondary to the weakened Gag-Gag interaction (50). In contrast to findings concerning the deletion mutants evaluated in those papers, we found here that virions produced by NC basic-residue mutants have a normal density. Since we have shown directly that the basic-residue mutants weaken the Gag-Gag interaction it appears that virion density is determined by some other property of Gag. Alternatively, the basic residues that remain in our most drastic mutant (M1-2/BR) may be sufficient for the minimal Gag-Gag interaction required to assemble virions of normal density.
Correlation between the effects of NC mutants on RNA binding and the effects on Gag-Gag interaction suggests that RNA is required for Gag polymerization. In fact, the NC assembly function can be performed instead by a heterologous RNA-binding protein (53). RNA promotes the formation of virion core structures from recombinant protein in vitro. RNase disrupts these structures (9) and disrupts the interaction between Gags in a GST pull-down as well (7).
RNA might promote Gag multimerization via any of a number of mechanisms. By binding to the basic residues in NC, RNA might neutralize charge repulsion between Gag polyprotein monomers. This would then permit protein-protein contacts involving NC or perhaps the CA-dimer interface (7, 22). Though this model is supported by the observation that virion cores assemble in vitro in the absence of RNA (albeit inefficiently) as long as the salt concentration is very high, no protein-protein contacts in the structure of HIV-1 NC bound to the HIV-1 SL3 RNA stem-loop were observed (16). This indicates that if direct protein-protein interactions do occur among Gag molecules, the contacts do not involve NC residues. We favor the hypothesis that RNA acts as a scaffold for Gag multimerization. The binding of Gag (via NC) to RNA would drive Gag accumulation and create a favorable environment for the formation of protein-protein contacts among CA and possibly MA domains, thus driving virion assembly.
ACKNOWLEDGMENTS
We thank Anna Aldovini, Heinrich Göttlinger, and Flossie Wong-Staal for generously providing plasmid DNAs. We thank Cagan Gurer for critical reading of the manuscript.
This work was supported by grant AI 41857 (J.L.) and by shared core facilities of the Columbia-Rockefeller Center for AIDS Research (P30 AI42848), both from the National Institutes of Health.
REFERENCES
- 1.Aldovini A, Young R. Mutations of RNA and protein sequences involved in human immunodeficiency virus type 1 packaging result in production of noninfectious virus. J Virol. 1990;64:1920–1926. doi: 10.1128/jvi.64.5.1920-1926.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bennett R P, Nelle T D, Wills J W. Functional chimeras of the Rous sarcoma virus and human immunodeficiency virus Gag proteins. J Virol. 1993;67:6487–6498. doi: 10.1128/jvi.67.11.6487-6498.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berkowitz R, Fisher J, Goff S P. RNA packaging. Curr Top Microbiol Immunol. 1996;214:177–218. doi: 10.1007/978-3-642-80145-7_6. [DOI] [PubMed] [Google Scholar]
- 4.Berthoux L, Pechoux C, Ottmann M, Morel G, Darlix J L. Mutations in the N-terminal domain of human immunodeficiency virus type 1 nucleocapsid protein affect virion core structure and proviral DNA synthesis. J Virol. 1997;71:6973–6981. doi: 10.1128/jvi.71.9.6973-6981.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowzard J B, Bennett R P, Krishna N K, Ernst S M, Rein A, Wills J W. Importance of basic residues in the nucleocapsid sequence for retrovirus Gag assembly and complementation rescue. J Virol. 1998;72:9034–9044. doi: 10.1128/jvi.72.11.9034-9044.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braaten D, Franke E K, Luban J. Cyclophilin A is required for an early step in the life cycle of human immunodeficiency virus type 1 prior to the initiation of reverse transcription. J Virol. 1996;70:3551–3560. doi: 10.1128/jvi.70.6.3551-3560.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burniston M T, Cimarelli A, Colgan J, Curtis S P, Luban J. Human immunodeficiency virus type 1 Gag polyprotein multimerization requires the nucleocapsid domain and RNA and is promoted by the capsid-dimer interface and the basic region of matrix protein. J Virol. 1999;73:8527–8540. doi: 10.1128/jvi.73.10.8527-8540.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campbell S, Rein A. In vitro assembly properties of human immunodeficiency virus type 1 Gag protein lacking the p6 domain. J Virol. 1999;73:2270–2279. doi: 10.1128/jvi.73.3.2270-2279.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campbell S, Vogt V M. Self-assembly in vitro of purified CA-NC proteins from Rous sarcoma virus and human immunodeficiency virus type 1. J Virol. 1995;69:6487–6497. doi: 10.1128/jvi.69.10.6487-6497.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carriere C, Gay B, Chazal N, Morin N, Boulanger P. Sequence requirements for encapsidation of deletion mutants and chimeras of human immunodeficiency virus type 1 Gag precursor into retrovirus-like particles. J Virol. 1995;69:2366–2377. doi: 10.1128/jvi.69.4.2366-2377.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cimarelli A, Luban J. Translation elongation factor 1-alpha interacts specifically with the human immunodeficiency virus type 1 Gag polyprotein. J Virol. 1999;73:5388–5401. doi: 10.1128/jvi.73.7.5388-5401.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clavel F, Orenstein J M. A mutant of human immunodeficiency virus with reduced RNA packaging and abnormal particle morphology. J Virol. 1990;64:5230–5234. doi: 10.1128/jvi.64.10.5230-5234.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Colgan J, Yuan H E H, Franke E K, Luban J. Binding of the human immunodeficiency virus type 1 Gag polyprotein to cyclophilin A is mediated by the central region of capsid and requires Gag dimerization. J Virol. 1996;70:4299–4310. doi: 10.1128/jvi.70.7.4299-4310.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Darlix J L, Lapadat-Tapolsky M, de Rocquigny H, Roques B P. First glimpses at structure-function relationships of the nucleocapsid protein of retroviruses. J Mol Biol. 1995;254:523–537. doi: 10.1006/jmbi.1995.0635. [DOI] [PubMed] [Google Scholar]
- 15.Dawson L, Yu X F. The role of nucleocapsid of HIV-1 in virus assembly. Virology. 1998;251:141–157. doi: 10.1006/viro.1998.9374. [DOI] [PubMed] [Google Scholar]
- 16.De Guzman R N, Wu Z R, Stalling C C, Pappalardo L, Borer P N, Summers M F. Structure of the HIV-1 nucleocapsid protein bound to the SL3 psi-RNA recognition element. Science. 1998;279:384–388. doi: 10.1126/science.279.5349.384. [DOI] [PubMed] [Google Scholar]
- 17.De Rocquigny H, Gabus C, Vincent A, Fournie-Zaluski M C, Roques B, Darlix J L. Viral RNA annealing activities of human immunodeficiency virus type 1 nucleocapsid protein require only peptide domains outside the zinc fingers. Proc Natl Acad Sci USA. 1992;89:6472–6476. doi: 10.1073/pnas.89.14.6472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dorfman T, Luban J, Goff S P, Haseltine W A, Gottlinger H G. Mapping of functionally important residues of a cysteine-histidine box in the human immunodeficiency virus type 1 nucleocapsid protein. J Virol. 1993;67:6159–6169. doi: 10.1128/jvi.67.10.6159-6169.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Franke E K, Yuan H E H, Bossolt K L, Goff S P, Luban J. Specificity and sequence requirements for interactions between various retroviral Gag proteins. J Virol. 1994;68:5300–5305. doi: 10.1128/jvi.68.8.5300-5305.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Freed E O. HIV-1 gag proteins: diverse functions in the virus life cycle. Virology. 1998;251:1–15. doi: 10.1006/viro.1998.9398. [DOI] [PubMed] [Google Scholar]
- 21.Fu X, Katz R A, Skalka A M, Leis J. Site-directed mutagenesis of the avian retrovirus nucleocapsid protein, pp12. J Biol Chem. 1988;263:2140–2145. [PubMed] [Google Scholar]
- 22.Gamble T R, Yoo S, Vajdos F F, von Schwedler U K, Worthylake D K, Wang H, McCutcheon J P, Sundquist W I, Hill C P. Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein. Science. 1997;278:849–853. doi: 10.1126/science.278.5339.849. [DOI] [PubMed] [Google Scholar]
- 23.Ganser B K, Li S, Klishko V Y, Finch J T, Sundquist W I. Assembly and analysis of conical models for the HIV-1 core. Science. 1999;283:80–83. doi: 10.1126/science.283.5398.80. [DOI] [PubMed] [Google Scholar]
- 24.Gheysen D, Jacobs E, de Foresta F, Thiriart C, Francotte M, Thines D, DeWilde M. Assembly and release of HIV-1 precursor Pr55gag virus-like particles from recombinant baculovirus-infected insect cells. Cell. 1989;59:103–112. doi: 10.1016/0092-8674(89)90873-8. [DOI] [PubMed] [Google Scholar]
- 25.Gross I, Hohenberg H, Krausslich H G. In vitro assembly properties of purified bacterially expressed capsid proteins of human immunodeficiency virus. Eur J Biochem. 1997;249:592–600. doi: 10.1111/j.1432-1033.1997.t01-1-00592.x. [DOI] [PubMed] [Google Scholar]
- 26.Huang M, Orenstein J M, Martin M A, Freed E O. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J Virol. 1995;69:6810–6818. doi: 10.1128/jvi.69.11.6810-6818.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jowett J B, Hockley D J, Nermut M V, Jones I M. Distinct signals in human immunodeficiency virus type 1 Pr55 necessary for RNA binding and particle formation. J Gen Virol. 1992;73:3079–3086. doi: 10.1099/0022-1317-73-12-3079. . (Erratum, 74:943, 1993.) [DOI] [PubMed] [Google Scholar]
- 28.Krausslich H G, Welker R. Intracellular transport of retroviral capsid components. Curr Top Microbiol Immunol. 1996;214:25–63. doi: 10.1007/978-3-642-80145-7_2. [DOI] [PubMed] [Google Scholar]
- 29.Lapadat-Tapolsky M, De Rocquigny H, Van Gent D, Roques B, Plasterk R, Darlix J L. Interactions between HIV-1 nucleocapsid protein and viral DNA may have important functions in the viral life cycle. Nucleic Acids Res. 1993;21:831–839. doi: 10.1093/nar/21.4.831. . (Erratum, 21:2024.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee Y M, Liu B, Yu X F. Formation of virus assembly intermediate complexes in the cytoplasm by wild-type and assembly-defective mutant human immunodeficiency virus type 1 and their association with membranes. J Virol. 1999;73:5654–5662. doi: 10.1128/jvi.73.7.5654-5662.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee Y M, Yu X F. Identification and characterization of virus assembly intermediate complexes in HIV-1-infected CD4+ T cells. Virology. 1998;243:78–93. doi: 10.1006/viro.1998.9064. [DOI] [PubMed] [Google Scholar]
- 32.Leis J, Jentoft J. Characteristics and regulation of interaction of avian retrovirus pp12 protein with viral RNA. J Virol. 1983;48:361–369. doi: 10.1128/jvi.48.2.361-369.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lever A, Göttlinger H, Haseltine W, Sodroski J. Identification of a sequence required for efficient packaging of human immunodeficiency virus type 1 RNA into virions. J Virol. 1989;63:4085–4087. doi: 10.1128/jvi.63.9.4085-4087.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luban J, Bossolt K A, Franke E K, Kalpana G V, Goff S P. Human immunodeficiency virus type 1 gag protein binds to cyclophilins A and B. Cell. 1993;73:1067–1078. doi: 10.1016/0092-8674(93)90637-6. [DOI] [PubMed] [Google Scholar]
- 35.Luban J, Goff S P. Binding of human immunodeficiency virus type 1 (HIV-1) RNA to recombinant HIV-1 gag polyprotein. J Virol. 1991;65:3203–3212. doi: 10.1128/jvi.65.6.3203-3212.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mak J, Khorchid A, Cao Q, Huang Y, Lowy I, Parniak M A, Prasad V R, Wainberg M A, Kleiman L. Effects of mutations in Pr160gag-pol upon tRNA(Lys3) and Pr160gag-pol incorporation into HIV-1. J Mol Biol. 1997;265:419–431. doi: 10.1006/jmbi.1996.0742. [DOI] [PubMed] [Google Scholar]
- 37.McDermott J, Farrell L, Ross R, Barklis E. Structural analysis of human immunodeficiency virus type 1 Gag protein interactions, using cysteine-specific reagents. J Virol. 1996;70:5106–5114. doi: 10.1128/jvi.70.8.5106-5114.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Myers G, Korber B, Wain-Hobson S, Jeang K-T, Henderson L E, Pavlakis G N. Human retroviruses and AIDS. Los Alamos, N.Mex: Los Alamos National Laboratory; 1994. [Google Scholar]
- 39.Ottmann M, Gabus C, Darlix J L. The central globular domain of the nucleocapsid protein of human immunodeficiency virus type 1 is critical for virion structure and infectivity. J Virol. 1995;69:1778–1784. doi: 10.1128/jvi.69.3.1778-1784.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Platt E J, Haffar O K. Characterization of human immunodeficiency virus type 1 Pr55gag membrane association in a cell-free system: requirement for a C-terminal domain. Proc Natl Acad Sci USA. 1994;91:4594–4598. doi: 10.1073/pnas.91.10.4594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poon D T, Wu J, Aldovini A. Charged amino acid residues of human immunodeficiency virus type 1 nucleocapsid p7 protein involved in RNA packaging and infectivity. J Virol. 1996;70:6607–6616. doi: 10.1128/jvi.70.10.6607-6616.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prats A C, Housset V, de Billy G, Cornille F, Prats H, Roques B, Darlix J L. Viral RNA annealing activities of the nucleocapsid protein of Moloney murine leukemia virus are zinc independent. Nucleic Acids Res. 1991;19:3533–3541. doi: 10.1093/nar/19.13.3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 44.Sandefur S, Varthakavi V, Spearman P. The I domain is required for efficient plasma membrane binding of human immunodeficiency virus type 1 Pr55Gag. J Virol. 1998;72:2723–2732. doi: 10.1128/jvi.72.4.2723-2732.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmalzbauer E, Strack B, Dannull J, Guehmann S, Moelling K. Mutations of basic amino acids of NCp7 of human immunodeficiency virus type 1 affect RNA binding in vitro. J Virol. 1996;70:771–777. doi: 10.1128/jvi.70.2.771-777.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schwartz S, Campbell M, Nasioulas G, Harrison J, Felber B K, Pavlakis G N. Mutational inactivation of an inhibitory sequence in human immunodeficiency virus type 1 results in Rev-independent gag expression. J Virol. 1992;66:7176–7182. doi: 10.1128/jvi.66.12.7176-7182.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sheng N, Erickson-Viitanen S. Cleavage of p15 protein in vitro by human immunodeficiency virus type 1 protease is RNA dependent. J Virol. 1994;68:6207–6214. doi: 10.1128/jvi.68.10.6207-6214.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sheng N, Pettit S C, Tritch R J, Ozturk D H, Rayner M M, Swanstrom R, Erickson-Viitanen S. Determinants of the human immunodeficiency virus type 1 p15NC-RNA interaction that affect enhanced cleavage by the viral protease. J Virol. 1997;71:5723–5732. doi: 10.1128/jvi.71.8.5723-5732.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith A J, Srinivasakumar N, Hammarskjold M L, Rekosh D. Requirements for incorporation of Pr160gag-pol from human immunodeficiency virus type 1 into virus-like particles. J Virol. 1993;67:2266–2275. doi: 10.1128/jvi.67.4.2266-2275.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Swanstrom R, Wills J W. Synthesis, assembly and processing of viral proteins. In: Coffin J M, Hughes S, Varmus H E, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 263–334. [PubMed] [Google Scholar]
- 51.Tanchou V, Gabus C, Rogemond V, Darlix J L. Formation of stable and functional HIV-1 nucleoprotein complexes in vitro. J Mol Biol. 1995;252:563–571. doi: 10.1006/jmbi.1995.0520. [DOI] [PubMed] [Google Scholar]
- 52.Weiss A, Wiskocil R, Stobo J. The role of T3 surface molecules in the activation of human T cells: a two stimulus requirement for IL-2 production reflects events occurring at a pretranslational level. J Immunol. 1984;133:123–128. [PubMed] [Google Scholar]
- 53.Zhang Y, Barklis E. Effects of nucleocapsid mutations on human immunodeficiency virus assembly and RNA encapsidation. J Virol. 1997;71:6765–6776. doi: 10.1128/jvi.71.9.6765-6776.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]