Abstract
Deep learning methods are a class of machine learning techniques capable of identifying highly complex patterns in large datasets. Here, we provide a perspective and primer on deep learning applications for genome analysis. We discuss successful applications in the fields of regulatory genomics, variant calling and pathogenicity scores. We include general guidance for how to effectively use deep learning methods as well as a practical guide to tools and resources. This primer is accompanied by an interactive online tutorial.
Deep learning has made impressive recent advances in applications ranging from computer vision to natural-language processing. This primer discusses the main categories of deep learning methods and provides suggestions for how to effectively use deep learning in genomics. The primer is intended for bioinformaticians who are interested in applying deep learning approaches, and for genomicists and general biomedical researchers who seek a high-level understanding of this rapidly evolving field. Computer scientists may also use the primer as an introduction to the exciting applications of deep learning in genomics. However, we do not provide a survey of deep learning in the biomedical field, which has been broadly covered in recent reviews1–5. This paper is accompanied by an interactive tutorial that we have created for interested readers to build a convolutional neural network to discover DNA-binding motifs (see URLs).
Deep learning as a class of machine learning methods
Machine learning techniques have been extensively used in genomics research3,6. Machine learning tasks fall within two major categories: supervised and unsupervised. In supervised learning, the goal is predicting the label (classification) or response (regression) of each data point by using a provided set of labeled training examples. In unsupervised learning, such as clustering and principal component analysis, the goal is learning inherent patterns within the data themselves.
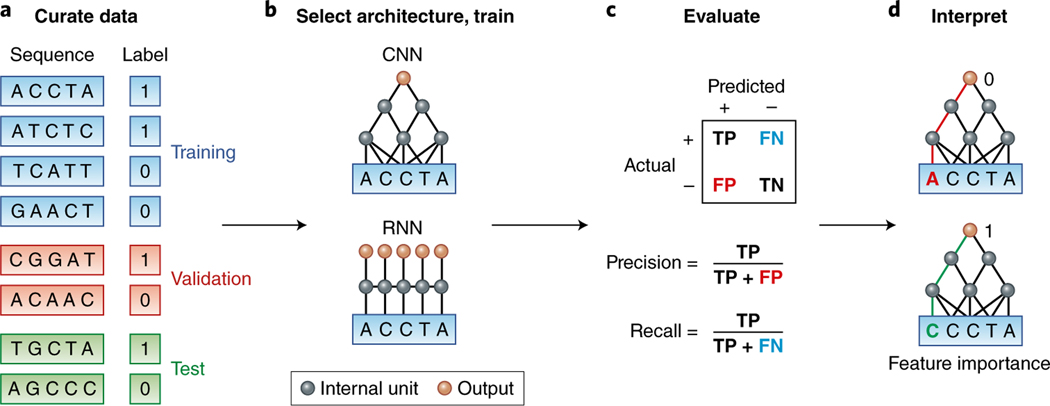
The ultimate goal in many machine learning tasks is to optimize model performance not on the available data (training performance) but instead on independent datasets (generalization performance). With this goal, data are randomly split into at least three subsets: training, validation and test sets. The training set is used for learning the model parameters (detailed discussion on parameter optimization in ref. 1), the validation set is used to select the best model, and the test set is kept aside to estimate the generalization performance (Fig. 1). Machine learning must reach an appropriate balance between model flexibility and the amount of training data. An overly simple model will underfit and fail to let the data ‘speak’. An overly flexible model will overfit to spurious patterns in the training data and will not generalize.
Fig. 1 |. Deep learning workflow in genomics.
a, A dataset should be randomly split into training, validation and test sets. The positive and negative examples should be balanced for potential confounders (for example, sequence content and location) so that the predictor learns salient features rather than confounders. b, The appropriate architecture is selected and trained on the basis of domain knowledge. For example, CNNs capture translation invariance, and RNNs capture more flexible spatial interactions. c, True positive (TP), false positive (FP), false negative (FN) and true negative (TN) rates are evaluated. When there are more negative than positive examples, precision and recall are often considered. d, The learned model is interpreted by computing how changing each nucleotide in the input affects the prediction. The interactive tutorial illustrates the four steps of this workflow (see URLs).
Large neural networks, a main form of deep learning, are a class of machine learning algorithms that can make predictions and perform dimensionality reduction. The key difference between deep learning and standard machine learning methods used in genomics—e.g., support vector machine and logistic regression—is that deep learning models have a higher capacity and are much more flexible. Typical deep learning models have millions of trainable parameters. However, this flexibility is a double-edged sword. With appropriately curated training data, deep learning can automatically learn features and patterns with less expert handcrafting. It also requires greater care to train on and to interpret the underlying biology. Box 1 summarizes the main messages of this primer on how to effectively use deep learning in genomics.
Box 1 |. Deep learning in genomics: key elements and guidance.
• Large training datasets (typically thousands of examples), curated to remove confounders, are required.
• The main architectures—feed-forward, convolutional and recurrent—correspond to different assumptions about data.
• Most genomics data do not require very deep networks.
• Researchers must be wary of high accuracy due to data imbalance or bias that makes classification too easy.
• A good practice is to compare against simpler machine learning models on the same dataset.
• Deep learning can achieve high accuracy, but the interpretation of results is more challenging than for standard statistical models.
Setting up deep learning
Deep learning is an umbrella term that refers to the recent advances in neural networks and the corresponding training platforms (e.g., TensorFlow and PyTorch). The starting point of a neural network is an artificial neuron, which takes as input a vector of real values and computes the weighted average of these values followed by a nonlinear transformation, which can be a simple threshold7. The weights are the parameters of the model that are learned during training. The power of neural networks stems from individual neurons being highly modular and composable, despite their simplicity8. The output of one neuron can be directly fed as input into other neurons. By composing neurons together, a neural network is created.
The input into a neural network is typically a matrix of real values. In genomics, the input might be a DNA sequence, in which the nucleotides A, C, T and G are encoded as [1,0,0,0], [0,1,0,0], [0,0,1,0] and [0,0,0,1]. Neurons that directly read in the data input are called the first, or input, layer. Layer two consists of neurons that read in the outputs of layer one, and so on for deeper layers, which are also referred to as hidden layers. The output of the neural network is the prediction of interest, e.g., whether the input DNA is an enhancer. Box 2 describes key terms and concepts in deep learning.
Box 2 |. Vocabulary and concepts.
Artificial neuron
A simple mathematical function that takes a vector as input and outputs a transformed weighted sum of the vector. The weights are the neuron’s parameters.
Deep learning
Neural networks with multiple layers of artificial neurons. The output of one layer is fed as input into the next layer to achieve greater flexibility.
Cross-validation
A common machine learning strategy wherein the dataset is split multiple times into training and validation sets. The average validation performance across the multiple splits is used to select the final model.
Back-propagation
A common method to train neural networks by updating its parameters (i.e., weights) by using the derivative of the network’s performance with respect to the parameters.
Feed-forward neural network
The most flexible class of neural networks, wherein each neuron can have arbitrary weights.
Convolutional neural network
A class of neural networks in which groups of artificial neurons are scanned across the input to identify translation-invariant patterns.
Recurrent neural network
A class of neural networks with cycles that can process inputs of varying lengths.
Autoencoder
A class of neural networks that performs nonlinear dimensionality reduction.
Feature importance
A saliency score for each input feature (for example, each nucleotide) that measures the extent to which changes in that feature affect the prediction.
There are three common families of architectures for connecting neurons into a network: feed-forward, convolutional and recurrent. Feed-forward is the simplest architecture7. Every neuron of layer i is connected only to neurons of layer i + 1, and all the connection edges can have different weights. Feed-forward architecture is suitable for generic prediction problems when there are no special relations among the input data features.
In a convolutional neural network (CNN), a neuron is scanned across the input matrix, and at each position of the input, the CNN computes the local weighted sum and produces an output value9. This procedure is very similar to taking the position weight matrix of a motif and scanning it across the DNA sequence. CNNs are useful in settings in which some spatially invariant patterns in the input are expected.
Recurrent neural networks (RNN) are designed for sequential or time-series data7. At each point in the sequence, a neural network, which could be feed-forward or convolutional, is applied to generate an internal signal, which is also fed to the next step of the RNN. Hidden layers of the RNN can be viewed as memory states that retain information from the sequence previously observed and are updated at each time step.
In addition, there are neural network architectures used for unsupervised learning. The most common are autoencoders that perform nonlinear dimensionality reduction10, in contrast to principal component analysis, which is linear. In an autoencoder, the output is set to be the input, and the network is encouraged to find a low-dimensional space that compresses the original information and reconstructs the inputs.
Training a neural network starts with a labeled dataset of (, ), where each is the th input, and is its output label. Each training point is fed into the network, and the network’s output is evaluated against the true label to produce a loss . Loss is the sum of the errors made for each example. The lower the loss, the better the model. Commonly used loss functions include squared error and cross-entropy. Squared error measures the difference between predicted and actual output values, which is especially relevant when the output is a continuous value. Cross-entropy measures the difference between two probability distributions over the same set of underlying events or classes, as is appropriate when the output is categorical7.
To train the network, the derivative of is computed with respect to the parameters of the network, which are the collection of neuron weights. By updating the weights in a small step in the direction of the derivative, the network’s prediction loss can be decreased, and its accuracy can be increased. The derivative can be efficiently computed for each training point via the chain rule from calculus; this process is commonly called back-propagation. In parallel, the network’s accuracy is also evaluated on the validation data, which are not used to update the weights. A good practice to avoid overfitting is to stop the training updates when the validation accuracy begins to decrease. More sophisticated techniques such as L2 regularization on the weights and dropout are also effective in controlling overfitting. L2 regularization encourages simpler, and thus more generalizable, models by penalizing models with large weights7. Dropout is a training process that randomly ignores nodes to mitigate overfitting11.
How to use deep learning effectively
The most important step in building an effective deep learning model is to first curate an appropriate training dataset and to select a suitable evaluation metric. The training set should be constructed to ensure that confounding biases that may artificially inflate performance are not introduced. For example, known pathogenic genetic variants may cluster in certain regions of the genome, i.e., exons or promoters, whereas known neutral variants may be more broadly distributed throughout the genome. A neural network naively applied to these unbalanced data may appear to perform well, but in reality it would probably learn to identify regions of the genome enriched in pathogenic variants without actually being able to distinguish neutral from pathogenic variants within these important genomic regions. Thus, it is important to design training datasets that are appropriately balanced for confounders that would detrimentally affect performance when applied to real-world-use cases12.
Furthermore, genomics data are often highly imbalanced13. For example, there are many more variants that are not disease causing than are disease causing, or only a small fraction of the population may develop a particular disease to be predicted. Therefore it is often more meaningful to assess precision and recall, measures of classifier performance that account for the imbalance of classes in a dataset rather than simple accuracy14.
Successful application of deep learning also requires domain knowledge, as with all other machine learning methods. For example, in support-vector-machine classification and logistic regression, domain knowledge is used to construct features from data, and in Bayesian models, expertise is incorporated into the prior distributions. In deep learning, domain knowledge is built into the design of the network architecture. The performance of the network crucially depends on understanding the assumptions and limitations behind different architectures.
As an illustration, suppose we want to build a model to predict whether a DNA sequence is an active enhancer8. Biological knowledge indicates that regulatory elements may be effective even after small spatial translations, thus suggesting that CNN might model them effectively. Moreover, the regulatory motifs are known to have a tendency to be relatively short (< 20 nt) thus suggesting that convolution filters should also be small (< 20 nt). Finally, if enhancers are being modeled, most of the activity is known to have a tendency to be clustered in regions from several hundred base pairs to two kilobases. Consequently, a reasonable design would be to limit inputs to the network to <2 kb. Any choice of the network architecture places an implicit prior over the model, and, as with any other machine learning, mismatch between the model’s prior and the underlying biology can lead to poor performance.
In computer vision, researchers have observed that performance can improve with very deep networks (more than 100 layers). In most genomics applications, fewer than five layers are sufficient13,15. Even relatively shallow networks can still have millions of parameters, and the most important factor that determines the success of a model is the availability of a large corpus of labeled training data. Most successful biological applications of deep learning have at least several thousand labeled examples1. It is always good practice to train simpler machine learning models—linear regression, least absolute shrinkage and selection operator (LASSO), gradient boosting or random forest—in parallel on the same dataset to compare against the deep learning models. Simpler models have fewer parameters and can work better when fewer training datasets are available16.
interpreting deep learning models
In many genomic applications, researchers are more interested in the biological mechanisms revealed by the predictive model rather than the prediction accuracy itself13,17. For example, the main motivation for building accurate deep learning models to predict chromatin patterns is a hope to learn new gene-regulation grammar by interpreting the trained model. Although deep learning can achieve state-of-the-art accuracy, it is more challenging to interpret than the more standard statistical models.
The simplest method to interpret a neural network is analogous to in silico mutagenesis. Given a particular data point X, each feature of X (for example, mutating each nucleotide) can be systematically varied while the rest of the features are fixed, and how the network’s output changes can be tracked. This approach is easy to implement but can be computationally expensive—the network recomputes the output for every mutation of X. A computationally tractable approximation to mutagenesis is to take the derivative of the network output with respect to each feature of X. This derivative can be computed in one back-propagation pass, and it conveys the sensitivity of the output to small perturbations in input features. Features with large positive or negative derivatives may be more influential to the outcome.
In strict terms, the derivative is a valid measure of influence for only infinitesimally small perturbations to the input, whereas in practice, researchers are interested in larger changes (for example, mutation of A to C). Several variations of the derivative-based interpretation methods have been developed to partially address this limitation—for example, integrated gradient18 and DeepLift19—and this goal is still currently an active area of research. Other interpretation methods, such as LIME20 select a small number of features to explain why a prediction is made.
For a CNN, it is also possible to visualize each convolution filter as a heat map or position weight matrix–style logo image. These visualizations are useful to obtain a sense of what local features the network might be learning. A caveat is that multiple convolution filters might be learning partially redundant features, and how the local features interact is less clear, because such interaction depends on the higher layers of the network.
The interpretation methods discussed here should not be confused with causal models that attempt to pinpoint cause–effect relationships. Interpreting a prediction model can identify salient features and generate hypotheses, but inferring actual causality requires experimental perturbation.
Applications in genomics
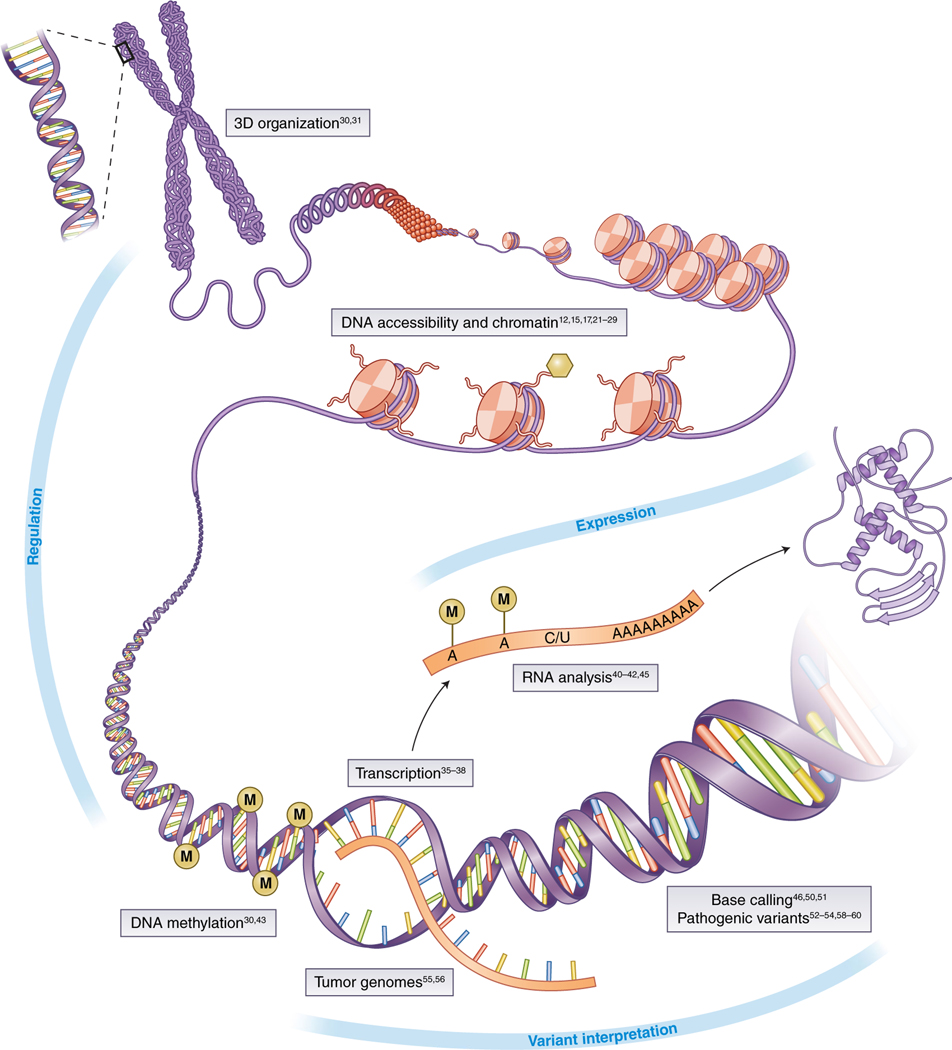
A growing number of publications are presenting deep learning approaches and tools to study the genome (Fig. 2). Functional genomics is a leading application domain of deep learning21. Examples include predicting the sequence specificity of DNA- and RNA-binding proteins and of enhancer and cis-regulatory regions, methylation status, gene expression and control of splicing. These tools are based on data generated by DNase I sequencing, assay for transposase-accessible chromatin using sequencing (ATAC–seq), chromatin immunoprecipitation (ChIP) sequencing, ChIP-on-chip, RNA immunoprecipitation sequencing, transcription-factor datasets and chromatin state15,17,22–24. Similarly, the identification of transcription start sites as well as cis-regulatory/enhancer elements25–29 can be executed with the addition of features from the Encyclopedia of DNA Elements (ENCODE) project, as well as transcription-start-site sequencing and RNA-seq signals. The methylation state of DNA, which also influences the expression of genes, has been inferred from three-dimensional genome topology (on the basis of Hi-C) and DNA sequence patterns30. Nucleotide sequence and DNase I assay signals predict Hi-C contacts at 1-kb resolution31.
Fig. 2 |. Applications of deep learning in genomics.
The boxes highlight several application domains and references discussed in the text. Image adapted with permission from ref. 65, Springer Nature.
Deep learning has been especially successful when applied to regulatory genomics, by using architectures directly adapted from modern computer vision and natural-language-processing applications32. Most approaches use CNN or RNN, which are well suited for the tasks of modeling regulatory elements, though genomics-specific modifications to the deep learning architecture can be useful. For example, Shrikumar et al.33 have addressed the phenomenon in which, in double-stranded DNA, the same pattern may appear identically on one strand and its reverse complement, owing to complementary base-pairing. Conventional deep learning models that do not explicitly model this property can produce different predictions on forward- and reverse-complement versions of the same DNA sequence.
Different tools are able to extract transcriptome patterns from large sets of gene expression data with the goal of estimating how much RNA is produced from a DNA template in a particular cell or condition34–37. Deep learning can build predictive models of gene expression from genotype data38 and can be used for studying the splicing-code model39 as well as for the identification of long noncoding RNAs40–42. Finally, deep learning has been used for the interpretation of regulatory control in single cells; for example, the detection of DNA methylation in single cells30,43, and for the identification of subgroups of cells by improving the representation of single-cell RNA-seq data44,45.
Predicting phenotypes from genetic data is also a major area of interest of deep learning. A first step in performing these types of predictions is to specify what genetic variants are present in an individual genome. This problem has been addressed by DeepVariant, which applies a CNN to make variant calls from short-read sequencing. The method treats DNA alignments as an image with a performance that appears to exceed that of standard variant callers46. Other methods for variant calling using more traditional DNA representations have also been developed47–49. Long-read-sequencing technologies also use deep learning for base calling50,51.
Prioritizing variants on the basis of the likelihood that they are pathogenic is important. Several methods that predict the pathogenicity of coding variants have been proposed52–54. These approaches are essentially aggregators that combine prior non–deep learning predictors as well as features known to be useful for the prediction of variant pathogenicity. A more challenging problem, the prediction of functional consequences of noncoding variants, has seen some success via DeepSEA13, a CNN trained to predict functional genomics features that can also be adapted to predict variant effects. In cancer genomics, deep learning can extract the high-level features between combinatorial somatic mutations and cancer types55 and learn prognostic information from multicancer datasets56. Preliminary phenotype prediction in agricultural applications appears promising57. However, a demonstration of the prediction of common complex human disease phenotypes is only now emerging: Zhou et al. have recently reported the extension of DeepSEA to the study of regulatory variants in autism spectrum disorder58. The same team has published ExPecto, the ab initio prediction of gene expression levels and variant effects from sequences from more than 200 tissues and cell types59. Also recently, Sundaram et al. have trained a DNN by using hundreds of common variants from population sequencing of nonhuman primate species to identify pathogenic variants in rare human diseases60.
Resources
Implementation of deep learning approaches requires familiarity with new tools and resources. These include access to computing power and general and genomics-specific software libraries (Box 3).
Box 3 |. Deep learning resources.
All the resources, except for those listed in the ‘specific for genomics’ block, are relevant for deep learning in general. Because the field is developing rapidly, this information is likely to be considerably different in the future.
| resource type | Name | urL | comment |
|---|---|---|---|
| Cloud platform | Amazon EC2 | https://aws.amazon.com/ec2/ | Most popular cloud platform |
| Microsoft Azure | https://azure.microsoft.com/ | Second-largest cloud platform | |
| Plug-and-play cloud GPU services | FloydHub | https://www.floydhub.com/ | All startups in the GPU service space; pay-by-the-hour model on top of basic monthly subscriptions |
| PaperSpace | https://www.paperspace.com/ | ||
| Valohai | https://valohai.com/ | ||
| Google CloudML | https://cloud.google.com/ml-engine/ | Can run your own models on Google’s hardware, including tensor processor units | |
| Google Colaboratory | https://colab.research.google.com/ | Notebook environment with free GPUs (during 12 h) | |
| Design services for deep learning models | Fabrik | https://github.com/Cloud-CV/Fabrik/ | Model export to Keras code; no training |
| IBM Data Cloud | https://datascience.ibm.com/ | Model export to Keras, PyTorch, TensorFlow or Caffe | |
| DeepCognition | http://deepcognition.ai/ | Training and evaluation included | |
| Prebuilt images with CUDA support | Docker Hub | https://hub.docker.com/r/nvidia/cuda/ | Docker images from NVIDIA with CUDA/cuDNN GPU support |
| Amazon Deep Learning AMIs | https://aws.amazon.com/machine-learning/amis/ | Amazon Machine Images (AMIs) with GPU support | |
| Software libraries | Keras | https://keras.io/ | More high-level than TensorFlow(general) (below) but can be integrated with it in many ways |
| TensorFlow | https://www.tensorflow.org/ | Developed by Google; most popular deep learning framework | |
| PyTorch | http://pytorch.org/ | Developed by Facebook | |
| Software libraries (specific for genomics) | DragoNN | https://kundajelab.github.io/dragonn/ | Tutorials included |
| Kipoi | http://kipoi.org/ | Model zoo for deep learning in genomics | |
| Educational resources | fast.ai | http://www.fast.ai/ | E.g., Deep Learning for Coders 1 and 2 |
| Coursera | https://www.coursera.org/specializations/deep-learning/ | Deep-learning-specialization course package | |
| Textbook | http://neuralnetworksanddeeplearning.com/ | Free online textbook with example code | |
| Fast.ai tips on configuring a deep learning environment | https://github.com/reshamas/fastai_deeplearn_part1/blob/master/README.md#platforms-for-using-fastai-gpu-required/ | Instructions for configuring deep learning frameworks for a variety of platforms; from the fast.ai course but general; the details of these procedures change quickly | |
| Setting up TensorFlow with GPU on Google Cloud Engine | https://medium.com/google-cloud/jupyter-tensorflownvidia-gpu-docker-google-compute-engine-4a146f085f17/ | Recipe for Docker-based setup of Google Cloud instance with TensorFlow, GPU support and Jupiter Notebooks |
Cloud platforms.
Recent advances in deep learning have, to a considerable degree, been driven by developments in graphical processing unit (GPU) technology. GPUs can significantly increase training speed because the way in which neural networks are trained lends itself well to their architecture, thus allowing for fast vector and matrix multiplications, owing to the large number of processing units and high memory bandwidth in GPUs. Some academic high-performance computing clusters offer access to GPUs, but because progress in GPU technology is rapid, those processors are at risk of becoming outdated. The major cloud-computing platforms, such as Amazon EC2, Google Cloud Engine, Microsoft Azure and IBM Cloud, can be more flexible in offering on-demand GPU access. However, even these cloud platforms require some degree of configuration from the user, such as installing and compiling an appropriate version of CUDA (a popular parallel computing platform and programming toolkit) for general GPU programming, upon which many deep learning frameworks depend for GPU acceleration.
For users wishing to avoid semimanual setup procedures, there are specialized platforms offering ‘plug-and-play’ GPU access, for instance FloydHub, Valohai, Paperspace and Google CloudML. Typically, these platforms provide configuration-free access to GPUs on Amazon or Google cloud infrastructures for a modest markup compared with running ‘raw’ cloud instances directly. Perhaps the simplest alternative at the time of writing of this paper is Google Colaboratory, a Python notebook environment that provides free use of a K80 GPU during 12 consecutive hours. The tutorial accompanying this paper (see URLs) is built on this platform. In some cases, using lightweight virtual machines such as Docker or Singularity containers is convenient for packaging software and dependencies so that they can be directly run on a cloud instance with minimal setup. All these solutions, however, still require users to write the code for the models.
Some emerging platforms are offering the possibility to design deep learning models without coding. For example, IBM Data Cloud has recently introduced a neural network designer in which users can build a model from building blocks and export the finished model to runnable code. The open-source Fabrik framework offers a similar capability. DeepCognition offers a platform in which users can design, train and evaluate models in the same framework.
Software libraries.
Many software libraries are commonly used for deep learning. Whereas earlier models were often written in frameworks developed at universities, such as Theano, Torch and Caffe, recent years have seen a strong showing of open-source Python libraries developed in corporate laboratories, for example TensorFlow (Google) and PyTorch (Facebook). Some frameworks were initially developed independently but subsequently received explicit backing from a company, for example, Keras (Google) and MXNet (Amazon). Keras has gained its popularity from being designed to be at a higher level of abstraction than most other frameworks; consequently, it allows for model construction in larger conceptual blocks such as network layers without requiring users to keep track of all the details.
Because of the relatively recent entry of deep learning into genomics, there are few genomics-specific software libraries (Box 3). One example is DragoNN, a toolkit to teach and learn about deep learning for genomics, which includes tutorials in Python notebook format, a command-line interface, tools for running models on cloud CPU or GPU, and a model-interpretation module. Kipoi is a repository of ready-to-use trained models in genomics. It currently contains 2,031 different models (‘model zoo’), covering canonical predictive tasks in transcriptional and post-transcriptional gene regulation61. Kipoi uses Conda virtual environments to install the correct dependencies for each model.
conclusions and perspectives
Although deep learning has demonstrated impressive potential in genomics, a number of outstanding issues remain62. First is the challenge of how to design deep learning systems that best augment and complement human experience in making medical decisions (for example, genome interpretation). Second is the challenge of how to avoid biases in training sets and how to interpret predictions. Interpretation and robustness are two important directions of method development63. Finally, there is a need for iterative experimentation, in which deep learning predictions can be validated by functional laboratory tests or by formal clinical assessment.
The most successful applications of deep learning in genomics to date has been in supervised learning, i.e., making predictions8. It is important to not confuse high prediction accuracy with the ultimate objective of extracting biomedical insights from data and making robust assessment in diverse settings that might be different from the training data. Beyond making predictions, deep learning can potentially become a powerful tool for synthetic biology by learning to automatically generate new DNA sequences and new proteins with desirable properties64. Such generative models are an exciting new frontier of research.
URLs. Interactive tutorial to build a convolutional neural network to discover DNA-binding motifs, https://colab.research.google.com/drive/17E4h5aAOioh5DiTo7MZg4hpL6Z_0FyWr.
Acknowledgements
We thank N. Wineinger, R. Dias, J. di Iulio and D. Evans for comments on the paper. The work of A. Telenti, A. Torkamani and P.M. is supported by the Qualcomm Foundation and the NIH Center for Translational Science Award (CTSA, grant UL1TR002550). Further support to A. Torkamani is from U54GM114833 and U24TR002306. J.Z. is supported by a Chan-Zuckerberg Biohub Investigator grant and National Science Foundation (NSF) grant CRII 1657155.
Footnotes
competing interests
M.H. is an employee of Peltarion.
Additional information
Reprints and permissions information is available at www.nature.com/reprints.
References
- 1.Angermueller C, Pärnamaa T, Parts L. & Stegle O. Deep learning for computational biology. Mol. Syst. Biol. 12, 878 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ching T. et al. Opportunities and obstacles for deep learning in biology and medicine. J. R. Soc. Interface 15, 20170387 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Telenti A, Lippert C, Chang PC & DePristo M. Deep learning of genomic variation and regulatory network data. Hum. Mol. Genet. 27, R63–R71 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yue T. & Wang H. Deep learning for genomics: a concise overview. Preprint at https://arxiv.org/abs/1802.00810 (2018). [Google Scholar]
- 5.Camacho DM, Collins KM, Powers RK, Costello JC & Collins JJ Next-generation machine learning for biological networks. Cell 173, 1581–1592 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Libbrecht MW & Noble WS Machine learning applications in genetics and genomics. Nat. Rev. Genet. 16, 321–332 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goodfellow I, Bengio Y, Courville A. & Bengio Y. Deep Learning Vol. 1 (MIT Press, Cambridge, 2016). [Google Scholar]
- 8.LeCun Y, Bengio Y. & Hinton G. Deep learning. Nature 521, 436–444 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Krizhevsky A, Sutskever I. & Hinton GE Imagenet classification with deep convolutional neural networks. Adv. Neural Inf. Process. Syst. 1, 1097–1105 (2012). [Google Scholar]
- 10.Hinton GE & Salakhutdinov RR Reducing the dimensionality of data with neural networks. Science 313, 504–507 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Srivastava N, Hinton G, Krizhevsky A, Sutskever I. & Salakhutdinov R. Dropout: a simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 15, 1929–1958 (2014). [Google Scholar]
- 12.Khodabandelou G, Mozziconacci J. & Routhier E. Genome functional annotation using deep convolutional neural network. Preprint at https://www.biorxiv.org/content/early/2018/05/25/330308 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou J. & Troyanskaya OG Predicting effects of noncoding variants with deep learning-based sequence model. Nat. Methods 12, 931–934 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Powers DMW Evaluation: from precision, recall and F-measure to ROC, informedness, markedness and correlation. J. Mach. Learn. Technol. 2, 37–63 (2011). [Google Scholar]
- 15.Kelley DR, Snoek J. & Rinn JL Basset: learning the regulatory code of the accessible genome with deep convolutional neural networks. Genome Res. 26, 990–999 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hastie T, Tibshirani R. & Friedman JH The Elements of Statistical Learning Vol. 1 (Springer Science+ Business Media, New York, 2001). [Google Scholar]
- 17.Quang D. & Xie X. DanQ: a hybrid convolutional and recurrent deep neural network for quantifying the function of DNA sequences. Nucleic Acids Res. 44, e107 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sundararajan M, Taly A. & Yan Q. Axiomatic attribution for deep networks. Preprint at https://arxiv.org/abs/1703.01365v2 (2017). [Google Scholar]
- 19.Shrikumar A, Greenside P. & Kundaje A. Learning important features through propagating activation differences. Proc. Int. Conf. Mach. Learn. 70, 3145–3153 (2017). [Google Scholar]
- 20.Ribeiro MT, Singh S. & Guestrin C. “Why should I trust you?”: explaining the predictions of any classifier. in KDD 1135–1144 (AAAI Press, Menlo Park, CA, USA, 2016). [Google Scholar]
- 21.Park Y. & Kellis M. Deep learning for regulatory genomics. Nat. Biotechnol. 33, 825–826 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Alipanahi B, Delong A, Weirauch MT & Frey BJ Predicting the sequence specificities of DNA- and RNA-binding proteins by deep learning. Nat. Biotechnol. 33, 831–838 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Lanchantin J, Singh R, Wang B. & Qi Y. Deep motif dashboard: visualizing and understanding genomic sequences using deep neural networks. Pac. Symp. Biocomput. 22, 254–265 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zeng H, Edwards MD, Liu G. & Gifford DK Convolutional neural network architectures for predicting DNA-protein binding. Bioinformatics 32, i121–i127 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu F, Li H, Ren C, Bo X. & Shu W. PEDLA: predicting enhancers with a deep learning-based algorithmic framework. Sci. Rep. 6, 28517 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kleftogiannis D, Kalnis P. & Bajic VB DEEP: a general computational framework for predicting enhancers. Nucleic Acids Res. 43, e6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Min X. et al. Predicting enhancers with deep convolutional neural networks. BMC Bioinformatics 18 (Suppl. 13), 478 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eser U. & Stirling Churchman L. FIDDLE: an integrative deep learning framework for functional genomic data inference. Preprint at https://www.biorxiv.org/content/early/2016/10/17/081380 (2016). [Google Scholar]
- 29.Li Y, Shi W. & Wasserman WW Genome-wide prediction of cis-regulatory regions using supervised deep learning methods. BMC Bioinformatics 19, 202 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Y. et al. Predicting DNA methylation state of CpG dinucleotide using genome topological features and deep networks. Sci. Rep. 6, 19598 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schreiber J, Libbrecht M, Bilmes J. & Noble W. Nucleotide sequence and DNaseI sensitivity are predictive of 3D chromatin architecture. Preprint at https://www.biorxiv.org/content/early/2017/01/30/103614 (2017). [Google Scholar]
- 32.Zeng W, Wu M. & Jiang R. Prediction of enhancer-promoter interactions via natural language processing. BMC Genomics 19 (Suppl. 2), 84 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shrikumar A, Greenside P. & Kundaje A. Reverse-complement parameter sharing improves deep learning models for genomics. Preprint at https://www.biorxiv.org/content/early/2017/01/27/103663 (2017). [Google Scholar]
- 34.Tan J, Hammond JH, Hogan DA & Greene CS ADAGE-based integration of publicly available Pseudomonas aeruginosa gene expression data with denoising autoencoders illuminates microbe-host interactions. mSystems 1, e00025–15 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen Y, Li Y, Narayan R, Subramanian A. & Xie X. Gene expression inference with deep learning. Bioinformatics 32, 1832–1839 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen L, Cai C, Chen V. & Lu X. Learning a hierarchical representation of the yeast transcriptomic machinery using an autoencoder model. BMC Bioinformatics 17 (Suppl. 1), 9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cui H. et al. Boosting gene expression clustering with system-wide biological information: a robust autoencoder approach. Preprint at https://www.biorxiv.org/content/early/2017/11/05/214122 (2017). [Google Scholar]
- 38.Xie R, Wen J, Quitadamo A, Cheng J. & Shi X. A deep auto-encoder model for gene expression prediction. BMC Genomics 18 (Suppl. 9), 845 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jha A, Gazzara MR & Barash Y. Integrative deep models for alternative splicing. Bioinformatics 33, i274–i282 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tripathi R, Patel S, Kumari V, Chakraborty P. & Varadwaj PK DeepLNC, a long non-coding RNA prediction tool using deep neural network. Netw. Model. Anal. Health Inform. Bioinform. 5, 21 (2016). [Google Scholar]
- 41.Yu N, Yu Z. & Pan Y. A deep learning method for lincRNA detection using auto-encoder algorithm. BMC Bioinformatics 18 (Suppl. 15), 511 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hill ST et al. A deep recurrent neural network discovers complex biological rules to decipher RNA protein-coding potential. Nucleic Acids Res. 46, 8105–8113 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Angermueller C, Lee HJ, Reik W. & Stegle O. DeepCpG: accurate prediction of single-cell DNA methylation states using deep learning. Genome Biol. 18, 67 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shaham U. et al. Removal of batch effects using distribution-matching residual networks. Bioinformatics 33, 2539–2546 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin C, Jain S, Kim H. & Bar-Joseph Z. Using neural networks for reducing the dimensions of single-cell RNA-Seq data. Nucleic Acids Res. 45, e156 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Poplin R. et al. Creating a universal SNP and small indel variant caller with deep neural networks. Preprint at https://www.biorxiv.org/content/early/2018/03/20/092890 (2017). [DOI] [PubMed] [Google Scholar]
- 47.Luo R, Sedlazeck FJ, Lam T-W & Schatz M. Clairvoyante: a multi-task convolutional deep neural network for variant calling in single molecule sequencing. Preprint at https://www.biorxiv.org/content/early/2018/09/26/310458 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Luo R, Lam T-W & Schatz M. Skyhawk: an artificial neural network-based discriminator for reviewing clinically significant genomic variants. Preprint at https://www.biorxiv.org/content/early/2018/05/01/311985 (2018). [Google Scholar]
- 49.Torracinta R. et al. Adaptive somatic mutations calls with deep learning and semi-simulated data. Preprint at https://www.biorxiv.org/content/early/2016/10/04/079087 (2016). [Google Scholar]
- 50.Boža V, Brejová B. & Vinař T. DeepNano: deep recurrent neural networks for base calling in MinION nanopore reads. PLoS One 12, e0178751 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Teng H, Hall MB, Duarte T, Cao MD & Coin L. Chiron: translating nanopore raw signal directly into nucleotide sequence using deep learning. Preprint at https://www.biorxiv.org/content/early/2017/08/23/179531 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Qi H. et al. MVP: predicting pathogenicity of missense variants by deep neural networks. Preprint at https://www.biorxiv.org/content/early/2018/02/02/259390 (2018). [Google Scholar]
- 53.Quang D, Chen Y. & Xie X. DANN: a deep learning approach for annotating the pathogenicity of genetic variants. Bioinformatics 31, 761–763 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Korvigo I, Afanasyev A, Romashchenko N. & Skoblov M. Generalising better: applying deep learning to integrate deleteriousness prediction scores for whole-exome SNV studies. PLoS One 13, e0192829 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yuan Y. et al. DeepGene: an advanced cancer type classifier based on deep learning and somatic point mutations. BMC Bioinformatics 17, 476 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yousefi S. et al. Predicting clinical outcomes from large scale cancer genomic profiles with deep survival models. Sci. Rep. 7, 11707 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ma W, Qiu Z, Song J, Cheng Q. & Ma C. DeepGS: predicting phenotypes from genotypes using deep learning. Preprint at https://www.biorxiv.org/content/early/2017/12/31/241414 (2017). [Google Scholar]
- 58.Zhou J. et al. Whole-genome deep learning analysis reveals causal role of noncoding mutations in autism. Preprint at https://www.biorxiv.org/content/early/2018/05/11/319681 (2018). [Google Scholar]
- 59.Zhou J. et al. Deep learning sequence-based ab initio prediction of variant effects on expression and disease risk. Nat. Genet. 50, 1171–1179 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sundaram L. et al. Predicting the clinical impact of human mutation with deep neural networks. Nat. Genet. 50, 1161–1170 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Avsec Z. et al. Kipoi: accelerating the community exchange and reuse of predictive models for genomics. Preprint at https://www.biorxiv.org/content/early/2018/07/24/375345 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Webb S. Deep learning for biology. Nature 554, 555–557 (2018). [DOI] [PubMed] [Google Scholar]
- 63.Ghorbani A, Abid A. & Zou J. Interpretation of neural networks is fragile. Preprint at https://arxiv.org/abs/1710.10547 (2017). [Google Scholar]
- 64.Gupta A. & Zou J. Feedback GAN (FBGAN) for DNA: a novel feedback-loop architecture for optimizing protein functions. Preprint at https://arxiv.org/abs/1804.01694 (2018).
- 65.Stranger B. et al. ; eGTEx Project. Enhancing GTEx by bridging the gaps between genotype, gene expression, and disease. Nat. Genet. 49, 1664–1670 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]