Abstract
Although evolution is driven by changes in how regulatory pathways control development, we know little about the molecular details underlying these transitions. The TRA-2 domain that mediates contact with TRA-1 is conserved in Caenorhabditis. By comparing the interaction of these proteins in two species, we identified a striking change in how sexual development is controlled. Identical mutations in this domain promote oogenesis in Caenorhabditis elegans but promote spermatogenesis in Caenorhabditis briggsae. Furthermore, the effects of these mutations involve the male-promoting gene fem-3 in C. elegans but are independent of fem-3 in C. briggsae. Finally, reciprocal mutations in these genes show that C. briggsae TRA-2 binds TRA-1 to prevent expression of spermatogenesis regulators. By contrast, in C. elegans TRA-1 sequesters TRA-2 in the germ line, allowing FEM-3 to initiate spermatogenesis. Thus, we propose that the flow of information within the sex determination pathway has switched directions during evolution. This result has important implications for how evolutionary change can occur.
Keywords: evolution of gene regulation, sex determination, nematodes
Introduction
Nematode sex Determination
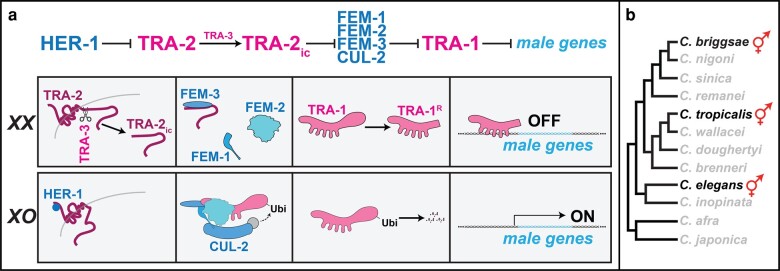
Because most animal species have two sexes, each individual needs to determine which sex to become, and regulate development to select the appropriate cell fates. For example, the nematode Caenorhabditis elegans uses a complex pathway that responds to the ratio of X chromosomes to autosomes, so that XX animals develop as hermaphrodites and XO animals as males (reviewed by Zarkower 2006). This pathway controls the activity of the Gli transcription factor TRA-1, which in turn regulates the expression of hundreds of target genes throughout the body (Berkseth et al. 2013).
How does this pathway work? In the embryo, the X/A ratio acts through xol-1 and three sdc genes to control the expression of her-1, which produces a male sex hormone. In XO animals, HER-1 binds the TRA-2 receptor to prevent it from promoting female fates (Fig. 1a). By contrast, in XX animals the absence of HER-1 allows the TRA-3 protease to cleave TRA-2 (Barnes and Hodgkin 1996; Sokol and Kuwabara 2000), releasing an intercellular fragment that promotes female development. This TRA-2ic fragment binds FEM-3, preventing a FEM-1/FEM-2/FEM-3 complex from ubiquitinating TRA-1 (Mehra et al. 1999; Starostina et al. 2007). Finally, TRA-1 is cleaved to produce a transcription factor (Zarkower and Hodgkin 1992; Schvarzstein and Spence 2006) that represses male genes (Berkseth et al. 2013), including fog-1 and fog-3, which direct spermatogenesis (Ellis and Kimble 1995; Chen and Ellis 2000; Jin et al. 2001).
Fig. 1.
The core sex determination pathway in Caenorhabditis nematodes. a) The top line summarizes the core regulatory interactions that control sexual fates. In XX animals, the TRA-2 receptor is free to be cleaved by TRA-3, releasing an intracellular fragment that binds FEM-3, preventing the FEM complex from working. This allows the TRA-1 transcription factor to be cleaved, forming a repressor of male genes. In XO animals, the male sex hormone HER-1 binds to and inactivates TRA-2. This leaves the FEM complex free to assemble with CUL-2 and ubiquitinate TRA-1, leading to its degradation. Hence, male genes are expressed. b) The phylogeny shows that ancestral Caenorhabditis species were male/female, and three species independently evolved XX animals that reproduce as self-fertile hermaphrodites.
Evolution of Self-Fertile Hermaphrodites
This regulatory process has been altered during recent evolution, during the production of new mating systems. Although most Caenorhabditis species are male/female, the phylogeny shows that C. elegans, Caenorhabditis briggsae, and Caenorhabditis tropicalis independently evolved XX animals that can reproduce as self-fertile hermaphrodites (Fig. 1b, Kiontke et al. 2011). Since a key feature of these hermaphrodites is the ability to make sperm as well as oocytes, each of these three species has altered the germ line to allow a brief period of XX spermatogenesis (Ellis 2022, 2016). So far, we only know about these changes in C. elegans and C. briggsae.
One of the key modifications was the recruitment of novel genes to modify the sex determination pathway in the XX germ line. For example, C. elegans uses fog-2 to regulate the activity of tra-2 in germ cells (Schedl and Kimble 1988). The fog-2 gene encodes an F-box protein that binds GLD-1, which regulates the expression of tra-2 mRNA (Clifford et al. 2000). However, because fog-2 arose through a series of gene duplications and is unique to C. elegans, it is not part of this pathway in other species (Nayak et al. 2005). Instead, the she-1 gene is needed for hermaphrodite spermatogenesis in C. briggsae (Guo et al. 2009). It also arose through a series of gene duplications and acts through TRA-2, but she-1 is unique to C. briggsae.
Other changes to the pathway involve the core genes themselves. For example, fem-1, fem-2, and fem-3 encode members of an E3 ubiquitin ligase complex that regulates TRA-1 stability (Starostina et al. 2007). In C. elegans, these genes are also required downstream of tra-1 to initiate spermatogenesis (Hodgkin 1986; Schedl et al. 1989). By contrast, fem-2 and fem-3 are not needed for spermatogenesis in C. briggsae (Hill et al. 2006).
Finally, some mutations that affect tra-2 or fem-3 regulation in C. elegans have major effects on hermaphrodite development. Gain-of-function mutations in fem-3 result in animals that make only sperm (Barton et al. 1987), whereas gain-of-function mutations in tra-2 cause them to produce only oocytes (Doniach 1986; Schedl and Kimble 1988). These mutations all affect the 3′-untranslated regions of their messages, so they only alter expression, not protein structure (Ahringer and Kimble 1991; Okkema and Kimble 1991; Kuwabara et al. 1992). Analyses of these mutations suggest that one way to achieve hermaphrodite spermatogenesis is by modulating the competition between TRA-2 and FEM-3 to control germ cell fates.
The tra-2(mx) Alleles
A separate group of tra-2 mutations shows more complex effects in C. elegans. These tra-2(mx) alleles are all missense mutations that alter a conserved region near the C-terminus of the protein (Lum et al. 2000; Wang and Kimble 2001). The mx alleles prevent TRA-2 from directly binding the TRA-1 transcription factor (Lum et al. 2000; Wang and Kimble 2001), and they cause most hermaphrodites to produce only oocytes, transforming them into true females (Doniach 1986). Besides this strong effect on germ cells, the mx mutations have only weak effects on the soma, which sometimes result in the partial retraction of the hermaphrodite tail, or in the deaths of the HSN neurons. Thus, they strongly promote oogenesis in the germ line, but weakly promote male fates in the soma. Hence, the tra-2(mx) alleles seem likely to reveal key aspects of hermaphrodite gene regulation. But how they work and why they have such different effects in the soma and germ line have remained mysteries. We used comparative evolution to answer these questions.
Results
Caenorhabditis briggsae tra-2(mx) Alleles Increase Hermaphrodite Spermatogenesis
Analyzing the sex determination pathway in C. elegans germ cells is tricky, because the FEM complex has multiple roles in the germ line, and is absolutely required for spermatogenesis (Hodgkin 1986). By contrast, neither the fem-2 nor the fem-3 gene is needed for spermatogenesis in C. briggsae (Hill et al. 2006). Since Wang and Kimble (2001) showed that interaction between TRA-2 and TRA-1 is conserved in C. briggsae, the fact that the fem genes are not needed to produce sperm makes C. briggsae an ideal system for studying tra-2(mx) mutations.
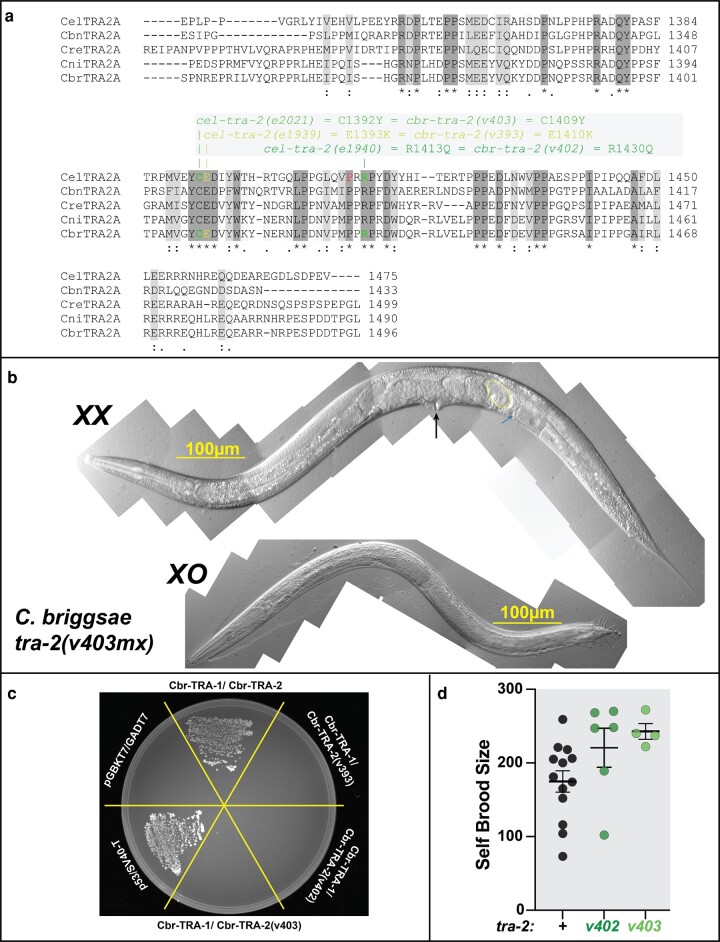
We began by preparing an alignment of five Caenorhabditis TRA-2 proteins, which revealed extensive conservation near the C-terminus, in the mx region (Fig. 2a). This part of the protein is released from the membrane following cleavage by the calpain protease TRA-3 (Barnes and Hodgkin 1996) and interacts with TRA-1 in both C. elegans (Lum et al. 2000) and C. briggsae (Wang and Kimble 2001). There are five missense mutations that cause the special tra-2(mx) phenotype in C. elegans (Kuwabara et al. 1998). Four of the affected residues are identical in all five species, while the fifth is partially conserved. Thus, we suspected that the role of the TRA-2(mx) domain in sex determination might also be conserved.
Fig. 2.
Caenorhabditis briggsae tra-2(mx) alleles do not feminize the germ line. a) Alignment of the C-termini of five Caenorhabditis TRA-2 proteins, prepared using MUSCLE. Identical residues are shaded dark gray, and conservative substitutions are shaded light gray. All C. elegans tra-2(mx) alleles affect one of five conserved sites in this region. We made identical C. briggsae mutations for three of the C. elegans alleles (marked in green shades). b) Differential interference contrast photomicrographs of a Cbr-tra-2(v403) hermaphrodite and male. The black arrow indicates the vulva, and one of the self-embryos in the uterus is outlined in yellow. The small blue arrow marks two self-sperm. c) Yeast two-hybrid results, with bait constructs listed before prey constructs. d) Self-brood size of hermaphrodites, with error bars indicating the mean and 95% confidence limits.
To learn the effect of mx mutations in C. briggsae, we made mutations identical to three of the C. elegans tra-2(mx) alleles, as shown in Fig. 2a. One of these three alleles, Cbr-tra-2(v393), showed more extensive masculinization than its C. elegans ortholog, tra-2(e1939) (supplementary fig. S1, Supplementary Material online). The v393 tail is blunt with several male characteristics, the gonad usually has a single arm ending in the cloaca, and the germ cells all form sperm. These traits suggest that this substitution causes more extensive harm to general TRA-2 function in C. briggsae than it does in C. elegans.
However, the other two alleles, cbr-tra-2(v402) and cbr-tra-2(v403), have only minor effects on the soma, since XX animals usually form typical hermaphrodite bodies and XO animals are normal males (Fig. 2b). In this respect, these mutants resemble their C. elegans orthologs.
We used the yeast two-hybrid assay to confirm that these C. briggsae mutations prevent TRA-2 from binding TRA-1 (Fig. 2c). Our assay can detect the normal interaction of TRA-2 with TRA-1 in this species, and control experiments using C. elegans proteins confirmed that we can also detect the disruption of binding by the mx alleles in that species (data not shown). Thus, the role of the MX domain in TRA-2/TRA-1 interactions is conserved, and this function generally plays little role in somatic sex determination.
Surprisingly, none of the C. briggsae tra-2(mx) mutants showed the characteristic Fog phenotype of their C. elegans counterparts. Although cbr-tra-2(v393) has the opposite phenotype—the animals make only sperm—it also causes XX animals to develop intersexual gonads (supplementary fig. S1, Supplementary Material online), which might influence germ cell fates. These somatic effects could be the result of broader problems in TRA-2ic folding, which might also impinge on its ability to bind FEM-3.
Thus, we focused our analyses on the other two cbr-tra-2(mx) alleles. To learn whether they reduced sperm number without eliminating it altogether, we measured the self-broods of these animals (Fig. 2d). If these mutants made fewer sperm than normal, they should make fewer self-progeny. Instead, we observed an increase in brood size for both alleles. This result suggested that v402 and v403 cause extra spermatogenesis in C. briggsae, a phenotype opposite that caused by the C. elegans mx alleles.
Caenorhabditis briggsae tra-2(mx) Alleles Restore Spermatogenesis to she-1 Mutants
In C. elegans, the fog-2 gene is required for hermaphrodite spermatogenesis, since fog-2 XX mutants develop as females rather than as hermaphrodites, whereas the XO mutants are normal males (Schedl and Kimble 1988). By contrast, mutations in the 3′-UTR of fem-3 increase hermaphrodite spermatogenesis (Barton et al. 1987; Rosenquist and Kimble 1988) and suppress fog-2 (Schedl and Kimble 1988). This suppression of fog-2 has been a key diagnostic tool for studying the control of spermatogenesis in C. elegans hermaphrodites.
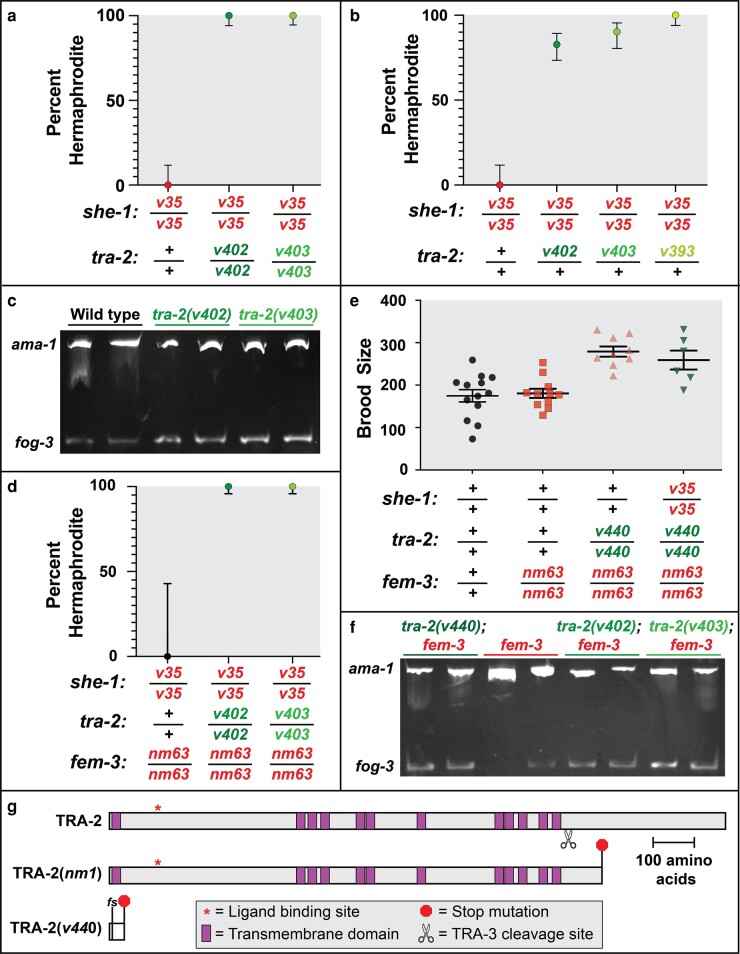
Although C. briggsae has no homolog of fog-2 (Nayak et al. 2005), mutations in she-1 have a similar effect, transforming XX animals into females (Guo et al. 2009). Thus, we tested the ability of Cbr-tra-2(v402mx) and Cbr-tra-2(v403mx) to suppress she-1 (Fig. 3a). Both alleles completely restored normal hermaphrodite development to she-1 XX animals. These results confirm the idea that Cbr-tra-2(mx) mutations promote spermatogenesis in XX hermaphrodites. In addition, tra-2 null mutations are dominant suppressors of fog-2 in C. elegans (Schedl and Kimble 1988) and of she-1 in C. briggsae (Guo et al. 2009), demonstrating the conserved importance of tra-2 dosage in the XX germ line. Although the C. briggsae tra-2(mx) alleles are not null, we tested them for dominant suppression of she-1 in the germ line (Fig. 3b). Our data revealed that they suppress she-1 almost as well as a null allele does. Furthermore, we included the Cbr-tra-2(v393mx) allele in these experiments, since it has no effect on somatic development in heterozygotes, and found that it behaves like the other mx alleles do.
Fig. 3.
Caenorhabditis briggsae tra-2(mx) mutations suppress she-1 independent of fem-3 function. a, b, d) Graphs showing the percent of XX animals of each genotype that develop as self-fertile hermaphrodites, rather than as females. The error bars show 95% confidence intervals. a) N = 29, 62, 67, respectively. b) N = 29, 87, 62, 60. d) N = 7, 87, 88. c, f) Semi-quantitative RT-PCR results showing fog-3 expression (lower band) and control ama-1 expression (upper band). These two products were produced in separate PCRs (using equivalent amounts of the cDNA template) and run on a single gel. Each lane used cDNA template produced from a mixture of 5 mid-L4 XX larvae. e) Self-brood size of hermaphrodites, with error bars indicating the mean and 95% confidence limits. g) Diagram showing the locations of the reference mutation Cbr-tra-2(nm1) and the null allele Cbr-tra-2(v440). The v440 allele truncates the protein prior to the crucial HER-1-binding site and removes the entire intracellular domain, so it represents a molecular null.
In both C. elegans and C. briggsae, spermatogenesis requires fog-3 (Ellis and Kimble 1995; Chen et al. 2001), which is a major target for regulation by the transcription factor TRA-1 (Chen and Ellis 2000). We used reverse transcription PCR (RT-PCR) to measure fog-3 expression in XX L4 larvae, during the normal period of spermatogenesis, and found that it was significantly higher in the cbr-tra-2(mx) mutants than in the wild type (Fig. 3c, supplementary fig. S2, Supplementary Material online). Thus, these alleles appear to affect spermatogenesis by increasing expression of the key regulator fog-3.
Caenorhabditis elegans mutations in tra-2 and its target fem-3 have opposite effects on the germ line, which implies that they compete with each other to regulate spermatogenesis (Barton et al. 1987). Hence, the predominant model in the field is that TRA-2 controls sex determination by inactivating FEM-3, which would otherwise direct male development. Although C. briggsae FEM-3 is not required for spermatogenesis, it remained possible that C. briggsae tra-2(mx) mutations promote spermatogenesis by increasing the activity of FEM-3 in germ cells. Thus, we tested their ability to suppress she-1 in a fem-3 null mutant background (Fig. 3d). As shown previously, a she-1 mutation feminizes the germ line of Cbr-fem-3 mutants (Guo et al. 2009). Surprisingly, Cbr-tra-2(mx) alleles suppress this feminization. Thus, C. briggsae tra-2(mx) alleles do not act through FEM-3 to promote spermatogenesis. Instead, they appear to alter the direct regulation of TRA-1 by TRA-2.
We also measured the effects of tra-2 mutations on fog-3 transcript levels in a fem-3 null mutant background (Fig. 3f, supplementary fig. S3, Supplementary Material online). Even so, the mx mutations of tra-2 caused an increase in fog-3 expression. Taking these results together, we infer that SHE-1 regulates TRA-2 activity in C. briggsae germ cells and that TRA-2 physically interacts with TRA-1 to regulate fog-3 expression and spermatogenesis, bypassing the need for FEM-3.
These studies suggested that null alleles of C. briggsae tra-2 should also promote spermatogenesis by preventing an interaction with TRA-1. To test this possibility (Fig. 3, e and f), we began by using gene editing to produce an early frameshift in C. briggsae tra-2 (Fig. 3g), since other Cbr-tra-2 alleles are located near the C-terminus and might have residual function. The Cbr-tra-2(v440) allele transforms all XX animals into pseudo-males, just like Cbr-tra-2(mn1) (Kelleher et al. 2008). These XX mutants have defective male tails, but normal male gonads and only produce sperm. As expected, this somatic effect is suppressed by a fem-3 null mutation, which restores normal hermaphrodite development. Thus, tra-2(v440) defines the null phenotype.
When we measured brood sizes to assay the number of hermaphrodite sperm that were produced (Singson 2001), we found that the wild type and fem-3 null mutants made the same number of sperm, but that tra-2(v440); fem-3 double mutants made extra sperm, confirming that TRA-2 regulates germ cell sexual development even in the complete absence of FEM-3 (Fig. 3e). This increased brood size is also observed in tra-2(v440); she-1 fem-3 triple mutants (Fig. 3e). As these results predict, tra-2(v440) also causes an increase in the expression of fog-3 (Fig. 3f). We conclude that C. briggsae TRA-2 regulates germ cell fates independent of FEM-3. However, SHE-1 cannot regulate germ cell fates without TRA-2 being present and functional.
Blocking the Ability of C. briggsae TRA-1 to Bind TRA-2 Also Promotes Spermatogenesis
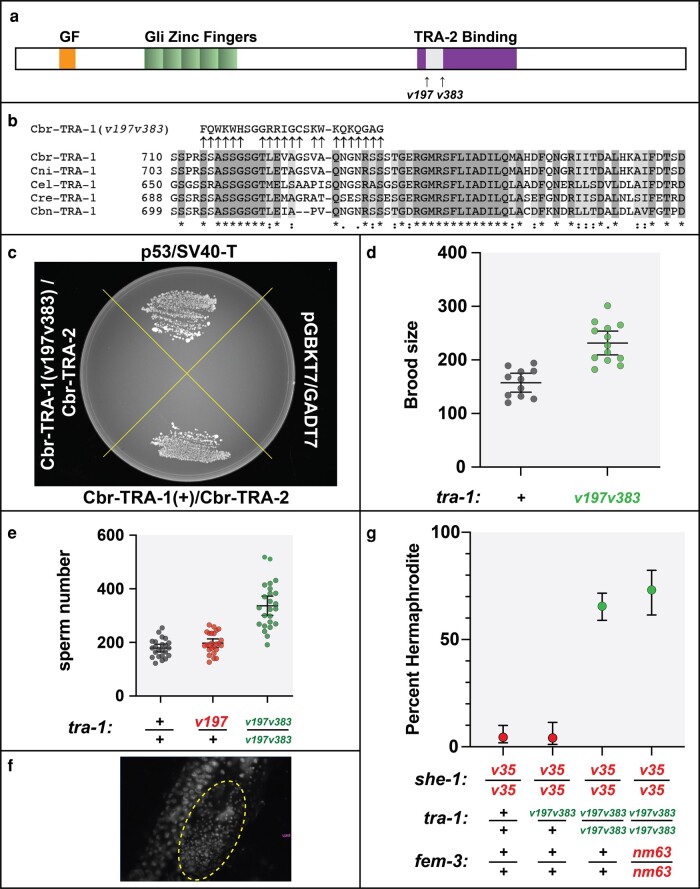
The TRA-1 transcription factor has a large conserved domain C-terminal to the five zinc fingers (de Bono and Hodgkin 1996), which is known to bind TRA-2 in both C. elegans (Lum et al. 2000) and C. briggsae (Wang and Kimble 2001). Since we did not know which residues were critical to its function, we made sequential frameshift mutations in C. briggsae tra-1, which alter the region of 24 amino acids that lies between them (Fig. 4, a and b). These Cbr-tra-1(v197 v383) mutants develop as XX hermaphrodites and XO males, which indicates that TRA-1's normal function of repressing male development must be unaffected. However, a yeast two-hybrid assay shows that this double mutation does prevent Cbr-TRA-1 from binding TRA-2 (Fig. 4c).
Fig. 4.
A C. briggsae tra-1 mutation that blocks TRA-2-binding favors spermatogenesis. a) Diagram of the structure of C. briggsae TRA-1, indicating the location of the double frameshift mutant v197 v383. Orange marks the conserved gain-of-function domain, green the five conserved zinc fingers, and purple the conserved TRA-2-binding domain. b) Alignment of five Caenorhabditis TRA-1 proteins, showing part of the TRA-2-binding domain. It was prepared using MUSCLE. Identical residues are shaded dark gray, and residues with conservative substitutions are shaded light gray. c) Yeast two-hybrid results, with bait constructs listed before prey constructs. d) The number of self-progeny for animals of the indicated genotypes. The thick line indicates the mean, and the error bars represent 95% confidence intervals. e) The number of sperm in individuals of the indicated genotypes, observed following DAPI staining. f) Extensive sperm in one ovotestis of a tra-1(v197 v383) XX animal. g) Graph showing the percent of XX animals of each genotype that develop as self-fertile hermaphrodites, rather than as females. The error bars show 95% confidence intervals. N = 113, 73, 212, and 67, respectively.
Much like the Cbr-tra-2(mx) alleles, this tra-1 mutation increases hermaphrodite brood size (Fig. 4d). To confirm its effect on hermaphrodite germ cell fates, we directly counted sperm and saw a significant increase in number (Fig. 4e), which is even apparent simply by looking at the gonad (Fig. 4f). As a control, we studied tra-1(v197)/+ animals, which should make half the normal amount of TRA-1, and they had the wild-type amount of sperm.
As with tra-2, we also built double and triple mutants using she-1, to confirm that this tra-1 allele promotes spermatogenesis. As expected, the tra-1(v197 v383) mutation suppresses she-1 (Fig. 4g), and this effect is independent of fem-3. However, this suppression is mostly recessive, whereas that by Cbr-tra-2(mx) alleles is dominant.
When we build Cbr-tra-2(mx); tra-1(v197 v383) animals, some produced many extra sperm, and others had completely masculinized germ lines (supplementary fig. S4, Supplementary Material online). Thus, disrupting the ability of C. briggsae TRA-2 to bind TRA-1 causes excess spermatogenesis in hermaphrodites. Since double mutants show a more severe effect, we infer that none of the tra-2 or tra-1 single mutants completely disrupts binding on its own.
The C. elegans tra-2(mx) Alleles Prevent Sequestration of TRA-2 by TRA-1
These results imply a simple model for how TRA-2 works in C. briggsae hermaphrodites. First, TRA-2ic binds FEM-3 in the soma; this interaction protects TRA-1, leaving it free to turn off male genes. Second, TRA-2ic directly binds TRA-1 in germ cells, to help turn off genes that promote spermatogenesis. Although null alleles of tra-2 block both functions, the mx alleles only block its germline activity.
By contrast, the role of TRA-2 and its mx mutations in C. elegans has always been confusing. Although C. elegans tra-2(mx) alleles disrupt the interaction between TRA-2 and the master transcription factor TRA-1 (Lum et al. 2000; Wang and Kimble 2001), they result in XX animals that make only oocytes (Doniach 1986). By contrast, tra-2(null) alleles, which also prevent interaction with TRA-1, cause XX animals to make only sperm (Hodgkin and Brenner 1977). This comparison implies that something complex is going on in this part of the C. elegans sex determination pathway.
This complexity has been hard to address with classical epistasis, since many of the genes that control sex determination act at multiple points in the pathway (Ellis 2022). To address this problem, we carried out experiments looking at the effects of heterozygosity for mutations in the sex determination pathway on tra-2(mx) phenotypes. Wang and Kimble (2001) used this approach to study how decreasing TRA-1 dosage affected the developing germ cells of tra-2(mx) heterozygotes. Their studies suggest that TRA-1 behaves like a repressor of TRA-2(mx) activity, since lower levels of TRA-1 lead to an increase in the fraction of XX females.
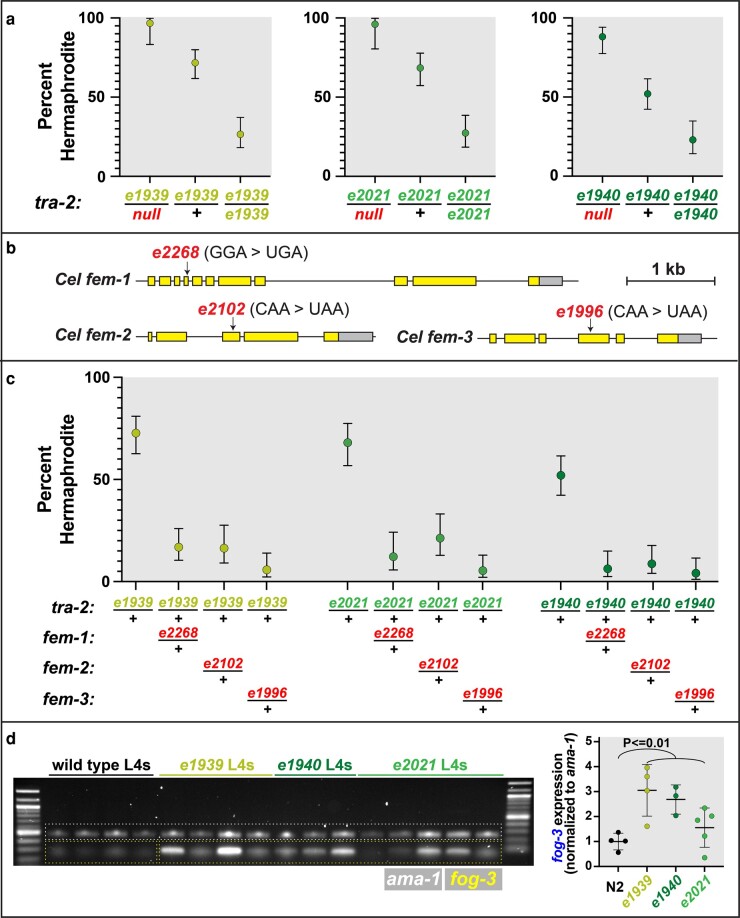
We began by studying tra-2 gene dosage, to learn how mx alleles altered TRA-2 activity itself. These experiments were inspired by the small number of tra-2(mx)/tra-2(null) animals observed by Doniach (1986). By using PCR or a marker mutation to identify these heterozygotes, we were able to amass large datasets for three tra-2(mx) alleles (Fig. 5a). For all three alleles, a tra-2(mx) allele in trans to a wild-type allele is significantly more likely to be female than one in trans to a null mutation. Thus, C. elegans tra-2(mx) alleles increase the normal germline function of TRA-2.
Fig. 5.
Caenorhabditis elegans tra-2(mx) alleles increase TRA-2's ability to target the FEM complex. a, c) Animals of the indicated genotypes were generated from crosses using tra-2(mx) fathers as described in the Materials and Methods and raised at 20 °C. On its own, each fem mutation is recessive. The error bars show 95% confidence intervals. a) N = 30, 92, 79, 25, 76, 73, 59, 100, and 61, respectively. c) N = 88, 89, 61, 69, 75, 49, 61, 75, 100, 64, 69, and 72, respectively. b) Location of the nonsense alleles we used in C. elegans fem genes. d) Semi-quantitative RT-PCR results showing fog-3 expression (lower band) and control ama-1 expression (upper band). These two products were produced in separate PCRs (using equivalent amounts of the cDNA template) and run on a single gel. Each lane used cDNA template produced from single mid-L4 XX larvae. The quantitation on the right was done using Carestream MI software. The fog-3 values are normalized to the average wild-type values for ama-1. (All calculations were done in arbitrary units.) Each genotype shows bars for the mean and standard deviation. The P-value was computed for a comparison of wild type with combined mutant data using the Mann–Whitney U test.
In addition, we studied interactions between tra-2(mx) alleles and null alleles of fem-1, fem-2, or fem-3, which encode components of the FEM complex. For these experiments, we used nonsense alleles (Fig. 5b) that are recessive and act zygotically. (By contrast, fem-1 deletions have a maternal effect due to the absence of germline licensing [Johnson and Spence 2011].) We observed strong genetic interactions between tra-2(mx) mutations and lowered expression of each fem gene (Fig. 5c). Moreover, these effects were particularly strong for fem-3, the direct target of TRA-2. We conclude that tra-2(mx) alleles increase inhibition of the FEM complex and that this effect is enhanced by reducing fem gene dosage, which is likely to result in fewer FEM complexes to compete with TRA-2.
These results suggest that in C. elegans TRA-1 negatively regulates TRA-2, resulting in higher activity of the FEM complex. Since the FEM complex not only targets TRA-1, but also acts downstream of it to promote spermatogenesis (reviewed by Ellis 2022), when tra-2(mx) mutations decrease FEM activity, they should favor oogenesis. Putting all these results together, we suggest that TRA-1 plays an important role in sequestering TRA-2 in the C. elegans germline, to facilitate hermaphrodite spermatogenesis.
Since this model depends on the downstream activity of the FEM complex, it does not require fog-3 transcript levels to be low to cause oogenesis in the tra-2(mx) mutants. We wondered whether the absence of an interaction with TRA-1 caused an increase in these transcripts, as is observed in C. briggsae. RT-PCR analyses show that this is in fact the case (Fig. 5d). Hence, TRA-2 binds TRA-1 in both species to lower the expression of the sperm regulator fog-3. However, in C. elegans the most important aspect of this interaction is that TRA-1 is sequestering TRA-2 in germ cells, allowing the FEM proteins to direct hermaphrodite spermatogenesis.
Discussion
Evolutionary biologists generally assume that if the genes in a regulatory pathway are conserved, their relationships within that pathway will have been conserved as well. For example, the identification of ras genes in C. elegans and Drosophila led to the hope that they could be used to elucidate the ras signal transduction pathway for other animals (Han 1992), a hope that was soon realized (Treisman 1996).
These types of studies often focus on long time scales and distantly related species, but a detailed picture of how evolutionary change occurs requires a narrower focus. Sex determination is a particularly useful trait, because it changes more rapidly than many other traits. In nematodes, these changes not only involve secondary sexual characteristics, but often reproductive traits and even entire mating systems (reviewed by Ellis and Lin 2014; Ellis 2016). To date, evolutionary studies using nematodes have focused on C. elegans and C. briggsae, because of the ease of using isogenic strains and hermaphrodite genetics to dissect the regulation of sexual traits.
In both species, tra-2 (Hodgkin and Brenner 1977; Kuwabara et al. 1992; Kuwabara 1996; Kelleher et al. 2008), fem-3 (Hodgkin 1986; Rosenquist and Kimble 1988; Haag et al. 2002; Hill and Haag 2009), and tra-1 (Hodgkin and Brenner 1977; Zarkower and Hodgkin 1992; de Bono and Hodgkin 1996; Kelleher et al. 2008) are sex determination genes with conserved structures. Moreover, their somatic functions are strongly conserved. However, fem-3 is absolutely required for spermatogenesis in C. elegans, but not in C. briggsae (Hill et al. 2006).
Not only is each gene conserved, the binding interactions between TRA-2 and FEM-3 (Mehra et al. 1999; Haag et al. 2002) and between TRA-2 and TRA-1 (Lum et al. 2000; Wang and Kimble 2001) are conserved as well. But although the TRA-2/TRA-1 interaction involves conserved residues near the C-terminus of TRA-2 (Fig. 2a), we show that the function of this interaction is exactly opposite in the two species. Identical tra-2(mx) mutations cause hermaphrodites to make oocytes instead of sperm in C. elegans, but sperm instead of oocytes in C. briggsae. How has this remarkable difference come to be?
The regulation in C. briggsae appears straightforward (Fig. 6). In this species, TRA-2 binds FEM-3 to promote female development throughout the soma, by preventing FEM-3 and its partners from causing TRA-1 degradation. Our data imply that TRA-2 also binds TRA-1 to promote oogenesis in the germ line, since the Cbr-tra-2(mx) mutants make extra sperm, and do so even in fem-3(null) mutants. Thus, C. briggsae TRA-2 promotes female development in both soma and germ line.
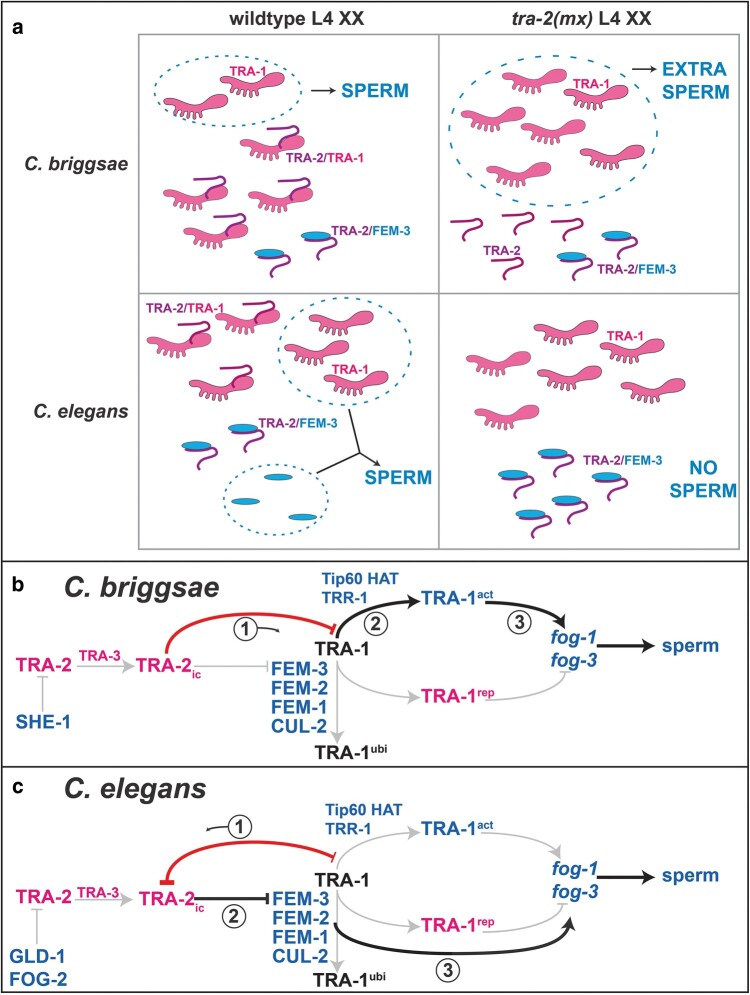
Fig. 6.
Recent evolutionary changes have rewired Caenorhabditis sex determination. a) Models for how the relative concentrations of TRA-2ic (purple loop), TRA-1 (pink), and FEM-3 (blue) specify germ cell fates. The stippled ovals enclose factors that promote spermatogenesis. In C. briggsae, TRA-2ic binds TRA-1 to prevent spermatogenesis. We present one possible mechanism by which it might act, in which full-length TRA-1 promotes the expression of sperm genes and TRA-2ic blocks this activation step. (Although TRA-1 repressor also participates, we omit it for clarity.) By contrast, C. elegans FEM-3 plays an essential role in spermatogenesis downstream of TRA-1. Thus, if TRA-2ic fails to bind TRA-1, it is free to inactivate FEM-3 and prevent spermatogenesis. b) Model for sex determination in the germ cells of larval C. briggsae hermaphrodites. Genes or proteins that promote male fates are in blue, and those that promote female fates in red. Proteins are in capitals and genes in lowercase italics. Arrows denote positive interactions, and “—|” denotes negative ones. Critical relationships in the regulation of spermatogenesis are highlighted in black and the key TRA-2/TRA-1 interaction in red. c) Similar model for C. elegans.
We previously suggested that TRA-1, like other Gli transcription factors, makes an activator as well as a cleaved repressor (Guo et al. 2013). Although TRA-1 repressor seems to turn off male genes throughout the body and germ line (Conradt and Horvitz 1999; Chen and Ellis 2000; Yi et al. 2000; Berkseth et al. 2013), our data suggested that TRA-1 activator specifically promotes spermatogenesis. Thus, one simple model is that TRA-2 binds TRA-1 activator in C. briggsae to stop it from inducing spermatogenesis. However, other models, such as TRA-2 facilitating the repression of fog-3 by TRA-1, also remain possible.
The situation in C. elegans is more complex, which is why these special tra-2 alleles were named mixomorphic—the mutations cause weak masculinization in the soma, but strongly promote oogenesis in the germ line. Hence, they have opposite effects on sexual development in different tissues. Based on several experiments, we propose that C. elegans TRA-1 binds to and sequesters TRA-2 in the germ line (Fig. 5, a and c). (i) Lowering the amount of TRA-1 increases the ability of tra-2(mx) alleles to promote oogenesis (Wang and Kimble 2001). This result suggests that TRA-1 could be a negative regulator of TRA-2 in germ cells. (ii) Increasing wild-type TRA-2 function in tra-2(mx) heterozygotes increases the likelihood that XX animals only make oocytes, which implies that the mx alleles augment the normal ability of TRA-2 to promote this fate. (iii) Decreasing the activity of the FEM complex increases the ability of tra-2(mx) alleles to promote oogenesis, which suggests that C. elegans TRA-2 regulates germ cells fates by inhibiting FEM-3 activity. And (iv) the tra-2(mx) mutants produce elevated levels of the sperm regulator fog-3, even though they make only oocytes. This final result suggests that the key factor that determines phenotype is low fem-3 activity, which is needed downstream of fog-3 transcription to allow spermatogenesis (Chen and Ellis 2000). Thus, we suggest that in C. elegans germ cells, TRA-1 regulates TRA-2 activity by sequestration and that this interaction prevents TRA-2 from binding FEM-3.
This situation would not be stable in C. elegans, if the FEM complex regulated germ cell fates only by controlling TRA-1. However, the three fem genes also have an essential spermatogenesis function downstream of TRA-1. In C. elegans, tra-1; fem double mutants make male bodies, the tra-1 phenotype, but only produce oocytes, the fem germ line phenotype (Hodgkin 1986). Furthermore, these double mutants have elevated expression of the critical spermatogenesis gene fog-3, a direct TRA-1 target, even though they make oocytes (Chen and Ellis 2000). Thus, the FEM complex regulates a downstream spermatogenesis factor, in addition to controlling TRA-1. This essential role for the C. elegans FEM complex in spermatogenesis means that regulation of TRA-2 levels by TRA-1 could indeed act through the FEM complex to control spermatogenesis.
Some evidence suggests that C. briggsae fem-3 retains the ability to promote spermatogenesis downstream of Cbr-tra-1 (Hill and Haag 2009), but it is not essential. Indeed, null mutants of Cbr-fem-2 or Cbr-fem-3 make sperm normally (Hill et al. 2006), and Cbr-fem-3 hermaphrodites produce the correct number of sperm before switching to oogenesis (Fig. 3e).
To date, the only molecular function known for the FEM complex is regulation of TRA-1 stability. Thus, an important goal for future research is identifying the downstream target of these proteins in C. elegans, and determining how the FEM complex controls its activity and why this control has been altered during recent evolution.
Our data show that C. briggsae she-1 controls hermaphrodite spermatogenesis by acting through tra-2 to control tra-1, since this regulation can occur even in fem-3 null mutants. By contrast, C. elegans fog-2 primarily acts through tra-2 to control fem-3 activity. These differences show that pathways with multiple branches are prone to changes that decrease or even eliminate the function of one of those branches. Despite the important changes we have observed, the sex determination pathway still retains multiple roles for the key genes tra-2, fem-3, and tra-1 in both species. However, the directionality and significance of their interactions have changed dramatically.
If TRA-1 indeed regulates the activity of its binding partner TRA-2 by sequestration, this result opens up new possibilities for the control of development by transcription factors. In some situations, they might determine cell fates not only by regulating the expression of target genes, but also by directly altering the levels of other binding factors. This would allow for multiple regulatory branches in a heretofore unanticipated manner.
Taken together, our results and those of previous studies suggest that hermaphrodite development is based on fundamentally different approaches in C. elegans and C. briggsae.
In C. elegans, the levels of TRA-2 and FEM-3 in the germ cells are tightly balanced, so that a change in either one can alter the decision to produce sperm or oocytes (Doniach 1986; Barton et al. 1987; Schedl and Kimble 1988). Furthermore, recent studies suggest that TRA-2 protein levels in the germ line are very low, so low as to be almost undetectable (Hu et al. 2019). These observations point to a model in which the germline expression of both tra-2 and fem-3 was lowered and balanced during the evolution of self-fertility. This arrangement would make it easier for the same germ line to produce sperm early in life, and later switch to oogenesis.
By contrast, C. briggsae hermaphrodites appear to have low or nonexistent expression of fem-3 in germ cells, and the regulation of spermatogenesis is instead controlled by the modulation of TRA-2 and TRA-1. As a consequence, fem-3 null mutations have no effect on hermaphrodite reproduction in this species (Hill et al. 2006). Indeed, their effects can only be discerned if TRA-2 is completely absent.
Although we now have a good picture of the differences between C. elegans and C. briggsae, we need to dissect the process of sex determination in their male/female relatives to be able to figure out the times and directions of the major events that shaped these species. Such studies had previously been impractical, but the advent of rapid gene editing should soon lead to results.
Materials and Methods
Strains
The following strains were used: C. elegans: N2, dpy-10(e128) (Brenner 1974) dpy-10(cn64) (Levy et al. 1993) tra-2(e1939mx), tra-2(e1940mx) and tra-2(e2021mx) II (Doniach 1986), tra-1(e1099) III (Hodgkin and Brenner 1977), fem-1(e2268) (Johnson and Spence 2011), fem-2(e2102) (Hodgkin 1986), and fem-3(e1996) (Hodgkin 1986). C. briggsae: AF16 (Fodor et al. 1983), tra-2(v440) II, tra-2(v393mx) II, tra-2(v402mx) II and tra-2(v403mx), tra-1(v197v383) III (this paper), dpy-18(mf104) III (Winter et al. 2007), she-1(v35) IV (Guo et al. 2009), and fem-3(nm63) IV (Hill et al. 2006).
Genetics
Animals were maintained at 20 °C on plates seeded with AMA1004, unless otherwise indicated. Procedures for raising animals and building double mutants were based on work by Brenner (1974). If necessary, genotypes were tested using single-worm PCR. For Fig. 5a, we studied C. elegans tra-2(mx) homozygotes from a male/female strain, heterozygotes from a cross of tra-2(mx) males by dpy-18 unc-32 hermaphrodites, and tra-2(mx)/tra-2(e1095null) animals from crosses of tra-2(mx) males by tra-2(e1095)/dpy-10 hermaphrodites. All animals were raised at 20 °C. For Fig. 5c, we crossed tra-2(mx) males by null mutants in the fem genes and observed the heterozygous progeny for enhancement of feminization.
Genome Editing
Mutations were made using gene editing with TALENs (Wood et al. 2011; Wei et al. 2014) or CRISPR (Arribere et al. 2014; Farboud et al. 2019). Specific edits were made using single-strand oligos as repair templates. All solutions were injected into the germline syncytium of young adult hermaphrodites, and F1 progeny screened for mutations by the PCR on acrylamide minigels, after they had successfully reproduced. Homozygotes were isolated from among their progeny.
Phenotype Assays
To identify hermaphrodites or females, individual L4 larvae were picked onto separate plates and scored as hermaphrodite by the production of eggs, or as female by the absence of eggs and the presence of stacked oocytes in the gonad. If necessary, genotypes were determined by single-worm PCR analysis. To assess dominant suppression of she-1 female phenotypes, she-1(v35) males were crossed with the appropriate double mutant XX animals, after which the F1 were raised and scored at 25 °C. If necessary, genotypes were determined by single-worm PCR analysis after their phenotypes had been determined.
Brood Counts
Individual L4 hermaphrodites were plated at 20 °C and passed to new plates every 8 to 14 h. After transfer, eggs and larvae were counted. Animals that did not survive through the period of sperm exhaustion were not included in the assays.
Sperm Counts
Young adults were stained with DAPI, their germ lines were photographed at four separate focal planes using fluorescent microscopy, and then, sperm were counted.
Quantitative RT-PCR
For C. briggsae, groups of five animals of the indicated ages were picked into 2 µl of sterile water in the lid of a microcentrifuge tube and then spun into a mixture of 50 µl of RNAzol (MRC Inc.), 16 µl of water, and 3 µl of precipitation carrier (MRC Inc.). The worms were lysed by freezing at −70 °C, thawing at 65 °C, and 30″ sonication on power 9 with a Misonix cup-horn sonicator. Afterward, we followed the MRC protocol for RNA preparation. The final pellet was resuspended in 20 µl of water, and 10 µl was used for reverse transcription, yielding 20 µl of cDNA. From this total, 1.5 µl of template was used for individual PCRs. The number of cycles was limited so that one-third and one-ninth dilutions of template were clearly distinguishable.
For C. elegans, we pushed the technology to study samples from individual worms. The only change we made was to add a proteinase K lysis step (Ly et al. 2015) before proceeding with the RNAzol procedure, as described above. For single animals, we resuspended the RNA in 10 µl and used all of it for reverse transcription.
Yeast Two-Hybrid Assays
The C-terminus of cDNA encoding TRA-1 was cloned into the bait vector pGBKT7 (Promega), to be fused with the GAL4 DNA-binding domain. The C-termini of cDNAs encoding different alleles of TRA-2 were cloned into the prey vector pGADT7 (Promega), to be fused with the GAL4 DNA activation domain. Each insertion was confirmed by DNA sequencing. Yeast transformations were performed using standard protocols (Lundblad 2001) with bait constructs into strain AH109 and prey into strain Y187. Afterward, the transformed AH109 and Y187 strains were crossed to generate progeny that contained both bait and prey vectors, using SD-Trp/-Leu dropout selection. The pGBKT7-p53/pGADT7-SV40-T–antigen pair was used as a positive control and empty bait/prey vector as a negative control. To test TRA-1/TRA-2 interactions, the established strains containing bait and prey were streaked on SD-Trp/-Leu dropout plates and incubated at 30 °C for 2 d, until the clones had grown to 1 to 2 mm2. Next, the appropriate clones were picked into H2O and 5 µl of 105/µl cells were streaked on SD-Trp/-Leu/-His/-Ade dropout plates and incubated at 30 °C for 3 to 5 d. The end-point was when colonies had grown up for the positive but not negative controls.
For Cbr-TRA-1, we used the C-terminus (residues 661-1146 of TRA-1A), and for Cel-TRA-1 we used the corresponding region (residues 624-1110). For Cbr-TRA-2, we used residues 1102-1496 of TRA-2A, and for Cel-TRA-2, we used residues 1089-1475.
Table of Primers
| Cbr-fog-3 | cbr-fog-3RT-F2 | TTCCACTCGCGTTGGAGAAG |
| … | cbr-fog-3RT-R2 | CGGATGTTGGCTTGAACGTG |
| Cbr-ama-1 | RE1041 | CGACAACCCACTCTCCATAA |
| … | RE1042 | GCCAATCGATGAAGATGTCAC |
| Cel-fog-3 | RE1033 | TTTGGCGCTGAACTTGGAAA |
| … | RE1034 | CATCGCAGTTCACATCTCCA |
| Cel-ama-1 | RE1049 | CCGACTCTCCACAAAATGTCA |
| … | RE1050 | GGACGGCGCAGAGAGTATC |
Supplementary Material
Acknowledgments
The authors thank D. Pilgrim and J. Hodgkin for strains. Additional strains were provided by the CGC, which was funded by the NIH Office of Research Infrastructure Programs (P40 OD010440). The authors also thank NIH Grants R01GM118836 and R01GM121688 for funding and the American Cancer Society for a Postdoctoral Award to support Shin-Yi Lin (126627-PF-15-228-01-DDC).
Contributor Information
Yongquan Shen, Department of Molecular Biology, Rowan-Virtua School of Translational Biomedical Engineering and Science, Rowan-Virtua School of Osteopathic Medicine, Stratford, NJ 08084, USA.
Shin-Yi Lin, Department of Molecular Biology, Rowan-Virtua School of Translational Biomedical Engineering and Science, Rowan-Virtua School of Osteopathic Medicine, Stratford, NJ 08084, USA.
Jonathan Harbin, Department of Molecular Biology, Rowan-Virtua School of Translational Biomedical Engineering and Science, Rowan-Virtua School of Osteopathic Medicine, Stratford, NJ 08084, USA.
Richa Amin, Department of Molecular Biology, Rowan-Virtua School of Translational Biomedical Engineering and Science, Rowan-Virtua School of Osteopathic Medicine, Stratford, NJ 08084, USA.
Allison Vassalotti, Department of Molecular Biology, Rowan-Virtua School of Translational Biomedical Engineering and Science, Rowan-Virtua School of Osteopathic Medicine, Stratford, NJ 08084, USA.
Joseph Romanowski, Department of Molecular Biology, Rowan-Virtua School of Translational Biomedical Engineering and Science, Rowan-Virtua School of Osteopathic Medicine, Stratford, NJ 08084, USA.
Emily Schmidt, Department of Molecular Biology, Rowan-Virtua School of Translational Biomedical Engineering and Science, Rowan-Virtua School of Osteopathic Medicine, Stratford, NJ 08084, USA.
Alexis Tierney, Department of Molecular Biology, Rowan-Virtua School of Translational Biomedical Engineering and Science, Rowan-Virtua School of Osteopathic Medicine, Stratford, NJ 08084, USA.
Ronald E Ellis, Department of Molecular Biology, Rowan-Virtua School of Translational Biomedical Engineering and Science, Rowan-Virtua School of Osteopathic Medicine, Stratford, NJ 08084, USA.
Supplementary Material
Supplementary material is available at Molecular Biology and Evolution online.
Data Availability
The data underlying this article are available in the article and in its online supplementary material. Strains are available upon request.
References
- Ahringer J, Kimble J. Control of the sperm-oocyte switch in Caenorhabditis elegans hermaphrodites by the fem-3 3′ untranslated region. Nature. 1991:349(6307):346–348. 10.1038/349346a0. [DOI] [PubMed] [Google Scholar]
- Arribere JA, Bell RT, Fu BX, Artiles KL, Hartman PS, Fire AZ. Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics. 2014:198(3):837–846. 10.1534/genetics.114.169730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes TM, Hodgkin J. The tra-3 sex determination gene of Caenorhabditis elegans encodes a member of the calpain regulatory protease family. EMBO J. 1996:15(17):4477–4484. 10.1002/j.1460-2075.1996.tb00825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton MK, Schedl TB, Kimble J. Gain-of-function mutations of fem-3, a sex-determination gene in Caenorhabditis elegans. Genetics. 1987:115(1):107–119. 10.1093/genetics/115.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkseth M, Ikegami K, Arur S, Lieb JD, Zarkower D. TRA-1 ChIP-seq reveals regulators of sexual differentiation and multilevel feedback in nematode sex determination. Proc Natl Acad Sci USA. 2013:110(40):16033–16038. 10.1073/pnas.1312087110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974:77(1):71–94. 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PJ, Cho S, Jin SW, Ellis RE. Specification of germ cell fates by FOG-3 has been conserved during nematode evolution. Genetics. 2001:158(4):1513–1525. 10.1093/genetics/158.4.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PJ, Ellis RE. TRA-1A regulates transcription of fog-3, which controls germ cell fate in C. elegans. Development. 2000:127(14):3119–3129. 10.1242/dev.127.14.3119. [DOI] [PubMed] [Google Scholar]
- Clifford R, Lee MH, Nayak S, Ohmachi M, Giorgini F, Schedl T. FOG-2, a novel F-box containing protein, associates with the GLD-1 RNA binding protein and directs male sex determination in the C. elegans hermaphrodite germline. Development. 2000:127(24):5265–5276. 10.1242/dev.127.24.5265. [DOI] [PubMed] [Google Scholar]
- Conradt B, Horvitz HR. The TRA-1A sex determination protein of C. elegans regulates sexually dimorphic cell deaths by repressing the egl-1 cell death activator gene. Cell. 1999:98(3):317–327. 10.1016/S0092-8674(00)81961-3. [DOI] [PubMed] [Google Scholar]
- de Bono M, Hodgkin J. Evolution of sex determination in Caenorhabditis: unusually high divergence of tra-1 and its functional consequences. Genetics. 1996:144(2):587–595. 10.1093/genetics/144.2.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doniach T. Activity of the sex-determining gene tra-2 is modulated to allow spermatogenesis in the C. elegans hermaphrodite. Genetics. 1986:114(1):53–76. 10.1093/genetics/114.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis RE. “The persistence of memory”—hermaphroditism in nematodes. Mol Reprod Dev. 2016:84(2):144–157. 10.1002/mrd.22668. [DOI] [PubMed] [Google Scholar]
- Ellis RE. Sex determination in nematode germ cells. Sex Dev. 2022:16(5-6):1–18. 10.1159/000520872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis RE, Kimble J. The fog-3 gene and regulation of cell fate in the germ line of Caenorhabditis elegans. Genetics. 1995:139(2):561–577. 10.1093/genetics/139.2.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis RE, Lin SY. The evolutionary origins and consequences of self-fertility in nematodes. F1000Prime Rep. 2014:6:62. 10.12703/P6-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farboud B, Severson AF, Meyer BJ. Strategies for efficient genome editing using CRISPR-Cas9. Genetics. 2019:211(2):431–457. 10.1534/genetics.118.301775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fodor A, Riddle DL, Nelson FK, Golden JW. Comparison of a new wild-type Caenorhabditis briggsae with laboratory strains of C. briggsae and C. elegans. Nematologica. 1983:29(2):203–217. 10.1163/187529283X00456. [DOI] [Google Scholar]
- Guo Y, Chen X, Ellis RE. Evolutionary change within a bipotential switch shaped the sperm/oocyte decision in hermaphroditic nematodes. PLoS Genet. 2013:9(10):e1003850. 10.1371/journal.pgen.1003850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Lang S, Ellis RE. Independent recruitment of F box genes to regulate hermaphrodite development during nematode evolution. Curr Biol. 2009:19(21):1853–1860. 10.1016/j.cub.2009.09.042. [DOI] [PubMed] [Google Scholar]
- Haag ES, Wang S, Kimble J. Rapid coevolution of the nematode sex-determining genes fem-3 and tra-2. Curr Biol. 2002:12(23):2035–2041. 10.1016/S0960-9822(02)01333-7. [DOI] [PubMed] [Google Scholar]
- Han M. Ras proteins in developmental pattern formation in Caenorhabditis elegans and Drosophila. Semin Cancer Biol. 1992:3(4):219–228. [PubMed] [Google Scholar]
- Hill RC, de Carvalho CE, Salogiannis J, Schlager B, Pilgrim D, Haag ES. Genetic flexibility in the convergent evolution of hermaphroditism in Caenorhabditis nematodes. Dev Cell. 2006:10(4):531–538. 10.1016/j.devcel.2006.02.002. [DOI] [PubMed] [Google Scholar]
- Hill RC, Haag ES. A sensitized genetic background reveals evolution near the terminus of the Caenorhabditis germline sex determination pathway. Evol Dev. 2009:11(4):333–342. 10.1111/j.1525-142X.2009.00340.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin J. Sex determination in the nematode C. elegans: analysis of tra-3 suppressors and characterization of fem genes. Genetics. 1986:114(1):15–52. 10.1093/genetics/114.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin JA, Brenner S. Mutations causing transformation of sexual phenotype in the nematode Caenorhabditis elegans. Genetics. 1977:86(2):275–287. 10.1093/genetics/86.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S, Skelly LE, Kaymak E, Freeberg L, Lo TW, Kuersten S, Ryder SP, Haag ES. Multi-modal regulation of C. elegans hermaphrodite spermatogenesis by the GLD-1-FOG-2 complex. Dev Biol. 2019:446(2):193–205. 10.1016/j.ydbio.2018.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin SW, Kimble J, Ellis RE. Regulation of cell fate in Caenorhabditis elegans by a novel cytoplasmic polyadenylation element binding protein. Dev Biol. 2001:229(2):537–553. 10.1006/dbio.2000.9993. [DOI] [PubMed] [Google Scholar]
- Johnson CL, Spence AM. Epigenetic licensing of germline gene expression by maternal RNA in C. elegans. Science. 2011:333(6047):1311–1314. 10.1126/science.1208178. [DOI] [PubMed] [Google Scholar]
- Kelleher DF, de Carvalho CE, Doty AV, Layton M, Cheng AT, Mathies LD, Pilgrim D, Haag ES. Comparative genetics of sex determination: masculinizing mutations in Caenorhabditis briggsae. Genetics. 2008:178(3):1415–1429. 10.1534/genetics.107.073668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiontke KC, Félix MA, Ailion M, Rockman MV, Braendle C, Penigault JB, Fitch DH. A phylogeny and molecular barcodes for Caenorhabditis, with numerous new species from rotting fruits. BMC Evol Biol. 2011:11(1):339. 10.1186/1471-2148-11-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwabara PE. Interspecies comparison reveals evolution of control regions in the nematode sex-determining gene tra-2. Genetics. 1996:144(2):597–607. 10.1093/genetics/144.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwabara PE, Okkema PG, Kimble J. tra-2 encodes a membrane protein and may mediate cell communication in the Caenorhabditis elegans sex determination pathway. Mol Biol Cell. 1992:3(4):461–473. 10.1091/mbc.3.4.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwabara PE, Okkema PG, Kimble J. Germ-line regulation of the Caenorhabditis elegans sex-determining gene tra-2. Dev Biol. 1998:204(1):251–262. 10.1006/dbio.1998.9062. [DOI] [PubMed] [Google Scholar]
- Levy AD, Yang J, Kramer JM. Molecular and genetic analyses of the Caenorhabditis elegans dpy-2 and dpy-10 collagen genes: a variety of molecular alterations affect organismal morphology. Mol Biol Cell. 1993:4(8):803–817. 10.1091/mbc.4.8.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum DH, Kuwabara PE, Zarkower D, Spence AM. Direct protein-protein interaction between the intracellular domain of TRA-2 and the transcription factor TRA-1A modulates feminizing activity in C. elegans. Genes Dev. 2000:14(24):3153–3165. 10.1101/gad.853700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundblad V. Yeast cloning vectors and genes. Curr Protoc Mol Biol. 2001. 10.1002/0471142727.mb1304s21. Chapter 13 Unit13.4. [DOI] [PubMed] [Google Scholar]
- Ly K, Reid SJ, Snell RG. Rapid RNA analysis of individual Caenorhabditis elegans. MethodsX. 2015:2:59–63. 10.1016/j.mex.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehra A, Gaudet J, Heck L, Kuwabara PE, Spence AM. Negative regulation of male development in Caenorhabditis elegans by a protein-protein interaction between TRA-2A and FEM-3. Genes Dev. 1999:13(11):1453–1463. 10.1101/gad.13.11.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak S, Goree J, Schedl T. fog-2 and the evolution of self-fertile hermaphroditism in Caenorhabditis. PLoS Biol. 2005:3(1):e6. 10.1371/journal.pbio.0030006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okkema PG, Kimble J. Molecular analysis of tra-2, a sex determining gene in C.elegans. EMBO J. 1991:10(1):171–176. 10.1002/j.1460-2075.1991.tb07933.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenquist TA, Kimble J. Molecular cloning and transcript analysis of fem-3, a sex-determination gene in Caenorhabditis elegans. Genes Dev. 1988:2(5):606–616. 10.1101/gad.2.5.606. [DOI] [PubMed] [Google Scholar]
- Schedl T, Graham PL, Barton MK, Kimble J. Analysis of the role of tra-1 in germline sex determination in the nematode Caenorhabditis elegans. Genetics. 1989:123(4):755–769. 10.1093/genetics/123.4.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schedl T, Kimble J. fog-2, a germ-line-specific sex determination gene required for hermaphrodite spermatogenesis in Caenorhabditis elegans. Genetics. 1988:119(1):43–61. 10.1093/genetics/119.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schvarzstein M, Spence AM. The C. elegans sex-determining GLI protein TRA-1A is regulated by sex-specific proteolysis. Dev Cell. 2006:11(5):733–740. 10.1016/j.devcel.2006.09.017. [DOI] [PubMed] [Google Scholar]
- Singson A. Every sperm is sacred: fertilization in Caenorhabditis elegans. Dev Biol. 2001:230(2):101–109. 10.1006/dbio.2000.0118. [DOI] [PubMed] [Google Scholar]
- Sokol SB, Kuwabara PE. Proteolysis in Caenorhabditis elegans sex determination: cleavage of TRA-2A by TRA-3. Genes Dev. 2000:14(8):901–906. 10.1101/gad.14.8.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starostina NG, Lim JM, Schvarzstein M, Wells L, Spence AM, Kipreos ET. A CUL-2 ubiquitin ligase containing three FEM proteins degrades TRA-1 to regulate C. elegans sex determination. Dev Cell. 2007:13(1):127–139. 10.1016/j.devcel.2007.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treisman R. Regulation of transcription by MAP kinase cascades. Curr Opin Cell Biol. 1996:8(2):205–215. 10.1016/S0955-0674(96)80067-6. [DOI] [PubMed] [Google Scholar]
- Wang S, Kimble J. The TRA-1 transcription factor binds TRA-2 to regulate sexual fates in Caenorhabditis elegans. EMBO J. 2001:20(6):1363–1372. 10.1093/emboj/20.6.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Q, Shen Y, Chen X, Shifman Y, Ellis RE. Rapid creation of forward-genetics tools for C. briggsae using TALENs: lessons for nonmodel organisms. Mol Biol Evol. 2014:31(2):468–473. 10.1093/molbev/mst213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter AD, Keskiaho K, Kukkola L, McCormack G, Felix MA, Myllyharju J, Page AP. Differences in collagen prolyl 4-hydroxylase assembly between two Caenorhabditis nematode species despite high amino acid sequence identity of the enzyme subunits. Matrix Biol. 2007:26(5):382–395. 10.1016/j.matbio.2007.01.008. [DOI] [PubMed] [Google Scholar]
- Wood AJ, Lo TW, Zeitler B, Pickle CS, Ralston EJ, Lee AH, Amora R, Miller JC, Leung E, Meng X, et al. Targeted genome editing across species using ZFNs and TALENs. Science. 2011:333(6040):307. 10.1126/science.1207773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi W, Ross JM, Zarkower D. mab-3 is a direct tra-1 target gene regulating diverse aspects of C. elegans male sexual development and behavior. Development. 2000:127(20):4469–4480. 10.1242/dev.127.20.4469. [DOI] [PubMed] [Google Scholar]
- Zarkower D (2006), ‘Somatic sex determination’, in C. elegans Research Community The (ed.), WormBook (Wormbook.org), doi/ 10.1895/wormbook.1.84.1. [DOI] [PMC free article] [PubMed]
- Zarkower D, Hodgkin J. Molecular analysis of the C. elegans sex-determining gene tra-1: a gene encoding two zinc finger proteins. Cell. 1992:70(2):237–249. 10.1016/0092-8674(92)90099-X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in the article and in its online supplementary material. Strains are available upon request.