Abstract
Embryo quality is an important determinant of successful implantation and a resultant live birth. Current clinical approaches for evaluating embryo quality rely on subjective morphology assessments or an invasive biopsy for genetic testing. However, both approaches can be inherently inaccurate and crucially, fail to improve the live birth rate following the transfer of in vitro produced embryos. Optical imaging offers a potential non-invasive and accurate avenue for assessing embryo viability. Recent advances in various label-free optical imaging approaches have garnered increased interest in the field of reproductive biology due to their ability to rapidly capture images at high resolution, delivering both morphological and molecular information. This burgeoning field holds immense potential for further development, with profound implications for clinical translation. Here, our review aims to: (1) describe the principles of various imaging systems, distinguishing between approaches that capture morphological and molecular information, (2) highlight the recent application of these technologies in the field of reproductive biology, and (3) assess their respective merits and limitations concerning the capacity to evaluate embryo quality. Additionally, the review summarizes challenges in the translation of optical imaging systems into routine clinical practice, providing recommendations for their future development. Finally, we identify suitable imaging approaches for interrogating the mechanisms underpinning successful embryo development.
Keywords: embryo assessment, optical imaging, autofluorescence, label-free imaging, metabolic imaging, non-invasive, hyperspectral imaging, fluorescent lifetime microscopy, light sheet microscopy
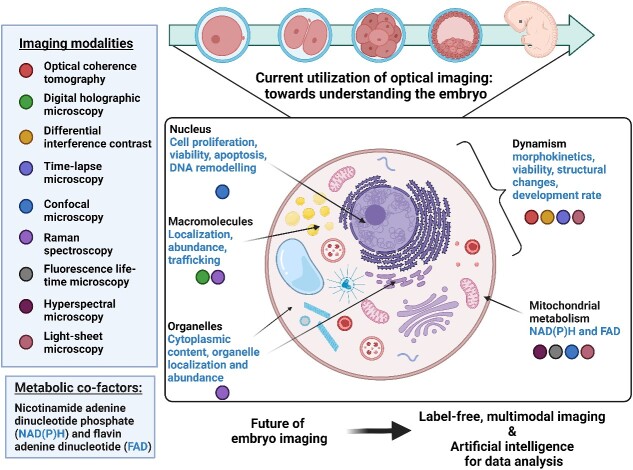
This review describes some of the most commonly used optical imaging approaches and explores their advantages, disadvantages, and potential use for viewing the embryo in the absence of exogenous tags.
Graphical Abstract
Graphical Abstract.
Introduction
Globally, one in every six individuals is affected by infertility [1]. This is defined as the inability to establish a pregnancy following 12 months of unprotected sexual intercourse [2, 3]. Assisted reproductive technologies, such as in vitro fertilization (IVF), are frequently used by those struggling with infertility. There is an apparent year-on-year increase in the demand for IVF with the number of cycles performed rising by ~8% each year since 1999 (overall four-fold increase in number of cycles from 1999 to 2021; Australia/New Zealand data; [4]). However, the success rate has remained at ~20% for more than a decade (live births per initiated cycle; [4]). This poses significant challenges—including financial, emotional, and physical burdens—for those who use IVF to overcome infertility. Embryo quality is a key determinant of pregnancy success in IVF [5]. Presently, the predominant method for assessing embryo quality involves morphological inspection by an embryologist. While many studies have sought to correlate embryo morphology with IVF outcomes (reviewed in [6]), the accuracy of this approach is low. This is likely due to morphological assessments being highly subjective with inherent inaccuracies in the evaluation process [7, 8]. An alternative approach for assessing embryo quality is preimplantation genetic testing (PGT). This involves genetic analysis of a small number of cells (biopsy) taken from the placenta cell lineage of a blastocyst-stage embryo [9]. However, in some embryos, this biopsy will fail to diagnose the presence of genetic aberrations within the same embryo due to mosaicism (a mixture of cells: a proportion with the expected number of chromosomes and a proportion with a deviation in this number) [10, 11]. Additionally, the transfer of biopsied embryos has been associated with an increased risk of developing preeclampsia, a pregnancy complication that predisposes the mother and child to a lifetime increased risk of developing cardiovascular disease [12]. Therefore, the pursuit of an accurate and non-invasive method for assessing embryo quality remains elusive: light-based imaging can fulfill this need.
Light offers an unprecedented route for imaging. The ability to select wavelengths means we can tune to the excitation of specific molecules within the sample, offering a high degree of selectivity. Furthermore, light can reflect and refract at boundaries between different substrates (or cells/tissues) in a manner indicative of the physical properties of the layers in question. As a result, harnessing and manipulating light presents an opportunity to investigate both morphological and molecular changes within living cells, revealing dynamic and diagnostic information on cellular health. This inherent quality has piqued significant interest in the field of reproductive biology. In particular, the potential application of light-based techniques for the assessment of embryo developmental potential. Historically, embryo imaging has been achieved through label-based methods, involving fixation and subsequent staining or labeling (reviewed in [13]). This includes both traditional histology as well as the employment of exogenous fluorescent molecules (immunohistochemistry), where different cellular components can be visualized. For live embryo imaging, the employment of genetically modified organisms expressing endogenous fluorescence labels, such as green fluorescence protein (GFP), is commonly used [14, 15]. All of the above-mentioned methodologies require the use of exogenous labels or stains, which may alter the innate biological processes within cells [16]. Importantly, such approaches are not suitable for the early embryo as they may negatively affect viability and downstream development potential. Consequently, there has been a shift toward label-free imaging to visualize the preimplantation embryo.
Recent advancements in label-free imaging have enabled the rapid acquisition of high-resolution images of the embryo, providing detailed information on morphology as well as molecular and bio-physical properties [17]. However, reproductive biologists and clinicians may not be aware of the available optical approaches and their potential for revealing mechanisms underpinning successful preimplantation embryo development. In this review, we describe the principles of the most commonly used optical imaging modalities, evaluate their merits and limitations, and importantly, assess their potential to develop into a powerful tool for label-free assessment of embryos. Additionally, we explore how these imaging techniques may inspire novel research questions previously hindered by the lack of suitable tools.
Principle of label-free imaging approaches
Label-free imaging techniques exploit the interplay between light and matter [18]. This interaction can provide insight into intricate biological processes within cells that are often concealed when using standard light microscopy. Consequently, label-free imaging presents an enticing avenue for the evaluation of cellular health and function. Label-free techniques can be divided into two broad categories based on the information they reveal: morphological or molecular. These mainly refer to the source of contrast in the respective imaging techniques employed. In this review, we will focus on label-free imaging technologies such as optical coherence tomography (OCT), digital holographic microscopy (DHM), and differential interference contrast (DIC) microscopy that generate a high-contrast image of morphological changes by utilizing the interaction between underlying cellular structures and the illuminating light. In essence, the path of light can be altered as it traverses through the sample. The change in the light path is dependent on the inherent physical properties of the sample. Furthermore, we also describe fluorescence microscopy such as confocal microscopy, fluorescence lifetime microscopy (FLIM), hyperspectral microscopy, and light sheet microscopy that achieve high-contrast images based on the fluorescence emitted by exogenous (labels) or endogenous molecules (autofluorescence). Raman spectroscopy also relies on endogenous fluorophores but uses the inelastic scattering of light (a shift in the energy of photons as light interacts with intracellular molecules) to provide a unique fingerprint of the chemical composition of cells.
Historically, autofluorescence from cells was considered background fluorescence, contributing to and contaminating the signal captured from specific fluorescence tags or stains [19]. More recently, researchers have taken advantage of such autofluorescence by exploiting a wide range of endogenous fluorophores that are associated with cellular function, such as amino acids, vitamins, lipids, and metabolic cofactors [20]. To date, advancements in microscopy techniques have shed new light on the potential for autofluorescence to reveal the inner biochemistry of living cells in a spatio-temporal manner [21, 22]. For example, the naturally fluorescent metabolic cofactors—nicotinamide adenine dinucleotide (NADH), nicotinamide adenine dinucleotide phosphate (NADPH) and flavin adenine dinucleotide (FAD)—may be used to non-invasively measure cellular metabolism [23]. As embryo metabolism is intimately associated with viability [20], exploiting the fluorescence signals from these metabolic cofactors has shown to be a valuable and non-invasive indicator of embryo quality [22, 24]. Here, we review existing studies that use label-free optical approaches to image embryos, categorizing them based on the type of information they provide—morphology or molecular (summarized in Graphical Abstract and Table 1).
Table 1.
Summary of existing imaging modalities used in reproductive biology
| Imaging system | Main application | Speed, spatial resolution, field of view (FOV)/depth of imaging | Pros & cons | Adaptability to clinic/field, compatibility for embryo imaging | Citations |
|---|---|---|---|---|---|
| Morphological information | |||||
| Optical coherence tomography (OCT) | Tissue structure |
Speed – Real-time, milliseconds Spatial resolution – High axial resolution (micron level), variable lateral resolution ~10–15 μm Field of view: – Limited to a few mm in depth |
Pros: – High-resolution, cross-sectional imaging – Non-invasive and real-time imaging – Suitable for 3D reconstruction Cons: – Limited penetration in highly scattering tissue – Limited contrast in some cases – Motion artifacts due to sample movement |
Medium—compatible for embryo imaging due to low risk of phototoxicity | [25–37, 157] |
| Differential interference contrast microscopy (DIC) | Cell structure |
Speed – Real time Spatial resolution – Good lateral resolution (sub-μm) but limited depth information Field of view: – Moderate, up to a few hundred μm |
Pros: – Enhanced contrast for transparent samples – Real-time imaging of live, unstained embryos Cons: – Limited depth information, 3D capabilities – Sensitive to focus adjustments and vibrations |
Low—compatible with embryo imaging due to low risk of phototoxicity | [38–44] |
| Digital holographic microscopy (DHM) | Bio-physical parameters (e.g., refractive index) |
Speed – Real time; several frames per second Spatial resolution – Submicron lateral resolution (~100–200 μm) Field of view: – Moderate; up to a few hundred μm |
Pros: – Quantitative phase imaging – Label-free and non-destructive – High spatial resolution Cons: – Limited depth penetration – Complex setup and data analysis – Susceptible to vibrations and environmental changes |
High—compatible for embryo imaging due to low risk of phototoxicity | [45–50] |
| Time-lapse microscopy | Morphological changes and developmental rate |
Speed – Real time or time-lapse; various frame intervals (seconds to hours) Spatial resolution – Lateral resolution varies based on setup, from sub-μm to μm Field of view: – Varies, often limited to small regions of cell |
Pros: – Dynamic observation of development – Real-time monitoring of cellular processes – Longitudinal, observational studies of living cells Cons: – Limited to surface imaging in some cases – Require special equipment and culture conditions – Limited depth of field |
High—compatible for embryo imaging due to low risk of phototoxicity | [51–63] |
| Molecular information | |||||
| Raman spectroscopy | Selective molecules structure such as proteins, lipids, and DNA |
Speed – Relatively slow, due to point-by-point acquisition Spatial resolution – Limited spatial resolution (μm-level) due to diffraction limited spot size Field of view: – Varies, but typically small due to mode of acquisition |
Pros: – Chemical composition analysis – Label-free and non-destructive – Can provide molecular information Cons: – Relatively slow imaging speed – Low signal-to-noise ratio in some cases (fluorescence interference) – Limited spatial resolution – Expensive and complex |
Low—not compatible for embryo imaging due to high risk of phototoxicity | [71–79] |
| Confocal microscopy | Cell and tissue structure, autofluorescence |
Speed – Moderate, can capture optical sections relatively quickly Spatial resolution – High axial and lateral resolution (sub-μm) Field of view: – Limited to a few hundred μm |
Pros: – High-resolution optical sections – 3D imaging and optical sectioning – Suitable for fluorescent labeling Cons: – Limited penetration in thick samples – Photobleaching and phototoxicity – Complex instrumentation and data analysis |
Low—not compatible for embryo imaging due to high risk of phototoxicity | [80–93] |
| Fluorescence lifetime microscopy (FLIM) | Cell structure, autofluorescence |
Speed – Moderate, requires time-resolved measurements Spatial resolution – Diffraction-limited Field of view: – Varies based on setup, typically limited to moderate-sized regions |
Pros: – Quantitative measurement of fluorescence lifetimes – Can provide insights into molecular interactions – Useful for studying metabolic processes Cons: – Require fluorescent labeling or intrinsic fluorophores – Limited to specific fluorophores (cannot do >3 at once) – Complex instrumentation and data analysis |
Low—not compatible for embryo imaging due to high risk of phototoxicity | [94–100] |
| Hyperspectral microscopy | Cell structure, autofluorescence |
Speed – Moderate to slow, depends on spectral channels acquisition Spatial resolution – Diffraction-limited Field of view: – Varies, often limited to small- to moderate-sized regions |
Pros: – Multispectral analysis of endogenous fluorophores – Potential for non-invasive metabolic assessment – Simultaneous detection of multiple fluorophores Cons: – Limited spatial resolution, depth penetration in some cases – Complex data analysis and interpretation |
High—compatible for embryo imaging due to low risk of phototoxicity | [24, 101–113] |
| Light sheet microscopy | Cell and tissue structure, autofluorescence |
Speed – Rapid 3D imaging, can capture volumes (entire cell) within seconds to minutes Spatial resolution – Good lateral and axial resolution (sub-μm). Diffraction-limited. Field of view: – Can be large, capturing entire embryos or tissues |
Pros: – Rapid, 3D imaging of large samples – Minimal photodamage and photobleaching – Low light exposure to samples Cons: – Require specialized sample preparation – Limited penetration in thick, opaque tissues – Complex setup with orthogonal illumination and detection |
High—compatible for embryo imaging due to low risk of phototoxicity | [114–135] |
Morphology-based imaging
Optical coherence tomography
Optical coherence tomography (OCT) is a low-coherence imaging modality that operates on the principle of interferometric detection of near-infrared light (the interaction of light waves to measure miniscule changes) [25]. It sends light into cells/tissue and captures the light that is reflected back from within the sample (backscattered light; Figure 1). Prior to light being sent into the sample, it is split into two beams: one passes through the sample, while the other (reference beam) does not. The interference patterns resulting from the combination of the reference and sample beams can be used to construct high-resolution cross-sectional images of the sample, revealing subsurface structures up to a few millimeters in depth [26]. This can be achieved in a number of ways (time and Fourier domain OCT) with the chosen approach dependent upon the sample being imaged (reviewed in [27]). Each approach has different advantages such as ease of implementation, imaging speed, penetration depth, and detection sensitivity [27]. Clinically, OCT has been widely employed for detecting structural abnormalities in biological tissues. It has seen exceptional success not only in the area of ophthalmology (retinal imaging) [28] but also for cardiology (vascular imaging) [29–31] and dermatology [32, 33]. The use of OCT is ideal for these applications as it does not require the addition of any contrast agents, allowing this system to image in real time without causing tissue damage.
Figure 1.
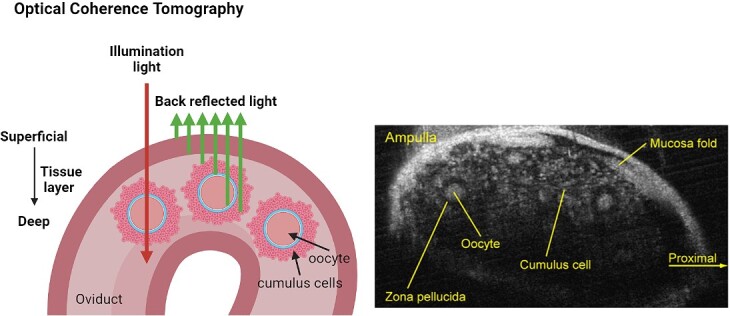
Optical coherence tomography records backscattered light from different cell/tissue layers to form a 3D morphological image. It can work in the temporal or Fourier domain. Image on the right shows ovulated murine cumulus oocyte complexes within the oviduct. Schematic was created using BioRender.com. Image on the right adapted from [157] [CC-BY-NC-ND 4.0 License].
Furthermore, in addition to structural information, OCT may capture functional information. A prime functional extension is Doppler OCT, which detects blood flow velocity [34]. In the context of developmental biology, OCT has been used to study cardiac, vascular, and neural development in post-implantation embryos (reviewed in [35, 36]). However, as OCT primarily relies on the backscattering of light from the sample, a challenge often arises due to insufficient contrast between different tissue types or structures, which may then require contrast agents. Near-infrared light can be used to increase imaging depth, though this may still remain problematic for highly scattering specimens where the light is deviated from its straight path. This limits the information obtained from imaging to the first couple of millimeters of tissue [33]. Excitingly, imaging depth may be increased using new approaches that illuminate the sample and subsequently collect the backscattered light from a separate path [37].
Differential interference contrast microscopy
Differential interference contrast (DIC) microscopy, also known as Nomarski interference contrast microscopy, is an approach that produces images with pseudo-relief, creating the appearance of three-dimensional (3D) structures with enhanced contrast [38]. The pseudo-relief effect is achieved with bright and dark regions generated in the final image that correspond to variations in the optical path length, which reveals underlying surface topography. More specifically, this works by using polarized light to illuminate the specimen. Polarization is a property of a transverse wave such as light. In the case of light, it denotes the plane of oscillation of the electric and magnetic fields, which is perpendicular to the direction of propagation of the beam. In DIC, light is separated into two orthogonally polarized beams which are sheared (displaced) with respect to one another. Both beams then pass through the sample, adjacent to one another, and will traverse different optical path lengths where the specimen differs in refractive index or thickness. The beams are subsequently recombined (interfered) to create an image with enhanced contrast and depth perception, though typically, this is not quantitative. This technique is well established and may be implemented on brightfield microscopes. This technique is most commonly used for observing live, unstained, and transparent biological specimens [39]. Therefore, the label-free nature of DIC gives promising indications of suitability for observing detailed cellular changes over time, such as cellular growth. Indeed, DIC has been widely utilized to study the development of rat neurons [40], mouse stem cells [41], plant cells [42], breast cancer cells [43], and mouse embryonic cells [44]. However, although DIC can be employed to study cell biology without the need for exogenous tags, it cannot provide further details about underlying molecular changes within cells, thereby limiting its utility for widespread adaptation of this technique for assessing embryo quality. Thus, DIC may be informative when integrated with other imaging techniques for a comprehensive understanding of embryo development, beyond observing morphological changes alone.
Digital holographic microscopy
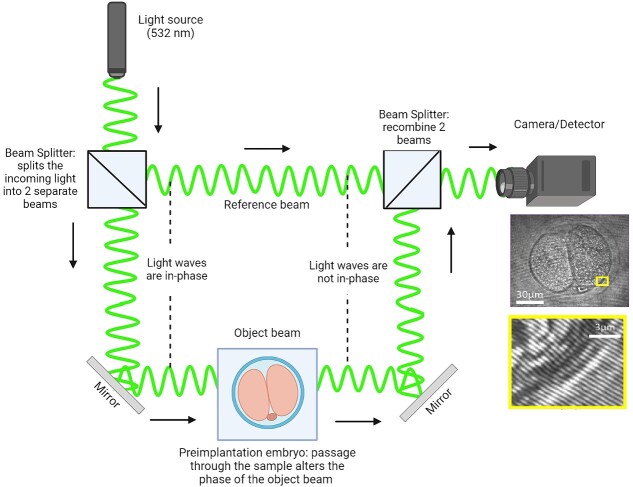
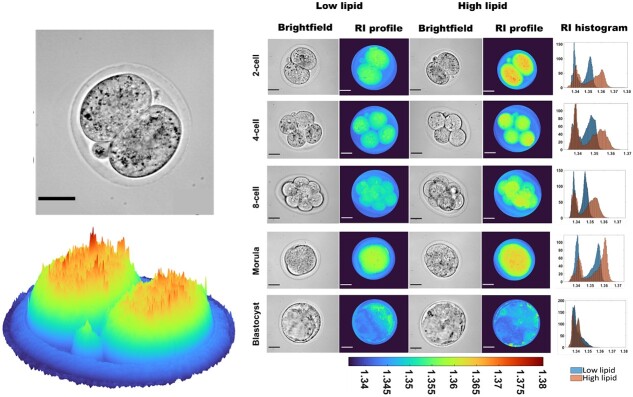
Digital holographic microscopy (DHM) is an advanced imaging technique that utilizes the principles of holography to capture detailed 3D information about samples with high precision [45]. The holograms are generated by recording the interference pattern from two interacting laser beams: one that passes through a specimen (object beam) and another that does not (reference beam) (Figure 2). Holograms contain information about both the shift in amplitude (intensity) and phase (linked to the optical path length/delay) of the light waves as they pass through the specimen [46]. The additional phase information acquired offers information for a more detailed understanding of the specimen and even applies to biological objects that appear transparent. This approach is quantitative and allows for the investigation of bio-physical information such as the dynamics of cellular growth, measurement of cell thickness, and refractive index [47]. The measurement of refractive index (indicative of how much light slows down and changes direction when it passes through a sample) is particularly intriguing for evaluating the biological health of a sample. In contrast to the use of DHM, the conventional refraction of light has been used to determine the refractive index of discarded human oocytes when placed between optical fibers [48]. However, this study reported exceedingly high refractive index values (average refractive index = 1.8), well beyond what might be expected in cells or tissues (range of 1.3–1.5) [49]. Further, this approach failed to provide spatial and temporal information of the oocyte, potentially omitting developmentally important information. On the other hand, we recently demonstrated the capability for DHM to spatiotemporally determine refractive index within mouse preimplantation embryos in response to variable intracellular lipid content (Figure 3; [50]). The presence of such lipids altered the refractive index of the embryo. As intracellular lipids act as an energy reservoir during embryo development, the ability of DHM to detect the associated changes in refractive index may be indicative of embryo viability. This demonstrates the potential of DHM to be a label-free diagnostic for embryo quality.
Figure 2.
Digital holographic microscopy can take a phase image of the object. The phase is recorded by interfering a beam passing through the sample with a reference beam.
Figure 3.
Refractive index profile of mouse embryos cultured in low- or high-lipid-containing culture media during preimplantation development. The refractive index was determined through digital holographic microscopy. Digital holographic microscopy was able to detect a higher refractive index in embryos cultured in high-lipid-containing media, particularly from the two-cell to the morula stage of development. These changes in refractive index were reflective of intracellular lipid abundance. Scale bar = 30 μm. Color coding indicates the determined refractive index. Figure adapted from [50] [CC-BY 4.0 License].
Time-lapse microscopy
Time-lapse microscopy is an imaging technique that employs brightfield microscopy equipped with a camera to capture images of a specimen at pre-defined time intervals. It is typically performed within a controlled environment to maintain optimal culture conditions for the viability of the specimen, including temperature, humidity, and gas concentrations. Time-lapse microscopy is well documented in the literature, playing a pivotal role in the study of various aspects of cell behavior [51, 52], morphological changes [53, 54], and responses to drug treatment [55–57]. Notably, time-lapse microscopy removes the need for human intervention during the culture process and obviates the need for the removal of the specimen from its culture environment for visual inspection. Through uninterrupted recording, time-lapse microscopy can capture and analyze dynamic morphological changes of an embryo during preimplantation development. Time-lapse microscopy is perhaps the most well-known and widely utilized imaging technique for assessing preimplantation development of human embryos [58, 59]. However, there is a lack of high-quality evidence that the use of time-lapse microscopy to select embryos for transfer improves the live birth rate [60–62]. This may stem from limitations in the morphological and developmental information obtained, such as the timing of cleavage-stage divisions and blastocyst formation. These may not be reliable indicators of successful implantation or live birth post-embryo transfer. It could also be due to differences in embryo culture conditions/media formulations, limited datasets, and the metrics (e.g. pregnancy rate vs. live birth rate) used to develop algorithms for predicting IVF success [63]. As such, accurately predicting embryo developmental potential using time-lapse microscopy alone remains a challenge.
Molecular-based optical imaging
Standard wide-field fluorescence microscopy
The term “fluorescence” originated from a discovery first made by Sir George Stokes in 1852 [64]. Stokes observed that certain substances, particularly a solution of quinine had the remarkable property of absorbing ultraviolet (UV) light and subsequently emitting visible light [65]. This pivotal observation laid the foundation for the field of fluorescence microscopy, where light emission from fluorescent molecules can be used to form an image depicting the localization and quantity of those molecules. This basic principle underlies all forms of fluorescence imaging. For fluorescent imaging of biological processes, there are a plethora of methods that either fluorescently tag recombinant proteins of interest (e.g., green fluorescent protein) or use exogenous fluorescent molecules (e.g., immunohistochemistry or fluorescent dyes).
While these methods have been advantageous in biological discovery, they are limited when considering their application to living embryos: the exogenous labeling may negatively impact subsequent development. It was not until the late 20th century that the potential of autofluorescence, the intrinsic fluorescence exhibited by particular molecules within cells, began to be explored and recognized for its informative value. In this case, light emitted from endogenous fluorophores in the cell could be measured directly. Early investigations into autofluorescence aimed to measure the dynamic aspects of metabolic reactions using a fluorometer to detect blue fluorescence in cells [66], which we now know to be NAD(P)H. However, this early one-photon approach had limitations, notably its reliance on blue or UV light for excitation, which is not ideal for imaging biological specimens due to the potential risk of photodamage [67]. Alternatively, the use of two-photon excited fluorescence (TPEF) microscopy overcomes this limitation [68, 69]: by using near-infrared light, the risk of photodamage is reduced. TPEF has the added benefit of increasing light penetration into the sample, allowing investigation into deeper layers of tissues. It works by allowing molecules to be excited through the simultaneous absorption of two near-infrared photons, each with half the energy and double the wavelength of traditional UV light. While TPEF microscopy has exciting possibilities, it also presents challenges as high-resolution 3D images are difficult to obtain due to weak signals from the use of longer wavelengths [70]. The inability to obtain high-resolution images in 3D impedes the use of this modality for imaging the developing embryo—itself a 3D structure.
Raman spectroscopy
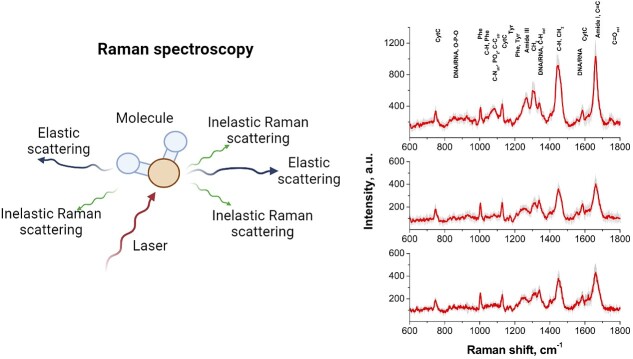
Raman spectroscopy is an advanced analytical technique that provides detailed information about the molecular composition and vibrational characteristics of a sample [71]. This technique relies on the principle of inelastic scattering, a phenomenon that occurs when monochromatic light interacts with a sample and the energy shift is indicative of the molecular composition of the sample (Figure 4). This interaction allows Raman spectroscopy to capture a unique “molecular fingerprint” corresponding to the alterations in biomolecules such as lipids, carbohydrates, deoxyribonucleic acid (DNA), and proteins [72]. Consequently, Raman spectroscopy enables direct evaluation of organelle structures and dynamic biochemical processes within living cells [73]. With the well-established knowledge that metabolism is linked to embryo quality, Raman spectroscopy may be an innovative technique to investigate the relationship between metabolomic profile and embryo quality. Confirming this, previous studies have demonstrated the ability of Raman spectroscopy to predict the developmental potential of human embryos through the analysis of metabolic by-products in spent culture media [74–76].
Figure 4.
The principle of Raman scattering (left). Some light from the laser pulse is inelastically scattered following its interaction with molecules. The change in energy manifests itself as a change in the wavelength of the scattered light leading to a spectrum [denoted as the Raman shift, measured in wavenumbers (cm−1)]. The peaks of the Raman signal can be attributed to specific proteins, nucleic acids, etc., in the sample. The spectra on the right show characteristic Raman bands for a living cell from a two-cell stage embryo. The position of the bands and relative intensities differ depending on the location within the cell investigated [top spectra from a lipid-rich (dark cytoplasm) area; middle spectra taken from a (light) cytoplasm region; and the bottom from within the nucleus]. Schematic was generated using BioRender.com. Figures adapted from [79] [CC-BY 4.0 License].
The use of Raman spectroscopy to assess embryo quality via spent culture media may appear indirect, but this approach avoids potential damage inflicted to the developing embryo during the imaging process. Raman spectroscopy, as with many optical approaches, is limited in terms of penetration depth and has issues with regard to signal strength [77]. Specifically, the probability of light undergoing Raman scattering is very low, with typically only 1 in 106 or 107 photons being Raman-scattered. Thus, in order to generate sufficient signal, high illumination powers or increasing acquisition time is required. However, the use of high illumination power may induce photodamage to the embryo, negatively impacting its viability. One study demonstrated that Raman spectroscopy could distinguish between low- and high-quality mouse embryos based on the distinct variations in their Raman spectral signals [78]. However, despite efforts to minimize phototoxicity, the study did not put forward a comprehensive experimental validation that Raman imaging did not affect embryo viability. Conversely, another study that performed Raman spectroscopy directly on mouse embryos reported a null impact on the ability of embryos to develop to the blastocyst-stage [79]. Having said that, the authors observed a significant reduction in the number of cells within resultant blastocyst-stage embryos from the Raman-imaged group compared to the non-imaged, control group. A reduction in cell number may adversely impact implantation potential and pregnancy outcomes, which was not assessed in the study. This suggests that in its current form, Raman spectroscopy may not be compatible with live embryo imaging and warrants further investigation to address its inherent limitations.
Laser scanning confocal microscopy
Laser scanning confocal microscopy, a variant of optical imaging, relies on point-by-point illumination, typically delivered by a high-power laser. This laser-induced point excitation generates intense fluorescence along the focal profile [80]. Fundamentally, confocal microscopy achieves sharp, focused information, at one point of illumination through spatial filtering [80]. This process effectively eliminates out-of-focus light, thereby enhancing the signal-to-noise ratio in the final image (Figure 5A). This fluorescence microscopy approach facilitates rapid, non-invasive exploration of changes in cellular morphology and the underlying biochemistry of the cell.
Figure 5.
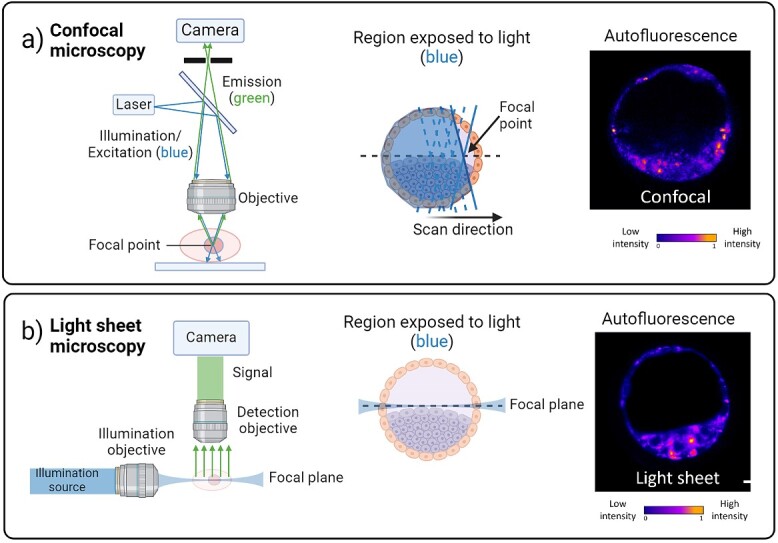
(A) shows the principle of epifluorescence microscopy, specifically the case for confocal microscopy. A laser is focused to a point on a sample. The resultant fluorescence from the focal point is captured by the very same microscope objective, passes through a pinhole, and is collected on a camera. An image in 3D can be captured in this way by point scanning the beam. At each point the light field passes through the sample above and below the plane of interest (compare to light sheet imaging). (B) Light sheet imaging illuminates a given plane of a sample with scattered or fluorescent light collected by a second microscope objective (detection objection) placed orthogonally to the one used for illumination. Light only illuminates the (focal) plane of interest [contrast this with confocal microscopy in (A)]. As such, the light dose is much lower than that for epifluorescence/confocal microscopy. The sample or illumination beam may be scanned to generate a 3D image. Representative autofluorescence images of murine blastocyst-stage embryos taken at an illumination wavelength of 405 nm are shown on the right and are false-colored for clarity. Schematic was generated using BioRender.com.
Historically, confocal microscopy stands as one of the earliest and most commonly used imaging modalities adapted for embryonic imaging, alongside methods such as ultrasound biomicroscopy, micro-magnetic resonance imaging, and micro-computed tomography [35]. However, all of these imaging systems struggle with limitations in imaging speed, spatial resolution, and image contrast, particularly when capturing dynamic biological processes, such as metabolism and morphokinetics [81–83]. Despite these limitations, confocal microscopy has remained a widely employed approach for studying developmental processes in different cell types, including human cornea [84, 85] and lung tissue [86], as well as mouse and hamster embryonic cells [87–89]. Many of these studies employed exogenous tags or molecular probes as direct indicators of cellular architecture or biochemistry, such as Mitotracker for mitochondria, Lysotracker for lysosomes, and 4′,6-diamidino-2-phenylindole (DAPI) for the nucleus. However, reliance on staining and fluorescence tags can render the labeled cells non-viable for long-term assessments due to the potential toxicity of these agents or the need for fixation [16].
Furthermore, the application of fluorescence tags raises a fundamental issue—the risk of photobleaching due to sensitivity of the tags to light exposure. This sensitivity can lead to fluctuating or degrading signal levels, potentially resulting in false-positive or false-negative outcomes. Additionally, phototoxicity is another concern typically associated with most confocal imaging systems [90] due to exposure to high-energy light sources and relatively slow imaging acquisition speeds, which can have adverse effects on embryo health [91–93]. Past studies have demonstrated that exposure to laser scanning confocal microscopy negatively impacts embryo viability by: (1) inhibiting embryo development to the blastocyst stage; (2) increasing the production of reactive oxygen species, which damages DNA integrity; and (3) increasing the number of apoptotic cells [89]. Importantly, the detrimental effects of laser exposure on cellular architecture and fluorescence tag detection hinge on the duration, intensity, and wavelength of the laser employed [90]. Overcoming these significant challenges related to fluorescent probes and laser power usage in visualization is paramount for the development of laser scanning confocal microscopy that is a safe and accurate diagnostic tool for embryo quality.
Fluorescence lifetime microscopy
Fluorescence lifetime microscopy (FLIM) is an advanced imaging modality that provides quantitative information about cellular and molecular processes [94, 95]. It operates on a different principle than simply recording fluorescence intensity. FLIM operates via the excitation of fluorescent molecules, typically with a laser, and measures the time taken for the molecules to transition from the excited state back to the ground state—their decay rate [94, 95]. This information can then be used to construct the fluorescence decay curve (lifetime; Figure 6), which is unique to the individual fluorescent molecules present within a sample that could not be elucidated with intensity-based fluorescence microscopy alone, such as laser scanning confocal microscopy. The contrast for imaging is based on the lifetime of individual fluorophores rather than their emission spectra, which is affected by the environment of the fluorophore. This allows for a more sensitive approach to detect autofluorescence as it enables discrimination between fluorescent molecules integral to embryo metabolism that share similar spectral characteristics such as bound- and free-NADH, enabling differentiation of NADH from NADPH [96]. Therefore, this imaging approach has the potential for label-free imaging of dynamic molecular processes in the embryo. Indeed, past studies have shown that FLIM was able to detect spatio-temporal changes in autofluorescence originating from the cytoplasm and mitochondrial regions in human [97, 98] and mouse embryos [99, 100] (Figure 6; [98]). However, FLIM requires a measurement of the time between sample excitation by a pulsed laser and the arrival of the emitted photon at the detector that may lead to increased complexity and cost of the system, which requires specialized lasers, detectors, and software [94]. Therefore, this may limit the accessibility of FLIM to researchers with limited resources and hinder adoption into clinical practice. Furthermore, as FLIM relies on the detection of individual photons emitted as excited molecules decay back to their ground state, the signal level detected is often very weak, this impacts image quality and requires longer acquisition times [94]. As such, like confocal microscopy, FLIM is prone to inducing photodamage and the photobleaching of fluorophores and may limit its ability to perform long-term monitoring of dynamic processes in live embryos [94].
Figure 6.
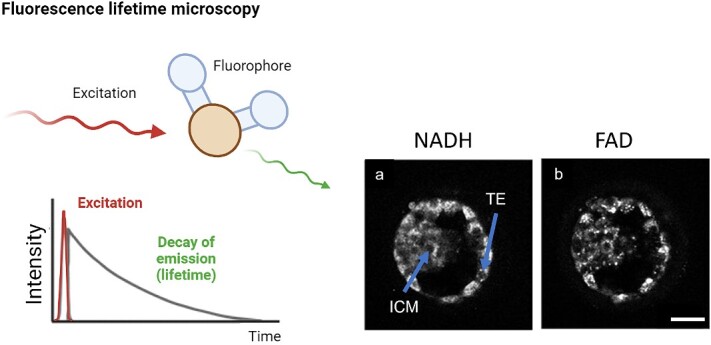
Fluorescence lifetime microscopy. Following excitation with a laser pulse the decay of the signal is measured to yield a fluorophore lifetime. The decay constant (lifetime) is dependent on the environment of the fluorophore and/or its conformational state. Importantly, is not dependent on the absolute intensity of the signal, and complements intensity-based imaging approaches. Images on the right are of a human blastocyst-stage embryo showing abundance of NADH and FAD following FLIM. ICM = inner cell mass, TE = trophectoderm. Scale bar = 40 μm. Schematic was generated using BioRender.com. Images adapted with permission from [98].
Hyperspectral microscopy
Hyperspectral microscopy, an emerging imaging modality, holds significant promise for exploiting cellular autofluorescence to assess embryos. This approach adopts a multispectral strategy, utilizing multiple low-power excitation photodiodes to capture fluorescent signals. Hyperspectral microscopy has the unique capability to detect light signals spanning the visible light spectrum, ranging from 380 to 780 nm (from UV to near-infrared) [101]. This contrasts with traditional confocal imaging, which typically employs two-channel excitation wavelengths, capable of capturing fluorescence from two molecules with distinct spectral properties (e.g. endogenous metabolic cofactors NAD(P)H and FAD; [102]). Hyperspectral microscopy has the capacity to comprehensively explore cellular metabolism [103] due to its unique capability to simultaneously capture a broad range of spectral information or “fingerprint” from multiple endogenous fluorescent molecules (Figure 7). Indeed, extensive studies have been conducted demonstrating that autofluorescence imaging with hyperspectral spectroscopy can discern subtle differences in metabolic signatures within and between cells. Notable examples include its successful detection of collagen, flavin, free NADH, and retinoids in different biological samples such as olfactory cells [104], articular cartilage tissue [105], benign and invasive melanoma [106], and pathological hallmarks of Alzheimer's disease in retinal tissue [107–109]. Given its ability to detect autofluorescence and its use of low-power photodiodes, hyperspectral imaging holds promise as a non-invasive means to assess embryo health. Indeed, hyperspectral microscopy has been effectively employed to detect changes in the endogenous metabolic co-factors NAD(P)H and FAD in bovine [110–112] and mouse [24] embryos. Recently, we demonstrated the capability of hyperspectral microscopy to spatially detect changes in autofluorescence related to metabolic activity within mouse blastocyst-stage embryos [24]. Excitingly, we showed that such changes in cell autofluorescence could be used to discriminate between euploid and aneuploid embryos (cells with an expected vs. unexpected number of chromosomes, respectively). These studies demonstrate the potential of label-free imaging to not only assess embryo viability but also obtain information on their ploidy status. Therein lies the intriguing opportunity to explore the capacity for such label-free imaging approaches to predict downstream development outcomes in the same embryo.
Figure 7.
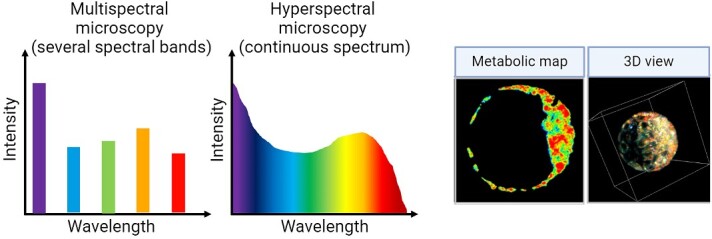
Hyperspectral and multispectral microscopy. The graphs on the left show the principal of multispectral and hyperspectral imaging. In both, spectral information is recorded. In multispectral imaging, several discrete wavelengths are captured, but in hyperspectral imaging, this is performed in a continuous fashion. On the right, hyperspectral light sheet microscopy was used to record cell autofluorescence from a murine blastocyst-stage embryo. From this we can create metabolic maps across the embryo and generate a 3D image. Schematic was generated using BioRender.com. Images adapted from [135] [CC-BY 4.0 License].
Despite the benefits of hyperspectral microscopy for imaging the embryo, there are concerns regarding the potential for light-induced photodamage. These apprehensions were addressed in our recent study, which demonstrated the safety of hyperspectral imaging for embryos [24]. Using a mouse model, we found no adverse impact on embryo developmental competence following exposure to hyperspectral imaging, specifically in their ability to develop to the blastocyst stage and generate healthy offspring following their transfer to pseudopregnant recipients. Our follow-up study on imaging the mouse oocyte demonstrated equivalent levels of safety: imaged and non-imaged oocytes had comparable rates of fertilization, development to the blastocyst-stage and post-transfer outcomes [113].
One noteworthy limitation of hyperspectral microscopy when using an epifluorescence approach [24] is the difficulty in generating a 3D image of the sample. This limitation is substantial when considering the imaging of embryos. As embryonic development is a highly dynamic process occurring in 3D, assessments limited to static 2D images may fail to capture information on critical developmental events. Therein lies the need for further development of hyperspectral spectroscopy. Employing an integrative strategy could be the key to addressing this need. Considering this, an alternative approach might involve the integration of hyperspectral imaging with a modality that enables rapid, volumetric imaging of the embryo.
Light sheet fluorescence microscopy
Recent advances in fluorescence-based microscopy have highlighted the growing superiority of light sheet microscopy (sometimes termed selective plane illumination microscopy). This approach offers exceptional advantages such as high image acquisition speed, low background-to-signal ratio, and minimal photobleaching and photodamage [114, 115]. The underlying principle of light sheet imaging involves illumination and detection through two separate and distinct objectives placed in an orthogonal orientation [116]. As a result, the excitation of the sample is restricted only to the imaging plane, using a thin sheet of light for illumination [117, 118] (Figure 5B). Consequently, light sheet imaging is well suited for fast, volumetric imaging of large specimens while minimizing light exposure [119]. This contrasts with other 3D imaging approaches such as confocal microscopy, where image acquisition is slow and the entire sample is illuminated for each optical section [120], thus making the light dose delivered to the sample prohibitively high. Light sheet microscopy has demonstrated its capability to capture the dynamics of small molecules; protein localization [121]; calcium ion signaling [122]; and oligonucleotide molecules within large, living organisms [123]. The application of light-sheet microscopy in the field of embryology is an emerging area of interest [124]. Studies have previously demonstrated the ability of light sheet microscopy to elucidate the dynamics of morphogenesis and embryogenesis in mouse [117, 125, 126], drosophila [127], and zebrafish [128, 129] with remarkable spatial and temporal resolution. However, it is important to note that these studies relied on genetically modified species expressing exogenous fluorescence reporters, such as GFP, and thus are not considered label-free imaging. More recently, there has been an increased interest in exploring the potential of light sheet microscopy for label-free imaging of cellular autofluorescence.
Several studies have now demonstrated the ability for label-free light sheet microscopy to assess metabolic activity in numerous pathological conditions including cancer [130–132] and neurological diseases [133, 134]. Recently, we demonstrated the first use of light sheet microscopy to detect a dynamic shift in metabolism during the preimplantation development of mouse embryos (Figure 8; [135]). This approach enabled rapid volumetric imaging of cellular autofluorescence from the embryo, which was achieved using a single excitation wavelength of 375 nm. This is in contrast with confocal microscopy, which requires dual wavelengths to excite NAD(P)H and FAD separately. Due to the judicious choice of wavelength, we were able to co-excite NAD(P)H and FAD equally. Further, due to the use of a hardware-based approach to separate the fluorescence signals emitted by NAD(P)H and FAD, we obviate the need for large post-processing power normally required in conventional hyperspectral microscopy [24]. Collectively, the use of light sheet metabolic imaging provides opportunities for high-throughput imaging of embryos to study development spatio-temporally and may serve as a valuable diagnostic tool for embryo quality.
Figure 8.
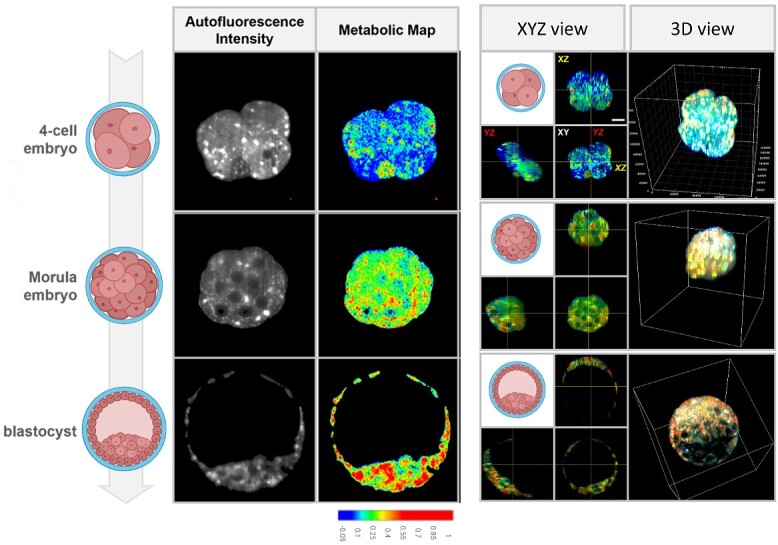
Autofluorescence profile of preimplantation mouse embryos as detected by hyperspectral light-sheet microscopy. Hyperspectral light sheet microscopy was able to detect dynamic changes in metabolism during preimplantation embryo development and the distribution of highly metabolically active sites within embryos. Scale bar = 20 μm. Images adapted from [135] [CC-BY 4.0 License].
Future directions
Use of advanced optical imaging together with artificial intelligence
One significant challenge associated with label-free optical imaging is the vast amount of data that it generates. Large amounts of data and the capacity to analyze may impede clinical adoption of label-free optical imaging methods, particularly in clinics or medical facilities with limited resources. Artificial intelligence (AI) has the potential to mitigate this challenge. In the context of optical imaging, AI offers the capacity to process and analyze large amounts of data within a short time frame, obviating the need for long-term storage. Examples of combining AI with optical imaging include: (1) its use with optical coherence tomography to accurately and automatically detect diabetic retinopathy [136]; (2) integrating AI with Raman spectroscopy to accurately diagnose and classify brain tumors [137, 138]; and (3) the use of AI with hyperspectral imaging to accurately and rapidly diagnose esophageal cancer [139]. In clinical IVF, there are multiple instances where AI might aid in improving the ability to predict live birth, such as gamete and embryo selection, and the development of personalized fertility medicine (reviewed in [63]). In recent years, the use of AI in analyzing time-lapse images has shown promise for sperm selection [140], assessing oocyte viability [141], and in predicting embryo developmental competence and pregnancy [142–148]. However, time-lapse microscopy together with AI does not improve live birth rate [149]. Consequently, more studies are warranted where much larger data sets are included to improve accuracy.
Additionally, there remains the potential of employing AI alongside optical imaging approaches. This integrative strategy could offer unique advantages, from improving the resolution of captured images to automated tracking of cellular features during development [150]. This may enhance accuracy in predicting embryo developmental potential when using optical imaging and uncover subtle biological phenomena that might otherwise remain undetected. Collectively, the rapidly growing and pervasive field of AI represents a powerful and exciting opportunity for the reproductive biology field and the IVF industry.
Utilization of label-free imaging for clinical in vitro fertilization
Label-free imaging techniques have the potential to revolutionize IVF. Specifically, such imaging approaches may improve the selection and ranking of developmentally competent embryos for transfer, thus improving the chance of a live birth outcome. These non-invasive approaches also reduce the risks associated with an invasive biopsy, a current clinical gold-standard assessment that does not improve, and may even decrease, live birth rate [151]. For example, optical imaging techniques such as DHM offer a non-invasive means to assess the biophysical properties of an embryo and in the absence of exogenous tags, revealing subtle changes in refractive index that may be indicative of developmental potential. Additionally, the use of hyperspectral and light sheet microscopy to measure cellular autofluorescence and metabolism in situ may provide additional information on developmental potential. An important consideration in the development of any of these imaging technologies is their suitability for the clinical environment. In considering these novel technologies, these systems will need to reach a competitive price point and have a relatively small footprint due to the limited bench space available within clinics. Currently, time-lapse microscopy is the only imaging modality integrated into and used by IVF clinics. It provides a stable culture environment while offering capacity for long-term monitoring of preimplantation embryo development within a confined space on the bench. Despite its integration, whether time-lapse microscopy alone can improve IVF outcomes—implantation and live birth– remains debatable [152]. Therein lies an opportunity to explore other forms of advanced imaging modality for improving IVF outcomes. In the future, these imaging modalities in isolation, or in suitable combination, may aid embryologists in ranking and selecting embryos for transfer, ultimately improving the success rate from IVF.
Application of label-free imaging to select in vitro-produced embryos in agriculturally important species
Apart from clinical IVF, non-invasive assessments of embryo quality by optical imaging could also benefit the livestock industry. The International Embryo Transfer Society recently reported a 31.5% increase in the transfer of in vitro-produced (IVP) cattle embryos in 2021 compared to 2020 [153]. However, the efficiency of IVP is still far from optimal, with pregnancy rates remaining below 50% [154]. One of the key hurdles faced by the livestock industry is the lack of accurate technologies to select good-quality embryos for transfer to recipient cows. To date, the critical assessment of cattle embryos prior to transfer continues to rely on the subjective analysis of morphology by trained embryologists. This reliance on traditional techniques could be due to the cost and challenging logistics of adopting modern analytical technologies in the livestock industry [155]. Therefore, there exists an untapped opportunity to provide the field with a cost-effective, simple-to-use, and accurate selection tool for predicting which embryos will result in a live birth. Moving forward, a more extensive evaluation of optical imaging as an assessment tool for cattle IVF is warranted and will require additional refinement and validation to demonstrate the association between developmental potential and post-transfer outcomes.
Summary of the potential of label-free optical imaging approaches
Label-free imaging approaches may play a crucial role in various biological research contexts, each offering unique advantages depending on the specific requirements of the study. Raman spectroscopy is particularly useful when detailed information about molecular composition is needed without the introduction of exogenous labels. It is employed in diverse applications such as studying cell and tissue composition, detecting changes in biomolecular structures, and investigating cellular responses to various stimuli. Light sheet microscopy, with its ability to provide optical sectioning and reduced phototoxicity, is ideal for live imaging of large specimens or dynamic processes. This technique is frequently used in developmental biology, neuroscience, and embryology, capturing high-resolution 3D images of biological samples without the need for labels. Optical coherence tomography excels in imaging biological tissues with high resolution and depth penetration, making it valuable for visualizing structural details in tissues and monitoring changes over time. Digital holographic microscopy, on the other hand, offers label-free, quantitative, and non-invasive imaging, making it suitable for studying the biophysical properties of living cells. Each of these label-free imaging approaches contributes uniquely to the biologist’s toolkit, allowing for a comprehensive understanding of diverse biological phenomena. Separately, there is a strong case for moving to multimodality using optical imaging approaches. As an example, Raman can be combined with optical coherence tomography to offer both molecular and morphological imaging in unison [156]. Although the concept of using multimodal imaging has been used in other areas of biology, the exciting prospect of using this in the field of reproductive biology would provide an unprecedented view of developing embryos in their natural state.
Conclusion
The field of label-free optical imaging in the context of embryo assessment is poised for exciting developments and discoveries. These advancements hold the promise of revolutionizing our understanding of embryo development and improving the success rate of IVF. One of the most exciting prospects is the potential to develop an integrated label-free imaging approach. By combining different imaging modalities, such as hyperspectral imaging with light sheet microscopy, scientists could simultaneously obtain information on dynamic changes in morphology together with cellular metabolism in a spatiotemporal manner. Further, the merit for using AI to analyze and handle large imaging datasets holds great potential to augment or replace current data analysis methods, thus reducing subjectivity and enhancing the accuracy of assessing embryo quality.
Label-free imaging has the potential to bridge the gap between biologists, engineers, and clinicians, fostering new collaborative efforts across diverse fields. These cooperative ventures may lead to the exploration of novel avenues for not only the embryo but also across a wide range of other cell types, such as other reproductive cell types and tissues. This could lead to the discovery of novel mechanisms and factors influencing gamete/embryo development that were previously hindered by the lack of suitable tools. Excitingly, the use of label-free optical imaging to reveal the inner workings of the embryo may subsequently inform the development of novel and accurate diagnostics for clinical IVF, ultimately aiding those affected by infertility.
Acknowledgment
We would like to thank Dr Humphrey Hung-Chang Yao for the opportunity to contribute to this special issue. We also acknowledge the scientific contributions of the many researchers who have utilized optical imaging for embryos that have not been referenced in this review due to content constraints.
Footnotes
† Grant Support: KRD is supported by Future Making Fellowship (The University of Adelaide) and the National Health and Medical Research Council (APP2003786 and APP2029067). KD is supported by Engineering and Physical Sciences Research Council (EP/P030017/1, EP/R004854/1) and Australian Research Council (FL210100099).
Contributor Information
Darren J X Chow, Robinson Research Institute, School of Biomedicine, The University of Adelaide, Adelaide, Australia; Institute for Photonics and Advanced Sensing, The University of Adelaide, Adelaide, Australia; Centre of Light for Life, The University of Adelaide, Adelaide, Australia.
Tiffany C Y Tan, Robinson Research Institute, School of Biomedicine, The University of Adelaide, Adelaide, Australia; Institute for Photonics and Advanced Sensing, The University of Adelaide, Adelaide, Australia.
Avinash Upadhya, Institute for Photonics and Advanced Sensing, The University of Adelaide, Adelaide, Australia; Centre of Light for Life, The University of Adelaide, Adelaide, Australia; School of Biological Sciences, The University of Adelaide, Adelaide, Australia.
Megan Lim, Robinson Research Institute, School of Biomedicine, The University of Adelaide, Adelaide, Australia; Institute for Photonics and Advanced Sensing, The University of Adelaide, Adelaide, Australia; Centre of Light for Life, The University of Adelaide, Adelaide, Australia; School of Biological Sciences, The University of Adelaide, Adelaide, Australia.
Kishan Dholakia, Centre of Light for Life, The University of Adelaide, Adelaide, Australia; School of Biological Sciences, The University of Adelaide, Adelaide, Australia; Scottish Universities Physics Alliance, School of Physics and Astronomy, University of St Andrews, St Andrews, United Kingdom.
Kylie R Dunning, Robinson Research Institute, School of Biomedicine, The University of Adelaide, Adelaide, Australia; Institute for Photonics and Advanced Sensing, The University of Adelaide, Adelaide, Australia; Centre of Light for Life, The University of Adelaide, Adelaide, Australia.
Conflict of interest: The authors have declared that no conflict of interest exists.
Author contributions
KRD and KD conceived the review content. DJXC, TCYT, AU, ML, KD, and KRD wrote the manuscript. All authors critically reviewed and approved the final version of the manuscript.
Data availability
No new data were generated or analysed in support of this review article.
References
- 1. Infertility prevalence estimates: 1990-2021 . Geneva: World Health Organization; 2023. Licence: CC BY-NC-SA 3.0 IGO. [Google Scholar]
- 2. Zegers-Hochschild F, Nygren KG, Adamson GD, de Mouzon J, Lancaster P, Mansour R, Sullivan E, International Committee Monitoring Assisted Reproductive Technologies . The international committee monitoring assisted reproductive technologies (ICMART) glossary on ART terminology. Fertil Steril 2006; 86:16–19. [DOI] [PubMed] [Google Scholar]
- 3. Inhorn MC, Patrizio P. Infertility around the globe: new thinking on gender, reproductive technologies and global movements in the 21st century. Hum Reprod Update 2015; 21:411–426. [DOI] [PubMed] [Google Scholar]
- 4. Newman JE, Paul RC, Chambers GM. Assisted reproductive technology in Australia and New Zealand 2021. Sydney, Australia: National Perinatal Epidemiology and Statistics Unit, The University of New South Wales; 2023. [Google Scholar]
- 5. van Loendersloot LL, van Wely M, Limpens J, Bossuyt PM, Repping S, van der Veen F. Predictive factors in in vitro fertilization (IVF): a systematic review and meta-analysis. Hum Reprod Update 2010; 16:577–589. [DOI] [PubMed] [Google Scholar]
- 6. Pirkevi Centinkaya C, Kahmaran S. Morphokinetics of embryos - where are we now? J Reprod Biotechnol Fertil 2016; 5:1–8. [Google Scholar]
- 7. Fordham DE, Rosentraub D, Polsky AL, Aviram T, Wolf Y, Perl O, Devir A, Rosentraub S, Silver DH, Gold Zamir Y, Bronstein AM, Lara Lara M, et al. Embryologist agreement when assessing blastocyst implantation probability: is data-driven prediction the solution to embryo assessment subjectivity? Hum Reprod 2022; 37:2275–2290. [DOI] [PubMed] [Google Scholar]
- 8. Takahashi S, Johnston J, Patrizio P. Lessons from the premature adoption of preimplantation embryo testing. Genet Med 2019; 21:1038–1040. [DOI] [PubMed] [Google Scholar]
- 9. Cimadomo D, Capalbo A, Ubaldi FM, Scarica C, Palagiano A, Canipari R, Rienzi L. The impact of biopsy on human embryo developmental potential during preimplantation genetic diagnosis. Biomed Res Int 2016; 2016:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sadeghi MR. The disputable discourse on accuracy and effectiveness of PGT-A in light of advancements in genetic tools. J Reprod Infertil 2021; 22:149–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gleicher N, Patrizio P, Brivanlou A. Preimplantation genetic testing for aneuploidy - a castle built on sand. Trends Mol Med 2021; 27:731–742. [DOI] [PubMed] [Google Scholar]
- 12. Ginod P, Dahan MH. Polygenic embryo screening: are there potential maternal and fetal harms? Reprod Biomed Online 2023; 47:103327–103310. [DOI] [PubMed] [Google Scholar]
- 13. Javaeed A, Qamar S, Ali S, Mustafa MAT, Nusrat A, Ghauri SK. Histological stains in the past, present, and future. Cureus 2021; 13:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Amsterdam A, Lin S, Hopkins N. The Aequorea victoria green fluorescent protein can be used as a reporter in live zebrafish embryos. Dev Biol 1995; 171:123–129. [DOI] [PubMed] [Google Scholar]
- 15. Ju B, Xu Y, He J, Liao J, Yan T, Hew CL, Lam TJ, Gong Z. Faithful expression of green fluorescent protein (GFP) in transgenic zebrafish embryos under control of zebrafish gene promoters. Dev Genet 1999; 25:158–167. [DOI] [PubMed] [Google Scholar]
- 16. Jensen EC. Use of fluorescent probes: their effect on cell biology and limitations. Anat Rec (Hoboken) 2012; 295:2031–2036. [DOI] [PubMed] [Google Scholar]
- 17. Wang S, Larina IV, Larin KV. Label-free optical imaging in developmental biology [invited]. Biomed Opt Express 2020; 11:2017–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wax A, Giacomelli MG, Matthews TE, Rinehart MT, Robles FE, Zhu Y. Optical Spectroscopy of biological cells. Adv Opt Photon 2012; 4:322–378. [Google Scholar]
- 19. Gao M, Lewis G, Turner GM, Soubret A, Ntziachristos V. Effects of background fluorescence in fluorescence molecular tomography. Appl Opt 2005; 44:5468–5474. [DOI] [PubMed] [Google Scholar]
- 20. Thompson JG, Brown HM, Sutton-McDowall ML. Measuring embryo metabolism to predict embryo quality. Reprod Fertil Dev 2016; 28:41–50. [DOI] [PubMed] [Google Scholar]
- 21. Croce AC, Bottiroli G. Autofluorescence spectroscopy and imaging: a tool for biomedical research and diagnosis. Eur J Histochem 2014; 58:320–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tan TCY, Dunning KR. Non-invasive assessment of oocyte developmental competence. Reprod Fertil Dev 2022; 35:39–50. [DOI] [PubMed] [Google Scholar]
- 23. Georgakoudi I, Quinn KP. Label-free optical metabolic imaging in cells and tissues. Annu Rev Biomed Eng 2023; 25:413–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tan TCY, Mahbub SB, Campbell JM, Habibalahi A, Campugan CA, Rose RD, Chow DJX, Mustafa S, Goldys EM, Dunning KR. Non-invasive, label-free optical analysis to detect aneuploidy within the inner cell mass of the preimplantation embryo. Hum Reprod 2022; 37:14–29. [DOI] [PubMed] [Google Scholar]
- 25. Popescu DP, Choo-Smith LP, Flueraru C, Mao Y, Chang S, Disano J, Sherif S, Sowa MG. Optical coherence tomography: fundamental principles, instrumental designs and biomedical applications. Biophys Rev 2011; 3:155–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Huang D, Swanson EA, Lin CP, Schuman JS, Stinson WG, Chang W, Hee MR, Flotte T, Gregory K, Puliafito CA, Fujimoto JG. Optical coherence tomography. Science 1991; 254:1178–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gao W, Wu X. Differences between time domain and Fourier domain optical coherence tomography in imaging tissues. J Microsc 2017; 268:119–128. [DOI] [PubMed] [Google Scholar]
- 28. Aumann S, Donner S, Fischer J, Muller F. Chapter 3 Optical Coherence Tomography (OCT): Principle and Technical Realization. In: Bille JF (ed.), High Resolution Imaging in Microscopy and Opthalmology: New Frontiers in Biomedical Optics [Internet]. Cham, Switzerland: Springer; 2019. [PubMed] [Google Scholar]
- 29. Gupta A, Shrivastava A, Vijayvergiya R, Chhikara S, Datta R, Aziz A, Singh Meena D, Nath RK, Kumar JR. Optical coherence tomography: an eye into the coronary artery. Front Cardiovasc Med 2022; 9:1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Araki M, Park SJ, Dauerman HL, Uemura S, Kim JS, Di Mario C, Johnson TW, Guagliumi G, Kastrati A, Joner M, Holm NR, Alfonso F, et al. Optical coherence tomography in coronary atherosclerosis assessment and intervention. Nat Rev Cardiol 2022; 19:684–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Terashima M, Kaneda H, Suzuki T. The role of optical coherence tomography in coronary intervention. Korean J Intern Med 2012; 27:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wan B, Ganier C, Du-Harpur X, Harun N, Watt FM, Patalay R, Lynch MD. Applications and future directions for optical coherence tomography in dermatology. Br J Dermatol 2021; 184:1014–1022. [DOI] [PubMed] [Google Scholar]
- 33. Sattler E, Kastle R, Welzel J. Optical coherence tomography in dermatology. J Biomed Opt 2013; 18:061224–061227. [DOI] [PubMed] [Google Scholar]
- 34. Braaf B, Grafe MGO, Uribe-Patarroyo N, Bouma BE, Vakoc BJ, de Boer JF, Donner S, Weichsel J. In: Bille JF (ed.), OCT-Based Velocimetry for Blood Flow Quantification, in High Resolution Imaging in Microscopy and Ophthalmology: New Frontiers in Biomedical Optics. Cham, Switzerland: Springer; 2019: 161–179. [PubMed] [Google Scholar]
- 35. Raghunathan R, Singh M, Dickinson ME, Larin KV. Optical coherence tomography for embryonic imaging: a review. J Biomed Opt 2016; 21:1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Scully DM, Larina IV. Mouse embryo phenotyping with optical coherence tomography. Front Cell Dev Biol 2022; 10:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Untracht GR, Chen M, Wijesinghe P, Mas J, Yura HT, Marti D, Andersen PE, Dholakia K. Spatially offset optical coherence tomography: leveraging multiple scattering for high-contrast imaging at depth in turbid media. Sci Adv 2023; 9:eadh5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lang W. Nomarski differential interference-contrast microscopy. Carl Zeiss: Oberkochen; 1982. [Google Scholar]
- 39. Ziv NE, Schiller J. Differential Interference Contrast (DIC) Imaging of Living Cells, vol. 2007. USA: CSH Protoc; 2007: pdb.prot4787. [DOI] [PubMed] [Google Scholar]
- 40. Dodt HU, Zieglgansberger W. Visualizing unstained neurons in living brain slices by infrared DIC-videomicroscopy. Brain Res 1990; 537:333–336. [DOI] [PubMed] [Google Scholar]
- 41. Nishimura K, Ishiwata H, Sakuragi Y, Hayashi Y, Fukuda A, Hisatake K. Live-cell imaging of subcellular structures for quantitative evaluation of pluripotent stem cells. Sci Rep 2019; 9:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bajer A, Allen RD. Structure and Organization of the Living Mitotic Spindle of Haemanthus endosperm. Science 1966; 151:572–574. [DOI] [PubMed] [Google Scholar]
- 43. Wang X, Wang H, Wang J, Liu X, Hao H, Tan YS, Zhang Y, Zhang H, Ding X, Zhao W, Wang Y, Lu Z, et al. Single-shot isotropic differential interference contrast microscopy. Nat Commun 2023; 14:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Newmark JA, Warger WC 2nd, Chang C, Herrera GE, Brooks DH, DiMarzio CA, Warner CM. Determination of the number of cells in preimplantation embryos by using noninvasive optical quadrature microscopy in conjunction with differential interference contrast microscopy. Microsc Microanal 2007; 13:118–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Balasubramani V, Kujawinska M, Allier C, Anand V, Cheng CJ, Depeursinge C, Hai N, Juodkazis S, Kalkman J, Kus A, Lee M, Magistretti PJ, et al. Roadmap on digital holography-based quantitative phase imaging. J Imaging 2021; 7:1–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Javidi B, Carnicer A, Anand A, Barbastathis G, Chen W, Ferraro P, Goodman JW, Horisaki R, Khare K, Kujawinska M, Leitgeb RA, Marquet P, et al. Roadmap on digital holography [invited]. Opt Express 2021; 29:35078–35118. [DOI] [PubMed] [Google Scholar]
- 47. Rappaz B, Marquet P, Cuche E, Emery Y, Depeursinge C, Magistretti P. Measurement of the integral refractive index and dynamic cell morphometry of living cells with digital holographic microscopy. Opt Express 2005; 13:9361–9373. [DOI] [PubMed] [Google Scholar]
- 48. Wacogne B, Ivascu I, Zeggari R, Pieralli C, Amiot C, Pazart L, Roux C. Microsensors and image processing for single oocyte qualification: toward multiparametric determination of the best time for fertilization. Laser Phys Lett 2013; 10:105601. [Google Scholar]
- 49. Gul B, Ashraf S, Khan S, Nisar H, Ahmad I. Cell refractive index: models, insights, applications and future perspectives. Photodiagn Photodyn Ther 2021; 33:102096. [DOI] [PubMed] [Google Scholar]
- 50. Dwapanyin GO, Chow DJX, Tan TCY, Dubost NS, Morizet JM, Dunning KR, Dholakia K. Investigation of refractive index dynamics during in vitro embryo development using off-axis digital holographic microscopy. Biomed Opt Express 2023; 14:3327–3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Fotos JS, Patel VP, Karin NJ, Temburni MK, Koh JT, Galileo DS. Automated time-lapse microscopy and high-resolution tracking of cell migration. Cytotechnology 2006; 51:7–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Huth J, Buchholz M, Kraus JM, Schmucker M, von Wichert G, Krndija D, Seufferlein T, Gress TM, Kestler HA. Significantly improved precision of cell migration analysis in time-lapse video microscopy through use of a fully automated tracking system. BMC Cell Biol 2010; 11:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Soukup J, Cisar P, Sroubek F. Segmentation method of time-lapse microscopy images with the focus on biocompatibility assessment. Microsc Microanal 2016; 22:497–506. [DOI] [PubMed] [Google Scholar]
- 54. Ascione F, Caserta S, Perris R, Guido S. Investigation of cell dynamics in vitro by time lapse microscopy and image analysis. Chem Eng Trans 2014; 38:517–522. [Google Scholar]
- 55. Orth JD, Kohler RH, Foijer F, Sorger PK, Weissleder R, Mitchison TJ. Analysis of mitosis and antimitotic drug responses in tumors by in vivo microscopy and single-cell pharmacodynamics. Cancer Res 2011; 71:4608–4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Jiang Q, Sudalagunta P, Silva MC, Canevarolo RR, Zhao X, Ahmed KT, Alugubelli RR, DeAvila G, Tungesvik A, Perez L, Gatenby RA, Gillies RJ, et al. CancerCellTracker: a brightfield time-lapse microscopy framework for cancer drug sensitivity estimation. Bioinformatics 2022; 38:4002–4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. D'Orazio M, Murdocca M, Mencattini A, Casti P, Filippi J, Antonelli G, Di Giuseppe D, Comes MC, Di Natale C, Sangiuolo F, Martinelli E. Machine learning phenomics (MLP) combining deep learning with time-lapse-microscopy for monitoring colorectal adenocarcinoma cells gene expression and drug-response. Sci Rep 2022; 12:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kovacs P. Embryo selection: the role of time-lapse monitoring. Reprod Biol Endocrinol 2014; 12:124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Minasi MG, Greco P, Varricchio MT, Barillari P, Greco E. The clinical use of time-lapse in human-assisted reproduction. Ther Adv Reprod Health 2020; 14:263349412097692–263349412097615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Chen M, Wei S, Hu J, Yuan J, Liu F. Does time-lapse imaging have favorable results for embryo incubation and selection compared with conventional methods in clinical in vitro fertilization? A meta-analysis and systematic review of randomized controlled trials. PLoS One 2017; 12:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ahlstrom A, Lundin K, Lind AK, Gunnarsson K, Westlander G, Park H, Thurin-Kjellberg A, Thorsteinsdottir SA, Einarsson S, Astrom M, Lofdahl K, Menezes J, et al. A double-blind randomized controlled trial investigating a time-lapse algorithm for selecting day 5 blastocysts for transfer. Hum Reprod 2022; 37:708–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Racowsky C, Kovacs P, Martins WP. A critical appraisal of time-lapse imaging for embryo selection: where are we and where do we need to go? J Assist Reprod Genet 2015; 32:1025–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Chow DJX, Wijesinghe P, Dholakia K, Dunning KR. Does artificial intelligence have a role in the IVF clinic? Reprod Fertil 2021; 2:C29–C34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Valeur B, Berberan-Santos MN. A brief history of fluorescence and phosphorescence before the emergence of quantum theory. J Chem Educ 2011; 88:731–738. [Google Scholar]
- 65. Stokes GG. On the change of refrangibility of light. Philos Trans R Soc 1852; 142:463–562. [Google Scholar]
- 66. Chance B, Thorell B. Localization and kinetics of reduced pyridine nucleotide in living cells by microfluorometry. J Biol Chem 1959; 234:3044–3050. [PubMed] [Google Scholar]
- 67. Waldchen S, Lehmann J, Klein T, van de Linde S, Sauer M. Light-induced cell damage in live-cell super-resolution microscopy. Sci Rep 2015; 5:15348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Quinn KP, Sridharan GV, Hayden RS, Kaplan DL, Lee K, Georgakoudi I. Quantitative metabolic imaging using endogenous fluorescence to detect stem cell differentiation. Sci Rep 2013; 3:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. So PT, Dong CY, Masters BR, Berland KM. Two-photon excitation fluorescence microscopy. Annu Rev Biomed Eng 2000; 2:399–429. [DOI] [PubMed] [Google Scholar]
- 70. Benninger RKP, Piston DW. Two-photon excitation microscopy for the study of living cells and tissues. Curr Protoc Cell Biol 2013; Chapter 4.1124; 1–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Smith R, Wright KL, Ashton L. Raman spectroscopy: an evolving technique for live cell studies. Analyst 2016; 141:3590–3600. [DOI] [PubMed] [Google Scholar]
- 72. Smith E, Dent G. Introduction, Basic Theory and Principles. In: Modern Raman Spectroscopy - A Practical Approach. England: John Wiley & Sons, Ltd.; 2004: 1–21. [Google Scholar]
- 73. Windom BC, Hahn DW, Spectroscopy R. In: Wang QJ, Chung Y-W (eds.), Encyclopedia of Tribology. Boston, MA: Springer US; 2013: 2742–2747. [Google Scholar]
- 74. Meng H, Huang S, Diao F, Gao C, Zhang J, Kong L, Gao Y, Jiang C, Qin L, Chen Y, Xu M, Gao L, et al. Rapid and non-invasive diagnostic techniques for embryonic developmental potential: a metabolomic analysis based on Raman spectroscopy to identify the pregnancy outcomes of IVF-ET. Front Cell Dev Biol 2023; 11:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Ding J, Xu T, Tan X, Jin H, Shao J, Li H. Raman spectrum: a potential biomarker for embryo assessment during in vitro fertilization. Exp Ther Med 2017; 13:1789–1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Zheng W, Zhang S, Gu Y, Gong F, Kong L, Lu G, Lin G, Liang B, Hu L. Non-invasive Metabolomic profiling of embryo culture medium using Raman Spectroscopy with deep learning model predicts the blastocyst development potential of embryos. Front Physiol 2021; 12:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Lohumi S, Kim MS, Qin J, Cho BK. Raman imaging from microscopy to macroscopy: quality and safety control of biological materials. TrAC Trends Anal Chem 2017; 93:183–198. [Google Scholar]
- 78. Ishigaki M, Hashimoto K, Sato H, Ozaki Y. Non-destructive monitoring of mouse embryo development and its qualitative evaluation at the molecular level using Raman spectroscopy. Sci Rep 2017; 7:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Perevedentseva E, Krivokharchenko A, Karmenyan AV, Chang HH, Cheng CL. Raman spectroscopy on live mouse early embryo while it continues to develop into blastocyst in vitro. Sci Rep 2019; 9:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Paddock SW. Principles and practices of laser scanning confocal microscopy. Mol Biotechnol 2000; 16:127–150. [DOI] [PubMed] [Google Scholar]
- 81. Phoon CK. Imaging tools for the developmental biologist: ultrasound biomicroscopy of mouse embryonic development. Pediatr Res 2006; 60:14–21. [DOI] [PubMed] [Google Scholar]
- 82. Horga G, Kaur T, Peterson BS. Annual research review: current limitations and future directions in MRI studies of child- and adult-onset developmental psychopathologies. J Child Psychol Psychiatry 2014; 55:659–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Keklikoglou K, Arvanitidis C, Chatzigeorgiou G, Chatzinikolaou E, Karagiannidis E, Koletsa T, Magoulas A, Makris K, Mavrothalassitis G, Papanagnou ED, Papazoglou AS, Pavloudi C, et al. Micro-CT for biological and biomedical studies: a comparison of imaging techniques. J Imaging 2021; 7:1–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Tavakoli M, Hossain P, Malik RA. Clinical applications of corneal confocal microscopy. Clin Ophthalmol 2008; 2:435–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Chiang JCB, Roy M, Kim J, Markoulli M, Krishnan AV. In-vivo corneal confocal microscopy: imaging analysis, biological insights and future directions. Commun Biol 2023; 6:652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Taylor MD, Roberts JR, Hubbs AF, Reasor MJ, Antonini JM. Quantitative image analysis of drug-induced lung fibrosis using laser scanning confocal microscopy. Toxicol Sci 2002; 67:295–302. [DOI] [PubMed] [Google Scholar]
- 87. Zucker RM. Whole insect and mammalian embryo imaging with confocal microscopy: morphology and apoptosis. Cytometry A 2006; 69A:1143–1152. [DOI] [PubMed] [Google Scholar]
- 88. Heppert JK, Dickinson DJ, Pani AM, Higgins CD, Steward A, Ahringer J, Kuhn JR, Goldstein B. Comparative assessment of fluorescent proteins for in vivo imaging in an animal model system. Mol Biol Cell 2016; 27:3385–3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Squirrell JM, Wokosin DL, White JG, Bavister BD. Long-term two-photon fluorescence imaging of mammalian embryos without compromising viability. Nat Biotechnol 1999; 17:763–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Icha J, Weber M, Waters JC, Norden C. Phototoxicity in live fluorescence microscopy, and how to avoid it. Bioessays 2017; 39:1–15. [DOI] [PubMed] [Google Scholar]
- 91. Takenaka M, Horiuchi T, Yanagimachi R. Effects of light on development of mammalian zygotes. Proc Natl Acad Sci U S A 2007; 104:14289–14293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Bognar Z, Csabai TJ, Pallinger E, Balassa T, Farkas N, Schmidt J, Gorgey E, Berta G, Szekeres-Bartho J, Bodis J. The effect of light exposure on the cleavage rate and implantation capacity of preimplantation murine embryos. J Reprod Immunol 2019; 132:21–28. [DOI] [PubMed] [Google Scholar]
- 93. Campugan CA, Lim M, Chow DJX, Tan TCY, Li T, Saini AA, Orth A, Reineck P, Schartner EP, Thompson JG, Dholakia K, Dunning KR. The effect of discrete wavelengths of visible light on the developing murine embryo. J Assist Reprod Genet 2022; 39:1825–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Datta R, Heaster TM, Sharick JT, Gillette AA, Skala MC. Fluorescence lifetime imaging microscopy: fundamentals and advances in instrumentation, analysis, and applications. J Biomed Opt 2020; 25:1–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Chang CW, Sud D, Mycek MA. Fluorescence lifetime imaging microscopy. Methods Cell Biol 2007; 81:495–524. [DOI] [PubMed] [Google Scholar]
- 96. Blacker TS, Mann ZF, Gale JE, Ziegler M, Bain AJ, Szabadkai G, Duchen MR. Separating NADH and NADPH fluorescence in live cells and tissues using FLIM. Nat Commun 2014; 5:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Venturas M, Shah JS, Yang X, Sanchez TH, Conway W, Sakkas D, Needleman DJ. Metabolic state of human blastocysts measured by fluorescence lifetime imaging microscopy. Hum Reprod 2022; 37:411–427. [DOI] [PubMed] [Google Scholar]
- 98. Shah JS, Venturas M, Sanchez TH, Penzias AS, Needleman DJ, Sakkas D. Fluorescence lifetime imaging microscopy (FLIM) detects differences in metabolic signatures between euploid and aneuploid human blastocysts. Hum Reprod 2022; 37:400–410. [DOI] [PubMed] [Google Scholar]
- 99. Seidler EA, Sanchez T, Venturas M, Sakkas D, Needleman DJ. Non-invasive imaging of mouse embryo metabolism in response to induced hypoxia. J Assist Reprod Genet 2020; 37:1797–1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Sanchez T, Venturas M, Aghvami SA, Yang X, Fraden S, Sakkas D, Needleman DJ. Combined noninvasive metabolic and spindle imaging as potential tools for embryo and oocyte assessment. Hum Reprod 2019; 34:2349–2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Lu G, Fei B. Medical hyperspectral imaging: a review. J Biomed Opt 2014; 19:1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Dumollard R, Ward Z, Carroll J, Duchen MR. Regulation of redox metabolism in the mouse oocyte and embryo. Development 2007; 134:455–465. [DOI] [PubMed] [Google Scholar]
- 103. Campbell JM, Mahbub SB, Habibalahi A, Agha A, Handley S, Anwer AG, Goldys EM. Clinical applications of non-invasive multi and hyperspectral imaging of cell and tissue autofluorescence beyond oncology. J Biophotonics 2023; 16:1–14. [DOI] [PubMed] [Google Scholar]
- 104. Gosnell ME, Anwer AG, Cassano JC, Sue CM, Goldys EM. Functional hyperspectral imaging captures subtle details of cell metabolism in olfactory neurosphere cells, disease-specific models of neurodegenerative disorders. Biochim Biophys Acta, Mol Cell Res 2016; 1863:56–63. [DOI] [PubMed] [Google Scholar]
- 105. Mahbub SB, Guller A, Campbell JM, Anwer AG, Gosnell ME, Vesey G, Goldys EM. Non-invasive monitoring of functional state of articular cartilage tissue with label-free unsupervised hyperspectral imaging. Sci Rep 2019; 9:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Paoli J, Polonen I, Salmivuori M, Rasanen J, Zaar O, Polesie S, Koskenmies S, Pitkanen S, Overmark M, Isoherranen K, Juteau S, Ranki A, et al. Hyperspectral imaging for non-invasive diagnostics of melanocytic lesions. Acta Derm Venereol 2022; 102:adv00815–adv00817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. More SS, Beach JM, Vince R. Early detection of amyloidopathy in Alzheimer's mice by hyperspectral endoscopy. Invest Ophthalmol Vis Sci 2016; 57:3231–3238. [DOI] [PubMed] [Google Scholar]
- 108. Hadoux X, Hui F, Lim JKH, Masters CL, Pebay A, Chevalier S, Ha J, Loi S, Fowler CJ, Rowe C, Villemagne VL, Taylor EN, et al. Non-invasive in vivo hyperspectral imaging of the retina for potential biomarker use in Alzheimer's disease. Nat Commun 2019; 10:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. More SS, Vince R. Hyperspectral imaging signatures detect amyloidopathy in Alzheimer's mouse retina well before onset of cognitive decline. ACS Chem Neurosci 2015; 6:306–315. [DOI] [PubMed] [Google Scholar]
- 110. Sutton-McDowall ML, Gosnell M, Anwer AG, White MA, Purdey M, Abell AD, Goldys EM, Thompson JG. Hyperspectral microscopy can detect metabolic heterogeneity within bovine post-compaction embryos incubated under two oxygen concentrations (7% versus 20%). Hum Reprod 2017; 32:2016–2025. [DOI] [PubMed] [Google Scholar]
- 111. Gosnell ME, Anwer AG, Mahbub SB, Menon Perinchery S, Inglis DW, Adhikary PP, Jazayeri JA, Cahill MA, Saad S, Pollock CA, Sutton-McDowall ML, Thompson JG, et al. Quantitative non-invasive cell characterisation and discrimination based on multispectral autofluorescence features. Sci Rep 2016; 6:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Santos Monteiro CA, Chow DJX, Leal GR, Tan TC, Reis Ferreira AM, Thompson JG, Dunning KR. Optical imaging of cleavage stage bovine embryos using hyperspectral and confocal approaches reveals metabolic differences between on-time and fast-developing embryos. Theriogenology 2021; 159:60–68. [DOI] [PubMed] [Google Scholar]
- 113. Tan TCY, Brown HM, Thompson JG, Mustafa S, Dunning KR. Optical imaging detects metabolic signatures associated with oocyte quality. Biol Reprod 2022; 107:1014–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Reynaud EG, Krzic U, Greger K, Stelzer EH. Light sheet-based fluorescence microscopy: more dimensions, more photons, and less photodamage. HFSP J 2008; 2:266–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Tomer R, Khairy K, Keller PJ. Shedding light on the system: studying embryonic development with light sheet microscopy. Curr Opin Genet Dev 2011; 21:558–565. [DOI] [PubMed] [Google Scholar]
- 116. Huisken J. Slicing embryos gently with laser light sheets. Bioessays 2012; 34:406–411. [DOI] [PubMed] [Google Scholar]
- 117. Strnad P, Gunther S, Reichmann J, Krzic U, Balazs B, de Medeiros G, Norlin N, Hiiragi T, Hufnagel L, Ellenberg J. Inverted light-sheet microscope for imaging mouse pre-implantation development. Nat Methods 2016; 13:139–142. [DOI] [PubMed] [Google Scholar]
- 118. Huisken J, Swoger J, Del Bene F, Wittbrodt J, Stelzer EH. Optical sectioning deep inside live embryos by selective plane illumination microscopy. Science 2004; 305:1007–1009. [DOI] [PubMed] [Google Scholar]
- 119. Weber M, Huisken J. Light sheet microscopy for real-time developmental biology. Curr Opin Genet Dev 2011; 21:566–572. [DOI] [PubMed] [Google Scholar]
- 120. Swoger J, Pampaloni F, Stelzer EH. Light-sheet-based fluorescence microscopy for three-dimensional imaging of biological samples. Cold Spring Harb Protoc 2014; 2014:1–8. [DOI] [PubMed] [Google Scholar]
- 121. Rieckher M. Light sheet microscopy to measure protein dynamics. J Cell Physiol 2016; 232:27–35. [DOI] [PubMed] [Google Scholar]
- 122. Ellefsen KL, Parker I. Dynamic Ca(2+) imaging with a simplified lattice light-sheet microscope: a sideways view of subcellular Ca(2+) puffs. Cell Calcium 2018; 71:34–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Ritter JG, Veith R, Veenendaal A, Siebrasse JP, Kubitscheck U. Light sheet microscopy for single molecule tracking in living tissue. PLoS One 2010; 5:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Tomer R, Khairy K, Amat F, Keller PJ. Quantitative high-speed imaging of entire developing embryos with simultaneous multiview light-sheet microscopy. Nat Methods 2012; 9:755–763. [DOI] [PubMed] [Google Scholar]
- 125. Udan RS, Piazza VG, Hsu CW, Hadjantonakis AK, Dickinson ME. Quantitative imaging of cell dynamics in mouse embryos using light-sheet microscopy. Development 2014; 141:4406–4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Reichmann J, Eguren M, Lin Y, Schneider I, Ellenberg J. Live imaging of cell division in preimplantation mouse embryos using inverted light-sheet microscopy. Methods Cell Biol 2018; 145:279–292. [DOI] [PubMed] [Google Scholar]
- 127. Khairy K, Lemon WC, Amat F, Keller PJ. Light sheet-based imaging and analysis of early embryogenesis in the fruit fly. Methods Mol Biol 2015; 1189:79–97. [DOI] [PubMed] [Google Scholar]
- 128. Keller PJ, Schmidt AD, Wittbrodt J, Stelzer EH. Reconstruction of zebrafish early embryonic development by scanned light sheet microscopy. Science 2008; 322:1065–1069. [DOI] [PubMed] [Google Scholar]
- 129. Bernardello M, Marsal M, Gualda EJ, Loza-Alvarez P. Light-sheet fluorescence microscopy for the in vivo study of microtubule dynamics in the zebrafish embryo. Biomed Opt Express 2021; 12:6237–6254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Xu HN, Tchou J, Feng M, Zhao H, Li LZ. Optical redox imaging indices discriminate human breast cancer from normal tissues. J Biomed Opt 2016; 21:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Sun N, Xu HN, Luo Q, Li LZ. Potential indexing of the invasiveness of breast cancer cells by mitochondrial redox ratios. Adv Exp Med Biol 2016; 923:121–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Skala MC, Riching KM, Gendron-Fitzpatrick A, Eickhoff J, Eliceiri KW, White JG, Ramanujam N. In vivo multiphoton microscopy of NADH and FAD redox states, fluorescence lifetimes, and cellular morphology in precancerous epithelia. Proc Natl Acad Sci U S A 2007; 104:19494–19499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Xu HN, Gourmaud S, Podsednik A, Li X, Zhao H, Jensen FE, Talos DM, Li LZ. Optical redox imaging of ex vivo hippocampal tissue reveals age-dependent alterations in the 5XFAD mouse model of Alzheimer's disease. Metabolites 2022; 12:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Jentsch S, Schweitzer D, Schmidtke KU, Peters S, Dawczynski J, Bar KJ, Hammer M. Retinal fluorescence lifetime imaging ophthalmoscopy measures depend on the severity of Alzheimer's disease. Acta Ophthalmol 2015; 93:e241–e247. [DOI] [PubMed] [Google Scholar]
- 135. Morizet JM, Chow DJX, Wijesinghe P, Schartner EP, Dwapanyin GO, Dubost NS, Bruce GD, Anckaert E, Dunning KR, Dholakia K. UVA hyperspectral light-sheet microscopy for volumetric metabolic imaging: application to pre-implantation embryo development. ACS Photonics 2023; 10:4177–4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Kapoor R, Whigham BT, Al-Aswad LA. Artificial intelligence and optical coherence tomography imaging. Asia Pac J Ophthalmol (Phila) 2019; 8:187–194. [DOI] [PubMed] [Google Scholar]
- 137. Hollon TC, Pandian B, Adapa AR, Urias E, Save AV, Khalsa SSS, Eichberg DG, D'Amico RS, Farooq ZU, Lewis S, Petridis PD, Marie T, et al. Near real-time intraoperative brain tumor diagnosis using stimulated Raman histology and deep neural networks. Nat Med 2020; 26:52–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Hollon T, Jiang C, Chowdury A, Nasir-Moin M, Kondepudi A, Aabedi A, Adapa A, Al-Holou W, Heth J, Sagher O, Lowenstein P, Castro M, et al. Artificial-intelligence-based molecular classification of diffuse gliomas using rapid, label-free optical imaging. Nat Med 2023; 29:828–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Tsai CL, Mukundan A, Chung CS, Chen YH, Wang YK, Chen TH, Tseng YS, Huang CW, Wu IC, Wang HC. Hyperspectral imaging combined with artificial intelligence in the early detection of Esophageal cancer. Cancers (Basel) 2021; 13:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. You JB, McCallum C, Wang Y, Riordon J, Nosrati R, Sinton D. Machine learning for sperm selection. Nat Rev Urol 2021; 18:387–403. [DOI] [PubMed] [Google Scholar]
- 141. Manna C, Nanni L, Lumini A, Pappalardo S. Artificial intelligence techniques for embryo and oocyte classification. Reprod Biomed Online 2013; 26:42–49. [DOI] [PubMed] [Google Scholar]
- 142. Berntsen J, Rimestad J, Lassen JT, Tran D, Kragh MF. Robust and generalizable embryo selection based on artificial intelligence and time-lapse image sequences. PLoS One 2022; 17:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Sawada Y, Sato T, Nagaya M, Saito C, Yoshihara H, Banno C, Matsumoto Y, Matsuda Y, Yoshikai K, Sawada T, Ukita N, Sugiura-Ogasawara M. Evaluation of artificial intelligence using time-lapse images of IVF embryos to predict live birth. Reprod Biomed Online 2021; 43:843–852. [DOI] [PubMed] [Google Scholar]
- 144. Hariharan R, He P, Meseguer M, Toschi M, Rocha JC, Zaninovic N, Malmsten J, Zhan Q, Hickman C. Artificial intelligence assessment of time-lapse images can predict with 77% accuracy whether a human embryo capable of achieving a pregnancy will miscarry. Fertil Steril 2019; 112:E38–E39. [Google Scholar]
- 145. Huang B, Tan W, Li Z, Jin L. An artificial intelligence model (euploid prediction algorithm) can predict embryo ploidy status based on time-lapse data. Reprod Biol Endocrinol 2021; 19:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Liao Q, Zhang Q, Feng X, Huang H, Xu H, Tian B, Liu J, Yu Q, Guo N, Liu Q, Huang B, Ma D, et al. Development of deep learning algorithms for predicting blastocyst formation and quality by time-lapse monitoring. Commun Biol 2021; 4:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Coticchio G, Fiorentino G, Nicora G, Sciajno R, Cavalera F, Bellazzi R, Garagna S, Borini A, Zuccotti M. Cytoplasmic movements of the early human embryo: imaging and artificial intelligence to predict blastocyst development. Reprod Biomed Online 2021; 42:521–528. [DOI] [PubMed] [Google Scholar]
- 148. Tran D, Cooke S, Illingworth PJ, Gardner DK. Deep learning as a predictive tool for fetal heart pregnancy following time-lapse incubation and blastocyst transfer. Hum Reprod 2019; 34:1011–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Kieslinger DC, Vergouw CG, Ramos L, Arends B, Curfs M, Slappendel E, Kostelijk EH, Pieters M, Consten D, Verhoeven MO, Besselink DE, Broekmans F, et al. Clinical outcomes of uninterrupted embryo culture with or without time-lapse-based embryo selection versus interrupted standard culture (SelecTIMO): a three-armed, multicentre, double-blind, randomised controlled trial. Lancet 2023; 401:1438–1446. [DOI] [PubMed] [Google Scholar]
- 150. Wijesinghe P, Dholakia K. Emergent physics-informed design of deep learning for microscopy. JPhys Photonics 2021; 3:1–10. [Google Scholar]
- 151. Kucherov A, Fazzari M, Lieman H, Ball GD, Doody K, Jindal S. PGT-A is associated with reduced cumulative live birth rate in first reported IVF stimulation cycles age </= 40: an analysis of 133,494 autologous cycles reported to SART CORS. J Assist Reprod Genet 2023; 40:137–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Wang J, Guo Y, Zhang N, Li T. Research progress of time-lapse imaging technology and embryonic development potential: a review. Medicine (Baltimore) 2023; 102:e35203.1–e35203.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Viana JHM. 2021 statistics of embryo production and transfer in domestic farm animals. Embryo Technology Newsletter 2022; 40:22–40. [Google Scholar]
- 154. Ealy AD, Wooldridge LK, McCoski SR. Post-transfer consequences of in vitro-produced embryos in cattle. J Anim Sci 2019; 97:2555–2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. McLennan HJ, Saini A, Dunning KR, Thompson JG. Oocyte and embryo evaluation by AI and multi-spectral auto-fluorescence imaging: livestock embryology needs to catch-up to clinical practice. Theriogenology 2020; 150:255–262. [DOI] [PubMed] [Google Scholar]
- 156. Ashok PC, Praveen BB, Bellini N, Riches A, Dholakia K, Herrington CS. Multi-modal approach using Raman spectroscopy and optical coherence tomography for the discrimination of colonic adenocarcinoma from normal colon. Biomed Opt Express 2013; 4:2179–2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Wang S, Larina IV. In vivo dynamic 3D imaging of oocytes and embryos in the mouse oviduct. Cell Rep 2021; 36:109382.1–109382.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No new data were generated or analysed in support of this review article.