Abstract
CST (BART BARF0) family viral RNAs are expressed in several types of Epstein-Barr virus (EBV) infection, including EBV-associated cancers. Many different spliced forms of these RNAs have been described; here we have clarified the structures of some of the more abundant splicing patterns. We report the first cDNAs representing a full-length CST mRNA from a clone library and further characterize the transcription start. The relative abundance of splicing patterns and genomic analysis of the open reading frames (ORFs) suggest that, in addition to the much studied BARF0 ORF, there may be important products made from some of the upstream ORFs in the CST RNAs. Potential biological functions are identified for two of these. The product of the RPMS1 ORF is shown to be a nuclear protein that can bind to the CBF1 component of Notch signal transduction. RPMS1 can inhibit the transcription activation induced through CBF1 by NotchIC or EBNA-2. The protein product of another CST ORF, A73, is shown to be a cytoplasmic protein which can interact with the cell RACK1 protein. Since RACK1 modulates signaling from protein kinase C and Src tyrosine kinases, the results suggest a possible role for CST products in growth control, perhaps consistent with the abundant transcription of CST RNAs in cancers such as nasopharyngeal carcinoma.
In several types of infection, in addition to the well-established EBNA, LMP and EBER genes, Epstein-Barr virus (EBV) has been found to express various spliced RNAs transcribed rightward from the region spanning 150,000 to 161,000 on the B95-8 EBV genetic map. These have been referred to as complementary strand transcripts (CSTs), BamHI A rightward transcripts (BARTs), or the BARF0 RNAs. RNAs of this type were originally identified in cDNA made from the C15 xenograft culture of nasopharyngeal carcinoma (NPC) tissue (17). Similar RNAs were subsequently found in various EBV-positive NPC tumor biopsies and xenografts, Burkitt's lymphoma, lymphoid cell lines (LCLs) (3, 6, 11, 15, 19, 31, 43), and biopsies of oral hairy leukoplakia (24). Expression of CST RNAs has also been demonstrated in peripheral blood of normal human carriers of EBV (5), sera from NPC patients have been found to immunoprecipitate a protein product of the BARF0 open reading frame (ORF) made in vitro (12), and cytotoxic T lymphocytes that respond to a peptide derived from BARF0 have been identified in EBV-infected people (22).
A very complicated picture of alternatively spliced CST RNAs has built up (36, 38), but some of the proposed structures have been deduced using very sensitive reverse transcription (RT)-PCR methods or have been only single isolates from cDNA libraries, and thus they may yet represent very minor species within the family of RNAs that can be expressed. Predominant sizes of the RNAs expressed have been deduced from Northern blots, but these have only partly been related to the spliced RNA structures. In addition, the coding content of the CST RNAs remains uncertain. Most attention has focused on the BARF0 ORF, particularly a spliced form called RK-BARF0, located near the common 3′ end of the RNAs, since this is the longest ORF. Protein expression has been reported for this ORF in cells infected by EBV (10), but these data have been complicated by a subsequent report (21) indicating that the antibody used cross-reacts with a cell protein similar in size to that predicted for the viral ORF, casting doubt on the specificity of the original results. No function has yet been reported for a product of the BARF0 ORF.
In this paper we report the structures of the first full-length cDNA clones for the CST family, compare by RNase protection assay (RPA) the abundance of various spliced forms of the RNA structures, and reexamine the coding potential of the RNAs. These results suggest that attention should now be directed to some of the smaller ORFs upstream of BARF0, the predicted products of which may have functions relevant to the observation that CST RNAs are frequently found to be expressed in human cancers containing EBV.
MATERIALS AND METHODS
Isolation of cDNA clones.
An oligo(dT)-primed cDNA library was constructed from 5 μg of polyadenylated RNA isolated from the C15 tumor using a TimeSaver cDNA synthesis kit (Pharmacia) and cloned into an EcoRI-digested λgt11 vector (Stratagene). To isolate large or full-length clones, duplicate lifts were taken from a total of 5 × 105 plaques. One set was then screened with an RsaI fragment from the 3′ end of the CSTs (EBV coordinates 160250 to 161063), and the other was screened with a BglII-SspI fragment containing exon I (EBV coordinates 150462 to 151009).
The library was separately probed with an XhoI restriction fragment (EBV genome coordinates 155939 to 156325) together with a duplicate set probed with the RsaI clone described above.
Positive clones were plaque purified, transferred into plasmids, and analyzed by sequencing and restriction digestion.
RPA.
RNA was extracted from cell lines and the C15 NPC xenograft, using RNAzol B (Tel-Test Inc.) RPAs were performed as described previously (37).
Primer extension.
HeLa cells were transfected using calcium phosphate (13) with a plasmid (SK) containing the CST promoter region (EBV coordinates 150201 to 151282) cloned upstream of the chloramphenicol acetyltransferase (CAT) reporter in pCAT Basic (Promega) or with pCAT Basic as a control. At 48 h posttransfection, the cells were harvested and total RNA was extracted using RNAzol B. Primer extension reactions were performed as described previously (38), using 25 μg of total RNA hybridized to a labeled oligonucleotide probe, GAAGAGGCTAGTGCCTACG (EBV positions 150762 to 150744), within CST exon I. Extension products were resolved on a 7.5% sequencing gel and visualized by autoradiography.
Expression of RPMS1.
ORFs within the full-length cDNA clones were cloned after PCR amplification into pcDNA3 vector (Invitrogen). Flag sequences were introduced into the RPMS1 ORF by PCR protocols. Similar Flag sequences were also introduced into the RPMS1 ORF within the full-length cDNA. cDNA clones containing either the tagged RPMS1 ORF alone or the full-length clone containing a tagged RPMS1 ORF were then transfected into 293 cells by using Superfect (Qiagen). Transfected cells were harvested and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) through a 12.5% gel, transferred to a polyvinylidene difluoride (PVDF) membrane, and probed with the anti-Flag monoclonal antibody M2 (Sigma).
In vitro transcription and translation.
Full-length cDNAs in pcDNA3 were linearized with XbaI; transcripts were synthesized using the T7 message machine kit (Ambion) and translated separately using a wheat germ translation system (Ambion). Products were separated by SDS-PAGE on a 12.5% gel and transferred to a PVDF membrane which was blocked (5% dry milk–phosphate-buffered saline [PBS]) and probed using a rabbit anti-RPMS1 peptide (SGQPRWWPWG) antibody, diluted 1:20 with milk-PBS. Following incubation with a goat anti-rabbit horseradish peroxidase conjugate (Dako), proteins were visualized using the Amersham ECL chemiluminescence system.
Immunofluorescence.
Epitope-tagged RPMS1 was transfected into HeLa cells by calcium phosphate precipitation, and expression was visualized using the anti-Flag antibody M2. Flag-tagged A73 was transfected into 293 cells by calcium phosphate precipitation and stained for expression as follows. Cells were fixed in 4% paraformaldehyde in PBS (30 min at 4°C), rinsed with PBS, and permeabilized with 0.5% Triton X-100 for 30 min at 4°C. Cells were stained with anti-Flag (1:100 dilution) or anti-RACK1 (1:100 dilution) for 60 min at 4°C. RACK1 staining was visualized with fluorescein isothiocyanate-conjugated anti-mouse immunoglobulin M (1:20; Sigma); Flag-tagged anti-A73 staining was visualized with tetramethyl rhodamine isothiocyanate-conjugated anti-mouse immunoglobulin G.
Fusion proteins.
RPMS1 ORF from the full-length cDNA clone was cloned into the pGEX-2 vector, using BamHI and EcoRI sites introduced by PCR amplification. RPMS1(34-103) was subcloned as a PstI-EcoRI fragment (PstI site at amino acid 34) into pBluescript vector and then recloned via the BamHI/EcoRI sites into pGEX-3.
A73 fusion protein constructions were generated by PCR, introducing BamHI and EcoRI sites at the N and C termini of A73 so that the products could be cloned into pGEX-4T. The resulting glutathione S-transferase (GST)–A73 fusion was then PCR amplified to introduce a HindIII site at the 5′ end of the GST protein and subcloned into pcDNA3.
In vitro CBF1 binding assays.
Linearized vector containing the CBF1 ORF was labeled in vitro with [35S]methionine by in vitro transcription and translation (Promega). Single bacterial colonies containing the relevant plasmids were picked into 5-ml cultures, grown overnight at 37°C, diluted (1:10) into 25-ml cultures, and grown, with shaking, for a further hour. Expression of fusion proteins was induced by the addition of 0.1 mM isopropyl-β-d-thiogalactopyranoside, and cultures were propagated for a further 3 h. Bacteria were harvested, resuspended in PBS containing protease inhibitors, and sonicated for 20 s on ice; debris was removed by centrifugation at 13,000 × g for 10 min, and the supernatant was incubated with glutathione beads (Sigma) for 30 min at 4°C. The resin was washed five times in 1 ml of PBS and then resuspended in 0.5 ml of PBS. Radiolabeled CBF1 (1 μl) was added to each sample and incubated for a further 30 min at 4°C. Complexes were washed a further five times with 1 ml of PBS containing 0.05% Tween and then eluted from the beads using 250 μl of protein sample buffer and heat (95°C for 2 min). Eluted complexes (50 μl) were separated by SDS-PAGE on 10% gels and dried, and bound CBF1 was visualized by autoradiography. After separation by SDS-PAGE, complexes were analyzed by Western blotting and probed with monoclonal anti-GST antibodies (a gift from S. Dilworth) to determine relative levels of fusion proteins in each reaction.
RPMS1-CBF1 interactions in yeast.
Saccharomyces cerevisiae and yeast plasmids pAS1-CYH2 and pACTII were obtained from S. Elledge (Baylor College of Medicine, Houston, Tex.). The RPMS1 ORF was fused to the Gal4 DNA binding domain in pAS-CYH2, and the various segments of the CBF1 ORF were fused to the Gal4 activation domain in pACTII. pAS-CYH2-RPMS1 and the different pACTII-CBF1 plasmids were cotransformed into S. cerevisiae Y190 (7). Each transformation mixture was plated on either SC medium lacking tryptophan and leucine (SD −Trp, −Leu; selection for the presence of both plasmids) or SC medium lacking tryptophan, leucine, and histidine but including 40 mM 3-aminotriazole (3-AT; Sigma) (selection for the presence of both plasmids and for interaction between RPMS1 and CBF1). Plates were incubated at 30°C for 5 days. Colonies from the −Trp, −Leu plates were further tested for β-galactosidase activity with a filter lift assay (2).
Yeast two-hybrid screen.
A73 was introduced into the pLEXA plasmid (Clontech) and was used as bait to screen a HeLa yeast library (Clontech) cloned into the pB42AD vector. Briefly, the bait and library plasmids were cotransfected into EGY48[p8op-lacZ] yeast cells and plated onto SD −Ura, −Trp, −His plates. All resulting colonies were collected and replated at high density on selection plates (SD −Ura, −His, −Trp plates additionally lacking Leu and containing 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside [X-Gal]). Positive colonies, as determined by growth and development of blue color, were collected. DNA was extracted from the yeast colonies, and activation domain library inserts were isolated by PCR amplification and sequenced.
To confirm interaction, library and bait plasmids were isolated, purified, and cotransfected back into yeast. The yeast cells were streaked on either SD −His, −Trp, −Ura plates to confirm the presence of both partner plasmids or on selection plates (SD −His, −Trp, −Ura, −Leu plus X-Gal) to test for interaction.
CAT assays.
HeLa cells (5 × 105) were transfected by calcium phosphate precipitation with plasmid pPDL83A (1 μg) containing four copies of the 100-bp EBNA-2-responsive region of the EBV C promoter (Cp) ligated upstream of the E1b promoter of E1b-CAT (pGH262). Activation of Cp was induced by cotransfection of either 1 μg of pPDL151 (26), encoding EBNA-2 under the control of the simian virus 40 (SV40) promoter/enhancer in vector pSG5 (Stratagene), or 1 μg of pJH197 (identical to mNotchIC-E2TANLS [18]), encoding NotchIC under the control of the SV40 promoter/enhancer in plasmid pSG5. The effect of RPMS1 was assessed by cotransfection of 5 μg of RPMS1 plasmid, RPMS1(34-103), RPMS1(SR), or pcDNA3 (vector without insert control), each under the control of the cytomegalovirus (CMV) immediate-early promoter (pcDNA3 vector; Invitrogen). In RPMS1(SR), the WW amino acids have been altered to SR by site-directed mutagenesis. Two days after transfection, CAT activity was determined (18); acetylated chloramphenicol levels were quantitated by fluorimetry. All experiments were carried out three times, and the results are shown as means of three experiments, normalized to the values for EBNA-2 or NotchIC activation.
Luciferase assays.
The reporter plasmid pJH26A (6 μg), containing eight CBF1 binding sites cloned upstream of a luciferase gene, was transfected by calcium phosphate precipitation into HeLa cells together with 1 μg of pPDL151 (EBNA2) or 1 μg of pJH197 (NotchIC) and 5 μg of either RPMS1, RPMS1(34-103), RPMS1(SR), or pcDNA3 (vector control). Expression of luciferase was determined in cell extracts made 2 days after transfection, using a luciferase assay kit (Promega) and a Jade luminometer (Labtech). Assays were equalized for protein content by using a Bio-Rad protein assay kit, and results are shown as means of duplicate readings from three experiments, normalized to the values for EBNA-2 or NotchIC activation.
Coprecipitation of A73 and RACK1.
GST-A73, GST-RPMS1, or GST alone cloned in pcDNA3 vectors was transfected into 293 cells (106) by using Superfect (Qiagen). Thirty minutes prior to harvest, cells were treated either with 100 ng of phorbol myristate acetate (PMA) in dimethyl sulfoxide or with dimethyl sulfoxide alone. Cells were harvested, resuspended in 100 mM NaCl–25 mM Tris-Cl (pH 7.6)–2 mM MgCl2–0.5% NP-40–protease inhibitors, incubated at 4°C for 60 min, and then centrifuged at 13,000 × g for 15 min at 4°C. A portion was retained as a total extract control, and the remainder was incubated with glutathione-agarose beads (Sigma) for 3 h at 4°C. The beads were washed five times with PBS containing 0.25% NP-40; following the final wash, the beads were resuspended in SDS-gel sample buffer and boiled for 2 min. The eluted proteins were separated by SDS-PAGE on a 12.5% gel and transferred to PVDF membranes. The membranes were probed with anti-RACK1 or anti-GST monoclonal antibodies as described above.
Nucleotide sequence accession numbers.
The sequences of the A73 and RPMS1 cDNAs have been deposited in the EMBL data library (accession no. AJ251096 and AJ251097, respectively).
RESULTS
Analysis of CST family RNAs.
We previously proposed a promoter and first exon for the CST transcripts (38). This was confirmed by RPA (36, 38). cDNA structures were assembled by linking overlapping partial cDNAs or PCR fragments. However, no cDNAs representing full-length mRNA were isolated directly by cloning from cDNA libraries. The very complicated nature of the splicing of the CST RNAs, uncertainty about whether the transcription start site at about 150641 represents the only promoter for these RNAs, and the possibility of RT-PCR methods giving prominence to minor transcripts selected by the choice of primers made it important to establish a complete structure for a CST RNA. By screening a cDNA library from C15 with probes in the exon I and exon VII regions (Fig. 1), we isolated four clones hybridizing to both probes. Two of these were completely characterized by nucleotide sequencing (structures RPMS1 and RPMS1A [Fig. 1]). The 5′ ends of these clones were at 150640 and 150690 on the B95-8 EBV sequence. The other two clones were characterized by restriction digestion and partial sequencing and appeared to be independent clones of the RPMS1 cDNA; the most frequent structure detected using these probes on the C15 library was thus the RPMS1 type (Fig. 1). Since the library had been amplified, this might not be a true reflection of the relative abundance of this splicing pattern, but further RPA analysis (see below) suggests that this type of RNA is a significant component of the CST family in C15 cells. Further screening of the library using probes in the exon V and VII regions (Fig. 1) gave clones which either were consistent with the RPMS1 structure or were of the A73 type. The A73 structure has been isolated by several groups using cDNA from NPC biopsies (6), lymphoid tumors (44), or C15 (36), but the clone that we have found is the longest reported so far. It is probably still a partial clone; its 5′ end was at position 156007 in the B95-8 EBV sequence. All of the cDNAs had a poly(A) addition following base 160989 of the B95-8 EBV sequence and included a poly(A) tract. There were no sequences changes from B95-8 in the A73 clone and only three nucleotides different in the 5′ part of the RPMS1 clone.
FIG. 1.
Map of the principal features of the EBV genome in the CST region and structures of the cDNAs discussed. The standard ORF map (9) is shown beneath a scale in kilobases, the segment deleted in B95-8 being numbered separately (30). RNA structures are shown as horizontal arrowed lines, and the three cDNA structures described in this paper are labeled A73, RPMS1A, and RPMS1. The CST exons are marked, and their coordinates are shown below. Numbers refer to the end nucleotides present in the spliced RNA at exon boundaries and relate to the B95-8 EBV sequence (1), except for exons IB, II, III, IIIA, IIIB, and IV, which are from the Raji EBV sequence (30). The protein sequences of RPMS1 and A73 are shown below in the one-letter amino acid code.
Previously published structures for CST RNAs have exon I linked directly to exon II. Two further novel exons (IA and IB) are also shown in Fig. 1. These were detected by PCR analysis of the cDNA library using primers in exon 3 and the lambda flanking sequence. The structures contained exons I, IA, II, and III or I, IB, II, and III. The quantitative significance of these exons in the total RNA population is unknown, but they further underline the complexity of splicing possible in the CST RNAs. The section of the map in Fig. 1 between 149000 and 152000 has been drawn using the B95-8 EBV sequence, but exons I and IA are also conserved in the sequence of Raji EBV in that region (data not shown).
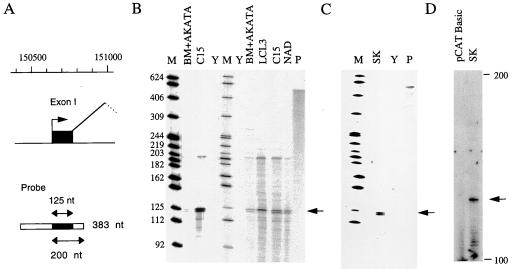
Previous cDNA analysis suggested multiple start sites for exon I in the range 150641 to 150648; the 5′ end of the RPMS1 cDNA was at 150640. Although there is considerable evidence to support exon I as the major start of CST RNAs, 5′ RACE (rapid amplification of 5′ cDNA ends) analysis indicated the existence of RNAs initiated at least 120 nucleotides (nt) upstream of this point (37). To further validate the proposed 5′ end of the CST RNAs, exon I was assayed by RPA using RNA from the C15 NPC xenograft and various cell lines. The genomic probe overlaps exon I at both ends (Fig. 2A); thus, when combined with the cDNA sequencing (Fig. 1) and primer extension data shown below, protection of the approximately 125-nt exon I fragment indicates initiation at about 150645 and splicing at position 150769. As expected, the 125-nt band characteristic of exon I was readily detected in C15 RNA (Fig. 2B). Expression of this exon was also found at a lower level in the LCLs NAD, BM+Akata, and LCL3. NAD contains the EBV from C15, BM+Akata contains Akata EBV, and LCL3 contains B95-8 EBV. Some of the RPAs also showed a longer band at about 200 nt; this was too small to correspond to RNA initiated far upstream of the mapped promoter and spliced at the exon I boundary but matched the size predicted for correctly initiated RNA that had not spliced at the 3′ end of exon I. There was thus no evidence in these assays for significant levels of RNA transcribed in the 200-bp region upstream of exon I.
FIG. 2.
Exon I RPAs and primer extension. (A) Illustration of the probe used for RPA of exon I. The probe contains 383 nt of EBV sequence (from 150844 to a BglII site at 150462), of which exon I protects 125 nt (marked by arrows in B and C). RNA initiated at the start of exon I but not spliced at the 3′ end of exon I would give a protected fragment of 200 nt. (B) RPA of RNA from cell lines BM+AKATA, LCL3, and NAD and RNA from the C15 NPC xenograft, using the exon I probe. Size markers (M; lengths shown in nucleotides) are an end-labeled MspI digest of pBR322 DNA. Yeast tRNA (Y) was used as a negative control in the RPA, and the undigested probe (1% of the amount used in the RPA) is shown in track P. (C) RNA from HeLa cells transfected with plasmid SK analyzed by RPA using the exon I probe as in panel B. Other symbols are as in panel B. (D) Primer extension analysis of RNA form HeLa cells transfected with SK plasmid or pCAT Basic negative control. The primer hybridized to exon I and was extended with reverse transcriptase to give a product about 122 nt long (arrow), consistent with a 5′ end at about 150640.
To confirm the promoter activity at this location, a plasmid (SK) was constructed that contained B95-8 EBV sequences 150199 to 151283 cloned upstream of a CAT reporter gene with the SV40 poly(A) addition signal at its 3′ end to permit the formation of stable RNA. When this was transfected into the epithelial cell line HeLa, exon I RNA was readily detected by RPA (Fig. 2C). Primer extension analysis of RNA from a further transfection of the same plasmid confirmed the initiation site as about 150645. These transfection experiments show that EBV sequences 150199 to 151283 are sufficient to allow substantial promoter activity, initiating at a position similar to that for RNA from EBV-infected cells. A more detailed analysis of sequences involved in transcription from this promoter will be published elsewhere.
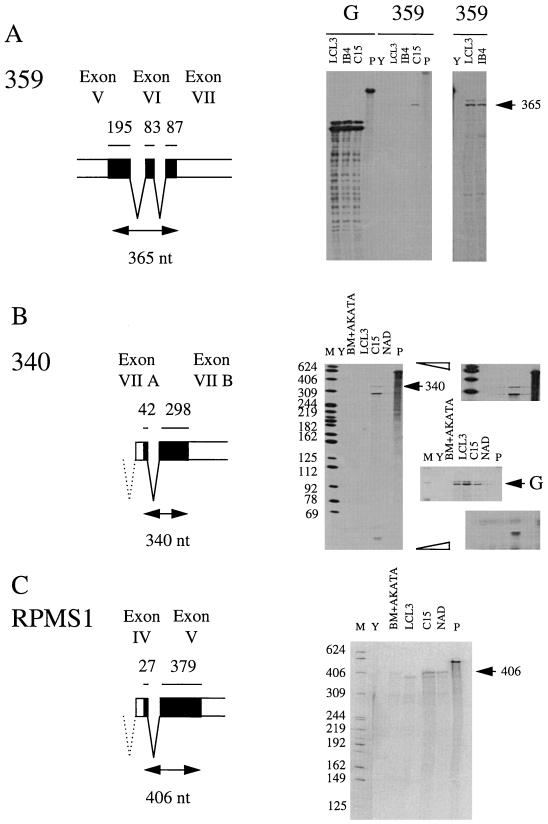
To further characterize the major CST RNAs and determine whether the splice patterns shown in Fig. 1 comprise a significant portion of the CST RNA, RPAs were performed with probes derived from the cDNAs spanning some of the splice junctions (Fig. 3). With these probes, full-length protection of the EBV content of the probe corresponds to splicing as in the probe; smaller protected fragments arise from differently spliced or unspliced RNA. The level of contaminating EBV DNA in the RNA preparations was so low as to be insignificant in these assays (for example, there was no full-length protection of the exon I probe in Fig. 2B). Probe 359 was tested on C15 and the B95-8 LCLs LCL3 and IB4 (Fig. 3A). In IB4, EBV DNA is present at only a few copies per cell and is integrated. In each case, the major band was at a size consistent with full-length protection of the EBV content of the probe, the signal being stronger in C15 than in the LCLs. In C15 some smaller, lower-level protected fragments were also detected. The sizes of these were consistent with individual CST exons, but it was clear that most of the RNA containing these exons was spliced in the A73 pattern in this region. Probe 340 (Fig. 3B) showed both full-length protection of the EBV content of the probe (i.e., spliced like the probe) and 298- and 41-nt fragments corresponding to discontinuity with the probe at the splice junction (for example, like the RPMS1 cDNA). This probe would conveniently also reveal use of the RK-BARF0 splice junction (10, 37), but there was no noticeable signal at the 235-nt position this would produce, suggesting it is only a minor component of the RNA in the cells tested. In C15 about 11% of the RNA had the A73-type splice in the 340 probe region (Fig. 3); in the LCLs, the proportion of A73 type splicing was higher, although the total amount of CST RNA was lower. The RPMS1 probe (Fig. 3C) showed mainly full-length protection of the EBV content of the probe (i.e., most of the RNA hybridizing to the probe contained the exon IV to exon V splice), but there was some signal probably corresponding to exon V alone. Exon IV is deleted in the B95-8 strain, and so LCL3 gave only the exon V band. There was no noticeable signal at the position that would be predicted for the RPMS1A splice, suggesting that only a minor fraction of the RNA has that structure.
FIG. 3.
RPA of splice junctions in CST RNAs. Markers (M), yeast negative control (Y), and probes (P) were loaded as in Fig. 2. (A) RPA of RNA from LCL3 and IB4 cell lines and C15 xenograft, using probe 359 spanning splices between exons V, VI, and VII. The 359 major protected fragment from the spliced RNA is indicated (arrow). The right-hand panel contains a longer exposure of tracks Y, LCL3, and IB4. (B) RPA as in panel A, using probe 340 spanning the exon VIIA/VIIB splice containing the A73 ORF. The spliced RNA gives a 340-nt protected fragment; individual exon protection gives the 298- and 42-nt fragments. Upper and lower right-hand panels are longer exposures. The middle right-hand panel is an RPA for GAPDH cell RNA to compare loadings; NAD is weaker than the other RNA samples (arrowed G indicates GAPDH-protected fragment). (C) RPA as in panel A, using probe RPMS1 covering the exon IV/V splice. Spliced RNA gives a 406-nt protected fragment (arrow), whereas individual exon protection gives fragments of 379 and 27 nt (not shown).
These results indicate that the splices shown in the RPMS1 and A73 cDNA structures covering parts of exons V, VI, and VII represent a substantial proportion of the CST RNA in the cell lines tested and will be relevant to interpretation of the potential coding content of the CST RNAs. Comparison of the RPA signals from the A73 (340) probe with the GAPDH signal suggests that this CST RNA is about 750-fold less abundant than GAPDH in C15, putting this RNA in the lower abundance class of RNAs. In LCL3, the A73 (340) signal was about 10-fold weaker than in C15.
Possible coding content of CST family RNAs.
The original interpretation of the EBV genome coding content (1) proposed a reading frame called BALF3, but no leftward RNA was detected that might encode this reading frame in B95-8 cells. The subsequent discovery of the CST RNAs (17) led to the possibility that the overlapping ORF on the opposite strand (BARF0) is the important gene in this region; attention has subsequently been focused on BARF0. However, it is now clear that BALF3 equivalents are present in all human herpesviruses that have been sequenced. The degree of protein sequence homology is substantial (Fig. 4A), particularly among the gammaherpesvirus group, and in herpes simplex virus (HSV) the equivalent gene has been shown to have an essential role in localization of capsids to intranuclear sites where DNA is cleaved and packaged (23, 39).
FIG. 4.
Genomic evaluation of BALF3 and BARF0. (A) Protein sequence comparison of two parts of the BALF3 equivalents of HHV8, herpesvirus saimiri, human CMV (HCMV), and HSV with the EBV BALF3 protein (amino acid numbers in the protein sequences shown at the ends of the lines). Identities in all five proteins (∗), identities in the three gammaherpesvirus (HHV8, HVS, and EBV) proteins (!), strong similarities in all five proteins (:), and identity of two of the three gammaherpesvirus sequences (.) are indicated. (B) Comparison of the EBV and HHV8 sequences at the position of the CST poly(A) addition signal (AATAAA underlined) and the poly(A) addition signal for the BALF2 RNA on the opposite strand (TTTATT underlined). The first A of the CST AATAAA is at position 160964 in the B95-8 EBV sequence and 7057 in the equivalent HHV8 sequence (opposite strand). The point at which poly(A) is added in the sequenced clones is shown by the left end of the AAAAAA motif aligned with the EBV sequence.
Another point against BARF0 expression from most of the CST RNAs is that the BARF0 ORF does not end with a termination codon in the cDNA clones that we have analyzed. Although the genomic sequence of EBV contains a termination codon at the end of the BARF0 ORF, in all of the sequenced cDNA clones, either from C15 or oral hairy leukoplakia biopsy cDNA, the point of poly(A) addition is just before the termination codon (Fig. 4B) so that BARF0, if translated, would apparently extend into a long run of AAA (lysine) codons [the poly(A) tail] without a termination codon. This has been discussed previously (36), and it seems unlikely that such BARF0 ORFs would be translated. RNA which terminates at a later position has been detected using 3′ RACE. It was concluded that up to 25% of CST RNA underwent poly(A) addition slightly further downstream, allowing proper termination (36). None of the C15 clones reported above or a further five partial clones analyzed by nucleotide sequencing (data not shown) or the five oral hairy leukoplakia cDNAs (24) contained the termination codon for BARF0. Our data thus suggest that the RNA containing a conventionally terminated BARF0 ORF can be only a minor fraction of the CST RNA in the cells we have studied.
Inspection of the EBV map shows that many of the genes of EBV are organized as 3′-coterminal groups of transcripts, each RNA usually encoding its most 5′ functional ORF. This would imply that ORFs upstream of BARF0 might be significant products of the CST RNAs. We have focused on two ORFs referred to here as RPMS1 (38) and A73. Systematic searching for ORFs in the sequences of the RPMS1 and A73 spliced RNAs (Fig. 1) identified these as the most likely to be significant, selecting on the basis of length (over 100 amino acids) and presence of an ATG initiator codon. Neither of these products has previously been identified at the protein level in EBV-infected cells. The limited quantities of antibody we have obtained so far against RPMS1 are of low titer and specificity and have not been suitable for screening cell lines and tissues for RPMS1 protein. However, we report below considerable evidence for functions of these proteins.
RPMS1.
Since full-length cDNAs that include the RPMS1 ORF had been isolated and our data show that RNA containing these splices is a significant proportion of the CST RNA in EBV-infected cells, we tested whether the RPMS1 protein could be expressed from this type of spliced cDNA. A Flag epitope tag was cloned in frame into the RPMS1 ORF, and a plasmid containing the tagged RPMS1 cDNA under the control of the CMV immediate-early promoter was transfected into cells. Western blotting of total cell lysates showed that RPMS1 was expressed from the tagged cDNA; the same-sized protein was produced when the RPMS1 ORF alone was cloned downstream of the promoter (Fig. 5A). Similar results were obtained using in vitro transcription and translation of analogous constructions without the Flag tag and Western blotting with an antibody to RPMS1 (Fig. 5B). These results showed that the presence of the leader sequences upstream of the RPMS1 ORF in the RPMS1 cDNA is compatible with RPMS1 expression. When a similar Flag tag was placed in the BARF0 ORF within the same cDNA structure, no BARF0-Flag expression was detected in transfected cells (data not shown).
FIG. 5.
Expression of RPMS1. (A) Cells were transfected with expression plasmids containing the Flag-tagged RPMS1 ORF, (lane 1), RPMS1 cDNA with no Flag tag (lane 2), or the RPMS1 cDNA with Flag-tagged RPMS1 (lane 3). Cell extracts were analyzed by Western blotting using an anti-Flag antibody. The position of the tagged RPMS1 protein is indicated (arrow). (B) In vitro translation of RPMS1. Proteins were derived from transcription and translation of RPMS1 cDNA (lane 1), 5′ deletion of RPMS1 cDNA commencing 11 bp prior to the start of the RPMS1 ORF (lane 2), RPMS1 ORF (lane 3), or negative control RPMS1A cDNA (lane 4). Proteins were analyzed by Western blotting using an antibody to RPMS1. The RPMS1 band is indicated (arrow). (C) The Flag-tagged RPMS1 expression construction was transfected into HeLa cells and tested for immunofluorescence using an anti-Flag antibody, demonstrating the nuclear location of the tagged RPMS1. Views i and ii are two fields of tagged RPMS1, view iii is a negative control omitting the Flag antibody, and view iv is the phase contrast image of the cells shown in view ii.
In the RPMS1A clone (Fig. 1), the RPMS1 ORF is modified by the splicing so that it is shorter and has a different C terminus. It is intriguing that the RPMS1A C terminus includes a consensus protein signal for isoprenylation, but we have no evidence at present for expression or function of RPMS1A, and only a very small proportion of the RNA is spliced in the RPMS1A pattern in the RNAs tested in Fig. 3C.
Inspection of the amino acid sequence of the RPMS1 ORF showed the presence of the amino acid sequence WWP (38), which is very rare in protein sequences but is also found in the EBV EBNA-2 sequence. The WW motif in EBNA-2 is essential for the binding of EBNA-2 to the cell CBF1 (RBP-Jk) protein and the regulation of transcription by EBNA-2 through CBF1 (16). The possibility that RPMS1 also binds to CBF1 was therefore considered. For binding to occur in cells, RPMS1 would have to be a nuclear protein. We therefore transfected the plasmid that expresses the Flag-tagged RPMS1 ORF (Fig. 5A) into 293 cells and assayed Flag-tagged RPMS1 expression by immunofluorescence. Clear nuclear expression of the protein was observed (Fig. 5C).
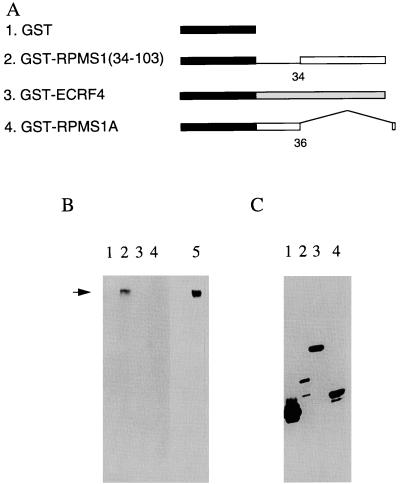
The RPMS1 protein was tested for its ability to bind to CBF1. Radiolabeled CBF1 was prepared by in vitro transcription and translation. The product was mixed with various GST fusion proteins (Fig. 6A) containing part of RPMS1 or control proteins. Complexes were isolated on glutathione-agarose beads, washed free of unbound CBF1, and analyzed by SDS-PAGE and fluorography (Fig. 6B). The input levels of the various GST fusion proteins into the assay were tested by Western blotting with an antibody to GST (Fig. 6C). GST-RPMS1 fusion protein was found to be very susceptible to degradation, even though protease inhibitors were present during the purification. Although binding to this protein could readily be observed (data not shown), it is not illustrated in Fig. 6 because most of the fusion protein containing full-length RPMS1 was degraded. The fusion protein containing amino acids 34 to 103 of RPMS1 was much more stable (Fig. 6C, track 2) and was found to bind to CBF1 (Fig. 6B, track 2). No binding was detected to GST alone, an unrelated GST fusion protein (ECRF4), or the RPMS1A protein.
FIG. 6.
Interaction of RPMS1 and CBF1. (A) Structures of GST fusion proteins. The GST portion is filled, and the amino acid numbers of RPMS1 are noted. The ECRF4 fusion protein (line 3) was a negative control, and the RPMS1A fusion (line 4) has the same sequence as RPMS1 up to amino acid 36 but then has a different C terminus. (B) Fluorograph of gel electrophoresis of radiolabeled CBF1 protein bound to the GST fusion proteins illustrated in panel A. Track 5 contains the input CBF1 protein (10% of the amount used in the assay), and the other tracks correspond to the construction numbers in panel A. The CBF1 band is marked by an arrow. (C) Western blot of input GST fusion proteins used in pull-down assays, probed with the anti-GST antibody. Tracks correspond to the construction numbers in panel A.
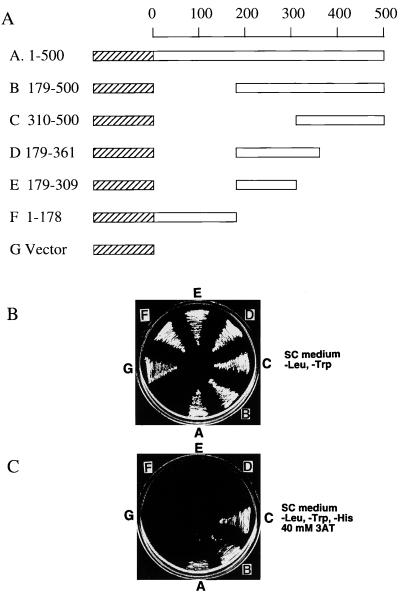
Because of the relatively high concentrations of target proteins that are used in the pull-down assay, we also tested whether the interaction between CBF1 and RPMS1 was sufficiently strong to be detected in a two-hybrid association experiment in yeast. Wild-type CBF1 and various deletion mutants (Fig. 7A) were constructed as fusion proteins with the activation domain of Gal4 in a vector suitable for expression in yeast. This vector confers growth on yeast plates lacking leucine. The plasmids were introduced into yeast in the presence of a plasmid expressing RPMS1 fused to the DNA binding domain of Gal4. This plasmid also encoded a gene allowing yeast cells to grow on medium lacking tryptophan. As expected, yeast containing both plasmid types grew on plates lacking Leu and Trp (Fig. 7B), but only constructions containing amino acids 310 to 500 of CBF1 were able to grow on plates also selecting for histidine auxotrophy (Fig. 7C). Growth on His-deficient plates (containing 3-AT to enhance the His selection) requires interaction between the Gal4 hybrid proteins to induce expression of the gene allowing synthesis of His, indicating that amino acids 310 to 500 of CBF1 are sufficient to interact with RPMS1. Since CBF1 amino acids 179 to 361 were not able to interact, amino acids 362 to 500 are likely to contribute to the sequence that interacts with RPMS1.
FIG. 7.
Two-hybrid interaction of CBF1 and RPMS1. (A) Structures of fusion proteins encoded by the plasmids used in a yeast two-hybrid assay. The portions of CBF1 protein (open boxes) fused to the Gal4 activation domain (hatched boxes) are shown below a scale of amino acid number of CBF1. (B) Presence of both activation domain and DNA binding domain plasmids in the yeast strains, shown by growth on −Leu, −Trp medium (letters refer to CBF1 fusion proteins in panel A). (C) Two-hybrid test of association on −Leu, −Trp, −His plates containing 40 mM 3-AT; strains are as in panel B.
Comparison of the RPMS1 protein sequence with sequences of other transcription factors did not suggest the presence of a transcription activation domain in RPMS1, but the binding data (Fig. 6 and 7) implied that RPMS1 might be able to compete with the binding of EBNA-2 or NotchIC to CBF1, preventing their transcription activation through CBF1. This was tested by cotransfection of an expression vector for RPMS1 in reporter assays on promoters induced by EBNA-2 or NotchIC (Fig. 8). The promoters used were either an artificial promoter composed of eight concatemerized CBF1 binding sites upstream of the HSV thymidine kinase promoter with a luciferase reporter or part of the EBV Cp promoter concatemerized with a CAT reporter. As expected, EBNA-2 and NotchIC both induced reporter gene expression. In both cases, this induction was prevented by RPMS1, but mutated RPMS1, in which the WW was changed to SR, was much less effective. A truncated RPMS1 (containing amino acids 34 to 103, just retaining the WW) still inhibited Notch1 transactivation (Fig. 8B) but was less effective at inhibiting EBNA-2 transactivation (Fig. 8A). The mutations in the RPMS1 protein did not prevent the localization of the protein to the nucleus, nor did RPMS1 reduce the expression of EBNA-2 in cotransfection assays (data not shown).
FIG. 8.
RPMS1 inhibition of transcription induced by EBNA-2 or NotchIC on reporter plasmids pJH26A (8 × CBF1-Luciferase) or pPDL83A (4 × CpE2RE-CAT). All transfections received 1 μg of reporter plasmid and 1 μg of either EBNA-2 or Notch expression vector, as indicated. Additionally, 5 μg of plasmids expressing RPMS1, RPMS1(34-103), or RPMS1(SR) was cotransfected as shown. Results are given as a mean of three experiments, normalized on the values for EBNA-2 or NotchIC activation.
These reporter assay results show that RPMS1 can act as an antagonist of EBNA-2 or Notch1 transcription activation. On the Cp promoter not all of the transactivation by EBNA-2 is mediated through the CBF1 interaction (8), perhaps accounting for the difference between Notch1 and EBNA-2 sensitivity to the RPMS1(34-103) mutant.
A73.
The second CST ORF we have studied is A73. Although our longest A73 clone extends farther in the 5′ direction than those reported previously (6, 36, 43), we suspect that even this is not a full-length cDNA for A73. The isolated A73 ORF was cloned into an expression vector with a Flag epitope tag at the C terminus of the protein. When this plasmid was transfected into cells, the tagged A73 protein was readily detected and was found to be cytoplasmic (Fig. 9A). The function of A73 being unknown, it was tested in a yeast two-hybrid screen using a library from HeLa cell RNA to search for potential binding partners. Eight strongly positive binding clones were isolated in this screen. When sequenced, five of them were found to be derived from human RACK1 (receptor for activated C kinase) (33), a 36-kDa WD repeat protein (29). Sequencing showed that at least four of the clones were independent isolates of RACK1 from the library. The structures of the RACK1 cDNAs are shown in Fig. 9B. The variant form of RACK1 that would be encoded by the clone starting at 152 has not been described previously. The two-hybrid interaction of A73 with RACK1 is illustrated in Fig. 9C; the left plate was under selection to show the presence of both plasmids in the yeast cells, and the right plate was under further selection so that only yeast cells in which the two-hybrid interaction was successful would grow. Sector 1 (A73/RACK1) and the positive control (sector 3) grew under these conditions, but the two negative controls did not. All of the RACK-1 cDNAs isolated by the two-hybrid procedure were partial, and the structures show that only amino acids 170 to 317 of RACK1 were required for the association with A73 in yeast.
FIG. 9.
A73 is a cytoplasmic protein and interacts with RACK1. (A) Detection of Flag-tagged A73 protein in transfected cells by immunofluorescence using an anti-Flag antibody. Upper left, anti-RACK1; upper right, anti-Flag; lower left, no antibody control; lower right, merge of upper panels. Scale bar is 20 μm. (B) Diagram of RACK1 clones isolated in a two-hybrid screen. The full-length human RACK1 cDNA (33) is 1,093 nt long, and the RACK1 ORF (filled box) is from positions 96 to 1046. All clones isolated were partial cDNAs starting at 302, 530, 497 (two clones isolated), or 152 and terminating at 1093, as indicated. The clone whose 5′ end was at 152 lacked 201 to 440 from the standard sequence and may represent an alternatively spliced form of RACK1 RNA. (C) Interaction with RACK1 in two-hybrid assay. Sectors: 1, A73 and RACK1; 2, pLexA vector and pB42AD (negative control); 3, pLexA-53 and pB42AD-T (positive control, p53 and SV40 large T antigen interaction); 4, pLexA and RACK1 (negative control). The left plate demonstrates the presence of both plasmids in each yeast strain allowing growth on the selective medium; the right plate is also under His selection for two-hybrid interactions. (D) Coprecipitation of A73 with RACK1. 293 cells containing endogenous RACK1 were transfected with expression constructions for GST-RPMS1 (track 1), GST-RPMS1(34-103) (track 2), GST-A73 (track 3), and A73 (track 4). The transfected cells (tracks 1 to 3) were treated with tetradecanoyl phorbol acetate for 30 min prior to protein extraction. Extracts of the cells were Western blotted for RACK1 (left) or used in a pull-down assay with glutathione-agarose beads, isolating proteins that bound to the beads. The bound proteins were then assayed (right) by Western blotting for RACK1. (E) The cell extracts in the left part of panel D were assayed for expression of the GST fusion proteins by Western blotting using an anti-GST antibody. Cell extracts: tracks 1 and 2, GST-A73; tracks 3 and 5, GST-RPMS1(34-103); tracks 4 and 6, GST-RPMS1. Tracks 1, 5, and 6 were from cells treated with PMA; in tracks 2 to 4, the cells were not treated with PMA.
Like A73, RACK1 is a cytoplasmic protein (33); in the two-hybrid experiment it was transferred to the yeast nucleus by a nuclear localization signal within the LexA fusion partner. To further validate the interaction between A73 and RACK1, plasmids expressing a GST-A73 fusion or two control proteins, GST-RPMS1, and GST-RPMS1(34-103), were transfected into 293 cells, which contain endogenous RACK1. The cells were treated with PMA for 30 min, and extracts were applied to glutathione-agarose beads; bound proteins were isolated and tested for the presence of RACK1 by Western blotting. All of the cell extracts contained RACK1 (Fig. 9D, left) but only the GST-A73 pulled down RACK1 protein (Fig. 9D). Control transfection of an A73 expression construction lacking GST did not bind RACK1 to the glutathione beads, supporting the specificity of the interaction. The lack of interaction between RACK1 and the control GST-RPMS1 and GST RPMS-1(34-103) proteins could not be attributed to a lack of expression since by Western blotting these control proteins were somewhat more abundant than the GST A73 fusion protein (Fig. 9E). The PMA treatment, which is frequently used to activate RACK1, did not affect the expression levels of the GST fusion proteins.
Two protein bands were detected on SDS gels using the RACK1 antibody. In this context it is interesting that one of the RACK1 clones was spliced differently from the published cDNA and might represent an alternative form of RACK1, but this has not been investigated.
The three remaining cDNAs isolated in the A73 two-hybrid screen encoded β5-integrin, protein KIAA0547 (28 (EMBL accession no. AB011119; of unknown function), and a further protein which was not detected in the EMBL, GenBank, or EST databases at the time of submission. The β5-integrin interaction with A73 is most probably direct, but it could also be an indirect interaction through RACK1. These three interactions have not yet been investigated.
DISCUSSION
Although they are not essential for immortalization of B cells in culture (20, 32), there is considerable evidence for expression of the CST RNAs in several types of human cancer (3, 6, 11, 15, 17, 19, 31, 43). Viral gene expression in tumor cells is usually very restricted, presumably to allow the tumor to avoid immune surveillance, perhaps suggesting a functional role for a product of the CST RNAs (or the RNAs themselves) in the tumor. By RNase protection of several exons, we have additionally confirmed that CST RNAs are also expressed at a low level in LCLs.
Many different spliced CST structures have been reported, but there has been little attention directed to the relative levels of expression of these different forms, and in some cases structures have been inferred by combining various overlapping partial cDNAs or PCR products, with no direct evidence for the combined structures. The emphasis in this paper has been on the most abundant CST RNA structures in the hope that this will direct attention to significant gene products. This allows an initial simplification of the complex pattern of transcripts. The only transcription start that we identified is at the previously mapped exon I, beginning between 150640 and 150645. Further mapping of exon I by RNase protection, primer extension, and sequencing of apparently full-length RPMS1 cDNAs indicates that this is a major initiation site for CST RNAs. We did not find evidence for significant transcription from the few hundred nucleotides upstream of exon I, although another study has indicated that there may be a low level of RNA initiated upstream of 150640 to 150645 (36). Our strategy for isolating full-length cDNAs (a library screen requiring hybridization with probes for exon I and exon VII) biased the selection of long clones to those starting at exon I. Nevertheless, further screening with probes in exons V and VII revealed only RNAs that were consistent with the RPMS1 structure or the A73 type. It is at present unclear whether the A73 RNA initiates at exon I or another promoter. In contrast to the dense packing of genes in the rest of the EBV genome, there are several kilobases of DNA containing no mapped features within the region covered by the CST transcription unit; thus, it is quite possible that other promoters, splicing patterns, ORFs, or unknown genetic elements remain to be discovered in this region of the genome.
RPAs using probes containing the spliced RNA sequence across CST RNA splice sites allowed measurement of their abundance as a proportion of the total RNA hybridizing to the probe. These assays confirmed that the RNAs covering the RPMS1 and A73 ORFs are significant proportions of the CST family of transcripts. RPMS1 and A73 are the longest ORFs with initiator ATG codons in the RNAs apart from BARF0, suggesting that they may be expressed as proteins. We propose that the BARF0 ORF, which has attracted the most attention, is not expressed from most of the CST RNA. It might be translated from a minor fraction of the RNA, perhaps as an RK-BARF0 form (10, 21). RNA that might express BARF0 with a functional termination codon was not detected in our screens.
BARF0 overlaps the BALF3 ORF on the opposite strand of the EBV genome. Although RNA encoding BALF3 has not yet been detected, the conservation of BALF3 equivalents in other herpesviruses (Fig. 4A) and the essential role of UL28 in HSV argue that BALF3 is likely to be a functional ORF. In human herpesvirus 8 (HHV8) and herpesvirus saimiri, there is no ORF equivalent to BARF0 on the strand opposite their BALF3 equivalents (orf7 and KRF2). Although the AATAAA sequence that acts as part of the polyadenylation signal for the CST RNAs is conserved in HHV8, in exactly the same position relative to the poly(A) site on the opposite strand for BALF2 (Fig. 4B), RNAs equivalent to the CST RNAs have not yet been found in HHV8-infected cells, and it might be that the presence of the CST AATAAA site in HHV8 is a coincidence since the protein sequence of BALF3 is conserved at that position between EBV and HHV8 and the amino acids at that position (FYC) have few possible codons in the genetic code.
Also, codon usage analysis supports the idea that BALF3 is an expressed gene in some circumstances. EBV has a relatively GC-rich DNA sequence (66.4% GC) in the BALF3 region, and the constraints imposed by the required amino acid sequence of a protein result in codon selection which concentrates the excess GC in the third position of the codons, where it has relatively little effect on the protein sequence. The presence of a systematic third-position enhanced GC constitutes strong evidence for the validity of the ORF and is particularly clear in BALF3, which has 67.0, 50.0, and 82.2% GC in the first, second, and third codon positions, respectively, a relative bias that is maintained throughout the ORF. Corresponding reciprocal analysis of BARF0 in the region overlapping BALF3 puts the highest GC (82%) in the first position of the codons, strongly skewing its potential amino acid content.
We have assembled a considerable amount of data suggesting that RPMS1 and A73 may be functionally significant and implying that it will be worthwhile to search further for these proteins. We showed that it is possible to express RPMS1 from the full-length cDNA both by in vitro translation and by transfection into cells. The RPMS1 protein was found to bind to CBF1, a mediator of Notch signaling and a target for the EBNA-2 and EBNA-3 proteins of EBV. Binding was demonstrated in vitro using a pull-down assay and in a yeast two-hybrid system. Epitope-tagged RPMS1 was shown to be located in the nucleus of transfected cells, like CBF1. RPMS1 antagonized Notch- or EBNA-2-mediated activation of transcription at promoters with CBF1 binding sites, suggesting a role for RPMS1 in virus-infected cells. The most obvious potential role for RPMS1 would thus be as a negative regulator of Notch pathway signal transduction, as has been proposed for EBNA-3 family proteins and suggested by the transcription assay we used. It should be noted that RPMS1 may be overexpressed relative to EBNA-2 and NotchIC in the experiment shown in Fig. 8 (the CMV promoter used for RPMS1 expression is about fivefold stronger in human cells than the SV40 promoter assayed on EBNA-2 expression), and so the significance of the effect of RPMS1 in EBV-infected cells remains to be determined. However, it is interesting that Notch expression is very low in undifferentiated epithelial cells (42), and so there might only be a very low level of NotchIC with which RPMS1 would have to compete. It might also be that RPMS1 has another effector function related to CBF1 that we have not measured in our assays. The slightly lesser inhibitory effect of RPMS1 on EBNA-2 transactivation of Cp-CAT (Fig. 8) than on the CBF1 luciferase target might reflect different affinities of EBNA-2 and RPMS1 for CBF1 in the context of the two promoters.
The A73 ORF is of particular interest since cDNAs containing this splicing pattern have been isolated by several groups from different types of tumor cell, including NPC (6). The association of A73 with RACK1 in the two-hybrid assay and transfected cells (Fig. 9) suggests a possible role for A73 in EBV-associated cancers. The region of EBV containing the A73 ORF is not required for immortalization of human B lymphocytes (20, 32), but a contribution of EBV to cell transformation, or a role in epithelial cells, might work by a different mechanism. Bearing in mind the expression of CST RNAs in various tumors, the ability of RACK1 to bind to certain protein kinase C isoforms, serving as a scaffold or anchor protein (27, 33–35), is of considerable interest. The ability of RACK1 also to interact with the β-integrin signaling protein (26) and cyclic AMP-specific phosphodiesterase PDE4D5 (41) is also suggestive of a functional role for A73. Furthermore, RACK1 interacts with the tyrosine kinases Src and Lck, inhibiting their activity (4), and RACK1 constitutive expression was reported to reduce the growth rate of NIH 3T3 cells, perhaps through inhibition of Src family kinases (4). The interaction that we have observed suggests that the EBV A73 protein might modulate the activity of RACK1. It is thus possible that the A73 protein might modify signaling through some of these proteins and mediate a contribution of EBV to tumor cell development. In view of the involvement of protein kinase C and the need for PMA treatment to allow association of RACK1 with some of its targets (25), it is interesting to recall earlier epidemiological studies (43) which suggested a link between the incidence of NPC and the presence of phorbol esters in the environment. It has also been proposed that EBV and phorbol esters might cooperate in the transformation of primary human epithelial cells (40). Transformation of monkey epithelial cells using an EBV cosmid which contains the CST gene region was reported (14, 19). Further understanding of the potential role of A73 will thus depend on whether the A73 protein is expressed in human tumor cells and on functional analysis of A73 in cell transformation assays.
The results shown in this paper provide a fresh perspective on the CST RNAs and give the first functional insights into potential protein products of CST RNAs other than the BARF0 ORF. The data suggest that investigation of the ORFs in the RNAs upstream of BARF0 may clarify the function of this poorly understood part of the EBV genome, which represents the EBV genomic region transcribed most abundantly into mRNA in NPC.
ACKNOWLEDGMENTS
The Cancer Research Campaign supported the work of P.R.S. and D.T. in this project. The work was also supported in part by NIH grant CA42245 to S.D.H. O.D. gratefully acknowledges a Ph.D. fellowship from Fundaca Para a Ciencia e Tecnologia (PRAXIS XXI/BD/13778/97).
REFERENCES
- 1.Baer R, Bankier A T, Biggin M D, Deininger P L, Farrell P J, Gibson T J, Hatfull G, Hudson G S, Satchwell S C, Seguin C, Tuffnell P, Barrell B. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature. 1984;310:207–211. doi: 10.1038/310207a0. [DOI] [PubMed] [Google Scholar]
- 2.Breeden L, Nasmyth K. Regulation of the yeast HO gene. Cold Spring Harbor Symp Quant Biol. 1985;50:643–650. doi: 10.1101/sqb.1985.050.01.078. [DOI] [PubMed] [Google Scholar]
- 3.Brooks L A, Lear A L, Young L S, Rickinson A B. Transcripts from the Epstein-Barr virus BamHI A fragment are detectable in all three forms of virus latency. J Virol. 1993;67:3182–3190. doi: 10.1128/jvi.67.6.3182-3190.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang B Y, Conroy K B, Machleder E M, Cartwright C A. RACK1, a receptor for activated C kinase and a homolog of the beta subunit of G proteins, inhibits activity of src tyrosine kinases and growth of NIH 3T3 cells. Mol Cell Biol. 1998;18:3245–3256. doi: 10.1128/mcb.18.6.3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen H, Smith P, Ambinder R F, Hayward S D. Expression of Epstein-Barr virus BamHI-A rightward transcripts in latently infected B cells from peripheral blood. Blood. 1999;93:3026–3032. [PubMed] [Google Scholar]
- 6.Chen H L, Lung M M, Sham J S, Choy D T, Griffin B E, Ng M H. Transcription of BamHI-A region of the EBV genome in NPC tissues and B cells. Virology. 1992;191:193–201. doi: 10.1016/0042-6822(92)90181-n. [DOI] [PubMed] [Google Scholar]
- 7.Durfee T, Becherer K, Chen P L, Yeh S H, Yang Y, Kilburn A E, Lee W H, Elledge S J. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- 8.Evans T J, Farrell P J, Swaminathan S. Molecular genetic analysis of Epstein-Barr virus Cp promoter function. J Virol. 1996;70:1695–1705. doi: 10.1128/jvi.70.3.1695-1705.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farrell P J. The Epstein-Barr virus genome. In: Klein I G, editor. Advances in viral oncology. New York, N.Y: Raven Press; 1989. pp. 103–132. [Google Scholar]
- 10.Fries K L, Sculley T B, Webster-Cyriaque J, Rajadurai P, Sadler R H, Raab-Traub N. Identification of a novel protein encoded by the BamHI A region of the Epstein-Barr virus. J Virol. 1997;71:2765–2771. doi: 10.1128/jvi.71.4.2765-2771.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilligan K, Sato H, Rajadurai P, Busson P, Young L, Rickinson A, Tursz T, Raab-Traub N. Novel transcription from the Epstein-Barr virus terminal EcoRI fragment, DIJhet, in a nasopharyngeal carcinoma. J Virol. 1990;64:4948–4956. doi: 10.1128/jvi.64.10.4948-4956.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gilligan K J, Rajadurai P, Lin J C, Busson P, Abdel-Hamid M, Prasad U, Tursz T, Raab-Traub N. Expression of the Epstein-Barr virus BamHI A fragment in nasopharyngeal carcinoma: evidence for a viral protein expressed in vivo. J Virol. 1991;65:6252–6259. doi: 10.1128/jvi.65.11.6252-6259.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Graham F L, van der Eb A. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973;52:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- 14.Griffin B E, Karran L. Immortalization of monkey epithelial cells by specific fragments of Epstein-Barr virus DNA. Nature. 1984;309:78–82. doi: 10.1038/309078a0. [DOI] [PubMed] [Google Scholar]
- 15.Griffin B E, Xue S A. Epstein-Barr virus infections and their association with human malignancies: some key questions. Ann Med. 1998;30:249–259. doi: 10.3109/07853899809005852. [DOI] [PubMed] [Google Scholar]
- 16.Henkel T, Ling P D, Hayward S D, Peterson M G. Mediation of Epstein-Barr virus EBNA2 transactivation by recombination signal-binding protein J kappa. Science. 1994;265:92–95. doi: 10.1126/science.8016657. [DOI] [PubMed] [Google Scholar]
- 17.Hitt M M, Allday M J, Hara T, Karran L, Jones M D, Busson P, Tursz T, Ernberg I, Griffin B E. EBV gene expression in an NPC-related tumour. EMBO J. 1989;8:2639–2651. doi: 10.1002/j.1460-2075.1989.tb08404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hsieh J J, Henkel T, Salmon P, Robey E, Peterson M G, Hayward S D. Truncated mammalian Notch1 activates CBF1/RBPJk-repressed genes by a mechanism resembling that of Epstein-Barr virus EBNA2. Mol Cell Biol. 1996;16:952–959. doi: 10.1128/mcb.16.3.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karran L, Gao Y, Smith P R, Griffin B E. Expression of a family of complementary-strand transcripts in Epstein-Barr virus-infected cells. Proc Natl Acad Sci USA. 1992;89:8058–8062. doi: 10.1073/pnas.89.17.8058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kempkes B, Pich D, Zeidler R, Sugden B, Hammerschmidt W. Immortalization of human B lymphocytes by a plasmid containing 71 kilobase pairs of Epstein-Barr virus DNA. J Virol. 1995;69:231–238. doi: 10.1128/jvi.69.1.231-238.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kienzle N, Buck M, Greco S, Krauer K, Sculley T. Epstein-Barr virus-encoded RK-BARF0 protein expression. J Virol. 1999;73:8902–8906. doi: 10.1128/jvi.73.10.8902-8906.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kienzle N, Sculley T B, Poulsen L, Buck M, Cross S, Raab-Traub N, Khanna R. Identification of a cytotoxic T-lymphocyte response to the novel BARF0 protein of Epstein-Barr virus: a critical role for antigen expression. J Virol. 1998;72:6614–6620. doi: 10.1128/jvi.72.8.6614-6620.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koslowski K M, Shaver P R, Casey II J T, Wilson T, Yamanaka G, Sheaffer A K, Tenney D J, Pederson N E. Physical and functional interactions between the herpes simplex virus UL15 and UL28 DNA cleavage and packaging proteins. J Virol. 1999;73:1704–1707. doi: 10.1128/jvi.73.2.1704-1707.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lau R, Sinclair A J, Brimmell M, Farrell P J. Epstein-Barr virus productive cycle gene expression in vivo. INSERM Colloq. 1993;225:203–209. [Google Scholar]
- 25.Liliental J, Chang D D. Rack1, a receptor for activated protein kinase C, interacts with integrin beta subunit. J Biol Chem. 1998;273:2379–2383. doi: 10.1074/jbc.273.4.2379. [DOI] [PubMed] [Google Scholar]
- 26.Ling P D, Rawlins D R, Hayward S D. The Epstein-Barr virus immortalizing protein EBNA-2 is targeted to DNA by a cellular enhancer-binding protein. Proc Natl Acad Sci USA. 1993;90:9237–9241. doi: 10.1073/pnas.90.20.9237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mochly-Rosen D. Localization of protein kinases by anchoring proteins: a theme in signal transduction. Science. 1995;268:247–251. doi: 10.1126/science.7716516. [DOI] [PubMed] [Google Scholar]
- 28.Nagase T, Ishikawa K, Miyajima N, Tanaka A, Kotani H, Nomura N, Ohara O. Prediction of the coding sequences of unidentified genes. IX. The complete sequences of 100 new cDNA clones from brain which can code for large proteins in vitro. DNA Res. 1998;5:31–39. doi: 10.1093/dnares/5.1.31. [DOI] [PubMed] [Google Scholar]
- 29.Neer E J, Schmidt C J, Nambudripad R, Smith T F. The ancient regulatory-protein family of WD-repeat proteins. Nature. 1994;371:297–300. doi: 10.1038/371297a0. [DOI] [PubMed] [Google Scholar]
- 30.Parker B D, Bankier A, Satchwell S, Barrell B, Farrell P J. Sequence and transcription of Raji Epstein-Barr virus DNA spanning the B95-8 deletion region. Virology. 1990;179:339–346. doi: 10.1016/0042-6822(90)90302-8. [DOI] [PubMed] [Google Scholar]
- 31.Raab-Traub N, Rajadurai P, Flynn K, Lanier A P. Epstein-Barr virus infection in carcinoma of the salivary gland. J Virol. 1991;65:7032–7036. doi: 10.1128/jvi.65.12.7032-7036.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robertson E, Kieff E. Reducing the complexity of the transforming Epstein-Barr virus genome to 64 kilobase pairs. J Virol. 1995;69:983–993. doi: 10.1128/jvi.69.2.983-993.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ron D, Chen C H, Caldwell J, Jamieson L, Orr E, Mochly-Rosen D. Cloning of an intracellular receptor for protein kinase C: a homolog of the beta subunit of G proteins. Proc Natl Acad Sci USA. 1994;91:839–843. doi: 10.1073/pnas.91.3.839. . (Erratum, 92:2016, 1995.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ron D, Luo J, Mochly-Rosen D. C2 region-derived peptides inhibit translocation and function of beta protein kinase C in vivo. J Biol Chem. 1995;270:24180–24187. doi: 10.1074/jbc.270.41.24180. [DOI] [PubMed] [Google Scholar]
- 35.Ron D, Mochly-Rosen D. An autoregulatory region in protein kinase C: the pseudoanchoring site. Proc Natl Acad Sci USA. 1995;92:492–496. doi: 10.1073/pnas.92.2.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sadler R H, Raab-Traub N. Structural analyses of the Epstein-Barr virus BamHI A transcripts. J Virol. 1995;69:1132–1141. doi: 10.1128/jvi.69.2.1132-1141.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sinclair A J, Farrell P J. Host cell requirements for efficient infection of quiescent primary B lymphocytes by Epstein-Barr virus. J Virol. 1995;69:5461–5468. doi: 10.1128/jvi.69.9.5461-5468.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith P R, Gao Y, Karran L, Jones M D, Snudden D, Griffin B E. Complex nature of the major viral polyadenylated transcripts in Epstein-Barr virus-associated tumors. J Virol. 1993;67:3217–3225. doi: 10.1128/jvi.67.6.3217-3225.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taus N S, Salmon B, Baines J D. The herpes simplex virus 1 UL 17 gene is required for localization of capsids and major and minor capsid proteins to intranuclear sites where viral DNA is cleaved and packaged. Virology. 1998;252:115–125. doi: 10.1006/viro.1998.9439. [DOI] [PubMed] [Google Scholar]
- 40.Tomei L D, Noyes I, Blocker D, Holliday J, Glaser R. Phorbol ester and Epstein-Barr virus dependent transformation of normal primary human skin epithelial cells. Nature. 1987;329:73–75. doi: 10.1038/329073a0. [DOI] [PubMed] [Google Scholar]
- 41.Yarwood S J, Steele M R, Scotland G, Houslay M D, Bolger G B. The RACK1 signaling scaffold protein selectively interacts with the cAMP-specific phosphodiesterase PDE4D5 isoform. J Biol Chem. 1999;274:14909–14917. doi: 10.1074/jbc.274.21.14909. [DOI] [PubMed] [Google Scholar]
- 42.Zagouras P, Stifani S, Blaumueller C M, Carcangui M L, Artavanis-Tsakonas S. Alterations in Notch signaling in neoplastc lesions of the human cervix. Proc Natl Acad Sci USA. 1995;92:6414–6418. doi: 10.1073/pnas.92.14.6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zeng Y, Miao X, Jaio B, Li H, Ni H, Ito Y. Epstein-Barr virus activation in Raji cells with ether extracts of soil from different areas in China. Cancer Lett. 1984;23:53–59. doi: 10.1016/0304-3835(84)90061-2. [DOI] [PubMed] [Google Scholar]
- 44.Zhang C X, Lowrey P, Finerty S, Morgan A J. Analysis of Epstein-Barr virus gene transcription in lymphoma induced by the virus in the cottontop tamarin by construction of a cDNA library with RNA extracted from a tumour biopsy. J Gen Virol. 1993;74:509–514. doi: 10.1099/0022-1317-74-3-509. [DOI] [PubMed] [Google Scholar]