Abstract
The Epstein-Barr virus (EBV) open reading frame BGLF4 was identified as a potential Ser/Thr protein kinase gene through the recognition of amino acid sequence motifs characteristic of conserved regions within the catalytic domains of protein kinases. In order to investigate this potential kinase activity, BGLF4 was expressed in Escherichia coli and the purified protein was used to generate a specific antiserum. Recombinant vaccinia virus vTF7-3, which expresses the T7 RNA polymerase, was used to infect 293 and 293T cells after transient transfection with a plasmid containing BGLF4 under the control of the T7 promoter. Autophosphorylation of the BGLF4 protein was demonstrated using the specific antiserum in an immune complex kinase assay. In addition, EBNA-1-tagged BGLF4 and EBNA-1 monoclonal antibody 5C11 were used to demonstrate the specificity of the kinase activity and to locate BGLF4 in the cytoplasm of transfected cells. Manganese ions were found to be essential for autophosphorylation of BGLF4, and magnesium can stimulate the activity. BGLF4 can utilize GTP, in addition to ATP, as a phosphate donor in this assay. BGLF4 can phosphorylate histone and casein in vitro. Among the potential viral protein substrates we examined, the EBV early antigen (EA-D, BMRF1), a DNA polymerase accessory factor and an important transactivator during lytic infection, was found to be phosphorylated by BGLF4 in vitro. Amino acids 1 to 26 of BGLF4, but not the predicted conserved catalytic domain, were found to be essential for autophosphorylation of BGLF4.
Protein kinases are known to be involved in the regulation of a wide variety of eukaryotic cellular functions including cell metabolism, cell cycle control, hormone response, and control of transcription and translation. Studying viral protein kinases might therefore lead to an understanding of the mechanisms of virus replication and virus-cell interactions. Most of the protein kinases of the retroviruses are Tyr protein kinases, such as v-src and v-erb, which may contribute to the growth transformation phenotype of the virally infected host cells (for a review, see reference 32). The first protein kinase gene demonstrated in a eukaryotic DNA virus was that contained in the unique short (US) regions of the related human and porcine alphaherpesviruses, herpes simplex virus type 1 (HSV-1), and pseudorabies virus (20). Other protein kinases have been reported in DNA viruses, including protein kinase B1 of the poxviruses (45, 46) and ORF9 of baculovirus (42).
Phosphorylation of cellular and viral proteins, which has been observed during lytic infection of cells by herpesviruses, seems to be a common phenomenon which involves a number of different protein kinase activities (21). Two groups of viral protein kinase activities, US3 and UL13, have been identified in alphaherpesviruses. The US3 gene of HSV-1 (37) and the VZV66 gene of varicella-zoster virus (VZV) (19) were predicted to encode protein kinases on the basis of their strong similarity to the family of eukaryotic serine/threonine protein kinases. Mutation of US3 seemed not to affect the replication of HSV-1 in vitro (44). However, UL13 is responsible for the posttranslational processing associated with phosphorylation of alpha-22 of HSV-1. In addition, it was demonstrated that eukaryotic elongation factor 1δ is hyperphosphorylated by the protein kinase encoded by the UL13 gene (27). This modification is believed to contribute to the shutoff of host cell functions during HSV-1 infection. In beta- and gammaherpesviruses, there is only one open reading frame that seems likely to encode a protein kinase. UL13 homologues identified by sequence homology searches include UL97 of cytomegalovirus (CMV), BGLF4 of Epstein-Barr virus (EBV) (5), 15R of human herpesvirus 6 (HHV-6) (31), and ORF36 of HHV-8 (47). This family of proteins is evolutionarily more distant from the cellular protein kinases than are the alphaherpesvirus US protein kinases. The homologue encoded by CMV, UL97, has been shown to phosphorylate ganciclovir (34). This finding illustrated the mechanism through which human CMV (HCMV) is sensitive to this nucleoside analogue despite lacking a thymidine kinase. It was found also that the resistance of certain strains of HCMV to ganciclovir was attributable to a mutation in UL97 (52). Recently, ORF36, the UL13 homologue of HHV-8, also was shown to phosphorylate ganciclovir in transfected cells (4). The functions of UL97 and ORF36 during virus infection have not been determined in these studies. However, a recent report indicated that a recombinant HSV, in which UL13 has been deleted and replaced by HCMV UL97, can restore the activity of modifying cellular elongation factor 1δ following virus infection (26).
Based on these observations, we hypothesize that the high degree of conservation, through the evolution of the herpesviruses, of these predicted kinases can be attributed to their importance for the replication of these viruses in their natural hosts and may contribute to their pathogenesis. The BGLF4 gene was identified as a Ser/Thr protein kinase-related gene using amino acid sequence alignment of regions conserved within the catalytic domains of protein kinases (5), and the BGLF4 kinase is the only potential protein kinase identified in the EBV genome. BGLF4 gene-containing RNA transcripts were detected by Northern blot analysis (9) and seemed to be expressed early in lytic EBV infection. Because a protein kinase activity has not yet been demonstrated for the BGLF4 protein, we expressed the protein in prokaryotic and eukaryotic systems to evaluate its potential kinase activity.
BGLF4 was detected by immunofluorescence in the cytoplasm of transfected cells using an EBNA-1 tag system for immunoprecipitation and was shown to autophosphorylate. Since many EBV proteins, such as the lytic protein Zta (BZFL1) and early antigen, diffuse type (EA-D, BMRF1), are reported to be phosphorylated in infected cells (15, 28), we also examined the ability of BGLF4 to phosphorylate other EBV proteins expressed in Escherichia coli. Specific phosphorylation of EA-D was observed in vitro. EA-D is a DNA polymerase accessory factor (33) and is also an important transactivator during lytic infection (55). We suggest that BGLF4 may be important for regulating EBV replication through the phosphorylation of EA-D.
MATERIALS AND METHODS
Plasmid construction.
The BGLF4 open reading frame is located between nucleotides 123614 and 122328 of the EBV genome (a BamHI site is located at 122313). Therefore, a T7 promoter primer (5′-TAATACGACTCACTATAGGG) and LMRC16 (GATCGGATCCATGGATGTGAATATG; containing the BGLF4 initiation codon) were used in PCR to amplify the BGLF4 sequence from pGEM3Z-EBVBamHI G. For expression in E. coli, the PCR product was then digested with BamHI and cloned into the vector pRSETA (Invitrogen), which encodes an N-terminally His-tagged protein under the control of the T7 promoter. Nucleotide sequences of this clone were confirmed, and it was designated pSJC1. The same BamHI DNA fragment was then cloned into pSG5 (Stratagene) to generate pSJC2 for expression of BGLF4 in mammalian cells.
The vector pGH254 was used for expression of BGLF4 using recombinant vaccinia virus. This is a derivative of pBD7 (18) and contains a T7 promoter sequence and a black beetle virus leader sequence to enhance protein expression levels. The BamHI DNA fragment containing BGLF4 was subcloned into the BglII site of pGH254, and then a PCR product, encoding amino acids 408 to 446 of EBNA-1, was cloned upstream of BGLF4 as a tag for detection and immunoprecipitation with monoclonal antibody 5C11 (8). This construct was designated pSJC12(E1/BGLF4). In order to examine BGLF4 protein expression in transfected cells, a similar tag strategy was used to clone the E1/BGLF4 DNA fragment into pSG5. The resultant clone, pCF4, expresses BGLF4 under the control of the simian virus 40 (SV40) promoter.
Generation of BGLF4 mutants.
All the primers used for generating point mutations or deletions within BGLF4 are summarized in Table 1. The oligonucleotide LMRC16, which anneals to the BGLF4 initiation codon, and T7 primer, which anneals to the vector, served as outer primers for PCR mutagenesis. Recombinant PCRs with these outer primers and a pair of overlapping inner primers were used to generate pSJC13(K102M) containing a change of codon 102 from a lysine to a methionine codon. The oligonucleotide LMRC36 (encoding a protein with a mismatch at amino acid 102) and BGLF4 5′ primer LMRC16 were used to generate a DNA fragment encoding the N terminus of BGLF4 using pGEM-BamHI G as the template and Pfu polymerase (Stratagene). LMRC 37 and the T7 primer were used to amplify the 3′ DNA fragment of BGLF4 whose product has a mutation at amino acid 102. The two PCR products were then purified, denatured, annealed, and again amplified with the outer primers. A similar strategy was used to generate pSJC14(H193A, G195A), pSJC15(D129A,G221A), and pSJC16(D297A), using LMRC39 and LMRC40, LMRC41 and LMRC42, and LMRC43 and LMRC44, respectively, as internal primers.
TABLE 1.
Oligonucleotide primers used in this study
| Primer | Sequencea | Amino acid coordinate to BGLF4 |
|---|---|---|
| T 7 | 5′-TAATAC GAC TCA CTA TAG GG | |
| LMRC 16 | 5′-GATCGGATCCATGGATGTGAATATG | ATG |
| LMRC 36 | 5′-CGGTCATGCTCTATGACTCTGTGAC | K102M(5′) |
| LMRC 37 | 3′-ATAGAGCATGACCGTGGCATTATCT | K102M(3′) |
| LMRC 39 | 5′-CGCTTCGGCCATTAGCCCCAGCAACAT | H193A/D195A(5′) |
| LMRC 40 | 3′-TGGCCGAAGCGAAGAGGCCGCAGTGCC | H193A/D195A(3′) |
| LMRC 41 | 5′-TGCTTATGCGACTGCTTCCCTCCAGA | D219/G221A(5′) |
| LMRC 42 | 3′-TCGCATAAGCAGTCAGGACCAGCCAT | D219/G221A(3′) |
| LMRC 43 | 5′-CCTGGCTCTGCAGTCGCTCGGCTA | D297A(5′) |
| LMRC 44 | 3′-GCAGAGCCAGGCGAAGGGCCGTCT | D297A(3′) |
| LMRC 73 | 3′-TTTTTGGAATTCCTGCCGGAGAACATGACG | VFΔ35-65(3′) |
| LMRC 74 | 5′-CGGCAGGAATTCCAAAAAGGCCTGGGTCTC | VFΔ35-65(5′) |
| LMRC 67 | 3′-CTGCTGCAGACGGAGCTGTATCACG | LQΔ85-107(3′) |
| LMRC 68 | 5′-CGTCTGCAGCAGATACTCGCAGGT | LQΔ85-107(5′) |
| LMRC 76 | 3′-CCGGCTGCAGTTTGAGCACCCTCATCTT | LQΔ367-403(3′) |
| LMRC 77 | 5′-CAAACTGCAGCCGGCACCACTGCAAGAA | LQΔ367-403(5′) |
| LMRC 75 | 5′-CGGGATCCCCTCGAGAGACCCAGGCC | GS27(5′) |
| LMRC 72 | 5′-CGGGATCCATGACGCGCTGTGATCAC | GS70(5′) |
| LMRC 26 | 5′-GAAGATCTATCCTGGTGGATTTCACA | RS201(5′) |
Sequences encoding relevant amino acids are underlined.
A second set of small internal deletions of BGLF4 also was generated by recombinant PCR using different sets of internal primers containing PstI restriction enzyme sites to loop out the desired codons. pHH5(VFΔ35-65) was generated using LMRC73 and LMRC74, pHH1(LQΔ85-107) was generated using LMRC67 and LMRC68, and pHH7(LQΔ367-403) was generated using LMRC76 and LMRC77. Double-deletion mutant pHH6(VFΔ35-65,LQΔ85-107) was generated using LMRC16 and LMRC68 to amplify the BGLF4 fragment which contained VFΔ35-65 from pHH5 and using LMRC67 and the T7 promoter primer to obtain the DNA fragment LQΔ85-107. These two fragments were then purified and used to generate pHH6(VFΔ35-65,LQΔ85-107).
Three N-terminal truncation mutants of BGLF4 were generated using pGEM3Z-EBV G as the template and 5′ primers containing BamHI or BglII sites which annealed to different codons of BGLF4 and T7 primer, which annealed to the vector as 3′ primers. The 5′ primer LMRC75 (GS27) annealed to codon 27 with additional nucleotide sequences encoding Gly and Ser for cloning. Another two 5′ primers used were LMRC72 (for GS70) and LMRC26 (for RS201). Individual DNA fragments were cloned into pDL118A to obtain pHH8(GS27-429), pHH4(GS70-429), and pHH2(RS201-429).
Expression and purification of recombinant BGLF4.
pRSETA-BGLF4(pSJC1) was transformed into BL21(DE3), which encodes T7 RNA polymerase under the control of the Tac promoter. The bacteria were cultured in ampicillin-Luria broth (100 mg/ml) until the optical density at 600 nm reached 0.6, and isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 0.1 mM. The cells were then cultured for another 3 h, pelleted by centrifugation, resuspended in lysis buffer (100 μg of lysozyme/ml, 5 mM imidazole, 100 mM NaCl, 8 M urea, 20 mM Tris-Cl, pH 7.4), and incubated on ice for 1 h. The cell lysate was sonicated, and the insoluble fraction was clarified by centrifugation before being loaded onto a nickel column. After the binding, the column was washed with 100 mM imidazole buffer (100 mM NaCl, 8 M urea, 20 mM Tris-Cl, pH 7.4) and eluted with 300 mM imidazole buffer (100 mM NaCl, 8 M urea, 20 mM Tris-Cl, pH 7.4). Total lysate and purified proteins were displayed on sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis (SDS–10% PAGE) gel, for analysis.
Antiserum and antibodies.
Purified BGLF4 was used to immunize New Zealand White rabbits to generate anti-BGLF4 specific antisera. The monoclonal antibody 5C11 specifically recognizes an epitope between amino acids 408 and 446 of EBNA-1 (8). Monoclonal antibodies antiphosphoserine (PSR-45, P3430) and antiphosphothreonine (pTR-8, P-3555) were from Sigma, and antiphosphotyrosine (4G10) was from Upstate Biotechnology.
In vitro coupled transcription and translation.
This reaction was carried out essentially according to the protocol suggested by the manufacturer (Promega). One microgram of template DNA was mixed with 25 μl of TNT rabbit reticulocyte lysate, 2 μl of reaction buffer, 1 μl of T7 RNA polymerase, 1 μl of amino acid mixture, and 4 μl of [35S]methionine (10 mCi/ml) to make a final volume of 50 μl. The reaction mixture was incubated at 30°C for 1.5 h.
Transfection and vaccinia virus infection.
Human 293 cells were grown in Dulbecco's modified Eagle minimal essential medium supplemented with 10% fetal calf serum. For transfection, 293 cells were plated in 100-mm-diameter dishes at 1.5 × 106 cells per well the day before transfection and 10 μg of each DNA was transfected using the calcium phosphate-BES [N′,N-bis(2-hydroxyethyl)-2-aminoethanesulfonic acid-buffered saline] procedure (6). Recombinant vaccinia virus vTF7-3 (ATCC UR-2153), which encodes the T7 RNA polymerase, was used to infect the cells 24 h posttransfection. After being rinsed with phosphate-buffered saline (PBS), vTF7-3 viruses (multiplicity of infection = 10) were resuspended in 1 ml of PBS–10 mM MgCl 2–0.01% bovine serum albumin and added to the transfected cells and the mixture was incubated for 15 min. Then 2 ml of complete medium was added, and the mixture was incubated for another 30 min, followed by a PBS rinse. The cells were incubated in complete medium at 37°C for 16 to 18 h and harvested by trypsinization. In later experiments, we found that the 293T cell line, a derivative of 293 cells, anchors on slides more efficiently than 293 cells. Therefore, 293T cells were used in the transfection experiment to determine the cellular location of the BGLF4 protein.
Immunoprecipitation.
The transfected cells were rinsed twice with methionine-free medium at 4 to 6 h before trypsin treatment and incubated in 3 ml of methionine-free RPMI 1640 containing [35S]methionine (100 μCi/ml) and 10% dialyzed fetal calf serum. The cells were then washed in PBS and lysed in 0.4 ml of radio immunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% Nonidet P-40, 0.5% deoxycholate, 0.1% SDS, 50 μg of aprotinin/ml, 50 μg of leupeptin/ml, 1 mM phenylmethylsulfonyl fluoride) on ice for 1 min and vortexed for 30 s. This was repeated three times. The lysate was spun at 14,000 rpm in a microcentrifuge to remove the debris and precleaned by incubation with 50 μl of 20% protein A-Sepharose beads at 4°C for 30 min. Two hundred microliters of lysate was then incubated with 5 μl of BGLF4-specific antiserum or EBNA-1 monoclonal antibody 5C11 at 4°C for 1.5 h; then 50 μl of 20% protein A-Sepharose beads was added, and the mixture was incubated at 4°C for another 1.5 h. The protein A beads were then spun and washed twice with 1 ml of high-salt buffer (1 M NaCl, 10 mM Tris-HCl [pH 8.0], 0.2% NP-40), once with 1 ml of low-salt buffer (0.1 M NaCl, 10 mM Tris-HCl [pH 8.0], 0.2% NP-40), and six times with RIPA buffer at 4°C. The immunocomplex was then boiled in 20 μl of SDS sample buffer for 3 min and displayed on an SDS–10% PAGE gel. After electrophoresis, the gel was dried for autoradiography. For immune complex kinase assays, 293 cells were transfected and infected with recombinant vTF7-3, as described previously, without the radiolabel.
Western immunoblotting.
For Western immunoblot analysis, cells were lysed 40 h after transfection in 2× sample buffer (50 mM Tris, 4% SDS, 20% glycerol, 0.04% bromophenol blue, 200 mM dithiothreitol) and sonicated briefly. Lysates were displayed on 10 or 12% polyacrylamide gels. After being blocked in Tris-buffered saline (TBS)–5% nonfat dry milk–0.1% Tween 20 for 1 h, the filter was incubated with 1:500-diluted rabbit anti-BGLF4 serum or EBNA-1 monoclonal antibody 5C11, diluted 1:50 in TBS–0.1% Tween 20 at room temperature for 1 h. The filter was washed three times with TBS–0.1% Tween 20 for 15 min each time and then incubated in a 1:5,000 dilution of horseradish peroxidase-conjugated secondary antibody. The filter was washed three times and developed with an ECL kit (Amersham Life Science) and then exposed to Kodak X-ray film.
Immune complex kinase assay.
Lysates containing 30 μg of total protein were used for each immunoprecipitation. Protein A-Sepharose beads containing immunoprecipitated protein kinase were washed with high-salt, low-salt, RIPA, and kinase buffers (once each) and resuspended in 50 μl of kinase buffer containing 5 μCi of [γ-32P]ATP. After incubation at 30°C for 30 min, the immunocomplexes were washed with RIPA buffer and then resuspended in 2× SDS sample buffer and boiled for 5 min before SDS-PAGE analysis. To optimize the conditions for kinase activity, conditions for various protein kinases, such as VZV ORF47 (25 mM HEPES [pH 7.4], 10 mM MnCl2, 50 mM KCl) (40), HSV UL13 (50 mM Tris [pH 8.0], 50 mM MgCl2, 0.5 M NaCl, 0.1% NP-40, 1 mM dithiothreitol) (14), and pp60c-src (50 mM HEPES [pH 7.2], 1 mM MgCl2, 1 mM MnCl2, 150 mM NaCl, 0.5% NP-40) (3), were tested first. The optimal buffer conditions determined for the BGLF4 autophosphorylation activity were 50 mM HEPES (pH 7.4)–10 mM MgCl2–10 mM MnCl2–300 mM KCl–0.5% NP-40. Casein kinase II (CK II; Promega) was included as a positive control in the heparin inhibition assay.
In the transphosphorylation assay, the partially purified BGLF4 immunocomplexes were washed with BGLF4 kinase buffer, without NP-40, twice and incubated with 1 μg of substrate in the presence of 5 μCi of [γ-32P]ATP in a volume of 70 μl at 30°C for 30 min. After incubation, 60 μl of supernatant was precipitated by adding 20 μl of 24% trichloroacetic acid (TCA) and 2 μl of 12 mM sodium deoxycholate, incubated on ice for 20 min, and centrifuged at 4°C for 10 min. The protein pellet was rinsed with 95% ethanol, dried, and resuspended in 2× SDS sample buffer for electrophoresis and autoradiography. Substrates used included histone (Sigma; H-7755, type II-As; from calf thymus), casein (Sigma; C-4765; from bovine milk), EBV proteins Zta and Rta (pRSETA-Rta; amino acids 8 to 357), DNase (54), glutathione S-transferase–EBNA-1 (amino acids 408 to 641) (7), major DNA binding protein and diffuse type early antigen EA-D (11).
IFA.
Forty-eight hours after transfection, cells were fixed for immunofluorescence assay (IFA) staining with 50% acetone and 50% methanol for 20 min at −20°C. EBNA-1 monoclonal antibody 5C11 or 1:100-diluted rabbit anti-BGLF4 serum was added to the smears, and they were incubated in a moist chamber at 37°C for 1 h. The smears were then washed in PBS for 5, 10, and 15 min. Fluorescein isothiocyanate-conjugated goat anti-mouse serum was diluted 1:100 and placed on the smears, and the smears were incubated at 37°C for 1 h. After incubation, the smears were washed as described above, mounted in a 90% phosphate-buffered glycerol solution, and examined under a UV microscope.
RESULTS
Expression of BGLF4 in E. coli and generation of a specific antiserum.
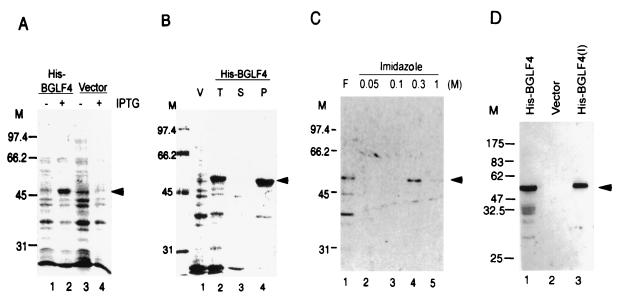
According to the EBV genome sequence deposited in GenBank (m80517.gb_vi), the BGLF4 open reading frame is located between nucleotides 123692 and 122328. However, examination of the sequence did not reveal an in-frame ATG within several hundred base pairs upstream of this region. Therefore, a PCR product containing the first in-frame ATG (nucleotide 123,614) to nucleotide 122328 was first cloned into pRSETA for expression in E. coli. The recombinant clone was then transformed into BL21(DE3), and expression was induced with IPTG. The recombinant protein (His-BGLF4) was visible on a Coomassie blue-stained SDS-PAGE gel. The additional band, which appeared following IPTG induction, migrated at the predicted size of 52 kDa (Fig. 1A, lane 2), since the cloning vector contains a 3.85-kDa polyhistidine coding sequence, which facilitates the purification of recombinant protein. However, the majority of the fusion protein appeared in insoluble form in the E. coli cell lysate (Fig. 1B, lane 4). Therefore, the bacterial pellet was solubilized in buffer containing 8 M urea and purified through a nickel column. The protein was eluted with buffer containing 300 mM imidazole (Fig. 1C, lane 4) and used to immunize a rabbit to generate a polyclonal antiserum. The reactivities and specificities of the polyclonal antibodies were examined by immunoblot analysis as shown in Fig. 1D. The rabbit anti-BGLF4 antiserum specifically recognized the 52-kDa protein in the E. coli cell lysate (Fig. 1D, lane 1), and no signal was observed in the lysate from the vector control (Fig. 1D, lane 2).
FIG. 1.
Expression of His-BGLF4 in E. coli BL21(DE3) and generation of BGLF4-specific antiserum. (A) Total cell lysate from bacteria carrying vector control (pRSETA) or pSJC1 (His-BGLF4). Lysate is shown uninduced (lanes 1 and 3) and after induction with IPTG (lanes 2 and 4) and was displayed on a 10% PAGE gel and stained with Coomassie blue. The predicted molecular mass of BGLF4 is approximately 52 kDa, as indicated by the arrowhead. (B) After lysis with buffer (lane 2; total lysate), the cell lysate was fractionated into soluble proteins (lane 3) and pellet. Most of the BGLF4 product appeared in insoluble form in E. coli (lane 4). V, vector control. (C) The recombinant BGLF4 protein was dissolved in 8 M urea–5 mM imidazole–100 mM NaCl and purified using a nickel column. After being washed with binding buffer (lane 2) and 100 mM imidazole buffer (lane 3), the BGLF4 protein was eluted with buffer containing 300 mM imidazole (lane 4). F, flowthrough. (D) Western immunoblotting demonstrates the specificity of rabbit anti-BGLF4 antiserum. Total cell lysates of E. coli carrying pSJC1 (lane 1) or pRSETA (lane 2) and purified BGLF4 (lane 3) were displayed on an SDS–10% PAGE gel, transferred onto a membrane, and probed with rabbit anti-BGLF4 antiserum as described in Materials and Methods. M, molecular weight markers.
Expression of BGLF4 in a eukaryotic system and autophosphorylation of BGLF4.
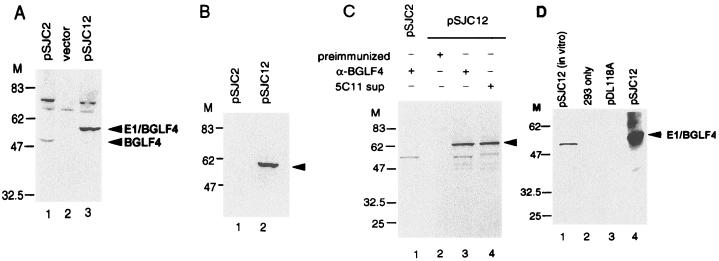
The purified, bacterially expressed protein was transferred onto a membrane and renatured for an in situ kinase assay (22), but the result was negative (data not shown). It is possible that the renaturation conditions used were not appropriate to restore enzyme activity or that some modification of the protein was required for kinase activity. We wished to express the BGLF4 protein under the control of the SV40 or CMV promoter following transient transfection in order to examine the kinase activity. However, the protein was barely detectable by immunoblotting using the anti-BGLF4 rabbit serum (data not shown). Since the construct pSJC2(pSG5-BGLF4) contains a T7 promoter upstream of the BGLF4 gene, it was transcribed and translated in a reticulocyte lysate system to check the gene product (Fig. 2D, lane 1). This product can be immunoprecipitated by the anti-BGLF4 specific antiserum (data not shown). We suggest that our inability to detect BGLF4 in transfected cells may be attributable to a low level of expression. In order to obtain sufficient protein, the recombinant vaccinia virus vTF7-3, which carries the T7 RNA polymerase gene, was used to infect 293 cells following transfection of pSJC2. Cell lysates harvested 16 h postinfection were found to contain a protein of approximately 48 kDa in a Western blot assay using polyclonal anti-BGLF4 serum (Fig. 2A, lane 1). The identity of some higher-molecular-mass protein species in this blot was not clear, which could be due to the cross-reaction to cellular proteins induced by the expression of BGLF4. Anti-BGLF4 antiserum can immunoprecipitate this product as shown in Fig. 2C, lane 1. Thus, the vTF-7 system may be used as a source of partially purified BGLF4 protein for kinase assays.
FIG. 2.
Expression of BGLF4 (pSJC2) and E1/BGLF4 (pSJC12) in 293 cells after transfection and infection with recombinant vaccinia virus vTF7-3, which carries a copy of the T7 RNA polymerase. (A) After transfection and infection, cell lysates of pSG5 (vector), pSJC2(pSG5-BGLF4), and pSJC12(pSG5-E1/BGLF4) were harvested, displayed on an SDS–10% PAGE gel, and reacted with BGLF4-specific antiserum in an immunoblotting assay. The 48-kDa product of BGLF4 and the 52-kDa product of E1/BGLF4 can be seen in lanes 1 and 3, respectively. (B) The cell lysates expressing BGLF4 and E1/BGLF4 were immunoblotted with EBNA-1 monoclonal antibody 5C11. Arrowhead, E1/BGLF4. (C) After transfection of BGLF4 or E1/BGLF4 expression plasmids and infection with vTF7-3, the cells were labeled with [35S]methionine for 4 h before lysis. The cell lysates were immunoprecipitated with preimmunized-rabbit sera, BGLF4-specific antisera, or the 5C11 monoclonal antibody. Arrowhead, E1/BGLF4. (D) Autophosphorylation of BGLF4. Protein A-Sepharose beads containing immunoprecipitated E1/BGLF4 were incubated in kinase buffer in the presence of [γ-32P]ATP. Autophosphorylation of BGLF4 may be seen in lane 4. Lane 1, in vitro transcription/translation product as a marker.
The immunoprecipitated BGLF4 obtained following vTF-7 infection was tested for autophosphorylation activity. Various experimental conditions used previously for other protein kinases, such as UL13 of HSV, VZV ORF47, and c-Src, were tested in the initial kinase assays. Autophosphorylation of BGLF4 can be detected using all of these conditions, but the strongest activity was observed in the buffer for VZV ORF47 (25 mM HEPES [pH 7.4], 10 mM MnCl2, 50 mM KCl) (40) (data not shown).
EBNA-1 tag system for expression and immune complex kinase assay.
Since most protein kinases share conserved motifs, it is possible that anti-BGLF4 antiserum could bring down cellular protein kinases in the immune complex kinase immunoprecipitation assay. An EBNA-1 monoclonal antibody tag system was therefore used to enhance the specificity of the immunoprecipitation reaction. 5C11 recognizes an epitope between amino acids 408 and 446 of EBNA-1, a region which contains no serine or threonine residues, and was shown to react with this sequence in both immunoprecipitation and Western immunoblot assays (8). Therefore, EBNA-1(408-446) and BGLF4 were expressed as a fusion protein using a derivative of pGH254 which contains a T7 RNA promoter and a black beetle virus leader sequence to enhance translation (18). Expression plasmid pSJC12 was transfected into 293 cells, followed by infection with vTF7-3. The expressed recombinant protein, E1/BGLF4, can be detected in Western immunoblot assays by anti-BGLF4 antiserum (Fig. 2A, lane 3) and by EBNA-1 monoclonal antibody 5C11 (Fig. 2B, lane 2). The E1/BGLF4 protein also can be immunoprecipitated by these two antibodies (Fig. 2C). The amount of protein expressed from pSJC12 was roughly 3- to 10-fold greater than that expressed from pSJC2 (Fig. 2A, lanes 1 and 3). The immunoprecipitated E1/BGLF4 also was shown to possess an autophosphorylation activity similar to that of BGLF4 (Fig. 2D).
Optimal conditions for BGLF4 kinase activity and use of GTP as a phosphate donor.
The E1/BGLF4 product and EBNA-1 monoclonal antibody were used to further characterize the BGLF4 kinase activity and distinguish it from cellular kinases in terms of optimal pH, dependence on monovalent and divalent cations, and the effect of detergent in the kinase buffer. We first examined the effect of pH in 50 mM HEPES buffer at a range between pH 6.2 and 8.3. The autophosphorylation products displayed on SDS-PAGE gel were quantitated with a phosphorimager (STORM 842; Captain Biolabtech Co.). Similar counts were obtained in the range from pH 6.5 to 8.0, whereas the activities were about 10% lower in the reactions at pH 6.2 or 8.3 (Fig. 3A). We then investigated the requirement of BGLF4 kinase activity for divalent cations (Fig. 3B). The buffer contained 50 mM HEPES (pH 7.4), 150 mM NaCl, and various concentrations of MgCl2 or MnCl2, as indicated. Manganese was found to be essential for BGLF4 activity (Fig. 3B, lane 6), and the activity can be stimulated further by magnesium ions (Fig. 3B, lane 5). The effect of monovalent cations also was examined; the presence of 300 mM KCl or NaCl gave increased activities (Fig. 3C, lanes 6 and 9). We also observed that the presence of 0.5% Triton X-100 or NP-40 can further enhance the autophosphorylation of BGLF4 (Fig. 3D). The final buffer conditions we chose for BGLF4 autophosphorylation were 50 mM HEPES (pH 7.4), 10 mM MgCl2, 10 mM MnCl2, 300 mM KCl, and 0.5% NP-40.
FIG. 3.
Optimal conditions for BGLF4 autophosphorylation. Protein A-Sepharose beads containing immunoprecipitated E1/BGLF4 were incubated in various kinase buffers in the presence of [γ-32P]ATP. (A) Effect of pH on BGLF4 kinase activity. The buffer containing 1 mM MnCl2, 1 mM MgCl2, 150 mM NaCl, 0.5% NP-40, and 50 mM HEPES was adjusted to different pHs as indicated. The autophosphorylation activities are similar in the range of pH 6.5 to 8.0, whereas the activities are lower at pH 6.2 and pH 8.3. (B) Effects of divalent cations on BGLF4 kinase activity. The buffer containing 50 mM HEPES (pH 7.4), 150 mM NaCl, and various amounts of MgCl2 and MnCl2, as indicated. Manganese is essential for BGLF4 activity (lane 6), which can be further stimulated by magnesium (lane 5). (C) Effects of monovalent cations on BGLF4 kinase activity. The buffer contained 50 mM HEPES (pH 7.4), 10 mM MgCl2, and 10 mM MnCl2, as indicated, and different concentrations of NaCl or KCl. The maximal activity was observed in the presence of 300 mM KCl (lane 6). (D) Effect of detergent on the BGLF4 kinase activity. In addition to 50 mM HEPES (pH 7.4), 10 mM MgCl2, 10 mM MnCl2, and 300 mM KCl, 0.5% NP-40 appeared to stimulate BGLF4 kinase activity. The in vitro transcription/translation product of BGLF4 (I) is shown as a marker. The immunoprecipitation product of pDL118-transfected cell lysate was also used in the kinase assay as a negative control (V) in each panel.
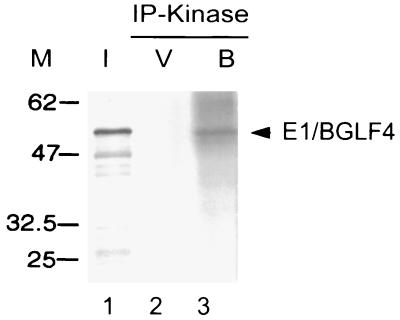
One of the important characteristics of UL13 of HSV and VZV 47 is that they can use both ATP and GTP as phosphate donors. Therefore, we tested [γ-32P]GTP instead of [γ-32P]ATP in the immune complex kinase reaction; autophosphorylation was observed using the authentic BGLF4 protein (data not shown) or EBNA-1 tag system (Fig. 4, lane 3).
FIG. 4.
Utilization of GTP as phosphate donor for BGLF4 autophosphorylation. The immunoprecipitation product of E1/BGLF4, as described for Fig. 3, was used for autophosphorylation in the kinase buffer containing [γ-32P]GTP instead of [γ-32P]ATP (lane 3). I, [35S]methionine-labeled in vitro transcription/translation product; V, vector (pPDL118) control.
Kinetics of BGLF4 autophosphorylation using high-salt buffer-washed immunocomplexes.
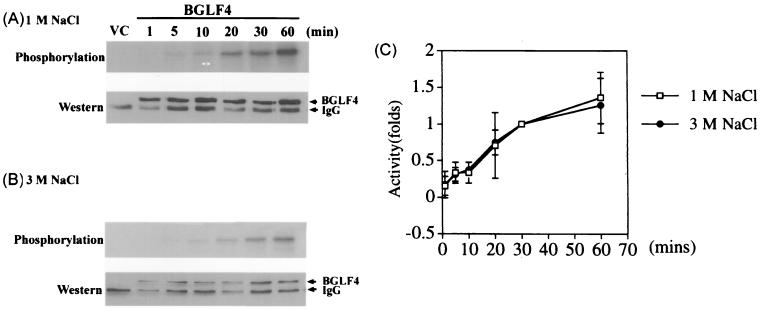
In addition to UL13 and US3, the large subunit of ribonucleotide reductase (R1) of HSV has been reported to have protein kinase activity (1, 12). The R1 protein kinase was demonstrated to utilize a catalytic domain different from those of other conserved protein kinases (36). However, the protein kinase activity of the HSV ribonucleotide reductase large subunit was recently claimed to be due to experimental contamination by casein kinase (30). The autophosphorylation activity of R1 was eliminated after washing the immune complexes with buffer containing 2 M NaCl. To eliminate potential contamination in our assay, we washed the anti-EBNA-1–E1/BGLF4 immunocomplexes with buffer containing 1.0, 1.5, 2.0, 2.5, or 3.0 M NaCl before switching to the kinase buffer for the assay. Similar relative activities were obtained after incubation for 30 min (data not shown). We then compared the kinetics of BGLF4 autophosphorylation using E1/BGLF4 immunocomplexes washed with 1 and 3 M NaCl, as shown in Fig. 5. After autophosphorylation with E1/BGLF4 complexes, the products were displayed on SDS–PAGE gel and transferred to a Hybond-C membrane. The radioactivity of the reaction mixtures in the upper portions of Fig. 5A and B were quantitated and divided by the relative amounts of protein detected by Western blotting as shown in the lower portions. Although the amount of E1/BGLF4 protein in the immunoprecipitation reaction mixtures decreased after the 3 M NaCl wash, the kinetics of autophosphorylation were very close to those of the immunocomplex washed with 1 M NaCl. This result indicated that the phosphorylation of BGLF4 was very likely to be autophosphorylation, unless there was a cellular enzyme strongly associated with BGLF4 which could tolerate washing with 3 M NaCl.
FIG. 5.
Kinetics of immunoprecipitated BGLF4 autophosphorylation using buffer containing 1 or 3 M NaCl to wash the immunocomplexes. Cell lysate (30 μg) harvested from pSJC12-transfected and vTF7-3-infected 293T cells was used for immunoprecipitation. The immunocomplexes were washed with 1 or 3 M NaCl buffer and incubated with [γ-32P]ATP in kinase buffer for various periods of time, as indicated. The reaction mixture containing vector control (VC) was incubated for 30 min. The products were displayed on SDS–10% PAGE gel, transferred to Hybond-C membranes, and quantitated by phosphorimager (A and B, top). The membranes were immunoblotted with 5C11 and developed with an ECL kit to demonstrate the relative amounts of BGLF4 in each reaction mixture (A and B, bottom), and the exposed films were scanned and quantitated with the Scion Imager (National Institutes of Health) program. (C) The kinase activities of the reaction mixtures were divided by the relative amount of protein in each reaction mixture, and the averages of three independent experiments were indicated as relative fold increases or decreases. The kinase activity at 30 min was counted as 1.
Autophosphorylation of BGLF4 was not affected by the presence of heparin or okadaic acid.
CK II is a well-known cellular kinase which also can use GTP as a phosphate donor. Although the kinase activity we observed in BGLF4 autophosphorylation is Mn2+ dependent, unlike that in CK II, we nonetheless examined the kinase activity of BGLF4 in the presence of heparin (Fig. 6A to C). As a positive control, 10 U of CK II was used to phosphorylate 10 μg of casein in BGLF4 buffer, and this activity was completely blocked by 1 μg of heparin/μl in the reaction mixture (Fig. 6A, lanes 7 and 8). However, the phosphorylation activity of BGLF4 was not affected by the presence of 0.25 to 2 μg of heparin/μl (Fig. 6C). In this property, BGLF4 is very similar to VZV47. These experiments excluded the possibility that the phosphorylation of BGLF4 we observed was due to the contamination of CK II.
FIG. 6.
Autophosphorylation of BGLF4 was not affected in the presence of heparin or okadaic acid. (A) Different concentrations of heparin, as indicated, were included in the autophosphorylation reaction mixture for E1/BGLF4 (lanes 2 to 6). Ten units of CK II phosphorylated 1 μg of casein, and this phosphorylation was completely blocked in the presence of heparin (lanes 7 and 8). VC, vector control. (D) The addition of phosphatase inhibitor okadaic acid (OA) did not increase the autophosphorylation of BGLF4. (B and E) The same blots as in panels A and D were probed with 5C11 to show the relative amounts of E1/BGLF4 in the reaction mixtures. (C and F) The kinase activities of each reaction mixture were divided by the relative amounts of protein in each reaction mixture, and the averages of three independent experiments were indicated as relative fold increases or decreases.
The kinetics of BGLF4 autophosphorylation were such that no plateau was reached by 60 min (Fig. 5). In order to examine whether a phosphatase activity is present in the BGLF4 autophosphorylation reaction, okadaic acid, a potent inhibitor of phosphatases A, 2A, and 2B (32), was added to the reaction mixture. Relative kinase activities, calculated by dividing the 32P activity by the amount of protein detected by immunoblotting (Fig. 6E and F), were not affected by the presence of 0.625 to 5 nM okadaic acid (Fig. 6D).
BGLF4 is phosphorylated on serine and threonine residues and is expressed in the cytoplasm of transfected cells.
In order to examine whether BGLF4 is a Ser/Thr protein kinase, as predicted by amino acid homology, the kinase immunoprecipitation products were displayed on SDS–10% PAGE gel, transferred to Hybond-C membranes, and probed with antiphosphoserine, antiphosphothreonine, or antiphosphotyrosine specific monoclonal antibodies. As shown in Fig. 7, only phosphoserine and phosphothreonine were detected in BGLF4. In an attempt to localize the cellular expression of BGLF4, anti-BGLF4 rabbit serum was used in an immunofluorescence assay of 12-O-tetradecanoylphorbol-13-acetate (TPA)-induced P3HR1 and B95-8 cells. Unfortunately, the signal was not obvious, probably due to the small amount of protein expressed in EBV-positive cells or the insensitivity of the antiserum (data not shown). To avoid the changes in cell morphology induced by vaccinia virus, E1/BGLF4 was further subcloned into a pSG5-based vector to place it under the control of the SV40 promoter. The amount of E1/BGLF4 expressed after transfection was about 1/10 to 1/20 that of the vaccinia virus system; however, the protein can be detected by 5C11 or anti-BGLF4 antiserum in transfected cells. BGLF4 was principally detected in the cytoplasm of pCF4 (E1/BGLF4)-transfected 293T cells (Fig. 8).
FIG. 7.
Analysis of the phosphoamino acids of BGLF4, using phosphoamino acid-specific monoclonal antibodies. After the immune complex kinase assay, the products were displayed on SDS–10% PAGE gel, transferred onto Hybond-C membranes, and immunoblotted with antiphosphoserine, antiphosphothreonine, or antiphosphotyrosine specific monoclonal antibodies. Arrowheads, positions of BGLF4 and immunoglobulin G (IgG). VC, vector control.
FIG. 8.

BGLF4 expressed in the cytoplasm of pCF4-transfected 293T cells. (A) 293T cells transfected with pCF4 were fixed with 50% methanol–50% acetone and reacted with 5C11 monoclonal antibody. (B) Photograph of cells reacted with anti-BGLF4 under lower magnification.
Transphosphorylation activity of BGLF4 in vitro.
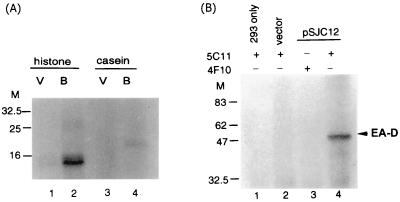
In addition to autophosphorylation, the ability of BGLF4 to catalyze the phosphorylation of heterologous proteins was examined. The immunocomplex containing E1/BGLF4 was incubated with 1 μg of casein or histone in kinase buffer without NP-40. After the reaction, the product was precipitated with TCA and displayed on SDS–10% PAGE gel. The phosphorylation of histone and casein was observed (Fig. 9A). We then examined which viral proteins can be phosphorylated by BGLF4. The EBV immediate-early gene product, Zta, is a phosphoprotein and plays a key role in the switch from latency to the lytic cycle. Serine-173 of Zta is required for DNA binding and is a target for CK II phosphorylation (15, 29). Early antigen (EA-D) is another EBV protein which is highly phosphorylated (33). In addition, UL12 of HSV-2 (alkaline phosphatase) is phosphorylated by US3 (16, 17). Therefore we tested a panel of E. coli-expressed recombinant proteins including Zta, Rta, EA-D, major DNA binding protein, DNase, and EBNA-1 in the transphosphorylation reaction. Specific phosphorylation was only observed for EA-D (Fig. 9B), which is an accessory factor of the EBV DNA polymerase.
FIG. 9.
Transphosphorylation of BGLF4. Protein A-Sepharose beads containing immunoprecipitated E1/BGLF4 were incubated with 1 μg of individual protein in the presence of 5 μCi of [γ-32P]ATP. After incubation, the products were precipitated with TCA and analyzed on an SDS–10% PAGE gel. (A) A stronger signal was observed in the reaction of histone (lane 2) than in that of casein (lane 4). V, cell lysate from vector control; B, cell lysate from E1/BGLF4-transfected cells. (B) Purified bacterially expressed EA-D was also phosphorylated by the 5C11-immunoprecipitated E1/BGLF4 product. 4F10 is a monoclonal antibody against EBV DNase which was used as a negative control in immunoprecipitation.
Amino acids 1 to 26 are important for BGLF4 autophosphorylation activity.
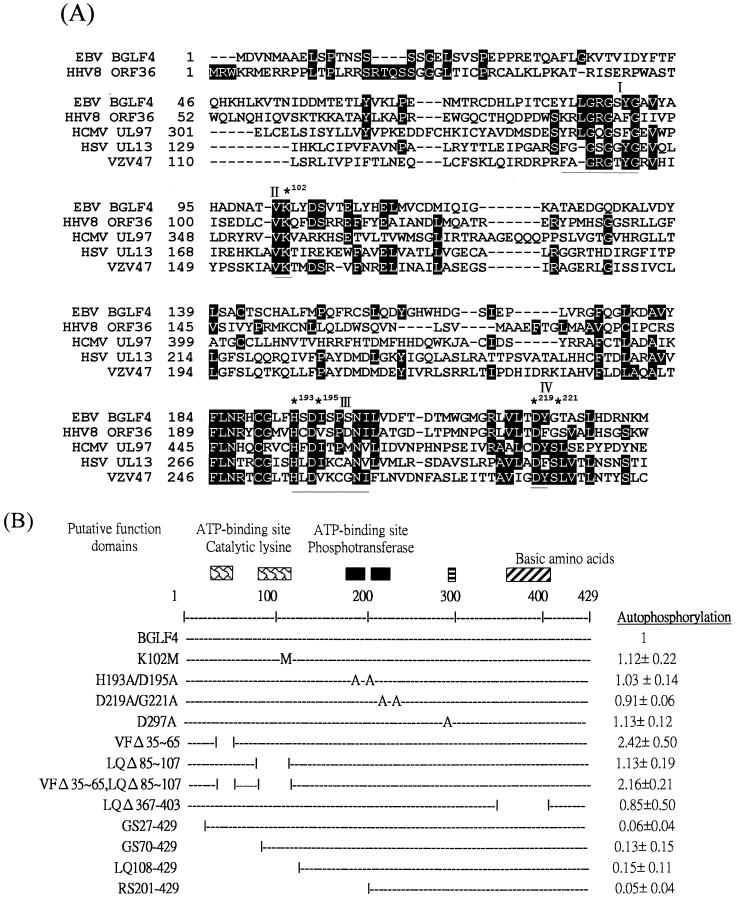
Computer-assisted analysis of the predicted amino acid sequence of EBV BGLF4 identified conserved catalytic motifs (5, 49), including motifs I and II (invariant Lys), which are implicated in the activity of many other protein kinases. In order to identify the functional domains for BGLF4 kinase activity, the amino acid sequence of BGLF4 was aligned with those of human herpesvirus homologues HHV8 ORF36, HCMV UL97, HSV UL13, and VZV 47 using CLUSTALW software (25, 53). Regions containing conserved boxes I to IV are shown in Fig. 10A. A mutant with a change of lysine to methionine (K102M) was generated to determine whether Lys-102 functions as an invariant catalytic Lys. In addition, H193A/D195A and D219A/G221A, which are changed in conserved box III or IV, and D297A, which carries a mutation within a nonconserved region, were generated and individual plasmids were transfected into 293T cells, followed by infection with vTF7-3. The cell lysates were harvested and examined for their abilities to autophosphorylate, as shown in Fig. 11A. To our surprise, these four mutants showed autophosphorylation activities very similar to that of wild-type BGLF4 in the immune complex kinase assay, as shown in Fig. 11C and 10B (right).
FIG. 10.
(A) Sequence alignment of BGLF4 and its homologues among other herpesviruses using the CLUSTALW program (25, 53). Amino acids conserved among herpesviruses relative to the N terminus at amino acid 231 of BGLF4 are shaded. Conserved boxes I to IV are underlined; ∗, amino acids chosen for mutation. (B) Summary of BGLF4 mutants and their relative activities. The BGLF4 coding region encodes 429 amino acids. The putative functional domains, based on alignment of different kinases, are indicated at the top. Different mutants were generated by PCR or recombinant PCR as described in Materials and Methods. Amino acid changes to methionine (M) or alanine (A) are indicated. Relative activities of autophosphorylation are summarized on the right.
FIG. 11.
Relative autophosphorylation activities of BGLF4 mutants. All the plasmids were expressed in 293T cells by transfection and infection of vTF7-3 and assayed for autophosphorylation activities as described for Fig. 6. (A) The immune complex kinase products were displayed on SDS–12% PAGE gel and transferred to a Hybond-C membrane and exposed to X-ray film. The autophosphorylation level was quantitated by STORM 842. (B) The Hybond-C membrane was then probed with the 5C11 monoclonal antibody, and the specific bands were quantitated with the Scion Imager program. (C) The kinase activities of individual lanes were divided by the relative amount of protein in each reaction mixture, and the averages of three independent experiments are indicated as relative fold increases or decreases.
In order to map the important functional domain within BGLF4, we generated mutants with small deletions consisting of the whole putative conserved catalytic domain of BGLF4 (LQΔ85-107), a domain which contains hydrophobic amino acids and a lysine conserved between BGLF4 and HHV8 ORF36 (VF35-65) or both regions (VF35-65,LQΔ85-107). Since some protein kinases contain a pseudosubstrate region which functions as an autoregulation domain (50), another mutant (LQΔ367-403) was generated to examine whether the stretch of basic amino acids within BGLF4 might suppress the protein kinase activity. In addition, four N-terminal deletion mutants, GS27-429, GS70-429, LQ108-429, and RS201-429, were generated and tested for their abilities to autophosphorylate. In conclusion, deletion of amino acids 85 to 107 did not affect the autophosphorylation of BGLF4 but deletion of amino acids 35 to 65 stimulated the phosphorylation signal approximately twofold (Fig. 10B and 11C). Deletion of amino acids 367 to 403 did not increase the kinase activity. An assay of N-terminal deletion mutants revealed that amino acids 1 to 26 are essential for the autophosphorylation of BGLF4.
DISCUSSION
This is the first report demonstrating a protein kinase activity of the EBV gene BGLF4, although the function of this gene was predicted several years ago. The optimal conditions for BGLF4-associated kinase activity, comprising 50 mM HEPES (pH 7.4), 10 mM MgCl2, 10 mM MnCl2, 300 mM KCl, and 0.5% NP-40, are very similar to those of VZV 47 (39). The ability of the BGLF4 protein to use both ATP and GTP as a phosphate donor is similar to that of HSV UL13, VZV 47, and ubiquitous cellular kinase CK II. Unlike the cellular enzyme, the viral kinases are not inhibited by heparin (32). By detecting phosphoamino acids, we found that BGLF4 is phosphorylated on serine and threonine residues (Fig. 7). BGLF4 protein appeared to be expressed principally in the cytoplasm of transfected cells (Fig. 8). We found difficulty in detecting the expression of the BGLF4 protein in TPA-induced B95-8 cells (data not shown), perhaps due to only a very small amount of protein being expressed for a short period of time during the viral replication cycle or to the insensitivity of the rabbit antiserum. Therefore it is still possible that authentic BGLF4 might be associated with other viral proteins in EBV-infected cells and might exhibit a different subcellular localization.
The BGLF4 expressed in E. coli was mostly in the insoluble fraction, and our inability to demonstrate phosphorylation activity using a renaturation gel activity assay could imply that certain posttranslational modifications or a particular conformation is required for enzyme activity. In our original experiments, we used BGLF4 expressed transiently in eukaryotic cells and BGLF4-specific antiserum in an immune complex kinase assay (data not shown). In order to increase further the specificity of immunoprecipitation, an EBNA-1 tag system was used to demonstrate BGLF4 phosphorylation. Because a recent study showed that herpesvirus ribonucleotide reductase R1 is a good substrate for host cell protein kinases but is not a protein kinase itself (30), we used immunocomplexes washed with 1 and 3 M NaCl in the kinase experiment and obtained similar kinetics of kinase activity. Although it is difficult to believe that a noncovalent bond between two molecules would tolerate a 3 M NaCl wash, we suggest that the kinase activity must be important for the virus regardless of whether BGLF4 is itself a protein kinase or whether the kinase activity is derived from a strongly linked cellular kinase. Indeed, this is the case for HSV R1. Although there are still arguments about the kinase activity of HSV R1, the PK domain of HSV-2 was demonstrated to be required for immediate-early gene expression and virus growth (48).
In addition to autophosphorylation of BGLF4, phosphorylation of EA-D in vitro supports our hypothesis of the significance of BGLF4 activity. EA-D is a phosphorylated protein, and the degree of phosphorylation increases as the lytic cycle of EBV replication progresses (28, 33). EA-D is a DNA polymerase accessory factor (33) and is also an important transactivator during lytic infection (55). Since the effect of phosphorylation on EA-D is still not clear, it will be interesting to determine the contribution of BGLF4 to the phosphorylation of EA-D in vivo.
The UL13 protein kinase of HSV-1 is expressed late in the replication cycle and, directly or indirectly, phosphorylates ICP22, as demonstrated by UL13 deletion mutants (13, 43). HSV UL13 protein is packaged into virions (41). Recently, it was found that HSV UL13 and ICP22, which is a substrate of UL13, are both required for HSV-induced modification of the large subunit of RNA polymerase II (Pol II) after viral infection (35). The altered phosphorylation state of Pol II was demonstrated to promote late viral transcription in some cell lines.
ORF47 and ORF66 of VZV were reported to be dispensable for propagation of VZV in vitro, but a double mutant lacking both proteins grows to a reduced titer (24, 51). ORF47 also phosphorylates ORF62, the major immediate-early transactivator (40). In addition, a recent study involving inoculation of knockout virus-infected human thymus/liver or skin implants into SCID-hu mice showed that the ORF47 protein was required for virus growth in human T cells and skin (38). In summary, the functions of UL13 homologs in herpesviruses include modifying viral transactivators and cellular macromolecular machinery.
For another DNA virus, vaccinia virus, B1, one of the two viral protein kinases, is required for viral DNA replication (45). One of the substrates for B1 kinase has been demonstrated in vitro to be the viral single-stranded DNA binding protein (2). It will be interesting to determine whether BGLF4 also plays a role in modifying cellular transcriptional or DNA replication machinery during EBV infection.
Definition of catalytic domains of nine motifs was achieved by alignment of 65 sequences by Hanks and Hunter (23). The nine conserved boxes of sequences were used by Chee et al. (5) in the identification of herpesvirus UL13 group protein kinases. The so-called catalytic lysine, in the second subdomain sequence AXKXO (where O denotes a hydrophobic residue), was found to be functionally irreplaceable in some protein kinases (10). The lysine residue of UL13 of pseudorabies virus was mutated by DeWind et al. (20), and this mutant was shown to lose kinase activity. This region is highly conserved among alphaherpesviruses, but the sequence of BGLF4 in this region, TVKLY, is less conserved. Furthermore, the distance between subdomain I (putative nucleotide binding site) and the lysine is four amino acids less in BGLF4 (Fig. 10A). The deletion mutant study demonstrated that amino acids 85 to 107 are dispensable for BGLF4 phosphorylation, whereas the sequence between amino acids 35 and 65 is a possible inhibition domain or autoregulatory domain. Deletion of amino acids 35 to 65 increased the phosphorylation of BGLF4 about twofold (Fig. 11). The N terminus, amino acids 1 to 26, was found to be essential for BGLF4 phosphorylation, the amino acid sequence in this region contains multiple serine residues and a possible N-linked glycosylation site (Fig. 12). Further experiments using purified BGLF4 in transphosphorylation assays need to be conducted to determine whether this region is the major phosphorylation site of BGLF4 or is required for catalytic activity.
FIG. 12.
The sequence of the first 26 amino acids of BGLF4 contains multiple serine residues and a possible glycosylation site (underlined).
During the preparation of this paper, we noted that ORF36 of HHV-8, another gammaherpesvirus, was reported to be more active than the thymidine kinase homologue in phosphorylation of ganciclovir and ganciclovir-mediated cell death (4). Since the amino acid sequences of BGLF4 and HHV-8 ORF36 have 50% homology, the possibility that BGLF4 can also phosphorylate ganciclovir is under investigation in our laboratory.
ACKNOWLEDGMENTS
We thank Tim J. Harrison of the Royal Free and University College School of Medicine (University College London) for critical reading of the manuscript. We also thank Li Wha Huang for the P3 laboratory facility for recombinant vaccinia virus infection and Yu-Fan Chen for photography.
This research was supported by the National Science Council, grants NSC88-2314-B-002-154 and NSC89-2320-B002-045.
REFERENCES
- 1.Ali M A, Prakash S S, Jariwalla R. Localization of the antigenic sites and intrinsic protein kinase domain within a 300 amino acid segment of the ribonucleotide reductase large subunit from herpes simplex virus type 2. Virology. 1992;187:360–367. doi: 10.1016/0042-6822(92)90328-m. [DOI] [PubMed] [Google Scholar]
- 2.Beaud G, Sharif A, Topa-Masse A, Leader D P. Ribosomal protein S2/Sa kinase purified from HeLa cells infected with vaccinia virus corresponds to the B1R protein kinase and phosphorylates in vitro the viral ssDNA-binding protein. J Gen Virol. 1994;75:283–293. doi: 10.1099/0022-1317-75-2-283. [DOI] [PubMed] [Google Scholar]
- 3.Branch D R, Mills G B. pp60c-src expression is induced by activation of normal human T lymphocytes. J Immunol. 1995;154:3678–3685. [PubMed] [Google Scholar]
- 4.Cannon J S, Hamzeh F, Moore S, Nicholas J, Ambinder R F. Human herpesvirus 8-encoded thymidine kinase and phosphotransferase homologues confer sensitivity to ganciclovir. J Virol. 1999;73:4786–4793. doi: 10.1128/jvi.73.6.4786-4793.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chee M S, Lawrence G L, Barrell B G. Alpha-, beta- beta- and gammaherpesviruses encode a putative phosphotransferase. J Gen Virol. 1989;70:1151–1160. doi: 10.1099/0022-1317-70-5-1151. [DOI] [PubMed] [Google Scholar]
- 6.Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen M-R, Yang J-F, Hsu T-Y, Liu M-Y, Chen J-Y, Yang C-S. Use of bacterially expressed GST/EBNA-1 fusion proteins for detection of antibodies in sera from patients with nasopharyngeal carcinoma and healthy donors. Chin J Microbiol Immunol (Taipei) 1996;29:65–79. [PubMed] [Google Scholar]
- 8.Chen M R, Tsai C H, Wu F F, Kan S H, Yang C S, Chen J Y. The major immunogenic epitopes of Epstin-Barr virus (EBV) nuclear antigen 1 are encoded by sequence domains which vary among nasopharyngeal carcinoma biopsies and EBV-associated cell lines. J Gen Virol. 1999;80:447–455. doi: 10.1099/0022-1317-80-2-447. [DOI] [PubMed] [Google Scholar]
- 9.Chen M R, Hsu T Y, Chen J Y, Yang C S. Molecular characterization of a cDNA clone encoding the Epstein-Barr virus (EBV) DNase. J Virol Methods. 1990;29:127–142. doi: 10.1016/0166-0934(90)90107-q. [DOI] [PubMed] [Google Scholar]
- 10.Chen W S, Lazar C S, Poenie M, Tsien R Y, Gill G N, Rosenfeld M G. Requirement for intrinsic protein tyrosine kinase in the immediate and late actions of the EGF receptor. Nature (London) 1987;328:820–823. doi: 10.1038/328820a0. [DOI] [PubMed] [Google Scholar]
- 11.Chow K C, Ma J, Lin L S, Chi K H, Yen S H, Liu S M, Liu W T, Chen W K, Chang T H, Chen K Y. Serum responses to the combination of Epstein-Barr virus antigens from both latent and acute phases in nasopharyngeal carcinoma: complementary test of EBNA-1 with EA-D. Cancer Epidemiol Biomarkers Prev. 1997;6:363–368. [PubMed] [Google Scholar]
- 12.Chung T D, Wymer J P, Smith C C, Kulka M, Aurelian L. Protein kinase activity associated with the large subunit of herpes simplex virus type 2 ribonucleotide reductase (ICP10) J Virol. 1989;63:3389–3398. doi: 10.1128/jvi.63.8.3389-3398.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coulter L J, Moss H W M, Lang J, McGeoch D J. A mutant of herpes simplex virus type 1 in which the UL13 protein kinase gene is disrupted. J Gen Virol. 1993;74:387–395. doi: 10.1099/0022-1317-74-3-387. [DOI] [PubMed] [Google Scholar]
- 14.Cunningham C, Davison A J, Dolan A, Frame M C, McGeoch D J, Meredith D M, Moss H W M, Orr A C. The UL13 virion protein of herpes simplex virus type 1 is phosphorylated by a novel virus-induced protein kinase. J Gen Virol. 1992;73:303–311. doi: 10.1099/0022-1317-73-2-303. [DOI] [PubMed] [Google Scholar]
- 15.Daibata M, Humphreys R E, Sairenji T. Phosphorylation of the Epstein-Barr virus BZLF1 immediate-early gene product ZEBRA. Virology. 1992;188:916–920. doi: 10.1016/0042-6822(92)90553-2. [DOI] [PubMed] [Google Scholar]
- 16.Daikoku T, Kurachi R, Tsurumi T, Nishiyama Y. Identification of a target protein of US3 protein kinase of herpes simplex virus type 2. J Gen Virol. 1994;75:2065–2068. doi: 10.1099/0022-1317-75-8-2065. [DOI] [PubMed] [Google Scholar]
- 17.Daikoku T, Yamashita Y, Tsurumi T, Nishiyama Y. The US3 protein kinase of herpes simplex virus type 2 is associated with phosphorylation of the UL12 alkaline nuclease in vitro. Arch Virol. 1995;140:1637–1644. doi: 10.1007/BF01322537. [DOI] [PubMed] [Google Scholar]
- 18.Dasmahapatra D B, Rozhan E J, Schwarz J. BD7, a novel cell free expression vector with an efficient translation initiation signal. Nucleic Acids Res. 1987;15:3933. doi: 10.1093/nar/15.9.3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davison A J, Scott J E. The complete DNA sequence of varicella-zoster virus. J Gen Virol. 1986;67:1759–1816. doi: 10.1099/0022-1317-67-9-1759. [DOI] [PubMed] [Google Scholar]
- 20.DeWind N, Domen J, Berns A. Herpesviruses encode an unusual protein-serine/threonine kinase which is nonessential for growth in cultured cells. J Virol. 1992;66:5200–5209. doi: 10.1128/jvi.66.9.5200-5209.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feighny R J, Farrell M P, Pagano J S. Polypeptide synthesis and phosphorylation in Epstein-Barr virus-infected cells. J Virol. 1980;34:455–463. doi: 10.1128/jvi.34.2.455-463.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Glover C V, Allis C D. Enzyme activity dot blot assaying protein kinases. Methods Enzymol. 1991;85:85–90. doi: 10.1016/0076-6879(91)00128-j. [DOI] [PubMed] [Google Scholar]
- 23.Hanks S K, Hunter T. Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J. 1995;9:576–596. [PubMed] [Google Scholar]
- 24.Heineman T C, Seidel K, Cohen J I. The varicella-zoster virus ORF66 protein induces kinase activity and is dispensable for viral replication. J Virol. 1996;70:7312–7317. doi: 10.1128/jvi.70.10.7312-7317.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Higgins D G, Bleasby A J, Fuchs R. CLUSTAL V: improved software for multiple sequence alignment. Comput Appl Biosci. 1992;8:189–191. doi: 10.1093/bioinformatics/8.2.189. [DOI] [PubMed] [Google Scholar]
- 26.Kawaguchi Y, Matsumura T, Roizman B, Hirai K. Cellular elongation factor 1δ is modified in cells infected with representative alpha-, beta-, or gammaherpesviruses. J Virol. 1999;73:4456–4460. doi: 10.1128/jvi.73.5.4456-4460.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kawaguchi Y, Van Sant C, Roizman B. Eukaryotic elongation factor 1δ is hyperphosphorylated by the protein kinase encoded by the UL13 gene of herpes simplex virus 1. J Virol. 1998;72:1731–1736. doi: 10.1128/jvi.72.3.1731-1736.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kiehl A, Dorsky D I. Cooperation of EBV DNA polymerase and EA-D(BMRF1) in vitro and colocalization in nuclei of infected cells. Virology. 1991;182:330–340. doi: 10.1016/0042-6822(91)90849-7. [DOI] [PubMed] [Google Scholar]
- 29.Kolman J L, Taylor N, Marshak D R, Miller G. Serine-173 of the Epstein-Barr virus ZEBRA protein is required for DNA binding and is a target for casein kinase II phosphorylation. Proc Natl Acad Sci USA. 1993;90:10115–10119. doi: 10.1073/pnas.90.21.10115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Langelier Y, Champoux L, Hamel M, Guilbault C, Lamarche C, Gaudreau P, Mawwie B. The R1 subunit of herpes simplex virus ribonucleotide reductase is a good substrate for host cell protein kinases but is not itself a protein kinase. J Biol Chem. 1998;273:1435–1443. doi: 10.1074/jbc.273.3.1435. [DOI] [PubMed] [Google Scholar]
- 31.Lawrence G L, Chee M, Craxton M A, Gompels U A, Honess R W, Barrell B G. Human herpesvirus 6 is closely related to human cytomegalovirus. J Virol. 1990;64:287–299. doi: 10.1128/jvi.64.1.287-299.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leader D P. Viral protein kinases and protein phosphatases. Pharmacol Ther. 1993;59:343–389. doi: 10.1016/0163-7258(93)90075-o. [DOI] [PubMed] [Google Scholar]
- 33.Li J S, Zhou B S, Dutschman G E, Grill S P, Tan R-S, Chen Y-C. Association of Epstein-Barr virus early antigen diffuse component and virus-specified DNA polymerase activity. J Virol. 1987;61:2947–2949. doi: 10.1128/jvi.61.9.2947-2949.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Littler E, Stuart A D, Chee M S. Human cytomegalovirus UL97 open reading frame encodes a protein that phosphorylates the antiviral nucleoside analog ganciclovir. Nature. 1992;358:160–162. doi: 10.1038/358160a0. [DOI] [PubMed] [Google Scholar]
- 35.Long M C, Leong V, Scharrer P A, Spencer C A, Rice S A. ICP22 and UL13 protein kinase are both required for herpes simplex virus-induced modification of the large subunit of RNA polymerase II. J Virol. 1999;73:5593–5604. doi: 10.1128/jvi.73.7.5593-5604.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luo J H, Aurelian L. The transmembrane helical segment but not the invariant lysine is required for the kinase activity of the large subunit of herpes simplex virus type 2 ribonucleotide reductase (ICP10) J Biol Chem. 1992;267:9645–9653. [PubMed] [Google Scholar]
- 37.McGeoch D J, Davison A J. Alphaherpesviruses possess a gene homologous to the protein kinase gene family of eukaryotes and retroviruses. Nucleic Acids Res. 1986;14:1765–1777. doi: 10.1093/nar/14.4.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moffat J F, Zerboni L, Sommer M H, Heineman T C, Cohen J I, Kaneshima H, Arvin A M. The ORF47 and ORF66 putative protein kinases of varicella-zoster virus determine tropism for human T cells and skin in the SCID-hu mouse. Proc Natl Acad Sci USA. 1998;95:11969–11974. doi: 10.1073/pnas.95.20.11969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ng T I, Grose C. Serine protein kinase associated with varicella-zoster virus ORF47. Virology. 1992;191:9–18. doi: 10.1016/0042-6822(92)90161-h. [DOI] [PubMed] [Google Scholar]
- 40.Ng T I, Keenan L, Kinchington P R, Grose C. Phosphorylation of varicella-zoster virus open reading frame (ORF) 62 regulatory product by viral ORF 47-associated protein kinase. J Virol. 1994;68:1350–1359. doi: 10.1128/jvi.68.3.1350-1359.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Overton H A, McMillan D J, Klavinskis L S, Hope L, Ritchie A J, Wong-kai-in P. Herpes simplex virus type 1 gene UL13 encodes a phosphoprotein that is a component of the virion. Virology. 1992;190:184–192. doi: 10.1016/0042-6822(92)91204-8. [DOI] [PubMed] [Google Scholar]
- 42.Possee R D, Sun T P, Howard S C, Ayres M D, Hill-Perkins M, Gearing K L. Nucleotide sequence of the Autographa californica nuclaer polyhedrosis 9.4 kbp EcoRI-I and -R (polyhedrin gene) region. Virology. 1991;185:229–241. doi: 10.1016/0042-6822(91)90770-c. [DOI] [PubMed] [Google Scholar]
- 43.Purves F C, Ogle W O, Roizman B. Processing of the herpes simplex virus regulatory protein α22 mediated by the UL13 protein kinase determines the accumulation of a subset of α and γ mRNAs and proteins in infected cells. Proc Natl Acad Sci USA. 1993;90:6701–6705. doi: 10.1073/pnas.90.14.6701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Purves F C, Roizman B. The UL13 gene of herpes simplex virus 1 encodes the functions for posttranslational processing associated with phosphorylation of the regulatory protein α22. Proc Natl Acad Sci USA. 1992;89:7310–7314. doi: 10.1073/pnas.89.16.7310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rempel R E, Anderson M K, Evans E, Traktman P. Temperature-sensitive vaccinia virus mutants identify a gene with an essential role in viral replication. J Virol. 1990;64:574–583. doi: 10.1128/jvi.64.2.574-583.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rempel R E, Traktman P. Vaccinia virus B1 kinase: phenotypic analysis of temperature-sensitive mutants and enzymatic characterization of recombinant proteins. J Virol. 1992;66:4413–4426. doi: 10.1128/jvi.66.7.4413-4426.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Russo J J, Bohenzky R A, Chien M C, Chen J, Yan M, Maddalena D, Parry J P, Peruzzi D, Edelamn I S, Chang Y, Moore P S. Nucleotide sequence of the Kaposi sarcoma-associated herpes virus (HHV8) Proc Natl Acad Sci USA. 1996;93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith C C, Peng T, Kulka M, Aurelian L. The PK domain of the large subunit of herpes simplex virus type 2 ribonucleotide reductase (ICP10) is required for immediate-early gene expression and virus growth. J Virol. 1998;72:9131–9141. doi: 10.1128/jvi.72.11.9131-9141.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith R F, Smith T F. Identification of new protein kinase-related genes in three herpesviruses, herpes simplex virus, varicella-zoster virus, and Epstein-Barr virus. J Virol. 1989;63:450–455. doi: 10.1128/jvi.63.1.450-455.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Soderling T R. Protein kinases and phosphatases: regulation by autoinhibitor domains. Biotechnol Appl Biochem. 1993;18:185–200. [PubMed] [Google Scholar]
- 51.Stevenson D, Colman K L, Davison A J. Characterization of the putative protein kinases specified by varicella-zoster virus genes 47 and 66. J Gen Virol. 1994;75:317–326. doi: 10.1099/0022-1317-75-2-317. [DOI] [PubMed] [Google Scholar]
- 52.Sullivan V, Talarico C L, Stanat S C, Davis M, Coen D M, Biron K K. Protein kinase homologue controls phosphorylation of ganciclovir in human cytomegalovirus-infected cells. Nature. 1992;358:162–164. doi: 10.1038/358162a0. [DOI] [PubMed] [Google Scholar]
- 53.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weigh matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tsai C-H, Liu M-T, Chen M-R, Lu J, Yang H-L, Chen J-Y, Yang C-S. Characterization of monoclonal antibodies to the Zta and DNase proteins of Epstein-Barr virus. J Biomed Sci. 1997;4:69–77. doi: 10.1007/BF02255596. [DOI] [PubMed] [Google Scholar]
- 55.Zhang Q, Holley-Guthrie E, Ge J-Q, Dorsky D, Kenney S. The Epstein-Barr virus (EBV) DNA polymerase accessory protein, BMRF1, activates the essential downstream component of the EBV oriLyt. Virology. 1997;230:22–34. doi: 10.1006/viro.1997.8470. [DOI] [PubMed] [Google Scholar]