Abstract
Equine infectious anemia virus (EIAV) infection of horses is characterized by recurring cycles of disease and viremia that typically progress to an inapparent infection in which clinical symptoms are absent as host immune responses maintain control of virus replication indefinitely. The dynamics of EIAV viremia and its association with disease cycles have been well characterized, but there has been to date no comprehensive quantitative analyses of the specific tissue sites of EIAV infection and replication in experimentally infected equids during acute disease episodes and during asymptomatic infections in long-term inapparent carriers. To characterize the in vivo site(s) of viral infection and replication, we developed a quantitative competitive PCR assay capable of detecting 10 copies of viral DNA and a quantitative competitive reverse transcription-PCR assay with a sensitivity of about 30 copies of viral singly spliced mRNA. Animals were experimentally infected with one of two reference viruses: the animal-passaged field isolate designated EIAVWyo and the virulent cell-adapted strain designated EIAVPV. Tissues and blood cells were isolated during the initial acute disease or from asymptomatic animals and analyzed for viral DNA and RNA levels by the respective quantitative assays. The results of these experiments demonstrated that the appearance of clinical symptoms in experimentally infected equids coincided with rapid widespread seeding of viral infection and replication in a variety of tissues. During acute disease, the predominant cellular site of viral infection and replication was the spleen, which typically accounted for over 90% of the cellular viral burden. In asymptomatic animals, viral DNA and RNA persisted in virtually all tissues tested, but at extremely low levels, a finding indicative of tight but incomplete immune control of EIAV replication. During all disease states, peripheral blood mononuclear cells (PBMC) were found to harbor less than 1% of the cellular viral burden. These quantitative studies demonstrate that tissues, rather than PBMC, constitute the predominant sites of virus replication during acute disease in infected equids and serve as resilient reservoirs of virus infection, even in the presence of highly effective immune responses that maintain a stringent control of virus replication in long-term inapparent carriers. Thus, these observations with EIAV, a predominantly macrophage-tropic lentivirus, highlight the role of tissues in sequestering lentiviral infections from host immune surveillance.
Equine infectious anemia virus (EIAV) is unique among lentiviruses in that the clinical course of infection in equids results initially in a rapid and dynamic series of clearly demarcated cycles of disease and associated viremia that begin by 3 weeks postinfection and continue at irregular intervals separated by weeks or months (reviewed in reference 25). Disease cycles last 3 to 5 days and are characterized by fever, diarrhea, lethargy, edema, anemia, and thrombocytopenia. This stage of disease, defined as chronic equine infectious anemia (EIA), typically lasts about 8 to 12 months postinfection, with the frequency and severity of clinical episodes decreasing with time. In contrast to the progressive degenerative disease associated with most lentiviral infections, horses infected with EIAV typically make a transition during the first year postinfection from chronic EIA to an inapparent infection in which clinical symptoms are absent and viremia is usually undetectable for the remainder of the animal's life span of up to about 20 years. Thus, the EIAV systems provides a novel model in which to examine the dynamics of lentivirus replication during clearly defined cycles of disease and during long-term asymptomatic infections.
Several lines of evidence indicate that the control of EIAV replication and disease in long-term inapparent carriers is mediated by virus-specific host immune responses that evolve during the first year postinfection to achieve an enduring effective suppression of virus replication. For example, experimental infection of foals with severe combined immunodeficiency results in a progressive infection leading to death, demonstrating the necessity of the host immune system in accomplishing the temporal control of virus replication associated with infection of immunocompetent horses (29). In addition, it has been shown that severe stress or treatment of long-term inapparent carriers with immunosuppressive drugs can cause recrudescence of viremia and disease, even after decades of clinical quiescence (16, 48). Finally, it has been demonstrated that transfer of whole blood from long-term inapparent carriers to naive horses results in EIAV infection and disease in the recipient horses (12). Taken together, these observations demonstrate the lack of attenuation of the infecting virus during persistent infection and the importance of host immune responses in establishing and maintaining control of EIAV replication and disease. The same experiments, however, also clearly demonstrate the persistence of virulent EIAV infection in infected horses during asymptomatic and nonviremic periods, either during chronic EIA or in long-term inapparent infections. This persistence of EIAV infection in the presence of enduring, robust, and highly suppressive host immunity raises a number of interesting questions about the reservoirs of EIAV infection that are able to escape host immune surveillance.
The primary target of EIAV in vivo are cells of the monocyte/macrophage lineage (25), although there has recently been a report of limited infection of macrovascular endothelial cells in the renal tissues of a long-term inapparent carrier horse (22). Infection of blood monocytes with EIAV results in a nonproductive infection, and differentiation of virus-infected monocytes to macrophages is required to activate virus replication (21, 43). This pattern of infection has led to the model that virus-infected monocytes may serve as “Trojan horses” that can widely disseminate EIAV infection to tissues without detection by the host immune system, suggesting that monocytes may sequester EIAV during asymptomatic infection. The identification of EIAV infection of macrovascular endothelial cells suggests that this cell type may also serve as a reservoir for virus infection, at least in long-term inapparent infections. The role, if any, of endothelial cell infection in acute disease remains to be defined.
The primary sites of EIAV replication in persistently infected horses has been examined by immunofluorescent techniques to detect viral antigens (23), by Southern blot analyses to detect proviral DNA (35), and more recently by qualitative PCR techniques to detect viral DNA or RNA (14, 27, 42). The results of these studies uniformly demonstrate that the high levels of viremia observed during acute EIA are associated with high levels of virus replication in macrophage-rich tissues, including liver, spleen, and kidney; peripheral blood mononuclear cells (PBMC) and other tissues appeared to contain only minor levels of virus infection, despite the high levels of virus in the blood. In contrast to the abundance of viral antigens and DNA observed in tissues from EIAV-infected horses during acute disease, qualitative analyses of EIAV infection sites by Southern blot (35) and PCR (14, 42) in a limited number of inapparent carrier horses indicate low levels of EIAV infection associated with tissue macrophages and typically undetectable levels of EIAV DNA in PBM. These general patterns of EIAV infection were confirmed recently in an informative study by Oaks et al. (26) using a combination of PCR and in situ hybridization techniques to elucidate tissue macrophages as the predominant site of EIAV infection during clinical and subclinical infections.
The previous studies of the sites of EIAV infection in experimentally infected horses provide an important qualitative description of virus infection in vivo, but there has been to date no comprehensive quantitative analyses of EIAV DNA and RNA in experimentally infected horses to ascertain the relative importance of different tissues in virus production during acute disease or in persistence during inapparent infections. In addition, the majority of these published studies of EIAV infection during acute disease are based on experimental infections with the highly virulent EIAVWyo strain of virus that usually causes death within several weeks postinfection, in marked contrast to the chronic EIA observed in most EIAV infections in the field. The highly virulent nature of the EIAVWyo raises some concerns about the relevance of this experimental infection model to the more typical infections that result in chronic EIAV that progresses to a long-term inapparent infection. Finally, the characterization of EIAV replication in vivo has been limited by the use of only one or two animals in many published studies.
Therefore, we sought in this current study to characterize viremia levels and the tissue sites of EIAV infection and replication during symptomatic and asymptomatic infections by using newly developed sensitive and quantitative PCR and reverse transcriptase PCR (RT-PCR) techniques to measure viral DNA and RNA, respectively. For these studies we employed the well-characterized EIAVPV reference strain (25, 38), which has been used extensively in our laboratory in experimental infections to reliably produce in equids a clinical progression from chronic EIA to a long-term inapparent infection. For comparison to previous studies, we also examined virus infection and replication during acute disease in horses inoculated with the highly virulent EIAVWyo. The results of these studies provide for the first time a comprehensive quantitative analysis of the in vivo sites of viral infection and replication in various tissues from experimentally infected animals during the initial acute disease and during long-term inapparent infection.
MATERIALS AND METHODS
Virus strains and experimental animals.
The highly virulent wild-type EIAVWyo strain, an animal-passaged isolate, was produced as serum taken during acute disease in an experimentally infected horse, as described earlier (35). The titer of the EIAVWyo stock was 106 horse infectious doses per ml. The cell-adapted pathogenic EIAVPV strain was derived, and the titers were determined as described elsewhere (49). Infectious stocks of EIAVPV were grown, and titers were determined in primary fetal equine kidney cell cultures. The titer of EIAVPV stock was 106.5 50% tissue culture infectious doses (TCID50) per ml.
Two thoroughbred horses (298 and 94-07) and four mixed-breed ponies (112, 123, 18, and 72) were used in these studies. Prior to infection, animals were shown to be seronegative for EIAV-reactive antibodies by the agar immunodiffusion test (IDEXX Laboratories, Westbrook, Mass.), and viral RNA was not detected in the plasma by RT-PCR analyses (see below). The two horses were intravenously inoculated with 1 ml of EIAVWyo serum (Table 1). Ponies 112 and 123 were inoculated intravenously with 106 TCID50 of the EIAVPV strain, while ponies 18 and 72 were inoculated intravenously with 300 and 104 TCID50 of the EIAVPV strain, respectively. All animals were monitored daily via physical exams and rectal temperatures for clinical signs of EIA.
TABLE 1.
Clinical profiles and levels of EIAV in the plasma of experimentally infected animals
| Animal no. | Virus strain | Infectious dosea | Euthanized (days postinfection) | Clinical status | Plasma viremia (TCID50/ml)b | Plasma RNAc |
|---|---|---|---|---|---|---|
| 298 | EIAVWyo | 1 × 106 | 12 | Acute | 105.0 | 3.1 × 108 |
| 94-07 | EIAVWyo | 1 × 106 | 11 | Acute | 103.9 | 3.6 × 107 |
| 112 | EIAVPV | 1 × 106 | 9 | Acute | 103.6 | 2.1 × 107 |
| 123 | EIAVPV | 1 × 106 | 13 | Acute | 104.7 | 4.2 × 107 |
| 18 | EIAVPV | 1 × 104 | 538 | Asymptomatic | Negd | Neg |
| 72 | EIAVPV | 3 × 102 | 506 | Asymptomatic | Neg | 1.2 × 102 |
EIAVWyo, horse infectious doses; EIAVPV, TCID50.
TCID50 per ml as determined in primary MDM cultures from plasma obtained the day of euthanization.
Copies of virion genomic RNA per ml of plasma as determined by RT-PCR from samples obtained the day of euthanization.
Neg, negative for detectable infectious virus or viral RNA.
Clinical samples.
Heparinized whole blood was collected by jugular venipuncture from experimentally infected animals during defined febrile episodes and from asymptomatic animals. Plasma and PBMC were isolated by discontinuous density gradient centrifugation. Serum samples were collected. Six animals were euthanized by administration of a lethal intravenous injection of Beuthanasia, at which time replicate tissues samples were collected and flash frozen immediately in liquid nitrogen. Tissue samples from the liver, spleen, medulla and cortex of the kidney, peripheral lymph nodes including the sublingual and mandibular nodes, and bone marrow were collected from all animals. The cerebrum, medulla, and cerebellum from the brain were collected from three animals (horse 94-07 and ponies 112 and 123). Tissue, cellular, plasma, and serum samples were stored at −80°C until isolation of nucleic acids.
Nucleic acid purification from tissue samples.
Total cellular DNA was purified by standard techniques (40). Briefly, tissues were immersed in liquid nitrogen and immediately pulverized. Samples were then suspended in 1 ml of 0.1% sodium dodecyl sulfate (SDS)–10 mM EDTA per 100 mg of tissue or per 106 PBMC, digested with 100 μg of proteinase K per ml at 37°C for a minimum of 5 h, followed by phenol-chloroform extraction and ethanol precipitation. Pelleted DNA was resuspended in 10 mM Tris-HCl–1 mM EDTA by gentle rocking at room temperature overnight. The concentration of the DNA was determined spectrophotometrically, and the quality of the DNA was assayed by gel electrophoresis. Samples were stored at −20°C until quantification of the cellular genomic and EIAV viral DNA content.
RNA was extracted from pulverized tissue samples and PBMC using either 1 ml of RNAzol (Biotecz) or TRIzol (BRL) per 100 mg of sample or per 106 PBMC according to the manufacturer's protocol. RNA was resuspended in diethyl pyrocarbonate (DEPC)-treated water. The concentration of the RNA was determined by spectrophotometry, and the integrity of the RNA was monitored by formaldehyde denaturing gel electrophoresis. Samples were stored at −70°C until assayed for viral and cellular RNAs.
Virion-associated genomic RNA was recovered from plasma samples by ultracentrifugation to pellet virus and extracted as described earlier (19). Briefly, a minimum of 1 ml of each plasma sample was centrifuged at 100,000 × g for 45 min in a type 55.1 rotor (Beckman Instruments, Palo Alto, Calif.) at 4°C. The supernatant was aspirated, and the pellet resuspended in 0.1% SDS. RNA was extracted using 1 ml of TRIzol as described above. The RNA was precipitated in the presence of 20 μg of glycogen as carrier. RNA was pelleted by centrifugation at 12,000 × g for 15 min at 4°C and resuspended in 10 to 20 μl of DEPC-treated water. All samples were stored at −70°C until quantification of the viral RNA.
Quantitative PCR analyses.
The viral DNA burden was determined by using a competitive PCR (QC-PCR) assay to quantify viral DNA (30). To optimize detection of variant quasispecies, highly conserved gag-specific primers were chosen based on alignment of all of the EIAV sequences in the database using the Genetics Computer Group (Madison, Wis.) package analyses software (10). A SmaI-XbaI fragment of the gag gene was cloned into a trpLE vector (3). The competitor template containing a 55-bp deletion at the unique ApaI site was generated by Bal31 exonuclease digestion. The competitor template was quantified by spectrophotometry and limiting-dilution analyses (data not shown). Amplification of wild-type and competitor templates by using the sense gag11 (5′-ATGTATGCTTGCAGAGACATTG-3′) and antisense gag34 (5′-GCTGACTCTTCTGTTGTATCG-3′) primers yielded 310- and 255-bp products, respectively. The PCR reaction contained 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl, 0.001% gelatin, 0.05 mM concentrations of each deoxyribonucleoside triphosphate, 12.5 pmol of each primer, 1 μCi of [α-32P]dATP (3,000 Ci/mmol; NEN, Boston, Mass.), and 2.5 U of AmpliTaq DNA polymerase (Perkin-Elmer, Norwalk, Conn.) in a final volume of 25 μl. The thermal cycling conditions included an initial hot start at 99°C, followed by 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min for 30 cycles, with a final 10-min extension at 72°C. PCR products were separated by electrophoresis on 6% acrylamide gels containing 8 M urea. Gels were fixed in a 10% methanol–10% acetic acid solution and dried. The radioactivity incorporated in each band was quantified by using phosphorimaging technologies (Molecular Dynamics, Sunnyvale, Calif.). Preliminary experiments were performed to determine the amount of sample DNA that was within the linear range of the amplification reaction. Addition of carrier DNA to maintain a constant mass had a minimal effect on the amplification efficiency; therefore, carrier DNA was not included in the reactions.
Independent amplification of a single-copy cellular genomic sequence was performed to control for sample loss or DNA degradation and to normalize the cellular DNA content. A 94-bp fragment of the equine sodium channel gene was amplified by using the sense (5′-GGGAGTGTGTGCTCAAGATGT-3′) and antisense (5′-AATGGACAGGATGACAACCAC-3′) primers, as described earlier (37). The concentrations of the PCR reagents were the same as those described above, except that 6.25 pmol of each primer was used. Half-log dilutions from 10 to 100 ng of the sample DNA were used. The thermal cycling conditions were 94°C for 45 s and 65°C for 4 min for 30 cycles, followed by a 10-min extension at 72°C. PCR products were electrophoretically separated and quantified as described above. A standard curve was generated by linear regression analysis of known amounts of cloned DNA. The cellular equivalents (CE) of all samples were calculated by using the equation of the line, as described previously (44).
Quantitative RT-PCR analyses.
To determine the levels of EIAV replication in various tissues, the amount of singly spliced viral mRNA was quantified by competitive RT-PCR (QC-RT-PCR [30]). A 910-bp cDNA fragment from singly spliced EIAV mRNA (33) was subcloned into a low-copy vector, pLG338 (6), modified to contain a T7 RNA promoter site. A 104-bp deletion was generated by ligation-mediated PCR techniques. Wild-type and competitor transcripts were generated by in vitro transcription using a commercially available T7 RNA polymerase kit (Ambion) according to the manufacturer's directions. Transcripts were subjected to a second RNase-free DNase (Promega) digestion, organic extraction, and ethanol precipitation. RNA was resuspended in DEPC-treated water and quantified by spectrophotometry. Reverse transcription of defined quantities of sample RNA and titered quantities of competitor transcripts was performed with Superscript II (BRL) according to the manufacturer's recommendations. cDNA was synthesized by using 12.5 pmol of an EIAV env-specific primer (5′-GTATTCCTCCAGTAGTTC-3′). Radiolabeled amplification of 2 μl of the RT reaction was performed by using the sense (5′-TGAACCTGGCTGATCGTAGGATCC-3′) and antisense (5′-GTATTCCTCCAGTAGTTCCTGCTAAGC-3′) primers. The concentration of the PCR reagents was as stated above for amplification of viral DNA. A 510-bp product and a 406-bp product were produced from the wild-type and competitor templates, respectively. PCR products were separated by polyacrylamide gel electrophoresis and quantified as described for viral DNA. Preliminary RT-PCR reactions were performed to determine the amount of sample RNA that fell within the linear range of the assay.
An equine glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNA clone was produced to provide a cellular marker gene. RT-PCR was performed using commercially available primers (Clontech) complementing highly homologous cross-species GAPDH-specific sequences. RT reactions were carried out as described above, except that 10 pmol of the antisense primer (5′-TCCACCACCCTGTTGCTGTA-3′) and 1 μg of cellular RNA from an EIAVWyo-infected animal was used. Then, 2 μl of the RT product was amplified with 10 pmol of each of the sense (5′-ACCACAGTCCATGCCATCAC-3′) and the above-described antisense primers and reagent concentrations analogous to the viral DNA reactions. The thermal cycling conditions were 94°C for 45 s, 45°C for 45 s, and 72°C for 2 min for 35 cycles, followed by a 10-min extension at 72°C. The resultant 452-bp product was purified by agarose gel electrophoresis and cloned into the pGEM T vector (Promega). The identity of an equine GAPDH cDNA was confirmed by gene sequencing. Tissue RNA samples were subjected to independent RT-PCR analyses for equine GAPDH transcripts to assess if the samples were amenable to RT-PCR analyses. The cDNAs were subjected to radiolabeled amplification and analyzed by denaturing acrylamide electrophoresis. The integrity of the tissue-derived RNAs was assayed by Northern blot hybridization with the equine GAPDH-specific probe (40).
The viral burden in the plasma was determined by quantitative RT-PCR. A synthetic gag-specific RNA was employed as the external standard. The reaction conditions were as described elsewhere (19). Briefly, 1 to 4 μl of virion RNA was reverse transcribed with 10 pmol of gag34 primer as described above. Radiolabeled hot start PCR was performed as described for viral DNA analyses by using 10 pmol of each of the gag11 and gag34 primers. PCR products were separated by polyacrylamide gel electrophoresis, and the amount of radiolabel incorporated was quantified as described above. The copies of virion RNA per milliliter of plasma were calculated by using the equation of the line. At least two independent assays were performed for each sample.
Measurement of infectious EIAV in plasma.
To complement the measurement of viral RNA levels in plasma, we also determined the levels of infectious EIAV by using primary equine monocyte-derived macrophage (MDM) cultures. Monocytes were isolated by overnight adherence, cultured, and frozen as previously described (32). Thawed monocytes were plated at 105 cells per well in 48-well plates in minimal essential medium-alpha medium (Gibco BRL) supplemented with 50% heat-inactivated adult bovine serum (HyClone), 2 mM glutamine, 100 U of penicillin per ml, and 100 μg of streptomycin per ml. Two days after thawing, the MDM cells were infected for 2 h at 37°C in Mg2+- and Ca2+-free Hanks balanced salt solution (Gibco BRL). Six wells of MDM cells per dilution of plasma were infected. Tenfold serial dilutions of plasma samples, starting at a 1:10 dilution, were used. Serum-free medium was added after 0.5 h to permit readherence of cells. Cells were washed once with serum-free medium. Cultures were maintained in minimal essential medium-alpha supplemented with 10% heat inactivated horse serum (HyClone), penicillin, streptomycin, and glutamine. Supernatants were removed weekly for micro-RT assays (19), and the TCID50 was calculated 14 days postinfection by using the Kärber formula. At least two independent infections of MDM cells were performed for each plasma sample.
RESULTS
Clinical profiles of EIAV experimentally infected animals.
Six animals were infected intravenously with either the EIAVWyo or EIAVPV strain (Table 1). All infected animals developed clinical disease symptoms, including febrile episodes, depression, and anorexia, within 1 month postinfection. Horses 298 and 94-07 infected with the highly virulent EIAVWyo strain became moribund and were euthanized 12 and 11 days postinfection, respectively. Four ponies were infected with the EIAVPV isolate. Ponies 112 and 123 were inoculated with EIAVPV and developed a fever associated with acute EIA at 9 and 13 days postinfection, respectively. The ponies were sacrificed at this time. Ponies 18 and 72 experienced multiple, irregularly spaced disease cycles typical of chronic EIA within 6 months of infection. Pony 18 resolved the chronic disease and remained clinically quiescent for more than 1 year. In contrast, pony 72 was clinically quiescent for 6 months and subsequently experienced two additional late disease cycles. These recrudescent febrile episodes were at 5 months and at 7 days prior to being euthanized, respectively. Ponies 18 and 72 were clinically normal (asymptomatic) at the time of sacrifice. Tissue samples and PBMC were isolated at the time of necropsy from all experimentally infected animals.
Quantification of EIAV in the plasma.
As an initial measure of the systemic levels of virus replication associated with the various experimental infections, we assayed the levels of plasma viremia by using infectivity assays in cultured MDM cells. The results of these assays (Table 1) indicated similar plasma viremia levels ranging from 103.6 to 105.0 TCID50 per ml during acute disease caused by either the EIAVWyo (animals 298 and 94-07) or EIAVPV (animals 112 and 123) infections. In marked contrast to the levels of infectious EIAV in plasma during acute disease, no infectious virus was detected in repeated assays of plasma taken during the asymptomatic stage of EIA (animals 18 and 72). The levels of infectious virus associated with acute and asymptomatic EIA were in general agreement with previous studies from this laboratory and others using infectivity assays in FEK cells (11, 18, 31). However, these experiments represented the first direct comparison of plasma samples from EIAVWyo- and EIAVPV-infected animals made possible by the use of MDM cells.
As noted above, EIAV plasma viremia has to date been measured using virus infectivity assays in cell culture. With the development of a quantitative RT-PCR technique (19), we were able to also compare plasma viral RNA levels in EIAVWyo and EIAVPV infections in parallel (Table 1). The results of these assays indicated high levels of plasma viral RNA in the two EIAVWyo-infected horses with 3.1 × 108 and 3.6 × 107 copies per ml in animals 298 and 94-07, respectively. The EIAVPV acutely infected ponies were found to have 2.1 × 107 and 4.2 × 107 copies per ml for ponies 112 and 123, respectively. The amounts of viral RNA detected in these experimentally infected animals were within the range of RNA levels observed during acute EIA in our historical panel of experimental infections (32). Using both the measurements of infectious virus and plasma RNA levels, it can be calculated for the first time that one infectious unit of either the EIAVWyo or EIAVPV in blood corresponded to about 3,000 copies of viral RNA as measured by RT-PCR.
In contrast to acute EIA, the asymptomatic animals had either undetectable or barely detectable levels of plasma viral RNA (Table 1). Interestingly, the level of suppression of EIAV replication was apparently similar in pony 18, which remained asymptomatic for over a year, and in pony 72, which had just recovered from a recrudescence of disease. One week prior to the euthanization of pony 72, the amount of viral RNA was determined to be about 4.2 × 107 copies of per ml of plasma. The precipitous drop of over 5 orders of magnitude in plasma RNA levels over the course of 1 week in an animal that had been infected for more than 1 year indicated the maintenance of rapid, potent host clearance mechanisms.
Taken together, these quantitative viremia studies demonstrate that acute disease was associated with very high plasma virus levels as measured by plasma viremia in MDM cells and by quantitation of viral RNA. While the sample size was small, no association between disease severity and virus levels were observed, confirming previous observations (31). Additionally, the lack of detectable infectious virus and the low levels of viral RNA indicated that production of EIAV is tightly controlled in asymptomatic animals.
Identification of the in vivo sites of EIAV infection.
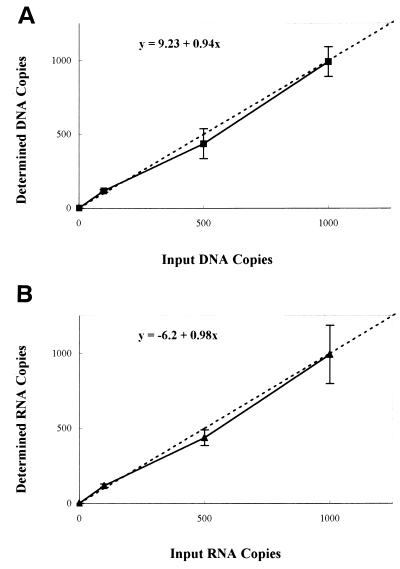
To determine the viral DNA burden in tissues and thus identify the in vivo sites of viral infection, we developed a sensitive and specific assay for quantifying viral DNA. To ascertain the specificity of the QC-PCR, DNA from infected and uninfected samples were amplified with viral gag-specific primers. A 310-bp product was detected only in samples from infected animals, indicating that the amplified DNA was of viral origin (data not shown). Amplification of viral sequences was additionally confirmed by restriction enzyme digestion, by Southern blot hybridization using EIAV gag-specific probes, and by sequencing (data not shown). To determine the sensitivity of the quantitative reactions, competitive amplification experiments were performed using specific known quantities of cloned viral DNA. The calculated copies were in close agreement with the expected values (Fig. 1, R = 0.99), and the sensitivity of the assay was determined to be 10 copies of viral DNA.
FIG. 1.
Detection and quantitation of known copy number of EIAV proviral DNA and singly spliced RNA. Wild-type and deleted cloned DNA fragments from the EIAV gag gene and cloned, in vitro-transcribed singly spliced RNA were quantified by spectrophotometry. Known template inputs of these DNA and RNA standards were subjected to direct 32P-labeled QC-PCR (A) or QC-RT-PCR (B) as described in Materials and Methods. Signal quantitation was performed with a storage phosphorimaging system (Molecular Dynamics). Values on the ordinate representing the determined copy number were calculated as described previously (30) and plotted as a function of the input copies of plasmid copy number (A) or in vitro-transcribed RNA copy number (B). The competitive amplifications gave a linear response over the range of templates studied.
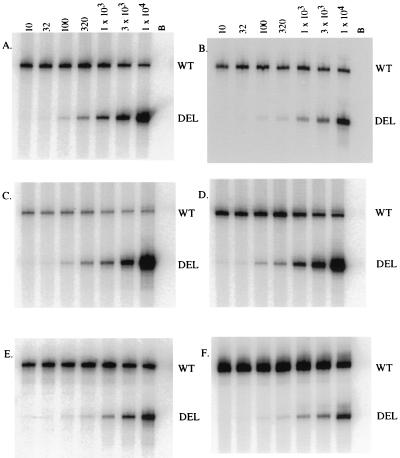
The levels of EIAV DNA were quantified from a panel of tissue samples isolated from the experimentally infected animals described in Table 1. Figure 2 summarizes representative data from a QC-PCR analysis using tissues taken during acute disease the EIAVPV-infected pony 123. These data qualitatively demonstrate markedly different levels of viral DNA in the various tissues.
FIG. 2.
Quantitation of viral DNA in the tissue samples from an EIAVPV-infected animal during acute disease. A representative QC-PCR was performed on equine tissue DNA samples from pony 123 amplified with gag-specific primers. Aliquots (2.5 μl) of direct 32P-labeled coamplification products from kidney, cortex (1 μg of DNA [A]), PBMC (1 μg of DNA [B]), bone marrow (1 μg of DNA [C]), mandibular lymph node (1 μg of DNA [D]), liver (500 ng [E]), and spleen (50 ng [F]) and increasing copy number (10 to 10,000) of competitor DNA were analyzed by denaturing 8% acrylamide electrophoresis. The amplified wild-type (WT) product was seen as the upper 310-bp band, and the competitor (DEL) was seen as the lower 255-bp band. The copies of competitor template were indicated above each lane. Lane B, amplification of buffer containing no tissue DNA.
Quantification of QC-PCR data for each of the six experimentally infected animals is summarized in Table 2. The results of the QC-PCR analyses revealed that the two EIAVWyo-infected animals had high cellular levels of viral DNA in most tissues, up to 290,000 copies/100,000 CE, indicating extensive disseminated infection. A broad range of copies of viral DNA that varied by up to a 100-fold (range, 2,700 to 290,000 and 1,200 to 120,000 copies/100,000 CE for horses 298 and 94-07, respectively) was observed, indicating preferential sites of infection. Interestingly, a different pattern of infection in the tissues of both animals was observed. Among the tissues examined from both animals, the spleen contained the highest concentration of cellular viral DNA at 290,000 and 120,000 copies/100,000 CE in horses 298 and 94-07, respectively. In horse 298, the burden was reduced to 45,000 copies/100,000 CE in the liver and to less than 8,000 copies/100,000 CE in the remaining samples. In contrast, the liver, lymph node, bone marrow, and PBMC from horse 94-07 harbored from 20,000 to 60,000 copies of viral DNA/100,000 CE, while the brain and kidney contained less than 2,000 copies.
TABLE 2.
QC-PCR quantification of tissue-specific EIAV DNA in experimentally infected equids
| Tissue sample | Mean copies of viral DNA/100,000 CEc
|
|||||
|---|---|---|---|---|---|---|
| EIAVWyo, acute, in animal:
|
EIAVPV, acute, in animal:
|
EIAVPV, asymptomatic, in animal:
|
||||
| 298 | 94-07 | 112 | 123 | 18 | 72 | |
| Braina | ND | 1,200 | <10 | 70 | ND | ND |
| Kidneyb | 5,500 | 1,400 | 10 | 500 | 30 | 80 |
| PBMC | 2,700 | 16,000 | 60 | 2,000 | 40 | 20 |
| Bone marrow | 3,500 | 62,000 | 90 | 160 | 10 | 80 |
| Lymph node | 7,800 | 17,000 | 40 | 430 | 10 | 80 |
| Liver | 45,000 | 23,000 | 110 | 3,200 | 180 | 300 |
| Spleen | 290,000 | 120,000 | 8,000 | 53,000 | 170 | 350 |
The cerebrum, medulla, and cerebellum were not significantly different from each other and were combined as the brain.
The cortex and medulla from the kidney were not significantly different from each other and were combined as the kidney.
ND, not determined. The values represent the mean copies of viral DNA per 100,000 CE. All data represent the results of at least two independent assays. The standard error was <15%.
A decreased amount of cellular viral DNA in all tissues in the EIAVPV acutely infected animals was observed compared to the EIAVWyo acutely infected animals (Table 2). Viral DNA was detectable in all samples in the EIAVPV acutely infected animals, once again indicating a rapid dissemination of viral infection. The cellular viral DNA burden in the two EIAVPV acutely infected ponies had viral DNA loads that varied by up to a 1,000-fold between samples within an animal (range, <10 to 8,000 and 70 to 53,000 copies/100,000 CE for ponies 112 and 123, respectively), revealing preferential sites of infection. Among the tissues, the spleen was the primary site of infected cells in both animals with 8,000 and 53,000 copies/100,000 CE in ponies 112 and 123, respectively. For pony 112, all of the remaining samples contained less than 100 copies of viral DNA/100,000 CE. In contrast, the liver and the PBMC from pony 123 harbored similar amounts of cellular viral DNA, up to 3,000 copies/100,000 CE. The remaining samples contained less than 500 copies/100,000 CE. Interestingly, the viral DNA burden differed between the two animals by approximately 10-fold, suggesting variation in the sensitivity of individual animals to infection.
In contrast to the acute infections described above, the two asymptomatic animals contained a greatly decreased cellular viral DNA load, with a maximal level of 350 copies/100,000 CE (Table 2). However, viral DNA was detectable in all tissues examined from both animals, reflecting widespread distribution of low levels of infected cells in asymptomatic animals. The cellular viral DNA levels varied only 10- to 20-fold between the samples (range, 10 to 180 copies for pony 18; 20 to 350 copies for pony 72), indicating limited partitioning of virally infected cells. The splenic and hepatic burdens were equal to each other in both animals. Pony 18 had approximately 200 copies of viral DNA/100,000 CE in these tissues, while the other samples harbored fewer than 40 copies. Interestingly, the cellular viral DNA levels in pony 72 were similar to those of pony 18 despite the difference in the recent clinical histories, containing approximately 300 copies/100,000 CE in the spleen and liver and fewer than 80 copies/100,000 CE in the remaining samples.
Comparison of the viral burden in particular tissues among the different infections and disease states revealed that the number of viral DNA copies in the EIAVWyo-infected animals was approximately 10-fold higher than in the EIAVPV acutely infected ponies, which in turn was in general 10-fold greater than the asymptomatic animals. These results suggested there was an association between the infecting viral strain and the viral DNA burden. However, regardless of the infecting strain, the appearance of clinical symptoms coincided with rapid widespread seeding of infection, although preferential sites of infection could be identified. During clinical quiescence, both long term and recent, persistent infection and widespread active viral replication was observed, suggesting reservoirs of infected cells apparently resistant to immune clearance.
Determination of the tissue sites of viral replication.
To determine sites of active viral replication during acute disease and of continuous virus production in acutely infected and asymptomatic animals, the amount of singly spliced EIAV mRNA was quantified by directly radiolabeled QC-RT-PCR. The active sites of lentivirus replication have been previously identified by quantification of either genomic or singly spliced mRNA species (2, 24, 34, 39). Studies performed by our laboratory observed high levels of cell-free virus, up to 108 particles per ml of plasma, during disease episodes (Table 1 and reference 32). Thus, levels of EIAV replication were assayed based on quantification of a singly spliced mRNA species to avoid any contribution of viral genomic RNA to these measurements. Amplification of viral singly spliced mRNA sequences was detected only in infected samples containing reverse transcriptase, indicating that the reaction was specific for this viral RNA (Fig. 3). No amplified product was detected in plasma RNA samples from febrile animals, indicating that virions do not contain viral singly spliced mRNA (data not shown). Restriction enzyme digestion and sequencing of the amplified products confirmed the specificity of this assay (data not shown). To determine the sensitivity of the QC-RT-PCR, experiments analogous to those described for viral DNA were performed. There was excellent agreement between the calculated copies and the determined copies of in vitro-transcribed RNAs (Fig. 1, R = 100). The assay was sensitive (limit of detection, 30 copies of viral singly spliced mRNA) and reproducible (coefficient of variation, <30%).
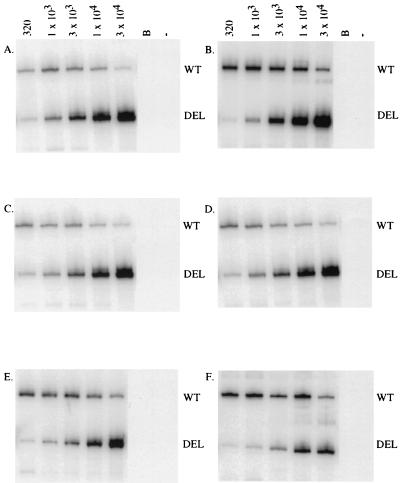
FIG. 3.
Quantitation of viral singly spliced mRNA in the tissue samples from an EIAVPV-infected animal during acute disease. A representative QC-RT-PCR was performed on equine tissue RNA samples from pony 123 amplified with primers specific for EIAV singly spliced RNA species. Aliquots (2.5 μl) of direct 32P-labeled coamplification products from the kidney, cortex (1 μg of RNA [A]), kidney, medulla (1 μg of RNA [B]), mandibular lymph node (1 μg of RNA [C]), bone marrow (1 μg of RNA [D]), liver (500 ng of RNA [E]), and spleen (50 ng of RNA [F]) and increasing copy number (320 to 32,000) of in vitro-synthesized competitor RNA were analyzed by denaturing 8% acrylamide electrophoresis. The amplified wild-type (WT) product was seen as the upper 510-bp band, and the deleted competitor (DEL) was seen as the lower 406-bp band. The copies of competitor RNA were indicated above each lane. Lane B, RT-PCR of buffer containing no tissue RNA; lane −, RT-PCR of RNA without reverse transcriptase.
Quantification of the amount of viral singly spliced mRNA was determined on the same panel of tissues as that used for the viral DNA analyses. A radiolabeled QC-RT-PCR experiment from pony 123, acutely infected with EIAVPV, is shown in Fig. 3. The abundance of viral singly spliced mRNA from the experimentally infected animals was measured (Table 3). Similar to the DNA results, there were various levels of replicating virus throughout these animals. A pattern reminiscent of the viral infection emerged when the distribution of viral replication was examined.
TABLE 3.
QC-RT-PCR quantification of tissue-specific EIAV singly spliced mRNA in experimentally infected equids
| Tissue samplea | Mean copies of viral singly spliced mRNA/μg of total RNAb
|
|||||
|---|---|---|---|---|---|---|
| EIAVWyo, acute, in animal:
|
EIAVPV, acute, in animal:
|
EIAVPV, asymptomatic, in animal:
|
||||
| 298 | 94-07 | 112 | 123 | 18 | 72 | |
| Brain | ND | 7,000 | <30 | 100 | ND | ND |
| Kidney | 650 | 27,000 | <30 | 650 | <30 | <30 |
| PBMC | 700 | 6,000 | <30 | 17,000 | <30 | 190 |
| Bone marrow | 1,500 | 6,000 | 130 | 700 | <30 | <30 |
| Lymph node | 130,000 | 90,000 | 90 | 1,600 | <30 | 30 |
| Liver | 110,000 | 76,000 | 3,600 | 23,000 | 740 | 140 |
| Spleen | 7,300,000 | 92,000 | 185,000 | 510,000 | 50 | 750 |
The brain and the kidney are as described in Table 2.
ND, not determined. The values represent the mean copies of viral singly spliced mRNA per μg of total RNA. All data represent the results of at least two independent assays. The standard error was <25%.
The EIAVWyo-infected animals harbored high levels, up to several million copies, of viral singly spliced mRNA in most of the samples, indicating abundant, rapid disseminated viral replication (Table 3). A broad range of copies of viral singly spliced mRNA in various tissues was observed (700 to 7,300,000 and 6,000 to 92,000 copies/μg of total RNA for horses 298 and 94-07, respectively), reflecting that virus replicated in preferential sites. The spleen of horse 298 contained the highest amount of replicating virus with 7,300,000 copies/μg of total RNA. The liver and the lymph nodes had comparable cellular viral RNA loads, harboring approximately 100,000 copies/μg of total RNA. The remaining samples had less than 1,500 copies/μg of total RNA. By comparison, similar levels of cellular viral singly spliced mRNA were observed in the spleen, liver, and lymph node from horse 94-07, with approximately 90,000 copies/μg of total RNA in each tissue. The other samples contained less than 30,000 copies/μg of total RNA. Interestingly, for both animals the lymph nodes contained a relatively high amount of viral singly spliced (Table 3) mRNA, despite containing low levels of viral DNA (Table 2).
The level of cellular viral singly spliced mRNA in most samples in the EIAVPV acutely infected animals was similar or 10-fold less than in the EIAVWyo infections (Table 3). A wide range of copies of viral singly spliced mRNA was determined among the different tissues (undetectable to 200,000 and 100 to 500,000 copies/μg of total RNA for ponies 112 and 123, respectively), again indicating the preferred sites of virus replication. In both ponies, the spleen contained the highest cellular levels of replicating virus, up to 500,000 copies/μg of total RNA. In pony 112, the cellular viral singly spliced mRNA load was reduced to 4,000 copies and less than 100 copies/μg of total RNA in the liver and the rest of the samples, respectively. By comparison, the liver and the PBMC in pony 123 contained equivalent levels of cellular viral singly spliced mRNA (20,000 copies/μg of total RNA), which was approximately 10-fold less than the spleen. Interestingly, relatively high levels of EIAV singly spliced mRNA were also detected in the PBMC from one of the EIAVWyo acutely infected animals (horse 94-07), suggesting that viral replication in this case was not confined to the tissues. This observation was in contrast to previous analyses, which report a lack of productive EIAV infection of blood monocytes (21, 43). The remaining samples in pony 123 harbored fewer than 2,000 copies of viral singly spliced mRNA/μg of total RNA, which was in general 100- to 1,000-fold less than the spleen. The viral singly spliced mRNA load in the two acute EIAVPV infections varied by approximately 10-fold between the two animals, indicating variation between animals.
In asymptomatic animals, the levels of viral singly spliced mRNA were dramatically reduced compared to the EIAVWyo-infected animals, with a maximal load of 750 copies/μg of total RNA (Table 3). In some tissue samples, viral singly spliced RNA was undetectable in repeated analyses. Within an animal, the difference in the number of copies of viral singly spliced mRNA between samples was limited compared to the acutely infected animals (range, undetectable to 750 copies/μg of total RNA for both animals). A predominant site of viral replication appeared to be either the hepatic or splenic compartments for ponies 18 and 72, respectively. The remaining samples from pony 18 harbored fewer than 50 copies and from pony 72 fewer than 150 copies of viral singly spliced mRNA/μg of total RNA, respectively. Together, these results indicated that viral replication was suppressed and restricted during clinical quiescence.
In comparison to the viral DNA analyses, the levels of singly spliced viral RNA failed to show a clear gradient when the different disease states and the infecting strains were compared (Table 3). The EIAVWyo-infected animals had similar or greater levels of viral replication relative to the EIAVPV acutely infected animals. In asymptomatic animals, the level of viral RNA expression was clearly reduced. These data suggested that during acute disease the amount of viral replication was similar in some samples between EIAV strains. During acute disease, replication was in general detected throughout infected animals, although the spleen was a predominant cellular site of highly active replication. During clinical quiescence, tight but not absolute control of viral replication was evident, indicating that a continuous low level of replication persisted.
Determination of the overall EIAV nucleic acid load.
The above assays determine the concentration of viral DNA and RNA in various tissues but do not reflect the total organ burden. To determine the relative contribution of each organ to the total viral nucleic acid burden, the amount of viral nucleic acids was calculated from generalized measurements of equid organs and body weights, the number of copies of viral nucleic acids, and the nucleic acid yields from each specimen. The kidney, peripheral lymph nodes, bone marrow, liver, spleen, and brain were estimated to account for approximately 0.25, 0.1, 3.5, 1, 0.4, and 0.1%, respectively, of the total body weight in horses (15, 45) and were assumed to be similar in ponies. The viral DNA loads in each organ revealed that the bone marrow, liver, and spleen were the principal sites of infection in the EIAVWyo and EIAVPV acutely infected and asymptomatic animals, containing an average of 1011, 109, and 108 copies in each organ, respectively (Table 4). Taken together, the spleen, bone marrow, and liver accounted for 98% of the viral DNA burden in all infected animals regardless of the disease state. In contrast, the liver and spleen each had approximately 1012, 1011, and 109 copies of viral singly spliced mRNA in the animals acutely infected with EIAVWyo and EIAVPV and in the asymptomatic animals, respectively (Table 4). Together, the spleen and liver accounted for up to 98% of the viral singly spliced mRNA and were the major producers of virus in all animals. In contrast to the abundance of viral DNA and RNA localized to tissues, the PBMC contained less than 0.1% of the total viral DNA or RNA during acute disease or during asymptomatic infections. These data clearly demonstrate that tissues constitute the predominant in vivo sites of EIAV infection during symptomatic and asymptomatic infections.
TABLE 4.
Determination of the total viral nucleic acid burden
| Tissue samplea | Total viral nucleic acid burden (copies)b
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EIAVWyo, acute, in animal:
|
EIAVPV, acute, in animal:
|
EIAVPV, asymptomatic, in animal:
|
||||||||||
| 298
|
94-07
|
112
|
123
|
18
|
72
|
|||||||
| DNA | RNA | DNA | RNA | DNA | RNA | DNA | RNA | DNA | RNA | DNA | RNA | |
| Brain | ND | ND | 1.1 × 108 | 2.6 × 109 | 9.5 × 104 | Neg | 4.5 × 106 | 1.4 × 107 | ND | ND | ND | ND |
| Kidney | 2.1 × 109 | 5.8 × 108 | 6.1 × 108 | 5.0 × 1010 | 1.6 × 106 | 9.1 × 106 | 5.2 × 107 | 2.1 × 108 | 5.5 × 106 | 4.6 × 106 | 3.0 × 107 | 1.9 × 106 |
| PBMC | 1.7 × 107 | 1.1 × 108 | 9.1 × 108 | 4.9 × 108 | 1.8 × 106 | 6.5 × 105 | 4.3 × 107 | 3.6 × 108 | 1.2 × 106 | 1.3 × 105 | 1.4 × 105 | 3.3 × 106 |
| Bone marrow | 6.1 × 1010 | 3.2 × 1010 | 5.5 × 1011 | 9.2 × 1010 | 1.5 × 108 | 8.3 × 108 | 2.7 × 107 | 2.1 × 108 | 9.9 × 107 | Neg | 1.1 × 109 | 3.2 × 108 |
| Lymph node | 2.9 × 1010 | 1.5 × 1011 | 1.1 × 1010 | 1.2 × 1011 | 2.9 × 106 | 2.4 × 107 | 3.0 × 107 | 3.0 × 108 | 5.3 × 106 | Neg | 1.6 × 107 | 8.9 × 106 |
| Liver | 8.6 × 1010 | 5.6 × 1011 | 8.8 × 1010 | 1.1 × 1012 | 1.3 × 108 | 1.1 × 1010 | 5.0 × 109 | 1.0 × 1011 | 2.5 × 108 | 1.8 × 109 | 5.1 × 108 | 4.8 × 108 |
| Spleen | 7.6 × 1010 | 4.2 × 1012 | 6.8 × 1010 | 1.1 × 1011 | 1.7 × 109 | 1.0 × 1011 | 6.7 × 109 | 3.1 × 1011 | 9.5 × 107 | 3.3 × 107 | 2.2 × 108 | 4.8 × 108 |
| Total | 2.3 × 1011 | 4.9 × 1012 | 7.2 × 1011 | 1.5 × 1012 | 2.0 × 109 | 1.1 × 1011 | 1.2 × 1010 | 4.1 × 1011 | 4.6 × 108 | 1.8 × 109 | 1.9 × 109 | 1.3 × 109 |
The brain and the kidney are as described in Table 2.
ND, not determined; Neg, no detectable RNA by RT-PCR.
To obtain a measure of the overall whole-body viral load, the total viral DNA and singly spliced mRNA burden of all of the organs was calculated, excluding the brain samples (Table 4). The whole-body viral DNA load was 1011 copies in the EIAVWyo acutely infected animals. In comparison, the EIAVPV acutely infected animals harbored 109 to 1010 copies of viral DNA, representing a 10- to 100-fold decrease. The viral DNA burden was decreased by up to an additional 10-fold in the asymptomatic animals, containing 108 copies of viral DNA. In contrast, the whole-body viral singly spliced RNA level was 1012 copies in the EIAVWyo acutely infected animals. In comparison, the overall viral singly spliced mRNA burden in the EIAVPV acutely infected animals was reduced approximately 10-fold to 1011 copies. The asymptomatic animals harbored 109 copies of viral singly spliced mRNA, which represented an additional 100-fold decrease. These data suggested that the virulence of the EIAVWyo strain was related to an increased rate of infection and replication in the tissues.
Calculation of the mean transcriptional level (the ratio of the total viral singly spliced RNA load to the total viral DNA load) has been used to compare human immunodeficiency virus type 1 (HIV-1) expression levels in various organs (1, 24). In a similar manner, the mean transcriptional level (viral transcripts per proviral genome) for the various organs in the EIAV-infected animals was calculated from the quantitative DNA and RNA data summarized in Table 4. In the EIAVWyo and EIAVPV acutely infected animals the spleen, liver, and lymph nodes consistently had transcriptional indices up to 85, indicating the highest levels of virus expression per proviral genome. While transcription was not limited to these sites, the levels in the remaining samples were typically less than 10. Notable exceptions were the brain and kidney in horse 94-07, which had indices of 25 and 80, respectively. In contrast to the acute infections, the highest transcriptional levels from the clinically quiescent animals were in the liver and the spleen; however, the indices were only 7 and 2. These results provided additional support for the suppression of viral transcription in tissues of inapparent carriers. Interestingly, the PBMC in three animals (298, 123, and 72) had transcriptional indices of 6, 8, and 24, respectively, suggesting low levels of EIAV proviral expression in blood monocytes during both acute and chronic infection.
DISCUSSION
The clearly demarcated disease cycles associated with EIAV infection provide a unique in vivo model for characterization of the tissue reservoirs of productive and latent lentiviral infection during clinical episodes and during long-term asymptomatic infections. The experiments described here present for the first time a rigorous quantitative analysis of the levels of both EIAV viral DNA as a measure of infection and singly spliced viral RNA as a measure of virus replication in various tissue compartments of experimentally infected equids. The results of these studies identified preferential sites of virus infection, replication, and latency associated with persistent EIAV infections.
The levels of EIAV DNA and singly spliced viral RNA measured in the experimentally infected equids differed markedly depending on the clinical status of the infected animals and the strain of virus used for the infection. In terms of plasma viremia, the current studies clearly demonstrate that acute EIAV infection is associated with very high levels of virus in the blood, as measured by infectivity in a recently developed quantitative equine macrophage assay (32). In marked contrast, infectious virus was undetectable in blood during asymptomatic EIAV infection. The current experiments for the first time extend the measurements of blood viral burden to include quantification of plasma viral RNA by RT-PCR. These data revealed plasma RNA levels of 107 to 108 copies of virion RNA per ml of plasma during acute disease and fewer than 100 copies per ml during asymptomatic stages of infection. Thus, the new combination of in vitro infectivity and PCR assays clearly illustrate the characteristic wave of viremia associated with EIAV disease episodes and the rigorous control of virus replication during clinical latency.
While these blood virus load measurements in general confirm previously reported viremia data derived from infectivity assays in equine fibroblastic cells, the new assays also provide new insights into the pattern of viremia during persistent EIAVPV infection. First, the viremia levels measured in equine macrophages are similar to viremia levels reported previously by infectivity assays in equine fibroblastic cells (11, 18, 31). This agreement indicates that the latter assays in fact provide a relevant measure of EIAVPV viremia, eliminating a lingering concern about the validity of using equine fibroblastic cells to measure EIAVPV infectivity. Second, the quantitative measurements of plasma viral RNA closely parallel viremia measurements in assessing viral load during acute disease and clinical latency. However, the viral RNA measurements demonstrate for the first time that the negligible levels of infectious EIAV associated with clinical latency is in fact due to an absence of viral particles in the blood and not to an inactivation of virus by neutralizing serum antibodies. This observation appears to indicate the efficacy of the host immune control of viral replication, presumably by the reduction of infected target cells. Finally, the viremia data summarized here indicates that the typically fatal outcome of EIAVWyo infection compared to the chronic EIA induced by EIAVPV is not reflected in substantial differences in plasma viremia levels, since the two infections produced similar plasma virus loads during the initial acute disease. As discussed below, however, the differences in disease severity may be more accurately associated with the tissue viral load rather than the levels of virus in blood.
During the initial stages of acute disease within 2 weeks postinfection, viral infection and replication were shown to be widely disseminated among all of the tissues examined in both the EIAVWyo- and EIAVPV-infected animals (Tables 2 to 4). In this regard, the results of the current studies are in agreement with previous analyses of EIAV replication in infected horses during acute disease using cultured cells to detect virus (17), fluorescent antibody (23), Southern blot (36), in situ hybridization (41), or PCR (13, 14, 26) procedures and with the pathological findings characteristic of EIAV infection (41). The results of these earlier qualitative analyses established an association between sites of virus infection and host tissues rich in macrophages. However, the quantitative assays employed here extend these studies to clearly identify the spleen as the predominant cellular site of EIAV infection and replication during acute disease. The concentration of proviral DNA and singly spliced viral RNA in the splenic tissues was typically 10 to 20 times greater than the concentration of EIAV DNA or RNA measured in any other tissue. In contrast, the asymptomatic animals had equivalent cellular loads of viral DNA and mRNA in the spleen and the liver (Tables 2 and 3). In all animals, estimation of total organ viral loads (Table 4) indicates further that the spleen and liver tissues contain up to 85% of the total viral DNA or mRNA that was detected in the tissues examined. It is assumed that the high levels of virus infection and replication in the splenic and hepatic tissues reflect the abundance of target macrophages in these organs. In addition, these quantitative analyses revealed that during the initial acute disease the EIAVWyo-infected animals contained 10- to 100-fold more viral DNA and singly spliced RNA in tissues compared to the levels measured in the EIAVPV-infected animals. This differential in tissue levels of viral DNA and singly spliced viral RNA is in contrast to the similar levels of plasma viral genomic RNA measured during acute disease in animals infected by the two viral strains. These observations indicate that the levels of tissue-associated viral DNA and singly spliced viral RNA, especially in the spleen and liver, constitute a more accurate correlate of disease severity than plasma viral RNA load.
Although the quantitative viral DNA and singly spliced viral RNA analyses in general identified macrophage-rich tissues as the predominant sites of viral infection during acute EIA, in agreement with pathological findings, these detailed analyses also revealed some unexpected aspects of EIAV infections. For example, a number of studies have reported that EIAV DNA and RNA is either very low or undetectable in PBMC isolated from horses infected with EIAV (14, 21, 42, 43). These observations support the widely accepted concept that infection of blood monocytes with EIAV results in latent infection and that cellular differentiation to macrophages is necessary for the production of virus, as proposed for other lentiviruses (8). However, in the current studies we observed substantial levels of viral DNA (2,000 to 16,000 copies per 100,000 CE) and viral singly spliced RNA (700 to 17,000 copies per μg of RNA) in PBMC in three of the four infected animals during acute disease. These findings suggest that blood monocytes can in fact serve as targets of EIAV infection and of virus production during waves of viremia associated with disease episodes, perhaps reflecting widespread activation of blood monocytes during cycles of EIAV disease.
A second unexpected observation is the relatively high level of viral DNA (Table 2) accompanied by a low level of viral singly spliced RNA (Table 3) in the bone marrow of the EIAVWyo acutely infected and the asymptomatic EIAVPV-infected animals. These observations extend previous findings from visna virus (9), simian immunodeficiency virus (4), and HIV-1 (7, 28) indicating that the less-differentiated cells of the bone marrow can serve as a reservoir of latently infected cells. Recent studies detected increased levels of inflammatory cytokines in the bone marrow during EIAV infection in vivo (47) and in bone marrow derived macrophages in vitro (46), which are hypothesized to suppress platelet production (5). Thus, cytokine dysregulation could result in thrombocytopenia, a clinical finding consistently associated with recrudescence of disease. Additionally, severe hemorrhaging is the principal contributing pathology in lethal EIAVWyo infection. Therefore, the organ-specific viral loads may directly or indirectly account for the pathological features manifest during disease cycles.
In striking contrast to the high levels of viral DNA and viral singly spliced RNA found in tissues from animals during acute disease, asymptomatic EIAV infections were characterized by extremely low levels of viral DNA (150 to 350 copies per 100,000 CE) and viral singly spliced RNA (50 to 750 copies per μg of RNA) concentrated predominantly in the liver and spleen. Other tissues examined contained fewer than 100 copies of DNA and usually <30 copies of singly spliced viral RNA. These observations demonstrate a rigorous control of EIAV replication that severely limits the number of EIAV-infected cells and the levels of virus production. However, it is important to note that despite the lifelong asymptomatic state usually maintained by EIAV-infected animals following 6 to 12 months of chronic EIA, the viral infection is never completely eliminated since tissue reservoirs of productively infected cells are sustained despite the high levels of immune control established by the host.
The ability of animals to routinely establish immunological control of EIAV infection and disease is considered the best example of natural immune control of a virulent lentivirus infection (25). The concept of immunologic control of EIAV replication in infected asymptomatic animals has been developed mostly based on measurements of viremia that indicate the absence of detectable virus in the blood of asymptomatic animals in marked contrast to the viremia levels of 105 to 106 TCID50 per ml of blood during clinical episodes (25). However, it has been shown that transfer of as little of 7 ml of whole blood from asymptomatic animals to naive equids produces infection of as least 80% of the recipient animals (12). Therefore, it has not been clear from these studies whether the lack of infectious virus in the blood actually reflected a lack of virus particles or inactivation of plasma virus by the high levels of EIAV-specific neutralizing antibodies present in asymptomatic animals (11). The RT-PCR assays described here demonstrate for the first time that the lack of infectious virus in the blood in asymptomatic animals is indeed reflective of a virtual absence of detectable viral genomic RNA in the plasma from these animals. Thus, the infectivity of blood from asymptomatic animals is most likely associated with the approximate 0.04% of PBMC containing EIAV proviral DNA.
The results described in the current study define the tissue sites of EIAV infection but do not specifically define the target cells within these tissues. In agreement with the well-established macrophage tropism of EIAV, the predominant sites of EIAV infection and replication during symptomatic and asymptomatic infections were determined to be tissues rich in macrophages, such as the spleen and liver. While a recent study (22) indicates that renal macrovascular endothelial cells may also serve as a reservoir of EIAV infection in a long-term inapparent carrier, the relative contributions of macrophages and endothelial cells in different tissues to viral persistence were not determined in this study. In addition, there has been no characterization of endothelial cell infection in experimentally infected horses during acute disease to assess the potential role of these cells in pathogenesis. The results of the current study seem to support the predominant importance of monocytes and macrophages in viral pathogenesis and persistence, since EIAV infection and replication correlated with the abundance of tissue macrophages in the respective organs.
These quantitative studies of EIAV infection and replication in vivo provide a detailed description of the patterns of virus replication associated with clinical and asymptomatic infections. While the data are specific for EIAV, the results have implications for broader issues related to the role of tissue reservoirs in lentiviral persistence, the potential for immune control or ultimate clearance of lentiviral infections, and for understanding the mechanisms of lentiviral pathogenesis. Based on the studies with EIAV, a predominantly macrophage-tropic lentivirus, it is clear that tissues can provide a widespread and resilient reservoir for lentiviral infection, even in the presence of potent host immune clearance mechanisms. Moreover, it appears that the extent of viral pathogenesis can be most closely associated with the levels of virus replication in tissues rather than plasma virus burden. Thus, it will be important to examine in more detail the potential role of tissue macrophages in the persistence of lentiviruses that are also lymphotropic and to define the mechanisms by which tissue macrophage infections lead to characteristic lentiviral pathologies such as fever, diarrhea, anemia, thrombocytopenia, and encephalitis.
ACKNOWLEDGMENTS
We thank Drew Lichtenstein for stimulating discussions, Caroline Leroux for thoughtful review of the manuscript, and Gary Thomas and Brian Meade for excellent animal care.
This work was supported by National Institutes of Health grant R01 CA49296, by funds from the Lucille P. Markey Charitable Trust and the Kentucky Agricultural Experimental Station, and in part by a grant from the Pittsburgh Supercomputer Center through the NIH National Center for Research Resources (resource grant 2 P41 RR06009). S.M.H. was supported by NIH AIDS training grant 5T32 AI07487.
REFERENCES
- 1.Bagnarelli P, Menzo S, Valenza A, Manzin A, Giacca M, Ancarani F, Scalise G, Varaldo P E, Clementi M. Molecular profile of human immunodeficiency virus type 1 infection in symptomless patients and in patients with AIDS. J Virol. 1992;66:7328–7335. doi: 10.1128/jvi.66.12.7328-7335.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bagnarelli P, Valenza A, Menzo S, Sampaolesi R, Varaldo P E, Butini L, Montroni M, Perno C-F, Balotta C, Clementi M. Dynamics and modulation of human immunodeficiency virus type 1 transcripts in vitro and in vivo. J Virol. 1996;70:7603–7613. doi: 10.1128/jvi.70.11.7603-7613.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chong Y-H, Payne S L, Issel C J, Montelaro R C, Rushlow K E. Characterization of the antigenic domains of the major core protein (p26) of equine infectious anemia virus. J Virol. 1991;65:1007–1012. doi: 10.1128/jvi.65.2.1007-1012.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clements J E, Zink M C. Molecular biology and pathogenesis of animal lentivirus infections. Clin Microbiol Rev. 1996;9:100–117. doi: 10.1128/cmr.9.1.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crawford T B, Wardrop K J, Tornquist S J, Reilich E, Meyers K M, McGuire T C. A primary production deficit in the thrombocytopenia of equine infectious anemia virus. J Virol. 1996;70:7842–7850. doi: 10.1128/jvi.70.11.7842-7850.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cunningham T P, Montelaro R C, Rushlow K E. Lentivirus envelope sequences and proviral genomes are stabilized in Escherichia coli when cloned in low-copy-number plasmid vectors. Gene. 1993;124:93–98. doi: 10.1016/0378-1119(93)90766-v. [DOI] [PubMed] [Google Scholar]
- 7.Embretson J, Zupancic M, Ribas J L, Burke A, Racz P, Tenner-Racz K, Haase A T. Massive covert infection of helper T lymphocytes and macrophages by HIV during the incubation period of AIDS. Nature. 1993;362:359–362. doi: 10.1038/362359a0. [DOI] [PubMed] [Google Scholar]
- 8.Gendelman H E, Narayan O, Kennedy-Stoskopf S, Kennedy P G E, Ghoti Z, Clements J E, Stanley J, Pezeshkpour G. Tropism of sheep lentiviruses for monocytes: susceptibility to infection and virus gene expression increase during maturation to macrophages. J Virol. 1986;58:67–74. doi: 10.1128/jvi.58.1.67-74.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gendelman H E, Narayan O, Moplineaux S, Clements J E, Ghotbi Z. Slow, persistent replication of lentiviruses: role of tissue macrophages and macrophage precursors in bone marrow. Proc Natl Acad Sci USA. 1985;82:7086–7090. doi: 10.1073/pnas.82.20.7086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Genetics Computer Group. Program manual for the Wisconsin Package. Madison, Wis: Genetics Computer Group; 1994. [Google Scholar]
- 11.Hammond S A, Cook S J, Lichtenstein D L, Issel C J, Montelaro R C. Maturation of the cellular and humoral immune responses to persistent infection in horses by equine infectious anemia virus is a complex and lengthy process. J Virol. 1997;71:3840–3852. doi: 10.1128/jvi.71.5.3840-3852.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Issel C J, Adams J W V, Meek L, Ochoa R. Transmission of equine infectious anemia virus from horses without clinical signs of disease. J Am Vet Med Assoc. 1982;180:272–275. [PubMed] [Google Scholar]
- 13.Kim C H, Casey J W. Genomic variation and segregation of equine infectious anemia virus during acute infection. J Virol. 1992;66:3879–3882. doi: 10.1128/jvi.66.6.3879-3882.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim C H, Casey J W. In vivo replication status and envelope heterogeneity of equine infectious anemia virus in an inapparent carrier. J Virol. 1994;68:2777–2780. doi: 10.1128/jvi.68.4.2777-2780.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klaus G G B, editor. Lymphocytes: a practical approach. Oxford, England: IRL Press, Ltd.; 1987. [Google Scholar]
- 16.Kono Y, Hirasawa K, Fukunaga Y, Taniguchi T. Recrudescence of equine infectious anemia by treatment with immunosuppressive drugs. Natl Inst Anim Health Q. 1976;16:8–15. [PubMed] [Google Scholar]
- 17.Kono Y, Kobayashi K, Fukunaga Y. Distribution of equine infectious anemia virus in horses infected with the virus. Natl Inst Anim Health Q. 1971;11:11–20. [PubMed] [Google Scholar]
- 18.Langemeier J L, Cook S J, Cook R F, Rushlow K E, Montelaro R C, Issel C J. Detection of equine infectious anemia viral RNA in plasma samples from recently infected and long-term inapparent carrier animals by PCR. J Clin Microbiol. 1996;34:1481–1487. doi: 10.1128/jcm.34.6.1481-1487.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lichtenstein D L, Rushlow K E, Cook R F, Raabe M L, Swardson C J, Kociba G J, Issel C J, Montelaro R C. Replication in vitro and in vivo of an equine infectious anemia virus mutant deficient in dUTPase activity. J Virol. 1995;69:2881–2888. doi: 10.1128/jvi.69.5.2881-2888.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lifson J D, Nowak M A, Goldstein S, Rossio J L, Kinter A, Vasquez G, Wiltrout T A, Brown C, Schneider D, Wahl L, Lloyd A L, Williams J, Elkins W R, Fauci A S, Hirsch V M. The extent of early viral replication is a critical determinant of the natural history of simian immunodeficiency virus infection. J Virol. 1997;71:9508–9514. doi: 10.1128/jvi.71.12.9508-9514.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maury W. Monocyte maturation controls expression of equine infectious anemia virus. J Virol. 1994;68:6270–6279. doi: 10.1128/jvi.68.10.6270-6279.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maury W, Oaks J L, Bradley S. Equine endothelial cells support productive infection of equine infectious anemia virus. J Virol. 1998;72:9291–9297. doi: 10.1128/jvi.72.11.9291-9297.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McGuire T C, Crawford T B, Henson J B. Immunofluorescent localization of equine infectious anemia virus in tissue. Am J Pathol. 1971;62:283–294. [PMC free article] [PubMed] [Google Scholar]
- 24.Michael N L, Vahey M, Burke D S, Redfield R R. Viral DNA and mRNA expression correlate with the stage of human immunodeficiency virus (HIV) type 1 infection in humans: evidence for viral replication in all stages of HIV disease. J Virol. 1992;66:310–316. doi: 10.1128/jvi.66.1.310-316.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Montelaro R C, Ball J M, Rushlow K E. Equine retroviruses. In: Levy J A, editor. The Retroviridae. Vol. 2. New York, N.Y: Plenum Press; 1993. pp. 257–360. [Google Scholar]
- 26.Oaks J L, McGuire T C, Ulibarri C, Crawford T C. Equine infectious anemia virus is found in tissue macrophages during subclinical infection. J Virol. 1998;72:7263–7269. doi: 10.1128/jvi.72.9.7263-7269.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O'Rourke K I, Besola M L, McGuire T C. Proviral sequences detected by polymerase chain reaction in peripheral blood cells of horses with equine infectious anemia virus. Arch Virol. 1991;117:109–119. doi: 10.1007/BF01310496. [DOI] [PubMed] [Google Scholar]
- 28.Pantaleo G, Graziosi C, Demarest J F, Butini L, Montroni M, Fox C H, Orenstein J M, Kotler D P, Fauci A S. HIV infection is active and progressive in lymphoid tissue during the clinically latent stage of disease. Nature. 1993;362:355–358. doi: 10.1038/362355a0. [DOI] [PubMed] [Google Scholar]
- 29.Perryman L E, O'Rourke K I, McGuire T C. Immune responses are required to terminate viremia in equine infectious anemia lentivirus infection. J Virol. 1988;62:3073–3076. doi: 10.1128/jvi.62.8.3073-3076.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Piatak M, Saag M S, Yang L C, Clark S J, Knappes J C, Luk K-C, Hahn B H, Shaw G M, Lifson J D. High levels of HIV-1 in plasma during all stages of infection determined by competitive PCR. Science. 1993;259:1749–1754. doi: 10.1126/science.8096089. [DOI] [PubMed] [Google Scholar]
- 31.Raabe M L, Issel C J, Cook S J, Cook R F, Woodson B, Montelaro R C. Immunization with a recombinant envelope protein (rgp90) of EIAV produces a spectrum of vaccine efficacy ranging from lack of clinical disease to severe enhancement. Virology. 1998;245:151–162. doi: 10.1006/viro.1998.9142. [DOI] [PubMed] [Google Scholar]
- 32.Raabe M L, Issel C J, Montelaro R C. Equine monocyte derived macrophage cultures and their application for infectivity and neutralization studies of equine infectious anemia virus. J Virol Methods. 1998;71:87–104. doi: 10.1016/s0166-0934(97)00204-8. [DOI] [PubMed] [Google Scholar]
- 33.Rasty S, Dhruva B R, Schlitz R L, Shih D S, Issel C J, Montelaro R C. Proviral DNA integration and transcriptional patterns of equine infectious anemia virus during persistent and cytopathic infections. J Virol. 1990;64:86–95. doi: 10.1128/jvi.64.1.86-95.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reinhart T A, Rogan M J, Huddleston D, Rausch D M, Eiden L E, Haase A T. Simian immunodeficiency virus burden in tissues and cellular compartments during clinical latency and AIDS. J Infect Dis. 1997;176:1198–1208. doi: 10.1086/514113. [DOI] [PubMed] [Google Scholar]
- 35.Rice N R, Lequarre A-S, Casey J W, Lahn S, Stephens R M, Edwards J. Viral DNA in horses infected with equine infectious anemia virus. J Virol. 1989;63:5194–5200. doi: 10.1128/jvi.63.12.5194-5200.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rice N R, Simek S, Ryder O A, Coggins L. Detection of proviral DNA in horse cells infected with equine infectious anemia virus. J Virol. 1978;16:577. doi: 10.1128/jvi.26.3.577-583.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rudolph J A, Spier S J, Byrns G, Rojas C V, Bernoco D, Hoffman E P. Periodic paralysis in quarter horses: a sodium channel mutation disseminated by selective breeding. Nat Genet. 1992;2:144–147. doi: 10.1038/ng1092-144. [DOI] [PubMed] [Google Scholar]
- 38.Rwambo P M, Issel C J, Hussain K A, Montelaro R C. In vitro isolation of a neutralization escape mutant of equine infectious anemia virus (EIAV) Arch Virol. 1990;111:275–280. doi: 10.1007/BF01311062. [DOI] [PubMed] [Google Scholar]
- 39.Saltarelli M J, Hadziyannis E, Hart C E, Harrison J V, Felber B K, Spira T J, Pavlakis G N. Analysis of human immunodeficiency virus type 1 mRNA splicing patterns during disease progression in peripheral blood mononuclear cells from individuals. AIDS Res Hum Retrovir. 1996;12:1443–1456. doi: 10.1089/aid.1996.12.1443. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 41.Sellon D C, Fuller F J, McGuire T C. The immunopathogenesis of equine infectious anemia virus. Virus Res. 1994;32:111–138. doi: 10.1016/0168-1702(94)90038-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sellon D C, Perry S T, Coggins L, Fuller F J. Wild-type equine infectious anemia virus replicates in vivo predominantly in tissue macrophages, not in peripheral blood monocytes. J Virol. 1992;66:5906–5913. doi: 10.1128/jvi.66.10.5906-5913.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sellon D C, Walker K M, Russell K E, Perry S T, Covington P, Fuller F J. Equine infectious anemia virus replication is upregulated during differentiation of blood monocytes from acutely infected horses. J Virol. 1996;70:590–594. doi: 10.1128/jvi.70.1.590-594.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simmonds P, Balfe P, Peutherer J, Ludlam C A, Bishop J O, Brown A J L. Human immunodeficiency virus-infected individuals contain provirus in small numbers of peripheral mononuclear cells and at low copy numbers. J Virol. 1990;64:864–872. doi: 10.1128/jvi.64.2.864-872.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith B P. Large animal internal medicine. Diseases of horses, cattle, sheep, and goats. St. Louis, Mo: C.V. Mosby Co.; 1990. [Google Scholar]
- 46.Swardson C J, Lichtenstein D L, Wang S, Montelaro R C, Kociba G J. Infection of bone marrow macrophages by equine infectious anemia virus. Am J Vet Res. 1997;58:1402–1407. [PubMed] [Google Scholar]
- 47.Tornquist S J, Oakes J L, Crawford T B. Elevation of cytokines associated with the thrombocytopenia of equine infectious anemia virus. J Gen Virol. 1997;78:2541–2548. doi: 10.1099/0022-1317-78-10-2541. [DOI] [PubMed] [Google Scholar]
- 48.Tumas D B, Hines M T, Perryman L E, Davis W C, McGuire T C. Corticosteroid immunosuppression and monoclonal antibody-mediated CD5+ T lymphocyte depletion in normal and equine infectious anemia virus-carrier horses. J Gen Virol. 1994;75:959–968. doi: 10.1099/0022-1317-75-5-959. [DOI] [PubMed] [Google Scholar]