Abstract
The Rep78 protein of adeno-associated virus (AAV) contains amino acid sequence motifs common to rolling-circle replication (RCR) initiator proteins. In this report, we describe RCR initiator-like activities of Rep78. We demonstrate that a maltose-binding protein (MBP)–Rep78 fusion protein can catalyze the cleavage and ligation of single-stranded DNA substrates derived from the AAV origin of replication. Rep-mediated single-stranded DNA cleavage was strictly dependent on the presence of certain divalent cations (e.g., Mn2+ or Mg2+) but did not require the presence of a nucleoside triphosphate cofactor. Electrophoretic mobility shift assays demonstrated that binding of single-stranded DNA by MBP-Rep78 was influenced by the length of the substrate as well as the presence of potential single-stranded cis-acting sequence elements. Site-directed mutagenesis was used to examine the role of specific tyrosine residues within a conserved RCR motif (motif 3) of Rep78. Replacement of Tyr-156 with phenylalanine abolished the ability of MBP-Rep78 to mediate the cleavage and ligation of single-stranded DNA substrates but not the ability to stably bind single-stranded DNA. The cleaving-joining activity of Rep78 is consistent with the mechanism of replicative intermediate dimer resolution proposed for the autonomous parvoviruses and may have implications for targeted integration of recombinant AAV vectors.
Adeno-associated virus type 2 (AAV-2), a member of the Parvoviridae family, is a nonenveloped, icosahedral virus with a linear, single-stranded DNA (ssDNA) genome of 4,680 nucleotides (nt) (reviewed in reference 3). The approximately 4.4-kb unique coding region of AAV-2 is flanked by inverted terminal repeat (ITR) sequences of 145 nt each. The ITRs serve as the viral origins of replication and are believed to form stem-loop structures that prime synthesis of the double-stranded, replicative form of the viral genome (11, 21, 35). The AAV genome contains two genes termed rep and cap. The cap gene encodes the viral capsid proteins. The rep gene encodes four overlapping nonstructural proteins (known as Rep78, Rep68, Rep52, and Rep40) from a single open reading frame (Fig. 1) via a combination of alternative promoter usage and differential splicing (17, 18, 26). The two largest Rep proteins, Rep78 and Rep68, recognize a cognate binding site within the AAV origin of replication, nick the origin in a site- and strand-specific fashion, and covalently attach to the 5′ end of the cleaved DNA substrate via a phosphotyrosine linkage (13, 19, 30, 33). In addition, the Rep proteins have been shown to possess ATPase and 3′-to-5′ helicase activities in vitro (14, 28, 37, 38). The ability to bind, cleave, and covalently attach to double-stranded ori sequences in a site- and strand-specific manner is a trait common to initiator proteins encoded by replicons employing rolling-circle DNA replication (RCR) (reviewed in references 5 and 22).
FIG. 1.
(A) Diagrammatic representation of the AAV genome. Relative positions of the rep and cap genes are indicated by open rectangles. The 145-nt ITRs, which serve as the viral origins of replication, are indicated by closed rectangles. (B) Schematic representation of the rep gene products. The 224-residue amino-terminal domain common to Rep78 and Rep68 contains a site-specific DNA-binding domain, as well as amino acid motifs conserved among RCR initiator proteins (12, 23). RCR motif 1 has not been identified within the parvovirus Rep proteins. An approximately 130-amino-acid domain containing motifs associated with helicase activity is indicated by the stippled rectangle. The helicase domain is common to all four rep gene products, as is the nuclear localization signal (NLS). Sequences encoding an approximately 90-amino-acid zinc finger domain (striped rectangle) are removed from Rep68 and Rep40 transcripts by a splicing event. (C) Comparison of the AAV Rep78/68 RCR motifs with equivalent motifs of RCR initiator proteins. Invariant amino acid residues are indicated by asterisks. A representative replicon is indicated at the left, with the associated initiator protein in parentheses. pUB110, an RCR plasmid of gram-positive bacteria; φX174, ssDNA bacteriophage; TYLCV, tomato yellow leaf curl geminivirus; B19, human parvovirus. (D) Representation of the AAV origin of replication. The trs is the site of strand-specific cleavage. RBS, Rep-binding site core.
RCR is an asymmetric replication mechanism in which leading-strand DNA synthesis is uncoupled from that of the lagging strand (5, 7, 22). The RCR mechanism has been described for a number of prokaryotic and eukaryotic replicons (5, 15, 22, 31), including ssDNA bacteriophage (e.g., φX174), various plasmids of gram-positive bacteria (e.g., pT181), and, more recently, members of the Geminiviridae (small ssDNA viruses infecting mono- and/or dicotyledonous plants). Moreover, an RCR-like mechanism is believed to play a role in the conjugative transfer of numerous bacterial plasmids, as well as the interspecies transfer of the Ti (tumor-inducing) plasmid of Agrobacterium tumefaciens (24, 25).
The Parvoviridae have been proposed to replicate via an RCR-like mechanism known as the rolling-hairpin model (32). The rolling-hairpin model, which utilizes a linear template with self-priming terminal palindromes, shares with the rolling-circle model the properties of asymmetric, leading-strand DNA synthesis resulting in the displacement of ssDNA, as well as initiation and termination of replication at a repetitively encountered ori sequence. Initiator-like proteins encoded by the Parvoviridae family (known as NS1 or Rep proteins) possess at least two RCR-associated amino acid sequence motifs (known as motifs 2 and 3) (12). Motif 2 contains the sequence HuHuuu (where u is any hydrophobic amino acid) and is believed to bind a divalent cation required for DNA strand cleavage. Motif 3 contains the sequence uxxYux1-2K (where u is any hydrophobic amino acid and x is any amino acid) and contains the catalytic tyrosine residue involved in nucleophilic attack and covalent attachment to the phosphodiester backbone of the DNA substrate. A motif 1 equivalent has not been identified within the parvovirus Rep proteins.
In this paper, we describe RCR initiator protein-like activities of the Rep78 protein of AAV-2. We demonstrate that a maltose-binding protein (MBP)–Rep78 fusion protein can specifically cleave ssDNA substrates derived from the AAV origin of replication. Cleavage of ssDNA is dependent on a divalent cation cofactor (Mn2+ supported single-stranded endonuclease activity much more efficiently than Mg2+) but does not require nucleoside triphosphate (NTP) hydrolysis. In addition, it is demonstrated that Rep78 can catalyze the ligation of single-stranded oligonucleotides derived from the AAV origin of replication. Site-directed mutagenesis demonstrated that tyrosine residue 156 of Rep78 (occurring within RCR motif 3) is essential for the cleavage-ligation activity.
MATERIALS AND METHODS
Plasmids and recombinant proteins.
Plasmids expressing an MBP-Rep78 fusion protein, pMBP-Rep78, and an MBP-Rep78(K340H) mutant, pMBP-Rep78(NTP), have been described previously (28, 29). MBP-LacZα is encoded by plasmid pMAL-c2 (New England Biolabs, Beverly, Mass.) MBP fusion proteins were expressed in Escherichia coli DH5α and purified by amylose affinity chromatography as described elsewhere (28, 29) except that the bacterial cultures were supplemented with 50 μM ZnSO4 prior to induction.
Site-directed mutagenesis.
Bacterial expression vectors encoding site-directed mutants of MBP-Rep78, pMBP-Rep78(Y152F), and pMBP-Rep78(Y156F) were constructed by high-fidelity, PCR-based mutagenesis of plasmid pMBP-Rep78 (28, 29), using a QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, Calif.) according to the manufacturer's instructions. Complementary mutagenic oligonucleotides used in the construction of pMBP-Rep78(Y152F) were 5′-GATGAGTGCTTTATCCCCAATTAC-3′ and 5′-GTAATTGGGGATAAAGCACTCATC-3′. The complementary oligonucleotide pair used in the construction of pMBP-Rep78(Y156F) was 5′-ATCCCCAATTTCTTGCTCCCCA-3′ and 5′-TGGGGAGCAAGAAATTGGGGAT-3′. Base changes relative to the wild-type rep sequence are underlined. Mutations were confirmed by DNA sequencing.
In vitro cleavage and ligation reactions.
Oligodeoxynucleotide substrates were 5′-end labeled with [γ-32P]ATP (>5,000 Ci/mmol; Amersham Pharmacia Biotech) in the presence of T4 polynucleotide kinase. Unincorporated radiolabel was removed by Sephadex G-25 chromatography. Labeled substrates were added to cleavage reaction buffer (14 μl) containing 30 mM Tris-HCl (pH 8.0), 4 mM MnCl2, 30 mM NaCl, 1 mM dithiothreitol (DTT), and various amounts of MBP fusion protein. Following incubation at 37°C for 1 h, each reaction mixture received 0.5 volume of loading buffer (0.15% bromophenol blue and 0.15% xylene cyanol in 100% formamide) and was heated to 90°C for 10 min prior to gel loading. Cleavage products were analyzed by electrophoresis in denaturing polyacrylamide gels containing 7 M urea and 1× Tris-borate-EDTA (TBE) buffer.
For Rep-mediated ligation reactions, approximately 50 pmol of unlabeled 26-mer (Fig. 2) was mixed with 2 μg of MBP fusion protein (approximately 18 pmol) and 0.1 pmol of 5′-end-labeled AAV ori oligonucleotide (55-mer [Fig. 2]) in the cleavage reaction buffer described above. Reactions were incubated at 37°C for 1 h. Following the addition of formamide loading buffer, samples were heated to 90°C for 10 min and then resolved by denaturing electrophoresis on a 13% polyacrylamide–7 M urea gel.
FIG. 2.
Oligodeoxynucleotide probes used in this study. Nucleotides 4536 to 4590 of the AAV ori region are shown at the top. The core Rep-binding motif (GCTC repeat) is boxed. The double-stranded ori nick site (known as the trs) is indicated. Sequences of various oligodeoxynucleotide probes are shown below. Non-AAV-derived nucleotides of the 63-mer and 33-mer probes are underlined. The 44-mer is a synthetic control oligonucleotide bearing a GACT repeat.
Electrophoretic mobility shift assays.
For ssDNA mobility shift assays, 0.5 μg of MBP-Rep78(Y156F) fusion protein was incubated for 30 min at 30°C with approximately 40,000 cpm of 5′-end-labeled oligonucleotide probe in a 14-μl reaction volume containing buffer A [30 mM Tris-HCl (pH 8.0), 50 mM NaCl, 4 mM MnCl2, 1 mM DTT, 0.1 mg of bovine serum albumin/ml, 25 μg of poly(dI-dC)/ml]. Following the addition of loading buffer (3.5 μl of 50% glycerol–0.5% bromophenol blue), each reaction was loaded onto a native 4.5% polyacrylamide gel and resolved by electrophoresis in 0.5× TBE running buffer.
For double-stranded DNA (dsDNA) mobility shift assays, a 5′-end-labeled 63-bp dsDNA oligonucleotide ori probe (80 fmol) containing sequences spanning AAV nt 4538 to 4584 (flanked by EcoRI and PstI sites) was incubated for 30 min at 30°C with 100 ng of MBP-Rep78(Y156F) or MBP-LacZα per reaction in buffer A (14 μl). For cold competition experiments, unlabeled ssDNA competitors were added in either 10- or 100-fold molar excess prior to addition of MBP fusion protein. Following incubation, the binding reactions were separated by electrophoresis on a native 5% polyacrylamide gel in 0.5× TBE buffer.
Gel elution and DNA sequencing.
A large-scale cleavage-ligation reaction (140 μl) containing 40 μg of MBP-Rep78, 1.7 × 107 cpm of 5′-end-labeled 55-mer (Fig. 2), and 10 μg of unlabeled 26-mer (Fig. 2) in reaction buffer (30 mM Tris-HCl [pH 8.0], 4 mM MnCl2, 30 mM NaCl, 1 mM DTT) was incubated in a 30°C waterbath for 1 h. After the addition of loading buffer and brief heating, the sample was separated on a preparative 8% polyacrylamide–6 M urea gel. Following exposure of the wet gel to a storage phosphor screen to identify the radiolabeled bands, Rep78-mediated ligation products were excised from the gel. Gel slices were finely diced and eluted in 10 ml of 1× TE buffer (10 mM Tris-HCl [pH 7.4], 1 mM EDTA) by soaking overnight at 37°C with constant agitation. Following volume reduction by sec-butanol extraction and ethanol precipitation, the 5′-end-labeled ligation products were sequenced using a Maxam-Gilbert DNA sequencing kit (Sigma, St. Louis, Mo.) as instructed by the manufacturer. M13 sequences were generated by cycle sequencing of single-stranded M13mp18 using a Taq DNA sequencing kit (Boehringer Mannheim) and 5′-end-labeled forward primer (17-mer) according to the manufacturer's instructions.
RESULTS
Site-specific cleavage of a single-stranded AAV ori oligonucleotide by MBP-Rep78.
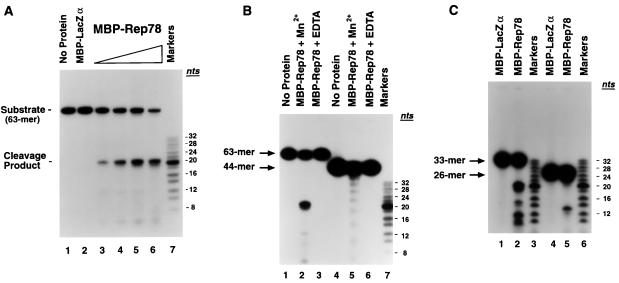
A common trait of RCR initiator proteins is the ability to specifically cleave ssDNA substrates derived from their origins of replication (5, 10). To determine whether the AAV Rep78 gene product (which contains conserved RCR motifs [Fig. 1]) is capable of site-specific cleavage of a single-stranded AAV ori substrate, an oligodeoxynucleotide (63-mer [Fig. 2]) derived from the AAV origin of replication (nt 4538 to 4584 flanked by EcoRI and PstI sites) was 5′-end labeled and incubated with purified fusion protein in an in vitro cleavage assay (Fig. 3A). The AAV ori oligonucleotide remained intact when incubated with an MBP-LacZα control protein (Fig. 3A, compare lanes 1 and 2). In contrast, incubation of the single-stranded ori substrate with increasing amounts of MBP-Rep78 fusion protein resulted in the dose-dependent production of a 21-nt cleavage product (lanes 3 to 6), indicating that the major ssDNA cleavage site maps to the same cleavage site utilized in Rep-mediated nicking of double-stranded AAV ori substrates (6, 13). This result indicates that Rep78 can specifically recognize its cognate, dsDNA cleavage site, known as the terminal resolution site (trs), in the form of ssDNA.
FIG. 3.
MBP-Rep78 fusion protein specifically cleaves a single-stranded AAV ori oligodeoxynucleotide. (A) A 5′-end-labeled, single-stranded AAV ori substrate (63-mer [Fig. 2]) was incubated with either no fusion protein (lane 1), MBP-LacZα (500 ng; lane 2), or an increasing amount of MBP-Rep78 (60, 125, 250, and 500 ng; lanes 3 to 6, respectively) for 1 h at 37°C. The reaction products were resolved by electrophoresis on a denaturing 18% polyacrylamide–7 M urea gel and visualized by autoradiography. Markers, 5′-end-labeled 2-nt oligodeoxynucleotide ladder (range, 8 to 32 nt; Pharmacia). (B) Cleavage of nonspecific ssDNA by MBP-Rep78. Oligodeoxynucleotides (63-mer and 44-mer [Fig. 2]) were 5′-end labeled and incubated with either no protein (lanes 1 and 4) or 0.8 μg of MBP-Rep78 in the presence (lanes 2 and 5) or absence (lanes 3 and 6) of 4 mM MnCl2. Cleavage products were separated on a denaturing 17% polyacrylamide–7 M urea gel. Markers, 2-nt oligodeoxynucleotide ladder (Pharmacia). (C) 5′-end-labeled oligonucleotides (33-mer and 26-mer [Fig. 2]) were incubated with either MBP-LacZα (0.8 μg; lanes 1 and 4) or MBP-Rep78 (0.8 μg; lanes 2 and 5) for 1 h at 37°C, then resolved in a denaturing 16% polyacrylamide–7 M urea gel, and visualized by autoradiography. Lanes 3 and 6 received a 5′-end-labeled 2-nt oligonucleotide ladder (Pharmacia).
To characterize further the specificity of MBP-Rep78 endonuclease activity on ssDNA, Rep-mediated cleavage of the 63-nt AAV ori substrate was compared to that observed with a non-AAV-derived, heterologous oligonucleotide substrate (Fig. 3B). As observed previously, cleavage of the 63-mer ori substrate by MBP-Rep78 occurred predominantly at the trs (Fig. 3B, lane 2), notable levels of cleavage also occurred between residues 12 and 13 relative to the 5′ end of the 63-mer substrate. Cleavage required the presence of a divalent cation cofactor (lane 3). Manganese supported ssDNA cleavage much more efficiently than did magnesium. Calcium and copper ions were ineffective as cofactors (data not shown). In contrast to requirements for the cleavage of dsDNA substrates (13, 14), Rep-mediated cleavage of ssDNA did not require (and was not enhanced by) the presence of ATP. To examine the effects of MBP-Rep78 on a nonspecific ssDNA substrate, a 44-mer consisting of a repeating GACT motif (and therefore lacking potential secondary structure) (44-mer [Fig. 2]) was 5′-end labeled and incubated with MBP-Rep78 in the presence or absence of MnCl2 (Fig. 3B, lanes 5 and 6, respectively). Although much less efficient that the cleavage of AAV ori sequences, cleavage of the 44-mer did occur (Fig. 3B, lane 5), resulting in the appearance of a 4-nt ladder. This result indicated that Rep-mediated cleavage occurred 5′ of each thymidine base within the GACT repeat sequence. This is consistent with cleavage at the double-stranded DNA trs, which occurs between the thymidine residues of a 5′-GTTG-3′ motif.
When a 33-nt substrate (33-mer [Fig. 2]) representing the left half of the 63-mer ori oligonucleotide (thus containing the trs but lacking the top strand of the dsDNA Rep-binding site) was incubated with MBP-Rep78, additional sites of DNA cleavage were more readily observed (Fig. 3C, lane 2). In addition to the trs, MBP-Rep78 cleaved the 33-mer 5′ of thymidine residues occurring at positions 10, 13, and 16 relative to the 5′ end of the oligonucleotide substrate. A radiolabeled, 26-nt ori-derived substrate (26-mer [Fig. 2]) was cleaved by MBP-Rep78 on the 5′-side of the first thymidine of the 5′-GTTG-3′ trs motif (Fig. 3C, lane 5). Rep-mediated cleavage at this position has been observed previously (6, 32a). Cleavage of the 26-mer at additional sites seen with the 63- and 33-nt substrates would result in fragments of 8 nt or less and therefore would not be visible on the gel.
Cleavage of single-stranded AAV ori sequences by Rep78 mutant proteins.
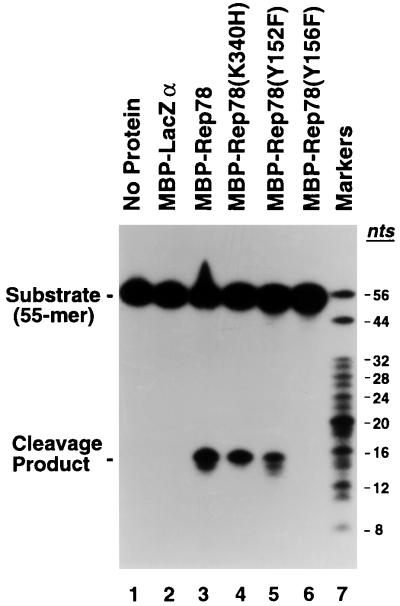
One of the conserved amino acid motifs (motif 3) associated with RCR initiator proteins is a uxxYxx1-2K motif (where u represents hydrophobic amino acid residues and x represents any amino acid) which contains a catalytic tyrosine residue involved in cleavage of (and covalent attachment to) the target DNA substrate (12). Certain RCR initiator proteins possess a second catalytic tyrosine four amino acid residues from the primary tyrosine and thus located on the same face of a potential α helix. One such initiator is the gene A protein of bacteriophage φX174 (Fig. 1C). Cleavage of ssDNA has been demonstrated for both gene A protein tyrosine residues (10, 34). Alternating cleavage activity between the tyrosine pair is believed to couple the termination of one round of strand displacement synthesis with reinitiation of the next, thus resulting in “runaway” replication of the phage DNA. As diagrammed in Fig. 1, AAV Rep78 also possesses two appropriately spaced tyrosine residues (Tyr-152 and Tyr-156) within its RCR motif 3 equivalent. To examine the role of Tyr-152 and Tyr-156 of Rep78 in ssDNA cleavage, site-directed mutagenesis was used to replace each tyrosine residue with phenylalanine. These mutant Rep proteins [Rep78(Y152F) and Rep78(Y156F)], as well as a previously described Rep78(K340H) mutant (28, 29), were expressed in E. coli as MBP-Rep fusion proteins. The purified fusion proteins were incubated with a 5′-end-labeled AAV ori substrate (55-mer [Fig. 2]) in an in vitro ssDNA cleavage assay (Fig. 4). MBP-LacZα failed to cleave the ssDNA oligonucleotide (lane 2). In contrast, MBP-Rep78 cleaved the 55-mer substrate in a site-specific fashion, resulting in the production of a 15-nt cleavage product (lane 3). An ATPase-deficient MBP-Rep78 mutant protein, MBP-Rep78(K340H), cleaved the ssDNA substrate as efficiently as wild-type MBP-Rep78 (lane 4), confirming that NTP hydrolysis is not required for Rep-mediated cleavage of ssDNA. MBP-Rep78(Y152F) retained the ability to specifically cleave ssDNA (lane 5). Mutation of the tyrosine residue at position 156 to phenylalanine, however, completely abolished the endonuclease activity of MBP-Rep78 (lane 6).
FIG. 4.
Single-stranded DNA cleavage by Rep78 mutant proteins. A radiolabeled oligonucleotide (55-mer [Fig. 2]) spanning AAV nt 4536 to 4590 was incubated with either no protein (lane 1), 2 μg of MBP-LacZα (lane 2), or 2 μg of the indicated MBP-Rep78 fusion protein (lanes 3 to 6). Following incubation, the cleavage products were resolved by electrophoresis in a denaturing 13% polyacrylamide–7 M urea gel. Markers, 5′-end-labeled oligonucleotide ladder (range, 8 to 32 nt; Pharmacia) supplemented with 44- and 56-nt markers.
Rep78 can stably bind single-stranded AAV ori sequences.
In light of the ability of MBP-Rep78 to specifically cleave a trs substrate presented in single-stranded form, we sought to determine the ability of Rep78 to stably bind single-stranded AAV ori sequences via an electrophoretic mobility shift assay using radiolabeled, oligodeoxynucleotides. One concern in these assays was the ability of Rep78 to cleave and covalently attach to the ssDNA probes. Binding experiments were therefore performed with the endonuclease-negative MBP-Rep78(Y156F) mutant protein.
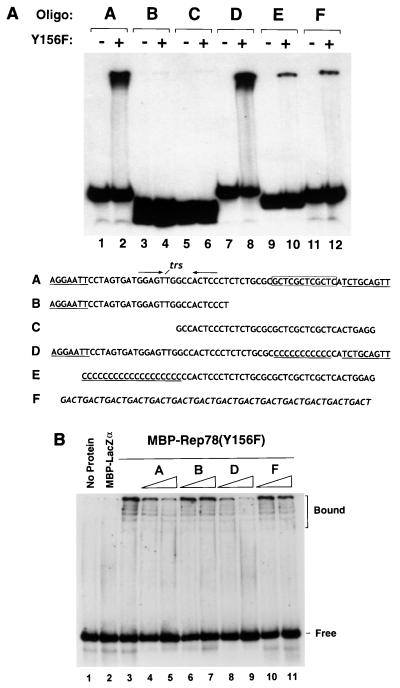
Characterization of the double-stranded AAV origin of replication has identified two important cis-acting elements: (i) the trs, consisting of a GTTG element flanked by a short palindrome, and (ii) the Rep-binding site (11, 19, 33, 35). We therefore sought to determine whether either of these known cis-acting elements plays a role in recognition of a single-stranded AAV ori substrate by Rep78. As shown in Fig. 5A, comparison of the binding levels observed with a non-AAV-derived control oligonucleotide to those observed with a 63-nt ori probe indicated that MBP-Rep78 was able to bind with high affinity to a single-stranded AAV ori substrate bearing the trs motif and top-strand Rep-binding site (compare lanes 2 and 12). However, when the trs motif and top strand of the Rep-binding site were presented separately in overlapping probes, only low levels of binding to either probe were observed (lanes 3 to 6). To address the possibility that binding of MBP-Rep78 to the radiolabeled ssDNA probes was subject to a minimal size requirement, the ability of MBP-Rep78 to bind a control oligonucleotide (in which the core GCTC triplet of the dsDNA Rep-binding site was replaced with cytosine residues) was determined. As shown in Fig. 5A, lane 8, MBP-Rep78 was able to bind the single-stranded GCTC-substituted substrate at levels comparable to those seen with the 63-nt parental probe. In contrast, a top-strand GCTC triplet-bearing probe extended at the 5′ end with cytosine residues was bound by MBP-Rep78 at levels equivalent to those seen with a non-AAV-derived probe (compare lanes 10 and 12). Taken together, these results indicate that Rep78 possesses the ability to bind with a relatively low affinity to nonspecific ssDNA and to bind with a much higher affinity to single-stranded AAV-derived sequences bearing the trs region.
FIG. 5.
Rep78 stably binds ssDNA oligonucleotides derived from the AAV origin of replication. (A) Single-stranded DNA mobility shift. MBP-Rep78(Y156F) (0.5 μg) was incubated with the indicated 5′-end-labeled single-stranded oligonucleotide probe as described in Materials and Methods. MBP-LacZα (0.5 μg; lanes 1, 3, 5, 7, 9, and 11) was tested as a control. The sequences and letter designations of the various oligonucleotide probes used are indicated below. Non-AAV sequences of probes A to E are underlined. The non-AAV-derived GACT-repeat control oligonucleotide (probe F) is italicized. (B) Cold competition experiment. Radiolabeled dsDNA probe (80 fmol) spanning the AAV ori region (nt 4538 to 4584) was incubated with either no fusion protein (lane 1), 100 ng of MBP-LacZα (lane 2), or 100 ng of MBP-Rep78(Y156F) (lane 3). Prior to the addition of fusion protein, samples resolved in lanes 4 to 11 received either a 10-fold (lanes 4, 6, 8, and 10) or 100-fold (lanes 5, 7, 9, and 11) molar excess of the indicated unlabeled, single-stranded competitor DNA. Positions of the bound and free probe are indicated at the right.
To confirm the ability of Rep78 to bind single-stranded AAV ori sequences with high affinity, we performed a cold competition experiment in which various unlabeled ssDNA oligonucleotides were tested for the ability to compete for binding of MBP-Rep78(Y156F) to a radiolabeled dsDNA ori probe consisting of the 63-mer oligonucleotide (probe A) annealed to its complementary strand. As shown in Fig. 5B, incubation of MBP-Rep78(Y156F) fusion protein with the dsDNA substrate resulted in the shift of the double-stranded ori probe (lane 3). Both the single-stranded 63-mer (probe A) and a 63-nt probe bearing a substitution of the GCTC triplet of the top-strand Rep-binding site (probe D), however, could compete for binding of MBP-Rep78 to double-stranded AAV ori sequences (lanes 4, 5, 8, and 9). In contrast, a 33-mer ori probe (probe B) and non-ori-derived 56-mer (probe F) failed to significantly compete for the binding of MBP-Rep78(Y156F) to dsDNA (lanes 6, 7, 10, and 11).
Cleavage-joining activities of MBP-Rep78.
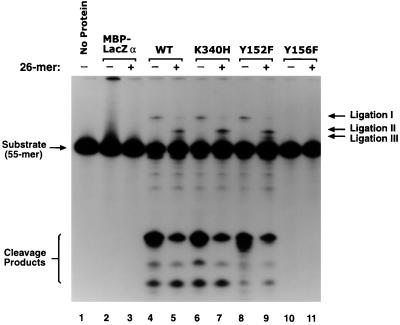
RCR initiator proteins have been shown to mediate in vitro recombination of oligonucleotide substrates bearing appropriate cleavage signals (10). As shown in Fig. 6, prolonged exposure of the MBP-Rep78 cleavage reaction gels to X-ray film indicated that novel Rep-dependent recombination products did indeed occur (note also that less abundant ssDNA cleavage products are more readily observed). Rep-mediated recombination of a radiolabeled input 55-mer oligodeoxynucleotide substrate (Fig. 2) resulted in a specific recombination product (ligation I) (Fig. 6, compare lanes 2 and 4). Addition of a second unlabeled oligodeoxynucleotide (26-mer [Fig. 2]) bearing the trs resulted in the appearance of a novel ligation product (ligation II) (Fig. 6, lane 5) resulting from Rep78-mediated recombination of the 55-mer and the 26-mer oligonucleotides. Rep-mediated recombination took place in the absence of NTP cofactors and could be observed with an ATPase-deficient Rep fusion protein (Fig. 6, lanes 6 and 7). An MBP-Rep78(Y152F) mutant protein, which retains the ability to cleave ssDNA, mediated recombination of the oligonucleotide substrates, resulting in the production of ligation products I and II, as well as an increased amount of a minor ligation product (ligation III) (lanes 8 and 9). The MBP-Rep78(Y156F) mutant, which binds ssDNA but lacks single-stranded endonuclease activity, failed to catalyze recombination of the oligonucleotide substrates. This result provides evidence that the ligation activity demonstrated by Rep78 is mediated by the catalytic tyrosine residue at position 156 within RCR motif 3 and not by a separate ligation domain within the protein.
FIG. 6.
MBP-Rep78 cleavage-ligation activity. The indicated MBP-Rep fusion protein (2 μg) was incubated with approximately 0.1 pmol of a 5′-end-labeled AAV ori oligonucleotide probe (55-mer) as described in Materials and Methods. Where indicated, an unlabeled 26-mer (50 pmol) bearing the trs (Fig. 2) was added to the reaction. After heating to 90°C for 10 min, samples were resolved on a denaturing 13% polyacrylamide–7 M urea gel in 1× TBE buffer. WT, wild type.
Characterization of Rep78-mediated ligation products.
To characterize further the Rep-mediated oligonucleotide ligation products, portions of samples used in Fig. 6 were separated on a DNA sequencing gel and compared to an M13 sequence ladder generated by the dideoxynucleotide chain termination method. Reactions which had received MBP-Rep78 and radiolabeled 55-mer (Fig. 7A, lane 1) contained a ligation product (ligation I) of approximately 104 nt. In addition to a small amount of ligation product I, reactions which had received a combination of MBP-Rep78, radiolabeled 55-mer, and unlabeled 26-mer (Fig. 7A, lane 2) contained a novel ligation product of approximately 76 nt (ligation II), as well as two less abundant molecular species of approximately 68 and 73 nt. To define more clearly the structure of the recombinant oligonucleotides, ligation products I and II were eluted from a preparative polyacrylamide gel and subjected to Maxam-Gilbert sequencing. The sequencing result for ligation product I is shown in Fig. 7B. Although hampered by the small amount of available starting material, the sequencing reactions clearly showed that ligation product I resulted from intermolecular recombination of the input 55-mer, while ligation product II arose from recombination of the 55-mer and 26-mer oligonucleotide substrates. The exact recombination junction for products I and II could not be determined from the sequencing gel, but based on size and flanking sequence information it was found that in each case the donor substrate (26-mer or 55-mer) was cleaved at a position six to eight bases from the 5′ end of the oligonucleotide and ligated to the 3′ end of an intact radiolabeled 55-mer acceptor molecule (Fig. 7C). The major ligation products therefore involved a donor substrate cleaved at a minor cleavage site upstream of the trs. One possible explanation for this observation is that when cleaved at the trs, intramolecular base pairing within the oligonucleotide substrate (see Fig. 7C) may cause the cleaved 5′ end of the oligonucleotide to be retained, thus favoring intramolecular (rather than intermolecular) rejoining. Alternatively, the retained 5′ cleavage product may block interactions between the acceptor molecule and the Rep-donor-substrate complex.
FIG. 7.
Characterization of Rep78-mediated ligation products. (A) The size of each Rep-mediated ligation product was determined by comparison to a Sanger sequencing reaction of M13 DNA (lanes G, A, T, and C). Portions of the samples analyzed in Fig. 6, lanes 2, 4, and 5 were separated on an 8% polyacrylamide sequencing gel. Lane 1, 5′-end-labeled 55-mer incubated with MBP-Rep78; lane 2, 5′-end-labeled 55-mer incubated with MBP-Rep78 and unlabeled 26-mer; lane 3, 5′-end-labeled 55-mer incubated with MBP-LacZα. (B) Ligation product I (ca. 104 nt) was eluted from a preparative polyacrylamide gel and subjected to Maxam-Gilbert sequencing as described in Materials and Methods. The sequence flanking the junction of the ligated oligonucleotide substrates is indicated. G, R, Y, and C, guanine-, purine-, pyrimidine-, and cytosine-specific cleavage reactions, respectively. (C) Schematic representation of the 55-mer ori substrate in its hairpin conformation. Rep78-mediated recombination occurred between the 3′ end of the 55-mer acceptor molecule and the cleaved 5′ end of a donor molecule (as indicated by the thin line). The major (large arrowhead) and minor (small arrowheads) single-stranded cleavage sites are indicated.
DISCUSSION
In this report, we demonstrate that the AAV Rep78 protein, which bears conserved amino acid motifs common to RCR initiator proteins, possesses the ability to cleave and ligate ssDNA substrates derived from the AAV origin of replication. Rep-mediated cleavage of non-ori substrates could be detected but was significantly less efficient than with ori-derived substrates. Cleavage of ssDNA by MBP-Rep78 required the presence of metal cations but did not require the presence of a NTP cofactor. Im and Muzyczka (13, 14) have shown that Rep-mediated cleavage of double-stranded AAV ori substrates requires the presence of both a divalent cation and ATP. One possible explanation for this contrast is that the preferred cleavage substrate of Rep78 is ssDNA and that NTP hydrolysis is required by Rep to partially unwind a dsDNA substrate prior to cleavage. Alternatively, the existence of a region of dyad symmetry centered about the trs (Fig. 2) raises the possibility that NTP hydrolysis may cause a conformational change in Rep that induces the formation of a stem-loop structure at the trs, thus exposing the cleavage site as a single-stranded region at the end of a hairpin. Rep-mediated ligation did not require NTP and was greatly enhanced when Mn2+, rather than Mg2+, was provided as a divalent cation cofactor. Site-directed mutagenesis of Tyr-156 within Rep78 RCR motif 3 abolished the ability of MBP-Rep78 to cleave and ligate ssDNA substrates.
The role of ssDNA ligation within the AAV replication cycle is speculative. Current models of AAV replication do not utilize a ligation step. However, replication models for the autonomous parvoviruses propose that resolution of the replicative intermediate form dimer requires a nicking and ligation step at the junction of the two monomeric units of the dimer (1, 2), and experimental evidence for the ligation event in infected cells has been reported (4). Applying the autonomous parvovirus dimer resolution model to AAV replication accounts for all of the known enzymatic activities of Rep78, including those reported in this paper. In this model, the viral ITR at the junction of the genomic dimer exists in its fully base-paired conformation (i.e., as linear DNA) (Fig. 8B). Rep78 binds its cognate dsDNA-binding site and uses the energy of ATP hydrolysis to either partially unwind the double-stranded ori sequence or induce the formation of a stem-loop structure, thereby exposing the Rep cleavage site (trs) in the form of ssDNA (Fig. 8C). Upon strand cleavage and covalent attachment to the 5′ end of the DNA, Rep78 helicase activity unwinds the viral DNA in a 3′-to-5′ direction (Fig. 8D), resulting in strand displacement and asymmetric DNA replication mediated by the cellular replication machinery and primed by the 3′-hydroxyl group generated by Rep-mediated strand cleavage. After traversing the ITR region, a second single-stranded ori element is encountered in the proper orientation. Cleavage of the newly encountered origin, either by a subunit of a Rep78 multimer or possibly by the second tyrosine residue occurring at position 152 within RCR motif 3, results in transfer of the covalently attached DNA strand of Rep78 to the newly cleaved trs, thus resolving the genomic dimer into replicative-form monomers (Fig. 8E). This model is consistent with the notion that rather than possessing a viral replication strategy that is unique to the Dependovirus genus, AAV may utilize a replication mechanism similar to that of the autonomous parvoviruses. The dimer resolution model resembles RCR replication in several ways: (i) both utilize a replicon-encoded initiator protein that recognizes the double-stranded form of an origin of replication; (ii) both initiate asymmetric, strand displacement DNA synthesis primed by a strand-specific DNA cleavage event; (iii) both terminate one round of DNA synthesis upon encountering a second origin of replication; and (iv) both utilize an initiator protein-mediated strand transfer mechanism to resolve the replication intermediates.
FIG. 8.
Genomic dimer resolution model. Details of the dimer resolution model (1, 2) as applied to AAV are described in the text. (A) Double-stranded AAV genomic dimer. Filled rectangles represent the viral ITR sequences. (B to E) Enlargement of the dimer junction ITR element. The left and right Rep-binding sites are designated by stippled and striped rectangles, respectively. The trs is represented by a filled circle. Rep78 (filled oval) is depicted as a monomer for simplicity. Cellular replication components are not indicated. RFD, replicative-form genomic dsDNA dimer; RFM, replicative-form genomic dsDNA monomer.
AAV is unique among the animal viruses in its ability to integrate within a specific chromosomal locus (16, 27). The chromosomal integration site on human chromosome 19 (known as AAVS1) resembles the AAV origin of replication (8, 33). The ability of Rep78 to covalently attach to its DNA substrate (30, 33) and to mediate recombination of AAV ori-derived sequences (this report) raises the possibility that Rep78 may mediate recombination between an AAV replication intermediate and the chromosomal AAVS1 locus by a strand cleavage and exchange mechanism. Interestingly, characterization of recombinant junctions formed in vivo between AAV DNA and the flanking integration site sequences revealed that the majority of integration events involved the viral ITR sequences (9, 20). Moreover, Rep78 has been demonstrated to interact with AAVS1 sequences in vitro (36). Understanding the relationship of Rep function to AAV integration may aid in the development of AAV gene therapy vectors.
ACKNOWLEDGMENTS
We thank Brian Safer for reviewing the manuscript, Sandra Afione for sharing unpublished observations, and Michael Schmidt for helpful technical advice.
REFERENCES
- 1.Astell C R, Chow M B, Ward D C. Sequence analysis of the termini of virion and replicative forms of minute virus of mice DNA suggests a modified rolling hairpin model for autonomous parvovirus DNA replication. J Virol. 1985;54:171–177. doi: 10.1128/jvi.54.1.171-177.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Astell C R, Liu Q, Harris C E, Brunstein J, Jindal H K, Tam P. Minute virus of mice cis-acting sequences required for genome replication and the role of the trans-acting protein, NS-1. Prog Nucleic Acid Res Mol Biol. 1996;55:245–285. doi: 10.1016/s0079-6603(08)60196-8. [DOI] [PubMed] [Google Scholar]
- 3.Berns K I, Bohenzky R A. Adeno-associated viruses: an update. Adv Virus Res. 1987;32:243–306. doi: 10.1016/s0065-3527(08)60479-0. [DOI] [PubMed] [Google Scholar]
- 4.Cotmore S F, Gunther M, Tattersall P. Evidence for a ligation step in the DNA replication of the autonomous parvovirus minute virus of mice. J Virol. 1989;63:1002–1006. doi: 10.1128/jvi.63.2.1002-1006.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Espinosa M, del Solar G, Rojo F, Alonso J C. Plasmid rolling circle replication and its control. FEMS Microbiol Lett. 1995;130:111–120. doi: 10.1111/j.1574-6968.1995.tb07707.x. [DOI] [PubMed] [Google Scholar]
- 6.Fife K H, Berns K I, Murray K. Structure and nucleotide sequence of the terminal regions of adeno-associated virus. Virology. 1977;78:475–487. doi: 10.1016/0042-6822(77)90124-6. [DOI] [PubMed] [Google Scholar]
- 7.Gilbert W, Dressler D. The rolling circle model. Cold Spring Harbor Symp Quant Biol. 1968;33:473–484. doi: 10.1101/sqb.1968.033.01.055. [DOI] [PubMed] [Google Scholar]
- 8.Giraud C, Winocour E, Berns K I. Site-specific integration by adeno-associated virus is directed by a cellular DNA sequence. Proc Natl Acad Sci USA. 1994;91:10039–10043. doi: 10.1073/pnas.91.21.10039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giraud C, Winocour E, Berns K I. Recombinant junctions formed by site-specific integration of adeno-associated virus into an episome. J Virol. 1995;69:6917–6924. doi: 10.1128/jvi.69.11.6917-6924.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanai R, Wang J C. The mechanism of sequence-specific DNA cleavage and strand transfer by φX174 gene A* protein. J Biol Chem. 1993;268:23830–23836. [PubMed] [Google Scholar]
- 11.Hong G, Ward P, Berns K I. In vitro replication of adeno-associated virus DNA. Proc Natl Acad Sci USA. 1992;89:4673–4677. doi: 10.1073/pnas.89.10.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ilyina T V, Koonin E V. Conserved sequence motifs in the initiator proteins for rolling circle DNA replication encoded by diverse replicons from eubacteria, eucaryotes and archaebacteria. Nucleic Acids Res. 1992;20:3279–3285. doi: 10.1093/nar/20.13.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Im D-S, Muzyczka N. The origin binding protein Rep68 is an ATP-dependent site-specific endonuclease with DNA helicase activity. Cell. 1990;61:447–457. doi: 10.1016/0092-8674(90)90526-k. [DOI] [PubMed] [Google Scholar]
- 14.Im D-S, Muzyczka N. Partial purification of adeno-associated virus Rep78, Rep52, and Rep40 and their biochemical characterization. J Virol. 1992;66:1119–1128. doi: 10.1128/jvi.66.2.1119-1128.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koonin E V, Ilyina T V. Geminivirus replication proteins are related to prokaryotic plasmid rolling circle DNA replication initiator proteins. J Gen Virol. 1992;73:2763–2766. doi: 10.1099/0022-1317-73-10-2763. [DOI] [PubMed] [Google Scholar]
- 16.Kotin R M, Siniscalco M, Samulski R J, Zhu X, Hunter L, Laughlin C A, McLaughlin S, Muzyczka N, Rocchi M, Berns K I. Site-specific integration by adeno-associated virus. Proc Natl Acad Sci USA. 1990;87:2211–2215. doi: 10.1073/pnas.87.6.2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laughlin C A, Westphal H, Carter B J. Spliced adenovirus-associated virus RNA. Proc Natl Acad Sci USA. 1979;76:5567–5571. doi: 10.1073/pnas.76.11.5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lusby E W, Berns K I. Mapping of the 5′ termini of two adeno-associated virus RNAs in the left half of the genome. J Virol. 1982;41:518–526. doi: 10.1128/jvi.41.2.518-526.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCarty D M, Ryan J H, Zolotukhin S, Zhou X, Muzyczka N. Interaction of the adeno-associated virus Rep protein with a sequence within the A palindrome of the viral terminal repeat. J Virol. 1994;68:4998–5006. doi: 10.1128/jvi.68.8.4998-5006.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakai H, Iwaki Y, Kay M A, Couto L B. Isolation of recombinant adeno-associated virus vector-cellular DNA junctions from mouse liver. J Virol. 1999;73:5438–5447. doi: 10.1128/jvi.73.7.5438-5447.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ni T-H, Zhou X, McCarty D M, Zolotukhin I, Muzyczka N. In vitro replication of adeno-associated virus DNA. J Virol. 1994;68:1128–1138. doi: 10.1128/jvi.68.2.1128-1138.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Novick R P. Staphylococcal plasmids and their replication. Annu Rev Microbiol. 1989;43:537–565. doi: 10.1146/annurev.mi.43.100189.002541. [DOI] [PubMed] [Google Scholar]
- 23.Owens R A, Weitzman M D, Kyostio S R M, Carter B J. Identification of a DNA-binding domain in the amino terminus of adeno-associated virus Rep proteins. J Virol. 1993;67:997–1005. doi: 10.1128/jvi.67.2.997-1005.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pansegrau W, Lanka E. Enzymology of DNA transfer by conjugative mechanisms. Prog Nucleic Acid Res Mol Biol. 1996;54:197–251. doi: 10.1016/s0079-6603(08)60364-5. [DOI] [PubMed] [Google Scholar]
- 25.Pansegrau W, Schoumacher F, Hohn B, Lanka E. Site-specific cleavage and joining of single-stranded DNA by VirD2 protein of Agrobacterium tumefaciens Ti plasmids: analogy to bacterial conjugation. Proc Natl Acad Sci USA. 1993;90:11538–11542. doi: 10.1073/pnas.90.24.11538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Redemann B E, Mendelson E, Carter B J. Adeno-associated virus Rep protein synthesis during productive infection. J Virol. 1989;63:873–882. doi: 10.1128/jvi.63.2.873-882.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Samulski R J, Zhu X, Xiao X, Brook J D, Housmann D E, Epstein N, Hunter L A. Targeted integration of adeno-associated virus (AAV) into human chromosome 19. EMBO J. 1991;10:3941–3950. doi: 10.1002/j.1460-2075.1991.tb04964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith R H, Kotin R M. The Rep52 gene product of adeno-associated virus is a DNA helicase with 3′-to-5′ polarity. J Virol. 1998;72:4874–4881. doi: 10.1128/jvi.72.6.4874-4881.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith R H, Spano A J, Kotin R M. The Rep78 gene product of adeno-associated virus (AAV) self-associates to form a hexameric complex in the presence of AAV ori sequences. J Virol. 1997;71:4461–4471. doi: 10.1128/jvi.71.6.4461-4471.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Snyder R O, Im D-S, Muzyczka N. Evidence for covalent attachment of the adeno-associated virus (AAV) Rep protein to the ends of the AAV genome. J Virol. 1990;64:6204–6213. doi: 10.1128/jvi.64.12.6204-6213.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stenger D C, Revington G N, Stevenson M C, Bisar D M. Replicational release of geminivirus genomes from tandemly repeated copies: evidence for rolling-circle replication of a plant viral DNA. Proc Natl Acad Sci USA. 1991;88:8029–8033. doi: 10.1073/pnas.88.18.8029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tattersall P, Ward D C. Rolling hairpin model for replication of parvovirus and linear chromosomal DNA. Nature (London) 1976;263:106–109. doi: 10.1038/263106a0. [DOI] [PubMed] [Google Scholar]
- 32a.Urabe M, Hasumi Y, Kume A, Surosky R T, Kurtzman G J, Tobita K, Ozawa K. Charged-to-alanine scanning mutagenesis of the N-terminal half of adeno-associated virus type 2 Rep78 protein. J Virol. 1999;73:2682–2693. doi: 10.1128/jvi.73.4.2682-2693.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Urcelay E, Ward P, Wiener S M, Safer B, Kotin R M. Asymmetric replication in vitro from a human sequence element is dependent on adeno-associated virus Rep protein. J Virol. 1995;69:2038–2046. doi: 10.1128/jvi.69.4.2038-2046.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Mansfeld A D M, van Teeffelen H A A M, Baas P D, Jansz H S. Two juxtaposed tyrosyl-OH groups participate in φX174 gene A protein catalysed cleavage and ligation of DNA. Nucleic Acids Res. 1986;14:4229–4238. doi: 10.1093/nar/14.10.4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ward P, Berns K I. Minimum origin requirements for linear duplex AAV DNA replication in vitro. Virology. 1995;209:692–695. doi: 10.1006/viro.1995.1306. [DOI] [PubMed] [Google Scholar]
- 36.Weitzman M D, Kyostio S R M, Kotin R M, Owens R A. Adeno-associated virus (AAV) Rep proteins mediate complex formation between AAV DNA and its integration site in human DNA. Proc Natl Acad Sci USA. 1994;91:5808–5812. doi: 10.1073/pnas.91.13.5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wonderling R S, Kyostio S R M, Owens R A. A maltose-binding protein/adeno-associated virus Rep68 fusion protein has DNA-RNA helicase and ATPase activities. J Virol. 1995;69:3542–3548. doi: 10.1128/jvi.69.6.3542-3548.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou X, Zolotukhin I, Im D-S, Muzyczka N. Biochemical characterization of adeno-associated virus Rep68 DNA helicase and ATPase activities. J Virol. 1999;73:1580–1590. doi: 10.1128/jvi.73.2.1580-1590.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]