Significance
Hepatitis B virus (HBV) infections are the leading cause of chronic hepatitis, which may lead to cirrhosis and hepatocellular carcinoma. We found the HBV-encapsidated pregenomic RNA (pgRNA) to be decorated with high amounts of small chemical modifications. Among which, the cytidine methylation, m5C is found on the HBV pgRNA packaging signal, epsilon. Depletion of the methyltransferase NSUN2 from host cells results in decreased m5C on epsilon, a corresponding decrease in viral core protein expression and a block in the reverse transcription of viral pgRNA into DNA. This study uncovers a host-aided RNA modification required for the production of genomic DNA-containing infectious viral particles and suggests a unique mechanism that may be an antiviral target.
Keywords: hepatitis B virus, reverse transcription, m5C, NSUN2, RNA modifications
Abstract
Epitranscriptomic RNA modifications have emerged as important regulators of the fate and function of viral RNAs. One prominent modification, the cytidine methylation 5-methylcytidine (m5C), is found on the RNA of HIV-1, where m5C enhances the translation of HIV-1 RNA. However, whether m5C functionally enhances the RNA of other pathogenic viruses remains elusive. Here, we surveyed a panel of commonly found RNA modifications on the RNA of hepatitis B virus (HBV) and found that HBV RNA is enriched with m5C as well as ten other modifications, at stoichiometries much higher than host messenger RNA (mRNA). Intriguingly, m5C is mostly found on the epsilon hairpin, an RNA element required for viral RNA encapsidation and reverse transcription, with these m5C mainly deposited by the cellular methyltransferase NSUN2. Loss of m5C from HBV RNA due to NSUN2 depletion resulted in a partial decrease in viral core protein (HBc) production, accompanied by a near-complete loss of the reverse transcribed viral DNA. Similarly, mutations introduced to remove the methylated cytidines resulted in a loss of HBc production and reverse transcription. Furthermore, pharmacological disruption of m5C deposition led to a significant decrease in HBV replication. Thus, our data indicate m5C methylations as a critical mediator of the epsilon elements’ function in HBV virion production and reverse transcription, suggesting the therapeutic potential of targeting the m5C methyltransfer process on HBV epsilon as an antiviral strategy.
Epitranscriptomic RNA modifications have been found on the RNA of diverse virus families, where they mostly enhance viral replication, with a few exceptions (1, 2). In particular, we previously found the RNA of HIV-1 highly enriched with multiple modifications, with ~20% more N6-methyladenosine (m6A), 14x more 5-methylcytidine (m5C), and ~20x more 2′-O-methylations (Nm) than that on host mRNAs (3). Functionally, m5C boosts HIV-1 mRNA translation, m6A enhances the stability of HIV-1 RNA, N4-acetylcytidine (ac4C) prevents viral RNA premature degradation, and Nm is reported to protect HIV-1 RNA from innate immune detection (3–7). Thus, HIV-1, if not other viruses as well, may have evolved to have its RNA enriched with modifications that enhance viral replication through divergent mechanisms.
Hepatitis B virus (HBV) is a highly pathogenic blood-borne virus whose DNA genome, similar to HIV-1, is produced through the reverse transcription of capsid-packaged viral RNA, making RNA an essential intermediate in the replication of the viral genome (8). The RNA modification, m6A has been identified on the pregenomic RNA (pgRNA) of HBV, with m6A reported to decrease viral RNA stability and yet enhance pgRNA encapsidation and reverse transcription (9–11). However, the presence/function of any other of the >150 modifications on HBV RNA remains enigmatic (12). In particular, the cytidine methylation m5C is prominently found on mammalian mRNAs (13–15), with its’ function diverse and seemingly context dependent. m5C abundance on mRNAs negatively correlates with ribosome occupancy on ~61% of transcripts, yet enhances the translation of specific transcripts such as p21 & CDK1, and mildly enhances mRNA nuclear export (16–19). In addition, m5C was also reported to stabilize mRNAs in cancer cells (20, 21). For viral transcripts, m5C was first found on mRNAs of Sindbis virus (22), with its’ presence enhancing viral gene translation of HIV-1 as well as the replication of the model retrovirus murine leukemia virus (MLV) (3, 23, 24).
Here, we hypothesize that the RNA of HBV can be decorated and functionally regulated by m5C, in analogy to other reverse-transcribing viruses. In support of our hypothesis, we found that HBV RNA is substantially more enriched with multiple RNA modifications than host mRNAs, including m5C. HBV RNA m5C is mostly deposited by the host methyltransferase NSUN2, the methyltransferase responsible for ~75% of the m5C on host mRNAs (14, 25). These viral m5C are located on the viral RNA terminal “epsilon” elements, which are essential for initiating viral reverse transcription while encompassing the viral capsid (HBc) start codon. Consistent with the function of epsilon, depletion of NSUN2 leads to a partial defect in HBc translation and an almost complete loss of viral reverse transcription products. A similar defect was observed when point mutants were introduced to remove the methylated C residues without disrupting the epsilon structure, while a small molecule drug disruptor of m5C methylation can significantly inhibit HBV replication. Overall, our data suggest that HBV RNA is highly enriched with diverse RNA modifications, with NSUN2-deposited m5Cs on the epsilon RNA element enhancing viral particle production, a host-mediated mechanism that may be targeted for future antiviral development.
Results
We first ask whether the cytidines on HBV RNA are m5C methylated. As encapsidated pgRNA is degraded during reverse transcription, for detection of m5C on pgRNA, we used an HBV replicon with a Y63D-mutant pol gene (reverse transcriptase, RT) to preserve pgRNA (26, 27). Supernatants were collected from human hepatocyte HuH-7 cells transfected with the RT-mutant replicon, and the purified viral RNA dotted on a membrane. The membrane was stained with an antibody previously used to detect m5C (3, 28) and counterstained with an HBV-specific Northern blot probe (SI Appendix, Fig. S1A). m5C signals were observed on the same dots as detected by HBV probes, but not in the virus-free (mock) controls, suggesting that HBV pgRNA may contain m5C. We next profiled the RNA modification landscape of HBV pgRNA using ultra-high-performance liquid chromatography linked to tandem mass spectrometry (UPLC–MS/MS). Viral capsids within lysates of RT-mutant replicon transfected cells were fractionated over a sucrose gradient (SI Appendix, Fig. S1B). Fractions with viral RNA-containing capsids were collected, the capsids pelleted, and the RNA extracted. This highly purified capsid-associated RNA contained the full-length pgRNA (~3.2 kb) along with a range of spliced viral transcripts (29), while devoid of the distinctive sizes of host ribosomal RNAs (rRNAs) (SI Appendix, Fig. S1 E and F). For comparison, HuH-7 cellular RNA was also prepared, poly(A) selected, and rRNA depleted, also resulting in minimal residual rRNA (SI Appendix, Fig. S1 C, D, and F). These purity-verified viral and cellular RNA were digested to single nucleotides and subjected to UPLC–MS/MS identification of nucleotides, using a panel of commercially available nucleotide standards as reference (SI Appendix, Table S1) (30). We were able to quantify thirteen modified nucleosides in HBV RNA and host mRNAs (Fig. 1A, detailed in Table 1). Interestingly, HBV is highly enriched with multiple modifications, with viral RNA m5C abundances ~11-fold higher than that on host cellular mRNAs, and most other modifications between 2 and 20x more abundant on HBV RNA as compared to cellular mRNAs, with the exception of 2’O-methyluridine (Um) and inosine (I). Among cytidine modifications, the modification rate of m5C on HuH-7 mRNAs is roughly half of that previously found in T lymphocytes, while 2’O-methylcytidine (Cm) rates are comparable (3). Notably, 0.22% of HBV C residues are m5C methylated (Fig. 1B), compared to 0.02% m5C on HuH-7 mRNAs. Given that there are 883 cytidines in the 1.1mer pgRNA (ayw strain), we estimate roughly two m5C residues on every pgRNA molecule. Other modifications including 2’O-methylcytidine (Cm at 0.77%), N4-acetylcytidine (ac4C at 0.037%), as well as pseudouridine (ψ at 1.025%) were also detected and may be of future interest. All in all, two orthogonal detection methods suggest that HBV pgRNA contains m5C-methylated cytidines.
Fig. 1.
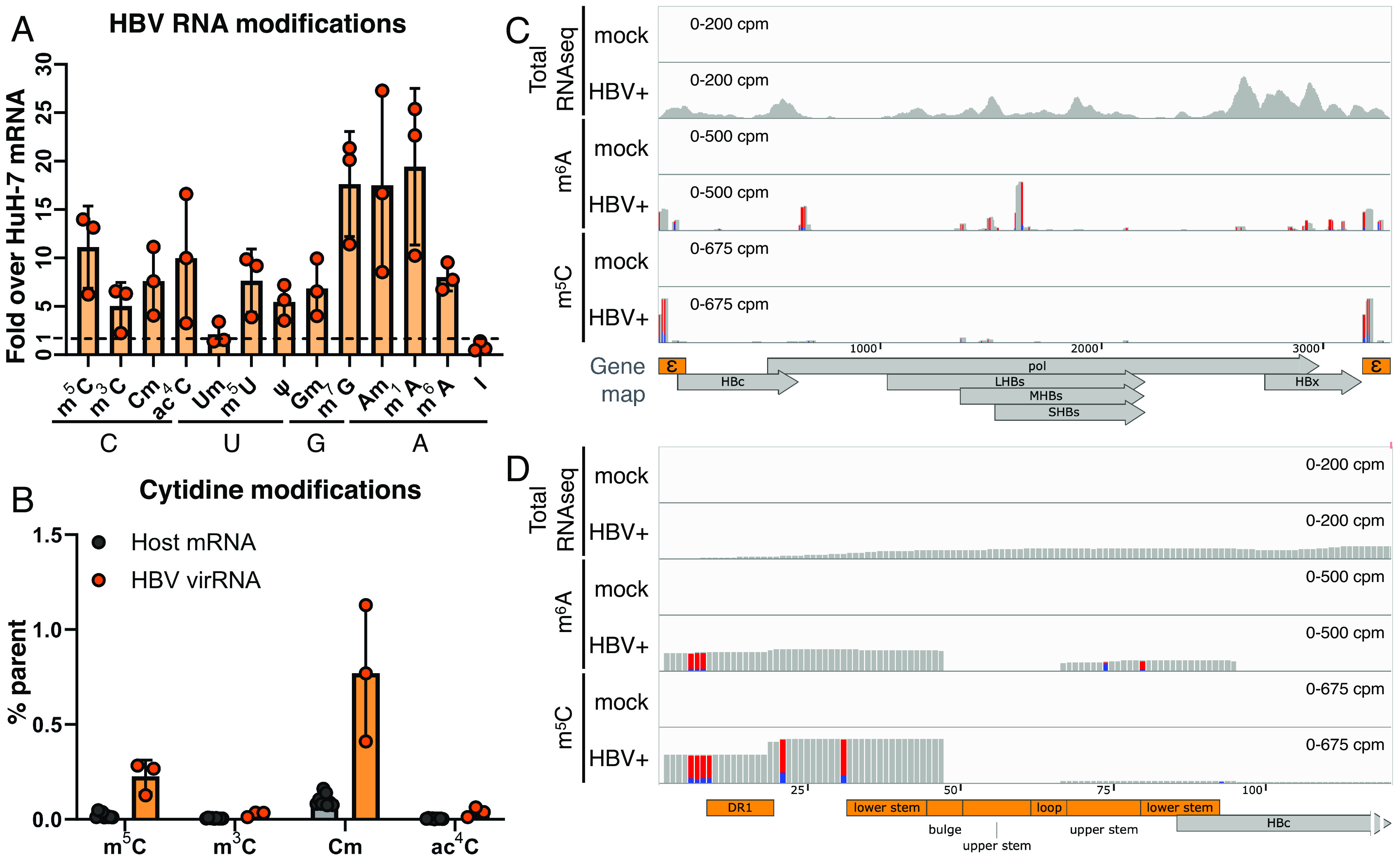
HBV RNA is m5C methylated. (A and B) Quantification of RNA modifications on HBV virion RNA by UPLC–MS/MS, purified from HBV pol Y63D (RT mut.) virions, shown normalized to the levels of that on HuH-7 mRNA (A), with cytidine modifications also presented as percent total cytidine (B). (C and D) m6A and m5C sites on HBV RNA were mapped using PA-m6A/m5C-seq on HuH-7 cells transfected with or without (mock) the wildtype ayw strain HBV replicon. Accompanying total RNA-seqs shown in the top track as the pulldown input control. The whole HBV pgRNA depicted in (C), with the epsilon region enlarged in (D). Track heights scaled as the shown counts per million total reads (cpm), red-blue bars depict T>C conversions from 4SU-crosslinking.
Table 1.
Relative amount of RNA modifications detected on purified HBV core-associated RNA and host (HuH-7) poly(A)+ mRNA by UPLC–MS/MS
| Residue | % Parent nucleotide on HBV RNA | % Parent nucleotide on host mRNA | HBV RNA /host mRNA |
|---|---|---|---|
| m5C | 0.226 | 0.020 | 11.12 |
| m3C | 0.027 | 0.005 | 5.04 |
| Cm | 0.769 | 0.101 | 7.59 |
| ac4C | 0.037 | 0.003 | 9.96 |
| Um | 0.046 | 0.021 | 2.12 |
| m5U | 0.035 | 0.004 | 7.64 |
| ψ | 1.025 | 0.187 | 5.46 |
| Gm | 0.147 | 0.021 | 6.96 |
| m7G | 1.643 | 0.093 | 17.63 |
| Am | 0.841 | 0.048 | 17.5 |
| m1A | 1.693 | 0.087 | 19.43 |
| m6A | 4.109 | 0.513 | 8.00 |
| I | 0.348 | 0.419 | 0.83 |
We previously utilized a photoactivatable-ribonucleoside-enhanced cross-linking and immunoprecipitation (PAR-CLIP)-derived strategy to map RNA modifications including m6A, m5C, ac4C, and ψ, on the RNA of HIV-1 and other viruses (3, 4, 6, 31, 32), originally developed as photo-crosslinking-assisted m6A sequencing (PA-m6A-seq) (33). To map m5C sites using an m5C antibody (PA-m5C-seq) (3), we extracted RNA from wildtype HBV replicon-transfected HuH-7 cells that were pulsed with the uridine analog 4-thiouridine (4SU) and crosslinked the 4SU+ RNA to m5C antibodies with 365 nm UV. The crosslinked and immunoprecipitated RNA was trimmed with RNase to the width of the antibody (~32 nt), reverse transcribed, and sequenced. Antibody-crosslinked 4SU results in characteristic T>C mutations in the sequencing reads, allowing for the identification of true antibody-crosslinked reads. As a control, PA-m6A-seq was also done in parallel, confirming the previously reported m6A sites on the epsilon element and HBx (9, 11, 34), with a few more peaks on pol and HBs (Fig. 1C) (35). To ensure 4SU-crosslinking wasn’t biased toward U-rich sequences, PA-m5C-seq was repeated using the alternative photoreactive nucleoside, 6-thioguanosine (6SG) (36). Regardless of 4SU or 6SG-crosslinking, m5C predominantly mapped to a cluster overlapping the epsilon element, with a few minor peaks in HBs (Fig. 1C and SI Appendix, Fig. S1G). Zooming in, the major m5C peak ranges from direct repeat 1 (DR1) to the lower stem of the epsilon hairpin (Fig. 1D and SI Appendix, Fig. S6A). Epsilon on the 5′ end of pgRNA acts as the RNA packaging signal and contains the reverse transcription initiation site, while every HBV transcript also includes a copy of epsilon in the 3′UTR. Due to the short read nature of Illumina sequencing, we could not differentiate the 5′ and 3′ epsilon -aligning reads and these reads were thus equally assigned to both ends.
If HBV RNA is indeed m5C methylated, we should be able to coimmunoprecipitate HBV RNA with the methyltransferase that deposits these m5Cs. m5C can be deposited on RNAs either by the NSUN family of methyltransferases (NSUN1-NSUN7) or DNMT2 (37). We cloned FLAG-tagged m5C methyltransferase candidates and yet were only able to obtain clones of NSUN2 and NSUN4 through NSUN6 from HuH-7 cDNA. Nevertheless, cells were cotransfected with these NSUN plasmids as well as an HBV replicon and pulsed with the cytidine analog 5-azacytidine (5-AzaC), which upon incorporation into RNA leads to m5C methyltransferases covalently stuck on the substrate RNA (38). Upon immunoprecipitating the 5-AzaC-stabilized NSUN-RNA complexes, NSUN2 showed the strongest copurification with HBV RNA (Fig. 2 A and B), suggesting that HBV RNA interacts with NSUN2, the primary host mRNA m5C methyltransferase (14, 25).
Fig. 2.
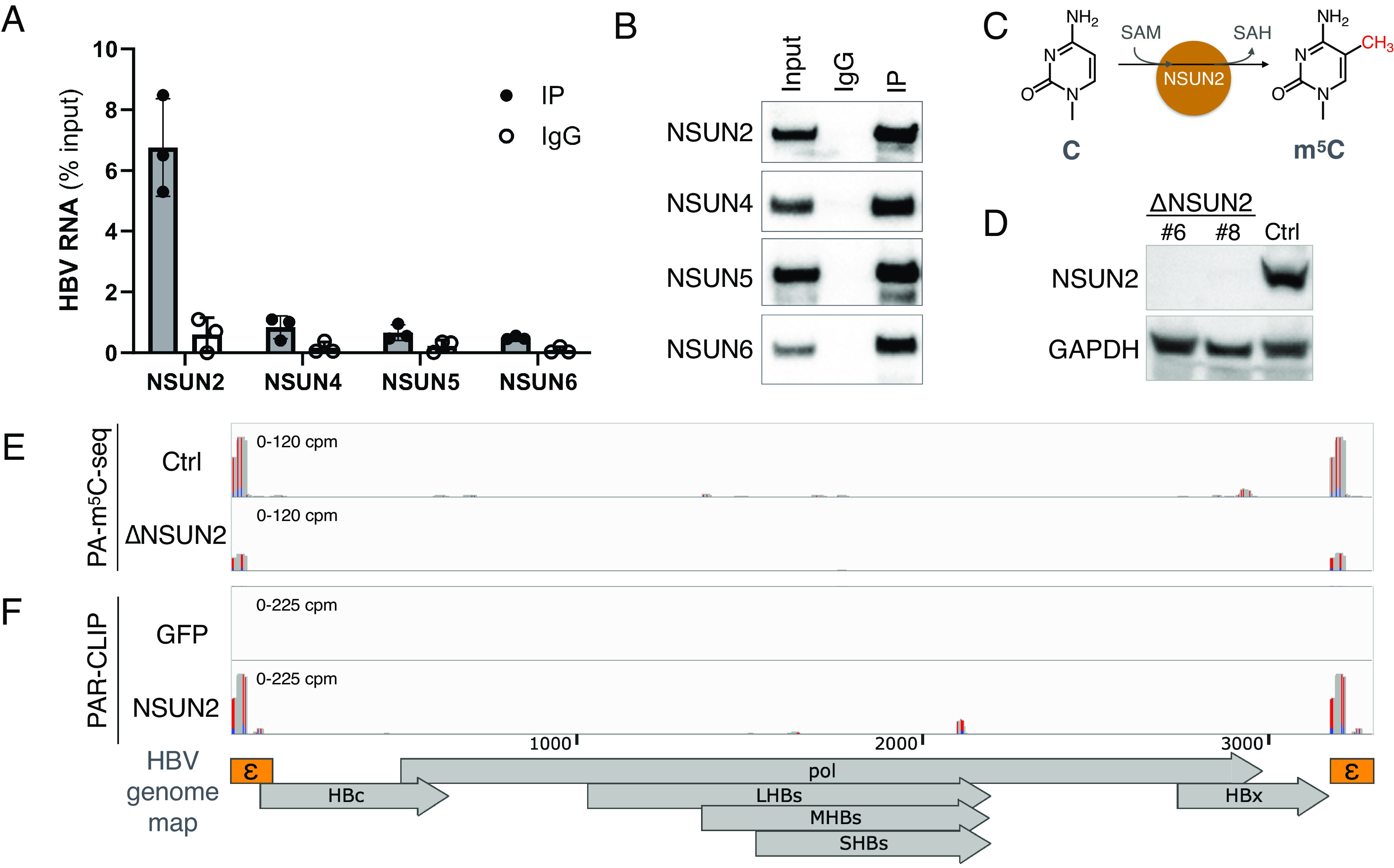
HBV RNA m5C is deposited by the host methyltransferase NSUN2. (A and B) 5-AzaC-mediated RNA immunoprecipitation (Aza-IP) of NSUN proteins, the pulled down HBV RNA quantified by qRT-PCR (n = 3, error bars = SD) (A), with Western blot validation of pulldown (B). (C) Depiction of NSUN2-mediated cytidine (C) methylation to produce 5-methylcytidine (m5C). (D) NSUN2 was depleted from HuH-7 cells by CRISPR-Cas9 (ΔNSUN2), with a GFP-targeted Cas9+ cell line produced alongside as a control (Ctrl). NSUN2 depletion in two single-cell clones (#6 & #8) of ΔNSUN2 and Ctrl cells validated by Western blot. (E) PA-m5C-seq of HBV RNA from Ctrl or ΔNSUN2 HuH-7 cells (clone #8). (F) NSUN2 or GFP (RNA-nonbinding control) binding sites on HBV RNA in HuH-7 cells mapped via PAR-CLIP. RNA-seq tracks scaled as the cpm shown, red-blue bars denote T>C conversions from 4SU-crosslinking.
If NSUN2 is indeed the methyltransferase depositing m5C on HBV RNA (Fig. 2C), we may predict that a) removal of NSUN2 should lead to the loss of m5C from HBV RNA, and b) NSUN2 should specifically coimmunoprecipitate with m5C+ regions of HBV RNA. To this end, we first utilized CRISPR/Cas9 to edit out NSUN2 from HuH-7 cells (ΔNSUN2, Fig. 2D and SI Appendix, Fig. S2A). ΔNSUN2 cells showed a slight impact to cell viability yet no impact on transfection efficiency (SI Appendix, Fig. S2 B, D, and E). Performing PA-m5C-seq on HBV-transfected ΔNSUN2 cells, we indeed observed ~75% shorter m5C peaks in ΔNSUN2 cells as compared to that in nontargeting CRISPR control cells (Ctrl) (Fig. 2E) (35). Next, we tested whether NSUN2 copurifies with HBV RNA at m5C sites, by performing PAR-CLIP on cells cotransfected with the HBV replicon and plasmids encoding either FLAG-tagged NSUN2 or green fluorescent protein (GFP). As in Fig. 2F, NSUN2, but not GFP, binds HBV RNA at the epsilon elements, colocalizing with the PA-m5C-seq peaks (35).
We next ask whether viral RNA m5C methylation affects the overall production of secreted HBV virions. If m5C enhances HBV replication as it does with HIV-1, and m5C deposition is NSUN2-dependent (Fig. 2E), then we would expect to see lower HBV viral production upon NSUN2 depletion. We transfected control and ΔNSUN2 HuH-7 cells with equal amounts of the wildtype HBV replicon (SI Appendix, Fig. S2E) and quantified the viral products in the supernatant. Indeed, we found almost undetectable amounts of virion-packaged HBV genomic DNA (Fig. 3A) and a ~60 to 90% loss of HBV surface antigen protein (HBs) in the supernatants of two independent single-cell clones of ΔNSUN2 HuH-7 cells as compared to control (Fig. 3 B and C). The loss of virion-packaged DNA could not be fully explained by the loss of HBs+ virions as quantifying HBV DNA from equal amounts of HBs+ virions still revealed a ~60% loss of secreted DNA from ΔNSUN2 cells (SI Appendix, Fig. S3 A and B). This loss of secreted viral products was also phenocopied in NSUN2 shRNA knocked down HuH-7 cells (Fig. 3 D and E). To ensure the above observations from replicon transfections were consistent in actual infections, we next turned to HepG2-NTCP cells which stably express the HBV receptor NTCP and thus can be infected (39, 40). Upon infecting NSUN2 CRISPR-depleted HepG2-NTCP cells (SI Appendix, Fig. S2 A and C) with wild-type HBV, we observed slower replication kinetics across 3 to 12 days postinfection (dpi, Fig. 3F), as well as a 60% drop in secreted HBc and HBs proteins and >90% loss in viral DNA at 12 dpi (Fig. 3 G and H). Thus, our observed NSUN2-dependent HBV replication phenotypes were consistent across NSUN2 CRISPR or RNAi-depletion, in both replicon transfection and HBV infection systems. Next, as a proof of concept, we treated HBV+ HuH-7 cells with the nucleoside analog 5-AzaC to test whether an m5C methyltransfer-disrupting drug could impede HBV replication. As discussed for NSUN Aza-IP, 5-AzaC incorporation in RNA may lead to NSUN2 covalently stuck on substrate RNA (38). If HBV were replicating in the presence of 5-AzaC, the resulting HBV RNA–NSUN2 complexes may impede viral replication. Indeed, when HBV+ HuH-7 cells were treated with 5-AzaC at minimally toxic dosages (Fig. 3I, gray lines), we observed a significant drop in secreted viral HBs protein and HBc capsids (Fig. 3I and SI Appendix, Fig. S3C), as well as viral DNA (SI Appendix, Fig. S3D). This antiviral activity is virus strain agnostic, as the antiviral effect was seen with other HBV genotypes as well (Fig. 3J).
Fig. 3.
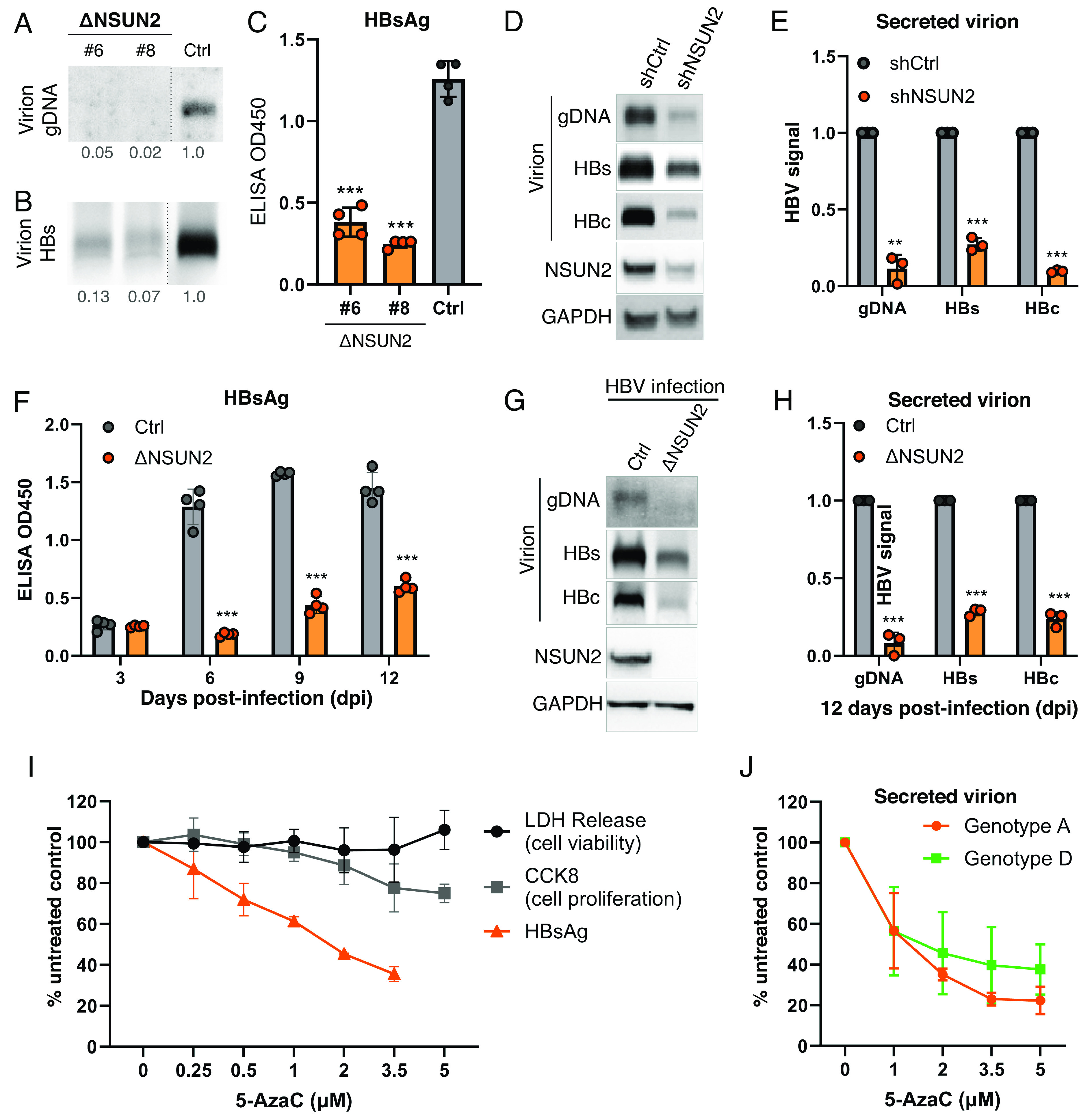
NSUN2 depletion diminishes virion secretion. (A–C) CRISPR-mediated NSUN2-depleted clones (ΔNSUN2 #6 & #8) or Cas9 control (Ctrl) HuH-7 cells were transfected with a wild-type HBV replicon. The supernatants harvested at 5 days posttransfection (dpt) were assayed for genomic DNA (gDNA) by Southern blot (A) and secreted HBs proteins by Western blot (B) and ELISA (C). Dashed lines between ΔNSUN2 and Ctrl lanes in (A and B) represent lanes removed from between these lanes. (D and E) HuH-7 cells transduced with shRNAs against NSUN2 (shNSUN2) or scrambled control (shCtrl) were transfected with wild-type HBV replicon, and the supernatant virions assayed 5dpt for genomic DNA (gDNA) by Southern blot and secreted HBs and HBc proteins by Western blot, with NSUN2 knockdown verified by cell lysate Western, repeats quantified in (E). (F–H) HepG2-NTCP-C4 cells with or without NSUN2-depletion by CRISPR (ΔNSUN2 or Cas9 control, Ctrl) were infected with wild-type HBV, the supernatant HBsAg measured by ELISA at 3 to 12 dpi (F). (G) Viral replication levels at 12 dpi were assayed for secreted virion gDNA, HBs, and HBc protein, with repeats quantified (H). (I) HBV-transfected HuH-7 cells were treated with the m5C methyltransfer disrupter 5-AzaC, secreted HBsAg assayed by ELISA at 3dpt. 5-AzaC toxicity at each dose assayed by lactate dehydrogenase (LDH) and CCK-8 assays to assess cell viability and proliferation. (J) 5-AzaC tested on HuH-7 cells transfected with HBV genotypes A & D, secreted virion HBc assayed by Western blot at 3 dpt with band intensities quantified. Error bars = SD with n = 3 to 4; ∗∗P < 0.01; ∗∗∗P < 0.001.
If NSUN2 depletion diminishes viral replication, then overexpression of NSUN2 may lead to increased HBV replication. Upon cotransfection of HuH-7 cells with the HBV replicon along with empty vector or an NSUN2 expression vector, we observed a 1.5 to 1.7x increase of intracellular and extracellular assembled capsid particles, as well as a 1.6x increase of HBc protein expression (SI Appendix, Fig. S4 A–C), supporting our finding that NSUN2 enhances viral replication. While this suggests that it may be advantageous for HBV to evolve to increase host NSUN2 expression, we only observed a modest ~1.1x increase in NSUN2 protein in HBV-transfected HuH-7 cells (SI Appendix, Fig. S4 D and E), which interestingly matches a ~1.1x higher NSUN2 expression in HBV+ hepatocellular carcinoma (HCC) tissues as compared to surrounding nontumor tissues in a published human patient RNA-seq dataset (SI Appendix, Fig. S4F) (41).
With NSUN2 depletion leading to the loss of secreted viral products, we next searched for the intracellular viral replication step(s) affected by NSUN2 and m5C that may explain the loss of secreted virions (Fig. 4A). Transfecting ΔNSUN2 HuH-7 cells with HBV replicons, we first tested the total intracellular RNA and protein levels. We observed a minor drop in viral pgRNA levels (Fig. 4B) and a ~50% drop in capsid (HBc) protein levels (Fig. 4C and SI Appendix, Fig. S5A). This loss of HBc protein expression in ΔNSUN2 cells is rescued upon exogenous NSUN2 add back in both HuH-7 and 293T cells (Fig. 4D and SI Appendix, Fig. S5B). With the drop of neither RNA nor protein production as pronounced as the loss of secreted DNA seen in Fig. 3 A, D, E, G, and H, and no evidence of NSUN2 affecting HBV RNA splicing or stability (SI Appendix, Fig. S5 C and D), we next probed the steps of capsid assembly. HBV virion assembly starts with HBV pol binding the pgRNA epsilon; the pgRNA–pol complex is then packaged into capsids assembled of HBc proteins. Once pgRNA–pol complexes are encapsidated, reverse transcription can commence, producing encapsidated viral DNA. Upon polyethylene glycol (PEG) precipitation of capsid particles from HBV+ ΔNSUN2 cell lysates, we observed a ~50% drop in pgRNA and HBc proteins in assembled capsids (Fig. 4 E and F), which can be fully explained by the 50% loss in HBc translation (Fig. 4C). However, the capsid-associated viral DNA in ΔNSUN2 cells are almost undetectable, similar to that seen in the ΔNSUN2 supernatants as well as with an RT-mutant control (Fig. 4G). While one may speculate that the removal of m5C may lead to changes in other modifications, we have not observed any change in HBV m6A levels in ΔNSUN2 cells (SI Appendix, Fig. S5E). All in all, depletion of NSUN2, which m5C-methylates epsilon, leads to a 50% loss of HBc protein production, and a ~95% loss of viral reverse transcription products. These phenotypes were orthogonally confirmed in shRNA-mediated NSUN2-knockdown HuH-7 cells (shNSUN2), which also showed diminished HBc protein production as well as a profound loss of HBV DNA (SI Appendix, Fig. S5 F and G).
Fig. 4.
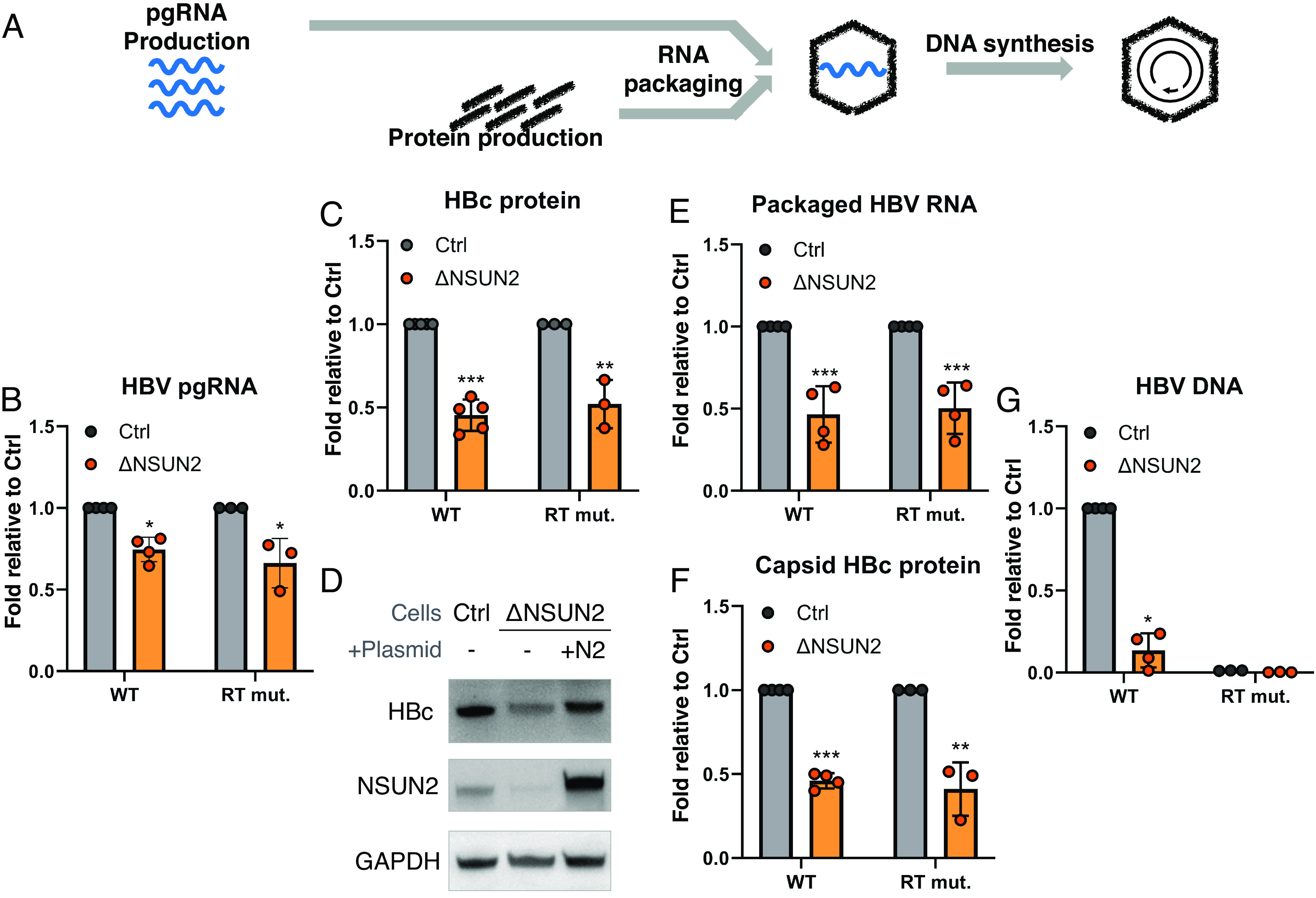
NSUN2 enhances HBV translation and is essential for reverse transcription. (A) Schematic of HBV replication cycle. ΔNSUN2 and control (Ctrl) HuH-7 cells were transfected with a wild-type HBV replicon (WT) or the pol Y63D mutant (RT mut.), and the following HBV products were measured. (B) Total intracellular HBV pgRNA levels measured by qRT-PCR at 2 dpt. (C) Intracellular core (HBc) protein at 3 dpt measured by Western blot, with band intensities quantified. (D) Intracellular HBc protein production from wild-type HBV in Ctrl and ΔNSUN2 cells, compared with NSUN2 add-back rescue in ΔNSUN2 cells (+N2). (E–G) Intracellular assembled capsid particles purified and the packaged core-associated viral RNA quantified by qRT-PCR (E), assembled particle capsid proteins assayed via band-quantified Western blotting (F), and capsid-associated viral DNA assayed by qPCR at 5 dpt (G). Error bars = SD with n > 3; ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001.
As depletion of NSUN2 may also impact nonviral m5C sites, we directly mutated the viral m5C sites to test whether these sites regulate viral pgRNA epsilon function in cis. PA-m5C-seq produce 32+nts wide peaks, leaving it unclear which exact cytidines are methylated. Our m5C peak covers from DR1 and the left strand of the epsilon lower stem up to the bulge (Fig. 1 C and D and SI Appendix, Fig. S6A), yet DR1 is required for the strand transfer step of reverse transcription while the bulge is critical for reverse transcription initiation (42). We thus mutated five C residues covered by our peak while avoiding DR1 and the bulge, introducing C>G or C>A mutations to remove potentially methylated Cs, with compensating G>C or G>U mutations on the opposing base pairing strand to conserve the epsilon hairpin structure (SI Appendix, Fig. S6A). The C opposite from the HBc start codon was avoided from mutation as well. With both (5′ or 3′ end) copies of epsilon potentially methylated, we made separate mutant replicons with C>G mutants on the 5′ epsilon (5′CG), C>A on the 5′ epsilon (5′CA), and C>G or C>A on the 3′ epsilon (3′CG or 3′CA, Fig. 5A). In the supernatants of HuH-7 cells transfected with these constructs, we observed minimal impact on secreted HBs levels, yet a >90% loss of secreted HBc and DNA levels with the 5′ mutants but not the 3′ mutants (Fig. 5B, repeats quantified in Fig. 5C). Upon testing intracellular viral products, we observed a negligible loss in pgRNA levels (Fig. 5D), yet a ~75% loss in packaged pgRNA and >90% loss in DNA and capsid HBc protein levels with the 5′ mutants but not the 3′ mutants (Fig. 5 E–G). Loss of HBc expression from the 5′CA mutant HBV is consistent in wild-type and ΔNSUN2 HuH-7 cells, as would be expected if our mutants and ΔNSUN2 both remove m5C (SI Appendix, Fig. S6B). While HBV epsilon is also modified with m6A, our mutations are distinct from the known m6A site and do not impact pgRNA m6A levels (SI Appendix, Fig. S6 A and C) (9). The >90% HBc protein loss in the 5′ mutants is more pronounced than the ~50% HBc loss with ΔNSUN2 cells (Fig. 4C), which makes sense as our base pairing strand “hairpin-maintaining” mutations were very close to the HBc start codon and may affect HBc translation in more than one way. To test whether our m5C-mutants also impact reverse transcription, we complemented our 5′CA mutant with an HBc expression vector to add-back HBc protein (Fig. 5H). Yet, despite the recovery of HBc protein to WT levels, the 5′CA mutant still showed a 50% deficiency in viral DNA production (Fig. 5I). Thus, our results suggest that mutational removal of methylated cytidine on the 5′ epsilon indeed confirmed our observation from ΔNSUN2 cells, where m5C/NSUN2 is required for HBc protein production and reverse transcription, both critical steps in HBV virion production.
Fig. 5.
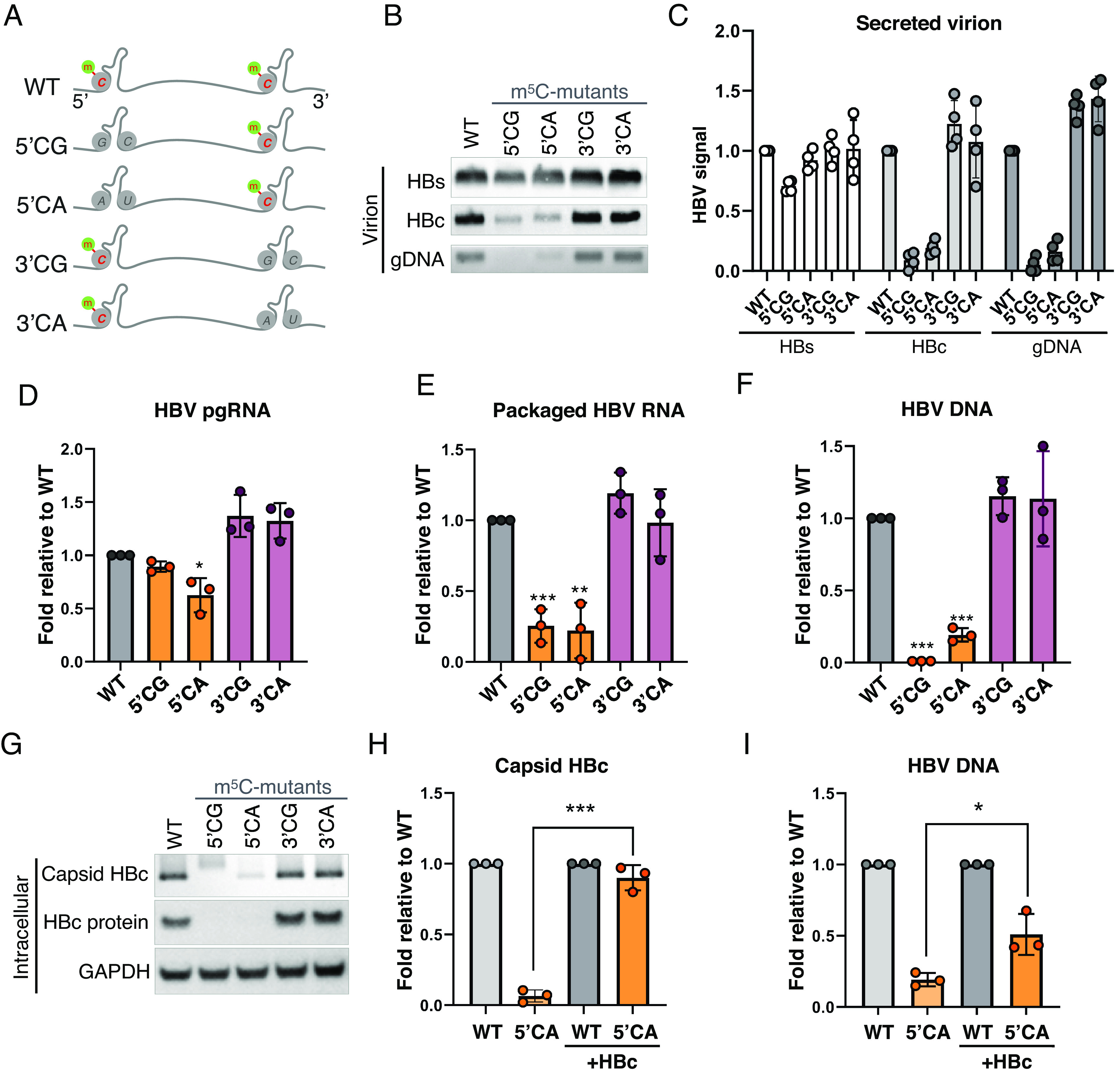
m5C at the 5′end epsilon is critical for HBV replication. (A) Schematic of m5C sites on HBV pgRNA epsilon hairpins. Lower four lanes depict replicons with the putative methylated cytidines (C1842, 1845, 1847, 1858, and 1859 in GenBank J02203 coordinates, SI Appendix, Fig. S6A) on the 5′ epsilons mutated to G or A (5′CG or 5′CA), or the 3′ epsilons (3′CG or 3′CA). G residues on the complimentary strand of the hairpin (G1899 & G1900, SI Appendix, Fig. S6A) were mutated to C or U to preserve the hairpin structure. (B–G) WT and mutant replicons were transfected into HuH-7 cells, with the secreted virion HBs and HBc proteins measured via Western blot, and gDNA assayed via Southern blot at 5 dpt (B), repeats of panel B quantified in (C). (D) Intracellular pgRNA quantified by qRT-PCR at 2 dpt. (E) Capsid-packaged viral RNA quantified via qRT-PCR at 3 dpt. (F) Capsid-associated viral DNA assayed via qPCR at 5 dpt. (G) Intracellular capsid HBc and total HBc proteins assayed 3 dpt via native agarose gel electrophoresis and SDS-PAGE followed by Western blotting. (H and I) WT and mutant replicons were transfected into HuH-7 cells with or without HBc expression vector supplementation, with the capsid HBc protein assayed via band-quantified Western blots (H) and viral DNA quantified by qPCR (I). Error bars = SD, n > 3; ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001.
Discussion
Recent progress in epitranscriptomics has highlighted the fact that the four-nucleotide sequence alone may not fully inform the function of the RNA. With pathogenic viruses, resolving this knowledge gap may unveil unrealized therapeutic potential. In this report, we found the pgRNA of HBV to be decorated with diverse modifications at stoichiometries significantly higher than cellular mRNA (Fig. 1 A and B and Table 1). Analogous to the highly modified RNA of HIV-1 and MLV, this may represent convergent evolution among reverse transcribing viruses, if not with other viruses as well (3, 23). Evolutionary traits would only make sense if it provides advantages to the production of offspring, as was shown with m5C enhancement of HIV-1 and MLV replication (3, 23). Here, we found NSUN2-mediated m5C methylation of HBV RNA on the terminal epsilon element (Figs. 1 and 2). Notably, upon NSUN2 depletion, we still found ~25% remaining m5C on epsilon elements (Fig. 2E). While host mRNAs can be methylated by both NSUN2 and NSUN6 (43, 44), we only observe strong copurification of HBV RNA with NSUN2 (Fig. 2A), thus we chose to focus on NSUN2, though we cannot rule out the role of other methyltransferases. Consistent with epsilon being essential for reverse transcription initiation and containing the HBc start codon, we found m5C to be critical for the production of DNA+ viral particles (Fig. 3). m5C provides a 2x boost to capsid protein (HBc) translation (Fig. 4C) and is essential for viral genome reverse transcription (Fig. 4G). m5C enhancement of translation mirrors its’ role in HIV-1. Although the mechanism for this translation boost remains unclear, we note that m5C is located close to not only the HBV HBc translation start site (SI Appendix, Fig. S6A) but also to the translation start site of several HIV ORFs including gag, env, and nef (3), making m5C enhancement of translation initiation an interesting possibility. Furthermore, the requirement of epsilon-located m5C for reverse transcription is particularly interesting. HBV and duck HBV studies have demonstrated that the first step of reverse transcription: pol-epsilon binding, is functional with in vitro transcribed, unmodified epsilon (42, 45). In the subsequent pgRNA encapsidation step, as the magnitude of encapsidation defect in ΔNSUN2 cells is equivalent to the loss in capsid protein (HBc) translation (Fig. 4 E and F), m5C is unlikely to impact encapsidation. With m5C not found in the epsilon bulge, where the first 5 nt of DNA is reverse transcribed off of (42), it may thus be speculated that NSUN2 and m5C may be critical for the subsequent first strand transfer step, which may be further tested in future studies.
During the preparation of this manuscript, another report was published supporting our finding that NSUN2 enhances HBV replication (46). However, through differing mapping techniques, Feng et al. reported two m5C sites that were not detected by m5C antibody (Fig. 1C) nor NSUN2 PAR-CLIP (Fig. 2F) and did not report any epsilon sites. We hope future advances in direct RNA sequencing may resolve this discrepancy (47). Mechanistically, Feng et al. found that NSUN2 depletion led to a modest drop in viral RNA stability, though consistent with our 10 to 20% decrease in pgRNA levels in ΔNSUN2 cells (Fig. 4B), we observed no decrease of pgRNA stability in our ΔNSUN2 HuH-7 cells (SI Appendix, Fig. S5D). However, we observed a larger loss of HBc translation and reverse transcription with NSUN2 depletion through RNAi and CRISPR in replicon transfection and infection studies (Figs. 3 and 4 and SI Appendix, Figs. S3 and S5). Finally, we have similarly noted a modest 1.1x increase in NSUN2 expression in HBV-transfected cells as well as in HBV+ patient samples (SI Appendix, Fig. S4 D and E), yet this is nowhere as pronounced as reported by Feng et al. Nevertheless, both studies confirm that NSUN2 deposition of m5C on viral RNA enhances HBV replication.
We also tested the concept that RNA modifications can be antiviral targets. This was best demonstrated with the m6A inhibitor STM2457 exhibiting antiviral activity against human coronaviruses (48). Extending this concept to m5C, we found the m5C methyltransfer disruptor, 5-AzaC to be antiviral against HBV replication (Fig. 3 I and J). While 5-AzaC is a widely used cancer drug with side effects that render it unsuitable for HBV treatment in its’ current form (49), we note that derivatives of this nucleotide analog or other NSUN2 inhibitors could be developed as alternatives to current reverse transcription inhibitors. Finally, our m5C site mutations on the 5′ epsilon but not the 3′, resulted in a loss of viral DNA production as well as a loss in HBc translation (Fig. 5). This makes sense as only the 5′ epsilon is required for reverse transcription and contains the HBc translation start site (which was avoided in our mutations, SI Appendix, Fig. S6A). This also implicates that at least the 5′epsilon (if not the 3′) must be m5C methylated (50–52). While these mutations had a stronger impact on HBc translation than NSUN2 depletion, trans-complementation of these mutants with HBc only rescued DNA production up to 50%, consistent with the interpretation that NSUN2/m5C enhances both HBc translation and reverse transcription.
Materials and Methods
Cell Lines and Viral Replicons.
HuH-7 cells were obtained from Michael Lai (Academia Sinica, Taiwan) (53). HepG2-NTCP-C4 cells were kindly gifted by Koichi Watashi, National Institute of Infectious Diseases, Japan (39). The HBV replicon pCHT-9/3091(54), a CMV promoter-driven 1.1mer HBV ayw genome, was used for all experiments unless specifically stated. The HBV 1.1mer replicon expressing an RT-defective Y63D mutant pol was used for m5C detection assays (26). HBV clinical isolates, genotypes A and D, were provided by Pei-Jer Chen (55). Epsilon mutant replicons 5′CG, 5′CA, 3′CG, and 3′CA were generated by QuickChange Lightning Site-Directed Mutagenesis Kit (Agilent Technologies) using a parental HBV replicon pCHT-9/3091.
Generation of Gene Knock Out HuH-7 and HepG2-NTCP-C4 Cells.
The ΔNSUN2 HuH-7 (#6 and #8) and ΔNSUN2 HepG2-NTCP-C4 cell lines were generated through CRISPR/Cas9 genome editing (56). A LentiCRISPRv2 lentiviral vector (provided by Feng Zhang, Addgene #52961) expressing an NSUN2-targeting guide RNA (GeCKO v2 library A #32780) was packaged in 293T cells with the packaging plasmids dR8.74 and pMD2.G. Naive HuH-7 and HepG2-NTCP-C4 cells were transduced with this lentivirus, puromycin selected, and single-cell cloned through limiting dilution. Clones were confirmed for NSUN2 depletion by Western blot and the edit site genotyped (SI Appendix, Fig. S2A).
PCR and Other Oligos.
All PCR primers and oligos are listed in SI Appendix, Table S2.
RNA Preparation for RNA Modification Profiling by UPLC–MS/MS.
Trizol-extracted cellular mRNA was purified with the Poly(A)Purist MAG Kit (Invitrogen). Capsid-associated viral RNA was obtained from the HBV Y63D (RT) mutant. Lysates of HuH-7 cells transfected with the RT-mut HBV, were digested with 3 units of DNase I and 30 units of Micrococcal Nuclease (NEB) at 37 °C 1 h to remove noncapsidated nucleic acids. The intracellular capsids were then pelleted over a 20% sucrose cushion at 100,000 rpm for 1 h (TLA110, Beckman) and fractionated on a 10 to 50% sucrose gradient at 35,000 rpm for 4 h (SW41, Beckman). Fractions containing HBV capsids and pgRNA were pooled (SI Appendix, Fig. S1B) and pelleted once again over a 20% sucrose cushion, subject to a repeat of DNase and micrococcal nuclease digestion, and the viral RNA was extracted with Trizol. The cellular mRNA and viral RNA were then ribo-depleted (RiboMinus Eukaryote System v2, Invitrogen), and the composition was verified by Fragment Analyzer (Agilent) microfluidic electrophoresis. Highly purified RNA samples were then digested to single nucleotides with the Nucleoside Digestion Mix (NEB, M0649S) for 2 h at 37 °C and cleaned using a Microcon-10 KDa filter (Millipore, MRCPRT010).
PA-m5C/m6A-seq.
As previously described (3, 4, 31, 33), HuH-7 cells were transfected with the wildtype HBV replicon and spiked with 100 mM 4-thiouridine (4SU, Biosynth NT06186) in fresh media 48 h posttransfection and harvested 18 h later. RNA was extracted with TRIzol, and poly(A) was selected [Poly(A)Purist MAG Kit, Invitrogen]. Then, 10 to 14 μg poly(A)+ RNA was incubated with 10 µg m6A antibody (SySy #202003) or 15 µg m5C antibody (Diagenode #C15200081) overnight, the antibodies crosslinked to RNA with 365 nm UV, and the coimmunoprecipitated complexes subject to a brief RNase T1 (Thermo EN0541) digestion. After rigorous washes, the m6A/m5C-antibody-bound RNA was eluted by proteinase K digestion and purified with Trizol LS (Invitrogen).
Secreted Virion Detection.
The extracellular HBV virion was precipitated using 10% PEG, as previously described (57). One mL of harvested cell culture supernatant was combined with 400 μL of 35% PEG-8000 to achieve a final concentration of 10% and incubated overnight at 4 °C. After centrifugation at 10,000 × g 5 min at 4 °C, the resulting viral pellet was resuspended in TNE buffer (10 mM Tris pH 7.4, 150 mM NaCl, 1 mM EDTA) and subjected to 1% native agarose gel electrophoresis. Gel-separated viral particles were transferred onto a nitrocellulose membrane by capillary transfer for Western blot and an N+ Nylon membrane for Southern blot analysis.
Northern and Southern Blot Analyses.
As previously described (29), DNA- or RNA-containing N+ Nylon membranes were prehybridized with DIG Easy Hyb (Roche) for 1 h at 42 °C and then hybridized with an HBV probe overnight at 42 °C. The probe consists of full-length HBV DNA, labeled with a DIG labeling kit (Roche). Following two 10-min washes with low stringency buffer (2X SSC, 0.1% SDS) at room temperature and two 15-min washes with high stringency buffer (0.5X SSC, 0.1% SDS) at 65 °C, the membrane was incubated with anti-DIG antibody (Roche). After three washes with wash buffer (0.1M Maleic acid, 0.15M NaCl, pH 7.5, 0.3% Tween20), the HBV genome signal was detected using the CSPD substrate (Roche), with band intensities quantified using ImageJ (NIH).
Native Agarose Gel and Western Blot of Capsid Particles.
As previously described (29), purified capsid particles were subjected to 1% native agarose gel electrophoresis. Rabbit anti-HBc polyclonal antibody [lab-generated (58)] and Rabbit anti-HBs polyclonal antibody (Novus) was used at 1/2,000x; goat anti-rabbit-IgG-HRP antibody (Sigma) at 1/10,000x.
HBV pgRNA, Capsid-Associated RNA, and DNA Detection.
Encapsidated viral RNA and DNA were extracted as reported (59). SuperScript III (Invitrogen) was used for cDNA synthesis, and specific primers designed for HBV pgRNA qPCR assay were utilized (SI Appendix, Table S2) (60).
HBV Infection.
HBV virions were collected from supernatants of HBV-transfected cells. HepG2-NTCP-C4 cells were maintained in DMEM with 2.5% DMSO for 72 h to induce cell differentiation. The differentiated HepG2-NTCP-C4 cells were then infected with HBV at 1,000 genome equivalents (GE) per cell in DMEM medium containing 2% FBS, 4% PEG-8000, and 2.5% DMSO (39, 61). After removing free virus by washing with 1X PBS, the HBV-infected cells were maintained for 12 d.
Quantification and Statistical Analysis.
All averaged data include error bars that denote SD, with single data points shown. Statistical significance was determined by Student’s t test, comparing test samples to control unless otherwise specified. Results were labeled as follows: *P < 0.05, **P < 0.01, and ***P < 0.001.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We would like to thank the generosity of Jianming Hu for sharing plasmids and Koichi Watashi for sharing HepG2-NTCP-C4 cells, as well as John Tavis for insightful discussions at the 2023 HBV meeting. Special thanks to Fu-An Li and the Institute of Biomedical Sciences (IBMS) Proteomics Core Facility for initial condition testing and optimizing UPLC–MS/MS analysis conditions, as well as Yun-Hua Lin in P.-J.C.’s lab for technical assistance. We also thank the following Academia Sinica core facilities: Agricultural Biotechnology Research Center Metabolomics Core, IBMS DNA Sequencing (AS-CFII-111-211), Institute of Molecular Biology and Biodiversity Research Center Next Generation Sequencing cores. This research was funded by the National Science and Technology Council (NSTC) grants 112-2320-B-001-023 to K.T., 112-2320-B-039-061 to C.S. 112-2326-B-002-001 to P.-J.C. and Academia Sinica career development award AS-CDA-112-L02 to K.T.
Author contributions
P.-Y.(A.)S., C.-H.C., S.-C.B.Y., M.-H.L., and K.T. designed research; P.-Y.(A.)S., C.-H.C., S.-C.B.Y., H.-Y.W., W.-J.T., Y.-P.H., Y.-Y.I.C., M.-H.L., and K.T. performed research; C.S., and P.-J.C. contributed new reagents; P.-Y.(A.)S., C.-H.C., S.-C.B.Y., H.-Y.W., M.-H.L., and K.T. analyzed data; and P.-Y.(A.)S. and K.T. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission. J.-h.J.O. is a guest editor invited by the Editorial Board.
Data, Materials, and Software Availability
All deep sequencing data have been deposited at the NCBI GEO database under accession number GSE250109 (35). All other study data are included in the manuscript and/or SI Appendix.
Supporting Information
References
- 1.Tsai K., Cullen B. R., Epigenetic and epitranscriptomic regulation of viral replication. Nat. Rev. Microbiol. 18, 559–570 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McIntyre W., et al. , Positive-sense RNA viruses reveal the complexity and dynamics of the cellular and viral epitranscriptomes during infection. Nucleic Acids Res. 46, 5776–5791 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Courtney D. G., et al. , Epitranscriptomic addition of m(5)C to HIV-1 transcripts regulates viral gene expression. Cell Host Microbe 26, 217–227.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kennedy E. M., et al. , Posttranscriptional m(6)A editing of HIV-1 mRNAs enhances viral gene expression. Cell Host Microbe 19, 675–685 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsai K., et al. , Epitranscriptomic addition of m(6)A regulates HIV-1 RNA stability and alternative splicing. Genes Dev. 35, 992–1004 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsai K., et al. , Acetylation of cytidine residues boosts HIV-1 gene expression by increasing viral RNA stability. Cell Host Microbe 28, 306–312.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ringeard M., Marchand V., Decroly E., Motorin Y., Bennasser Y., FTSJ3 is an RNA 2’-O-methyltransferase recruited by HIV to avoid innate immune sensing. Nature 565, 500–504 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Tsukuda S., Watashi K., Hepatitis B virus biology and life cycle. Antiviral Res. 182, 104925 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Imam H., et al. , N6-methyladenosine modification of hepatitis B virus RNA differentially regulates the viral life cycle. Proc. Natl. Acad. Sci. U.S.A. 115, 8829–8834 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Imam H., Kim G. W., Mir S. A., Khan M., Siddiqui A., Interferon-stimulated gene 20 (ISG20) selectively degrades N6-methyladenosine modified Hepatitis B Virus transcripts. PLoS Pathog. 16, e1008338 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim G. W., Siddiqui A., Hepatitis B virus X protein expression is tightly regulated by N6-methyladenosine modification of its mRNA. J. Virol. 96, e0165521 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boccaletto P., et al. , MODOMICS: A database of RNA modification pathways. 2021 update. Nucleic Acids Res. 50, D231–D235 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dubin D. T., Taylor R. H., The methylation state of poly A-containing messenger RNA from cultured hamster cells. Nucleic Acids Res. 2, 1653–1668 (1975). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang T., Chen W., Liu J., Gu N., Zhang R., Genome-wide identification of mRNA 5-methylcytosine in mammals. Nat. Struct. Mol. Biol. 26, 380–388 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Squires J. E., et al. , Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 40, 5023–5033 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Q., et al. , NSUN2-mediated m5C methylation and METTL3/METTL14-mediated m6A methylation cooperatively enhance p21 translation. J. Cell Biochem. 118, 2587–2598 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schumann U., et al. , Multiple links between 5-methylcytosine content of mRNA and translation. BMC Biol. 18, 40 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xing J., et al. , NSun2 promotes cell growth via elevating cyclin-dependent kinase 1 translation. Mol. Cell Biol. 35, 4043–4052 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang X., et al. , 5-methylcytosine promotes mRNA export - NSUN2 as the methyltransferase and ALYREF as an m(5)C reader. Cell Res. 27, 606–625 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen X., et al. , 5-methylcytosine promotes pathogenesis of bladder cancer through stabilizing mRNAs. Nat. Cell Biol. 21, 978–990 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Chen T., et al. , NSUN2 is a glucose sensor suppressing cGAS/STING to maintain tumorigenesis and immunotherapy resistance. Cell Metab. 35, 1782–1798.e8 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dubin D. T., Stollar V., Methylation of Sindbis virus “26S” messenger RNA. Biochem. Biophys. Res. Commun. 66, 1373–1379 (1975). [DOI] [PubMed] [Google Scholar]
- 23.Courtney D. G., et al. , Extensive epitranscriptomic methylation of A and C residues on murine leukemia virus transcripts enhances viral gene expression. mBio 10, e01209-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eckwahl M., et al. , 5-methylcytosine RNA modifications promote retrovirus replication in an ALYREF reader protein-dependent manner. J. Virol. 94, e00544-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hussain S., et al. , NSun2-mediated cytosine-5 methylation of vault noncoding RNA determines its processing into regulatory small RNAs. Cell Rep. 4, 255–261 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lanford R. E., Notvall L., Lee H., Beames B., Transcomplementation of nucleotide priming and reverse transcription between independently expressed TP and RT domains of the hepatitis B virus reverse transcriptase. J. Virol. 71, 2996–3004 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chang C. H., Shih C., Significance of hepatitis B virus capsid dephosphorylation via polymerase. J. Biomed. Sci. 31, 34 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Edelheit S., Schwartz S., Mumbach M. R., Wurtzel O., Sorek R., Transcriptome-wide mapping of 5-methylcytidine RNA modifications in bacteria, archaea, and yeast reveals m5C within archaeal mRNAs. PLoS Genet. 9, e1003602 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Su P. Y., et al. , HBV maintains electrostatic homeostasis by modulating negative charges from phosphoserine and encapsidated nucleic acids. Sci. Rep. 6, 38959 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Basanta-Sanchez M., Temple S., Ansari S. A., D’Amico A., Agris P. F., Attomole quantification and global profile of RNA modifications: Epitranscriptome of human neural stem cells. Nucleic Acids Res. 44, e26 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cullen B. R., Tsai K., Mapping RNA modifications using photo-crosslinking-assisted modification sequencing. Methods Mol. Biol. 2298, 123–134 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martinez Campos C., et al. , Mapping of pseudouridine residues on cellular and viral transcripts using a novel antibody-based technique. RNA 27, 1400–1411 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen K., et al. , High-resolution N(6) -methyladenosine (m(6) A) map using photo-crosslinking-assisted m(6) A sequencing. Angew. Chem. Int. Ed. Engl. 54, 1587–1590 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murata T., et al. , N6-methyladenosine modification of hepatitis B virus RNA in the coding region of HBx. Int. J. Mol. Sci. 24, 2265 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Su P. Y., et al. , Data from “Epitranscriptomic cytidine methylation of the Hepatitis B viral RNA encapsidation signal ensure the reverse transcription of viral RNA.” NCBI Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE250109. Deposited 13 December 2023.
- 36.Hafner M., et al. , Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141, 129–141 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bohnsack K. E., Hobartner C., Bohnsack M. T., Eukaryotic 5-methylcytosine (m(5)C) RNA methyltransferases: Mechanisms, cellular functions, and links to disease. Genes (Basel) 10, 102 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khoddami V., Cairns B. R., Identification of direct targets and modified bases of RNA cytosine methyltransferases. Nat. Biotechnol. 31, 458–464 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Iwamoto M., et al. , Evaluation and identification of hepatitis B virus entry inhibitors using HepG2 cells overexpressing a membrane transporter NTCP. Biochem. Biophys. Res. Commun. 443, 808–813 (2014). [DOI] [PubMed] [Google Scholar]
- 40.Yan H., et al. , Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. Elife 1, e00049 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yoo S., et al. , A pilot systematic genomic comparison of recurrence risks of hepatitis B virus-associated hepatocellular carcinoma with low- and high-degree liver fibrosis. BMC Med. 15, 214 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tavis J. E., Perri S., Ganem D., Hepadnavirus reverse transcription initiates within the stem-loop of the RNA packaging signal and employs a novel strand transfer. J. Virol. 68, 3536–3543 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu J., et al. , Sequence- and structure-selective mRNA m(5)C methylation by NSUN6 in animals. Natl. Sci. Rev. 8, nwaa273 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Selmi T., et al. , Sequence- and structure-specific cytosine-5 mRNA methylation by NSUN6. Nucleic Acids Res. 49, 1006–1022 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jones S. A., Boregowda R., Spratt T. E., Hu J., In vitro epsilon RNA-dependent protein priming activity of human hepatitis B virus polymerase. J. Virol. 86, 5134–5150 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Feng J., et al. , NSUN2-mediated m5C modification of HBV RNA positively regulates HBV replication. PLoS Pathog. 19, e1011808 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leger A., et al. , RNA modifications detection by comparative Nanopore direct RNA sequencing. Nat. Commun. 12, 7198 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Burgess H. M., et al. , Targeting the m(6)A RNA modification pathway blocks SARS-CoV-2 and HCoV-OC43 replication. Genes Dev. 35, 1005–1019 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Christman J. K., 5-Azacytidine and 5-aza-2’-deoxycytidine as inhibitors of DNA methylation: Mechanistic studies and their implications for cancer therapy. Oncogene 21, 5483–5495 (2002). [DOI] [PubMed] [Google Scholar]
- 50.Hirsch R. C., Loeb D. D., Pollack J. R., Ganem D., Cis-acting sequences required for encapsidation of duck hepatitis B virus pregenomic RNA. J. Virol. 65, 3309–3316 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kawamoto S., Ueda K., Mita E., Matsubara K., The packaging signal in hepatitis B virus pregenome functions only at the 5’ end. J. Virol. Methods 49, 113–127 (1994). [DOI] [PubMed] [Google Scholar]
- 52.Ryu D. K., Ahn B. Y., Ryu W. S., Proximity between the cap and 5’ epsilon stem-loop structure is critical for the suppression of pgRNA translation by the hepatitis B viral polymerase. Virology 406, 56–64 (2010). [DOI] [PubMed] [Google Scholar]
- 53.Nakabayashi H., Taketa K., Miyano K., Yamane T., Sato J., Growth of human hepatoma cells lines with differentiated functions in chemically defined medium. Cancer Res. 42, 3858–3863 (1982). [PubMed] [Google Scholar]
- 54.Nassal M., The arginine-rich domain of the hepatitis B virus core protein is required for pregenome encapsidation and productive viral positive-strand DNA synthesis but not for virus assembly. J. Virol. 66, 4107–4116 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu H. L., Chen P. J., Lin M. H., Chen D. S., Temporal aspects of major viral transcript expression in Hep G2 cells transfected with cloned hepatitis B virus DNA: With emphasis on the X transcript. Virology 185, 644–651 (1991). [DOI] [PubMed] [Google Scholar]
- 56.Sanjana N. E., Shalem O., Zhang F., Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yan R., Cai D., Liu Y., Guo H., Detection of hepatitis B virus particles released from cultured cells by particle gel assay. Methods Mol. Biol. 1540, 193–202 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li H. C., et al. , Nuclear export and import of human hepatitis B virus capsid protein and particles. PLoS Pathog. 6, e1001162 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Su P. Y., et al. , Hepatitis B virus virion secretion is a CRM1-spike-mediated late event. J. Biomed. Sci. 29, 44 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Qi Y., et al. , DNA polymerase kappa is a key cellular factor for the formation of covalently closed circular DNA of hepatitis B virus. PLoS Pathog. 12, e1005893 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Choijilsuren G., et al. , Heparin at physiological concentration can enhance PEG-free in vitro infection with human hepatitis B virus. Sci. Rep. 7, 14461 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
All deep sequencing data have been deposited at the NCBI GEO database under accession number GSE250109 (35). All other study data are included in the manuscript and/or SI Appendix.


