Abstract
The levels of population diversity of three related Sindbis-like plant viruses, Tobacco mosaic virus (TMV), Cucumber mosaic virus (CMV), and Cowpea chlorotic mottle virus (CCMV), in infections of a common host, Nicotiana benthamiana, established from genetically identical viral RNA were examined. Despite probably having a common evolutionary ancestor, the three viruses maintained different levels of population diversity. CMV had the highest levels of diversity, TMV had an intermediate level of diversity, and CCMV had no measurable level of diversity in N. benthamiana. Interestingly, the levels of diversity were correlated to the relative host range sizes of the three viruses. The levels of diversity also remained relatively constant over the course of serial passage. Closer examination of the CMV and TMV populations revealed biases for particular types of substitutions and regions of the genome that may tolerate fewer mutations.
The error-prone replication, large populations, and rapid replication times associated with RNA viruses result in the potential for genetically diverse populations, termed quasispecies, arising within a single host (16). Developed as a model for early forms of life (14), the theoretical quasispecies describes a steady-state collection of genetic mutants that vary around a consensus sequence. Viral quasispecies are complex and dynamic distributions where the level of population variation (quasispecies cloud size) reacts to changes in selection pressures (8). There are a number of biological and evolutionary implications associated with highly diverse populations (7). Maintaining a large quasispecies cloud size could allow a virus ready access to a pool of mutants which could become the selectively advantaged dominant RNA species in a shift to a new environment. Conversely, highly diverse populations that are subjected to repeated bottlenecks have been shown to lose fitness through a process known as Muller's ratchet (4, 9, 10, 25, 26).
Both DNA (15, 39) and RNA plant viruses (6, 23, 30, 32) can maintain highly diverse populations in collections of field isolates, but the quasispecies variation of single plant virus isolates has not been examined. Plant viruses can use several mechanisms to generate quasispecies diversity, including replication error, recombination, and, for multipartite viruses, reassortment (for a review see reference 33). However, the extent of population variation is limited by selection pressure for variants that interact successfully with different host and viral proteins necessary for completion of the infection cycle.
The Sindbis-like virus group includes a number of plant and animal viruses with similarities in their genome organizations and nonstructural proteins. The three virus species chosen for this study were Tobacco mosaic virus (TMV; genus Tobamovirus), Cucumber mosaic virus (CMV; genus Cucumovirus, family Bromoviridae), and Cowpea chlorotic mottle virus (CCMV; genus Bromovirus, family Bromoviridae). TMV is a monopartite rod-shaped virus, and CMV and CCMV are tripartite icosahedral viruses. Although the genomic organization of regions producing similar functional domains for nonstructural proteins (Fig. 1A) indicates that TMV, CMV, and CCMV evolved from a common ancestor, these three viruses differ greatly in terms of the sizes of their host ranges. CMV infects about 1,000 plant species (12), TMV infects 80 to 100 plant species (17), and CCMV infects only a few plant species (22). A correlation between diversity in the viral quasispecies and the sizes of viral host ranges seems possible (33). However, there are few experimental examinations of plant virus population diversity, and no controlled comparisons between different viruses, related or otherwise, have been reported. The well-characterized cDNA clones and the availability of a common host (Nicotiana benthamiana) for TMV, CMV, and CCMV make it possible to do a controlled experimental comparison of quasispecies diversity for these related viruses. Here we examine CMV, TMV, and CCMV quasispecies, both in initial infections and following consecutive passages in N. benthamiana.
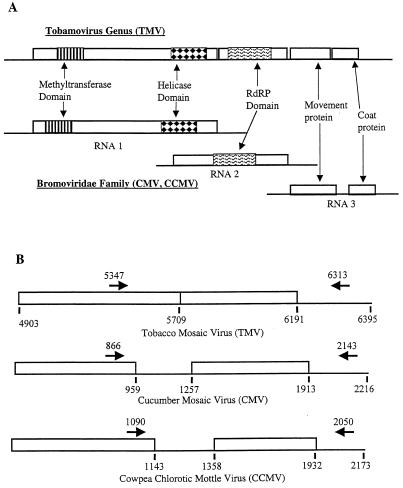
FIG. 1.
Genomic organization of the three viruses used in this study and locations of analyzed sequence regions. (A) Genomic maps of TMV, CMV, and CCMV with the common regions labeled. (B) Portions of viral genomes encoding the coat protein and flanking regions that were amplified and cloned for sequence analysis. Arrows, primer sites; numbers, nucleotide positions.
MATERIALS AND METHODS
Plants and viral inoculations.
N. benthamiana plants were grown under greenhouse conditions (16-h days at 24°C, 8-h nights at 20°C; supplemental light intensity, 500 μmol of photons m−2s−1) and inoculated at the three- or four-leaf stage. Plants were maintained in the greenhouse after inoculation. Viral RNAs were generated from cDNA clones of Fny-CMV (29, 34), CCMV (1, 11), and U1 TMV (36). For Fny-CMV and CCMV, transcripts were generated in vitro using SP6 or T7 RNA polymerase (Ambion). The TMV construct was inoculated as plasmid DNA, with viral RNA generated in planta from the Cauliflower mosaic virus 35S promoter (36). Transcript- or DNA-inoculated plants constituted passage zero. Subsequent passages (1 through 10) were at 14-day intervals, using sap extracts from the infected plant material. Two plants were inoculated at the passage zero level. One or two systemically infected leaves were removed from both plants, combined, and ground in 1 mM NaH2PO4 buffer (pH 7.0). The combined sap was used to inoculate both plants in the next passage.
Extraction of total RNA and cDNA synthesis.
Fourteen to 20 days postinoculation total RNA was extracted from systemically infected leaves of passage 0, passage 1, and passage 10 plants as previously described (34). One-fifth of the total RNA extraction was used as a template for reverse transcription (RT) with Superscript reverse transcriptase as prescribed by the manufacturer (Gibco). Two RT reactions were done for each infected plant. RT reactions were primed with primers 4144 (AACCTACTGCAGTGAGTCCGAGGATTA) for CMV, 4140 (AACCTACTGCAGGATAAAATCGCCGTAAC) for CCMV, and 4145 (TAATCGGAATTCAGGAAACAGCTATGACCCGCGCGATCCAGACAC) for TMV. The cDNAs were used as templates for thermal cycling reactions using the following primers: 4139 (AACCTACTGCAGTGAGTCCGAGGATTA) and 4144 for CMV, 4140 and 4146 (AACCTACTGCAGGATAAAATCGCCGTAAC) for CCMV, and 4143 (AACCTACTGCAGCGGGTTTCTGTCCGC) and 4145 for TMV (Fig. 1B). Thermal cycling reactions were carried out for 15 cycles (94°C denaturation for 30 s, 50°C annealing for 1 min, 72°C extension for 1 min), and included a polymerase with proofreading capability (Pfu; Stratagene). Between 82 and 123 nucleotides at the 3′ termini of the viral RNAs were not included in the cloned and sequenced segments. In addition to reactions using the N. benthamiana viral populations, RT and thermal cycling reactions were done using in vitro transcripts of all three viruses as templates, in order to establish the level of error introduced by the experimental method.
Analysis of viral clones.
The amplified products generated from the viral RNAs were cloned into the vector pBSKS− (Stratagene). The viral clones were sequenced using dideoxy sequencing and the Taq DyeDeoxy terminator cycle sequencing kit (Applied Biosystems). Sequencing gels were run on an ABI 377 sequencer as described by the manufacturer (Perkin-Elmer). Eleven to 16 clones were sequenced from each virus treatment, roughly half from each of the two infected plants used as source tissue. Changes between the sequence of the cDNA clone and the viral population clones were recorded as mutations. In cases where multiple mutations occurred in close proximity in the same clone, each mutated base was considered a unique mutation. The mutation frequency was calculated as the total number of mutations observed in all clones for a given viral population divided by the total number of bases sequenced for the population. To compare variation levels for the three viruses, the mutation frequencies and percentages of mutated clones from the passages were totaled. Comparisons of individual passages and comparisons between populations were tested for statistical significance using a χ2 test.
RESULTS AND DISCUSSION
Generation and analysis of quasispecies.
Infections were initiated for all three viruses using transcripts derived from cDNA clones. This ensured that the infections of each virus began with genetically identical sources of RNA. All three sets of viral clones used for in vitro transcription were previously thoroughly characterized, and transcript infections displayed no detectable phenotypic differences from wild-type viral infections. A minimum of two plants were used for each mechanical inoculation of N. benthamiana plants, and 14 to 20 days postinoculation total RNA was extracted from systemically infected leaves. A segment of viral RNA was cloned from total RNA pools of all three viruses. The cloned region represented the coat protein and flanking regions (Fig. 1), including both translated and nontranslated sequences (roughly a 1-kb segment for all three viruses). An excess of viral RNA was used in the cDNA reactions to prevent amplification of only a small subset of the viral population. Eleven to 16 clones (at least 10,000 nucleotides) were sequenced for each virus population. The sequences of all viral clones were compared to the sequences of the source infectious clones, and the percentage of clones with mutations and the mutation frequency were calculated for each population.
A number of steps were taken to minimize the level of error introduced by the experimental procedure. Eighty-two to 123 bases from the 3′ termini of the viral RNAs were excluded from the cloned region, because RNA polymerases used for in vitro transcription have a tendency to make errors at the ends of transcripts (24). Errors were reduced during cDNA synthesis by using a high-fidelity reverse transcriptase and by thermal cycling using a polymerase with proofreading capabilities. Only 15 cycles were completed for each thermal cycling reaction in order to prevent mutations introduced by fluctuations in nucleotide levels (38). In addition, control reactions were done using in vitro transcripts as the template RNA to estimate the level of variability introduced by transcription, RT, and thermal cycling. Only 1 of 22 control clones (5%) derived from in vitro transcripts contained a single mutation, establishing the background level of experimental mutation frequency at 4.5 × 10−5 mutations per nucleotide.
Quasispecies variation over serial passage.
The three viruses were passaged 10 times in N. benthamiana to analyze for fluctuations in the levels of population variation during serial passage. Clones representing the viral populations were analyzed from the construct- or transcript-inoculated plants (passage 0), 1st-passage plants, and 10th-passage plants, using the percentage of mutated clones and the mutation frequency as indicators of population variation. The percentage of mutated clones did not significantly rise or fall for any of the three viruses during serial passage. The mutation frequencies also remained consistent for TMV and CCMV, with the exception of the passage 10 CCMV populations (Table 1), where one cloned RNA had undergone a recombination event incorporating 24 nucleotides into the intercistronic region. This single mutational event artificially raised the mutation frequency, but if the percentage of mutated clones is considered or if the recombination event is considered as a single mutation, the levels of population diversity in CCMV populations remain statistically unchanged over the course of serial passage. The CMV populations showed a slightly significant increase in mutation frequency between passage 0 and passage 1 but then stabilized through passage 10.
TABLE 1.
Quasispecies variation in CMV, TMV, and CCMV populations after serial passage in N. benthamiana
| RNA | Passage | % Mutated clones (no. mutated/ total no.) | No. of mutations/no. of bases sequenced | Mutation frequency |
|---|---|---|---|---|
| RT-PCR controls | 5 (1/22) | 1/23,488 | 4.5 × 10−5 | |
| CMV | 0 | 43 (6/14) | 8/16,454 | 4.6 × 10−4 |
| 1 | 60 (9/15) | 15/18,496 | 8.1 × 10−4 | |
| 10 | 69 (9/13) | 11/15,827 | 7.0 × 10−4 | |
| Total | 57 (24/42) | 34/50,777 | 6.6 × 10−4 | |
| TMV | 0 | 29 (4/14) | 7/13,227 | 5.2 × 10−4 |
| 1 | 33 (4/12) | 5/11,350 | 4.4 × 10−4 | |
| 10 | 27 (3/11) | 3/10,234 | 2.9 × 10−4 | |
| Total | 30 (11/37) | 15/34,811 | 4.3 × 10−4 | |
| CCMV | 0 | 0 (0/14) | 0/13,454 | 0 |
| 1 | 7 (1/14) | 1/13,969 | 7.1 × 10−5 | |
| 10 | 6 (1/16) | 24/15,847a | 1.5 × 10−3 | |
| Total | 5 (2/44) | 25/43,270 | 5.7 × 10−4 |
The single mutant derived from the CCMV passage 10 plants was a recombinant which had incorporated a stretch of 24 bases in the nontranslated intercistronic region.
There are a number of possible scenarios that could affect diversity levels of RNA viruses in serial passage experiments. The levels of diversity could rise as the virus population expands exponentially and each new infection is started with an increasing spectrum of mutants. Alternatively, the levels of diversity could fall as selection pressures select for mutants that are better suited to infecting N. benthamiana and these mutants begin to dominate the population. However, these experiments seem to indicate that the viral populations rapidly reach a level of diversity in the initially inoculated plant which is maintained relatively constant over the course of passaging. This would imply that the sap passaging technique used is not a severe bottleneck, since the accumulation of deleterious mutants associated with Muller's ratchet (7) did not occur in the higher-diversity CMV and TMV populations. In addition, this raises the possibility of a threshold limit to population diversity for plant viruses. Quasispecies clouds for these three viruses appear to follow Eigen's prediction (13) that there is selection for a level of variation. Alternatively, these viruses might establish each subsequent infection in the passage with viral RNAs having sequences that are identical to the consensus sequence, in effect rendering infections in later passages the same as the initial-transcript infections. No changes to the consensus sequence were observed in any of the passaged populations; mutations arose and then disappeared without becoming fixed. However, it should be noted that for most plant viruses, especially those with divided genomes, there is nothing equivalent to a plaque assay. Hence, there is no way of accurately quantifying the amount of virus used in the inoculum or quantifying the number of viruses that initiate the infection in the new host.
Comparing quasispecies diversity.
Comparing the sequence data from the clones derived from the N. benthamiana viral populations demonstrated differences in the level of quasispecies variation between the three viruses (Table 1). Because the populations changed very little over the course of serial passage, the data from all three sampled passages were combined for purposes of comparison. CMV populations showed the highest level of variation, significantly higher than that of the controls (P < 0.05). Similarly, TMV populations were also significantly more diverse than the controls, but somewhat less diverse than CMV populations (P < 0.05). In contrast, CCMV populations had no more variation than the controls. Additionally, a comparison of the percentages of mutated clones shows that this trend is consistent within each individual passage. Interestingly, these results correlated with the relative host range sizes of the three viruses. No two clones had the same mutations, indicating that each mutant clone represented a unique viral RNA. Thus, we can infer that each clone with a wild-type sequence also represents a cDNA generated from a unique RNA template and that the experimental procedure provides a representative sampling of the population.
There are a number of possible explanations for the observed differences in the levels of genetic diversity of viral populations. Certainly the effects of host selection play a role in limiting error accumulation in the population, but here all three viruses were subjected to the same environment. The differences in diversity are likely not due to differences in population size, since all three viruses replicate to high levels in N. benthamiana. In fact, CMV, with the highest level of diversity, has the lowest viral titer (0.1 to 0.3 mg/gm) (28). CCMV accumulates to 0.3 to 0.5 mg/gm (22), and TMV accumulates to 1 mg/gm (41). The relative viral titers do not correlate with the levels of population diversity.
Another potential source of the different levels of diversity may be differences in the fidelity of replication. The differences in population variation may simply be due to differences in the error rates and recombination rates of the viral replicases. For example, the high mutation frequencies in the CMV populations may be due to higher error rates in replication. Certainly the replication fidelity of these viruses could be affected by the intracellular site of replication due to nucleotide levels, but there are no data on this for any of these viruses. Recombination, which can act as a purifying mechanism that reduces diversity (5), has been well documented in members of the Bromovirus genus such as CCMV (3, 37). Recombination may reduce diversity in the CCMV population. However, these viruses have similar origins and similar replication proteins. It seems likely that in a common host these three viruses utilize the same host factors and have replicating units with similar characteristics. While there are no studies of replicase error rates or recombination rates for any plant viruses to date, it is hard to imagine that these viruses vary greatly in their abilities to generate diversity through replication. Alternatively, the differences could be explained by a factor unique to the infection cycle of each particular virus. Perhaps the low-diversity CCMV populations are subjected to frequent bottlenecks, which would prevent the population from developing high diversity without losses of fitness. CMV and TMV quasispecies could also be limited by bottlenecks of various sizes.
Distribution and types of mutations.
None of the mutations in either the CMV or TMV populations became fixed during the passage experiments. All of the mutations observed in the passage 0 clones were not evident in the passage 1 clones, and the mutations observed in the passage 1 clones were subsequently lost by passage 10 (data not shown). There was no change in the consensus sequence, indicating that none of the mutations conferred any selective advantage to the virus and emphasizing the rare nature of adaptive mutations in viral populations. Occasionally multiple mutations were observed in close proximity to each other in the same viral clone (data not shown). These mutations may have arisen as a result of the same replication event, although this is difficult to test. When mutation frequencies were calculated, each altered nucleotide was considered an individual mutation, regardless of its proximity to other mutations in the same viral clone. If mutations in close proximity were to be considered as single mutational events, the mutation frequencies observed would be lowered slightly but the comparative mutation frequencies for the three viruses as well as the relative levels of diversity in passaged populations would remain the same.
An examination of the locations of the mutations observed suggests that mutations may not be completely random. The observed mutations in the CMV populations were distributed throughout the sequenced region, both in translated and nontranslated regions (Fig. 2), with a bias for nontranslated regions over translated regions. In addition, there were large areas where no mutations were observed, in particular the area between nucleotides 1577 and 1846 of the coat protein gene. This could represent a region where selection acts against mutation tolerance, but more mutations need to be mapped to confirm that it is not occurring by random chance. The distribution of TMV mutations also covered the majority of the sequenced portions of the genome (Fig. 3), although in TMV 14 of the 15 mutations observed were in the coding regions. However, less of the TMV cloned segment represents nontranslated regions. Both of the CCMV mutational events occurred in the nontranslated intercistronic region (data not shown). Any selection that might be in effect here must be working at the RNA level. All populations have more than enough functional copies of the genes in question to supply the appropriate proteins in trans. This is confirmed by examining the classes of mutations found in the coding regions, where there is no bias for synonymous mutations. Four of the 11 mutations in the CMV coding region were silent, and 5 of the 14 mutations in the TMV coding region were silent. Thus, any selection is likely to be selection for the capacities of the viral RNA in replication, movement, encapsidation, or stability.
FIG. 2.
Distribution of accumulated mutations observed in clones derived from CMV populations in N. benthamiana. The region sequenced extends from base 868 in the 3a open reading frame to base 2134 in the 3′ nontranslated region. Sites of mutations are indicated by lines below the map. A line with a number indicates a position where more than one mutation occurred at or near the same nucleotide position. Actual nucleotide positions of mutations are listed as base numbers below the map.
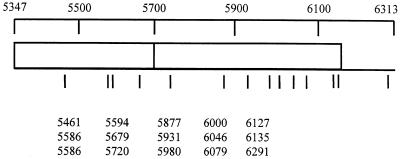
FIG. 3.
Distribution of accumulated mutations observed in clones derived from TMV populations in N. benthamiana. The region sequenced extends from base 5347 in the 30-kDa movement protein open reading frame to base 6313 in the 3′ nontranslated region. Sites of mutations are indicated by lines below the map. Actual nucleotide positions of mutations are listed as base numbers below the map.
The majority of observed mutations in the viral populations were substitutions: 31 of the 34 observed mutations in CMV populations were substitutions, all of the observed mutations in TMV populations were substitutions, and 1 of 2 mutations in the CCMV populations was a substitution. The three remaining mutations in CMV were single-base deletions. The CCMV recombination event was the only example of an addition mutation; no recombination events in the CMV and TMV populations were observed. Close examination of the specific changes indicates a bias for transitions, in particular G-to-A and C-to-U transitions. There was a strong bias for G-to-A transitions in the CMV populations (12 of 31 substitutions; Table 2). The TMV populations also demonstrated a slight preference for G-to-A and C-to-U transitions (7 of 15 substitutions; Table 3). This sort of transition bias, noted in other viral systems (21, 40), is higher than what is seen in DNA evolution (40). The bias may be due to the ability of guanosine to form a hydrogen bond with uridine in RNA base pairing. A phylogenetic study of CMV RNA 3 sequences has also indicated a strong bias for G-to-A and C-to-U transitions, although C-to-U transitions appear to be more common (35).
TABLE 2.
Types of substitution mutations observed in CMV populations
| Original base | No. of occurrences of mutated base:
|
|||
|---|---|---|---|---|
| G | A | U | C | |
| G | 12 | 2 | 0 | |
| A | 3 | 1 | 0 | |
| U | 3 | 3 | 4 | |
| C | 1 | 0 | 2 | |
TABLE 3.
Types of substitution mutations observed in TMV populations
| Original base | No. of occurrences of mutated base:
|
|||
|---|---|---|---|---|
| G | A | U | C | |
| G | 3 | 1 | 1 | |
| A | 2 | 1 | 0 | |
| U | 0 | 1 | 1 | |
| C | 0 | 1 | 4 | |
Conclusions.
Different levels of population diversity in field isolates of plant viruses have been observed, but until now there have been no controlled systematic studies of plant viral quasispecies and how they related to other biological properties. This study is also the first controlled comparison of related viral populations in a common host. CMV, CCMV, and TMV represent three viruses with a common evolutionary link that have distinctly different host range sizes. Previous studies on the population diversity of these three viruses suggest that CMV replicated-RNA populations (especially CMV satellite RNA populations) can be highly diverse (2, 20, 27, 30). Reports indicate that other strains of TMV maintain a lower level of diversity for genomic RNAs (31), even though TMV replicated satellite virus RNAs have been shown to have high levels of population diversity (19). A recent study of TMV suggested a mutation frequency of 3.1 × 10−4 per nucleotide in passaged TMV populations on a variety of hosts (18). CCMV populations have not been previously studied for diversity levels. A controlled study of these three viruses in a common host provides an opportunity to better understand the quasispecies nature of plant RNA viruses and how quasispecies may relate to host range.
Interestingly, the levels of quasispecies diversity for CMV, TMV, and CCMV in the common host N. benthamiana correlate directly with the relative sizes of the viral host ranges (Table 1). This has important evolutionary implications for the quasispecies structure of viral populations, since it suggests that highly diverse viruses such as CMV have a better chance of expanding into a new niche and thus pose a greater threat of emerging as new crop diseases. Several studies have examined the fitness effects of highly diverse viral populations in different circumstances. However, the overall effects of population diversity on the long-term evolutionary trajectories of viruses, particularly as they relate to the emergence of new diseases, are not well understood. The advantage of genetic diversity in populations that encounter new environmental challenges is a consistent theme that runs through all levels of biology. Theoretically, the ability to maintain genetic diversity in viral populations should enhance chances for adaptation to new selective regimes. Alternatively, if high diversity in the viral population resulted in fitness losses, the forces of selection would rapidly eliminate viruses that surpass viable limits of population diversity. Although a correlation between viral population diversity and expansion into new hosts is intriguing, only one host species and three viruses were used in this study. It will be important to analyze more hosts and more viruses to confirm this observation.
ACKNOWLEDGMENTS
We thank Rick Nelson and Shelly Carter for supplying U1 TMV; Robert Gonzales, Angela Scott, and Ann Harris for assistance in sequencing; Joachim de Miranda, Jim Bull, Holly Wichman, and Isabella Novella for helpful comments and discussion; and Greg May and Xin Shun Ding for critical reviews of the manuscript.
REFERENCES
- 1.Allison R F, Janda M, Ahlquist P. Infectious in vitro transcripts from cowpea chlorotic mottle virus cDNA clones and exchange of individual RNA components with brome mosaic virus. J Virol. 1988;62:3581–3588. doi: 10.1128/jvi.62.10.3581-3588.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aranda M A, Fraile A, García-Arenal F. Genetic variability and evolution of the satellite RNA of cucumber mosaic virus during natural epidemics. J Virol. 1993;67:5896–5901. doi: 10.1128/jvi.67.10.5896-5901.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bujarski J J, Nagy P D. Different mechanisms of homologous and nonhomologous recombination in brome mosaic virus: role of RNA sequences and replicase proteins. Semin Virol. 1996;7:363–372. [Google Scholar]
- 4.Chao L. Fitness of RNA virus decreased by Muller's ratchet. Nature. 1990;348:454–455. doi: 10.1038/348454a0. [DOI] [PubMed] [Google Scholar]
- 5.Chao L, Tran T, Matthews C. Muller's ratchet and the advantage of sex in the RNA virus ø6. Evolution. 1992;46:289–299. doi: 10.1111/j.1558-5646.1992.tb02038.x. [DOI] [PubMed] [Google Scholar]
- 6.deMiranda J R, Stevens M, deBruyne E, Smith H G, Bird C, Hull R. Beet luteovirus coat protein sequence variation. Ann Appl Biol. 1995;127:113–124. [Google Scholar]
- 7.Domingo E, Escarmís C, Sevilla N, Moya A, Elena S F, Quer J, Novella I, Holland J J. Basic concepts in RNA virus evolution. FASEB J. 1996;10:859–864. doi: 10.1096/fasebj.10.8.8666162. [DOI] [PubMed] [Google Scholar]
- 8.Domingo E, Holland J J. RNA virus mutations and fitness for survival. Annu Rev Microbiol. 1997;51:151–178. doi: 10.1146/annurev.micro.51.1.151. [DOI] [PubMed] [Google Scholar]
- 9.Duarte E, Clarke D, Moya A, Domingo E, Holland J. Rapid fitness losses in mammalian RNA virus clones due to Muller's ratchet. Proc Natl Acad Sci USA. 1992;89:6015–6019. doi: 10.1073/pnas.89.13.6015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duarte E A, Novella I S, Ledesma S, Clarke D K, Moya A, Elena S F, Domingo E, Holland J J. Subclonal components of consensus fitness in an RNA virus clone. J Virol. 1994;68:4295–4301. doi: 10.1128/jvi.68.7.4295-4301.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dzianott A M, Bujarski J J. The nucleotide sequence and genome organization of the RNA-1 segment in two bromoviruses: broad bean mottle virus and cowpea chlorotic mottle virus. Virology. 1991;185:553–562. doi: 10.1016/0042-6822(91)90525-g. [DOI] [PubMed] [Google Scholar]
- 12.Edwardson J R, Christie R G. CRC handbook of viruses infecting legumes. Boca Raton, Fla: CRC Press; 1991. Cucumoviruses; pp. 293–319. [Google Scholar]
- 13.Eigen M. The physics of molecular evolution. Chem Scri. 1986;26B:13–16. [Google Scholar]
- 14.Eigen M. Viral quasispecies. Sci Am. 1993;269:42–49. doi: 10.1038/scientificamerican0793-42. [DOI] [PubMed] [Google Scholar]
- 15.Gilbertson R L, Rojas M R, Russell D R, Maxwell D P. Use of the asymmetric polymerase chain reaction and DNA sequencing to determine genetic variability of bean golden mosaic geminivirus in the Dominican Republic. J Gen Virol. 1991;72:2843–2848. doi: 10.1099/0022-1317-72-11-2843. [DOI] [PubMed] [Google Scholar]
- 16.Holland J, Spindler K, Horodyski F, Grabau E, Nichol S, VandePol S. Rapid evolution of RNA genomes. Science. 1982;215:1577–1585. doi: 10.1126/science.7041255. [DOI] [PubMed] [Google Scholar]
- 17.Holmes F O. A comparison of the experimental host ranges of tobacco-etch and tobacco-mosaic viruses. Phytopathology. 1946;36:643–659. [Google Scholar]
- 18.Kearney C M, Thomson M J, Roland K E. Genome evolution of tobacco mosaic virus populations during long-term passaging in a diverse range of hosts. Arch Virol. 1999;144:1–4. doi: 10.1007/s007050050607. [DOI] [PubMed] [Google Scholar]
- 19.Kurath G, Heick J A, Dodds J A. RNase protection analyses show high genetic diversity among field isolates of satellite tobacco mosaic virus. Virology. 1993;194:414–418. doi: 10.1006/viro.1993.1278. [DOI] [PubMed] [Google Scholar]
- 20.Kurath G, Palukaitis P. RNA sequence heterogeneity in natural populations of three satellite RNAs of cucumber mosaic virus. Virology. 1989;173:231–240. doi: 10.1016/0042-6822(89)90239-0. [DOI] [PubMed] [Google Scholar]
- 21.Kurath G, Rey M E C, Dodds J A. Analysis of genetic heterogeneity within the type strain of satellite tobacco mosaic virus reveals several variants and a strong bias for G to A substitution mutants. Virology. 1992;189:233–244. doi: 10.1016/0042-6822(92)90699-p. [DOI] [PubMed] [Google Scholar]
- 22.Lane L C. Bromoviruses. In: Kurstack E, editor. Handbook of plant virus infections and comparative diagnosis. Amsterdam, The Netherlands: Elsevier Biomedical Press; 1981. pp. 333–376. [Google Scholar]
- 23.López C, Ayllón M A, Navas-Castillo J, Guerri J, Moreno P, Flores R. Molecular variability of the 5′- and 3′-terminal regions of citrus tristeza virus RNA. Phytopathology. 1998;88:685–691. doi: 10.1094/PHYTO.1998.88.7.685. [DOI] [PubMed] [Google Scholar]
- 24.Milligan J F, Groebe D R, Witherell G W, Uhlenbeck O C. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Novella I S, Elena S F, Moya A, Domingo E, Holland J J. Size of genetic bottlenecks leading to virus fitness loss is determined by mean initial population fitness. J Virol. 1995;69:2869–2872. doi: 10.1128/jvi.69.5.2869-2872.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Novella I S, Quer J, Domingo E, Holland J J. Exponential fitness gains of RNA virus populations are limited by bottleneck effects. J Virol. 1999;73:1668–1671. doi: 10.1128/jvi.73.2.1668-1671.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Palukaitis P, Roossinck M J. Variation in the hypervariable region of cucumber mosaic virus satellite RNAs is affected by the helper virus and the initial sequence context. Virology. 1995;206:765–768. doi: 10.1016/s0042-6822(95)80006-9. [DOI] [PubMed] [Google Scholar]
- 28.Palukaitis P, Roossinck M J, Dietzgen R G, Francki R I B. Cucumber mosaic virus. Adv Virus Res. 1992;41:281–348. doi: 10.1016/s0065-3527(08)60039-1. [DOI] [PubMed] [Google Scholar]
- 29.Rizzo T M, Palukaitis P. Nucleotide sequence and evolutionary relationships of cucumber mosaic virus (CMV) strains: CMV RNA 2. J Gen Virol. 1988;69:1777–1787. doi: 10.1099/0022-1317-69-8-1777. [DOI] [PubMed] [Google Scholar]
- 30.Rodríguez-Alvarado G, Kurath G, Dodds J A. Heterogeneity in pepper isolates of cucumber mosaic virus. Plant Dis. 1995;79:450–455. [Google Scholar]
- 31.Rodríguez-Cerezo E, García-Arenal F. Genetic heterogeneity of the RNA genome population of the plant virus U5-TMV. Virology. 1989;170:418–423. doi: 10.1016/0042-6822(89)90432-7. [DOI] [PubMed] [Google Scholar]
- 32.Rodríguez-Cerezo E, Moya A, García-Arenal F. Variability and evolution of the plant RNA virus pepper mild mottle virus. J Virol. 1989;63:2198–2203. doi: 10.1128/jvi.63.5.2198-2203.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roossinck M J. Mechanisms of plant virus evolution. Annu Rev Phytopathol. 1997;35:191–209. doi: 10.1146/annurev.phyto.35.1.191. [DOI] [PubMed] [Google Scholar]
- 34.Roossinck M J, Kaplan I, Palukaitis P. Support of a cucumber mosaic virus satellite RNA maps to a single amino acid proximal to the helicase domain of the helper virus. J Virol. 1997;71:608–612. doi: 10.1128/jvi.71.1.608-612.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roossinck M J, Zhang L, Hellwald K-H. Rearrangements in the 5′ nontranslated region and phylogenetic analyses of cucumber mosaic virus RNA 3 indicate radial evolution of three subgroups. J Virol. 1999;73:6752–6758. doi: 10.1128/jvi.73.8.6752-6758.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shintaku M H, Carter S A, Bao Y, Nelson R S. Mapping nucleotides in the 126-kDa protein gene that controls the differential symptoms induced by two strains of tobacco mosiac virus. Virology. 1996;221:218–225. doi: 10.1006/viro.1996.0368. [DOI] [PubMed] [Google Scholar]
- 37.Simon A E, Bujarski J J. RNA-RNA recombination and evolution in virus-infected plants. Annu Rev Phytopathol. 1994;32:337–362. [Google Scholar]
- 38.Smith D B, McAlister J, Casino C, Simmonds P. Virus ‘quasispecies’: making a mountain out of a molehill? J Gen Virol. 1997;78:1511–1519. doi: 10.1099/0022-1317-78-7-1511. [DOI] [PubMed] [Google Scholar]
- 39.Stenger D C. Genetic variability and the occurrence of less than genome-length viral DNA forms in a field population of beet curly top geminivirus. Phytopathology. 1995;85:1316–1322. [Google Scholar]
- 40.Vartanian J-P, Plikat U, Henry M, Mahieux R, Guillemot L, Meyerhans A, Wain-Hobson S. HIV genetic variation is directed and restricted by DNA precursor availability. J Mol Biol. 1997;270:139–151. doi: 10.1006/jmbi.1997.1104. [DOI] [PubMed] [Google Scholar]
- 41.Zaitlin M, Israel H W. Tobacco mosaic virus (type strain). CMI/AAB descriptions of plant viruses no. 151. Warwick, United Kingdom: Association of Applied Biologists; 1975. [Google Scholar]