Abstract
Due to various advantageous features there is current interest in retroviral vectors derived from primate foamy viruses (PFVs). Two PFV cis-acting sequences have been mapped in the 5′ region of the RNA (pre-)genome and in the 3′ pol genomic region. In order to genetically separate PFV packaging constructs from vector constructs, we investigated the effect of deletions in the 5′ untranslated region (UTR) of PFV packaging constructs and vectors on gene expression and RNA incorporation into viral particles. Our results indicate that the 5′ UTR serves different previously unknown functions. First, the R region of the long terminal repeat was found to be required for PFV gag gene expression. This regulation of gene expression appeared to be mainly posttranscriptional. Second, constructs with sequence deletions between the R region and the gag gene start codon packaged as much viral mRNA into particles as the undeleted construct, and RNA from such a 5′-UTR-deleted packaging construct was copackaged into vector-virus particles, together with vector RNA which was preferentialy packaged. Finally, in the U5 region a sequence was identified that was required to allow cleavage of the Gag precursor protein by the pol gene-encoded protease, suggesting a role of RNA in PFV particle formation. Taken together, the results indicate that complex interactions of the viral RNA, capsid, and polymerase proteins take place during PFV particle formation and that a clear separation of PFV vector and packaging construct sequences may be difficult to achieve.
Foamy viruses constitute the most divergent genus in the family Retroviridae. The primate foamy virus (PFV) replication strategy differs fundamentally from all other retroviruses and appears to bridge the retroviral and hepadnaviral replication pathway (for a recent review, see reference 31). PFVs have a genomic structure similar to other retroviruses and require provirus integration for replication (14, 42). However, the way in which polymerase protein is expressed (13, 57), the mode of viral capsid assembly and interaction with the cognate envelope (Env) glycoprotein (4, 18, 19, 41), and the time point of reverse transcription in the viral life cycle (35, 58) are unique among retroviruses and show functional analogies to hepadnaviruses (37).
PFVs have been suggested as good candidates from which to develop viral vectors, and several vectors are currently under construction for gene transfer purposes (9, 15, 21, 26, 38, 45, 47, 55, 56). A retroviral vector system consists of the vector proper, which harbors all elements in cis required for genome packaging, reverse transcription, integration, and transgene expression, and a packaging cell line, which supplies in trans the viral structural and enzymatic proteins required to execute these functions (34). Ideally, there is little genetic overlap between vector and packaging construct sequences to reduce the problem of generating replication-competent virus following recombination events (34).
In general, retroviruses harbor a cis-acting sequence (psi in murine retroviruses) in the 5′ untranslated region (UTR) of the genome which facilitates the dominant package of genomic viral RNA (8, 32). This sequence can usually be deleted from the packaging constructs expressing the Gag and Pol proteins (25, 34). In PFVs the Pol protein is expressed from a spliced RNA which uses the major splice donor (SD) located at position +53 following the start of transcription (+1) and a splice acceptor located in the gag gene (+1,850) (27, 57). Therefore, the deletion of the SD site from PFV packaging constructs and expression of Pol protein from a separate construct may disturb a balanced expression of Gag and Pol proteins, provided this is essential for optimal vector production. Furthermore, it has recently been shown that PFV vectors essentially require, besides a sequence in the 5′ region of the (pre-)genomic RNA (cis-acting sequence I [CASI]), essentially a second sequence located in the 3′ pol gene (CASII) in order to transfer successfully marker genes (15, 21, 55). Deletion of CASII from vector constructs largely reduced the viral RNA content of particles and also influenced the function of the pol-encoded protease to cleave the Gag precursor protein (21). This suggested complex interactions between the Gag and Pol proteins and the (pre-) genomic RNA in forming a functional PFV capsid. Since CASII is located in a coding region which cannot be deleted from packaging constructs, we aimed to investigate deletions in CASI to achieve separation of vector sequences from packaging sequences.
To do this, we used molecular clones of an isolate that was reported to be obtained from human material (1). Since there is a lack of evidence for authentic human foamy viruses (2, 44, 49), the origin of this isolate most probably related to an incidental trans-species transmission from chimpanzees harboring virtually identical PFVs (22, 23, 48).
MATERIALS AND METHODS
Cells.
Human fibrosarcoma (HT1080) and 293T (11) cells were cultivated in Dulbecco modified Eagle minimal essential medium supplemented with 10% fetal calf serum and antibiotics.
Recombinant DNA.
The plasmids pCgp-1, pCpol-2, and pCenv-1, directing the expression of PFV Gag and Pol, Pol, and Env proteins, respectively, under the transcriptional control of the human cytomegalovirus (CMV) enhancer-promoter, as well as the vector pMH5/M54 and the plasmid pSP/HFV-2, which was used to generate in vitro RNA transcripts, have been described previously (21). The pMH vectors contain the internal green fluorescent protein (GFP) expression cassette under transcriptional control of the spleen focus-forming virus U3 region (6). The plasmid pMH104 was derived from pMH5/M54. In addition to the pol gene ATG downmutation, in pMH104 the initiation sequences of the gag gene was changed from ATG′GCT′TCA to TTG′GCT′TAA, and thus the expression of Gag protein ablated. This mutation and all further mutants in the 5′ UTR in the background of the gag and pol expression plasmid pCgp-1 and the vector pMH5/M54 (as shown in detail in the figures) were generated by recombinant PCR (24). All mutations were verified by automated DNA sequence analysis of the amplicon by using AmpliTaq FS and the ABI 310 sequence analysis system (Perkin-Elmer) to exclude unwanted nucleotide exchanges.
The plasmid pSP/HFV-3 contains a part of the PFV gag gene (5′ of the NcoI restriction site) and the 5′ UTR inserted in antisense direction downstream of the SP6 promoter in the vector pSP65.
Transfections and analysis of vector transfer efficiency.
All transfections were carried out by using a calcium-phosphate cotransfection protocol, essentially as described previously (18, 21, 29) with a total of 27 μg of plasmid DNA per 1.8 × 106 cells. Virus from pMH104 vector (9 μg) was produced by cotransfection with gag and pol (pCgp) and env (pCenv-1) expression constructs (9 μg each) into 293T cells. When only one or two plasmids were transfected, the total DNA amount was adjusted to 27 μg by using pcDNA plasmid (Invitrogen). The vector transduction efficiency was determined as described previously (21). Briefly, 48 h after transfection 1 ml of cell-free supernatant (0.45-μm [pore-size] filtrate) was added to HT1080 recipient cells, which were seeded at a density of 3 × 103 cells per well in 24-well plates 1 day before. The cells were monitored for GFP expression 3 days after transduction by flow cytometry on a FACScan using the LysisII and CellQuest software packages (Becton Dickinson). The transduction efficiency was expressed as the percentage of GFP-positive cells in relation to the total number of cells analyzed.
Protein analysis.
Lysates from transfected cells or from cell-free virus were prepared by resuspension in detergent containing buffer, as described elsewhere (20). Cell-free virus was obtained by filtration of supernatant from transfected cells through a 0.45-μm (pore-size) filter (Schleicher & Schuell) and subsequent sedimentation through a 20% sucrose cushion in TNE (100 mM Tris-HCl, 100 mM NaCl, 1 mM EDTA; pH 8.0) for 3 h in an SW41 rotor (Beckman) at 25,000 rpm and 4°C. The samples were subjected to electrophoresis through sodium dodecyl sulfate–8% polyacrylamide gels and semi-dry blotted onto nitrocellulose membranes (Schleicher & Schuell). The blots were incubated with rabbit antisera raised against recombinant PFV Gag (20) and RNase H proteins (28) or, alternatively, with a mouse monoclonal antibody directed against PFV Pol protein (H. Imrich, M. Heinkelein, and A. Rethwilm, unpublished data), and were developed by using the ECL detection system (Amersham).
RPA.
All RNase protection analyses (RPAs) were performed by using the Direct Detection or the pTRI-GAPDH human kits from Ambion, essentially as described by the manufacturer. For the detection of RNA in virions, virus was produced as described previously (21). For the detection of cellular RNA by RPA, total RNA was prepared 48 h following transfection with the RNEasy Mini Kit (Qiagen), and 5 μg of RNA was used per assay. In some experiments virions were treated with 20 μg RNase A (Sigma) per ml at 25°C for 30 min prior to RNA extraction, and cellular RNA was treated with 0.5 U of RQ1 DNase (Promega) at 37°C for 25 min. Antisense RNA probes were generated in vitro from XbaI-linearized pSP/HFV-3 plasmid and HindIII-linearized pSP/HFV-2 plasmid (21).
RESULTS
Sequences in the R region of the long terminal repeat (LTR) are required for Gag expression.
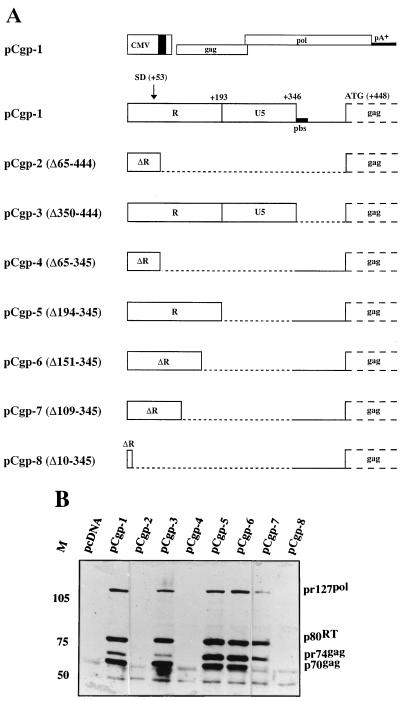
The 5′ SD of PFV, which is used to generate the subgenomic pol, env, and accessory gene transcripts, is located at position +53 following the transcriptional start (+1) of the viral RNA (33, 36). A difference from the previously published PFV sequence (33) is due to two additional nucleotides found upon resequencing our molecular clones. This revealed the sequence 5′-GAGCTCTCTTCACTA following the start of transcription. We assumed that sequences downstream from the SD and upstream of the gag gene (the gag ATG is located at position +448) may be dispensable for PFV gene expression. Deletion mutants in this region downstream of the SD (3′ of position +65) were generated in a CMV enhancer-promoter-directed expression construct, as shown in Fig. 1A. 293T cells were transfected with these constructs and analyzed for intracellular expression of Gag and Pol proteins. With the wild-type construct, pCgp-1, expression of the 127-kDa Pol precursor, the 80-kDa reverse transcriptase (RT) cleavage product, the 74-kDa Gag precursor, and the 70-kDa amino-terminal cleavage product were readily observed (Fig. 1B). Deletion of the sequences +65 to +444 in pCgp-2 completely abolished PFV protein expression (Fig. 1B). While the deletion of the leader sequences downstream of the R-U5 region in pCpg-3 (Δ350–444) had no influence on Gag and Pol protein expression, the deletion of sequences in R-U5 (Δ65–345) in pCgp-4 affected protein expression, as did the large deletion in pCgp-2 (Fig. 1B). To characterize further the element required to express PFV Gag and Pol proteins, we generated additional R-U5 deletion mutants (pCgp-5 to pCgp-8 in Fig. 1A). Absence of the U5 region in pCgp-5 (Δ194–345) and further deletion of 3′ R sequences in pCgp-6 (Δ151–345) allowed for Gag and Pol protein expression. However, with these constructs, and in particular with pCgp-7, a reduced level of cleaved p70gag was seen (Fig. 1B). Further truncation of R (pCgp-8; Δ10–345) abolished detectable PFV pr74 Gag protein expression, while due to the absence of the 5′ SD in this construct expression of Pol proteins could not be expected (Fig. 1B). We conclude from these experiments that sequences located in the R region downstream of the major 5′ SD (namely, in constructs pCgp-2 and pCgp-4) are required to express PFV Gag and Pol proteins.
FIG. 1.
Analysis of deletion mutants in the 5′ UTR for the expression of Gag and Pol proteins. (A) Wild-type pCgp-1 gag and pol expression construct and deletion mutants in the 5′ UTR. The major 5′ SD, the primer binding site (pbs), and the poly(A) signal (pA+), which was derived from the bovine growth hormone gene and is present in the pcDNA vector (Invitrogen), are indicated. The numbering of the deletions (Δ) is relative to the start of transcription (+1). The deletions in the constructs are marked by stippled lines. (B) Analysis of Gag and Pol protein expression levels in 293T cells transfected with the pCgp plasmids. Cellular lysates were reacted with PFV Gag and Pol antibodies in an immunoblot. M, relative molecular mass standards in kilodaltons.
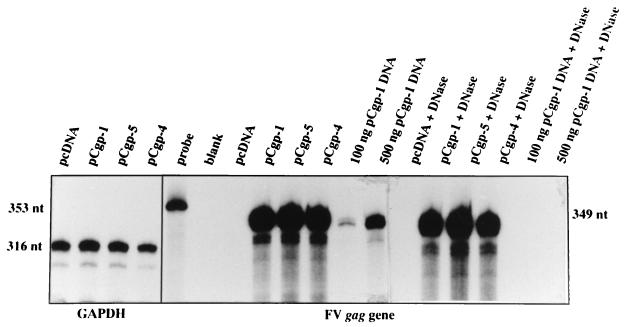
To investigate whether the lack of gag gene expression in the deletion mutants is reflected by the level of the respective mRNA in the transfected cells, RPA was performed. A representative experiment is shown in Fig. 2. No difference in gag mRNA expression levels in total cellular RNA extracted from the transfected cells was observed between Cpg-5 and pCgp-4; the former contains the complete R region and is able to generate Gag protein, while the latter is deleted in R and unable to express Gag (Fig. 1). Similar results were obtained when other R region-deleted constructs, such as pCgp-2 and pCgp-8, were analyzed (data not shown).
FIG. 2.
Analysis of gag-encoding RNA levels by RPA in 293T cells transfected with different gag and pol expression constructs from Fig. 1. A total of 5 μg of total cellular RNA was assayed per lane by using the 353-nt gag gene-derived pSP/HFV-3 probe. For control purposes, the levels of glyceraldehyde-3′-phosphate dehydrogenase (GAPDH) were analyzed. To investigate the possibility that contaminating plasmid DNA from the transfections was detected by RPA, we assayed pCgp-1 DNA at the indicated amounts in two lanes. DNase digestion prior to RPA completely eliminated detectable traces of plasmid DNA.
Deletion mutants in the 5′ UTR package viral RNA into particles.
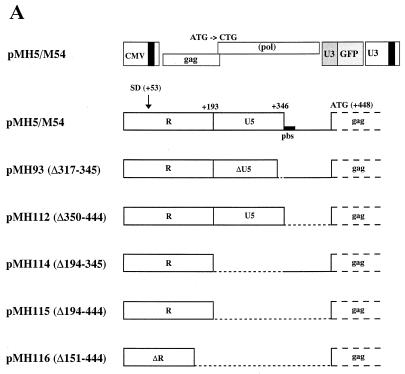
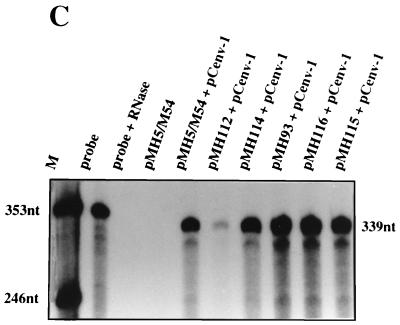
In retroviral packaging constructs the deletion of sequences within the 5′ UTR (the psi sequence) disables the packaging of the gag-pol mRNA and allows for the relatively selective packaging of psi sequence-containing vector RNA (34, 36). In order to investigate whether a similar approach is feasible to separate PFV packaging construct sequences from vector sequences, we generated the internal 5′ UTR deletion mutants shown in Fig. 3A. The constructs were made in the pMH5/M54 vector backbone (21). This vector has a wild-type gag gene and a pol gene ATG-to-CTG mutation (21). Since no Pol protein is expressed from this vector genome, immature viral pr74 Gag particles (18), which are released from cells following cotransfection with an env expression construct, only contain viral RNA and no viral DNA. Therefore, the vector RNA content of the particles can be analyzed quantitatively by RPA (21). All constructs harbored the R region-located cis-acting sequence essential for Gag protein expression, which was identified in the previous experiment. As shown in Fig. 3B, similar amounts of Gag protein were expressed upon transfection of the vectors into 293T cells. Cotransfection of an Env expression plasmid allowed the viral particles to egress from the cells and pellet through a 20% sucrose cushion (Fig. 3B). When the RNA content in the particles was determined by RPA with a gag gene-derived probe, which can hybridize only to the full-length transcript, all particle preparations, except those derived from pMH112 (Δ350–444), contained roughly similar amounts of viral RNA (Fig. 3C). Quantification based on six independent experiments revealed that in relation to pMH5/M54 the viral RNA content of particles derived from pMH112 transfection was reduced to 33%, whereas the values for pMH114, pMH115, and pMH93 were 124, 94, and 166%, respectively. A mild treatment of particles with RNase A prior to RNA extraction did not change this result significantly (data not shown).
FIG. 3.
Analysis of the viral RNA content in particulate material derived from vector genomes deleted in the 5′ UTR. (A) Schematic view of 5′ UTR deletion constructs. The deletions are marked by stippled lines. (B) Detection of pr74 Gag protein expressed by the pMH vectors shown in panel A and cotransfected with an envelope expression construct (pCenv-1) in cellular lysates and in cell-free particulate material sedimented through a sucrose cushion. M, relative molecular mass standard in kilodaltons. (C) RPA of viron RNA of the particles shown in panel B using a gag gene-derived probe of 353 nt. A representative example of six experiments is shown. The marker lane (M) contains in vitro-transcribed RNA of 353 and 246 nt.
These results show that even large deletions in the 5′ UTR of PFV do not abrogate viral RNA incorporation. Interestingly, pMH112 had a smaller deletion (Δ350–444) in the 5′ UTR and, nonetheless, packaged less viral RNA compared to the constructs pMH115 and pMH116, which had larger 5′ UTR deletions (Δ194–444 and Δ151–444, respectively).
Identification of a small sequence in the U5 region that is required for protease-mediated cleavage of the Gag protein precursor in virions.
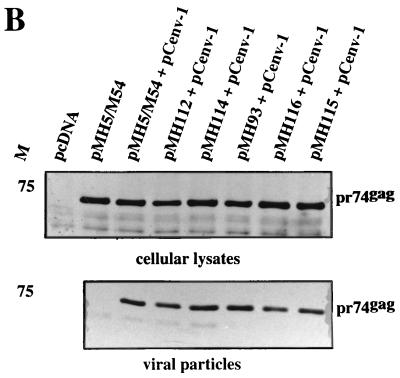
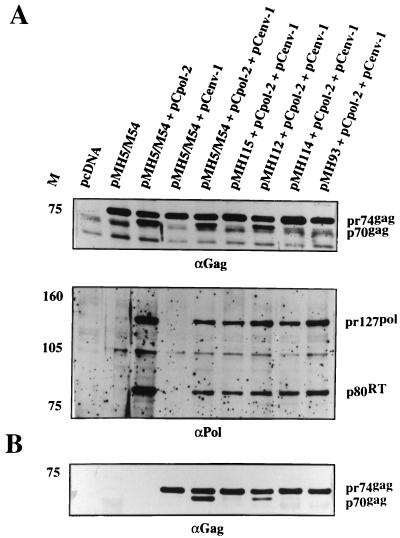
It has been shown previously that the incorporation of viral RNA into PFV particles affected the cleavage of the Gag protein by the pol gene-encoded protease (21). Therefore, the 5′ UTR deleted pMH vectors were, in addition to an env expression plasmid, also cotransfected with a pol expression plasmid, and the Gag proteins were analyzed in cellular extracts and extracellular particles. As shown in Fig. 4A, the triple transfection of the different vectors and pol and env encoding plasmids resulted in roughly similar levels of intracellular pr74 Gag precursor, pr127 Pol precursor and the p80 RT cleavage product, respectively. Intracellular cleavage of pr74 Gag was readily observed upon transfection of pMH5/M54 and pMH112 vectors together with pCpol-2 and pCenv-1. However, with the other constructs (pMH93, pMH114, and pMH115) the intracellular cleavage appeared to be reduced (Fig. 4A). The analysis of extracellular particles revealed a more drastic pattern. Only particles derived from pMH5/M54 and pMH112 transfections unequivocally showed the characteristic doublet of pr74/p70 Gag. pMH114 and pMH115 were found to be heavily defective in Gag cleavage, despite the presence of proteolytically active Pol protein in the transfected cells (Fig. 4B). Of particular interest was mutant pMH93 bearing only a 29-nucleotide (nt) deletion in the 3′ portion of the U5 region (Δ316–344) and being readily able to incorporate viral RNA (Fig. 3C). However, this mutant was also found to be unable to allow cleavage of the Gag precursor by the pol-encoded protease supplied in trans (Fig. 4B). In contrast, mutant pMH112, the RNA of which was found to have a reduced capacity to be packaged (Fig. 3), allowed for at least partial cleavage of Gag compared with wild-type pMH5/M54 (Fig. 4).
FIG. 4.
Analysis of the Gag and Pol expression profile in cell extracts and extracellular particulate material from cells cotransfected with 5′ UTR-deleted PFV vectors shown in Fig. 3A and pol and env expression constructs. A representative example of three experiments is shown. M, relative molecular mass standards in kilodaltons. (A) Detection of Gag and Pol proteins in cellular lysates from transfected cells. (B) Immunoblot analysis of Gag proteins in viral particles sedimented through a sucrose cushion.
Use of deleted packaging constructs in vector transfer experiments.
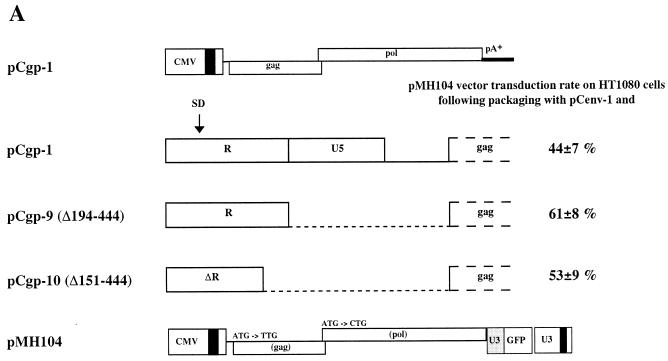
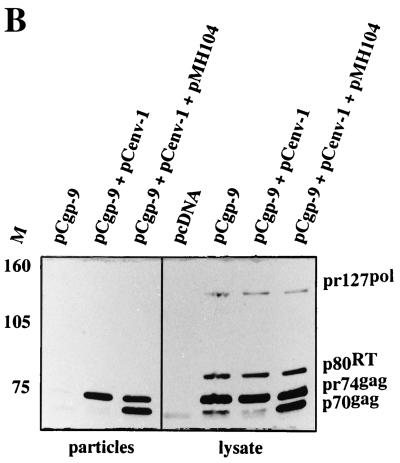
We next applied packaging constructs deleted in the 5′ UTR in vector transfer experiments. To do this, we used packaging constructs with maximal tolerable deletions in the 5′ UTR regardless of whether they package their own RNA. 293T cells were cotransfected with pCgp-derived gag and pol expression constructs, together with an env expression plasmid and the pMH104 vector, which is unable to generate Gag and Pol proteins from its own genome (Fig. 5A). Vector virus from the cell-free supernatant was subjected to recipient HT1080 cells, which were subsequently monitored for GFP expression. As shown in Fig. 5A, the 5′ UTR-deleted gag and pol expression constructs pCgp-9 (Δ192–442) and pCgp-10 (Δ149–442) gave rise to similar or even higher transduction rates than the wild-type pCgp-1 expression construct. When the PFV protein profile was analyzed in cellular extracts from cells transfected only with the packaging plasmids, pCgp-9 and pCenv-1, a reduced level of cleaved pr74 Gag precursor was detected despite the presence of proteolytically active Pol protein in the cells (Fig. 5B). This defect in Gag cleavage was much more pronounced in the corresponding viral particle preparations centrifuged through a sucrose cushion (Fig. 5B). Interestingly, addition of the vector genome which contained all cis-acting sequences to the transfection mix led to the activation of the proteolytic activity of Pol to cleave the Gag precursor and thus to the generation of infectious vector particles (Fig. 5B).
FIG. 5.
Analysis of the vector transduction rate and protein profile in cellular extracts and extracellular viruses from cells transfected with 5′ UTR-deleted packaging constructs, an env expression plasmid, and a vector genome. (A) Schematic view of pCgp packaging and pMH104 vector constructs and vector transduction rate on HT1080 target cells with cell-free vector virus produced by transient cotransfection, together with the pCenv-1 plasmid in 293T cells. The transduction rates are the means and standard variations of six experiments. (B) PFV Gag and Pol proteins in cellular lysates and extracellular particles centrifuged through a sucrose cushion. 293T cells were transfected as indicated, and the viral proteins were analyzed by immunoblot. M, relative molecular mass standards in kilodaltons.
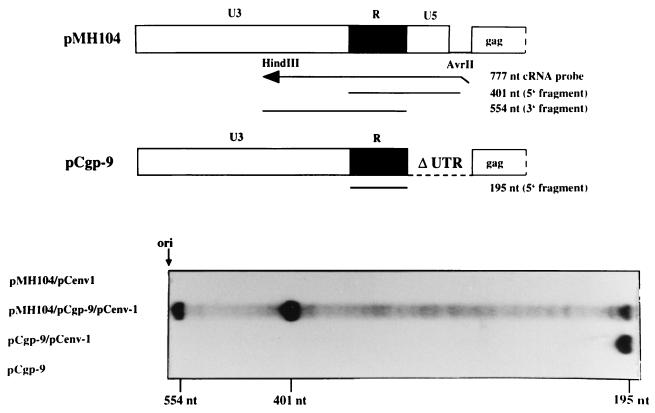
To investigate whether RNA from the packaging construct was copackaged into virions, RPA was performed. As shown in Fig. 6, the gag mRNA of the construct pCgp-9 was detected in particulate material when this packaging construct was cotransfected with an env expression plasmid. This finding corroborated the result shown in Fig. 3 on the packaging of 5′ UTR-deleted vectors. Upon triple transfection of packaging plasmids, together with the pMH104 vector, the majority of the viral RNA protected was derived from the vector genome (Fig. 6). However, the indicative band of 195 nt, which corresponds to the gag RNA from pCgp-9, was still present (Fig. 6), which indicated that this RNA was copackaged into vector genome containing virions. No particle-associated RNA was found when pCgp-9 was transfected in the absence of Env or the pMH104 vector was cotransfected together with pCenv-1 in the absence of a gag- and pol-encoding packaging construct.
FIG. 6.
Packaging of RNA from the pCgp-9 packaging constructs into PFV vector particles. 293T cells were transfected with the plasmids as indicated. Viral particles were prepared from the cell-free supernatant by centrifugation through sucrose, and the particle-associated RNA was analyzed by RPA by using the radiolabeled LTR-spanning pSP/HFV-2 probe (21).
DISCUSSION
For the establishment of a retroviral gene transfer vector and a corresponding packaging cell line, constructs with little or no genetic overlap are preferentially used (34). In PFVs, an essential component of the vector, the CASII element, resides in a coding sequence and is consequently present on both the vector and the packaging construct genomes (21). We therefore aimed to investigate the influence of deletions in CASI in PFV packaging constructs on gene expression and RNA packaging.
The R region of the LTR was found to be essential for Gag and Pol protein expression. Since the total gag mRNA levels were unaffected by constructs from which no Gag protein expression could be detected, the effect of the R region on gag gene expression is mainly posttranscriptional. PFVs are an exception to other complex regulated retroviruses (10) since they encode a transcriptional trans-activator (43) and lack a regulatory protein acting at the posttranscriptional level (7). PFVs make extensive use of splicing to generate at least nine major subgenomic mRNAs which encode viral proteins (30). However, a mechanism to regulate gene expression from unspliced PFV RNA has so far not been identified. We hypothesize that cis-acting sequences in the R region are required to serve this function and may act similarly to the recently identified R region-located elements in murine retroviruses (52).
While such a mechanism could explain the lack of gag gene expression from R region-deleted constructs, we also found a lack of pol gene expression from constructs deleted in the UTR from sequences 12 nt 3′ of the SD, which is more difficult to explain. Thus, pol expression was found to depend on intron sequences. The expression of functionally active Pol protein from a cDNA clone, pCpol-2 (21; Imrich et al., unpublished), directed from the CMV enhancer-promoter was not influenced by the presence or absence of the cis-acting element in question, and it was also found to be independent of simultaneous Gag protein expression (5; Imrich et al., unpublished). Pol protein expression directed by the R region-deleted constructs described in this study requires the generation of a spliced transcript. Usually only six nucleotides 3′ of the SD site are necessary for recognition by the splicing machinery (40, 50, 54). However, it is not known how the unusually spliced (27, 57) PFV pol transcript behaves in this respect. Therefore, a lack of spliced pol transcript following transfection of the 5′ UTR-deleted plasmids is one possible explanation for the observed phenomenon.
Clearly, the role of the R region in regulating gag and pol gene expression needs an in-depth analysis which is beyond the scope of this study. In this respect it may be worth reevaluating recently published data on the expression of structural proteins in packaging cell lines for simian foamy virus type 1 vectors (56). The constructs that were used to establish these packaging cells were devoid of most of the R region. The R region, however, is approximately 80% conserved amongst the PFVs (23), and we believe this sequence to be unlikely to serve different functions in these related viruses.
It has been described previously that a PFV vector genome, pMH38, which was severely impaired in packaging RNA because of the deletion of CASII, was also impaired with respect to cleavage of the Gag protein by the pol-encoded protease (21). This left the question as to whether viral RNA packaging was required for incorporation of the Pol protein into the viral particle (21). We now describe a mutant, pMH93, which packages large amounts of RNA but still fails to allow cleavage of Gag. Plasmid pMH93 has only a small deletion of 29 nt in the U5 region. This result indicates that it is not the incorporation of viral RNA as such that is required to mediate Gag cleavage but rather the presence of specific sequences. The deletion in pMH93 is located just 5′ of the pbs and may, therefore, influence reverse transcription of the RNA (pre-)genome which one could assume to be a prerequisite of Gag cleavage. However, pMH112 is deleted in most of the pbs, it packaged less of its RNA than pMH93 and allowed at least partial cleavage of Gag. This makes it unlikely that reverse transcription must precede Gag cleavage. Furthermore, the analysis of gag mutants has suggested that Gag cleavage must precede reverse transcription and not vice versa (12).
While in other retroviruses RT activation is initiated by cleavage of the Gag-Pol precursor molecule (54, 55), the way in which the polymerase is activated in PFV is still unresolved (5). We did not specifically address this question. However, we found that a vector containing the cis-acting sequences must be present to allow Gag cleavage by the pol-encoded protease. This indicates that at least the protease function of the PFV Pol precursor to cleave Gag requires specific RNA sequences or structures. These sequences are in part located in U5, and they can be deleted from packaging constructs, since they are present on the vector genome. With respect to the activation of the RT, it is worth mentioning that PFV pol gene expression in the vaccinia virus system revealed high and almost identical RT values for the wild-type pol gene and a protease active site mutant with heterologous added template (17; N. Fischer and A. Rethwilm, unpublished data). Reverse transcription of the PFV pregenome, however, appears to require specific interactions of Gag, Pol, and viral RNA. Based on this and previous studies (12, 18, 21), we suggest a role for the viral RNA in PFV particle formation. RNA appears to be essential to allow for the completion of the particle formation via pr74-p70 cleavage and this cleavage, in turn, is required to allow reverse transcription of the RNA.
With respect to viral RNA incorporation, our results show that even RNA from 5′ UTR-deleted packaging constructs was efficiently packaged. We also found packaging construct RNA in infectious vector particles together with the vector genome. The finding that a smaller 5′ UTR deletion was packaged less efficiently than the RNA of the larger deletions indicates that the RNA-protein interactions may be controlled, perhaps at the structural level, by positive- and negative-acting 5′ UTR sequences. In other retroviral vector systems the specificity of packaging genomic over subgenomic viral RNA or even unrelated RNA is by far less than 100% (8, 32). Furthermore, sequences outside the 5′ psi signal have been identified which support the packaging of genomic RNA and are probably responsible for a residual packaging of subgenomic RNA in these systems (3, 8, 32). However, other studies indicated that these effects may be due to sequences promoting RNA stability and nuclear export, rather than acting as additional genuine packaging sequences (25, 39, 52). It will, therefore, be interesting to investigate how the two foamy viral CAS elements act in concert to allow for efficient RNA packaging and how overlapping elements in CASI, such as the signal for RNA dimerization (16), can be distinguished. Since a definite separation of packaging construct sequences and vector sequence, which avoids the incorporation of packaging construct RNA, may be difficult to achieve in PFV vector systems, it is worth investigating the frequency of recombinations between suitable vector and packaging construct sequences during reverse transcription. However, even in a worst case scenario such recombination events of transcriptional trans-activator gene (tas)- and 3′ U3 region-deleted constructs are very unlikely to generate a replication-competent virus (46).
ACKNOWLEDGMENTS
We thank Ottmar Herchenröder for critical review of the manuscript and Rolf Flügel for the gift of Pol antiserum.
This work was supported by grants from Deutsche Forschungsgemeinschaft (Re627/6-1), Bundesministerium für Bildung und Forschung (01KV9817/0), Bayerische Forschungsstiftung, Sächsisches Staatsministerium für Umwelt und Landwirtschaft, EU (BMH4-CT97-2010), and The Wellcome Trust.
REFERENCES
- 1.Achong B G, Mansell P W A, Epstein M A, Clifford P. An unusual virus in cultures from a human nasopharyngeal carcinoma. J Natl Cancer Inst. 1971;46:299–307. [PubMed] [Google Scholar]
- 2.Ali M, Taylor G P, Pitman R J, Parker D, Rethwilm A, Cheingsong-Popov R, Weber J N, Bieniasz P D, Bradley J, McClure M O. No evidence of antibody to human foamy virus in widespread human populations. AIDS Res Hum Retrovir. 1996;12:1473–1483. doi: 10.1089/aid.1996.12.1473. [DOI] [PubMed] [Google Scholar]
- 3.Aschoff J M, Foster D, Coffin J M. Point mutations in the avian sarcoma/leukosis virus 3′ untranslated region result in a packaging defect. J Virol. 1999;73:7421–7429. doi: 10.1128/jvi.73.9.7421-7429.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baldwin D N, Linial M L. The roles of pol and env in the assembly pathway of human foamy virus. J Virol. 1998;72:3658–3665. doi: 10.1128/jvi.72.5.3658-3665.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baldwin D N, Linial M L. Proteolytic activity, the carboxy terminus of Gag, and the primer binding site are not required for Pol incorporation into foamy virus particles. J Virol. 1999;73:6387–6393. doi: 10.1128/jvi.73.8.6387-6393.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baum C, Ito K, Meyer J, Laker C, Ito Y, Ostertag W. The potent enhancer activity of the polycythemic strain of spleen focus-forming virus in hematopoietic cells is governed by a binding site for Sp1 in the upstream control region and by a unique enhacer core motif, creating an exclusive target for PEBP/CBF. J Virol. 1997;71:6323–6331. doi: 10.1128/jvi.71.9.6323-6331.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baunach G, Maurer B, Hahn H, Kranz M, Rethwilm A. Functional analysis of human foamy virus accessory reading frames. J Virol. 1993;67:5411–5418. doi: 10.1128/jvi.67.9.5411-5418.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berkowitz R, Fischer J, Goff S P. RNA packaging. Curr Top Microbiol Immunol. 1996;214:177–218. doi: 10.1007/978-3-642-80145-7_6. [DOI] [PubMed] [Google Scholar]
- 9.Bieniasz P D, Erlwein O, Aguzzi A, Rethwilm A, McClure M O. Gene transfer using replication-defective human foamy virus vectors. Virology. 1997;235:65–72. doi: 10.1006/viro.1997.8658. [DOI] [PubMed] [Google Scholar]
- 10.Cullen B R. Human immunodeficiency virus as a prototypic complex retrovirus. J Virol. 1991;65:1053–1056. doi: 10.1128/jvi.65.3.1053-1056.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DuBridge R B, Tang P, Hsia H C, Leong P-M, Miller J H, Calos M P. Analysis of mutation in human cells by using Epstein-Barr virus shuttle system. Mol Cell Biol. 1987;7:379–387. doi: 10.1128/mcb.7.1.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Enssle J, Fischer N, Moebes A, Mauer B, Smola U, Rethwilm A. Carboxy-terminal cleavage of the human foamy virus gag precursor molecule is an essential step in the viral life cycle. J Virol. 1997;71:7312–7317. doi: 10.1128/jvi.71.10.7312-7317.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Enssle J, Jordan I, Mauer B, Rethwilm A. Foamy virus reverse transcriptase is expressed independently from the gag protein. Proc Natl Acad Sci USA. 1996;93:4137–4141. doi: 10.1073/pnas.93.9.4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Enssle J, Moebes A, Heinkelein M, Panhuysen M, Mauer B, Schweizer M, Neumann-Haefelin D, Rethwilm A. An active human foamy virus integrase is required for viral replication. J Gen Virol. 1999;80:1445–1452. doi: 10.1099/0022-1317-80-6-1445. [DOI] [PubMed] [Google Scholar]
- 15.Erlwein O, Bieniasz P D, McClure M O. Sequences in pol are required for transfer of human foamy virus-based vectors. J Virol. 1998;72:5510–5516. doi: 10.1128/jvi.72.7.5510-5516.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Erlwein O, Cain D, Fischer N, Rethwilm A, McClure M O. Identification of sites that act together to direct dimerization of human foamy virus RNA in vitro. Virology. 1997;229:251–258. doi: 10.1006/viro.1997.8438. [DOI] [PubMed] [Google Scholar]
- 17.Fischer N, Enssle J, Müller J, Rethwilm A, Niewiesk S. Characterization of human foamy virus proteins expressed by recombinant vaccinia viruses. AIDS Res Hum Retrovir. 1997;13:517–521. doi: 10.1089/aid.1997.13.517. [DOI] [PubMed] [Google Scholar]
- 18.Fischer N, Heinkelein M, Lindemann D, Enssle J, Baum C, Werder E, Zentgraf H, Müller J G, Rethwilm A. Foamy virus particle formation. J Virol. 1998;72:1610–1615. doi: 10.1128/jvi.72.2.1610-1615.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goepfert P A, Shaw K L, Ritter G D, Mulligan M J. A sorting motif localizes the foamy virus glycoprotein to the endoplasmatic reticulum. J Virol. 1997;71:778–784. doi: 10.1128/jvi.71.1.778-784.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hahn H, Baunach G, Bräutigam S, Mergia A, Neumann-Haefelin D, Daniel M D, McClure M O, Rethwilm A. Reactivity of primate sera to foamy virus Gag and Bet proteins. J Gen Virol. 1994;75:2635–2644. doi: 10.1099/0022-1317-75-10-2635. [DOI] [PubMed] [Google Scholar]
- 21.Heinkelein M, Schmidt M, Fischer N, Moebes A, Lindemann D, Enssle J, Rethwilm A. Characterization of a cis-acting sequence in the pol region required to transfer human foamy virus vectors. J Virol. 1998;72:6307–6314. doi: 10.1128/jvi.72.8.6307-6314.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heneine W, Switzer W M, Sandstrom P, Brown J, Vedapuri S, Schable C A, Khan A S, Lerche N W, Schweizer M, Neumann-Haefelin D, Chapman L E, Folks T M. Identification of a human population infected with simian foamy viruses. Nat Med. 1998;4:403–407. doi: 10.1038/nm0498-403. [DOI] [PubMed] [Google Scholar]
- 23.Herchenröder O, Renne R, Loncar D, Cobb E K, Murthy K K, Schneider J, Mergia A, Luciw P A. Isolation, cloning, and sequencing of simian foamy viruses from chimpanzees (SFVcpz): high homology to human foamy virus (HFV) Virology. 1994;201:187–199. doi: 10.1006/viro.1994.1285. [DOI] [PubMed] [Google Scholar]
- 24.Higuchi R. Recombinant PCR. In: Innis M A, Gelfand D H, White T J, editors. PCR protocols: a guide to methods and applications. San Diego, Calif: Academic Press; 1990. pp. 177–183. [Google Scholar]
- 25.Hildinger M, Abel K L, Ostergat W, Baum C. Design of 5′ untranslated sequences in retroviral vectors developed for medical use. J Virol. 1999;73:4083–4089. doi: 10.1128/jvi.73.5.4083-4089.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hirata R K, Miller A D, Andrews R G, Russel D W. Transduction of hematopoietic cells by foamy virus vectors. Blood. 1996;88:3654–3661. [PubMed] [Google Scholar]
- 27.Jordan I, Enssle J, Güttler E, Mauer B, Rethwilm A. Expression of human foamy virus reverse transcriptase involves a spliced pol mRNA. Virology. 1996;224:314–319. doi: 10.1006/viro.1996.0534. [DOI] [PubMed] [Google Scholar]
- 28.Kögel D, Aboud M, Flügel R M. Molecular biological characterization of the human foamy virus reverse transcriptase and ribonuclease H domains. Virology. 1995;213:97–108. doi: 10.1006/viro.1995.1550. [DOI] [PubMed] [Google Scholar]
- 29.Lindemann D, Bock M, Schweizer M, Rethwilm A. Efficient pseudotyping of murine leukemia virus particles with chimeric human foamy virus envelope proteins. J Virol. 1997;71:4815–4820. doi: 10.1128/jvi.71.6.4815-4820.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lindemann D, Rethwilm A. Characterization of a human foamy virus 170-kDa Env-Bet fusion protein generated by alternative splicing. J Virol. 1998;72:4088–4094. doi: 10.1128/jvi.72.5.4088-4094.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Linial M L. Foamy viruses are unconventional retroviruses. J Virol. 1999;73:1747–1755. doi: 10.1128/jvi.73.3.1747-1755.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Linial M L, Miller A D. Retroviral RNA packaging: sequence requirements and implications. Curr Top Microbiol Immunol. 1990;157:125–152. doi: 10.1007/978-3-642-75218-6_5. [DOI] [PubMed] [Google Scholar]
- 33.Maurer B, Bannert H, Darai G, Flügel R M. Analysis of the primary structure of the long terminal repeat and the gag and pol genes of the human spumaretrovirus. J Virol. 1988;62:1590–1597. doi: 10.1128/jvi.62.5.1590-1597.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller A D. Development and applications of retroviral vectors. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 437–473. [PubMed] [Google Scholar]
- 35.Moebes A, Enssle J, Bieniasz P D, Heinkelein M, Lindemann D, Bock M, McClure M O, Rethwilm A. Human foamy virus reverse transcription that occurs late in the viral replication cycle. J Virol. 1997;71:7305–7311. doi: 10.1128/jvi.71.10.7305-7311.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muranyi W, Flügel R M. Analysis of splicing patterns of human spumaretrovirus by polymerase chain reaction reveals complex RNA structures. J Virol. 1991;65:727–735. doi: 10.1128/jvi.65.2.727-735.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nassal M, Schaller H. Hepatitis B virus replication. Trends Microbiol. 1993;1:221–228. doi: 10.1016/0966-842x(93)90136-f. [DOI] [PubMed] [Google Scholar]
- 38.Nestler U, Heinkelein M, Lücke M, Meixensberger J, Scheurlen W, Kretschmer A, Rethwilm A. Foamy virus vectors for suicide gene therapy. Gene Ther. 1997;4:1270–1277. doi: 10.1038/sj.gt.3300561. [DOI] [PubMed] [Google Scholar]
- 39.Ogert R A, Lee L H, Beemon K L. Avian retroviral RNA element promotes unspliced RNA accumulation in the cytoplasm. J Virol. 1996;70:3834–3843. doi: 10.1128/jvi.70.6.3834-3843.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Padgett R A, Grabowski P J, Konarska M M, Seiler S, Sharp P A. Splicing of messenger RNA precursors. Annu Rev Biochem. 1986;55:1119–1150. doi: 10.1146/annurev.bi.55.070186.005351. [DOI] [PubMed] [Google Scholar]
- 41.Pietschmann T, Heinkelein M, Heldmann M A, Zentgraf H, Rethwilm A, Lindemann D. Foamy virus capsids require the cognate envelope protein for particle export. J Virol. 1999;73:2613–2621. doi: 10.1128/jvi.73.4.2613-2621.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rethwilm A. Regulation of foamy virus gene expression. Curr Top Microbiol Immunol. 1995;193:1–23. doi: 10.1007/978-3-642-78929-8_1. [DOI] [PubMed] [Google Scholar]
- 43.Rethwilm A, Erlwein O, Baunach G, Maurer B, Meulen V T. The transcriptional transactivator of human foamy virus maps to the bel-1 genomic region. Proc Natl Acad Sci USA. 1991;88:941–945. doi: 10.1073/pnas.88.3.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rösener M, Hahn H, Kranz M, Heeney J, Rethwilm A. Absence of serological evidence for foamy virus infection in patients with amyotrophic lateral sclerosis. J Med Virol. 1996;48:222–226. doi: 10.1002/(SICI)1096-9071(199603)48:3<222::AID-JMV2>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 45.Russel D W, Miller A D. Foamy virus vectors. J Virol. 1996;70:217–222. doi: 10.1128/jvi.70.1.217-222.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schenk T, Enssle J, Fischer N, Rethwilm A. Replication of a foamy virus mutant with a constitutively active U3 promoter and deleted accessory genes. J Gen Virol. 1999;80:1591–1598. doi: 10.1099/0022-1317-80-7-1591. [DOI] [PubMed] [Google Scholar]
- 47.Schmidt M, Rethwilm A. Replicating foamy virus-based vectors directing high level expression of foreign genes. Virology. 1995;210:167–178. doi: 10.1006/viro.1995.1328. [DOI] [PubMed] [Google Scholar]
- 48.Schweizer M, Falcone V, Gänge J, Turek R, Neumann-Haefelin D. Siman foamy virus isolated from an accidentally infected human individual. J Virol. 1997;71:4821–4824. doi: 10.1128/jvi.71.6.4821-4824.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schweizer M, Turek R, Hahn H, Schliephake A, Netzer K-O, Eder G, Reinhardt M, Rethwilm A, Neumann-Haefelin D. Markers of foamy virus (FV) infections in monkeys, apes, and accidentally infected humans: appropriate testing fails to confirm suspected FV prevalence in man. AIDS Res Hum Retrovir. 1995;11:161–170. doi: 10.1089/aid.1995.11.161. [DOI] [PubMed] [Google Scholar]
- 50.Sharp P A. Split genes and RNA splicing. Cell. 1994;77:805–815. doi: 10.1016/0092-8674(94)90130-9. [DOI] [PubMed] [Google Scholar]
- 51.Swanstrom R, Wills J W. Synthesis, assembly, and processing of viral proteins. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 263–334. [PubMed] [Google Scholar]
- 52.Trubetskoy A M, Okenquist S A, Lenz J. R region sequences in the long terminal repeat of a murine retrovirus specifically increase expression of unspliced RNAs. J Virol. 1999;73:3477–3483. doi: 10.1128/jvi.73.4.3477-3483.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vogt V M. Proteolytic processing and particle maturation. Curr Top Microbiol Immunol. 1996;214:95–131. doi: 10.1007/978-3-642-80145-7_4. [DOI] [PubMed] [Google Scholar]
- 54.Wieringa B, Hofer E, Weissmann C. A minimal intron length but no specific internal sequence is required for splicing the large rabbit β-globin intron. Cell. 1984;37:915–925. doi: 10.1016/0092-8674(84)90426-4. [DOI] [PubMed] [Google Scholar]
- 55.Wu M, Chari S, Yanchis T, Mergia A. cis-Acting sequences required for simian foamy virus type 1 (SFV-1) vectors. J Virol. 1998;72:3451–3454. doi: 10.1128/jvi.72.4.3451-3454.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu M, Mergia A. Packaging cell lines for simian foamy virus type 1 vectors. J Virol. 1999;73:4498–4501. doi: 10.1128/jvi.73.5.4498-4501.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yu S F, Baldwin D N, Gwynn S R, Yendapalli S, Linial M L. Human foamy virus replication: a pathway distinct from that of retroviruses and hepadnaviruses. Science. 1996;271:1579–1582. doi: 10.1126/science.271.5255.1579. [DOI] [PubMed] [Google Scholar]
- 58.Yu S F, Sullivan M D, Linial M L. Evidence that the human foamy virus genome is DNA. J Virol. 1999;73:1565–1572. doi: 10.1128/jvi.73.2.1565-1572.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]