Abstract
The complete nucleotide sequence of hibiscus chlorotic ringspot virus (HCRSV) was determined. The genomic RNA (gRNA) is 3,911 nucleotides long and has the potential to encode seven viral proteins in the order of 28 (p28), 23 (p23), 81 (p81), 8 (p8), 9 (p9), 38 (p38), and 25 (p25) kDa. Excluding two unique open reading frames (ORFs) encoding p23 and p25, the ORFs encode proteins with high amino acid similarity to those of carmoviruses. In addition to gRNA, two 3′-coterminated subgenomic RNA (sgRNA) species were identified. Full-length cDNA clones derived from gRNA and sgRNA were constructed under the control of a T7 promoter. Both capped and uncapped transcripts derived from the full-length genomic cDNA clone were infectious. In vitro translation and mutagenesis assays confirmed that all the predicted ORFs except the ORF encoding p8 are translatable, and the two novel ORFs (those encoding p23 and p25) may be functionally indispensable for the viral infection cycle. Based on virion morphology and genome organization, we propose that HCRSV be classified as a new member of the genus Carmovirus in family Tombusviridae.
Hibiscus chlorotic ringspot virus (HCRSV) is an isometric monopartite plant virus which measures 28 nm in diameter. It was first identified in a hibiscus cultivar imported to the United States from El Salvador. The virus is found worldwide where hibiscus is cultivated (15, 36, 37). The symptoms on HCRSV-infected plants range from a generalized mottle to chlorotic ring spots and vein-banding patterns (37). Many hibiscus hybrids grown in the tropics as ornamental plants showed severe stunting and flower distortion when infected by HCRSV (41).
Serological evidence has demonstrated that HCRSV belongs to the genus Carmovirus (13). So far, the complete nucleotide sequences for seven carmoviruses have been determined. These include the type member carnation mottle virus (CarMV) (9), turnip crinkle virus (TCV) (3), melon necrotic spot virus (MNSV) (27), cardamine chlorotic fleck virus (CCFV) (33), cowpea mottle virus (CPMoV) (42), saguaro cactus virus (SCV) (38), and galinsoga mosaic virus (GaMV) (6).
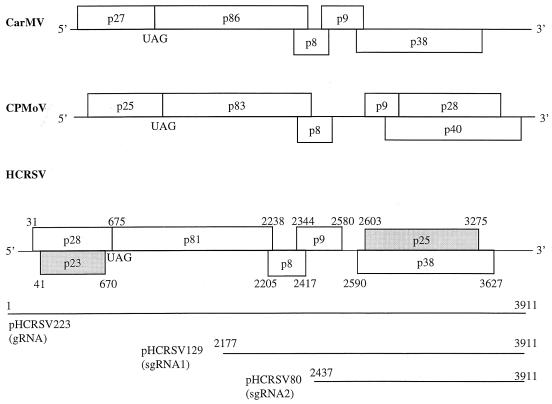
All carmoviruses have a genome organization similar to that of CarMV (Fig. 1). These viruses possess two open reading frames (ORFs), one of which results from an in-frame readthrough mechanism, with the potential to encode two polypeptides starting from the first AUG (5, 8, 22). Both proteins are putative subunits of the viral replicase. Two centrally located small ORFs encode proteins p8 and p9, which are required for virus movement (10, 17). The coat protein gene is located in the 3′ region of the genome. CPMoV also has a unique ORF that encodes a 28-kDa protein by in-frame readthrough of the ORF encoding p9 [ORF(p9)] (42). The function of p28 of CPMoV is still unknown.
FIG. 1.
Comparison of genome organization between HCRSV (this study), CarMV (9), and CPMoV (42). Open rectangles, predicted ORFs in each viral genome; shaded rectangles, novel ORFs. The potential readthrough and in-frame UAG amber termination codons for the first ORF of each virus and the locations of various HCRSV ORFs, the full-length gRNA, and sgRNA1 and 2 are shown. Numbers represent nucleotide positions.
Here we report the complete nucleotide sequence, genome organization, construction of a cDNA clone from which infectious transcripts can be derived, and expression of HCRSV in vitro. In addition to the five ORFs found in other carmoviruses, the HCRSV genomic RNA (gRNA) contains two novel ORFs, ORF(p23) and ORF(p25), located near the 5′- and 3′-terminal regions of the genome, respectively. Two 3′-coterminal subgenomic RNA (sgRNA) species were identified. In vitro expression of cDNA clones corresponding to viral gRNA and sgRNA showed that all the predicted ORFs except ORF(p8) were translated from transcripts derived from the cDNA clones. Mutagenesis studies confirmed that the in vitro-translated products were encoded by their corresponding ORFs, suggesting that these ORFs may encode genuine viral products. Furthermore, evidence showed that the two novel ORFs, ORF(p23) and ORF(p25), may encode products functionally important for viral replication and movement.
MATERIALS AND METHODS
Virus source, cDNA cloning, and nucleotide sequencing.
A single local lesion of HCRSV, serially transferred, was isolated in Singapore (41) and propagated in kenaf (Hibiscus cannabinus L.). Virions and viral RNA were purified as described by Hurtt (13) and Carrington and Morris (4), respectively. Viral RNA was analyzed by formaldehyde gel electrophoresis (16). Five micrograms of viral RNA was denatured and polyadenylated using poly(A) polymerase and ATP, according to the supplier's instructions (Life Technologies, Grand Island, N.Y.). First-strand cDNA was synthesized with an oligo(dT) primer and avian myeloblastosis virus reverse transcriptase, followed by second-strand synthesis using the Riboclone cDNA synthesis kit (Promega, Madison, Wis.). A library of cDNA clones was generated by ligation of the double-stranded cDNAs with EcoRI adapters and inserted into the EcoRI site of pBluescript SK+ and/or KS+ (Stratagene, San Diego, Calif.). Nucleotide sequences were determined from both strands of two independent cDNA clones using T7 or T3 universal primers or HCRSV sequence-specific primers by the dideoxynucleotide chain termination reaction (31) and automated sequencing using the ABI PRISM dye terminator cycle sequencing ready reaction kit (Perkin-Elmer, Norwalk, Conn.).
Construction of a biologically active cDNA clone.
The full-length cDNA clone was constructed from a nearly full-length cDNA clone, pHCRSV220, which is 11 nucleotides short at the 5′ end. A primer consisting of the T7 promoter sequence, the 11 missing nucleotides, and 22 nucleotides from the 5′ end of pHCRSV220 was synthesized. The sequence of the 11 missing nucleotides was determined using the 5′-AmpliFINDER rapid amplification of cDNA ends and PCR-Direct cloning kits (Clontech, Palo Alto, Calif.). cDNA synthesis was carried out using 3 μg of HCRSV RNA as the template and 10 pmol of primer P1 (5′-CAAGCTTCAGGTTCCTCATCAGTGGG-3′) annealing approximately 200 nucleotides from the 5′ end of pHCRSV220. RNA was hydrolyzed, and 10 pmol of AmpliFINDER anchor primer was ligated to the 5′ end of the first-strand cDNA by T4 RNA ligase. PCR amplification was carried out using the anchor primer and primer P2 (5′CTCGCTCGCCCATGGAGCGATACGGGGG-3′) priming approximately 130 nucleotides from the 5′ end. With a primer containing the 3′-terminal sequence of HCRSV plus a SmaI sequence, PCR was carried out to obtain the full-length HCRSV genome with pHCRSV220 as the template. The Taq polymerase-amplified PCR fragment was cloned into an SrfI site of PCRscript (Stratagene, San Diego, Calif.). Similarly, cDNA clones of sgRNA1 and sgRNA2 were constructed by the same method, and their sequences were verified by sequencing.
Verification of start, readthrough, and stop codons by sequencing.
The sequences around the start, readthrough, and stop codon regions of the predicted ORFs were verified by directly sequencing reverse transcription-PCR (RT-PCR) fragments (Access RT-PCR system; Promega). For each region, three RT-PCR fragments were synthesized from three independent RNA samples extracted from virions purified from three separate kenaf plants infected with HCRSV. Automated DNA sequencing was carried out using the same primer used for RT-PCR.
Primer extension and generation of subgenomic cDNA (sgcDNA) clones.
To identify the 5′ ends for the three HCRSV RNA species by primer extension analyses, 28-mer, 26-mer, and 18-mer oligonucleotides complementary to RNA (nucleotides 129 to 144, 2270 to 2296, and 2607 to 2625, respectively) were synthesized. Primer extension analyses as described by Boorstein and Craig (2) were performed using the primer extension system (Promega). Total viral RNA was extracted from purified virions and separated on a 1% low-melting-point agarose gel. The separated RNA bands corresponding to gRNA, sgRNA1, or sgRNA2 were excised and purified by phenol-chloroform extraction and ethanol precipitation. Viral RNAs (10 ng of each) were reverse transcribed to cDNAs by avian myeloblastosis virus reverse transcriptase at 58°C for 20 min using 100 fmol of 5′-end-labeled [γ-32P]ATP primers. Sizes of the primer extension products were determined by alignment with a pSK(+) sequence ladder encompassing the beta-galactosidase gene.
Northern blot analysis.
For Northern analysis, 3 μg of viral RNA was denatured, separated on a 1.2% formaldehyde–MOPS (morpholinepropanesulfonic acid) agarose gel and transferred to the Hybond-Nylon membrane (Amersham Pharmacia Biotech, Piscataway, N.J.) according to the method of Sambrook et al. (30). Three cDNA probes encompassing nucleotides 1 to 1027 (5′ end of HCRSV), nucleotides 2050 to 3212 (middle portion of genome), and nucleotides 2598 to 3911 (3′ end of HCRSV) were labeled with [32P]dCTP using the Prime-a-Gene system (Promega). The three 32P-labeled probes were hybridized separately at 65°C for 16 h (32). The RNA blots were washed and autoradiographed.
In vitro transcription and translation.
Viral gRNA and sgRNAs were prepared by the gel purification method as described above. Capped and uncapped RNA transcripts were synthesized in vitro using the RiboMAX T7 kit (Promega) according to the manufacturer's instructions. Purified viral RNAs at approximately 100 μg/ml were translated in wheat germ extracts or in the rabbit reticulocyte lysate in vitro translation system (Promega). In addition, cDNA clones at approximately 100 μg/ml were transcribed and translated in vitro using the TnT system (Promega). Two microliters each of [35S]methionine-labeled in vitro translation products were resolved in either sodium dodecyl sulfate (SDS)–8.5, –12.5, –17.5, or –20% PAGE gel and visualized by autoradiography or fluorography.
Site-directed mutagenesis.
All mutants were generated using the Quikchange site-directed mutagenesis kit (Stratagene) by following the manufacturer's instructions. The mutants were verified by sequencing.
Nucleotide sequence accession number.
The complete nucleotide sequence of HCRSV was deposited in GenBank under accession no. X86448.
RESULTS
Genome organization of HCRSV.
The complete nucleotide sequence of HCRSV and its deduced amino acid sequence were obtained. The sequences around the start and stop codons of each of the predicted ORFs were verified by RT-PCR and sequencing. The HCRSV genome consists of 3,911 nucleotides and contains seven ORFs (Fig. 1). ORF(p28) encodes a protein with the calculated molecular mass of 24 kDa. However, as the product encoded by this ORF showed an apparent molecular mass of 28 kDa by SDS-PAGE (see Fig. 3), this ORF is referred to as ORF(p28) in this report. Within ORF(p28) is ORF(p23). By in-frame readthrough of the stop codon of ORF(p28), the reading frame extends towards the 3′ terminus of the genome, giving rise to p81. Similar to what is found for all other carmoviruses, two centrally located and overlapping small ORFs, ORF(p8) and ORF(p9), were present. The 3′-proximal ORF(p38) encodes a protein with a molecular mass of 38 kDa, and an additional ORF, ORF(p25), is found within ORF(p38). Although the overall genome organization of HCRSV is similar to those of other members of the genus Carmovirus, including CarMV, TCV, MNSV, CCFV, CPMoV, SCV, and GaMV, the genome of HCRSV presents two additional ORFs, ORF(p23) and ORF(p25), that are unique to this virus (Fig. 1).
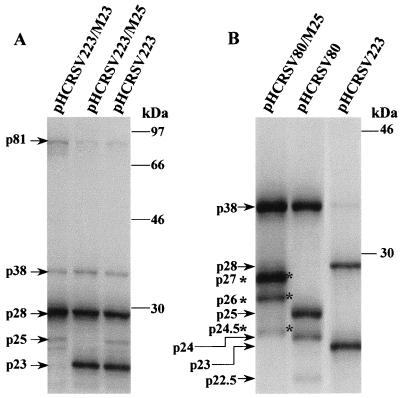
FIG. 3.
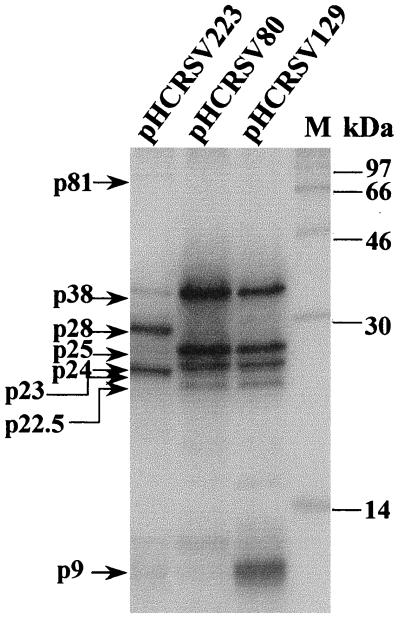
Analysis of the in vitro translation products of RNAs cotranscribed from pHCRSV223 (gRNA), pHCRSV129 (sgRNA1), and pHCRSV80 (sgRNA2) in wheat germ extracts with the TnT coupled translation system. [35S]methionine-labeled translation products were separated on SDS–17.5% polyacrylamide gel and visualized by autoradiography. Lane M, molecular mass markers in kilodaltons.
Determination of the gRNA and sgRNAs.
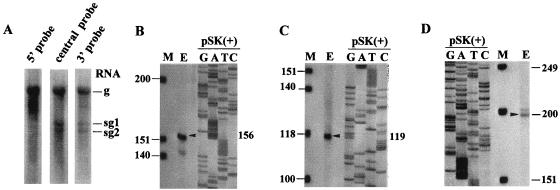
Purified virions were obtained from HCRSV-infected kenaf (H. cannabinus L.) leaves. Three distinct RNA species of approximately 3.9, 1.7, and 1.5 kb, corresponding to the gRNA and two putative sgRNAs, were observed in Northern blot analysis (Fig. 2A). Probes corresponding to the 5′-end, central, and 3′-end regions of the genome all hybridized to gRNA. Only the central and 3′-end probes hybridized to the two sgRNAs. These results suggest that HCRSV replication results in two 3′-coterminal sgRNA species in addition to the gRNA.
FIG. 2.
(A) Northern blot analysis of single-stranded HCRSV RNAs isolated from virion particles obtained from infected kenaf leaves. The lanes represent hybridization of probes to the 5′, central, and 3′ regions of the viral RNA. Also shown is a determination of the 5′-terminal ends of HCRSV gRNA (B), sgRNA1 (C), and sgRNA2 (D) by primer extension assay. The size of the primer extension product was analyzed on 8% polyacrylamide denaturing gel. Lane M, molecular weight marker provided by the primer extension kit. Positions of primer extension products (lane E) are indicated by arrowheads, and accurate sizes were determined through alignment with a pSK(+) sequence ladder encompassing the beta-galactosidase gene.
The 5′-terminal sequences of the three HCRSV RNA species were determined by sequencing the cDNA clones obtained using the 5′-AmpliFINDER rapid amplification of cDNA ends kit and primer extension analyses. The lengths of the primer extension products with sizes of 156, 119, and 200 nucleotides corresponded to the numbers of bases between the labeled nucleotides of the primers and the 5′ ends of the gRNA (Fig. 2B), sgRNA1 (Fig. 2C), and sgRNA2 (Fig. 2D).
Construction of a biologically active full-length cDNA clone.
A full-length cDNA clone of HCRSV (pHCRSV223) from which infectious transcripts could be derived and sgcDNA clones (pHCRSV129 and pHCRSV80) were constructed under the control of a T7 promoter. In vitro RNA transcripts derived from the full-length cDNA clone pHCRSV223 alone were infectious on inoculated kenaf. Symptoms that developed were identical to those induced by wild-type virus. Systemic chlorotic ring spots, vein-banding patterns, and stunted growth were observed 10 to 15 days postinoculation (Table 1).
TABLE 1.
Infectivity of transcripts derived from wild-type and mutant full-length cDNA constructs of HCRSV on inoculated kenaf plants
| cDNA construct | No. of inoculated plants showing viral symptoms/ 8 plants tested | Infectivitya (days) | Symptom severityb | RT-PCR resultc for:
|
|
|---|---|---|---|---|---|
| Inoculated leaves | Systemic leaves | ||||
| pHCRSV223 (capped) | 8 | 10–15 | +++ | + | + |
| pHCRSV223 (uncapped) | 8 | 10–15 | +++ | + | + |
| pHCRSV223/M28 | 0 | — | − | − | − |
| pHCRSV223/M23 | 0 | — | − | − | − |
| pHCRSV223/M81 | 8 | 14–17 | +++ | + | + |
| pHCRSV223/M25 | 8 | 30–35 | + | + | + |
Days required for symptom appearance after inoculation. —, uninfected.
Severity of systemic symptoms. +++, very severe, extensive spread of chlorotic ring spots (equivalent to wild type); +, sparsely scattered chlorotic ring spots; −, no symptoms.
All viable mutants and wild-type viruses were verified through RT-PCR using appropriate sequence-specific primers. +, presence of virus; −, absence of virus.
The infectivity of several members of Tombusviridae, e.g., TCV, tomato bushy stunt virus, and cucumber necrosis virus (12, 29, 39), does not require the cap structure at the 5′ end of the gRNA. However, uncapped brome mosaic virus and tobacco mosaic virus (TMV) were found to be largely noninfectious (1, 7, 19). Uncapped HCRSV transcripts were found to be as infectious as the capped transcripts, and the time taken for symptom development was the same as that required in plants inoculated with the capped transcripts (Table 1).
In vitro translation of HCRSV RNAs.
To investigate whether the predicted ORFs of HCRSV were translatable, plasmids pHCRSV223, pHCRSV129, and pHCRSV80 containing cDNA sequences corresponding to gRNA, sgRNA1, and sgRNA2, respectively (Fig. 1), were translated in vitro using the wheat germ transcription-coupled translation system (TnT system). Five proteins with apparent molecular masses of 81, 38, 28, 25, and 23 kDa were synthesized from pHCRSV223 in wheat germ extracts (Fig. 3). The putative p38 and p25 were also produced from pHCRSV129 and pHCRSV80 (Fig. 3). In addition, a protein with a molecular mass of approximately 9 kDa was observed from the translation of pHCRSV129 (Fig. 3), representing the product of ORF(p8) and/or ORF(p9). This product was later assigned to ORF(p9) by mutagenesis studies (see Fig. 4C). Unexpectedly, two other protein species with apparent molecular masses of 24 and 22.5 kDa were also observed from the translation of pHCRSV80 and pHCRSV129, respectively (Fig. 3). The possibility that these proteins were processed forms of p25 was excluded by a pulse-chase assay. Both p22.5 and p24 were detected after a 5-min chase and remained stable over the time course of 95 min (data not shown). Upon analysis of the sequence, we found that two AUG codons are located at nucleotide positions 2630 to 2632 and 2666 to 2668, respectively, within ORF(p25). These two AUG codons may be used as translation initiation signals for the 24- and 22.5-kDa proteins by a leaky translation mechanism. Translation of viral gRNA and sgRNA in wheat germ extracts yielded patterns identical to those of the corresponding cDNA clones (data not shown).
FIG. 4.
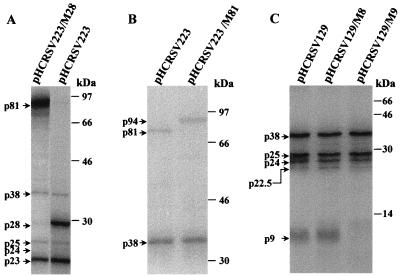
Analysis of the in vitro translation products of RNAs cotranscribed from mutants pHCRSV223/M28 (A), pHCRSV223/M81 (B), and pHCRSV129/M8 and pHCRSV223/M9 (C) and controls pHCRSV223 and pHCRSV129 in wheat germ extracts with the TnT coupled translation system. [35S]methionine-labeled translation products were separated on SDS–12.5% (A), –8.5% (B), and –17.5% (C) polyacrylamide gels and visualized by autoradiography. The molecular mass markers in kilodaltons are indicated at the right of each panel.
Verification of ORF(p28), ORF(p81), ORF(p8), and ORF(p9) by mutagenesis analysis.
Plant RNA viruses commonly exploit leaky translation termination signals in order to express internal protein-coding regions (43). Among carmoviruses, readthrough of the amber stop codon to produce a putative RdRp is a characteristic feature of their genome organization (22). Site-directed mutagenesis of TMV showed that the downstream cistron from the leaky amber codon is essential for regulating the expression of the putative viral RdRp (14).
To investigate if the same readthrough strategy is used by HCRSV in the synthesis of the 81-kDa protein, mutant construct pHCRSV223/M28 was made by mutating the amber stop codon, 673TAG, for ORF(p28) to 673GCT. Translation of transcripts derived from pHCRSV223/M28 showed that no p28 was detected, while p23, p25, and p38 were synthesized at levels similar to those translated from transcripts derived from pHCRSV223 (Fig. 4A). As expected, increased expression of the 81-kDa protein was observed from expression of pHCRSV223/M28 (Fig. 4A).
ORF(p81) was then confirmed by expression of mutant pHCRSV223/M81, in which the stop codon, 2236TAG, for ORF(p81) was mutated to 2236TAC. Translation of pHCRSV223/M81 showed a protein band with a molecular mass of 94 kDa; the 81-kDa protein was not detected (Fig. 4B). This is consistent with the predicted molecular mass of 94 kDa, which can be translated by an in-frame readthrough of the ORF(p81) stop codon and terminated at the ORF(p9) stop codon. This 94-kDa polypeptide, therefore, may be similar to the 98-kDa protein of CarMV that was assumed to be produced by a double-readthrough strategy (9).
Previous studies on TCV have suggested that the two small ORFs coding for 8- and 9-kDa proteins were conserved among all carmoviruses. Both proteins are required for cell-to-cell movement (10, 40) and can complement in trans (17). Only one small protein with a molecular mass of approximately 9 kDa was detected from the translation of sgcDNA1 (pHCRSV129) (Fig. 3). To verify whether only one or both ORFs [ORF(p8) and ORF(p9)] were translated in vitro, mutants pHCRSV129/M8 and pHCRSV129/M9 were used. They were generated by changing the start codon for ORF(p8) from 2205ATG to 2205GTG and the start codon for ORF(p9) from 2344ATG to 2344ACG and were translated in the TnT system. The small protein band observed from the expression of pHCRSV129 was not detected from the translation of pHCRSV129/M9, although p38, p25, p24, and p22.5 were translated at the same efficiency as those from transcripts derived from pHCRSV129 (Fig. 4C). Translation of pHCRSV129/M8 showed an expression pattern identical to that of pHCRSV129 (Fig. 4C). These results confirm that only ORF(p9) is translated to a detectable level. ORF(p8) may not be translated or may be translated at such a low level that it could not be detected. The amounts of CarMV p7 (18) and TCV p9 (17) have been shown to be very low in infected cells.
Detection of two novel ORFs in the HCRSV genome.
Both ORF(p23) and ORF(p25) were uniquely identified in the HCRSV genome. The authenticity of the start codon for ORF(p23) was confirmed by using the mutant pHCRSV223/M23. In this mutant, 41ATG was mutated to 41ACG. Translation of transcripts derived from pHCRSV223/M23 failed to produce p23, although other proteins were produced at levels similar to those translated from transcripts derived from pHCRSV223 (Fig. 5A).
FIG. 5.
Analysis of the in vitro translation products of RNAs cotranscribed from pHCRSV223, pHCRSV223/M23, and pHCRSV223/M25 (A) and from pHCRSV/M25 and pHCRSV80 (B) in wheat germ extracts with the TnT coupled translation system. [35S]methionine-labeled translation products were separated on SDS–12.5% polyacrylamide gel and were visualized by autoradiography. The molecular mass markers in kilodaltons are indicated at the right of each panel.
ORF(p25) is nested in ORF(p38), and its position in the genome is similar to that of ORF(p28) coding for p28 in the genome of CPMoV (42), a member of the genus Carmovirus. To authenticate the expression of ORF(p25), mutant pHCRSV223/M25, in which the stop codon 3275TGA for ORF(p25) was changed to 3275TGC, was generated. The 25-kDa protein could not be expressed from pHCRSV223/M25, but other products produced by pHCRSV223 were expressed (Fig. 5A). It was predicted based on the sequence that removal of the stop codon 3275TGA would generate a 27-kDa protein terminating at the next in-frame stop codon, 3352TAG. As the 27-kDa protein likely migrates to a position similar to that of the product of ORF(p28), it would be difficult to separate them. To clarify this ambiguity, mutant pHCRSV80/M25 was generated from pHCRSV80. Expression of this mutant produced the 27-kDa protein instead of the 25-kDa protein (Fig. 5B). In addition, two proteins with molecular masses of approximately 26 and 24.5 kDa instead of 24 and 22.5 kDa were observed (Fig. 5B). The translation efficiencies of these downstream proteins (p26 and p24.5) were also low (Fig. 5B). This is consistent with the previous observation that both p24 and p22.5 were inefficiently expressed from pHCRSV80 (Fig. 3), possibly by a leaky expression mechanism.
ORF(p38) coding for the viral coat protein.
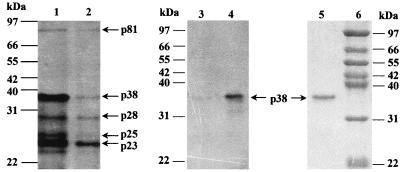
Comparison of p38 with the coat proteins of other carmoviruses showed 34 to 38% amino acid identity, indicating that ORF(p38) may encode the coat protein of HCRSV. To test this possibility, total viral RNA was extracted from purified virions and translated in vitro in wheat germ extracts. The in vitro-translated products were subjected to immunoprecipitation with anti-HCRSV antiserum. The 38-kDa protein was specifically immunoprecipitated by anti-HCRSV antiserum and was also detected exclusively from the purified virus particles (Fig. 6). These results confirm that p38 is the coat protein. This protein could be produced from both gcDNA and sgcDNAs (Fig. 3). However, its translation efficiency from the transcript of gcDNA was at least 10 times lower than that from sgcDNAs (Fig. 3). The coat protein gene can also be expressed from the gRNA of tobacco necrosis virus (20) but not from gRNAs of CarMV, TCV, and SCV (3, 5, 38).
FIG. 6.
Analysis of the in vitro translation products from total viral RNA (lane 1) and gel-purified gRNA (lane 2) in wheat germ extracts separated on SDS–12% polyacrylamide gel. The radiolabeled in vitro translation products from total RNA were immunoprecipitated with either preimmune serum (lane 3) or anti-HCRSV (lane 4), separated on SDS–12.5% polyacrylamide gels, and visualized by autoradiography. The coat protein was also detected by electrophoresis of the purified virus particles on SDS–12.5% polyacrylamide gel and visualized by staining with Coomassie brilliant blue (lane 5). Numbers indicate molecular markers in kilodaltons (lane 6).
Analysis of the infectivities of four HCRSV mutants.
The in vitro-transcribed transcripts derived from mutants pHCRSV223/M28, pHCRSV223/M23, pHCRSV223/M81, and pHCRSV223/M25 were inoculated on kenaf plants. Each transcript was inoculated into eight 2-week-old plants. The plants were regularly observed up to 60 days postinoculation for the development of viral symptoms (Table 1). All of the plants inoculated with RNA from pHCRSV223/M81 showed severe chlorotic ring spots and vein banding at 14 to 17 days postinoculation. All of the plants inoculated with RNA from pHCRSV223/M25 showed sparse systemic chlorotic ring spots at 30 to 35 days postinoculation. RT-PCR results indicated viral replication of mutants in inoculated kenaf leaves (Table 1). Sequencing the RT-PCR products from the leaves inoculated with pHCRSV223/M25 or pHCRSV223/M81 confirmed that the original mutations were unchanged. The plants inoculated with transcripts derived either from pHCRSV223/M28 or from pHCRSV223/M23 remained uninfected during the whole observation period. The viral RNA could not be detected from leaves inoculated with pHCRSV223/M23 and pHCRSV223/M28 (Table 1). These results demonstrate that mutation of the start codon for ORF(p23) and the amber stop codon for ORF(p28) completely abolishes the infectivity of HCRSV. The HCRSV p28 is similar to the p28 of TCV in that it is essential for genome replication (39). The mutation of the stop codon for ORF(p25) attenuates the infectivity of HCRSV. Extension of the 81-kDa protein to a putative 94-kDa protein by mutation of the stop codon for ORF(p81), however, does not affect the infectivity of HCRSV (Table 1). It is suggested that ORF(p28)-, ORF(p23)-, and ORF(p25)-encoded products may play essential roles in the viral life cycle.
Analysis of the 3′-UTR.
The 3′-untranslated region (3′-UTR) of HCRSV contains 286 nucleotides. A small stable hairpin loop composed of nucleotides 3876 to 3911 (5′-AAUAC C C UCACAACGGC CAUAGUUGUGAGG CAG C C C - 3 ′ ) was predicted using the sequence analysis program DNAsis (Hitachi Engineering Ltd.). Analysis of the 3′-terminal sequences of the genomic RNAs of CCFV, CPMoV, MNSV, CarMV, SCV, and TCV showed that similar structures could be formed. Conservation of this structure among members of the genus Carmovirus suggests that this cis element may be of functional importance in the replication of gRNA and sgRNAs (34).
DISCUSSION
Although HCRSV shares the general characteristics of the members of the genus Carmovirus, nucleotide sequence and in vitro translation data show that HCRSV exhibits distinct novel features in the genome organization. Two novel ORFs, ORF(p23) and ORF(p25), with the potential to encode polypeptides of 23 (p23) and 25 kDa (p25), respectively, were identified in the HCRSV genome. Expression and mutagenesis studies demonstrate that both ORFs are translatable in vitro. The functions of these two proteins are yet to be understood.
Mutation of the start codon for ORF(p23) totally abolished the infectivity of the viral RNA (Table 1). p23 possesses a leucine zipper motif and four potential phosphorylation sites, as shown by sequence analysis with a computer-aided program (SOSUI program, version 1.0, Mitaku Group, Department of Biotechnology, Tokyo University of Agriculture and Technology, Tokyo, Japan [http://azusa.proteome.bio.tuat.ac.jp/sosui]). It is unclear if this protein functions as a novel transcription factor that regulates HCRSV RNA replication. Further characterization of p23 in virus-infected plants is required to determine its function(s).
ORF(p25) is also unique in the genus Carmovirus although it has certain similarities to ORF(p28) of CPMoV (42). For example, both ORFs are located in the same position in the viral genomes (Fig. 1); sequence analysis shows that p25 and p28 are 52 and 28% similar at the nucleotide and amino acid levels, respectively, and that the sequence similarities are scattered. Although the function of p25 is unknown, a computer-aided program (SOSUI, version 1.0 [http://azusa.proteome.bio.tuat.ac.jp.sosui]) predicts that it may be a membrane protein with a 22-amino-acid transmembrane domain. As mutation of the stop codon for ORF(p25) attenuated the virus (Table 1), it is believed that p25 may be a functionally important product of HCRSV.
We have shown that p81 is expressed by readthrough of the ORF(p28) amber termination codon. This strategy is also utilized by other members of Tombusviridae to express replicase-associated proteins (32, 39) and by luteovirus to express coat protein (21). A number of other viruses, including TMV (25), tobacco rattle virus (26), lucerne transient streak virus (23), and tomato bushy stunt virus (12), also employ this strategy to express specific proteins. In some cases, a double-readthrough mechanism is used for expression of viral products. For example, a putative 100-kDa RdRp of CarMV could be expressed in vitro and in virus-infected protoplasts by such a strategy (11). However, there is no direct evidence to show that the protein is expressed in virus-infected plants. The 94-kDa polypeptide was not observed from the in vitro translation of the full-length cDNA, pHCRSV223 (Fig. 3 and 4A and B), or viral RNA, although the protein was clearly detected when the stop codon for ORF(p81) was abolished by site-directed mutagenesis. Interestingly, plants inoculated with transcripts derived from mutant pHCRSV223/M81 showed symptoms similar to those inoculated with wild-type transcripts. This indicates that termination of the C terminus of p81 is not necessary for virus infectivity.
Characterization of the sequence requirement for efficient readthrough shows that the consensus sequence, AA(A/G)-UAG-G(G/U)(G/A), is well conserved among several characterized plant viral RNAs including those of CarMV (9), maize chlorotic mottle virus (24), tomato bushy stunt virus (12), cucumber necrosis virus (28), barley yellow dwarf virus (21), and beet western yellow virus (35). Sequence analysis showed that the region surrounding the amber stop codon for ORF(p28) in HCRSV is 673AAA-UAG-GGG, which is consistent with the consensus sequence, while the sequence surrounding the amber stop codon for ORF(p81) is 673UCA-UAG-AAC, which exhibits low similarity to the consensus sequence. This is consistent with the observation that only p81 was detected when the full-length cDNA pHCRSV223 was expressed in vitro.
The unique genome structure of HCRSV not only adds new information to the diversity and complexity of the genus Carmovirus but also challenges our understanding of viral replication, gene expression, and RNA recombination in the family of Tombusviridae. Based on virion morphology and nucleotide sequence similarity of the major ORFs with those of other carmoviruses, we propose that HCRSV be classified as a new member of the genus Carmovirus.
ACKNOWLEDGMENTS
We thank S. S. Hurtt, USDA, for providing the kenaf seeds and D. X. Liu and S. W. Ding from the Institute of Molecular Agrobiology, Singapore, for helpful discussion and critical reading of the manuscript.
This research was supported by the National University of Singapore, research grant RP3920302.
REFERENCES
- 1.Ahlquist P, Janda M. cDNA cloning and in vitro transcription of the complete brome mosaic virus genome. Mol Cell Biol. 1984;4:2867–2882. doi: 10.1128/mcb.4.12.2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boorstein W R, Craig E A. Primer extension analysis of RNA. Methods Enzymol. 1989;180:347. doi: 10.1016/0076-6879(89)80111-9. [DOI] [PubMed] [Google Scholar]
- 3.Carrington J C, Heaton L A, Zuidema D, Hillman B I, Morris T J. The genome structure of turnip crinkle virus. Virology. 1989;170:219–226. doi: 10.1016/0042-6822(89)90369-3. [DOI] [PubMed] [Google Scholar]
- 4.Carrington J C, Morris T J. Complementary DNA cloning and analysis of carnation mottle virus RNA. Virology. 1984;139:22–31. doi: 10.1016/0042-6822(84)90326-x. [DOI] [PubMed] [Google Scholar]
- 5.Carrington J C, Morris T J. Characterization of cell-free translation products of carnation mottle genomic and subgenomic RNAs. Virology. 1985;144:1–10. doi: 10.1016/0042-6822(85)90299-5. [DOI] [PubMed] [Google Scholar]
- 6.Ciuffreda P, Rubino L, Russo M. Molecular cloning and the complete nucleotide sequence of galinsoga mosaic virus genomic RNA. Arch Virol. 1998;143:173–180. doi: 10.1007/s007050050277. [DOI] [PubMed] [Google Scholar]
- 7.Dawson W O, Beck K L, Knorr D A, Grantham G L. cDNA cloning of the complete genome of tobacco mosaic virus and production of infectious transcripts. Proc Natl Acad Sci USA. 1986;83:1832–1836. doi: 10.1073/pnas.83.6.1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dougherty W G, Kaesberg P. Turnip crinkle virus RNA and its translation in rabbit reticulocyte and wheat embryo extracts. Virology. 1981;42:45–56. doi: 10.1016/0042-6822(81)90087-8. [DOI] [PubMed] [Google Scholar]
- 9.Guilley H, Carrington J C, Balazs E, Jonard G, Richards K, Morris T J. Nucleotide sequence and genome organization of carnation mottle virus RNA. Nucleic Acids Res. 1985;13:6663–6677. doi: 10.1093/nar/13.18.6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hacker D L, Petty D T, Wei N, Morris T J. Turnip crinkle virus genes required for RNA replication and virus movement. Virology. 1992;186:1–8. doi: 10.1016/0042-6822(92)90055-t. [DOI] [PubMed] [Google Scholar]
- 11.Harbison S A, Davies J W, Wilson T M A. Expression of high molecular weight polypeptides by carnation mottle virus RNA. J Gen Virol. 1985;66:2597–2604. [Google Scholar]
- 12.Hearne P Q, Knorr D A, Hillman B I, Morris T J. The complete genome structure and synthesis of infectious RNA clones of tomato bushy stunt virus. Virology. 1990;177:141–151. doi: 10.1016/0042-6822(90)90468-7. [DOI] [PubMed] [Google Scholar]
- 13.Hurtt S S. Detection and comparison of electrophorotypes of hibiscus chlorotic ringspot virus. Phytopathology. 1987;77:845–850. [Google Scholar]
- 14.Ishikawa M, Meshi T, Ohno T, Okada Y. Specific cessation of minus-strand RNA accumulation at an early stage of tobacco mosaic virus infection. J Virol. 1991;65:861–868. doi: 10.1128/jvi.65.2.861-868.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones D R, Behncken G M. Hibiscus chlorotic ringspot, a widespread virus disease in the ornamental Hibiscus rosa-sinensis. Aust J Plant Pathol. 1980;9:4. [Google Scholar]
- 16.Lehrach H, Diamond D, Wozney J M, Boedtker H. RNA molecular weight determinations by gel electrophoresis under denaturing conditions, a critical reexamination. Biochemistry. 1977;16:4743–4751. doi: 10.1021/bi00640a033. [DOI] [PubMed] [Google Scholar]
- 17.Li W Z, Qu F, Morris T J. Cell-to-cell movement of turnip crinkle virus is controlled by two small open reading frames that function in trans. Virology. 1998;244:405–416. doi: 10.1006/viro.1998.9125. [DOI] [PubMed] [Google Scholar]
- 18.Marcos J F, Vilar M, Perez-Paya E, Pallas V. In vitro detection, RNA-binding properties and the characterisation of the RNA-binding domain of the p7 putative movement protein from carnation mottle carmovirus (CarMV) Virology. 1999;255:354–365. doi: 10.1006/viro.1998.9596. [DOI] [PubMed] [Google Scholar]
- 19.Meshi T, Ishikawa M, Motoyoshi F, Semba K, Okada Y. In vitro transcription of infectious RNA from full-length cDNAs of tobacco mosaic virus. Proc Natl Acad Sci USA. 1986;74:5463–5467. doi: 10.1073/pnas.83.14.5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meulewaeter F, Seurink J, Van Emmelo J. Genome structure of tobacco necrosis virus strain A. Virology. 1990;177:699–709. doi: 10.1016/0042-6822(90)90536-z. [DOI] [PubMed] [Google Scholar]
- 21.Miller W A, Waterhouse P M, Gerlach W L. Sequence and organization of barley yellow dwarf virus genomic RNA. Nucleic Acids Res. 1988;16:6097–6111. doi: 10.1093/nar/16.13.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morris T J, Carrington J C. Carnation mottle virus and viruses with similar properties. In: Koenig R, editor. The Plant Viruses. Vol. 3. New York, U.S.A: Plenum Press; 1988. pp. 73–112. [Google Scholar]
- 23.Morris-Krsinich B A M, Forster R L S. Lucerne transient streak virus RNA and its translation in rabbit reticulocyte lysate and wheat germ extract. Virology. 1983;128:176–185. doi: 10.1016/0042-6822(83)90328-8. [DOI] [PubMed] [Google Scholar]
- 24.Nutter R C, Scheets K, Panganiban L C, Lommel S A. The complete nucleotide sequence of the maize chlorotic mottle virus genome. Nucleic Acids Res. 1989;17:3163–3177. doi: 10.1093/nar/17.8.3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pelham H R B. Leaky UAG termination codon in tobacco mosaic virus RNA. Nature (London) 1978;272:469–471. doi: 10.1038/272469a0. [DOI] [PubMed] [Google Scholar]
- 26.Pelham H R B. Translation of tobacco rattle virus RNAs in vitro: four proteins from three RNAs. Virology. 1979;97:256–265. doi: 10.1016/0042-6822(79)90337-4. [DOI] [PubMed] [Google Scholar]
- 27.Riviere C J, Rochon D M. Nucleotide sequence and genomic organization of melon necrotic spot virus. J Gen Virol. 1990;71:1887–1896. doi: 10.1099/0022-1317-71-9-1887. [DOI] [PubMed] [Google Scholar]
- 28.Rochon D M, Tremaine J H. Complete nucleotide sequence of the cucumber necrosis virus genome. Virology. 1989;169:251–259. doi: 10.1016/0042-6822(89)90150-5. [DOI] [PubMed] [Google Scholar]
- 29.Rochon D M, Johnston J C. Infectious transcripts from the cloned cucumber necrosis virus cDNA: evidence for a bifunctional subgenomic mRNA. Virology. 1991;181:656–665. doi: 10.1016/0042-6822(91)90899-m. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. pp. 7.43–7.52. [Google Scholar]
- 31.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scholthof K B, Scholthof H B, Jackson A O. The tomato bushy stunt virus replicase proteins are coordinately expressed and membrane associated. Virology. 1995;208:365–369. doi: 10.1006/viro.1995.1162. [DOI] [PubMed] [Google Scholar]
- 33.Skotnicki M L, Mackenzie A M, Torronen M, Gibbs A J. The genomic sequence of cardamine chlorotic fleck carmovirus. J Gen Virol. 1993;74:1933–1937. doi: 10.1099/0022-1317-74-9-1933. [DOI] [PubMed] [Google Scholar]
- 34.Song C, Simon A E. Requirement of a 3′-terminal stem-loop in in vitro transcription by an RNA-dependent RNA polymerase. J Mol Biol. 1995;254:6–14. doi: 10.1006/jmbi.1995.0594. [DOI] [PubMed] [Google Scholar]
- 35.Veidt I, Lot H, Leiser M, Scheidecher D, Guilley H, Richards K, Jonard G. Nucleotide sequence of beet western yellow virus RNA. Nucleic Acids Res. 1988;16:9917–9932. doi: 10.1093/nar/16.21.9917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Waterworth H E. Hibiscus chlorotic ringspot virus. CMI/AAB descriptions of plant viruses no. 227. Warwick, United Kingdom: Association of Applied Biologists; 1980. [Google Scholar]
- 37.Waterworth H E, Lawson R H, Monroe R L. Purification and properties of hibiscus chlorotic ringspot virus. Phytopathology. 1976;64:570–575. [Google Scholar]
- 38.Weng Z, Xiong Z. Genome organization and gene expression of saguaro cactus carmovirus. J Gen Virol. 1997;78:525–534. doi: 10.1099/0022-1317-78-3-525. [DOI] [PubMed] [Google Scholar]
- 39.White K A, Skuzeski J M, Li W Z, Wei N, Morris T J. Immunodetection, expression strategy and complementation of turnip crinkle virus p28 and p88 replication components. Virology. 1995;211:525–534. doi: 10.1006/viro.1995.1434. [DOI] [PubMed] [Google Scholar]
- 40.Wobbe K K, Akgoz M, Dempsey D A, Klessig D F. A single amino acid change in turnip crinkle virus movement protein p8 affects RNA binding and virulence on Arabidopsis thaliana. J Virol. 1998;72:6247–6250. doi: 10.1128/jvi.72.7.6247-6250.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wong S M, Chng C G. Occurrence of hibiscus chlorotic ringspot virus in Singapore. Phytopathology. 1992;82:722. [Google Scholar]
- 42.You X J, Kim J W, Stuart G W, Bozarth R F. The nucleotide sequence of cowpea mottle virus and its assignment to the genus Carmovirus. J Gen Virol. 1995;76:2841–2845. doi: 10.1099/0022-1317-76-11-2841. [DOI] [PubMed] [Google Scholar]
- 43.Zaccomer B, Haenni A L, Macaya G. The remarkable variety of plant RNA virus genomes. J Gen Virol. 1995;76:231–247. doi: 10.1099/0022-1317-76-2-231. [DOI] [PubMed] [Google Scholar]