Abstract
Equine arteritis virus (EAV), the type member of the family Arteriviridae, is a single-stranded RNA virus with a positive-stranded genome of approximately 13 kb. EAV uses a discontinuous transcription mechanism to produce a nested set of six subgenomic mRNAs from which its structural genes are expressed. We have generated the first documented arterivirus defective interfering (DI) RNAs by serial undiluted passaging of a wild-type EAV stock in BHK-21 cells. A cDNA copy of the smallest DI RNA (5.6 kb) was cloned. Upon transfection into EAV-infected BHK-21 cells, transcripts derived from this clone (pEDI) were replicated and packaged. Sequencing of pEDI revealed that the DI RNA was composed of three segments of the EAV genome (nucleotides 1 to 1057, 1388 to 1684, and 8530 to 12704) which were fused in frame with respect to the replicase reading frame. Remarkably, this DI RNA has retained all of the sequences encoding the structural proteins. By insertion of the chloramphenicol acetyltransferase reporter gene in the DI RNA genome, we were able to delimitate the sequences required for replication/DI-based transcription and packaging of EAV DI RNAs and to reduce the maximal size of a replication-competent EAV DI RNA to approximately 3 kb.
Equine arteritis virus (EAV) is the type member of the family Arteriviridae (38), which was recently grouped together with the coronaviruses and the toroviruses in the newly established order of the Nidovirales (4, 13). Other members of the Arteriviridae are Porcine reproductive and respiratory syndrome virus, Lactate dehydrogenase-elevating virus, and Simian hemorrhagic fever virus.
EAV is a spherical, enveloped virus with a diameter of 50 to 60 nm (18, 30) and a positive-stranded RNA genome of about 12,700 nucleotides (nt) (8). The virion envelope is derived from intracellular host cell membranes and contains two major and three or four minor structural proteins (12, 38, 39). The envelope surrounds an isometric nucleocapsid of about 35 nm (18), which is composed of the genomic RNA and multiple copies of the nucleocapsid protein (N).
The EAV replicase is produced in the form of two large polyproteins: the open reading frame 1a (ORF1a) protein and the ORF1ab protein, the C-terminal part of which is expressed by ORF1a/1b ribosomal frameshifting (8). An apparently complete proteolytic processing scheme for the ORF1a and ORF1ab nonstructural polyproteins, which results in the generation of 12 end products (nsp1 to nsp12) and a large number of processing intermediates, was recently obtained (41, 46, 47, 50).
The EAV structural proteins are translated from a 3′-coterminal nested set of subgenomic (sg) mRNAs, which also carry a common 5′ leader sequence derived from the 5′ end of the genome (11). The mechanism of sg mRNA transcription, which resembles that of the coronaviruses in many aspects, involves a discontinuous step of which many details remain to be elucidated (4, 24, 38, 48).
Little is known about the genome replication of arteriviruses at the molecular level. The genomic 3′ end is polyadenylated and contains a nontranslated region (NTR) of 59 to 117 nt (38). A short, conserved sequence is present just upstream of the poly(A) tail, but its function remains to be elucidated (16). The 5′ end of the genomic RNA is capped (38) and contains an NTR of 156 to 224 nt. It was shown by Hwang and Brinton (19) that at least four host proteins from MA-104 cells interact with in vitro-transcribed RNA representing the 3′ end of the genomic negative strand of simian hemorrhagic fever virus, lactate dehydrogenase-elevating virus, and EAV. This region is the complement of the genomic leader sequence and is assumed to be involved in the initiation of plus-strand RNA synthesis.
Encapsidation of EAV sg mRNAs in virus particles has never been observed (R. Molenkamp, B. C. D. Rozier, and E. J. Snijder, unpublished data), which suggests that only genomic RNA is encapsidated and that a specific encapsidation signal that is lacking from the sg mRNAs could be located in the ORF1ab region. It has been shown that the same region in defective genomes of mouse hepatitis coronavirus (MHV) contains an encapsidation signal that is required for the specific encapsidation of MHV DI RNAs (2, 15, 35, 43). However, the infectious bronchitis coronavirus is able to encapsidate small amounts of sg mRNAs (54), suggesting that the determinant for specific encapsidation is not exclusively located in the ORF1ab region of this virus.
Recently, a full-length EAV cDNA clone from which infectious transcripts can be produced has been generated (45). Although many aspects of the viral life cycle can be investigated by using this tool, the study of replication and encapsidation signals would strongly benefit from the development of EAV replicons that can be replicated in trans. Defective interfering (DI) RNAs have been widely used for the analysis of cis-acting replication signals (6, 15, 20, 22, 26, 28, 36, 43, 52). DI RNAs are truncated, and in some cases rearranged, genomes that have usually lost the potential to replicate autonomously due to deletions in the viral replicase gene(s). They can be generated by serial undiluted passaging of viruses (3, 10, 31, 33, 34) during which their replication depends on the replicative enzymes encoded by the helper virus. DI RNAs have retained all replication signals and frequently also the sequences required for RNA encapsidation (15, 26, 36, 43, 52). Because DI RNAs are generally much smaller than the helper virus genome, they are replicated more efficiently. Together with competition for viral proteins, the replicative advantage of DI RNAs explains the interference with helper virus replication. Although arterivirus DI genomes would be very useful, the detection or isolation of arterivirus DI RNAs has not been reported.
In this paper, we describe the generation of the first documented EAV DI genomes by serial undiluted passaging of a virus stock. We characterized the smallest of the DI RNAs that were generated, the 5.6-kb DI-b, and constructed a DI-b cDNA clone. Transcripts derived from this construct (pEDI) were replicated and packaged in helper virus-infected cells. Sequence analysis revealed that in comparison with the genome, DI-b contains two large in-frame deletions in the replicase gene. Remarkably, the entire region encoding the structural proteins was retained in DI-b. Density gradients showed that virus particles containing EDI RNA are of the same density as virus particles containing only full-length EAV RNA. Finally, introduction of the chloramphenicol acetyltransferase (CAT) gene in the DI genome allowed us to identify sequences required for replication/transcription and encapsidation of EAV DI RNAs and to reduce the maximal size of a replication-competent EAV DI RNA to 3.0 kb.
MATERIALS AND METHODS
Cells and virus.
Baby hamster kidney cells (BHK-21 cells) were grown in BHK-21 medium (Life Technologies) supplemented with 5% fetal calf serum, 10% tryptose phosphate broth, and 10 mM HEPES. The EAV Bucyrus strain (14) was used in all experiments. All infections with EAV were carried out at 39.5°C.
Recombinant DNA techniques.
Standard recombinant DNA procedures were used (37). Restriction enzymes, T4 DNA ligase, and T7 RNA polymerase were obtained from Life Technologies. All enzyme incubations and biochemical reactions were performed according to the instructions of the manufacturers. Sequencing reactions were performed with the Big Dye Terminator kit (Perkin-Elmer) and analyzed using an ABI PRISM 310 genetic analyzer (Perkin-Elmer).
Serial undiluted passages.
BHK-21 cells were seeded in 10-cm2 dishes and infected with the EAV Bucyrus strain at a multiplicity of infection (MOI) of 50. Medium was harvested after incubation for 24 h at 39.5°C, at which time point complete cytopathogenic effect (CPE) was observed. For each virus passage, a similar dish with fresh BHK-21 cells was infected with one third (600 μl) of the culture medium from the previous passage. To preserve high helper virus titers during passaging, extra helper virus was added to the inoculum at an MOI of 10 whenever a delay of CPE was observed.
Isolation and analysis of viral RNAs.
Intracellular RNA was isolated at 12 h postinfection (p.i.) by using Trizol (Life Technologies) followed by isopropanol precipitation. Denaturing RNA electrophoresis was carried out using 1 to 1.5% agarose gels containing 10 mM MOPS (morpholinepropanesulfonic acid) and 2.2 M formaldehyde. Gels were dried and hybridized with appropriate 5′-end-labeled oligonucleotides (Table 1) as described by Meinkoth and Wahl (32).
TABLE 1.
Summary of hybridization experiments to map EAV sequences in DI-a and DI-b
| Oligonucleotide | Position | Sequence 5′-3′ | DI-a | DI-b | Genome |
|---|---|---|---|---|---|
| E274 | 228 | CCAGTAGCGGAGAAGGTTGC | + | + | + |
| E277 | 3752 | CGCCACTAGTAGTCCACGCC | − | − | + |
| E279 | 6452 | GCAGGCACGCAAACCATGGCC | − | − | + |
| E104 | 7534 | CCAGGATCGGGTGAGGC | + | − | + |
| E275 | 9192 | GGGCAACAGCGTCTGCGG | + | + | + |
| E280 | 11525 | GCGTAGCATAGGGTAGTACTG | + | + | + |
| E154 | 12680 | TTGGTTCCTGGGTGGCTAATAA CTACTT | + | + | + |
RT-PCR and cDNA cloning.
Intracellular poly(A)-containing RNA was purified using oligo(dT)-coupled magnetic beads (Dynal) according to the instructions of the manufacturer. cDNA synthesis was performed using Superscript II reverse transcriptase (Life Technologies), and subsequently cDNA was amplified by using the GeneAmp XL-PCR system (Perkin-Elmer). Oligonucleotides used as primers during reverse transcription (RT) and PCR are described in Table 2. The DI RNA-specific RT-PCR product was digested with MluI and BamHI and cloned between the MluI and BamHI sites in pEAV030 (EMBL/GenBank accession no. Y07862) (45), which are located at nt 589 and 9149, respectively. The resulting plasmid was named pEDI.
TABLE 2.
RT and PCR oligonucleotides
| Oligonucleotide | Polarity | Sequence 5′-3′a | Position (nt) |
|---|---|---|---|
| E266 | + | GACTCACTATC GCTCGAAGTGTG | 1 |
| E261 | + | GCAACCTTCTCCGCTACTGG | 228 |
| E275 | − | GGGCAACAGCGTCTGCGG | 9192 |
| E280 | − | GCGTAGCATAGGGTAGTACTG | 11525 |
| E154 | − | TTGGTTCCTGGGTGGCTAATAACTACTT | 12680 |
| E298 | + | CAACGTGC/GAAGTTCACC | 1679/8530 |
| E288 | − | GGTGAACTTC/GCACGTTG | 1679/8530 |
Non-EAV sequences are indicated in italics.
RNA transcription and transfection.
pEAV030, pEDI, and derivatives were linearized with XhoI. DNA templates were extracted with phenol-chloroform and ethanol precipitated. RNA was synthesized in vitro by using T7 RNA polymerase for 2 h at 37°C. Reactions contained 4 μg of linearized template DNA, 1 mM each GTP, CTP, ATP, and UTP, 5 mM dithiothreitol, 0.1 μg of bovine serum albumin per μl, 125 U of T7 RNA polymerase, 63.5 U of unmethylated capping analog [G(5′)ppp(5′)G; New England Biolabs], and 40 U of RnaseOut (Life Technologies) in a final volume of 50 μl of T7 transcription buffer (Life Technologies). For transfections with pEDI transcripts, BHK-21 cells were infected with EAV at an MOI of 10. After 1 h, the inoculum was removed and the cells were transfected with EDI RNA as previously described (45). Alternatively, BHK-21 cells were double transfected with EAV030 RNA and EDI RNA.
Sucrose gradient purification of EAV and EDI-containing particles.
BHK-21 cells were infected with EDI-containing EAV stocks at high MOI, and viral RNAs were labeled with [3H]uridine (100 μCi/ml) at 10 h p.i. in the presence of 10 μg of actinomycin D per ml. Medium from 106 infected cells was harvested at 24 h p.i., cleared by low-speed centrifugation, and loaded on a linear 20 to 50% (wt/wt) sucrose gradient in 10 mM Tris-HCl (pH 7.5)–1 mM EDTA–100 mM NaCl. The gradient was centrifuged at 4°C for 16 h at 40,000 rpm in a Beckman SW41 rotor. Subsequently, 600-μl fractions were collected from bottom to top and assayed for incorporation of label by trichloroacetic acid precipitation and liquid scintillation counting. RNA from sucrose gradient fractions was isolated as described by Spaan et al. (42).
Construction and analysis of pEDIC2, pEDIC7, and derivatives.
pEDI-based CAT expression constructs were created by introducing a BamHI-XhoI (nt 9149 to 12704) restriction fragment from the previously described constructs pEAVCAT2 and pEAVCAT7 (45) into BamHI-XhoI-digested pEDI. The resulting plasmids, pEDIC2 and pEDIC7, contained the CAT reporter gene downstream of the RNA2 and RNA7 sg mRNA promoters, respectively. An additional MluI restriction site was engineered at nt 435 to 440 by PCR mutagenesis (TGGCTT to ACGCGT). Truncated derivatives of pEDIC2 and pEDIC7 were generated by fusion of the restriction sites indicated in Table 3. With the exception of pEDIC2-1820, all deletions in the ORF1ab region of pEDIC2 were made in frame with respect to the replicase reading frame. In vitro-transcribed RNA from pEDIC2, pEDIC7, and derivatives was transfected into helper virus-infected cells as described above. As controls, pEDIC2 and pEDIC7 RNA was transfected into noninfected BHK-21 cells. At 12 h p.i., cell lysates were made by using the CAT enzyme-linked immunosorbent assay (ELISA) lysis buffer supplemented with the CAT ELISA kit (Boehringer Mannheim). At 16 h p.i., virus was harvested (P0 [passage 0] virus), mixed with helpervirus (MOI of 10), and used to infect a fresh monolayer of BHK-21 cells. Cell lysates were made at 12 h p.i., and P0 and P1 CAT expression was determined by CAT-ELISA (Boehringer Mannheim) by using an ELISA reader set at the recommended wavelength of 405 nm.
TABLE 3.
EDIC2 and EDIC7 deletion mutants
| Construct | Upstream site (nt) | Downstream site |
|---|---|---|
| EDIC2-0406 | MluI (435)a | MluI (589) |
| EDIC2-0613 | MluI (589) | NcoI (8566) |
| EDIC2-1318 | NcoI (8566) | SalI (8996) |
| EDIC2-1820 | SalI (8996) | BamHI (9149) |
| EDIC2-3457 | SphI (10403) | NspV (12350) |
| EDIC2-3450 | SphI (10403) | BalI (11636) |
| EDIC2-4150 | StuI (10723) | BalI (11636) |
| EDIC2-DD1 | MluI (589) | NcoI (8566) |
| SphI (10403) | BalI (11636) | |
| EDIC7-2050 | BamHI (9149) | StuI (10723) |
| EDIC7-DD2 | MluI (589) | NcoI (8566) |
| BamHI (9149) | BalI (11636) |
Position in the EAV genome.
Immunofluorescence assays.
Immunofluorescence assays were essentially performed as described before (47). A CAT-specific rabbit antiserum was obtained from 5 Prime → 3 Prime Inc. and used at a 1:500 dilution. A Cy3-conjugated donkey anti-rabbit immunoglobulin G (Jackson ImmunoResearch Laboratories) was used as the secondary antibody at a 1:1,000 dilution.
RESULTS
Generation of EAV DI RNAs during serial undiluted virus passaging.
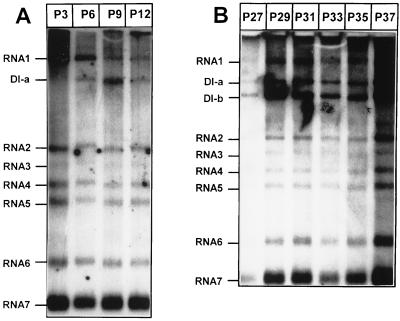
For many viruses, serial undiluted passaging in tissue culture has resulted in the generation of DI viruses (3, 10, 31, 33, 34). Likewise, in this study an EAV stock was passaged up to 40 times in BHK-21 cells. At each passage, intracellular RNA was isolated, subjected to denaturing gel electrophoresis, and hybridized to a radiolabeled oligonucleotide complementary to the 3′ end of the EAV genome and all sg mRNAs. At P6 we observed, the first new EAV RNA species, which migrated between the 12.7-kb genome (RNA1) and sg mRNA2 (3.2 kb) in denaturing agarose gels (Fig. 1A) and which appeared to be passaged from dish to dish. The size of this new RNA (DI-a) was estimated to be approximately 8 kb. Serial passaging was continued and at P27 a second new RNA species was observed (Fig. 1B). This RNA was approximately 6 kb long and was named DI-b. Both DI-a and DI-b accumulated in subsequent passages and remained present up to P40. During the passaging experiments, a decrease in CPE was regularly observed, suggesting that that at least one of the new RNA species interfered with helper virus replication.
FIG. 1.
Generation of natural EAV DI RNAs by serial undiluted passaging of a wt virus stock. Intracellular RNA was isolated from infected BHK-21 cells at P3 to P12 (A) and P27 to P37 (B) and subjected to gel electrophoresis and hybridization with an oligonucleotide recognizing the 3′ end of all viral mRNAs. The positions of the new RNA species DI-a and DI-b are indicated.
cDNA cloning and composition of an EAV DI RNA.
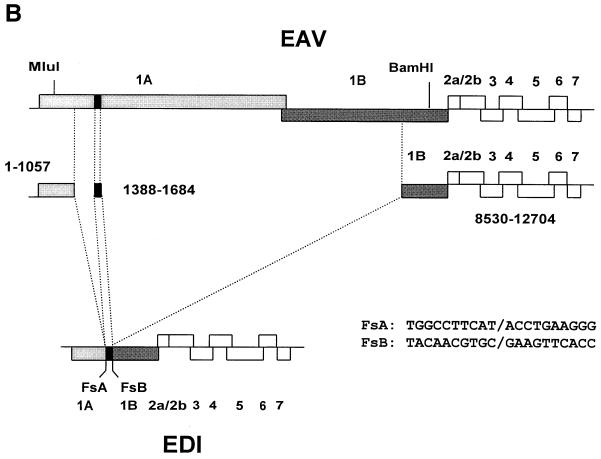
To determine which regions of the EAV genome were represented in DI-a and DI-b, intracellular RNA was isolated at P35 and subjected to gel electrophoresis and hybridization with a number of antisense oligonucleotides. The results of this analysis are summarized in Table 1. Apparently, DI-a and DI-b both lacked large portions of the 9.5-kb EAV replicase gene. Therefore, we attempted to generate and clone a cDNA copy of this region by using an RT-PCR approach based on a primer set derived from the terminal sequences of the replicase gene. cDNA was synthesized with oligonucleotide E275 (nt 9192 to 9209; Table 2) and a PCR product was generated using oligonucleotide E275 and E261 (nt 228 to 247; Table 2). An RT-PCR product (Fig. 2A) specific for DI RNA-containing intracellular RNA was obtained, and this approximately 1.9-kb fragment was cloned between the 5′- and 3′-terminal sequences of the infectious EAV cDNA clone, pEAV030 (45), using internal MluI (nt 589) and BamHI (nt 9149) restriction sites. This resulted in a plasmid containing a central replicase gene region that was presumably derived from a DI RNA and pEAV030-derived sequences representing nt 1 to 589 and 9149 to 12704 of the EAV genome. A schematic representation of this construct, which was named pEDI, is shown in Fig. 2B. The MluI-BamHI restriction fragment of pEDI was sequenced and found to consist of three segments of the EAV replicase gene (Fig. 2B). Two DI RNA-specific fusion sites, designated FsA (nt 1057 fused to 1388 of the EAV genome) and FsB (nt 1684 fused to 8530 of the EAV genome), were identified. These fusions were in frame with respect to the replicase gene and are also shown in Fig. 2B.
FIG. 2.
(A) RT and PCR on RNA isolated from P35 or wt-infected cells (EAV) using a primer set derived from the terminal sequences of the replicase gene. An approximately 1.9-kb DI RNA-specific PCR product was detected. Molecular weight markers are indicated. (B) Schematic representation of the EAV genome and EDI RNA. The replicase gene region is indicated by shaded boxes. The 296-nt middle segment of EDI is indicated as a black box. The internal MluI and BamHI restriction sites used for cloning of the DI replicase gene region and the sequences and locations of the two fusion sites FsA and FsB are also depicted. (C) RT-PCR approach to determine the sequence of the 5′- and 3′-terminal sequences of EDI RNA. An EDI RNA-specific RT-PCR was carried out on P35 RNA using a primer specific for FsA (primer E299) or FsB (primer E298). The specific RT-PCR products representing the 3′ (4 kb; lane 3)- and 5′ (1.1 kb; lane 5)-terminal sequences of the DI RNA were sequenced and found to contain sequences collinear with the wt EAV genome only. As a negative control, the same RT-PCR was performed on RNA isolated from wt EAV-infected cells (lanes 2 and 4).
On the basis of the estimated sizes of DI-a and DI-b, we expected no large deletions in the sequences outside the replicase gene. To confirm this, 5′- and 3′-end-specific RT-PCR analyses were carried out on RNA isolated at P35 or on RNA from wild-type (wt) EAV-infected cells. When an FsB-specific oligonucleotide and an primer complementary to the 3′ end of the genome were used, a specific 4-kb PCR product was obtained with intracellular RNA from P35 (Fig. 2C, lane 2) but not with intracellular RNA from wt EAV-infected cells (Fig. 2C, lane 1). Likewise, when an FsB-specific oligonucleotide and a primer complementary to the 5′ end of the genome were used, a specific 1.1-kb PCR product was obtained (Fig. 2C, lanes 3 and 4). Both DI RNA-specific RT-PCR products were sequenced entirely (data not shown) and contained only sequences that were identical to the published sequence of the corresponding regions of the EAV genome (8). From these data we concluded that the EDI cDNA sequence between nt 228 and 12680 (Fig. 2B) represents one of the DI RNAs generated during serial undiluted passaging of an EAV stock. The length of the EDI cDNA clone (5.6 kb) suggested that it is derived from the smaller DI-b. Remarkably, our results revealed that DI-b has retained the entire structural protein-coding region of the EAV genome.
In vitro-transcribed EDI RNA can be replicated and packaged by EAV helper virus.
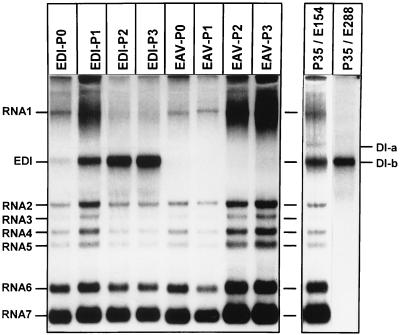
To show that pEDI transcripts are replication competent and behave similarly to the DI RNAs generated in our passaging experiments, we transfected in vitro-transcribed EDI RNA into EAV-infected cells and performed passaging experiments. BHK-21 cells were infected with helper virus at an MOI of 10. Half of the cells were transfected with EDI RNA, and the other half were mock transfected. Cells were plated and incubated for 16 to 20 h at 39.5°C. Culture supernatant (P0 virus) was harvested, and at the same time intracellular RNA was isolated (P0 RNA). One-third of the P0 virus was used to infect a fresh monolayer of BHK-21 cells, and at 16 to 20 h p.i. the medium was harvested (P1 virus). This procedure was repeated two more times. P0 to P2 virus was used to infect BHK-21 cells, and at 12 h p.i. intracellular RNA was isolated and analyzed. Figure 3 shows the results of such a transfection/passaging experiment. In lane 1 (EDI-P0 RNA), a small amount of EDI and a relatively large amount of helper virus genome could be observed. EDI accumulated in P2 and P3 (lane 3 and lane 4) at the expense of genomic RNA, which strongly suggested that EDI indeed interfered with the replication of the latter. The control experiment without EDI transcript (lanes 5 to 8) did not show the appearance of a DI RNA. Throughout this EDI passaging experiment, the amount of sg mRNAs produced in infected cells seemed to remain constant, while the amount of helper virus genomic RNA decreased drastically. This suggested that EDI was used in trans as a template for the synthesis of sg mRNAs. EDI comigrated with DI-b from P35 (lane 9), which again suggested that EDI is identical to DI-b. Hybridization of intracellular RNA from P35 with the EDI FsB-specific oligonucleotide (Table 2) supported this assumption.
FIG. 3.
Transfection and passaging of EDI RNA. In vitro-transcribed EDI RNA was transfected into EAV-infected BHK-21 cells. At 16 to 20 h p.i., medium was harvested and used to infect fresh BHK-21 cells. This procedure was repeated two times. RNA isolated from each passage (EDI-P0 to EDI-P3) was subjected to gel electrophoresis and hybridization with an oligonucleotide recognizing the 3′ end of all viral RNAs. As a control, BHK-21 cells were infected with wt EAV and passaged in the same manner (EAV-P0 to EAV-P3). RNA isolated from P35 of the initial passaging experiment was put on the same gel and hybridized with an oligonucleotide recognizing all viral mRNAs (P35/E154) or hybridized with an FsB-specific oligonucleotide (P35/E288).
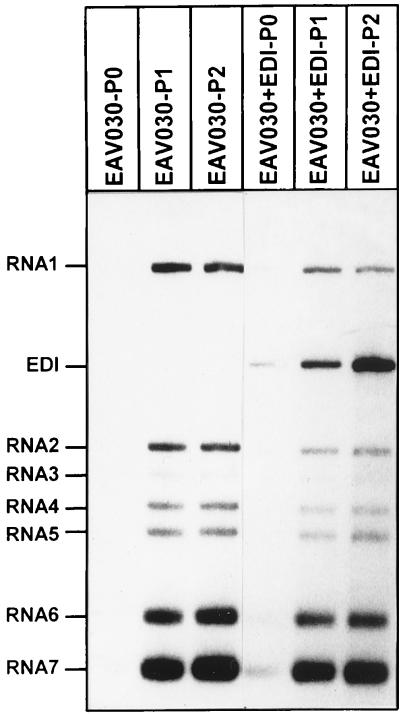
To show that the accumulation of DI RNA in the former experiment was not due to the accumulation of endogenous DI RNAs already present in the helper virus stock, and to investigate whether EDI could be replicated and packaged by cotransfection of full-length synthetic RNA transcribed from the infectious EAV cDNA clone pEAV030 (45), coelectroporations of EDI RNA and EAV030 RNA were performed. BHK-21 cells were double electroporated, and passaging experiments were carried out as described above. In double-transfected cells, a small amount of EDI was observed in P0 (Fig. 4, lane 4) and EDI accumulated during P1 and P2 (Fig. 4, lanes 5 and 6) at the expense of EAV030. Again, the levels of sg mRNAs remained constant during passaging. In cells transfected with EAV030 RNA only, a small amount of EAV030 was seen after P0 (Fig. 4, lane 1). During P1 and P2, EAV030 accumulated but DI RNAs were not detected (Fig. 4, lanes 2 and 3). These results indicate that EDI replication and packaging were driven by EAV030, in a fashion similar to replication and packaging of EDI by wt EAV helper virus. Furthermore, these data show that the accumulation of EDI in earlier experiments was not due to the accumulation of endogenous DI RNAs from the helper virus stock.
FIG. 4.
In vitro-transcribed EDI RNA was cotransfected with EAV030 RNA into BHK-21 cells. At 16 to 20 h p.i., medium was harvested and used to infect fresh BHK-21 cells. This procedure was repeated one more time. RNA isolated from each passage (P0 to P2; lanes 4 to 6) was subjected to gel electrophoresis and hybridization with an oligonucleotide recognizing the 3′ end of all viral mRNAs. As a control, BHK-21 cells were transfected with EAV030 RNA only, and virus was passaged in the same manner (lanes 1 to 3).
EDI RNA and EAV genomic RNA are packaged in virus particles of equal density.
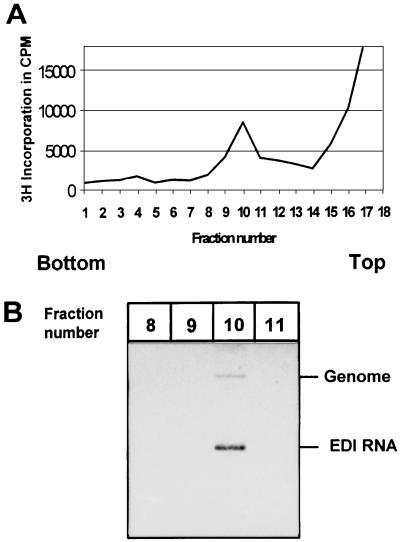
Since EDI RNA is much smaller than the EAV genome, we investigated whether EDI RNA is packaged into virus particles with a density different from that of regular EAV particles. BHK-21 cells were infected with EDI- and helper virus-containing P3 virus from the passaging experiment shown in Fig. 3. Viral RNAs were metabolically labeled by using [3H]uridine. The culture supernatant was loaded on a 20 to 50% (wt/wt) linear sucrose gradient, which was centrifuged to equilibrium. Subsequently, 600-μl fractions were collected from bottom to top and assayed for the presence of label by trichloroacetic acid precipitation and liquid scintillation counting.
This analysis revealed that the peak of radioactivity was present in fraction 10 (Fig. 5A). Subsequently, RNA was isolated from fractions 8 to 11 and subjected to denaturing agarose gel electrophoresis and hybridization with an antisense oligonucleotide recognizing the 3′ end of all EAV mRNAs. The peak amount of both EAV genomic RNA and EDI RNA was recovered from fraction 10 (Fig. 5B, lane 3). We therefore concluded that EDI RNA is packaged in virus particles with the same density as EAV helper virus particles.
FIG. 5.
Sucrose gradient analysis of an EDI-containing EAV stock. BHK-21 cells were infected at high MOI, and viral RNAs were labeled with [3H]uridine. Medium was loaded on a 20 to 50% (wt/wt) sucrose gradient which was centrifuged to equilibrium; 600-μl fractions were collected from bottom to top and analyzed for the presence of label. (A) Sucrose gradient profile with the peak of radioactivity in fraction 10. (B) RNA isolated from fractions 8 to 11 was subjected to gel electrophoresis and hybridization with an oligonucleotide recognizing all viral mRNAs. Both EAV genomic RNA and EDI RNA were found almost exclusively in fraction 10.
EDI replication and packaging monitored by expression of the CAT reporter gene.
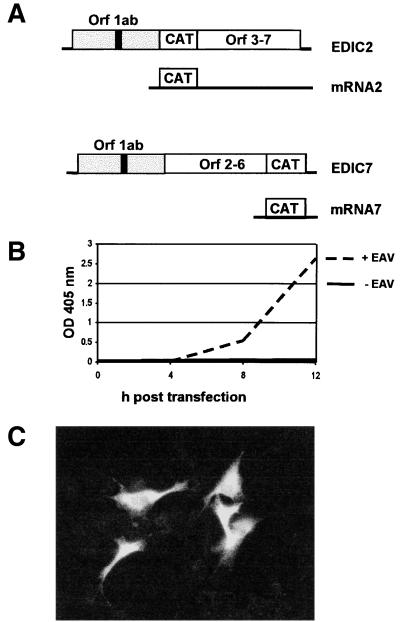
The analysis of the replication and packaging of an RNA replicon is, obviously, most straightforward by direct metabolic labeling of the replicon in transfected cells. However, in the case of EDI, such an approach was hampered by low transfection and replication efficiencies of the RNA replicon. We therefore engineered the expression of a reporter gene to monitor replication and packaging of EDI-based replicons in a sensitive and convenient manner. A similar approach has been used successfully during molecular studies on the replication, packaging, and transcription of, e.g., coronavirus and poliovirus subgenomic replicons (1, 20, 27, 29, 53). Clearly, this kind of analysis is most straightforward when the assay is based directly on translation of the replicating RNA itself. However, in the case of EDI, the presence of a truncated replicase ORF in the 5′ end of the replicon posed a potential problem, in particular because the importance of a similar ORF in certain coronavirus DI RNAs has been well established in our laboratory (7, 44). To avoid the risk of impairing EDI replication by inserting a reporter gene in its 5′-terminal region, we chose an indirect approach to study replication and packaging. The CAT reporter gene was inserted at the position normally occupied by ORF2b or ORF7 (Fig. 2B). Consequently, the CAT gene should be expressed from sg mRNA 2 or 7, both of which are normally used to translate EAV structural proteins. We have previously shown that in the EAV full-length cDNA clone pEAV030 efficient CAT expression from these positions is possible (45). Therefore, we engineered pEDI derivatives carrying the CAT gene at the same positions (pEDIC2 and pEDIC7) (Fig. 6A and 7), thereby allowing its expression from EDI-derived sg mRNAs.
FIG. 6.
(A) Schematic representation of EDIC2 and EDIC7. Subgenomic mRNAs from which the CAT reporter gene is expressed are indicated. (B) CAT protein expression from EDIC2 RNA transfected into EAV-infected BHK-21 cells during a 12-h time period. OD, optical density. (C) Immunofluorescence staining with a CAT-specific antiserum on EAV-infected BHK-21 cells, transfected with EDIC2 RNA. The transfection efficiency in this particular experiment was estimated to be approximately 10%.
FIG. 7.
Replication and packaging of EDIC2 and EDIC7 RNA deletion mutants. EDIC2, EDIC7, and restriction sites used for deletion mutagenesis are schematically depicted. The position of the CAT reporter gene behind the RNA2 sg mRNA promoter (EDIC2) or RNA7 sg mRNA promoter (EDIC7) is indicated. Deleted regions are indicated by dashed lines. CAT expression levels in P0 and P1 comparable to those observed for EDIC2 or EDIC7 are indicated by +; background CAT levels are indicated by −.
The level of CAT expression in transfected and infected cells was monitored using a sensitive CAT ELISA. We assumed that EDI replication and EDI-based sg mRNA synthesis would result in CAT expression during P0 and that its kinetics would be similar to that of EAV structural protein synthesis. Provided that the EDIC2- and EDIC7-based replicons would also be packaged into virus particles, CAT expression should be detectable during passaging experiments carried out in the presence of sufficient helper virus. A representative example of the analysis of EDIC2-driven CAT expression during P0 is shown in Fig. 6B. In helper virus-infected cells but not in mock-infected cells, significant CAT expression was detected at 4 h posttransfection and strongly increased between 8 and 12 h posttransfection, when EAV sg mRNA synthesis is known to reach its maximum (9). An immunofluorescence analysis using a CAT-specific antiserum revealed that (in this particular experiment) approximately 10% of the cells expressed the reporter gene at 12 h posttransfection (Fig. 6C). Essentially similar results were obtained with the EDIC7 replicon. Subsequently, we repeated this analysis during a standard passaging experiment with medium harvested from P0 at 16 h posttransfection. The virus harvested from both EDIC2 and EDIC7 transfections yielded P1 CAT protein expression levels that were comparable to those observed in P0. From these data, we concluded that EDIC2 and EDIC7 were replicated and packaged and could therefore be used as tools to delimitate the EDI sequences required for replication and packaging of EAV DI RNAs. It should, however, be noted that for reasons explained above, our assay does not discriminate between replication and transcription defects. Thus, lack of CAT protein expression by EDIC2 and EDIC7 derivatives can be explained by the abrogation of either DI RNA replication or DI RNA-based sg mRNA transcription.
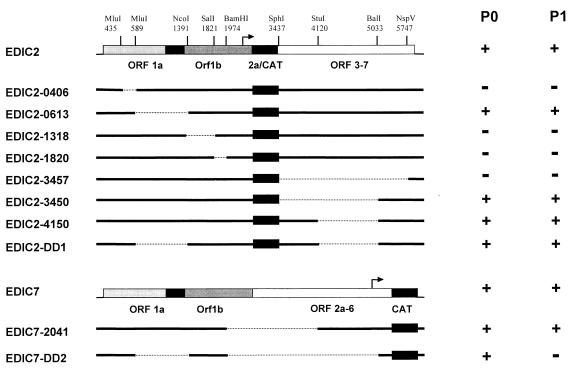
Delimitation of sequences required for replication and packaging of EDI.
Several deletions were made in pEDIC2 and pEDIC7 (Fig. 7), and in vitro-transcribed RNA from these derivatives was transfected into helper virus-infected BHK-21 cells. With the exception of EDIC2-1820, deletions in ORF1ab were in frame with respect to the replicase reading frame. As controls, EDIC2 and EDIC7 RNA were transfected into infected and mock-infected cells. Transfection efficiencies were determined by immunofluorescence assays (data not shown) and were judged to be similar for all CAT-expressing constructs. CAT protein expression in P0 and P1 was quantitated by using a sensitive CAT ELISA. The results of this analysis are summarized in Fig. 7.
EDIC2-0406, which lacks only a small portion (154 nt) of the 5′-terminal segment of EDI, did not express detectable levels of CAT in P0. EDIC2-0613, lacking an 802-nt region of EDI which included the middle, ORF1a-derived segment (nt 1388 to 1684 in the EAV genome), expressed the CAT protein in P0 and P1 in amounts similar to those observed for EDIC2 RNA. This implied that the region between nt 589 and 1391 in EDIC2 is dispensable for DI RNA replication and packaging. For EDIC2-1318 and EDIC2-1820, which both contained deletions in the ORF1b segment of EDI RNA, no CAT expression could be detected in P0.
EDIC2-3457, carrying a deletion of EAV ORF3 to ORF7, failed to express the CAT protein in P0. As stated above, the lack of CAT expression for EDIC2-0406, EDIC2-1318, EDIC2-1820, and EDIC2-3457 could be due to a defect in either replication or sg mRNA transcription. EDIC2-3457 still contains the 354 3′-terminal nucleotides of the EAV genome [not including the poly(A) tail]. When this region was extended to 1,066 nt in EDIC2-3450, CAT expression in P0 and P1 was restored and similar to that of EDIC2 RNA. A 913-nt deletion in the 3′-terminal segment of EDIC2 (EDIC2-4150) resulted in CAT expression levels in P0 and P1 that were comparable to those observed for EDIC2. When the deletions from EDIC2-0613 and EDIC2-4150 were combined in EDIC2-DD1, CAT expression levels in P0 and P1 were again comparable to those observed for EDIC2.
To analyze the region containing the promoter for sg mRNA2 synthesis (which is required for the expression of the CAT reporter gene in EDIC2), pEDIC7 was constructed, in which the CAT gene is expressed from sg mRNA7. EDIC7 expressed similar amounts of CAT as EDIC2 in both P0 and P1 (Fig. 7). EDIC7-2041, which lacked the 3′ end of ORF1b (including the sg mRNA2 promoter), and ORF2a, -2b, and -3, expressed similar amounts of CAT as EDIC7 in P0 and P1. The three deletions present in EDIC2-0613, EDIC2-4150, and EDIC7-2041, which apparently did not interfere with replication or packaging of the replicons, were then combined in EDIC7-DD2. This RNA expressed a significant amount of CAT in P0, but no expression of CAT protein was detected in P1, suggesting that EDIC7-DD2 was replicated but not packaged. EDIC7-DD2 is the smallest DI RNA in our studies. Based on these results, we concluded that the sequences required for EDI replication or EDI-based transcription have been reduced to 589 nt at the 5′ end, 1,066 nt at the 3′ end, and an internal ORF1b-derived segment of 583 nt.
Further characterization of EDIC2-DD1 and EDIC7-DD2.
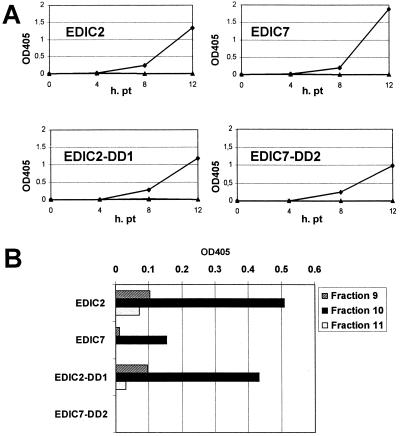
To further characterize the EDIC2-DD1 and EDIC7-DD2 replicons, CAT expression levels during P0 were monitored for EDIC2, EDIC7, EDIC2-DD1, and EDIC7-DD2. Transfection efficiencies were determined by immunofluorescence assay, and CAT values were corrected for transfection efficiency. For all constructs, significant CAT protein expression levels were detected at 4 h posttransfection (Fig. 8A) and CAT levels increased between 8 and 12 h posttransfection, when EAV sg mRNA synthesis reaches its maximum (9). No significant differences in replication efficiencies between the various EDI derivatives were found. To investigate whether the EDIC2, EDIC7, EDIC2-DD1, and EDIC7-DD2 replicons were packaged in virus particles, and to exclude the possibility that the RNA replicons could be passaged otherwise, a gradient analysis was performed. Medium harvested from transfected cells was passaged once, and the resulting P1 virus was loaded on a 20 to 50% linear sucrose gradient, which was centrifuged to equilibrium. Virus was isolated from the fraction that was earlier shown to contain the virus peak (Fig. 5, fraction 10) and fractions 9 and 11. Virus isolated from these fractions was mixed with helper virus (MOI of 10) and used to infect a fresh monolayer of BHK-21 cells. At 12 h postinfection, lysates were made and CAT expression was determined. Alternatively, immunofluorescence assays with a CAT-specific antibody were performed. For gradient-purified EDIC2, EDIC7, and EDIC2-DD1 virus, significant CAT expression (Fig. 8B) and a positive immunofluorescence signal were observed in a low percentage of cells (data not shown). In contrast, gradient-purified EDIC7-DD2 virus did not show CAT expression or a positive signal in immunofluorescence. From these data we conclude that the EDIC2, EDIC7, and EDIC2-DD1 replicons are indeed packaged into virus particles, while EDIC7-DD2 is not.
FIG. 8.
(A) Replication of EDIC2, EDIC7, EDIC2-DD1, and EDIC7-DD2 replicons during the first 12 h posttransfection (pt) as measured by the expression of the CAT reporter gene (optical density at 405 nm [OD 405]) in the presence (⧫) or absence (▴) of helper virus. (B) CAT expression in BHK-21 cells infected with sucrose gradient-purified virus from EDIC2, EDIC7, EDIC2-DD1, and EDIC7-DD2 replicons.
DISCUSSION
Generation of the first arterivirus DI RNA.
To our knowledge, the work presented in this paper constitutes the first characterization of a defective arterivirus genome. A DI RNA of approximately 8 kb (DI-a) was readily observed in P6 of our serial passaging experiment, but the 6-kb DI-b RNA used in this study surfaced only after 21 subsequent passages. No other new RNA species were observed up to P40, at which passage the experiment was terminated. DI-b cDNA was generated, and a presumably full-length DI-b cDNA clone (pEDI) was constructed and subjected to sequence analysis. Our preliminary hybridization analysis (Table 1) suggests that DI-a contains an additional part of ORF1b and may therefore be a precursor of DI-b. However, since the nature of DI-a was not further investigated, it remains unclear whether DI-b has evolved independently or is derived from DI-a. Synthetic pEDI transcripts were replicated and packaged in EAV-infected cells and also in cells transfected with transcripts derived from the full-length EAV cDNA clone. Density gradient centrifugation showed that virus particles containing EDI RNA were of the same density as virus particles containing only EAV genomic RNA. Finally, the insertion of the CAT reporter gene downstream of the RNA2 or RNA7 sg mRNA promoter allowed us to delimit the sequences required for replication/EDI-based transcription and packaging of EDI RNA. Based on the results obtained with deletion mutagenesis, these sequences are now reduced to 589 nt at the 5′ end, 1,066 nt at the 3′ end, and an internal sequence of 583 nt derived from ORF1b. However, the deletion mutant EDIC7-DD2, which contained these EAV sequences only, did replicate but was not packaged into virions.
Molecular characterization of DI-b.
Sequence analysis of the cloned DI-b RNA revealed some intriguing features. DI-b is composed of three genome segments joined in frame with respect to the replicase gene. The first segment (nt 1 to 1057) consists of the 5′ NTR (leader) sequence and part of replicase ORF1a. The second segment is a small (296-nt) region corresponding to nt 1388 to 1684 of the EAV genome, which encodes part of nsp2. The third segment represents the region from nt 8530 up to the 3′ end of the EAV genome. As a result, EDI contains a reading frame of 783 amino acids, starting at the ORF1a translation initiation codon and terminating at the ORF1b stop codon. Since the sequence upstream of the translation initiation codon of this fusion ORF is identical to the 5′ NTR of the EAV genome, we assume that this truncated replicase gene is indeed translated. The translation product consists of the entire nsp1 sequence, two segments of nsp2, the C-terminal domain of nsp10, nsp11, and nsp12. In EAV- and EDI-infected cells the proteolytic processing of the EDI 1ab fusion protein will probably be similar to that of the corresponding parts of the full-length EAV 1ab polyprotein (41, 46, 47, 50). The nsp1 autoprotease can be expected to cleave itself at the nsp1/2 site (40). The nsp10/11 and nsp11/12 sites will probably be processed in trans by the nsp4 protease encoded by the helper virus (46). The remainder of the EDI translation product would then be a small (185-amino-acid) fusion protein containing two segments of nsp2 and the C-terminal part of nsp10. It is unclear whether any of these EDI translation products has a function in DI RNA replication. It was proposed that for MHV DI RNAs, translation of a fusion reading frame might promote the stability and enhance the replication of DI RNAs (7). Studies to investigate whether the truncated EDI ORF is indeed translated and whether translation per se is required for efficient DI RNA replication are in progress.
One of our most surprising findings is that DI-b has retained all sequences encoding EAV structural proteins. During passaging experiments we observed that the levels of sg mRNAs remained more or less constant (Fig. 5), despite the fact that the amount of helpervirus genomic RNA was significantly reduced. This strongly suggested that EDI RNA can be used in trans as a template for the transcription of sg mRNAs, a finding which has very interesting implications. Most documented nidovirus DI RNAs contain only small parts of the sequences encoding the structural proteins. To our knowledge, there is no other example of a natural nidovirus DI RNA that has retained the entire region downstream of the replicase gene. We can envision different reasons why it might be advantageous for the EDI/DI-b genome to retain and/or express the viral structural genes. First of all, this region could contain important RNA signals that are crucial for replication or packaging of the DI RNA. This seems unlikely, however, since deletions leaving only 1,066 nt of the 3′-terminal region had no effect on DI RNA replication and packaging (EDIC2-3450 [Fig. 7]), suggesting that at least most of the region containing the structural genes is dispensable. A second explanation might be that sg mRNA transcription from the 3′-terminal part of the DI RNA enhances its stability. In the case of EDI/DI-b RNA, translation of the truncated replicase gene could stabilize the 5′-terminal part of the RNA, while sg mRNA synthesis could increase the stability of the remaining part.
Finally, the expression of structural proteins from DI RNA-derived sg mRNAs could provide a selective advantage. The interference of EDI/DI-b RNA with helper virus replication must result in a severe reduction of structural protein synthesis by the latter. Structural protein expression from the 3′-terminal part of EDI/DI-b may (partially) compensate for the interference with helper virus genome replication and structural protein expression and thereby promote EDI/DI-b packaging.
In vitro-transcribed EDI RNA was efficiently rescued by EAV helper virus after transfection into BHK-21 cells. In addition, synthetic EDI RNA could be rescued by cotransfection with RNA derived from the EAV full-length infectious cDNA clone. This cotransfection system of EDI and EAV030 RNAs may prove to be a very powerful tool in the studies on replication, transcription, recombination, packaging, and other aspects of the EAV life cycle. For instance, a complementation system in which the packaging of the helper virus genome is dependent on the structural proteins expressed from the DI RNA could be useful for the study of viral assembly.
Delimitation of sequences required for replication and packaging of EDI RNA.
On the basis of the results obtained with constructs EDIC2-0406 and EDIC2-0613, the 5′-terminal sequence required for efficient replication or transcription and packaging of EAV DI RNAs has been reduced to a maximum of 589 nt. Likewise, constructs EDIC2-3457 and EDIC2-3450 demonstrated that the 3′-terminal genomic sequence required for replication and packaging of EAV DI RNAs is at most 1,068 nt long. In addition to these 5′- and 3′-terminal sequences, a 583-nt sequence from the central part of replicase ORF1b (nt 8566 to 9149 in the EAV genome) was found to be essential for EDI-driven CAT expression (EDIC2-1318 and EDIC2-1820). Since our assay did not differentiate between replication and sg mRNA transcription defects, we cannot exclude that the minimal sequences that are required are in fact smaller. With the exception of EDIC2-1820, all deletions in the EDI ORF1ab region were made in frame with respect to the replicase reading frame. Thus, if the inability of EDIC2-0406 and EDIC2-1318 to express the CAT protein is due to an effect at the level of replication, this defect is not caused by the disruption of the truncated EDI ORF1ab.
The smallest replication-competent EDI RNA derivative was the 3.0-kb EDIC7-DD2, which contained a combination of the deletions present in EDIC2-0613, EDIC2-4150, and EDIC7-2041. This 3.0-kb sequence contained the CAT reporter gene behind the mRNA7 promoter and therefore only 2.2 kb of EAV sequence. EDIC7-DD2 was replicated but could not be rescued. Since the RNAs containing individual deletions (EDIC2-0613, EDIC2-4150, and EDIC7-2041) could be rescued efficiently, we believe that the size rather than the deletion of specific RNA encapsidation signals may have resulted in a packaging defect. Alternatively, the combination of sequences in EDIC7-DD2 might disrupt the structure of a specific encapsidation signal, resulting in a packaging-deficient RNA. The sucrose gradient analysis of EDIC2, EDIC2-DD1, EDIC7, and EDIC7-DD2 virions demonstrated that EDIC7-DD2 is indeed not packaged into virions, whereas EDIC2, EDIC2-DD1, and EDIC7 are.
The smallest EDI derivative that was still rescued efficiently was the 3.8-kb EDIC7-2050 RNA, which again contained the CAT reporter gene behind the mRNA7 promoter. Unfortunately, we were unable to identify a specific domain or sequence required for packaging within the remaining 2.2 kb of EAV sequences present in EDIC7-DD2, since other deletion mutants that expressed CAT during P0 but not during P1 were not obtained.
DI RNAs have proven to be helpful or even essential tools in the molecular studies of virus replication. Alphavirus DI RNAs have been used successfully to identify sequences involved in replication, encapsidation, and sg mRNA synthesis (17, 25, 26, 51). MHV DI RNAs have proved to be very useful to analyze regulation of discontinuous mRNA transcription (49), replication (5–7, 23), and packaging (2, 15, 43). A novel DI RNA complementation system, with one DI genome containing the structural genes and the second DI genome supporting replication and transcription allowed Kim et al. to study coronavirus assembly (21). By introducing the β-glucuronidase reporter gene in a transmissible gastroenteritis coronavirus (TGEV) DI RNA, Izeta et al. (20) were able to detect replication of the TGEV DI RNA at Po and identify sequences required for replication and encapsidation of TGEV DI RNAs. With the generation of the EAV DI-b cDNA clone (pEDI), described in this report, we now have obtained a powerful tool to study EAV replication, transcription, and packaging in more detail.
ACKNOWLEDGMENTS
We thank Guido van Marle, Marieke Tijms, and Peter Bredenbeek for stimulating discussions and Leonie van Dinten for assistance with the EAV infectious cDNA clone pEAV030 and constructs pEAVCAT2 and pEAVCAT7.
R.M. was supported by grant 700-31-020 from the Council for Chemical Sciences of the Netherlands Organisation for Scientific Research.
REFERENCES
- 1.Barclay W, Li Q, Hutchinson G, Moon D, Richardson A, Percy N, Almond J W, Evans D J. Encapsidation studies of poliovirus subgenomic replicons. J Gen Virol. 1998;79:1725–1734. doi: 10.1099/0022-1317-79-7-1725. [DOI] [PubMed] [Google Scholar]
- 2.Bos E C W, Dobbe J C, Luytjes W, Spaan W J M. A subgenomic mRNA transcript of coronavirus MHV-A59 DI RNA is packaged when it contains the DI-packaging signal. J Virol. 1997;71:5684–5687. doi: 10.1128/jvi.71.7.5684-5687.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burgyan J, Rubino L, Russo M. De novo generation of cymbidium ringspot virus defective interfering RNA. J Gen Virol. 1991;72:505–509. doi: 10.1099/0022-1317-72-3-505. [DOI] [PubMed] [Google Scholar]
- 4.Cavanagh D. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch Virol. 1997;142:629–633. [PubMed] [Google Scholar]
- 5.Chang R Y, Brian D A. cis-requirement for N-specific protein sequence in bovine coronavirus defective interfering RNA replication. J Virol. 1996;70:2201–2207. doi: 10.1128/jvi.70.4.2201-2207.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang R Y, Hofmann M A, Sethna P B, Brian D A. A cis-acting function for the coronavirus leader in defective interfering RNA replication. J Virol. 1994;68:8223–8231. doi: 10.1128/jvi.68.12.8223-8231.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Groot R J, van der Most R G, Spaan W J M. The fitness of defective interfering murine coronavirus DI-a and its derivatives is decreased by nonsense and frameshift mutations. J Virol. 1992;66:5898–5905. doi: 10.1128/jvi.66.10.5898-5905.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.den Boon J A, Snijder E J, Chirnside E D, de Vries A A F, Horzinek M C, Spaan W J M. Equine arteritis virus is not a togavirus but belongs to the coronaviruslike superfamily. J Virol. 1991;65:2910–2920. doi: 10.1128/jvi.65.6.2910-2920.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.den Boon J A, Spaan W J M, Snijder E J. Equine arteritis virus subgenomic RNA transcription: UV inactivation and translation inhibition studies. Virology. 1995;213:364–372. doi: 10.1006/viro.1995.0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Derdeyn C A, Frey T K. Characterization of defective interfering RNAs of rubella virus generated during serial undiluted passage. Virology. 1995;206:216–226. doi: 10.1016/S0042-6822(95)80036-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Vries A A F, Chirnside E D, Bredenbeek P J, Gravestein L A, Horzinek M C, Spaan W J M. All subgenomic mRNAs of equine arteritis virus contain a common leader sequence. Nucleic Acids Res. 1990;18:3241–3247. doi: 10.1093/nar/18.11.3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Vries A A F, Chirnside E D, Horzinek M C, Rottier P J M. Structural proteins of equine arteritis virus. J Virol. 1992;66:6294–6303. doi: 10.1128/jvi.66.11.6294-6303.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Vries A A F, Horzinek M C, Rottier P J M, de Groot R J. The genome organization of the Nidovirales: similarities and differences between arteri-, toro-, and coronaviruses. Semin Virol. 1997;8:33–47. doi: 10.1006/smvy.1997.0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doll E R, Bryans J T, McCollum W H, Crowe M E W. Isolation of a filterable agent causing arteritis of horses and abortion by mares. Its differentiation from the equine abortion (influenza) virus. Cornell Vet. 1957;47:3–41. [PubMed] [Google Scholar]
- 15.Fosmire J A, Hwang K, Makino S. Identification and characterization of a coronavirus packaging signal. J Virol. 1992;66:3522–3530. doi: 10.1128/jvi.66.6.3522-3530.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Godeny E K, Chen L, Kumar S N, Methven S L, Koonin E V, Brinton M A. Complete genomic sequence and phylogenetic analysis of the lactate dehydrogenase-elevating virus (LDV) Virology. 1993;194:585–596. doi: 10.1006/viro.1993.1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grakoui A, Levis R, Raju R, Huang H V, Rice C M. A cis-acting mutation in the Sindbis virus junction region which affects subgenomic RNA synthesis. J Virol. 1989;63:5216–5227. doi: 10.1128/jvi.63.12.5216-5227.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horzinek M C, Maess J, Laufs R. Studies on the substructure of togaviruses. II. Analysis of equine arteritis, rubella, bovine viral diarrhea, and hog cholera viruses. Arch Gesamte Virusforsch. 1971;33:306–318. [PubMed] [Google Scholar]
- 19.Hwang Y-K, Brinton M A. A 68-nucleotide sequence within the 3′ noncoding region of simian hemorrhagic fever virus negative-strand RNA binds to four MA104 cell protein. J Virol. 1998;72:4341–4351. doi: 10.1128/jvi.72.5.4341-4351.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Izeta A, Smerdou C, Alonso S, Penzes Z, Mendez A, Plana-Duran J, Enjuanes L. Replication and packaging of transmissible gastroenteritis coronavirus-derived synthetic minigenomes. J Virol. 1999;73:1535–1545. doi: 10.1128/jvi.73.2.1535-1545.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim K H, Narayanan K, Makino S. Assembled coronavirus from complementation of two defective interfering RNAs. J Virol. 1997;71:3922–3931. doi: 10.1128/jvi.71.5.3922-3931.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim Y N, Jeong Y S, Makino S. Analysis of cis-acting sequences essential for coronavirus defective interfering RNA replication. Virology. 1993;197:53–63. doi: 10.1006/viro.1993.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim Y N, Makino S. Characterization of a murine coronavirus defective interfering RNA internal cis-acting replication signal. J Virol. 1995;69:4963–4971. doi: 10.1128/jvi.69.8.4963-4971.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lai M M C, Cavanagh D. The molecular biology of coronaviruses. Adv Virus Res. 1997;48:1–100. doi: 10.1016/S0065-3527(08)60286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levis R, Schlesinger S, Huang H V. Promoter for Sindbis virus RNA-dependent subgenomic RNA transcription. J Virol. 1990;64:1726–1733. doi: 10.1128/jvi.64.4.1726-1733.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levis R, Weiss B G, Tsiang M, Huang H V, Schlesinger S. Deletion mapping of Sindbis virus DI RNAs derived from cDNAs defines the sequences essential for replication and packaging. Cell. 1986;44:137–145. doi: 10.1016/0092-8674(86)90492-7. [DOI] [PubMed] [Google Scholar]
- 27.Liao C L, Lai M M C. Requirement of the 5′-end genomic sequence as an upstream cis-acting element for coronavirus subgenomic mRNA transcription. J Virol. 1994;68:4727–4737. doi: 10.1128/jvi.68.8.4727-4737.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin Y J, Lai M M C. Deletion mapping of a mouse hepatitis virus defective interfering RNA reveals the requirement of an internal and discontiguous sequence for replication. J Virol. 1993;67:6110–6118. doi: 10.1128/jvi.67.10.6110-6118.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin Y J, Liao C L, Lai M M C. Identification of the cis-acting signal for minus-strand RNA synthesis of a murine coronavirus: implications for the role of minus-strand RNA in RNA replication and transcription. J Virol. 1994;68:8131–8140. doi: 10.1128/jvi.68.12.8131-8140.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maess J, Reczko E, Bohm H O. Equine arteritis virus: multiplication in BHK 21-cells buoyant density and electron microscopical demonstration. Arch Gesamte Virusforsch. 1970;30:47–58. [PubMed] [Google Scholar]
- 31.Makino S, Shieh C K, Soe L H, Baker S C, Lai M M C. Primary structure and translation of a defective interfering RNA of murine coronavirus. Virology. 1988;166:1–11. doi: 10.1016/0042-6822(88)90526-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meinkoth J, Wahl G. Hybridization of nucleic acids immobilized on solid supports. Anal Biochem. 1984;138:267–284. doi: 10.1016/0003-2697(84)90808-x. [DOI] [PubMed] [Google Scholar]
- 33.Mendez A, Smerdou C, Izeta A, Gebauer F, Enjuanes L. Molecular characterization of transmissible gastroenteritis coronavirus defective interfering genomes: packaging and heterogeneity. Virology. 1996;217:495–507. doi: 10.1006/viro.1996.0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Migliaccio G, Castagnola P, Leone A, Cerasuolo A, Bonatti S. mRNA activity of a Sindbis virus defective interfering RNA. J Virol. 1985;55:877–880. doi: 10.1128/jvi.55.3.877-880.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Molenkamp R, Spaan W J M. Identification of a specific interaction between the coronavirus mouse hepatitis virus A59 nucleocapsid protein and packaging signal. Virology. 1997;239:78–86. doi: 10.1006/viro.1997.8867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Penzes Z, Wroe C, Brown T D K, Britton P, Cavanagh D. Replication and packaging of coronavirus infectious bronchitus virus defective RNAs lacking a long open reading frame. J Virol. 1996;70:8660–8668. doi: 10.1128/jvi.70.12.8660-8668.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1989. , N.Y. [Google Scholar]
- 38.Snijder E J, Meulenberg J J M. The molecular biology of arteriviruses. J Gen Virol. 1998;79:961–979. doi: 10.1099/0022-1317-79-5-961. [DOI] [PubMed] [Google Scholar]
- 39.Snijder E J, van Tol H, Pedersen K W, Raamsman M J B, de Vries A A F. Identification of a novel structural protein of arteriviruses. J Virol. 1999;73:6335–6345. doi: 10.1128/jvi.73.8.6335-6345.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Snijder E J, Wassenaar A L M, Spaan W J M. The 5′ end of the equine arteritis virus replicase gene encodes a papainlike cysteine protease. J Virol. 1992;66:7040–7048. doi: 10.1128/jvi.66.12.7040-7048.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Snijder E J, Wassenaar A L M, Spaan W J M. Proteolytic processing of the replicase ORF1a protein of equine arteritis virus. J Virol. 1994;68:5755–5764. doi: 10.1128/jvi.68.9.5755-5764.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spaan W J M, Rottier P J M, Horzinek M C, van der Zeijst B A M. Isolation and identification of virus-specific mRNAs in cells infected with mouse hepatitis virus (MHV-A59) Virology. 1981;108:424–434. doi: 10.1016/0042-6822(81)90449-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van der Most R G, Bredenbeek P J, Spaan W J M. A domain at the 3′ end of the pol gene is essential for encapsidation of coronaviral defective interfering RNAs. J Virol. 1991;65:3219–3226. doi: 10.1128/jvi.65.6.3219-3226.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van der Most R G, Luytjes W, Rutjes S, Spaan W J M. Translation but not the encoded sequence is essential for the efficient propagation of the defective interfering RNAs of the coronavirus mouse hepatitis virus. J Virol. 1995;69:3744–3751. doi: 10.1128/jvi.69.6.3744-3751.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Dinten L C, den Boon J A, Wassenaar A L M, Spaan W J M, Snijder E J. An infectious arterivirus cDNA clone: identification of a replicase point mutation which abolishes discontinuous mRNA transcription. Proc Natl Acad Sci USA. 1997;94:991–996. doi: 10.1073/pnas.94.3.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Dinten L C, Rensen S, Spaan W J M, Gorbalenya A E, Snijder E J. Proteolytic processing of the open reading frame 1b-encoded part of the arterivirus replicase is mediated by the nsp4 serine protease and is essential for virus replication. J Virol. 1999;73:2027–2037. doi: 10.1128/jvi.73.3.2027-2037.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Dinten L C, Wassenaar A L M, Gorbalenya A E, Spaan W J M, Snijder E J. Processing of the equine arteritis virus replicase ORF1b protein: identification of cleavage products containing the putative viral polymerase and helicase domains. J Virol. 1996;70:6625–6633. doi: 10.1128/jvi.70.10.6625-6633.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Marle G, Dobbe J C, Gultyaev A P, Luytjes W, Spaan W J M, Snijder E J. Arterivirus discontinuous mRNA transcription is guided by base-pairing between sense and antisense transcription-regulating sequences. Proc Natl Acad Sci USA. 1999;96:12056–12061. doi: 10.1073/pnas.96.21.12056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van Marle G, Luytjes W, van der Most R G, van der Straaten T, Spaan W J M. Regulation of coronavirus mRNA transcription. J Virol. 1995;69:7851–7856. doi: 10.1128/jvi.69.12.7851-7856.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wassenaar A L M, Spaan W J M, Gorbalenya A E, Snijder E J. Alternative proteolytic processing of the arterivirus ORF1a polyprotein: evidence that nsp2 acts as a cofactor for the nsp4 serine protease. J Virol. 1997;71:9313–9322. doi: 10.1128/jvi.71.12.9313-9322.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weiss B G, Nitschko H, Ghattas I, Wright R, Schlesinger S. Evidence for specificity in the encapsidation of Sindbis virus RNAs. J Virol. 1989;63:5310–5318. doi: 10.1128/jvi.63.12.5310-5318.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.White C L, Thomson M, Dimmock N J. Deletion analysis of a defective interfering Semliki Forest virus RNA genome defines a region in the nsp2 sequence that is required for efficient packaging of the genome into virus particles. J Virol. 1998;72:4320–4326. doi: 10.1128/jvi.72.5.4320-4326.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang X, Lai M M C. A 5′-proximal RNA sequence of murine coronavirus as a potential initiation site for genomic-length mRNA transcription. J Virol. 1996;70:705–711. doi: 10.1128/jvi.70.2.705-711.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao X, Shaw K, Cavanagh D. Presence of subgenomic mRNAs in virions of coronavirus IBV. Virology. 1993;196:172–178. doi: 10.1006/viro.1993.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]