Abstract
Caveolin-1 (Cav1), the core structural and scaffolding protein of caveolae membrane domains, is highly expressed in many retinal cells and is associated with ocular diseases. Cav1 regulates innate immune responses and is implicated in neuroinflammatory and neuroprotective signaling in the retina. We have shown that Cav1 expression in Müller glia accounts for over 70% of all retinal Cav1 expression. However, the proteins interacting with Cav1 in Müller glia are not established. Here, we show that immortalized MIO-M1 Müller glia, like endogenous Müller glia, highly express Cav1. Surprisingly, we found that Cav1 in MIO-M1 cells exists as heat-resistant, high molecular weight complexes that are stable after immunoprecipitation (IP). Mass spectrometric analysis of high molecular weight Cav1 complexes after Cav1 IP revealed an interactome network of intermediate filament, desmosomes, and actin-, and microtubule-based cytoskeleton. These results suggest Cav1 domains in Müller glia act as a scaffolding nexus for the cytoskeleton.
Keywords: Caveolin-1, Caveolae, Müller glia, Immunoprecipitation, Mass spectrometry, Cav1 complexes, Cavin1/PTRF
1. Introduction
Caveolin-1 (Cav1), the signature structural protein of caveolae is known to play a role in cell signaling, lipid metabolism, endocytosis, and mechanotransduction [3, 24, 25]. Cav1 has been associated with ocular neuroinflammation, age-related macular degeneration, diabetic retinopathy, blood-retinal barrier function, and primary open-angle glaucoma [7, 9–11, 30]. It is expressed in a variety of cell types including retinal pigment epithelium (RPE) and choroidal and retinal vascular endothelium [8, 11]. Furthermore, Cav1 is highly expressed in Müller glia [11, 21, 26], and its expression correlates with Müller glia differentiation [8, 23]. We have shown that neuroretinal Cav1 expression overwhelmingly accounts for the majority of Cav1 expression in the retina, with most Cav1 being localized to Müller glia [11]. While Cav1 is highly expressed in Müller glia, its function in these cells is only beginning to be appreciated. Cav1 regulates cytokine secretion and immune cell influx into the retina, as global Cav1 knockout (KO) simultaneously suppresses cytokine secretion and increases immune cell influx into the retina [19]. Neuroretinal deletion of Cav1 suppresses both proinflammatory cytokine secretion and immune cell infiltration into the retina [11], further confirming a role for Müller glial Cav1 in innate immune responses. Cav1 is significantly upregulated in Müller glia in autoimmune uveitis [12]. The role of Cav1 as an immune modulator is likely cell-context dependent as it can either promote or suppress the inflammatory response depending on the cell type examined [19, 31, 32]. Müller glia express toll-like receptors (TLRs), whose activities can be enhanced or suppressed by interaction with Cav1 [22, 31]. Further, we and others have shown Cav1 to be an important regulator of blood-retinal barrier (BRB) function [2, 18, 19, 33].
While Müller glia abundantly express Cav1, it is unclear what proteins interact with Cav1 in these cells. The aim of this study was to identify the Cav1 interactome in MIO-M1 Müller glia by immunoprecipitating Cav1 and analyzing immune complexes by mass spectrometry. We show that Cav1 in MIO-M1 Müller glia exists as high molecular weight aggregates, which are resistant to heating in reducing SDS-PAGE buffer. Mass spectrometric analysis of Cav1 complexes revealed a network of cytoskeletal proteins that interact with Cav1.
2. Materials and Methods
2.1. Cell Lines and Culture Conditions
Immortalized MIO-M1 Müller glia were cultured in DMEM (1X) + GlutaMax™-I (ThermoFisher Scientific) supplemented with 10% fetal bovine serum (FBS), and 1% penicillin-streptomycin. Prostate cancer cells (PC3s) were cultured in F-12K medium (ATCC Cat#: 30–2004). Retinal microvascular endothelial cells (RMECs) were cultured in Endothelial Cell Basal Medium-2 (Lonza Cat#: CC-3156), supplemented with 2% FBS, human fibroblast growth factor, vascular endothelial growth factor, insulin-like growth factor-1, ascorbic acid, gentamicin-amphotericin B hydroxycortisone, and human endothelial growth factor. Cells were maintained in a humidified atmosphere of 5% CO2, at 37 °C.
2.2. Western Blotting
Cells were lysed in buffer containing 120 mM octylglucoside, 150 mM NaCl, 10 mM Tris–HCl pH 7.4, 0.5 mM EDTA, 0.1% Triton X-100 and protease inhibitor cocktail. Lysates were cleared by centrifugation and protein concentration was determined using a BCA reagent (ThermoFisher Scientific). Equal amounts of proteins were separated by reducing SDS-PAGE and were transferred to nitrocellulose membranes. Membranes were blocked for 1 h in 5% BSA and were probed with primary antibodies of choice: rabbit anti-Cav1 (Cell Signaling Technology, cat. #3267, 1:1000), rabbit anti-PTRF (Abcam, 1:1000) and mouse anti-β-actin (Abcam, 1:5000). Primary antibodies were detected using Horseradish peroxidase (HRP)-conjugated secondary antibodies. To visualize protein bands after SDS-PAGE electrophoresis, gels were stained for 1 h with SimplyBlue™ Safestain (Thermofisher Scientific). Western blot images were captured using the In Vivo F-Pro imaging system.
2.3. Immunoprecipitation
Immunoprecipitation was performed using the Dynabeads™ Protein G Immunoprecipitation Kit (ThermoFisher Scientific) according to the manufacturer’s instructions. Briefly, Cav1 primary antibody (Cell Signaling Technology, cat. #3267) was conjugated to magnetic beads for 20 min at room temperature (RT). Then, the beads-antibody conjugate was incubated with equal amounts of protein lysates for 20 min at RT. After several rounds of washing, beads were resuspended in Laemmli buffer and the immunoprecipitates were separated by reducing SDS-PAGE without heating. The gel was stained for 1 h using SimplyBlue™ SafeStain and washed several times with water. Visible Cav1 complexes were excised and used for mass spectrometry analysis (Fig. 1). A portion of the immune complexes was boiled in Laemmli buffer, separated by reducing SDS-PAGE, and transferred to nitrocellulose membranes for Western blotting.
Fig. 1.
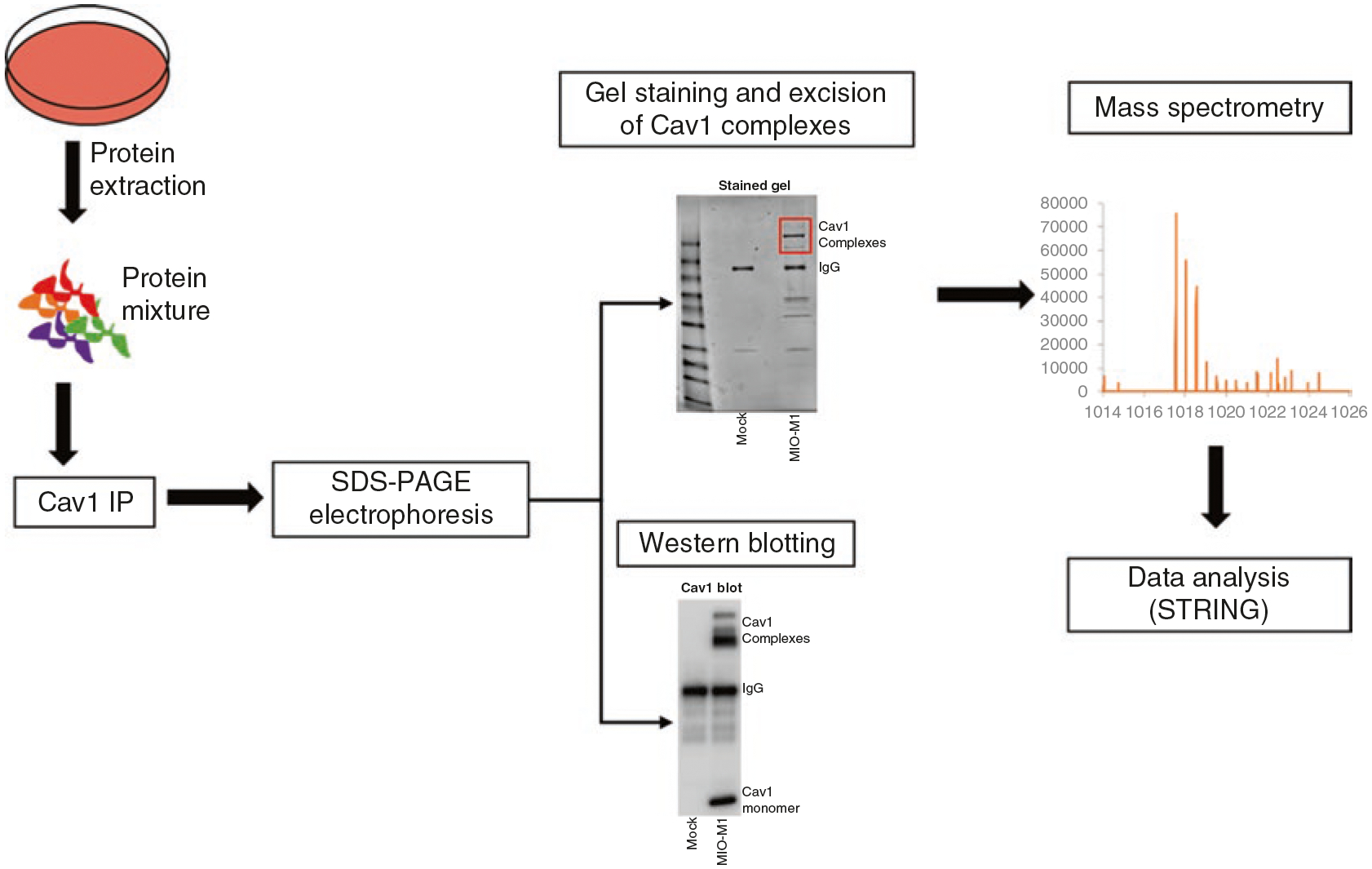
Schematic overview of Cav1 immunoprecipitation and mass spectrometry workflow. Proteins were extracted from untreated MIO-M1 Müller glia and were immunoprecipitated using Cav1 primary antibodies. Cav1 immunoprecipitates were separated by reducing SDS-PAGE without heating. Gels were stained for 1 h and visible Cav1 complexes were excised and analyzed by mass spectrometry. A portion of the immunoprecipitate was transferred to nitrocellulose membranes to evaluate interactions by Western blotting
2.4. Mass Spectrometry
Cav1 immunoprecipitates were separated by SDS-PAGE, gel-stained, and visible high molecular weight Cav1 complexes excised for mass spectrometry (Fig. 2d). Proteins were subjected to the FASP (filter-aided sample preparation) protocol [34] and digested overnight with Sequencing Grade Modified Trypsin (Promega, V5111) at 37 °C in 40 mM NH4HCO3. Peptides were desalted, concentrated, and loaded onto C18 sequencing columns (Acclaim™ PepMap™ 100 C18, ThermoFisher). Peptide elution was performed using a 90-min acetonitrile gradient for label-free quantification. Eluted peptides were analyzed by LC-MS/MS analysis using a Thermo Lumos Fusion tribrid Orbitrap mass spectrometer, coupled to an Ultimate 3000 RSLC nano ultra-high-performance liquid chromatography (UHPLC) system. Proteins were identified by Proteome Discoverer 2.4, with SEQUEST as the search engine. Protein identification required the detection of at least two peptides per protein. STRING open-source software was used to identify protein networks.
Fig. 2.
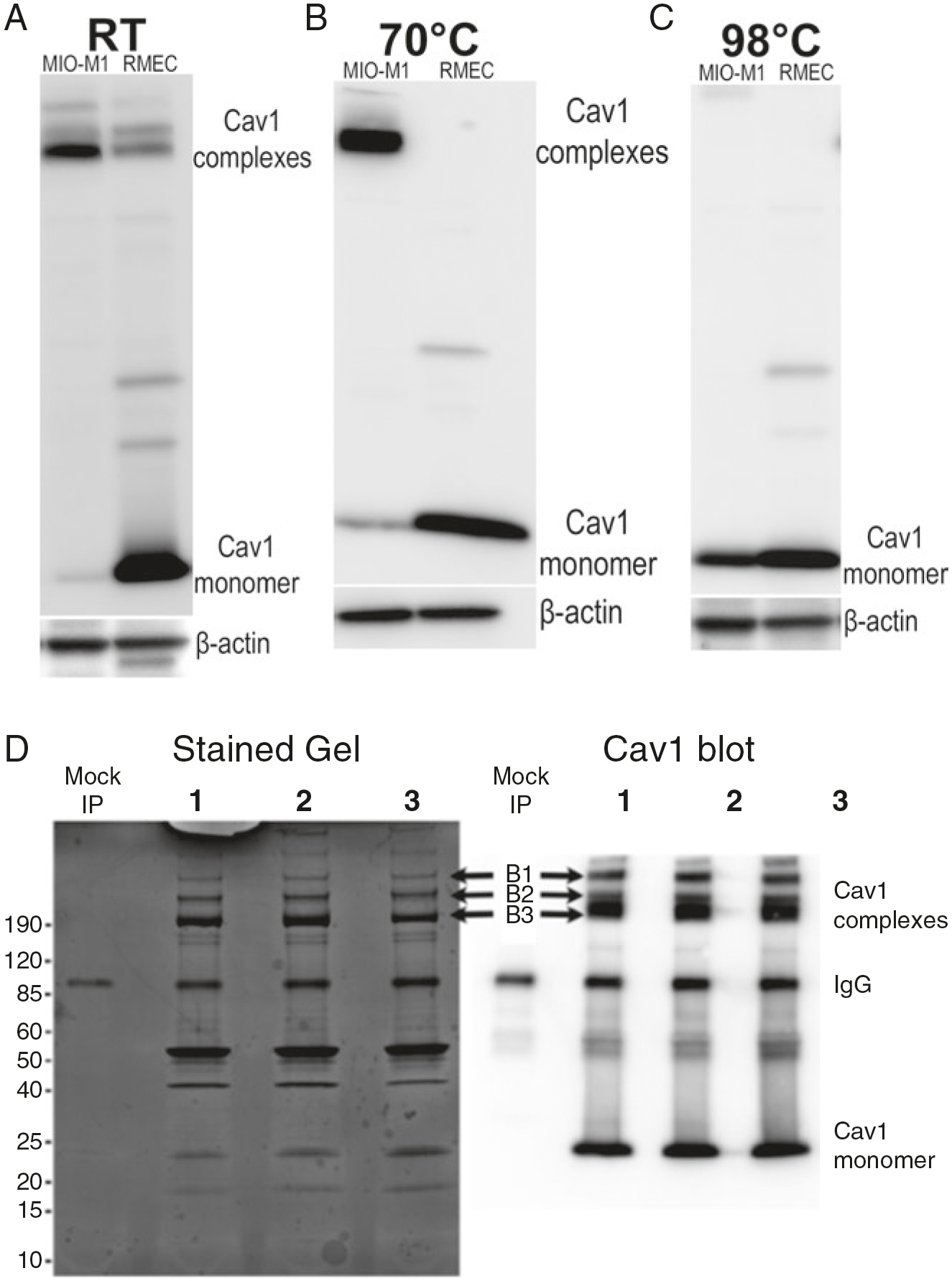
Cav1 in MIO-M1 cells exists in heat-resistant, high molecular weight complexes. (a) Representative Western blots showing expression of Cav1 in MIO-M1 and RMEC cells. MIO-M1 cells, like endogenous Müller glia, abundantly expresses Cav1. Interestingly, Cav1 in these cells exists in high molecular weight complexes. (b) Cav1 complexes are resistant to heating at 70 °C for 10 min in reducing SDS-PAGE. On the contrary, Cav1 in RMEC cells was mostly monomeric both at RT and after heating at 70 °C for 10 min. (c) Representative Western blots showing that Cav1 migrates as a monomer in reducing SDS-PAGE buffer only after rigorous heating at 98 °C for 10 min. (d) Stained gel and representative Western blot showing stable Cav1 complexes after Cav1 immunoprecipitation. Bands from the stained gel (B1, B2, and B3 indicated by arrows) were excised and analyzed by mass spectrometry
3. Results
3.1. Cav1 in MIO-M1 Müller Glia Exist as High Molecular Weight Complexes
To determine the protein composition of Cav1 complexes, we first evaluated the expression of Cav1 in authenticated MIO-Müller glia and compared the expression to retinal microvascular endothelial cells (RMEC), which also abundantly express Cav1 [29]. Our results show that MIO-M1 cells, like endogenous Müller glia, abundantly express Cav1 (Fig. 2). To our surprise, we observed that the majority of Cav1 in MIO-M1 cells exist as high molecular weight complexes (Fig. 2a) which are resistant to heating in reducing SDS-PAGE buffer (Fig. 2b). However, upon heating at 98 °C for 10 min, Cav1 complexes dissociate and migrate as monomers on reducing SDS-PAGE (Fig. 2c). High molecular weight Cav1 complexes in MIO-M1 cells remain relatively stable after Cav1 immunoprecipitation (Fig. 2d). On the contrary, Cav1 in RMECs migrates predominantly as monomers on SDS-PAGE gels without heating. While most Cav1 complexes in MIO-M1s remain stable after heating at 70 °C for 10 min, the small fraction of aggregated Cav1 in RMECs dissociates to monomers after heating at 70 °C. These data suggest that different proteins may be interacting with and stabilizing Cav1 complexes in MIO-M1 cells.
3.2. Proteins Associated with Cav1 Complexes Are Involved with the Cell Cytoskeleton
Next, we were interested in identifying the proteins that interact with Cav1 complexes in MIO-M1 cells. To identify the protein composition of Cav1 complexes, we immunoprecipitated Cav1 from MIO-M1 cells and analyzed the high molecular weight complexes by mass spectrometry. We analyzed the same complexes from three replicate samples (Fig. 2d). Thirty-three proteins were found to associate with Cav1 complexes after mass spectrometry. Interestingly, the majority of these proteins including β-actin (ACTB), myosin, vimentin (VIM), plectin (PLEC), and nestin (NES) have been shown to play a role in the cytoskeleton and cell-cell junction structure [4].
4. Discussion
In this study, we sought to identify the protein composition of Cav1 complexes in MIO-M1 cells. We show for the first time that Cav1 in MIO-M1 cells exists as heat-resistant, high molecular weight aggregates, which interact with important cytoskeletal proteins. Cav1 is the major protein component of caveolae [5, 6, 27], flask-shaped plasma membrane invaginations whose formation requires another protein called Cavin1 or PTRF (Polymerase I and Transcript Release Factor) [14, 20]. It is currently unclear why Cav1 exist in this aggregated form in MIO-M1 cells, as opposed to the monomeric form in RMEC cells. However, we speculate that the absence of Cavin1/PTRF expression in MIO-M1 cells may provide an explanation for this phenomenon. Cavin1/PTRF stabilizes Cav1 in multiple tissues, as Cavin1/PTRF deficiency downregulates Cav1 protein expression [20]. However, Cav1 is stably expressed in prostate cancer (PC3) cells without Cavin1/PTRF [1, 14, 15] in functional domains described as “Cav1 scaffolds” [16, 17]. Further, multiple studies have shown that approximately 150 Cav1 molecules are required per caveola formed [25]. However, during caveolae biogenesis, Cav1 form oligomers of 12–16 Cav1 molecules, which associate with lipid rafts in the Golgi and adopt detergent-resistant properties [13, 28], similar to Cav1 on the plasma membrane. Thus, it is intriguing to speculate that in the absence of Cavin1/PTRF in MIO-M1 Müller glial cells, these Cav1 high molecular weight oligomers represent non-caveolar Cav1 scaffolds previously described [16, 17]. Therefore, the expression of Cavin1/PTRF in MIO-M1 cells likely provides a mechanism to biochemically resolve Cav1 scaffolds (Fig. 3).
Fig. 3.
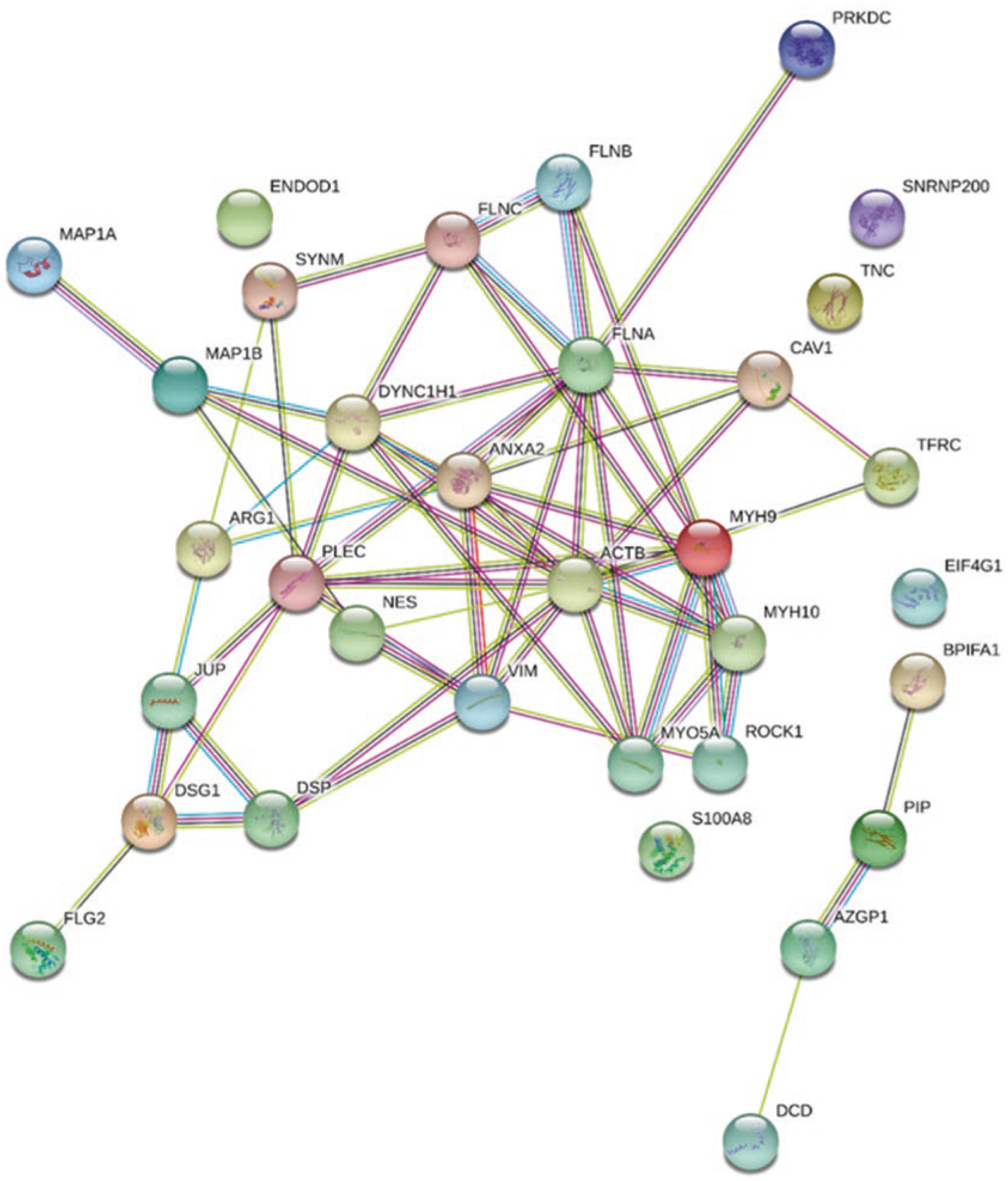
Proteins that interact with Cav1 complexes are associated with the cell cytoskeleton. Cav1 protein-protein interaction by STRING analysis. Cav1 complexes were analyzed by mass spectrometry and STRING open-source database was used to identify protein-protein interactions. A total of 33 proteins were found to interact with Cav1 complexes, most of which play a role in the cell cytoskeletal architecture and include β-actin (ACTB), myosin (MYO5A), vimentin (VIM) plectin (PLEC), and nectin (NEC)
Acknowledgments
This work was supported by NIH Grants R01EY019494 (MHE) and NEI Core Grant P30EY021725, by Presbyterian Health Foundation Research Scholar Award to Eric Enyong, and by an unrestricted grant to the OUHSC Department of Ophthalmology from Research to Prevent Blindness, Inc. Virginie Sjoelund was supported in part by the National Institute of General Medical Sciences of the National Institutes of Health under award number P20GM103447. We thank the Laboratory for Molecular Biology and Cytometry Research at OUHSC for the use of the Core Facility, which provided the proteomics services.
Contributor Information
Eric N. Enyong, Department of Physiology, Dean A. McGee Eye Institute, University of Oklahoma Health Sciences Center, Oklahoma City, OK, USA Department of Ophthalmology, Dean A. McGee Eye Institute, University of Oklahoma Health Sciences Center, Oklahoma City, OK, USA.
Jami Gurley, Department of Physiology, Dean A. McGee Eye Institute, University of Oklahoma Health Sciences Center, Oklahoma City, OK, USA; Department of Ophthalmology, Dean A. McGee Eye Institute, University of Oklahoma Health Sciences Center, Oklahoma City, OK, USA.
Virginie Sjoelung, Department of Cell Biology, University of Oklahoma Health Sciences Center, Oklahoma City, OK, USA.
Michael H. Elliott, Department of Physiology, Dean A. McGee Eye Institute, University of Oklahoma Health Sciences Center, Oklahoma City, OK, USA Department of Ophthalmology, Dean A. McGee Eye Institute, University of Oklahoma Health Sciences Center, Oklahoma City, OK, USA.
References
- 1.Aung CS, Hill MM, Bastiani M, Parton RG, Parat MO. PTRF-cavin-1 expression decreases the migration of PC3 prostate cancer cells: role of matrix metalloprotease 9. Eur J Cell Biol. 2011;90:136–42. [DOI] [PubMed] [Google Scholar]
- 2.Chow BW, Gu C. Gradual suppression of transcytosis governs functional blood-retinal barrier formation. Neuron. 2017;93:1325–1333 e1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cohen AW, Hnasko R, Schubert W, Lisanti MP. Role of caveolae and caveolins in health and disease. Physiol Rev. 2004;84:1341–79. [DOI] [PubMed] [Google Scholar]
- 4.Fletcher DA, Mullins RD. Cell mechanics and the cytoskeleton. Nature. 2010;463:485–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Glenney JR Jr. Tyrosine phosphorylation of a 22-kDa protein is correlated with transformation by Rous sarcoma virus. J Biol Chem. 1989;264:20163–6. [PubMed] [Google Scholar]
- 6.Glenney JR Jr, Soppet D. Sequence and expression of caveolin, a protein component of caveolae plasma membrane domains phosphorylated on tyrosine in Rous sarcoma virus-transformed fibroblasts. Proc Natl Acad Sci U S A. 1992;89:10517–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gu X, Fliesler SJ, Zhao YY, Stallcup WB, Cohen AW, Elliott MH. Loss of caveolin-1 causes blood-retinal barrier breakdown, venous enlargement, and mural cell alteration. Am J Pathol. 2014a;184:541–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gu X, Reagan A, Yen A, Bhatti F, Cohen AW, Elliott MH. Spatial and temporal localization of caveolin-1 protein in the developing retina. Adv Exp Med Biol. 2014b;801:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gu X, Reagan AM, McClellan ME, Elliott MH. Caveolins and caveolae in ocular physiology and pathophysiology. Prog Retin Eye Res. 2017;56:84–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gurley JM, Elliott MH. The role of caveolin-1 in retinal inflammation. Adv Exp Med Biol. 2019;1185:169–73. [DOI] [PubMed] [Google Scholar]
- 11.Gurley JM, Gmyrek GB, McClellan ME, Hargis EA, Hauck SM, Dozmorov MG, Wren JD, Carr DJJ, Elliott MH. Neuroretinal-derived caveolin-1 promotes endotoxin-induced inflammation in the murine retina. Invest Ophthalmol Vis Sci. 2020;61:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hauck SM, Dietter J, Kramer RL, Hofmaier F, Zipplies JK, Amann B, Feuchtinger A, Deeg CA, Ueffing M. Deciphering membrane-associated molecular processes in target tissue of autoimmune uveitis by label-free quantitative mass spectrometry. Mol Cell Proteom. 2010;9:2292–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hayer A, Stoeber M, Bissig C, Helenius A. Biogenesis of caveolae: stepwise assembly of large caveolin and cavin complexes. Traffic. 2010;11:361–82. [DOI] [PubMed] [Google Scholar]
- 14.Hill MM, Bastiani M, Luetterforst R, Kirkham M, Kirkham A, Nixon SJ, Walser P, Abankwa D, Oorschot VM, Martin S, Hancock JF, Parton RG. PTRF-Cavin, a conserved cytoplasmic protein required for caveola formation and function. Cell. 2008;132:113–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Inder KL, Zheng YZ, Davis MJ, Moon H, Loo D, Nguyen H, Clements JA, Parton RG, Foster LJ, Hill MM. Expression of PTRF in PC-3 Cells modulates cholesterol dynamics and the actin cytoskeleton impacting secretion pathways. Mol Cell Proteom. 2012;11:M111012245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khater IM, Aroca-Ouellette ST, Meng F, Nabi IR, Hamarneh G. Caveolae and scaffold detection from single molecule localization microscopy data using deep learning. PLoS One. 2019a;14:e0211659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khater IM, Liu Q, Chou KC, Hamarneh G, Nabi IR. Super-resolution modularity analysis shows polyhedral caveolin-1 oligomers combine to form scaffolds and caveolae. Sci Rep. 2019b;9:9888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klaassen I, Hughes JM, Vogels IM, Schalkwijk CG, Van Noorden CJ, Schlingemann RO. Altered expression of genes related to blood-retina barrier disruption in streptozotocin-induced diabetes. Exp Eye Res. 2009;89:4–15. [DOI] [PubMed] [Google Scholar]
- 19.Li X, Gu X, Boyce TM, Zheng M, Reagan AM, Qi H, Mandal N, Cohen AW, Callegan MC, Carr DJ, Elliott MH. Caveolin-1 increases proinflammatory chemoattractants and blood-retinal barrier breakdown but decreases leukocyte recruitment in inflammation. Invest Ophthalmol Vis Sci. 2014;55:6224–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu L, Brown D, McKee M, Lebrasseur NK, Yang D, Albrecht KH, Ravid K, Pilch PF. Deletion of Cavin/PTRF causes global loss of caveolae, dyslipidemia, and glucose intolerance. Cell Metab. 2008;8:310–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, Trombetta JJ, Weitz DA, Sanes JR, Shalek AK, Regev A, McCarroll SA. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161:1202–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mirza MK, Yuan J, Gao XP, Garrean S, Brovkovych V, Malik AB, Tiruppathi C, Zhao YY. Caveolin-1 deficiency dampens Toll-like receptor 4 signaling through eNOS activation. Am J Pathol. 2010;176:2344–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nelson BR, Ueki Y, Reardon S, Karl MO, Georgi S, Hartman BH, Lamba DA, Reh TA. Genome-wide analysis of Muller glial differentiation reveals a requirement for Notch signaling in postmitotic cells to maintain the glial fate. PLoS One. 2011;6:e22817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parton RG. Caveolae: structure, function, and relationship to disease. Annu Rev Cell Dev Biol. 2018;34:111–36. [DOI] [PubMed] [Google Scholar]
- 25.Parton RG, del Pozo MA. Caveolae as plasma membrane sensors, protectors and organizers. Nat Rev Mol Cell Biol. 2013;14:98–112. [DOI] [PubMed] [Google Scholar]
- 26.Roesch K, Jadhav AP, Trimarchi JM, Stadler MB, Roska B, Sun BB, Cepko CL. The transcriptome of retinal Muller glial cells. J Comp Neurol. 2008;509:225–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rothberg KG, Heuser JE, Donzell WC, Ying YS, Glenney JR, Anderson RG. Caveolin, a protein component of caveolae membrane coats. Cell. 1992;68:673–82. [DOI] [PubMed] [Google Scholar]
- 28.Sargiacomo M, Scherer PE, Tang Z, Kubler E, Song KS, Sanders MC, Lisanti MP. Oligomeric structure of caveolin: implications for caveolae membrane organization. Proc Natl Acad Sci U S A. 1995;92:9407–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stitt AW, Burke GA, Chen F, McMullen CB, Vlassara H. Advanced glycation end-product receptor interactions on microvascular cells occur within caveolin-rich membrane domains. FASEB J. 2000;14:2390–2. [DOI] [PubMed] [Google Scholar]
- 30.Thorleifsson G, Walters GB, Hewitt AW, Masson G, Helgason A, DeWan A, Sigurdsson A, Jonasdottir A, Gudjonsson SA, Magnusson KP, Stefansson H, Lam DS, Tam PO, Gudmundsdottir GJ, Southgate L, Burdon KP, Gottfredsdottir MS, Aldred MA, Mitchell P, St Clair D, Collier DA, Tang N, Sveinsson O, Macgregor S, Martin NG, Cree AJ, Gibson J, Macleod A, Jacob A, Ennis S, Young TL, Chan JC, Karwatowski WS, Hammond CJ, Thordarson K, Zhang M, Wadelius C, Lotery AJ, Trembath RC, Pang CP, Hoh J, Craig JE, Kong A, Mackey DA, Jonasson F, Thorsteinsdottir U, Stefansson K. Common variants near CAV1 and CAV2 are associated with primary open-angle glaucoma. Nat Genet. 2010;42:906–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang XM, Kim HP, Nakahira K, Ryter SW, Choi AM. The heme oxygenase-1/carbon monoxide pathway suppresses TLR4 signaling by regulating the interaction of TLR4 with caveolin-1. J Immunol. 2009;182:3809–18. [DOI] [PubMed] [Google Scholar]
- 32.Wang XM, Kim HP, Song R, Choi AM. Caveolin-1 confers antiinflammatory effects in murine macrophages via the MKK3/p38 MAPK pathway. Am J Respir Cell Mol Biol. 2006;34:434–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Z, Liu CH, Huang S, Fu Z, Tomita Y, Britton WR, Cho SS, Chen CT, Sun Y, Ma JX, He X, Chen J. Wnt signaling activates MFSD2A to suppress vascular endothelial transcytosis and maintain blood-retinal barrier. Sci Adv. 2020;6:eaba7457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wisniewski JR. Filter-aided sample preparation for proteome analysis. Methods Mol Biol. 2018;1841:3–10. [DOI] [PubMed] [Google Scholar]


