Abstract
Human immunodeficiency virus type 1 (HIV-1) patient isolates and molecular clones were used to analyze the determinants responsible for human CD4+ thymocyte depletion in SCID-hu mice. Non-syncytium-inducing, R5 or R3R5 HIV-1 isolates from asymptomatic infected people showed little or no human CD4+ thymocyte depletion in SCID-hu mice, while syncytium-inducing (SI), R5X4 or R3R5X4 HIV-1 isolates from the same individuals, isolated just prior to the onset of AIDS, rapidly and efficiently eliminated CD4-bearing human thymocytes. We have mapped the ability of one SI HIV-1 isolate to eliminate CD4+ human cells in SCID-hu mice to a region of the env gene including the three most amino-terminal variable regions (V1 to V3). We find that for all of the HIV-1 isolates that we studied, a nonlinear relationship exists between viral replication and the depletion of CD4+ cells. This relationship can best be described mathematically with a Hill-type plot indicating that a threshold level of viral replication, at which cytopathic effects begin to be seen, exists for HIV-1 infection of thymus/liver grafts in SCID-hu mice. This threshold level is 1 copy of viral DNA for every 11 cells (95% confidence interval = 1 copy of HIV-1 per 67 cells to 1 copy per 4 cells). Furthermore, while SI viruses more frequently achieve this level of replication, replication above this threshold level correlates best with cytopathic effects in this model system. We used GHOST cells to map the coreceptor specificity and relative entry efficiency of these early- and late-stage patient isolates of HIV-1. Our studies show that coreceptor specificity and entry efficiency are critical determinants of HIV-1 pathogenesis in vivo.
Progression to AIDS in human immunodeficiency virus type 1 (HIV-1)-infected individuals follows a long but highly variable asymptomatic period, suggesting that viral and/or host heterogeneity contribute to the development of disease. Despite the genetic diversity of HIV-1 isolates, few differences among HIV-1 strains have been linked to their pathogenic potential. Among these, variation in the region of the env gene which encodes the third variable region (V3) of gp120 has been most consistently correlated with HIV-1 pathogenesis in infected individuals (11, 12, 39–41). The sequence of the V3 loop determines whether HIV-1 is syncytium inducing (SI) in MT-2 cells. The SI phenotype is associated with advanced HIV-1 disease and with poor prognosis in patients and with increased cytopathic effects in thymic organ culture, hu-PBL-SCID mice, and SCID-hu mice (11, 12, 19, 25, 32, 33, 39–41). This region of the HIV-1 env gene has been shown to determine cellular tropism and replication properties as well as SI phenotype (7, 20, 30). Recently it has been shown that the NSI and SI phenotypes result from the inability and ability, respectively, of HIV-1 strains to utilize CXCR4 in entering cells (15, 17, 37). Furthermore, recent work has shown that variations in the V3 loop of gp120 determine the ability of HIV-1 to productively interact with one or more cell surface chemokine receptor during viral entry or viral glycoprotein-mediated cell-cell fusion (9, 10, 34).
For this study we used viral isolates from three individuals previously characterized as rapid progressors and one long-term nonprogressor (11). Each of the three rapid progressors, patients A, B, and C, went from normal levels of CD4+ peripheral blood T cells to less than 200 such cells per microliter of plasma, an AIDS-defining condition, in less than 2 years. In each case the rapid decline in CD4+ T cells was preceded by a phenotypic shift, from non-SI (NSI) to SI on MT-2 cells, in HIV-1 recovered from the patients. In contrast, the long-term nonprogressor, patient D, maintained a stable CD4+ cell count for over 7 years, and viral isolates from this patient were always NSI on MT-2 cells. We assayed the replication and thymocyte pathogenesis in the SCID-hu mouse of sequential isolates from these patients. For each of the three rapid progressors, we studied an early isolate obtained during the asymptomatic phase and a later isolate taken from the first time point at which SI virus was detected, just prior to the rapid decline in CD4+ T cells in each patient. Two isolates from the long-term nonprogressor, separated by 15 months in their time of isolation, were also studied. As controls, we used molecular clones of HIV-1 differing in tropism and NSI/SI phenotype. We have also constructed and assayed chimeric viruses bearing the V1 to V3 gp120-encoding region from four of the patient isolates described above.
SCID-hu mice, created by surgical implantation of human fetal thymus and liver tissue, develop a conjoint thymus/liver graft which in large part has the morphology of normal human thymus with small islands of fetal liver-derived hematopoietic tissue (29). Previous studies have shown that infection of human thymus/liver grafts in SCID-hu mice provides a model of HIV-1 infection in the human (1, 5). The SCID-hu mouse model of HIV-1 infection accurately reflects viral phenotypes seen in vivo. For example, the nef gene has a significant effect on replication and pathogenesis, and late-stage patient isolates are more pathogenic than earlier isolates from the same patients (21, 25). The data presented here confirm and extend previous reports that late-stage SI isolates are pathogenic in SCID-hu mice by genetically mapping a determinant of pathogenesis, showing that coreceptor utilization efficiency is an important pathogenic determinant and demonstrating that HIV-1 replication above a threshold level correlates with pathogenesis in the SCID-hu mouse.
MATERIALS AND METHODS
Construction of ΠSVNX.
The small simian virus 40 replication origin bearing plasmid ΠSVNX was constructed to facilitate bacterial production of plasmid containing the HIV-1 genome and the subsequent production of high titer HIV-1 stocks by transfection. CDM7-CD62L (6) was cut with SphI and PstI, the resulting overhangs were made blunt with T4 DNA polymerase, and the ends were ligated with T4 DNA ligase to delete the CD62L cDNA and the simian virus 40-derived intron and polyadenylation signal. An HIV-1 env DNA fragment flanked by several restriction sites and cut with MluI and XbaI was cloned into this vector. The resulting plasmid was cut with MluI and PstI, the overhangs were made blunt with T4 DNA polymerase, and the ends were ligated with T4 DNA ligase to delete the HIV-1 env gene and the MluI and PstI sites, leaving unique NotI and XbaI sites behind. Finally the M13 origin fragment was removed, since it contained a DraIII site, by cutting with NheI and SacII, making the ends blunt with T4 DNA polymerase, and ligating with T4 DNA ligase. All enzymes were from New England Biolabs, Beverly, Mass., and were used in buffers provided or recommended by the manufacturer.
Construction of recombinant infectious molecular clones of HIV-1.
HIV-1/JR-CSF proviral DNA without flanking cellular DNA was inserted into ΠSVNX in two steps. The HIV-1/JR-CSF long terminal repeat (LTR) was amplified from pYK-JRCSF (27), using a 5′ oligonucleotide primer bearing the restriction site NotI and a 3′ primer bearing XbaI and inserted into ΠSVNX to create ΠSVNX-CSF-LTR. The 9-kb SacI fragment of pYK-JRCSF was then inserted into ΠSVNX-CSF-LTR at the unique SacI site to create ΠSVNX-JRCSF. For the construction of env recombinant HIV-1 genomes based on JR-CSF, ΠSVNX-JRCSF was further modified by removing the 716-bp DraIII-to-BamHI fragment and replacing it with a 1,294-bp DraIII-to-BamHI fragment derived from pNL-Thy1-Δ-Bgl-II to create ΠSVNX-JRCSF-(D3/B stuffer) (35). Patient HIV-1 isolate or biological clone env fragments were isolated by nested PCR, from thymocyte DNA prepared 3 weeks after SCID-hu mouse infection, using a QIAamp blood kit (Qiagen, Valencia, Calif.). The first round of PCR was performed with oligonucleotides homologous to conserved regions of the HIV-1 genome, flanking the env gene. Products of the first round of PCR were used in a second PCR using a 5′ oligonucleotide primer homologous to the region of env including the HindIII site at nucleotide 6575 of the JR-CSF isolate of HIV-1 and a 3′ primer homologous to a region including the BamHI site at nucleotide 7325 of JR-CSF. PCR products were cut with HindIII and BamHI and inserted into pUC19 cut with the same restriction endonucleases. The env fragments were then removed from pUC19 by cleavage with DraIII and BamHI and ligated into ΠSVNX-JRCSF-(D3/B stuffer) which had been cut with the same enzymes. The resulting plasmids were used to create viral stocks.
Preparation and titration of HIV-1 stocks.
Viral stocks were prepared by transfection of COS-M6 cells with molecular clones of HIV-1 carried on the plasmid ΠSVNX or propagation of patient isolates. Cells were transfected by electroporation of 25 to 50 μg of plasmid DNA into 1.6 × 107 COS-M6 cells in 0.8 ml of Iscove's medium with 20% fetal bovine serum, using a Gene Pulser set at 250 V and 960 μF (Bio-Rad Laboratories, Hercules, Calif.). Transfected cells were returned to tissue culture flasks or plates with growth medium; 2 and 3 days later, the medium was harvested and replaced; on the fourth day the medium was again harvested, and the cells were discarded. These viral stocks were titrated by infection of 2-day phytohemagglutinin (PHA)-stimulated peripheral blood mononuclear cells (PBMC) for 24 h followed by harvest of the infected cells, preparation of cellular DNA and quantitative PCR as outlined below. Patient isolates were biologically cloned by limiting dilution of patient plasma which was added to 2-day PHA-stimulated PBMC and propagated for 14 days. Culture supernatant from wells containing the highest dilution of patient plasma at which HIV-1 replication was detected by p24 enzyme-linked immunosorbent assay (ELISA) were expanded by an additional week of culture in 10 to 20 ml of 2-day PHA-stimulated PBMC. These stocks were titrated by limiting dilution in 2-day PHA-stimulated PBMC followed by p24 ELISA.
HIV-1 infections in vitro.
HIV-1 infection of PBMC, MT-2 cells, and GHOST cells (NIH AIDS Research and Reference Reagent Program) was carried out by incubation of viral stocks with cells in the presence of Polybrene (4 or 8 μg/ml; Sigma Chemical, St. Louis, Mo.). For PBMC and MT-2 cells, the incubation was performed in a 15-ml centrifuge tube with gentle rocking at 37°C for 2 h; for GHOST cells, the incubation was done at 37°C overnight in 12-well plates. The plates were seeded the previous day with 2.5 × 104 cells per well, and the virus was added in a volume of 0.5 ml per well.
Preparation and maintenance of SCID-hu mice.
SCID-hu thymus/liver mice were created by implantation of human fetal thymus and liver fragments under the kidney capsule of C.B-17 SCID mice as originally described by McCune and colleagues (29). SCID and SCID-hu mice were maintained in microisolater cages on racks with HEPA-filtered air blown into each cage (Allentown Caging, Allentown, Pa.). The mice were implanted with 1-mm3 pieces of human fetal thymus and liver when they were 6 to 8 weeks old. Tissue at 16 to 24 weeks of gestational age was obtained from Advanced Bioscience Resources (Alameda, Calif.). One piece of fetal thymus and two of fetal liver were inserted under the left kidney capsule of each mouse, using a 16-gauge cancer implant needle set (Popper and Sons, New Hyde Park, N.Y.). The grafts were left undisturbed for 4 to 6 months prior to infection with HIV-1.
Infection of SCID-hu mice with HIV-1 and biopsy of infected grafts.
Mice were anesthetized with ketamine and xylazine (8 and 0.8 μg, respectively, per g of body weight) injected intraperitoneally prior to infection or biopsy. Methoxyfluorane was used if additional anesthesia was necessary, and buprenone was administered to minimize postoperative discomfort for all surgical procedures. Thymus/liver grafts were exteriorized and measured with a caliper. Only grafts larger than or equal to 0.5 cm in diameter were used. Freshly titered HIV-1 stocks were diluted to 4 × 103 or 2 × 104 50% tissue culture infective doses (TCID50) per ml in Iscove's medium with 2% fetal calf serum, and 200 or 1,000 TCID50 was injected directly into the thymus/liver grafts in a volume of 50 μl. SCID-hu mice were biopsied at 3, 6, 9, and 12 weeks postinfection. For each biopsy, the grafts were again exteriorized and one-fourth to one-half of the tissue, depending on the size of the graft, was removed. A single-cell suspension was made by mincing the tissue with two scalpels in Iscove's medium (Life Technologies, Rockville, Md.) supplemented with 2% fetal bovine serum (Omega Scientific, Tarzana, Calif.) and gentamicin (50 μg/ml; Life Technologies). The cells were filtered through 70-μm nylon mesh and transported on ice from the BL2+ mouse facility to the BL3 laboratory.
Flow cytometry.
Cells were washed twice in phosphate-buffered saline (PBS), counted, and aliquoted (5 × 105 cells per well) into 96-well V-bottom plates (Costar, Cambridge, Mass.). Fluorochrome-conjugated monoclonal antibodies (MAbs) were added to each well, and the plates were agitated and incubated 30 to 60 min in the dark at 4°C. MAbs used together were CD7-fluorescein isothiocyanate (FITC), CD4-phycoerythrin (PE) (CalTag, South San Francisco, Calif.), CD8-peridinin chlorophyll protein (PerCP) (BDIS, San Jose, Ca.), CD8-FITC (BDIS), CD4-PE, and CD3-PerCP (BDIS). Following incubation with MAb, the cells were washed twice with 200 μl of PBS, resuspended in 100 μl of PBS–2% formaldehyde, and incubated for 16 h at 4°C in the dark. Samples were diluted with PBS, and 104 cells, discriminated by their 90° and low-angle light scattering properties, were analyzed with a FACScan flow cytometer fitted with a helium-neon laser tuned to emit 488 nm light. Filters appropriate for the fluorochromes were used. CellQuest software was used to collect and analyze the flow cytometric data.
Quantitative PCR.
Genomic DNA was purified using a QIAamp blood kit (Qiagen) from approximately 107 cells from each biopsy or infected PBMC for titration of viral stocks. PCR amplification was performed by an initial denaturation step at 94°C for 2 min followed by 23 cycles of 94°C for 30 s and 65°C for 1 min with primers M667 and AA55, specific for the R/U5 region of the HIV-1 LTR (42), using a model PT-200 thermocycler (MJ Research, Watertown, Mass.). Primers specific for the human β-globin gene were used to detect cellular DNA (42). In each case, one of the two primers used was labeled on the free 5′ phosphate using T4 bacteriophage polynucleotide kinase (New England Biolabs) and [γ-32P]ATP. A standard curve for the number of HIV-1 copies was generated for each PCR with fivefold dilutions of EcoRI-digested ΠSVNX-JRCSF mixed with genomic DNA from 105 PBMC. A standard curve for the number of β-globin copies was generated for each PCR with fivefold dilutions of genomic DNA from PBMC. In both cases, the standard curve was used only for the range of values over which a linear regression gave an r2 value of greater than or equal to 0.98. Radiolabeled PCR products were resolved by electrophoresis on a 6% polyacrylamide–1× Tris-borate-EDTA gel. HIV-1 and β-globin copy numbers were obtained from the standard curve using a model 425 PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.).
Curve-fitting and statistical analysis.
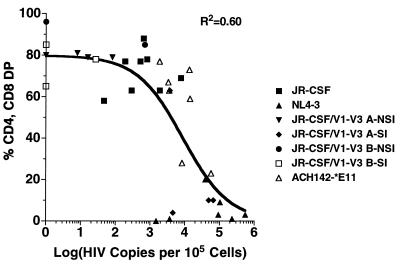
Both a straight line and a sigmoid curve were fit to the data plotted in Fig. 6, showing the relationship between the number of HIV-1 DNA copies per cell and the percentage of CD4 CD8 double-positive (DP) thymocytes, using linear and nonlinear least-squares programs, respectively (24). The sigmoid curve was derived from a modification of the Hill equation as follows: y = y1 − y1[(x/xmed)s/(1 + (x/xmed)s)], where y1 is the percentage CD4 CD8 DP cells when viral load was ≤10 copies/105 cells, xmed is the value of x at the half-maximal value of y, and s is the Hill coefficient which is related to the steepness of the curve at xmed. The variance of fit (ς2) for the straight line and sigmoid curve were calculated as follows: ς2 = Σt=1 (yi − f(xi))2/N−P, where N is the number of data points and P is the number of parameters fit. The 95% confidence intervals were evaluated by a bootstrap procedure (16). Prism software was used to plot the data and sigmoid curve (GraphPad, San Diego, Calif.).
FIG. 6.
HIV-1 DNA copies versus percentage of CD4 CD8 DP cells in human thymus/liver graft cells derived from SCID-hu mice. The number of copies of HIV-1 DNA per 105 cells derived from human thymus liver grafts in SCID-hu mice 3, 4, or 6 weeks postinfection was determined as described in the legend to Fig. 2. In each case, the number of viral DNA copies reached its maximum measured value at this time point. The percentage of CD4 CD8 DP cells recovered 6 or 7 weeks postinfection was determined as described in the legend to Fig. 1 and plotted against the viral load at 3. 4, or 6 weeks postinfection for each graft. Each symbol represents a separate human thymus/liver graft infected as defined in the key.
Nucleotide sequence accession numbers.
The nucleotide sequences of the entire cloned region from each patient isolate will be deposited with the Los Alamos database of HIV-1 sequences.
RESULTS
Previously characterized HIV-1 isolates and biological clones from three patients chosen for their rapid progression to AIDS and one long-term nonprogressor were used to infect SCID-hu mice (11).
These isolates and biological clones were derived from the same patients but are distinct from those used in a previous in vivo study (25). All viral isolates and biological clones used in this study replicated in normal human PHA-stimulated PBMC and in monocyte-derived macrophages in tissue culture; those termed NSI did not replicate or form syncytia in MT-2 cells, while those termed SI did (11). Human thymus/liver grafts implanted in SCID or SCID-beige mice (SCID-hu mice) were infected with 200 TCID50 of each HIV-1 stock, and the infections were monitored by biopsy at 3, 6, 9, and 12 weeks postinfection. At each time point the cells were incubated with CD3, CD4, CD7, and CD8 MAbs; at some time points cells were lysed and nucleic acids were prepared for assay of viral replication by quantitative PCR.
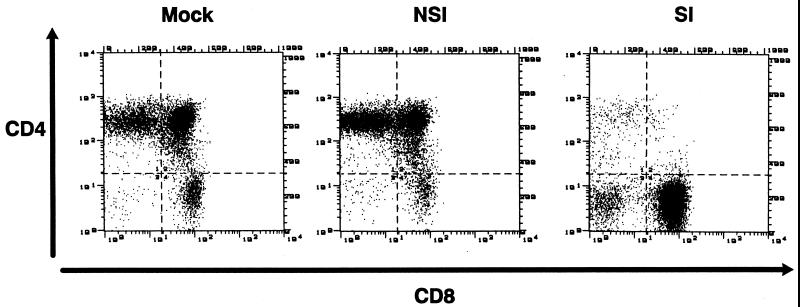
Infection by the early MT-2 cell NSI isolates and biological clones from the rapid progressors, patients A, B, and C, induced less perturbation of the thymocyte subset pattern seen in uninfected grafts than did later MT-2 cell SI isolates from the same patients at 6 weeks postinfection (Table 1 and Fig. 1). Infection by either isolate from the long term nonprogressor, patient D, disturbed thymopoiesis even less at this or any time point. Uninfected thymus liver grafts from a variety of tissue donors were consistent in the percentage of CD4 CD8 DP cells and, with the exception of one implant series, in the ratio of mature CD4+ CD8− (CD4 single-positive [SP]) cells to mature CD8+ CD4− (CD8 SP) cells. This ratio varied for tissue derived for the donor used in implant 23, however, which yielded few mature CD8+ cells. Nevertheless, the thymocyte populations were little affected by infection with the NSI primary HIV-1 isolates used. In contrast, later isolates and biological clones from patients A, B, and C which were SI for MT-2 cells in vitro induced a marked loss of CD4-bearing cells by 6 weeks postinfection (Table 1 and Fig. 1). In six of nine SI virus-infected grafts, both the immature CD4 CD8 DP cortical thymocytes and the more mature CD4 SP medullary thymocytes were depleted (Table 1 and Fig. 1). In one more graft, implanted in mouse 19-22, only the mature CD4 SP cells were depleted, resulting in an abnormal CD4 SP/CD8 SP ratio of 0.7. Therefore, in total seven of nine SI HIV-1-infected grafts showed CD4+ thymocyte depletion.
TABLE 1.
CD4 and CD8 phenotypes of human thymocytes in SCID-hu mice 6 weeks after infection with sequential patient isolates from four patientsa
| Patient-virus phenotype-date | Mouse | Cell distribution
|
||||
|---|---|---|---|---|---|---|
| % CD4+ | % CD4+ CD8+ | % CD4− CD8− | % CD8+ | % CD4+ % CD8+ | ||
| Mock (n = 9) | 15 ± 5 | 71 ± 12 | 8 ± 5 | 9 ± 5 | 2.1 ± 0.7 | |
| A-NSI-3/86 | 23-2 | 43 | 51 | 1 | 5 | 8.6 |
| A-NSI-3/86 | 23-4 | 47 | 46 | 1 | 6 | 7.6 |
| A-NSI-3/86 | 23-10 | 26 | 72 | 0 | 2 | 11.1 |
| A-SI-4/88 | 21-21 | 4 | 1 | 11 | 84 | 0.1 |
| A-SI-4/88 | 23-1 | 15 | 2 | 10 | 73 | 0.2 |
| A-SI-4/88 | 23-15 | 10 | 2 | 10 | 78 | 0.1 |
| B-NSI-11/86 | 17-3 (1) | 21 | 64 | 2 | 14 | 1.6 |
| B-NSI-11/86 | 17-6 (2) | 24 | 60 | 2 | 14 | 1.7 |
| B-NSI-11/86 | 17-15 (5) | 15 | 79 | 1 | 5 | 2.7 |
| B-SI-6/88 | 17-8 (3) | 25 | 2 | 30 | 46 | 0.5 |
| B-SI-6/88 | 17-14 (4) | 22 | 3 | 7 | 68 | 0.3 |
| B-SI-6/88 | 18-5 (6) | 12 | 82 | 1 | 5 | 2.4 |
| C-NSI-1/85 | 19-4 | 18 | 68 | 1 | 13 | 1.4 |
| C-NSI-1/85 | 20-20 | 37 | 38 | 3 | 23 | 1.6 |
| C-NSI-1/85 | 20-24 | 15 | 82 | 1 | 3 | 6.1 |
| C-SI-2/86 | 19-12 | 21 | 13 | 10 | 56 | 0.4 |
| C-SI-2/86 | 19-15 | 12 | 82 | 1 | 6 | 2.0 |
| C-SI-2/86 | 19-22 | 14 | 60 | 6 | 20 | 0.7 |
| D-NSI-3/86 | 23-19 | 31 | 65 | 0 | 4 | 7.1 |
| D-NSI-3/86 | 23-20 | 20 | 78 | 0 | 2 | 8.2 |
| D-NSI-3/86 | 23-29 | 19 | 78 | 0 | 2 | 7.9 |
| D-NSI-6/87 | 23-17 | 26 | 72 | 1 | 2 | 14.4 |
| D-NSI-6/87 | 23-21 | 28 | 69 | 0 | 4 | 7.9 |
| D-NSI-6/87 | 23-31 | 28 | 70 | 0 | 2 | 11.5 |
Each patient is designated by A to D, the viral phenotype in MT-2 cell syncytium assay is indicated as NSI or SI, and the month and year of isolation are indicated. Each mouse is identified by two numbers, the first identifying the tissue implantation and the second identifying the individual mouse. For patient B isolate-infected grafts, numbers in parentheses identify grafts in Fig. 3.
FIG. 1.
Two-color flow cytometric immunofluorescence assay of CD4 and CD8 expression on human thymocytes derived from SCID-hu mice 6 weeks after mock infection or infection with sequential NSI and SI patient A-derived biological clones of HIV-1. Cells were isolated from thymus/liver grafts in SCID-hu mice, incubated with CD4-PE and CD8-FITC, washed, and run on a FACScan flow cytometer. The data are representative of those presented in Table 1.
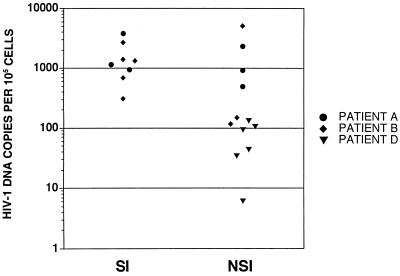
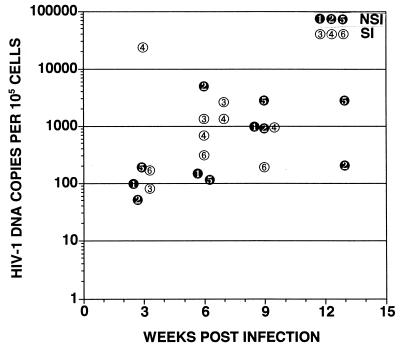
Viral replication was monitored by DNA isolation followed by quantitative PCR using primers specific for HIV-1 and for the cellular gene β-globin. At 6 weeks postinfection, the level of HIV-1 DNA found in thymus/liver grafts infected with NSI or SI isolates from two rapid progressors, patients A and B, was approximately 10-fold higher than the level achieved by the two isolates from a long-term nonprogressor, patient D (Fig. 2). The amount of HIV-1 DNA detected 6 weeks following infection with the SI patient A and B isolates, however, was not significantly higher than that detected at the same time point for the NSI isolates from these patients. Viral replication was therefore examined over the time course of infection with patient B NSI and SI biological clones. In this experiment, similar viral replication was observed for the NSI and SI clones except that in one of the three grafts infected with the SI clone, a peak level of viral DNA corresponding to more than one copy for every 10 cells was observed at 3 weeks postinfection (Fig. 3, graft 4). This graft along with graft 3 (Fig. 3), which were infected with the SI patient B clone, were almost completely depleted of CD4+ cells at 6 weeks postinfection. This finding of a peak of viral replication prior to the appearance of cytopathic effects in one graft was not sufficient to draw conclusions. This result, however, led us to carefully monitor the relationship between viral replication and cytopathic effects in subsequent thymus/liver graft infections.
FIG. 2.
HIV-1 DNA copies per 105 cells detected in thymus/liver graft cells derived from SCID-hu mice 6 weeks after infection with sequential NSI and SI isolates and biological clones derived from patients A, B, and D. Cells were isolated and lysed, nucleic acids were purified, and PCR was performed using a primer pair complementary to HIV-1 LTR DNA and separately with a primer pair complementary to the human β-globin gene. One oligonucleotide of each primer pair was labeled with 32P, and the PCR products were resolved on a 6% polyacrylamide gel and quantitated using a set of standards and a PhosphorImager as described in Materials and Methods.
FIG. 3.
Time course of HIV-1 DNA copies per 105 cells detected in cells derived from SCID-hu mice 3, 6, 9, and 13 weeks postinfection with sequential NSI and SI biological clones derived from patient B. Not all grafts were biopsied at time points after 6 weeks postinfection due to death of the mice. Only grafts 3 and 4 were also biopsied 7 weeks postinfection. The number of copies of HIV-1 DNA per 105 cells derived from human thymus liver grafts in SCID-hu mice at each time point was determined as described in the legend to Fig. 2.
Recombinant infectious molecular clones of HIV-1 were created to identify the determinants responsible for the depletion of CD4-bearing cells.
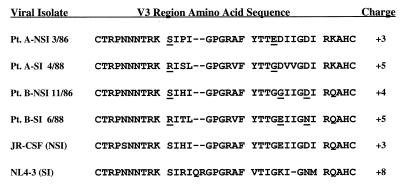
The region of the env gene encoding gp120 V1 to V3 was isolated by PCR from thymus/liver grafts infected with the biological clones of HIV-1 derived from patients A and B used above. These DNA fragments were inserted into plasmid vectors, sequenced, and then introduced into the genome of the NSI infectious molecular clone of HIV-1, JR-CSF, carried on plasmid ΠSVNX-JRCSF. The predicted 35-amino-acid V3 domain sequences of the patient isolates conform to what is known about residues which are required for interaction with CXCR4. Both SI patient isolates have an arginine residue (R) at position 11 which has been previously described to be a critical determinant of the SI phenotype in patient isolates (14, 18) (Fig. 4). In addition, the presence of a negatively charged residue (glutamic acid [E]) at position 24 in the patient A-NSI sequence and a neutral residue at this position in the patient A-SI sequence fits the pattern of substitutions seen by others in determining the SI or X4 phenotype. Similarly, the patient B-SI sequence has an asparagine (N) residue at position 29, while the B-NSI sequence has an aspartic acid (D) at this position. In this case, however, there is a compensatory charge for neutral substitution at position 25 (Fig. 4).
FIG. 4.
Alignment of predicted V3 regions of gp120 encoded by the env genes of HIV-1 strains used in this study. The patient (Pt.) from which the isolates are derived or clone name and MT-2 cell syncytium induction phenotype of each viral isolate is shown along with the predicted amino acid sequence and charge of the disulfide-bound V3 loop domain. For the primary isolates sequenced and characterized in this study, the date of viral isolation is also given. Amino acid positions where charge differences exist among the residues predicted for the two isolates from each patient are underlined. The sequences of JR-CSF and NL4-3 were obtained from the Los Alamos National Laboratory HIV sequence database.
Chimeric recombinant HIV-1 stocks were prepared by transfection of COSM6 cells, titered on PHA-stimulated PBMC, and used to infect MT-2 cells to confirm their syncytium induction phenotype. Infected MT-2 cell cultures were kept for 7 days, and syncytia were identified visually with an inverted microscope on the final day of culture. The recombinant viruses exhibited the same MT-2 cell phenotype, NSI or SI, as the biological clone from which they were derived (data not shown).
SCID-hu mice were infected with 200 TCID50 of the patient A and patient B NSI and SI recombinant viral stocks.
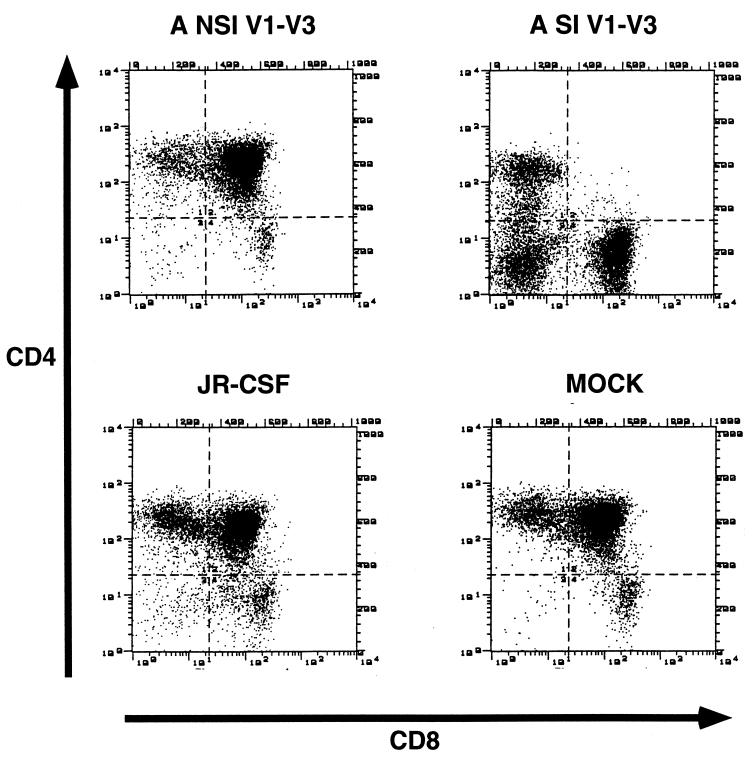
Additional thymus/liver grafts were infected with the HIV-1 molecular clone JR-CSF or NL4-3 or the late-stage R5 AIDS-associated patient clone ACH142-*E11, described in detail in the accompanying report (38), or left undisturbed as controls. Human tissue grafts were biopsied every 3 weeks beginning on week 3 or 4 postinfection, and a single-cell suspension was made. At each time point, cells were prepared for flow cytometry by incubation with CD7-FITC, CD4-PE, and CD8-PerCP and separately with CD8-FITC, CD4-PE, and CD3-PerCP. The remaining cells were lysed, and nucleic acids were prepared for analysis of viral DNA by quantitative PCR. Every NL4-3-infected graft analyzed was highly depleted of CD4+ cells by 6 weeks postinfection, as were three of the four grafts infected with JR-CSF/V1-V3 A-SI Env (Fig. 5 and 6) and two of seven grafts infected with ACH142-*E11. None of the grafts infected with JR-CSF or with the other chimeric viruses, JR-CSF/V1-V3 A-NSI Env, JR-CSF/V1-V3 B-NSI Env, and JR-CSF/V1-V3 B-SI Env, showed significant depletion of CD4+ cells. There was a strong nonlinear correlation between the level of viral replication measured at 3 or 4 weeks postinfection and the CD4 CD8 DP cell depletion observed at 6 or 7 weeks postinfection in these experiments (r2 = 0.60 [Fig. 6]). Bootstrap analysis indicated that the data were significantly better fit by a sigmoid curve derived from the Hill equation than by a straight line (P < 0.05). This indicates that HIV-1 replication in thymus/liver grafts exhibits a threshold characteristic with respect to cytopathic effects on CD4 CD8 DP thymocytes. This threshold level of viral replication, where 50% of the CD4 CD8 DP cells are depleted, is one copy of HIV-1 DNA for every 11 cells. The 95% confidence limits of this threshold level are one copy of HIV-1 DNA per 67 cells to one copy per 3.6 cells. In these experiments, NL4-3 and JR-CSF/V1-V3 A-SI Env, both SI and CXCR4 competent clones of HIV-1, were able to achieve viral replication above the threshold needed for CD4 CD8 DP cell depletion. In addition, the R5 AIDS-associated viral clone ACH-142 replicated close to or beyond this level in four of seven grafts and significantly depleted CD4 CD8 DP cells in two of seven grafts. The relationship between replication above the threshold value and cytopathic effects is not perfect in our data, likely because we could measure viral replication and CD4 CD8 DP cell depletion only every 3 weeks, and the peak level of HIV-1 DNA present in the grafts is not maintained following the depletion of target cells (i.e., graft 4 in Fig. 2 [23]). The other SI clone used, JR-CSF/V1-V3 B-SI Env, however, did not replicate to a sufficient level to induce depletion of CD4 CD8 DP cells. Patient B-derived recombinant viruses (1,000 TCID50) were subsequently used to infect thymus/liver grafts in SCID-hu mice. Again, the replication of these recombinant viruses did not achieve a high level and thymopoiesis was not noticeably disturbed (data not shown).
FIG. 5.
Two-color flow cytometric immunofluorescence assay of CD4 and CD8 expression on human thymocytes derived from SCID-hu mice 9 weeks after mock infection or infection with 200 TCID50 of JR-CSF, JR-CSF/V1-V3 A-NSI Env, and JR-CSF/V1-V3 A-SI Env molecular clones of HIV-1. Cells were isolated from thymus/liver grafts in SCID-hu mice, incubated with CD4-PE and CD8-FITC, washed, and run on a FACScan flow cytometer. The data are representative of those presented in Fig. 6.
GHOST cell infection assays were used to further determine the coreceptor specificity of the chimeric HIV-1 molecular clones that we derived.
A panel of 10 GHOST (3) indicator cell lines bearing an HIV-2 LTR-driven humanized green fluorescent protein (GFP) S65T gene and expressing CD4 along with a single coreceptor or related chemokine receptor was obtained from the NIH AIDS Research and Reference Reagent Program. Two infection experiments were performed using the protocol provided by the originators of the cell lines, V. KewalRamani and D. Littman. In the first infection, 50 μl of JR-CSF, 50 μl of the patient B-derived chimeric viral stocks, and 200 μl of the patient A chimeras were used to infect seven of the GHOST cell lines. In the second experiment, lesser doses of the four chimeric viruses were used to infect all 10 cell lines. Two days after each infection, the cells were removed from their plates and GFP fluorescence was measured with a FACScan flow cytometer. All of the viruses could most efficiently infect GHOST-CCR5 cells, the patient B-derived chimeric viruses exhibited the ability to infect GHOST-CCR3 cells, and both SI patient isolate-derived chimeric viruses could infect GHOST-CXCR4 cells (Table 2). The efficiency with which the SI patient A- and B-derived chimeric viruses infected GHOST-CXCR4 cells, however, was very different. The JR-CSF/V1-V3 B-SI Env viral stock had approximately a 10-fold higher titer on GHOST-CCR5 cells than the JR-CSF/V1-V3 A-SI Env viral stock, based on infections with the lowest dose of each used which gave approximately the same percentages of GFP+ cells (10% versus 13%). In contrast, the JR-CSF/V1-V3 B-SI Env viral stock had a much lower titer on GHOST-CXCR4 cells than the JR-CSF/V1-V3 A-SI Env viral stock. At the intermediate doses used, the JR-CSF/V1-V3 A-SI Env viral stock rendered 44% of the GHOST-CXCR4 cells GFP+, while the JR-CSF/V1-V3 B-SI Env viral stock gave only a background level of 1% GFP+ cells. Thus, JR-CSF/V1-V3 A-SI was at least 44-fold more efficient in infecting GHOST-CXCR4 than JR-CSF/V1-V3 B-SI Env when used at doses which gave similar high levels of infection of GHOST-CCR5 cells (Table 2).
TABLE 2.
Percentage of GHOST indicator cells expressing GFPa
| Viral inoculum | JR-CSF (50 μl) | JR-CSF V1–V3 A-NSI
|
JR-CSF V1–V3 A-SI
|
JR-CSF V1–V3 B-NSI
|
JR-CSF V1–V3 B-SI
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 200 μl | 50 μl | 5 μl | 200 μl | 50 μl | 5 μl | 50 μl | 5 μl | 0.5 μl | 50 μl | 5 μl | 0.5 μl | ||
| GHOST-CCR1 | 0 | 2 | 0 | 0 | 15 | 7 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| GHOST-CCR2b | ND | ND | 1 | 1 | ND | 6 | 0 | ND | 1 | 0 | ND | 1 | 0 |
| GHOST-CCR3 | 4 | 3 | 1 | 0 | 5 | 2 | 0 | 16 | 0 | 0 | 16 | 1 | 0 |
| GHOST-CCR4 | ND | ND | 0 | 0 | ND | 1 | 0 | ND | 0 | 0 | ND | 0 | 0 |
| GHOST-CCR5 | 49 | 42 | 28 | 4 | 64 | 60 | 13 | 45 | 88 | 29 | 70 | 77 | 10 |
| GHOST-CCR8 | ND | ND | 1 | 1 | ND | 2 | 0 | ND | 0 | 0 | ND | 0 | 0 |
| GHOST-CXCR4 | 0 | 1 | 1 | 1 | 36 | 44 | 3 | 0 | 1 | 1 | 8 | 1 | 1 |
| GHOST-V28 | 0 | 1 | 1 | 1 | 5 | 5 | 0 | 0 | 1 | 1 | 1 | 0 | 0 |
| GHOST-BONZO | 0 | 1 | 1 | 0 | 5 | 3 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| GHOST-BOB | 1 | 2 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 0 |
GHOST cell lines expressing the chemokine receptor or related molecule indicated were infected with chimeric HIV-1 stocks derived from molecular clones as described in Materials and Methods. Two days later, the cells were removed from their wells with trypsin, washed, and incubated in PBS–2% formaldehyde overnight. Cells fluorescence was measured with a FACScan flow cytometer using uninfected cells as a negative control. ND, not determined.
DISCUSSION
Our results show that the SI or X4 phenotype of HIV-1 patient isolates is an important determinant of pathogenesis in human thymocytes in SCID-hu thymus/liver mice. We have confirmed previous work showing that SI patient isolates and molecular clones are consistently more pathogenic and replicate more readily in human thymus/liver grafts in this in vivo model system than NSI isolates and clones (3, 22, 25). Furthermore, we found that an SI chimeric virus and the molecular clone NL4-3 reached higher peak levels of viral replication in thymus/liver graft cells, sufficient for CD4 CD8 DP cell depletion, which NSI chimeric clones did not reach. The late-stage R5 AIDS-associated clone ACH142-*E11, however, did replicate to high levels and occasionally depleted CD4 CD8 DP cells. Taking all of these data together, we found that the relationship between HIV-1 replication and cytopathic effects was nonlinear. The relationship was significantly better fit by a variation of the Hill equation than by a straight line (P < 0.05). This indicates that there is a threshold effect seen for depletion of CD4 CD8 DP cells with respect to HIV-1 copy number in thymus/liver grafts. Our data are imprecise in that viral load could not be continuously monitored and peak viral load is not stable (Fig. 2, graft 4) (23). We measured viral load every 3 weeks; therefore, the peak viral load that we measured may not correspond to the true maximum point of viral replication. Nevertheless, taken together the data shown in Fig. 6 indicate that replication above the threshold level of one copy of HIV-1 DNA for every 11 cells in thymus liver grafts in SCID-hu mice correlates with the depletion of CD4 CD8 DP thymocytes from thymus/liver grafts in SCID-hu mice. The inability of most NSI or R5 strains of HIV-1 to reach this threshold may result from the paucity of human thymocytes which express levels of CCR5 detectable by flow cytometry (4, 26, 31). The capability of late-stage AIDS-associated R5 HIV-1 clones to perturb thymopoiesis is described in detail in the accompanying report (38). The implication of Fig. 6 is that viral replication is an important determinant of cytopathic effects in SCID-hu thymus/liver graft infection and that its effect is manifest in a nonlinear way. These data do not, however, allow us to discern the mechanism of the HIV-1-mediated cytopathic effects that we observed.
Our results suggest that acquisition of the SI/X4 phenotype leads to the rapid loss of CD4+ T cells in patients and is not just a consequence of immune deficiency. We further found that two NSI HIV-1 isolates from a long-term nonprogressor replicated 10-fold less well in thymus/liver grafts in SCID-hu mice than any of the isolates from the rapid progressor. This implicates the particular HIV-1 quasispecies as at least one cause of the slow progression seen in this patient. Many other reports have documented that defective or relatively slowly replicating isolates of HIV-1 are found in long-term nonprogressors, consistent with our findings (13, 28, 36).
In this report, we have directly demonstrated that transfer of the V1 to V3 region of the env gene from an SI patient A isolate of HIV-1 to an NSI molecular clone conferred increased viral replication and pathogenesis. This finding directly implicates the SI/X4 phenotype in pathogenesis in this model of HIV-1-mediated disease. This likely recapitulates what is seen in patients, since the presence of SI HIV-1 in patient plasma has been correlated with more rapid disease progression and has been shown to arise just prior to a rapid decline in CD4+ cell numbers (11, 12, 39–41). In contrast, the transfer of V1 to V3 region of the env gene from an SI patient B isolate of HIV-1 to an NSI molecular clone resulted in a nonpathogenic, poorly replicating chimeric viral clone in SCID-hu mice. This may be explained if the resulting chimeric Env glycoprotein was not stable or was poorly functional as has been seen with similar chimeras in tissue culture studies (8).
The V1 to V3 regions that we sequenced from the NSI and SI patient isolates following infection of SCID-hu mice are unique and consistent with evolution within each patient. For both patients A and B, the sequences that we determined for each of their isolates are closely related to each other. Furthermore, the predicted V3 domain amino acid sequences of the sequential patient isolates of HIV-1 are consistent with the observed viral phenotype. In particular, the predicted presence of a positively charged amino acid residue at position 11 in the disulfide bound V3 loop and the absence of a negative charge at position 24 or 29 in the SI isolates are consistent with previous reports of the SI-determining genotype in patient isolates of HIV-1 (14, 18).
GHOST (3) cells were used to map the coreceptor specificity of the patient-derived chimeric molecular clones we constructed. Consistent with other reports, we found that all of these clade B isolates could infect GHOST-CCR5 cells, the isolates from patient B could also use CCR3 much less efficiently, and the SI patient isolate-derived chimeric molecular clones could infect GHOST-CXCR4 cells (2, 12, 37, 43, 44). The efficiency of infection of infect GHOST-CXCR4 cells, however, was strikingly different for the two SI patient isolate-derived chimeric molecular clones. We found that the patient A clone was at least 44-fold more efficient in infecting GHOST-CXCR4 than the patient B-derived clone. This difference likely accounts for the inability of the SI patient B-derived chimeric molecular clone to replicate to high levels in SCID-hu thymus/liver grafts and its correlated inability to deplete CD4+ thymocytes in this system. We do not yet know the precise step in viral entry into GHOST-CXCR4 cells which is less efficient. Further studies will be required to define this interesting phenotypic difference.
ACKNOWLEDGMENTS
We thank Ruth Connor for providing the patient-derived biological clones of HIV-1 used in this study. We also thank Grace Aldrovandi, Beth Jamieson, and Jayanand Vasudevan for useful discussions and help, Yongde Bao and Jay Fox of the University of Virginia Biomolecular Core Facility for DNA sequencing, and Lance Hultin and Ingrid Schmid of the UCLA FACS Core Facility and William Ross of the University of Virginia FACS Core Facility for flow cytometry.
This work was supported by NIH grant AI39943 and an AmFAR Scholar Award to D.C.
REFERENCES
- 1.Aldrovandi G M, Feuer G, Gao L, Jamieson B, Kristeva M, Chen I S, Zack J A. The SCID-hu mouse as a model for HIV-1 infection. Nature. 1993;363:732–736. doi: 10.1038/363732a0. [DOI] [PubMed] [Google Scholar]
- 2.Bazan H A, Alkhatib G, Broder C C, Berger E A. Patterns of CCR5, CXCR4, and CCR3 usage by envelope glycoproteins from human immunodeficiency virus type 1 primary isolates. J Virol. 1998;72:4485–4491. doi: 10.1128/jvi.72.5.4485-4491.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berkowitz R D, Alexander S, Bare C, Lindquist-Stepps V, Bogan M, Moreno M E, Gibson L, Wieder E D, Kosek J, Stoddart C A, McCune J M. CCR5- and CXCR4-utilizing strains of human immunodeficiency virus type 1 exhibit differential tropism and pathogenesis in vivo. J Virol. 1998;72:10108–10117. doi: 10.1128/jvi.72.12.10108-10117.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berkowitz R D, Beckerman K P, Schall T J, McCune J M. CXCR4 and CCR5 expression delineates targets for HIV-1 disruption of T cell differentiation. J Immunol. 1998;161:3702–3710. [PubMed] [Google Scholar]
- 5.Bonyhadi M L, Rabin L, Salimi S, Brown D A, Kosek J, McCune J M, Kaneshima H. HIV induces thymus depletion in vivo. Nature. 1993;363:728–732. doi: 10.1038/363728a0. [DOI] [PubMed] [Google Scholar]
- 6.Camerini D, James S P, Stamenkovic I, Seed B. Leu-8/TQ1 is the human equivalent of the Mel-14 lymph node homing receptor. Nature. 1989;342:78–82. doi: 10.1038/342078a0. [DOI] [PubMed] [Google Scholar]
- 7.Cheng-Mayer C, Quiroga M, Tung J W, Dina D, Levy J A. Viral determinants of human immunodeficiency virus type 1 T-cell or macrophage tropism, cytopathogenicity, and CD4 antigen modulation. J Virol. 1990;64:4390–4398. doi: 10.1128/jvi.64.9.4390-4398.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cho M W, Lee M K, Carney M C, Berson J F, Doms R W, Martin M A. Identification of determinants on a dualtropic human immunodeficiency virus type 1 envelope glycoprotein that confer usage of CXCR4. J Virol. 1998;72:2509–2515. doi: 10.1128/jvi.72.3.2509-2515.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, Mackay C R, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 10.Cocchi F, DeVico A L, Garzino-Demo A, Cara A, Gallo R C, Lusso P. The V3 domain of the HIV-1 gp120 envelope glycoprotein is critical for chemokine-mediated blockade of infection. Nat Med. 1996;2:1244–1247. doi: 10.1038/nm1196-1244. [DOI] [PubMed] [Google Scholar]
- 11.Connor R I, Mohri H, Cao Y, Ho D. Increased viral burden and cytopathicity correlate temporally with CD4+ T-lymphocyte decline and clinical progression in human immunodeficiency virus type 1-infected individuals. J Virol. 1993;67:1772–1777. doi: 10.1128/jvi.67.4.1772-1777.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Connor R I, Sheridan K E, Ceradini D, Choe S, Landau N R. Change in coreceptor use correlates with disease progression in HIV-1-infected individuals. J Exp Med. 1997;185:621–628. doi: 10.1084/jem.185.4.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deacon N J, Tsykin A, Solomon A, Smith K, Ludford-Menting M, Hooker D J, McPhee D A, Greenway A L, Ellett A, Chatfield C, et al. Genomic structure of an attenuated quasi species of HIV-1 from a blood transfusion donor and recipients. Science. 1995;270:988–991. doi: 10.1126/science.270.5238.988. [DOI] [PubMed] [Google Scholar]
- 14.De Jong J J, De Ronde A, Keulen W, Tersmette M, Goudsmit J. Minimal requirements for the human immunodeficiency virus type 1 V3 domain to support the syncytium-inducing phenotype: analysis by single amino acid substitution. J Virol. 1992;66:6777–6780. doi: 10.1128/jvi.66.11.6777-6780.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doranz B J, Rucker J, Yi Y, Smyth R J, Samson M, Peiper S C, Parmentier M, Collman R G, Doms R W. A dual-tropic primary HIV-1 isolate that uses fusin and the beta-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 16.Efron B, Tibshirani R J. An introduction to the bootstrap. Vol. 57. New York, N.Y: Chapman and Hall; 1993. [Google Scholar]
- 17.Feng Y, Broder C C, Kennedy P E, Berger E A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 18.Fouchier R A, Groenink M, Kootstra N A, Tersmette M, Huisman H G, Miedema F, Schuitemaker H. Phenotype-associated sequence variation in the third variable domain of the human immunodeficiency virus type 1 gp120 molecule. J Virol. 1992;66:3183–3187. doi: 10.1128/jvi.66.5.3183-3187.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glushakova S, Grivel J C, Fitzgerald W, Sylwester A, Zimmerberg J, Margolis L B. Evidence for the HIV-1 phenotype switch as a causal factor in acquired immunodeficiency. Nat Med. 1998;4:346–349. doi: 10.1038/nm0398-346. [DOI] [PubMed] [Google Scholar]
- 20.Hwang S S, Boyle T J, Lyerly H K, Cullen B R. Identification of envelope V3 loop as the major determinant of CD4 neutralization sensitivity of HIV-1. Science. 1992;257:535–537. doi: 10.1126/science.1636088. [DOI] [PubMed] [Google Scholar]
- 21.Jamieson B D, Aldrovandi G M, Planelles V, Jowett J B, Gao L, Bloch L M, Chen I S, Zack J A. Requirement of human immunodeficiency virus type 1 nef for in vivo replication and pathogenicity. J Virol. 1994;68:3478–3485. doi: 10.1128/jvi.68.6.3478-3485.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jamieson B D, Pang S, Aldrovandi G M, Zha J, Zack J A. In vivo pathogenic properties of two clonal human immunodeficiency virus type 1 isolates. J Virol. 1995;69:6259–6264. doi: 10.1128/jvi.69.10.6259-6264.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jamieson B D, Uittenbogaart C H, Schmid I, Zack J A. High viral burden and rapid CD4+ cell depletion in human immunodeficiency virus type 1-infected SCID-hu mice suggest direct viral killing of thymocytes in vivo. J Virol. 1997;71:8245–8253. doi: 10.1128/jvi.71.11.8245-8253.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson M L, Frasier S G. Nonlinear least-squares analysis. Methods Enzymol. 1985;117:301–341. [Google Scholar]
- 25.Kaneshima H, Su L, Bonyhadi M L, Connor R I, Ho D D, McCune J M. Rapid-high, syncytium-inducing isolates of human immunodeficiency virus type 1 induce cytopathicity in the human thymus of the SCID-hu mouse. J Virol. 1994;68:8188–8192. doi: 10.1128/jvi.68.12.8188-8192.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kitchen S G, Zack J A. Distribution of the human immunodeficiency virus coreceptors CXCR4 and CCR5 in fetal lymphoid organs: implications for pathogenesis in utero. AIDS Res Hum Retroviruses. 1999;15:143–148. doi: 10.1089/088922299311565. [DOI] [PubMed] [Google Scholar]
- 27.Koyanagi Y, Miles S, Mitsuyasu R T, Merrill J E, Vinters H V, Chen I S. Dual infection of the central nervous system by AIDS viruses with distinct cellular tropisms. Science. 1987;236:819–822. doi: 10.1126/science.3646751. [DOI] [PubMed] [Google Scholar]
- 28.Learmont J C, Geczy A F, Mills J, Ashton L J, Raynes-Greenow C H, Garsia R J, Dyer W B, McIntyre L, Oelrichs R B, Rhodes D I, Deacon N J, Sullivan J S, McPhee D A, Crowe S, Solomon A E, Chatfield C, Cooke I R, Blasdall S, Kuipers H. Immunologic and virologic status after 14 to 18 years of infection with an attenuated strain of HIV-1—a report from the Sydney Blood Bank Cohort. N Engl J Med. 1999;340:1715–1722. doi: 10.1056/NEJM199906033402203. [DOI] [PubMed] [Google Scholar]
- 29.McCune J M, Namikawa R, Kaneshima H, Shultz L D, Lieberman M, Weissman I L. The SCID-hu mouse: murine model for the analysis of human hematolymphoid differentiation and function. Science. 1988;241:1632–1639. doi: 10.1126/science.241.4873.1632. [DOI] [PubMed] [Google Scholar]
- 30.O'Brien W A, Koyanagi Y, Namazie A, Zhao J Q, Diagne A, Idler K, Zack J A, Chen I S. HIV-1 tropism for mononuclear phagocytes can be determined by regions of gp120 outside the CD4-binding domain. Nature. 1990;348:69–73. doi: 10.1038/348069a0. [DOI] [PubMed] [Google Scholar]
- 31.Pedroza-Martins L, Gurney K B, Torbett B E, Uittenbogaart C H. Differential tropism and replication kinetics of human immunodeficiency virus type 1 isolates in thymocytes: coreceptor expression allows viral entry, but productive infection of distinct subsets is determined at the postentry level. J Virol. 1998;72:9441–9452. doi: 10.1128/jvi.72.12.9441-9452.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Penn M L, Grivel J C, Schramm B, Goldsmith M A, Margolis L. CXCR4 utilization is sufficient to trigger CD4+ T cell depletion in HIV-1-infected human lymphoid tissue. Proc Natl Acad Sci USA. 1999;96:663–668. doi: 10.1073/pnas.96.2.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Picchio G R, Gulizia R J, Wehrly K, Chesebro B, Mosier D E. The cell tropism of human immunodeficiency virus type 1 determines the kinetics of plasma viremia in SCID mice reconstituted with human peripheral blood leukocytes. J Virol. 1998;72:2002–2009. doi: 10.1128/jvi.72.3.2002-2009.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pleskoff O, Sol N, Labrosse B, Alizon M. Human immunodeficiency virus strains differ in their ability to infect CD4+ cells expressing the rat homolog of CXCR-4 (fusin) J Virol. 1997;71:3259–3262. doi: 10.1128/jvi.71.4.3259-3262.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poon B, Jowett J B, Stewart S A, Armstrong R W, Rishton G M, Chen I S. Human immunodeficiency virus type 1 vpr gene induces phenotypic effects similar to those of the DNA alkylating agent, nitrogen mustard. J Virol. 1997;71:3961–3971. doi: 10.1128/jvi.71.5.3961-3971.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Premkumar D R, Ma X Z, Maitra R K, Chakrabarti B K, Salkowitz J, Yen-Lieberman B, Hirsch M S, Kestler H W. The nef gene from a long-term HIV type 1 nonprogressor. AIDS Res Hum Retroviruses. 1996;12:337–345. doi: 10.1089/aid.1996.12.337. [DOI] [PubMed] [Google Scholar]
- 37.Scarlatti G, Tresoldi E, Bjorndal A, Fredriksson R, Colognesi C, Deng H K, Malnati M S, Plebani A, Siccardi A G, Littman D R, Fenyo E M, Lusso P. In vivo evolution of HIV-1 co-receptor usage and sensitivity to chemokine-mediated suppression. Nat Med. 1997;3:1259–1265. doi: 10.1038/nm1197-1259. [DOI] [PubMed] [Google Scholar]
- 38.Scoggins R M, Taylor J R, Jr, Patrie J, van't Wout A B, Schuitemaker H, Camerini D. Pathogenesis of primary R5 human immunodeficiency virus type 1 clones in SCID-hu mice. J Virol. 2000;74:3205–3216. doi: 10.1128/jvi.74.7.3205-3216.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tersmette M, Goede R E Y D, Al B J M B, Winkel I M, Gruters R A, Cuypers H T, Huisman H G, Miedema F. Differential syncytium-inducing capacity of human immunodeficiency virus isolates: frequent detection of syncytium-inducing isolates in patients with acquired immunodeficiency syndrome (AIDS) and AIDS-related complex. J Virol. 1988;62:2026–2032. doi: 10.1128/jvi.62.6.2026-2032.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tersmette M, Gruters R A, de Wolf F, de Goede R E, Lange J M, Schellekens P T, Goudsmit J, Huisman H G, Miedema F. Evidence for a role of virulent human immunodeficiency virus (HIV) variants in the pathogenesis of acquired immunodeficiency syndrome: studies on sequential HIV isolates. J Virol. 1989;63:2118–2125. doi: 10.1128/jvi.63.5.2118-2125.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tersmette M, Lange J M, de Goede R E, de Wolf F, Eeftink-Schattenkerk J K, Schellekens P T, Coutinho R A, Huisman J G, Goudsmit J, Miedema F. Association between biological properties of human immunodeficiency virus variants and risk for AIDS and AIDS mortality. Lancet. 1989;i:983–985. doi: 10.1016/s0140-6736(89)92628-7. [DOI] [PubMed] [Google Scholar]
- 42.Zack J A, Arrigo S J, Weitsman S R, Go A S, Haislip A, Chen I S. HIV-1 entry into quiescent primary lymphocytes: molecular analysis reveals a labile, latent viral structure. Cell. 1990;61:213–222. doi: 10.1016/0092-8674(90)90802-l. [DOI] [PubMed] [Google Scholar]
- 43.Zhang L, He T, Huang Y, Chen Z, Guo Y, Wu S, Kunstman K J, Brown R C, Phair J P, Neumann A U, Ho D D, Wolinsky S M. Chemokine coreceptor usage by diverse primary isolates of human immunodeficiency virus type 1. J Virol. 1998;72:9307–9312. doi: 10.1128/jvi.72.11.9307-9312.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Y J, Dragic T, Cao Y, Kostrikis L, Kwon D S, Littman D R, KewalRamani V N, Moore J P. Use of coreceptors other than CCR5 by non-syncytium-inducing adult and pediatric isolates of human immunodeficiency virus type 1 is rare in vitro. J Virol. 1998;72:9337–9344. doi: 10.1128/jvi.72.11.9337-9344.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]