Abstract
The Prp19 complex (Prp19C), also named NineTeen Complex (NTC), is conserved from yeast to human and functions in many different processes such as genome stability, splicing, and transcription elongation. In the latter, Prp19C ensures TREX occupancy at transcribed genes. TREX, in turn, couples transcription to nuclear mRNA export by recruiting the mRNA exporter to transcribed genes and consequently to nascent mRNAs. Here, we assess the function of the nonessential Prp19C subunit Syf2 and the nonessential Prp19C-associated protein Cwc15 in the interaction of Prp19C and TREX with the transcription machinery, Prp19C and TREX occupancy, and transcription elongation. Whereas both proteins are important for Prp19C–TREX interaction, Syf2 is needed for full Prp19C occupancy, and Cwc15 is important for the interaction of Prp19C with RNA polymerase II and TREX occupancy. These partially overlapping functions are corroborated by a genetic interaction between Δcwc15 and Δsyf2. Finally, Cwc15 also interacts genetically with the transcription elongation factor Dst1 and functions in transcription elongation. In summary, we uncover novel roles of the Prp19C component Syf2 and the Prp19C-associated protein Cwc15 in Prp19C's function in transcription elongation.
Keywords: transcription elongation, Prp19 complex, NTC, Syf2, Cwc15
INTRODUCTION
For gene expression to culminate in protein synthesis, a series of dynamic and interconnected processes take place. The pre-mRNA is synthesized by RNA polymerase II (RNAPII) and is processed into mature mRNA by capping of the 5′ end, splicing and 3′ end formation. Furthermore, several proteins bind to the mRNA and package it into a messenger ribonucleoprotein particle (mRNP) (Mitchell and Parker 2014; Meinel and Sträßer 2015; Singh et al. 2015; Wegener and Müller-McNicoll 2019; Wende et al. 2019; Khong and Parker 2020). These events largely occur already cotranscriptionally. Only correctly processed and packaged mRNPs are exported from the nucleus to the cytoplasm (Mitchell and Parker 2014; Oeffinger and Montpetit 2015; Wende et al. 2019; Xie and Ren 2019). Here, the mRNA serves as the template for protein synthesis.
A key player of mRNP assembly is the TREX complex, which couples transcription to nuclear mRNA export and is conserved from yeast to human (Heath et al. 2016). In Saccharomyces cerevisiae, TREX is a heterononameric complex consisting of the THO complex (Tho2, Hpr1, Mft1, Thp2, and Tex1), the SR-like proteins Gbp2 and Hrb1, the DEAD-box RNA-helicase Sub2, and the export adaptor Yra1 (Sträßer et al. 2002). TREX is recruited to RNAPII-transcribed genes by several mechanisms. It binds to the nascent mRNA and interacts both with the S2-phosphorylated C-terminal domain (CTD) of Rpb1, the largest subunit of RNAPII, and with the Prp19 complex (Prp19C) (Abruzzi et al. 2004; Chanarat et al. 2011, 2012; Meinel et al. 2013; Meinel and Sträßer 2015). Prp19C, a complex best known for its function in splicing, in turn likewise interacts with RNAPII and furthermore with Mud2 (see Fig. 5; Chan et al. 2003; Chanarat et al. 2011; Minocha et al. 2018). Mud2, a protein that also functions in splicing, is recruited to the transcription machinery by the S2-phosphorylated CTD of Rpb1 (see Fig. 5; Kistler and Guthrie 2001; Minocha et al. 2018). Thus, TREX is recruited to the transcription machinery by several interactions.
FIGURE 5.
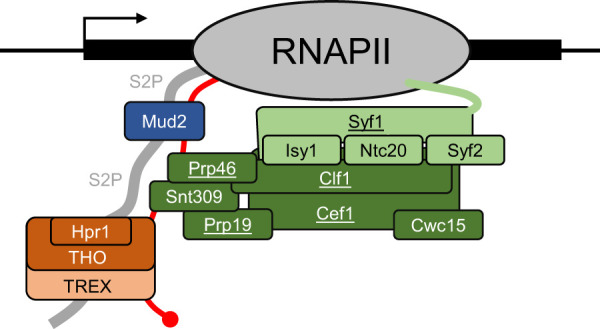
Model of Prp19C/NTC function in transcription elongation, nuclear mRNP assembly, and export. Prp19C/NTC (green) consists of the four essential proteins Prp19, Cef1, Syf1, and Clf1 (underlined) and the four nonessential proteins Snt309, Syf2, Isy1, and Ntc20. Isy1, Ntc20, Syf1, and Syf2 (light green) form a subcomplex of Prp19C/NTC. In addition, the essential protein Prp46 and the nonessential protein Cwc15 associate with Prp19C/NTC. Prp19C/NTC is present at transcribed genes through multiple interactions: It interacts directly with RNAPII (gray) via the C-terminal domain of its subunit Syf1. Moreover, the Prp19C/NTC subunits Snt309 and Cef1 interact with Mud2 (blue), which in turn directly binds to the S2-phosphorylated CTD of RNAPII (S2P). Prp19C/NTC facilitates efficient transcription elongation, most likely by its function in the occupancy of TREX (light orange), with which it interacts via its subunits Cwc15 and Syf2. Importantly, Cwc15 also mediates the interaction of Prp19C/NTC with RNAPII and is needed for Prp19C/NTC and TREX occupancy as well as efficient transcription elongation. Furthermore, THO (orange), a subcomplex of TREX, interacts directly with the CTD of RNAPII. Hpr1 (orange) is a THO subunit. TREX also binds to the nascent mRNA (red) and functions in mRNP assembly and nuclear mRNA export.
As mentioned above, Prp19C, which was also named NineTeen Complex (NTC), is important for TREX occupancy at transcribed genes and thus for efficient transcription elongation (see Fig. 5; Chanarat et al. 2012; Chanarat and Sträßer 2013). Prp19C and its associated proteins join the spliceosome during or after the dissociation of the U4 snRNP. Prp19C stabilizes the association of the U5 and U6 snRNPs by promoting RNA–RNA interaction between U5 and U6 in the activated spliceosomal B* complex, which catalyzes the first transesterification reaction and remains bound throughout the second step of splicing (Tarn et al. 1994; Chan et al. 2003; Chan and Cheng 2005; Fabrizio et al. 2009). In addition to splicing and transcription elongation, Prp19C is involved in genome stability (Idrissou and Maréchal 2022) and lipid droplet formation (Cho et al. 2007). Like the TREX complex, Prp19C is conserved from yeast to human. In humans, at least three different Prp19-like complexes exist, whereas only one Prp19C is known in yeast (Chanarat and Sträßer 2013). In S. cerevisiae, Prp19C consists of eight core subunits and 26 associated proteins (Ohi et al. 2002; Chanarat and Sträßer 2013). The core subunits are the four essential proteins Prp19, Cef1, Syf1, and Clf1 and the four nonessential proteins Snt309, Syf2, Isy1, and Ntc20 (see Fig. 5; Grote et al. 2010; Chanarat et al. 2012). In addition to these eight core proteins in S. cerevisiae, the human Prp19C contains additional core proteins—namely, PRL1/PRLG1, AD002/HSPC148, CTNNBL1/NAP, and HSP73. The yeast orthologs of PRL1/PRLG1 and AD002/HSPC148 are the Prp19C-associated proteins Prp46 and Cwc15, respectively, whereas the other two proteins do not have orthologs in yeast (Grote et al. 2010). Whereas Prp46 is essential, Cwc15 is not (Albers et al. 2003). Interestingly, Cwc15 is involved in pre-mRNA splicing by contributing to spliceosome assembly, activation, and regulation and interacts directly with the essential subunit Cef1 (Ohi et al. 2002). In addition, a loss-of-function mutation in Cwc15 is lethal in Schizosaccharomyces pombe and synthetically lethal with the prp19-1 mutation in S. cerevisiae indicating an important function of Cwc15 for Prp19C function (Ohi and Gould 2002). In Arabidopsis thaliana, Cwc15 is ubiquitously expressed and important in the developmental process (Slane et al. 2020). In humans, Cwc15 is essential for viability and exists as a stable heterodimer with CTNNBL1, a component that is lacking in yeast (van Maldegem et al. 2015). Structurally, Cwc15 is located on the surface of Prp8, where it associates with the PPIL4 and facilitates its anchoring to the spliceosome (Schmitzová et al. 2023).
Single deletion mutants of the nonessential Prp19C subunits do not show a strong phenotype (Chen et al. 1998, 2001, 2002; Ohi et al. 2002). Three of the four nonessential proteins—namely, Isy1, Ntc20, and Syf2—have overlapping functions and form a subcomplex together with the essential subunit Syf1 (Chen et al. 2002). The triple deletion of ISY1, NTC20, and SYF2 is lethal, whereas the different double deletions are not (Chen et al. 2001, 2002). Double deletion of ISY1 and NTC20, however, causes a severe growth defect and a high accumulation of nonspliced mRNAs (Chen et al. 2001). Deletion of SNT309, encoding the fourth nonessential core protein of Prp19C, leads to the accumulation of nonspliced mRNAs at elevated temperatures (Chen et al. 1998). Within Prp19C, Snt309 interacts exclusively with Prp19, and the lack of Snt309 results in the destabilization of the whole complex (Chen et al. 1999). Taken together, the nonessential proteins show no severe effects if deleted separately, but combinations of two or three deletions show more severe phenotypes. Therefore, while some of these proteins can compensate for the lack of other nonessential Prp19C components, collectively they perform essential functions.
Here, we elucidate the function of the nonessential Prp19C subunits in Prp19C and TREX occupancy at transcribed genes and in transcription elongation. In addition, we analyzed the function of the nonessential associated protein Cwc15, because it functions in pre-mRNA splicing and interacts with Cef1 (Ohi et al. 2002). We show that Syf2 plays a role in the interaction of Prp19C with TREX and in Prp19C occupancy. Cwc15 plays a role in the interaction between Prp19C and RNAPII, TREX occupancy, and efficient transcription elongation. Furthermore, the deletion of either CWC15 or SYF2 leads to a reduced interaction of Prp19C with TREX. This interaction is further decreased in a Δcwc15 Δsyf2 double deletion strain, and Δcwc15 and Δsyf2 are synthetically lethal at 37°C. Interestingly, Δcwc15 is also synthetically sick with Δdst1 at 37°C and in the presence of 6-azauracil (6-AU), indicating a function of Cwc15 in transcription elongation. Consistent with this conclusion, the mRNA synthesis rate is decreased in a Δcwc15 strain. Taken together, we show that Cwc15 and Syf2 are important for the interaction between TREX and Prp19C and that Cwc15 has a role in transcription elongation.
RESULTS
The nonessential Prp19C subunit Snt309 is needed for the interaction of Prp19C with Mud2
Prp19C is recruited to transcribed genes by Mud2 in an RNA-independent manner (Minocha et al. 2018), and the essential Prp19C subunit Clf1 most likely is important for this interaction (Chung et al. 1999). No further interactions between Prp19C components and Mud2 are known to date. We, therefore, investigated whether any of the nonessential Prp19C subunits are involved in the Prp19C–Mud2 interaction. To do so, we purified Prp19C from different strains expressing Syf1-TAP, each carrying a deletion of one of the nonessential Prp19C subunits, and assessed the copurification of hemagglutinin (HA)-tagged Mud2 by western blotting. To exclude any differences in the interaction of Mud2 and Prp19C due to changes in Syf1 or Mud2 total protein levels caused by one of the deletions, we determined the amount of Syf1 and Mud2 in whole cell lysates of each deletion mutant. Both Syf1 and Mud2 levels are constant in each of the deletion mutants (Supplemental Fig. S1A,B). Interestingly, deletion of ISY1 leads to an increased interaction between Prp19C and Mud2 (Fig. 1A,B). In contrast, the interaction between Prp19C and Mud2 is significantly reduced when SNT309 is deleted (Fig. 1A,B). A possible explanation for this weaker interaction could be the lower integrity of Prp19C in Δsnt309 cells (Chen et al. 1999), because a weaker association of Clf1 with Prp19C could, in turn, weaken the interaction of Prp19C with Mud2. Nevertheless, Snt309 does play a role in the interaction between Prp19C and Mud2.
FIGURE 1.
Snt309 is necessary for the interaction between Prp19C and Mud2, and Snt309 and Cwc15 are important for the interaction of Prp19C with RNAPII. (A) Coomassie gel of the EGTA eluates of Prp19C purified via TAP-tagged Syf1 from wild-type (WT) and the five deletion strains. The levels of copurifying HA-tagged Mud2 and Rpb1 were assessed by western blotting with an antibody directed against HA (αHA) or the antibody 8WG16 recognizing Rpb1, respectively. A strain expressing Mud2-HA only (i.e., lacking Syf1-TAP) served as negative control. (B,C) Exemplary western blot of three independent experiments using αHA (B) and 8WG16 (C) antibodies as described above (left panels) and quantification of the respective bands normalized to the essential Prp19C component Syf1 (right panels).
The Prp19C subunits Cwc15 and Snt309 mediate the interaction between Prp19C and RNAPII
The essential Prp19C subunit Syf1 interacts—directly or indirectly—with the large subunit of RNAPII (Chanarat et al. 2011). Thus, we were interested whether any of the nonessential Prp19C components are important for the interaction between these two complexes. The copurification of RNAPII with Prp19C was assessed in the five deletion mutants of each of the nonessential Prp19C components (Fig. 1A). To exclude any effects by reduced total RNAPII levels caused by deletion of one of these genes, we determined the total levels of the largest RNAPII subunit Rpb1 in whole cell extracts. A minor decrease of total Rpb1 levels occurs in Δntc20 cells (Supplemental Fig. S1C); however, we did not observe a significant difference in the interaction of Prp19C with Rpb1 in Δntc20 cells (Fig. 1A,C). In contrast, the interaction between Prp19C and Rpb1, and thus most likely the whole RNAPII complex, decreases in Δcwc15 and Δsnt309 cells (Fig. 1A,C). Thus, Cwc15 and Snt309 contribute to the interaction between Prp19C and RNAPII.
Cwc15, Syf2, and Snt309 play an important role in the interaction of Prp19C and TREX
Prp19C is important for the recruitment of TREX to actively transcribed genes and interacts with TREX in an RNA-independent manner in vivo (Chanarat et al. 2011). Thus, we determined, whether Prp19C and THO interact directly using an in vitro binding experiment. First, we purified THO and the heterotetrameric mRNA capping enzyme, the latter of which served as a negative control, bound to IgG beads. During the purification, the lysates were treated with RNaseA to prevent any RNA-mediated interactions. In addition, the IgG beads with bound THO or capping complex were washed with high salt buffer to obtain pure complexes. Equal amounts of Prp19C purified until the EGTA elution step were incubated with THO or the mRNA capping enzyme bound to beads, respectively. Bound proteins were eluted by cleavage with TEV protease. Prp19C indeed binds directly to the THO complex (Supplemental Fig. S2). In contrast, no interaction was observed between Prp19C and the mRNA capping enzyme (Supplemental Fig. S2). Thus, Prp19C binds directly to the THO complex in an RNA-independent manner.
Next, we determined which of the nonessential Prp19C components are involved in this direct interaction between Prp19C and TREX. To do this, we purified TREX using a strain expressing Hpr1-TAP in deletion mutants of each of the nonessential Prp19C subunits and assessed the copurification of Prp19C by western blotting for HA-tagged Syf1. The amount of copurified Prp19C increases when ISY1 is deleted, indicating that the interaction between the two complexes is stronger in the absence of Isy1 (Fig. 2A,B). In deletion mutants of CWC15, SYF2, and SNT309, the amount of copurified Syf1 and thus the interaction of TREX with Prp19C is reduced (Fig. 2A,B).
FIGURE 2.
Cwc15, Syf2, and Snt309 are needed for the interaction between TREX and Prp19C. (A) Coomassie gel of EGTA eluates from cells expressing Hpr1-TAP and HA-tagged Syf1 in a WT, Δcwc15, Δisy1, Δntc20, Δsyf2, and Δsnt309 background (upper panel). CBP, Calmodulin-binding protein; the faster migrating band of Hpr1-CBP in Δcwc15 and Δsyf2 cells was verified by mass spectrometry and is indicated by a star. Syf1-HA levels of extracts (Input) and eluates (IP) were assessed by western blotting using antibody against the HA-tag (lower panels). A strain expressing Syf1-HA only (i.e., lacking the TAP tag on Hpr1) served as negative control. (B) Quantification of western blots of three independent copurifications as shown in A. The intensity of the western band of copurified Syf1 was normalized to the amount of purified Tho2. (C) Coomassie gel of EGTA eluates of purified, N-terminally TAP-tagged Hpr1 from WT, Δcwc15, Δsyf2, and Δcwc15Δsyf2 cells (upper panel) and western blots assessing the corresponding Syf1-HA levels of extracts (Input) and eluates (IP) (lower panels). (D) Quantification of Syf1 copurified with TREX and normalized to Tho2 of three independent experiments as shown in C.
Interestingly, the total Hpr1-TAP levels in whole cell lysates also decrease in the absence of either Cwc15 or Syf2 as determined with an antibody directed against the protein A moiety of the C-terminal TAP tag (Supplemental Fig. S3A,B). These lower Hpr1 levels coincide with a faster migrating band of Hpr1-CBP visible in the TREX eluates from Δcwc15 and Δsyf2 cells (Fig. 2A, indicated by a star). To investigate whether the double deletion of CWC15 and SYF2 has additive effects on the amount of the faster migrating Hpr1-CBP band as well as the TREX–Prp19C interaction, we purified TREX from a Δcwc15 Δsyf2 double deletion strain. Only the faster migrating band of Hpr1-CBP is visible in the EGTA eluate from this Δcwc15 Δsyf2 strain (Supplemental Fig. S3C, indicated by a star). In addition, the interaction of TREX with Prp19C is further decreased (Supplemental Fig. S3C,D). Surprisingly, total levels of Hpr1-TAP are largely restored in the Δcwc15 Δsyf2 strain although several degradation bands are still visible (Supplemental Fig. S3E,F). Mass spectrometric analysis of the Hpr1-CBP bands purified from the wild-type and the double deletion strain revealed peptides corresponding to full-length Hpr1 in both cases (data not shown). As Hpr1 is ubiquitylated (Gwizdek et al. 2005; Babour et al. 2012), the size difference could be due to ubiquitylation. Detection of ubiquitylated Hpr1 in the Hpr1-TAP eluates by western blotting with an antibody directed against ubiquitin indeed revealed strongly decreased ubiquitylation of Hpr1 in the double mutant compared to wild-type cells (Supplemental Fig. S4A,B). Hpr1 is ubiquitinylated at its C terminus by the E3 ubiquitin ligase Rsp5, destabilizing Hpr1 at 37°C (Gwizdek et al. 2005). Indeed, Hpr1 is degraded more quickly at 37°C than at 30°C (Supplemental Fig. S4C; Gwizdek et al. 2005). Deletion of CWC15 and SYF2 increases the half-life of Hpr1-TAP at 37°C, consistent with its lower ubiquitylation level (Supplemental Fig. S4C). Thus, the C-terminal TAP tag impairs the ubiquitylation of Hpr1 when Cwc15 and/or Syf2 is lacking.
To assess the effect of Cwc15 and Syf2 on the interaction of TREX with Prp19C independent of changes in Hpr1 ubiquitylation, we tagged Hpr1 on its N terminus. No faster migrating band is visible in the EGTA eluates of TREX purifications from Δcwc15, Δsyf2, and Δcwc15 Δsyf2 cells expressing TAP-Hpr1 (Fig. 2C). Consistently, the total levels of TAP-Hpr1 in whole cell lysates of Δcwc15 and Δsyf2 cells are not reduced (Supplemental Fig. S3G,H). Importantly, as for the C-terminally TAP-tagged Hpr1 (Supplemental Fig. S3C,D), less Prp19C copurifies with TREX in Δcwc15 and Δsyf2 cells when purified via the N-terminally TAP- tagged Hpr1, and even less Prp19C copurifies with the N-terminally TAP-tagged Hpr1 in the Δcwc15 Δsyf2 double deletion strain (Fig. 2C,D). Thus, Cwc15 and Syf2 are important for the interaction of Prp19C and TREX.
Cwc15 and Syf2 function in Prp19C and TREX occupancy at transcribed genes
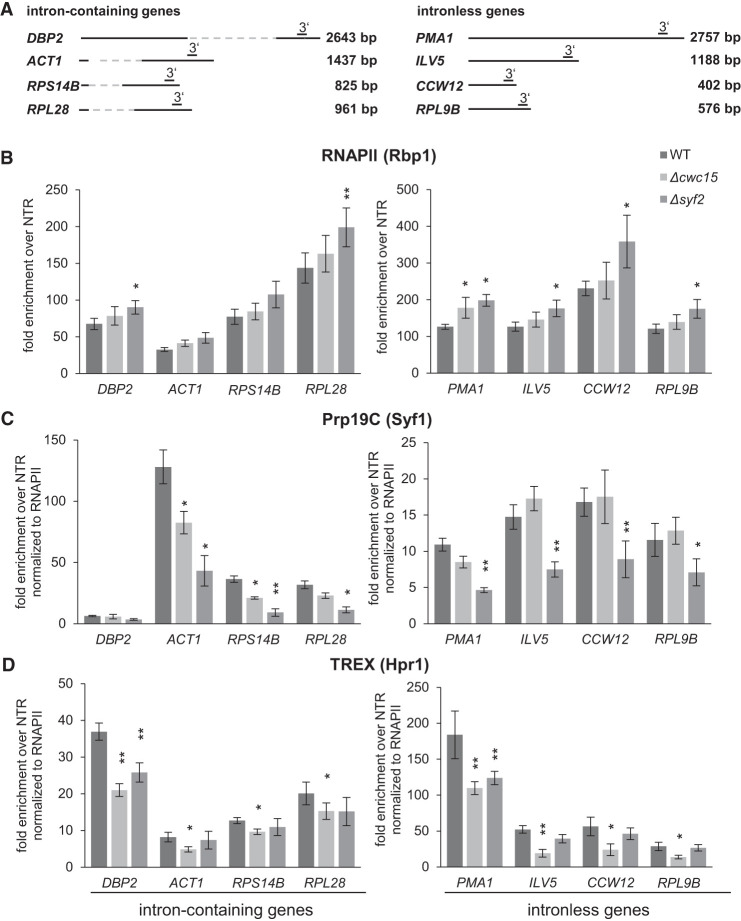
As Cwc15 and Syf2 are needed for the interaction of Prp19C with TREX and RNAPII, these two Prp19C components might also be important for the occupancy of Prp19C and TREX at transcribed genes. Thus, we performed chromatin immunoprecipitation (ChIP) experiments to determine the occupancy of TREX and Prp19C at four exemplary, highly transcribed intron-containing (DBP2, ACT1, RPS14B, RPL28) and four intronless (PMA1, ILV5, CCW12, RPL9B) genes (Fig. 3A) in deletion mutants of CWC15 or SYF2. As the recruitment of RNA-binding protein complexes to the site of transcription is at least partially dependent on RNA and thus on ongoing transcription (i.e., the presence of RNAPII), we first determined the occupancy of RNAPII. In Δcwc15 cells, the occupancy of RNAPII is unchanged compared to wild-type cells at the tested eight genes (Fig. 3B), indicating that the lack of Cwc15 does not cause a strong transcription defect. Interestingly, Prp19C occupancy decreases at half of the selected intron-containing genes, whereas it does not change at intronless genes in Δcwc15 cells, indicating that Cwc15 is needed for Prp19C occupancy at a subset of intron-containing genes (Fig. 3C; Supplemental Fig. S5A). In contrast, TREX occupancy decreases at all tested intron-containing and intronless genes in Δcwc15 cells (Fig. 3D; Supplemental Fig. S5B). This is consistent with our observation that Cwc15 is needed for a stable Prp19C–TREX interaction and the requirement of Prp19C for TREX occupancy (Fig. 2; Chanarat et al. 2011). In Δsyf2 cells, in contrast, RNAPII occupancy increases at both intron-containing and intronless genes (Fig. 3B) indicating that loss of Syf2 affects transcription. Prp19C occupancy decreases, whereas TREX occupancy is not affected in Δsyf2 cells (Fig. 3C,D). This result contradicts the function of Prp19C in TREX occupancy indicating that we do not fully understand the function of Prp19C and its subunit Syf2. Taken together, Syf2 is needed for Prp19C occupancy, and Cwc15 is needed for Prp19C occupancy at a subset of intron-containing genes and for TREX occupancy.
FIGURE 3.
Syf2 functions in Prp19C occupancy, and Cwc15 functions in TREX occupancy. (A) Scheme of four exemplary intron-containing genes (DBP2, ACT1, RPS14B, and RPL28) and four exemplary intronless genes (PMA1, ILV5, CCW12, and RPL9B) used for ChIP experiments. Open reading frames (ORFs) are represented by solid lines and introns by hatched lines. The bars above the genes indicate the positions of the primer pairs used for analysis by quantitative PCR. (B) RNAPII occupancy increases in Δsyf2 cells both at intron-containing and intronless genes. The occupancy of the RNAPII component Rpb1 was assessed in WT, Δcwc15, and Δsyf2 mutant cells by ChIP experiments at the four intron-containing (left panel) and the four intronless (right panel) genes depicted in A. (C) Prp19C occupancy decreases in Δsyf2 mutant cells at intron-containing genes. The occupancy of Prp19C was assessed by ChIP of Syf1-TAP normalized to the occupancy of RNAPII. (D) TREX occupancy decreases in Δcwc15 cells. The occupancy of TREX was assessed by ChIP of Hpr1-TAP normalized to the occupancy of RNAPII. Of note, total levels of Hpr1-TAP decrease in Δcwc15 and Δsyf2 cells. However, as the amount of beads is limiting in the ChIP protocol used, this should not influence the result. Moreover, Hpr1-TAP occupancy in Δsyf2 cells is only decreased at some genes.
Cwc15 is important for efficient transcription in vivo
Because deletion of SYF2 or CWC15 leads to similar phenotypes such as reduced TREX–Prp19C interaction, we also tested for a genetic interaction between these two genes as a genetic interaction indicates a function in the same cellular process. Interestingly, deletion of both SYF2 and CWC15 causes a strong additive growth defect at 37°C compared to the single deletions (Fig. 4A), indeed suggesting overlapping functions of Syf2 and Cwc15. However, SYF2 and CWC15 do not interact genetically in the presence of 6-AU (Fig. 4A). 6-AU leads to decreased intracellular GTP and UTP levels, thereby impeding RNA synthesis. Consequently, sensitivity to 6-AU indicates a function of the deleted gene in transcription elongation. Taken together, Syf2 and Cwc15 likely have overlapping functions, but this overlap appears not to be in transcription elongation.
FIGURE 4.
Cwc15 activity is needed for full transcriptional activity. (A) CWC15 and SYF2 interact genetically. Δcwc15 and Δsyf2 are synthetically lethal at 37°C. Tenfold serial dilutions WT, Δcwc15, Δsyf2, and Δcwc15 Δsyf2 cells were spotted on YPD plates and incubated for 3 d at 37°C (left panel) and on SDC(-ura) plates containing solvent (−6-AU) or 75 μg/mL 6-AU (+6-AU) and incubated for 2–3 d at 30°C (right panel). (B) Deletion of SYF2 does not cause a temperature-sensitive phenotype and does not interact genetically with Δdst1. Dot spots of WT, Δsyf2, Δdst1, and Δdst1 Δsyf2 cells as described in A. (C) Syf2 is not needed mRNA synthesis in vivo. Expression of the endogenous, intronless GAL10 gene and the plasmid-encoded, intron-containing ACT1 gene driven by the GAL10 promoter was induced for 5, 10, 20, and 30 min by the addition of galactose. Total RNA was extracted and the amount of GAL10 and ACT1 mRNA was determined by primer extension. The amount of GAL10 and ACT1 mRNA at the different time points of three independent experiments in WT and Δsyf2 cells was quantified and normalized to the levels of the RNAPIII transcript SCR1. (D) Deletion of CWC15 causes a temperature-sensitive phenotype and is synthetically sick with Δdst1 both at 37°C and on 6-AU plates. Dot spots of WT, Δcwc15, Δdst1, and Δdst1 Δcwc15 cells grown under the conditions described for A. (E) Cwc15 is required for efficient mRNA synthesis in vivo. Experiment as in C.
Nevertheless, Syf2 and Cwc15 are both needed for Prp19C and TREX occupancy, and these two complexes function in transcription elongation (Fig. 3; Chanarat et al. 2011). We, therefore, further investigated the potential function of either Syf2 or Cwc15 in transcription elongation. As a first indication, we tested for a genetic interaction between SYF2 and DST1, the gene encoding the transcription elongation factor TFIIS (Exinger and Lacroute 1992). However, the deletion of SYF2 does not lead to a growth defect at 37°C, and the double deletion of SYF2 and DST1 does not show any additive growth defect (Fig. 4B, left panel). Similarly, Δsyf2 cells are not sensitive to 6-AU and do not interact with Δdst1 in the presence of 6-AU (Fig. 4B, right panel). As an independent line of evidence, we assessed mRNA synthesis in an in vivo transcription assay using two reporter genes: the endogenous, intronless GAL10 gene and the intron-containing ACT1 gene encoded on a plasmid, likewise under control of the GAL10 promotor. Expression from these two GAL10 promoters was induced by the addition of galactose for 5, 10, 20, and 30 min, respectively, and the amount of synthesized GAL10 and ACT1 mRNA was quantified by primer extension. However, the amounts of synthesized GAL10 and ACT1 mRNA in Δsyf2 cells are similar to the respective mRNA levels in wild-type cells, also indicating that Syf2 function is dispensable for efficient mRNA synthesis (Fig. 4C).
In contrast to Δsyf2 cells, Δcwc15 cells have a growth defect comparable to that of Δdst1 cells at 37°C (Fig. 4D, left panel). Furthermore, the Δcwc15 Δdst1 double deletion strain has an additive growth defect at 37°C, i.e., CWC15 and DST1 interact genetically (Fig. 4D, left panel), indicating a function of Cwc15 in transcription elongation. Growth of Δcwc15 cells on 6-AU-containing plates is not impaired compared to wild-type cells; however, deletion of CWC15 exacerbates the growth defect of Δdst1 cells in the presence of 6-AU (Fig. 4D, right panel), again indicating a role of Cwc15 in transcription elongation. To substantiate this conclusion, we performed in vivo transcription assays with the two above-mentioned reporter genes. Consistent with the afore-mentioned data, both GAL10 and ACT1 mRNA levels are significantly lower in Δcwc15 cells compared to wild-type cells (Fig. 4E). Consequently, Cwc15 is needed for efficient mRNA synthesis or stability of intron-containing and intronless mRNA in vivo. Interestingly, the occupancy of RNAPII does not change in Δcwc15 cells (Fig. 3B). Moreover, the occupancy of RNAPII with an S2-phosphorylated CTD, which is the predominant state of RNAPII during transcription elongation, even increases (Supplemental Fig. S5C). As nevertheless mRNA levels decrease in Δcwc15 cells, this might indicate that RNAPII stalls during transcription elongation in Δcwc15 cells.
Taken together, our data show that the two nonessential Prp19C subunits Cwc15 and Syf2 play similar but different roles in the interactions of Prp19C with TREX, Mud2, and RNAPII. Importantly, Cwc15 is needed for efficient transcription elongation in vivo.
DISCUSSION
The TREX complex functions in transcription elongation and mRNA export (Sträßer et al. 2002). TREX is recruited to the transcription machinery by several mechanisms: (i) by direct interaction with the S2-phosphorylated CTD, (ii) by the nascent RNA, and (iii) by Prp19C (Chanarat et al. 2011; Meinel et al. 2013). Prp19C, in turn, is recruited to transcribed genes (i) by Mud2, which also interacts with the S2-phosphorylated CTD, and (ii) via direct interaction with RNAPII, mediated by the C-terminal domain of its subunit Syf1 (Chanarat et al. 2011; Minocha et al. 2018).
The aim of this study was to elucidate the role of the nonessential Prp19C components in the interaction with RNAPII, Mud2, and TREX, in Prp19C and TREX occupancy and in transcription elongation. Our data show that the Prp19C subunit Snt309 is important for the interaction between Prp19C and Mud2, RNAPII as well as TREX (Figs. 1, 2, and 5), which is reflected by the growth impairment caused by deletion of SNT309. Snt309 is required for the stable association of Cef1, an essential Prp19C core component, with Prp19C, and the deletion of SNT309 thus destabilizes the complex (Chen et al. 1999). In addition, the strong growth defect of Δsnt309 cells renders the analysis of Snt309's function difficult because of secondary effects.
Cwc15 is needed for the interaction between Prp19C and RNAPII as well as TREX (Figs. 1, 2, and 5). Consistently, TREX occupancy is reduced in Δcwc15 cells (Fig. 3). As we see similar effects on TREX occupancy at all eight exemplary genes, we believe that it is very similar for all or at least most genes. In addition, the transcriptional activity of two reporter genes is reduced in Δcwc15 compared to wild-type cells (Fig. 4E). This is also consistent with the observed genetic interaction between Δcwc15 and Δdst1 on 6-AU plates and at 37°C (Fig. 4D). In this respect, the phenotypes of Δcwc15 cells are similar to those of Δmud2 cells (Minocha et al. 2018). This is interesting as Cwc15 is known to be a loosely attached protein of Prp19C, and for S. cerevisiae it is not considered a core component of Prp19C (Ohi et al. 2002; van Maldegem et al. 2015).
The nonessential Prp19C component Syf2, in turn, stabilizes the Prp19C–TREX interaction and is needed for full Prp19C occupancy at transcribed genes (Figs. 2 and 3), most likely in a genome-wide manner as we observe the same effects at all eight tested genes. However, Syf2 forms a Prp19C subcomplex together with Syf1, Isy1, and Ntc20, and the reduced Prp19C occupancy in Δsyf2 cells could thus be an indirect effect caused by destabilization of this subcomplex and, consequently, of the interaction of Syf1 with the transcription machinery. Nevertheless, Cwc15 and Syf2 are both needed for Prp19C–TREX interaction, and this interaction is further reduced when both proteins are missing (Fig. 2C; Supplemental Fig. S3C,D). This is reflected by their genetic interaction: Δcwc15 and Δsyf2 are synthetically lethal at 37°C (Fig. 4A).
As determined by the recently published cryo-electron microscopy (cryo-EM) structures of activated spliceosomal complexes B and C, Syf2 helps to bridge Syf1 to the U2–U6 snRNA helix II during splicing, anchoring Prp19C to the active site through its component Cef1 (Bertram et al. 2020; Wilkinson et al. 2021). Syf2 thus helps to stabilize the catalytic core, and its deletion could disrupt this stable conformation, which may, in turn, render the whole Prp19C unstable. Cwc15, which was earlier thought to be a spliceosome-associated protein, has been regarded as a core component of the spliceosomal machinery in recent cryo-EM studies. It is in direct contact with all three snRNA elements—U4, U5, and U6—and interacts simultaneously with multiple components and other NTC-related proteins at the catalytic center of the spliceosome (Wan et al. 2016). Together with Prp45 and a C-terminal fragment of Cef1, Cwc15 acts as a rope-like structure providing flexible tethering to seal together the interactions between different components of Prp19C as well as the spliceosome (Yan et al. 2019). These structural data support our findings that the roles of Syf2 and Cwc15 in stabilizing Prp19C indeed overlap.
The total levels of C-terminal TAP-tagged Hpr1 decrease in Δcwc15 and Δsyf2 cells (Supplemental Fig. S3A,B), and a faster migrating band appears in the TREX purification (Supplemental Fig. S3C,D). Interestingly, total Hpr1-TAP levels are restored in Δcwc15 Δsyf2 double deletion strain (Supplemental Fig. S3E,F), and only the faster migrating band is visible (Supplemental Fig. S3C,D). As determined by mass spectrometric analysis, both the slower and the faster migrating band of Hpr1-CBP from wild-type and Δcwc15 Δsyf2 cells, respectively, contain full-length protein (data not shown). Hpr1 is ubiquitylated by the ubiquitin ligase Rsp5, degraded by the proteasome, and thus more unstable at 37°C (Gwizdek et al. 2005). However, the Hpr1 ubiquitylation does not only serve a degradative function: Ubiquitylated Hpr1 recruits the mRNA exporter Mex67 to transcribed genes, and, consistently, its ubiquitin ligase Rps5 is essential for nuclear mRNA export (Rodriguez et al. 2003; Gwizdek et al. 2006). As Hpr1-TAP is less ubiquitylated in the Δcwc15 Δsyf2 strain (Supplemental Fig. S4A), the faster migrating band most likely corresponds to nonubiquitylated Hpr1. This is consistent with our observation that Hpr1-TAP is more stable at 37°C in Δcwc15 Δsyf2 than in wild-type cells (Supplemental Fig. S4C), similar to the stabilization of Hpr1 observed in Δrsp5 cells (Gwizdek et al. 2005). Thus, the combination of a C-terminal TAP tag on Hpr1 with the deletion of CWC15 and/or SYF2 interferes with the ubiquitylation of Hpr1 and thus a longer half-life.
Δcwc15, Δsyf2, and Δcwc15 Δsyf2 cells do not show a nuclear mRNA export defect at 30°C or 37°C (data not shown). This is comparable to deletion of the C terminus of Syf1 in syf1–37 cells or deletion of MUD2, both of which also cause a decrease in TREX and Prp19C occupancy, but no nuclear mRNA export defect (Chanarat et al. 2011; Minocha et al. 2018). Most likely, the decrease in transcription elongation and thus in mRNA synthesis caused by the mutations alleviates the possibly existing delay in mRNA export.
In summary, Cwc15 and Syf2 mediate the interaction of TREX with Prp19C, whereas Cwc15 mediates the interaction of Prp19c with RNAPII (Fig. 5). In addition, Cwc15 and Syf2 function in Prp19C and TREX occupancy (Fig. 5). Importantly, Cwc15 is needed for efficient transcription elongation. Notably, this is the first time a specific function of these nonessential Prp19C subunits beyond their role in splicing has been determined.
MATERIALS AND METHODS
Quantification of total protein levels by western blot
To quantify total protein levels, 5 OD600 units of cells grown to the mid-log phase were harvested and lysed by the NaOH method and their proteins precipitated by TCA (Karakasili et al. 2014). Briefly, the cell pellet was resuspended in 500 µL of water, 150 µL of pretreatment solution (7.5% [v/v] β-mercaptoethanol, 1.85 M NaOH) was added, and the mixture was incubated for 20 min on ice. TCA was added to a final concentration of 10%, and the solution was incubated for 20 min on ice and centrifuged for 20 min at 15,000 rpm at 4°C. The supernatant was discarded, the pellet was resuspended in 90 µL of 1× SDS-loading buffer, and 10 µL 1 M Tris-base was added. Equal amounts of total protein corresponding to ∼6 µL of whole cell extract were separated by SDS-PAGE. Proteins tagged with the TAP or HA tag were detected with PAP or an anti-HA antibody (Sigma; R&D Systems) and a horseradish peroxidase (HRP)-coupled secondary antibody and CheLuminate-HRP ECL solution (Applichem) according to the manufacturer's instructions. Western blot signals were imaged using a ChemoCam Imager (Intas) and quantified using ImageJ. Each quantification is based on at least three biologically independent replicates.
Tandem affinity purification of native protein complexes
Purification of native protein complexes via a TAP-tagged subunit was performed as described previously (Puig et al. 2001). Briefly, 2 L of cell culture was harvested at an OD600 of 3.5 and lysed with a cryo-mill (Freezer/Mill 6870D, Spex Sample Prep). Lysis buffer (50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 1.5 mM MgCl2, 0.15% NP40, 1 mM DTT, 1.3 μg/mL pepstatin A, 0.28 μg/mL leupeptin, 170 μg/mL PMSF, 330 μg/mL benzamidine) was added to the cell powder and centrifuged (1 h, 164,700g, 4°C) and the supernatant was incubated for 1.5 h with IgG-coupled Sepharose beads (GE Healthcare). The beads were washed with lysis buffer and proteins were eluted by cleavage with TEV protease. The TEV eluates were incubated for 1 h with prewashed calmodulin Sepharose beads (Agilent Technologies) and washed with lysis buffer containing 2 mM CaCl2. Proteins were eluted with buffer containing 25 mM EGTA, precipitated with 10% TCA (v/v), and separated by SDS-PAGE. Copurifying proteins were analyzed by Coomassie staining or western blotting using antibodies against HA (Roche) or the CTD of Rpb1 (8WG16, Biolegend).
In vitro binding assay of Prp19C and THO
Prp19C was purified from 12 L of culture of an SYF1-TAP strain until EGTA elution (EGTA-E). THO and mRNA capping enzyme, a heterotetramer composed of two Ceg1 and two Cet1 molecules that served as a negative control, were purified from an HPR1-TAP strain and a CEG1-TAP strain, respectively. Lysates of the HPR1-TAP and CEG1-TAP strains were treated with 100 µg/mL RNase A and incubated with IgG Sepharose beads. Beads were washed with 10 mL high salt lysis buffer (50 mM Tris-HCl at pH 7.5, 1 M NaCl, 1.5 mM MgCl2, 0.15% NP40, 1 mM DTT, 1.3 μg/mL pepstatin A, 0.28 μg/mL leupeptin, 170 μg/mL PMSF, 330 μg/mL benzamidine) in order to obtain complexes. Equal amounts of purified and concentrated Prp19C were incubated with THO or the mRNA capping complex bound to IgG-coupled Sepharose beads for 1 h at 4°C on a turning wheel. After incubation, the beads were washed with 5 mL TAP lysis buffer, and the proteins were eluted by cleavage with TEV protease for 1 h at 4°C and separated by SDS-PAGE.
Chromatin immunoprecipitation experiments
ChIP experiments were performed as described in Reuter et al. (2015) with some modifications. Briefly, 100 mL yeast culture was grown to an OD600 of 0.8 and cross-linked with 1% formaldehyde. Cells were lysed with an equal volume of glass beads and sonicated three times for 15 min each with intermittent cooling, resulting in chromatin fragments of 200–250 bp. Endogenously TAP-tagged versions of the proteins of interest were immunoprecipitated by incubation of whole cell lysate with IgG-coupled Dynabeads (tosylactivated M280, Thermo Scientific) for 2.5 h at RT. For ChIP experiments of RNAPII and S2-phosphorylated RNAPII, 4 µL of the monoclonal antibody 8WG16 (Biolegend) or 2 µL of the antibody 3E10 (Abcam), respectively, were added for 1.5 h at RT followed by 1 h incubation with Protein G Dynabeads. After washing and elution, eluates, as well as input samples, were treated with proteinase K overnight at 65°C for the reversal of crosslinks. To determine the occupancy of each protein of interest at transcribed genes, eight paradigmatic genes were amplified from the precipitated DNA by quantitative PCR using specific primer pairs; a nontranscribed region (NTR1, 174,131–174,200 on Chr. V) served as negative control. The occupancy of each protein was calculated as the enrichment in the IP over the input (Input) sample of each transcribed gene normalized to NTR1: .
In vivo transcription assay
The in vivo transcription assays were performed as described in Minocha et al. (2018). Shortly, transcription of two genes, the endogenous intronless GAL10 gene and the intron-containing ACT1 gene, also under the control of the GAL10 promoter and encoded on a plasmid, was examined; the RNAPIII transcript SCR1 served as a standard. Cells were grown in media containing raffinose as a carbon source (2%, w/v), and expression from the GAL10 promotors was induced by the addition of 2% (w/v) galactose for 0–30 min. The amount of GAL10 and ACT1 mRNA and SCR1 was measured by primer extension using 5′-Cy5-labeled oligonucleotides specific for GAL10, ACT1, and SCR1, respectively. The cDNA was separated on a 7 M urea 7% polyacrylamide gel and quantified using ImageJ.
Statistical analysis
All data are presented as mean ± standard deviation (error bars) of at least three biologically independent experiments. Asterisks indicate the statistical significance (Student's t-test; *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank Günter Lochnit for the analysis of the Hpr1-CBP band purified from wild-type and Δcwc15 Δsyf2 cells. We thank Sittinan Chanarat for strain SYF1-TAP (BY4741), Susanne Röther for strain HPR1-TAP (BY4741), and Eleni Karakasili for strain Δdst1 (W303). We thank Vera Bettenworth for the critical reading of the manuscript. This work was supported by the European Union (European Research Council Consolidator Grant 772049 to K.S.) and the German Research Foundation (RTG2355 to K.S.).
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.079944.124.
Freely available online through the RNA Open Access option.
MEET THE FIRST AUTHORS
Laura Henke-Schulz.

Rashmi Minocha.

Meet the First Author(s) is an editorial feature within RNA, in which the first author(s) of research-based papers in each issue have the opportunity to introduce themselves and their work to readers of RNA and the RNA research community. Laura Henke-Schulz and Rashmi Minocha are co-first authors of this paper, “The Prp19C/NTC subunit Syf2 and the Prp19C/NTC-associated protein Cwc15 function in TREX occupancy and transcription elongation.” Laura completed her undergraduate and graduate studies in Biochemistry at Goethe University in Frankfurt. Subsequently, she pursued her doctoral degree in Biochemistry at Justus Liebig University in Giessen. Rashmi completed her undergraduate and postgraduate studies in India and then moved to Germany to pursue her doctorate in Biochemistry from the Ludwig Maximilian University of Munich. She also completed a part of her doctoral work at Justus Liebig University of Giessen, where the research for this published work was carried out. After her PhD, she moved back to India to pursue her interests in health-based research.
What are the major results described in your paper and how do they impact this branch of the field?
In our paper, we show that Cwc15 plays a pivotal role in mediating interactions between Prp19C and RNAPII/TREX, affecting transcription elongation, whereas Syf2 stabilizes Prp19C–TREX interaction, impacting mRNA export regulation. Deletion of Cwc15 and/or Syf2 interferes with Hpr1 ubiquitylation. These findings contribute to our understanding of the roles of nonessential Prp19C subunits beyond splicing, particularly in mediating interactions between transcription and mRNA export machinery.
What led you to study RNA or this aspect of RNA science?
LH-S: My interest in RNA science originated from its central role as a dynamic molecule in the complex mechanisms of life. I found its ability to modulate gene expression and participate in multiple cellular processes in various ways fascinating.
RM: I have always been intrigued by the complex molecular mechanisms underlying gene expression regulation. RNA, acting as a pivotal bridge, controls processes from transcription to mRNA processing and translation, shaping cellular activities and fate. I wanted to indulge deeper into understanding the multifaceted roles of RNA in these processes.
What are some of the landmark moments that provoked your interest in science or your development as a scientist?
LH-S: During my childhood, I developed a strong inclination toward exploration, often peppering my parents with inquiries about the inner workings of the world. This innate curiosity gradually evolved into a structured approach, characterized by the formulation of hypotheses followed by diligent testing and validation.
RM: My interest in science was sparked by my childhood curiosity, when I often found myself asking questions such as “How does this process work?” or “What scientific principles are at play here?” Biology always fascinated me, especially the complexities of a single cell shaping the entirety of life. It was this drive, along with witnessing new discoveries unfolding in the scientific community around me, that motivated me to pursue a career in science.
If you were able to give one piece of advice to your younger self, what would that be?
LH-S: If I could give just one piece of advice to my younger self, it would be to always trust your instincts. Although it may be difficult at times, it often leads to positive outcomes. However, do not hesitate to seek help when needed, and do not wait too long to ask for assistance.
RM: Embrace failure as an essential part of the learning process. Never let struggles demotivate you, both in science and in personal life! Setbacks and challenges are just opportunities for growth and resilience. If you feel stuck, take a step back, relax, and find joy in life's simple pleasures. Above all, trust that you have inside you the capability to achieve anything you desire!
How did you decide to work together as co-first authors?
LH-S: We decided to collaborate as co-first authors because although my colleague had completed her PhD program, we both desired to see the project through to its conclusion. This arrangement proved to be an excellent solution for both of us, allowing us to achieve our goals effectively.
REFERENCES
- Abruzzi KC, Lacadie S, Rosbash M. 2004. Biochemical analysis of TREX complex recruitment to intronless and intron-containing yeast genes. EMBO J 23: 2620–2631. 10.1038/sj.emboj.7600261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albers M, Diment A, Muraru M, Russell CS, Beggs JD. 2003. Identification and characterization of Prp45p and Prp46p, essential pre-mRNA splicing factors. RNA 9: 138–150. 10.1261/rna.2119903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babour A, Dargemont C, Stutz F. 2012. Ubiquitin and assembly of export competent mRNP. Biochim Biophys Acta 1819: 521–530. 10.1016/j.bbagrm.2011.12.006 [DOI] [PubMed] [Google Scholar]
- Bertram K, El Ayoubi L, Dybkov O, Agafonov DE, Will CL, Hartmuth K, Urlaub H, Kastner B, Stark H, Lührmann R. 2020. Structural insights into the roles of metazoan-specific splicing factors in the human step 1 spliceosome. Mol Cell 80: 127–139.e6. 10.1016/j.molcel.2020.09.012 [DOI] [PubMed] [Google Scholar]
- Chan SP, Cheng SC. 2005. The Prp19-associated complex is required for specifying interactions of U5 and U6 with pre-mRNA during spliceosome activation. J Biol Chem 280: 31190–31199. 10.1074/jbc.M505060200 [DOI] [PubMed] [Google Scholar]
- Chan SP, Kao DI, Tsai WY, Cheng SC. 2003. The Prp19p-associated complex in spliceosome activation. Science 302: 279–282. 10.1126/science.1086602 [DOI] [PubMed] [Google Scholar]
- Chanarat S, Sträßer K. 2013. Splicing and beyond: the many faces of the Prp19 complex. Biochim Biophys Acta 1833: 2126–2134. 10.1016/j.bbamcr.2013.05.023 [DOI] [PubMed] [Google Scholar]
- Chanarat S, Seizl M, Sträßer K. 2011. The Prp19 complex is a novel transcription elongation factor required for TREX occupancy at transcribed genes. Genes Dev 25: 1147–1158. 10.1101/gad.623411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanarat S, Burkert-Kautzsch C, Meinel DM, Sträßer K. 2012. Prp19C and TREX: interacting to promote transcription elongation and mRNA export. Transcription 3: 8–12. 10.4161/trns.3.1.19078 [DOI] [PubMed] [Google Scholar]
- Chen HR, Jan SP, Tsao TY, Sheu YJ, Banroques J, Cheng SC. 1998. Snt309p, a component of the Prp19p-associated complex that interacts with Prp19p and associates with the spliceosome simultaneously with or immediately after dissociation of U4 in the same manner as Prp19p. Mol Cell Biol 18: 2196–2204. 10.1128/MCB.18.4.2196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HR, Tsao TY, Chen CH, Tsai WY, Her LS, Hsu MM, Cheng SC. 1999. Snt309p modulates interactions of Prp19p with its associated components to stabilize the Prp19p-associated complex essential for pre-mRNA splicing. Proc Natl Acad Sci 96: 5406–5411. 10.1073/pnas.96.10.5406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CH, Tsai WY, Chen HR, Wang CH, Cheng SC. 2001. Identification and characterization of two novel components of the Prp19p-associated complex, Ntc30p and Ntc20p. J Biol Chem 276: 488–494. 10.1074/jbc.M006958200 [DOI] [PubMed] [Google Scholar]
- Chen CH, Yu WC, Tsao TY, Wang LY, Chen HR, Lin JY, Tsai WY, Cheng SC. 2002. Functional and physical interactions between components of the Prp19p-associated complex. Nucleic Acids Res 30: 1029–1037. 10.1093/nar/30.4.1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SY, Shin ES, Park PJ, Shin DW, Chang HK, Kim D, Lee HH, Lee JH, Kim SH, Song MJ, et al. 2007. Identification of mouse Prp19p as a lipid droplet-associated protein and its possible involvement in the biogenesis of lipid droplets. J Biol Chem 282: 2456–2465. 10.1074/jbc.M608042200 [DOI] [PubMed] [Google Scholar]
- Chung S, McLean MR, Rymond BC. 1999. Yeast ortholog of the Drosophila crooked neck protein promotes spliceosome assembly through stable U4/U6.U5 snRNP addition. RNA 5: 1042–1054. 10.1017/S1355838299990635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exinger F, Lacroute F. 1992. 6-Azauracil inhibition of GTP biosynthesis in Saccharomyces cerevisiae. Curr Genet 22: 9–11. 10.1007/BF00351735 [DOI] [PubMed] [Google Scholar]
- Fabrizio P, Dannenberg J, Dube P, Kastner B, Stark H, Urlaub H, Lührmann R. 2009. The evolutionarily conserved core design of the catalytic activation step of the yeast spliceosome. Mol Cell 36: 593–608. 10.1016/j.molcel.2009.09.040 [DOI] [PubMed] [Google Scholar]
- Grote M, Wolf E, Will CL, Lemm I, Agafonov DE, Schomburg A, Fischle W, Urlaub H, Lührmann R. 2010. Molecular architecture of the human Prp19/CDC5L complex. Mol Cell Biol 30: 2105–2119. 10.1128/MCB.01505-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwizdek C, Hobeika M, Kus B, Ossareh-Nazari B, Dargemont C, Rodriguez MS. 2005. The mRNA nuclear export factor Hpr1 is regulated by Rsp5-mediated ubiquitylation. J Biol Chem 280: 13401–13405. 10.1074/jbc.C500040200 [DOI] [PubMed] [Google Scholar]
- Gwizdek C, Iglesias N, Rodriguez MS, Ossareh-Nazari B, Hobeika M, Divita G, Stutz F, Dargemont C. 2006. Ubiquitin-associated domain of Mex67 synchronizes recruitment of the mRNA export machinery with transcription. Proc Natl Acad Sci 103: 16376–16381. 10.1073/pnas.0607941103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath CG, Viphakone N, Wilson SA. 2016. The role of TREX in gene expression and disease. Biochem J 473: 2911–2935. 10.1042/BCJ20160010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idrissou M, Maréchal A. 2022. The PRP19 ubiquitin ligase, standing at the cross-roads of mRNA processing and genome stability. Cancers (Basel) 14: 878. 10.3390/cancers14040878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karakasili E, Burkert-Kautzsch C, Kieser A, Sträßer K. 2014. Degradation of DNA damage-independently stalled RNA polymerase II is independent of the E3 ligase Elc1. Nucleic Acids Res 42: 10503–10515. 10.1093/nar/gku731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khong A, Parker R. 2020. The landscape of eukaryotic mRNPs. RNA 26: 229–239. 10.1261/rna.073601.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kistler AL, Guthrie C. 2001. Deletion of MUD2, the yeast homolog of U2AF65, can bypass the requirement for sub2, an essential spliceosomal ATPase. Genes Dev 15: 42–49. 10.1101/gad.851301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinel DM, Sträßer K. 2015. Co-transcriptional mRNP formation is coordinated within a molecular mRNP packaging station in S. cerevisiae. Bioessays 37: 666–677. 10.1002/bies.201400220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinel DM, Burkert-Kautzsch C, Kieser A, O'Duibhir E, Siebert M, Mayer A, Cramer P, Söding J, Holstege FC, Sträßer K. 2013. Recruitment of TREX to the transcription machinery by its direct binding to the phospho-CTD of RNA polymerase II. PLoS Genet 9: e1003914. 10.1371/journal.pgen.1003914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minocha R, Popova V, Kopytova D, Misiak D, Hüttelmaier S, Georgieva S, Sträßer K. 2018. Mud2 functions in transcription by recruiting the Prp19 and TREX complexes to transcribed genes. Nucleic Acids Res 46: 9749–9763. 10.1093/nar/gky640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell SF, Parker R. 2014. Principles and properties of eukaryotic mRNPs. Mol Cell 54: 547–558. 10.1016/j.molcel.2014.04.033 [DOI] [PubMed] [Google Scholar]
- Oeffinger M, Montpetit B. 2015. Emerging properties of nuclear RNP biogenesis and export. Curr Opin Cell Biol 34: 46–53. 10.1016/j.ceb.2015.04.007 [DOI] [PubMed] [Google Scholar]
- Ohi MD, Gould KL. 2002. Characterization of interactions among the Cef1p-Prp19p-associated splicing complex. RNA 8: 798–815. 10.1017/S1355838202025050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohi MD, Link AJ, Ren L, Jennings JL, McDonald WH, Gould KL. 2002. Proteomics analysis reveals stable multiprotein complexes in both fission and budding yeasts containing Myb-related Cdc5p/Cef1p, novel pre-mRNA splicing factors, and snRNAs. Mol Cell Biol 22: 2011–2024. 10.1128/MCB.22.7.2011-2024.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig O, Caspary F, Rigaut G, Rutz B, Bouveret E, Bragado-Nilsson E, Wilm M, Séraphin B. 2001. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24: 218–229. 10.1006/meth.2001.1183 [DOI] [PubMed] [Google Scholar]
- Reuter LM, Meinel DM, Sträßer K. 2015. The poly(A)-binding protein Nab2 functions in RNA polymerase III transcription. Genes Dev 29: 1565–1575. 10.1101/gad.266205.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez MS, Gwizdek C, Haguenauer-Tsapis R, Dargemont C. 2003. The HECT ubiquitin ligase Rsp5p is required for proper nuclear export of mRNA in Saccharomyces cerevisiae. Traffic 4: 566–575. 10.1034/j.1600-0854.2003.00115.x [DOI] [PubMed] [Google Scholar]
- Schmitzová J, Cretu C, Dienemann C, Urlaub H, Pena V. 2023. Structural basis of catalytic activation in human splicing. Nature 617: 842–850. 10.1038/s41586-023-06049-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh G, Pratt G, Yeo GW, Moore MJ. 2015. The clothes make the mRNA: past and present trends in mRNP fashion. Annu Rev Biochem 84: 325–354. 10.1146/annurev-biochem-080111-092106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slane D, Lee CH, Kolb M, Dent C, Miao Y, Franz-Wachtel M, Lau S, Maček B, Balasubramanian S, Bayer M, et al. 2020. The integral spliceosomal component CWC15 is required for development in Arabidopsis. Sci Rep 10: 13336. 10.1038/s41598-020-70324-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sträßer K, Masuda S, Mason P, Pfannstiel J, Oppizzi M, Rodriguez-Navarro S, Rondón AG, Aguilera A, Struhl K, Reed R, et al. 2002. TREX is a conserved complex coupling transcription with messenger RNA export. Nature 417: 304–308. 10.1038/nature746 [DOI] [PubMed] [Google Scholar]
- Tarn WY, Hsu CH, Huang KT, Chen HR, Kao HY, Lee KR, Cheng SC. 1994. Functional association of essential splicing factor(s) with PRP19 in a protein complex. EMBO J 13: 2421–2431. 10.1002/j.1460-2075.1994.tb06527.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Maldegem F, Maslen S, Johnson CM, Chandra A, Ganesh K, Skehel M, Rada C. 2015. CTNNBL1 facilitates the association of CWC15 with CDC5L and is required to maintain the abundance of the Prp19 spliceosomal complex. Nucleic Acids Res 43: 7058–7069. 10.1093/nar/gkv643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan R, Yan C, Bai R, Huang G, Shi Y. 2016. Structure of a yeast catalytic step I spliceosome at 3.4 Å resolution. Science 353: 895–904. 10.1126/science.aag2235 [DOI] [PubMed] [Google Scholar]
- Wegener M, Müller-McNicoll M. 2019. View from an mRNP: the roles of SR proteins in assembly, maturation and turnover. Adv Exp Med Biol 1203: 83–112. 10.1007/978-3-030-31434-7_3 [DOI] [PubMed] [Google Scholar]
- Wende W, Friedhoff P, Sträßer K. 2019. Mechanism and regulation of co-transcriptional mRNP assembly and nuclear mRNA export. Adv Exp Med Biol 1203: 1–31. 10.1007/978-3-030-31434-7_1 [DOI] [PubMed] [Google Scholar]
- Wilkinson ME, Fica SM, Galej WP, Nagai K. 2021. Structural basis for conformational equilibrium of the catalytic spliceosome. Mol Cell 81: 1439–1452.e9. 10.1016/j.molcel.2021.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Ren Y. 2019. Mechanisms of nuclear mRNA export: a structural perspective. Traffic 20: 829–840. 10.1111/tra.12691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C, Wan R, Shi Y. 2019. Molecular mechanisms of pre-mRNA splicing through structural biology of the spliceosome. Cold Spring Harb Perspect Biol 11: a032409. 10.1101/cshperspect.a032409 [DOI] [PMC free article] [PubMed] [Google Scholar]