Abstract
Endemic/epidemic dengue viruses (DEN) that are transmitted among humans by the mosquito vectors Aedes aegypti and Aedes albopictus are hypothesized to have evolved from sylvatic DEN strains that are transmitted among nonhuman primates in West Africa and Malaysia by other Aedes mosquitoes. We tested this hypothesis with phylogenetic studies using envelope protein gene sequences of both endemic/epidemic and sylvatic strains. The basal position of sylvatic lineages of DEN-1, -2, and -4 suggested that the endemic/epidemic lineages of these three DEN serotypes evolved independently from sylvatic progenitors. Time estimates for evolution of the endemic/epidemic forms ranged from 100 to 1,500 years ago, and the evolution of endemic/epidemic forms represents relatively recent events in the history of DEN evolution. Analysis of envelope protein amino acid changes predicted to have accompanied endemic/epidemic emergence suggested a role for domain III in adaptation to new mosquito and/or human hosts.
Dengue viruses (DEN) (Flaviviridae: Flavivirus) are serious human pathogens that occur nearly throughout the tropics, with ca. 100 million cases annually (16). DEN comprise four serotypes (DEN-1 to DEN-4); although epidemiologically similar, they are genetically and antigenically distinct. Infection with one serotype leads to lifelong protection against homologous reinfection but only brief protection against heterologous challenge (21, 38).
DEN cause dengue fever, a self-limited febrile illness lasting 2 to 10 days that has been known in the medical literature for over 200 years. Infrequent epidemics of dengue fever occurred in tropical areas until the 1950s. After War World II, this pattern of disease was disrupted by the emergence of dengue hemorrhagic fever and dengue shock syndrome, more severe diseases characterized by thrombocytopenia, hemorrhage, and excessive plasma leakage (16, 28). Two principal hypotheses have been proposed to explain the hemorrhagic form of disease: (i) the immune enhancement theory maintains that hemorrhage occurs in secondary infections when DEN-specific antibodies and memory T cells resulting from primary infection with another serotype enhance the binding of virus-immunoglobulin G complexes to FcY receptors on monocytic cells, and (ii) certain phenotypes of DEN are more virulent than others. Recent phylogenetic studies suggest that an Asian genotype of DEN-2 recently introduced into the New World may be associated with increased risk for hemorrhagic fever and shock in the presence of heterologous antibody (32). DEN strains of reduced virulence have also been described; endemic transmission on the South Pacific islands of Tonga, involving vectors other than Aedes aegypti, may result in less severe disease because of the lesser selection for high viremia imposed by more susceptible vectors, like Aedes polynesiensis (13).
Two distinct DEN transmission cycles occur: (i) endemic and epidemic DEN involving human hosts and transmission by A. aegypti, with Aedes albopictus and other Aedes mosquitoes serving as secondary vectors, and (ii) a zoonotic or sylvatic cycle in sylvatic habitats of Africa and Malaysia, involving nonhuman primate reservoir hosts and several different Aedes mosquitoes (12, 16). Although A. aegypti plays a greater role in urban transmission, A. albopictus and some other secondary Aedes vectors are generally more susceptible to experimental infection (12). Considerable variation in susceptibility and transmission efficiency among geographic populations of both A. aegypti and A. albopictus has also been demonstrated (14, 15). A. aegypti has reinfested most of the neotropics since its partial eradication earlier this century, resulting in reemergence of neotropical dengue (13).
The sylvatic transmission cycles of DEN have received little study. In West Africa, nonhuman primate cycles have been identified in several countries. DEN-2 has been isolated from Aedes (S.) africanus, Aedes (S.) leuteocephalus, Aedes (S.) opok, Aedes (D.) taylori, and Aedes (D.) furcifer (5, 33). African DEN-2 sylvatic isolates are genetically distinct from all endemic/epidemic isolates and are believed to be evolutionarily distinct (12, 16, 31). In Malaysia, all four serotypes of DEN are maintained in canopy-dwelling Aedes niveus mosquitoes and nonhuman primates (34–37). DEN-1, -2, and -4 were isolated from sentinel monkeys, and monkey seroconversion was demonstrated against DEN-1, -2, and -3. DEN-4 was also isolated from canopy collections of A. niveus mosquitoes (35).
Gubler (13) has hypothesized that endemic/epidemic DEN evolved from sylvatic forms of the viruses that utilize nonhuman primate hosts and gallery forest Aedes vectors (i.e., not A. aegypti or A. albopictus). To test this hypothesis, we have sequenced the complete envelope (E) protein gene of DEN-1, -2, and -4 strains isolated in primary tropical forest habitats in Malaysia by Albert Rudnick and colleagues during the 1960s and 1970s (36, 37), as well as DEN-2 strains from sylvatic locations in West Africa analyzed previously using other, smaller genome regions (31). Phylogenetic analyses indicated that the endemic/epidemic lineages of DEN-1, -2, and -4 evolved independently from sylvatic viruses circulating in the Asian-Oceania region. Estimates of divergence times suggested that emergences of endemic/epidemic DEN occurred on the order of 100 to 1,500 years ago.
MATERIALS AND METHODS
Viruses.
The DEN isolates we sequenced were obtained from reference center collections at the Centers for Disease Control and Prevention, Fort Collins, Colo., and the University of Texas Medical Branch, Galveston (Table 1). Sylvatic DEN from Malaysia included sentinel monkey isolates of DEN-1 (strain P72-1244), DEN-2 (P8-1407), and DEN-4 (P73-1120 and P75-514). Sylvatic DEN-4 strain P75-215 was isolated from A. niveus mosquitoes collected in the Malaysian forest canopy. Endemic/epidemic strains from Malaysia included mosquito isolates of DEN-2 (P7-863 and P8-377) and DEN-4 (P7-1006); DEN-4 strain 703-4 was isolated from Thailand. The DEN-2 African sylvatic strains sequenced were DAKAr A578, PM33974, and DAK HD 10674. The sources and years of isolation for these and other DEN strains whose sequences were used in our analysis are also listed in Table 1.
TABLE 1.
DEN isolates studied
| Strain | Epidemiological typea | Host | Yr isolated | Country where isolated | Serotype | GenBank accession no. | Reference |
|---|---|---|---|---|---|---|---|
| BR/90 | Endemic | Human | 1990 | Brazil | 1 | S64849 | 6 |
| S275/90 | Endemic | Human | 1990 | Singapore | 1 | M87512 | 10 |
| 45A25 | Endemic | Human | 1974 | Nauru | 1 | U88536 | 27 |
| 836-1 | Endemic | Human | 1984 | Philippines | 1 | D00503 | 4 |
| 1636 | Endemic | Human | 1977 | Jamaica | 1 | D00501 | 4 |
| AHF82 | Endemic | Human | 1980 | Thailand | 1 | D00502 | 4 |
| P72-1244b | Sylvatic | Sentinel monkey | 1972 | Malaysia | 1 | AF231721 | |
| H87 | Endemic | NRc | 1956 | Philippines | 3 | M93130 | 23 |
| 168-AP-2 | Endemic | NR | 1983 | Philippines | 3 | L11432 | 23 |
| 260698 | Endemic | NR | 1989 | Sri Lanka | 3 | L11437 | 23 |
| 1327 | Endemic | NR | 1965 | Tahiti | 3 | L11439 | 23 |
| 1340 | Endemic | NR | 1977 | Puerto Rico | 3 | L11434 | 23 |
| 2783 | Endemic | NR | 1991 | Sri Lanka | 3 | L11438 | 23 |
| 1594 | Endemic | NR | 1985 | Sri Lanka | 3 | L11436 | 23 |
| 1326 | Endemic | NR | 1981 | Sri Lanka | 3 | L11431 | 23 |
| 1416 | Endemic | NR | 1984 | India | 3 | L11424 | 23 |
| 1696 | Endemic | NR | 1986 | Samoa | 3 | L11435 | 23 |
| 1558 | Endemic | NR | 1985 | Mozambique | 3 | L11430 | 23 |
| 2167 | Endemic | NR | 1989 | Tahiti | 3 | L11619 | 23 |
| 29472 | Endemic | NR | 1992 | Fiji | 3 | L11422 | 23 |
| 1300 | Endemic | NR | 1974 | Malaysia | 3 | L11429 | 23 |
| 228761 | Endemic | NR | 1973 | Indonesia | 3 | L11425 | 23 |
| 159 | Endemic | NR | 1985 | Indonesia | 3 | L11428 | 23 |
| 29586 | Endemic | NR | 1981 | Malaysia | 3 | L11427 | 23 |
| CH3489D73-1 | Endemic | NR | 1973 | Thailand | 3 | L11620 | 23 |
| MK315 | Endemic | NR | 1987 | Thailand | 3 | L11442 | 23 |
| D86-007 | Endemic | NR | 1986 | Thailand | 3 | L11441 | 23 |
| 5987 | Endemic | NR | 1962 | Thailand | 3 | L11430 | 23 |
| 1280 | Endemic | NR | 1978 | Indonesia | 3 | L11426 | 23 |
| PR6 | Endemic | NR | 1963 | Puerto Rico | 3 | L11433 | 23 |
| ThNH-28/93 | Endemic | NR | 1993 | Thailand | 2 | U31950 | 43 |
| ThNH-52/93 | Endemic | NR | 1993 | Thailand | 2 | U31951 | 43 |
| ThNH-7/93 | Endemic | NR | 1993 | Thailand | 2 | U31959 | 43 |
| ThNH-p11/93 | Endemic | NR | 1993 | Thailand | 2 | U31952 | 43 |
| 200787 | Endemic | NR | 1983 | Mexico | 2 | L04561 | 41 |
| P8-377b | Sylvatic | A. albopictus | 1969 | Malaysia | 2 | AF231715 | |
| P7-863b | Endemic | A. aegypti | 1969 | Malaysia | 2 | AF231716 | |
| 16681 | Endemic | Human | 1964 | Thailand | 2 | U87411 | 18 |
| TH-36 | Endemic | NR | 1958 | Thailand | 2 | D10514 | 41 |
| PUO-218 | Endemic | Human | 1980 | Thailand | 2 | D00345 | 11 |
| New Guinea C | Endemic | Human | 1944 | New Guinea | 2 | AF038403 | 11 |
| N.1409 | Endemic | Human | 1983 | Jamaica | 2 | M20558 | 7 |
| P8-1407b | Sylvatic | Sentinel monkey | 1970 | Malaysia | 2 | AF231717 | |
| DAK Ar A578b | Sylvatic | A. taylori sensu lato | 1980 | Ivory Coast | 2 | AF231718 | |
| PM33974b | Sylvatic | A. africanus | 1981 | Guinea | 2 | AF231719 | |
| DAK HD10674b | Sylvatic | Human | 1970 | Senegal | 2 | AF231720 | |
| PR158 | Endemic | Human | 1969 | Puerto Rico | 2 | L10046 | 25 |
| P9122 | Endemic | NR | 1957 | India | 2 | L10043 | 25 |
| TR1751 | Endemic | Human | 1953 | Trinidad | 2 | L10053 | 25 |
| 1897 | Endemic | NR | 1987 | Taiwan | 2 | L10052 | 25 |
| 2088 | Endemic | NR | 1983 | Philippines | 2 | L10045 | 25 |
| #10 | Endemic | NR | 1984 | Somalia | 2 | L10051 | 25 |
| 0190 | Endemic | NR | 1983 | Burkina Faso | 2 | L10042 | 25 |
| 1051 | Endemic | NR | 1976 | Indonesia | 2 | L10044 | 25 |
| S-44554 | Endemic | Human | 1977 | Seychelles | 2 | L10048 | 25 |
| S-44552 | Endemic | Human | 1977 | Seychelles | 2 | L10047 | 25 |
| 1583 | Endemic | NR | 1985 | Sri Lanka | 2 | L10050 | 25 |
| 1592 | Endemic | NR | 1985 | Sri Lanka | 2 | L10040 | 25 |
| 271206 | Endemic | NR | 1990 | Sri Lanka | 2 | L10049 | 25 |
| 206714 | Endemic | NR | 1989 | Sri Lanka | 2 | L10055 | 25 |
| 271235 | Endemic | NR | 1990 | Sri Lanka | 2 | L10054 | 25 |
| 40274 | Endemic | NR | 1990 | Brazil | 2 | L10041 | 25 |
| M56309 | Endemic | Human | 1986 | Malaysia | 2 | X15433 | 40 |
| MS8455 | Endemic | Human | 1987 | Malaysia | 2 | X15434 | 39 |
| 1036 | Endemic | NR | 1976 | Indonesia | 4 | U18492 | 22 |
| 1132 | Endemic | NR | 1977 | Indonesia | 4 | U18431 | 22 |
| 1411 | Endemic | NR | 1983 | El Salvador | 4 | U18426 | 22 |
| 5489 | Endemic | NR | 1981 | New Caledonia | 4 | U18432 | 22 |
| 1385 | Endemic | NR | 1982 | Brazil | 4 | U18425 | 22 |
| S-44754 | Endemic | NR | 1979 | Tahiti | 4 | U18438 | 22 |
| 703-4b | Endemic | Human | 1994 | Thailand | 4 | AF231726 | |
| 1650 | Endemic | NR | 1986 | Puerto Rico | 4 | U18436 | 22 |
| 114-094 | Endemic | NR | 1985 | Tahiti | 4 | U18439 | 22 |
| BC6494 | Endemic | NR | 1994 | El Salvador | 4 | U18427 | 22 |
| P7-1006b | Endemic | A. aegypti | 1969 | Malaysia | 4 | AF231722 | |
| H241-P | Endemic | Human | 1956 | Philippines | 4 | S66064 | 17 |
| P73-1120b | Sylvatic | Sentinel monkey | 1973 | Malaysia | 4 | AF231724 | |
| P75-215b | Sylvatic | A. niveus | 1975 | Malaysia | 4 | AF231725 | |
| P75-514b | Sylvatic | Sentinel monkey | 1975 | Malaysia | 4 | AF231723 | |
| 1492 | Endemic | NR | 1984 | Mexico | 4 | U18431 | 22 |
| 30153 | Endemic | NR | 1973 | Indonesia | 4 | U18428 | 22 |
| D84-024 | Endemic | NR | 1984 | Thailand | 4 | U18442 | 22 |
| D78-017 | Endemic | NR | 1978 | Thailand | 4 | U18441 | 22 |
| 12123 | Endemic | NR | 1984 | Philippines | 4 | U18435 | 22 |
| 16598 | Endemic | NR | 1964 | Philippines | 4 | U18434 | 22 |
| TC2443 | Endemic | NR | 1963 | Thailand | 4 | U18440 | 22 |
| S44750 | Endemic | NR | 1978 | Sri Lanka | 4 | U18437 | 22 |
“Endemic” indicates human or A. aegypti isolates or those associated with peridomestic transmission; “sylvatic” indicates sentinel monkey or canopy-dwelling-mosquito isolates.
The sequence was determined in this study.
NR, not reported.
Virus growth, RNA extraction, and sequencing.
DEN stocks were prepared in C6/36 A. albopictus cell culture monolayers at 28°C with a multiplicity of infection of 0.1 to 1.0 PFU per cell. Culture media consisted of Eagle's minimal essential medium supplemented with 2% fetal bovine serum. Seven days after infection, supernatants were clarified by centrifugation at 1,000 × g for 5 min. RNA was extracted with Trizol LS (Bethesda Research Laboratories, Bethesda, Md.) according to the manufacturer's protocol. PCR primers were designed to amplify the E protein gene based on the genomic sequences of DEN previously published (Tables 1 and 2). RNA was denatured at 65°C for 5 min, and cDNA was synthesized in a 20-μl reaction volume with Superscript II reverse transcriptase (Bethesda Research Laboratories) at 42°C for 1 h using the antisense primers listed in Table 2. The PCR products were purified from 1% agarose gels and sequenced directly using an Applied Biosystems (Foster City, Calif.) Prism automated DNA sequencing kit and model 377 sequencer according to the manufacturer's protocol.
TABLE 2.
Oligonucleotides used for PCR amplification of DENa
| Primer (genetic sense) | Sequence (5′→3′) |
|---|---|
| DEN1-635 (+) | AGACACATGGGTGACCTAGG |
| DEN1-2472 (−) | CTCTGTCCAAGTGTGGACTTC |
| DEN2-835 (+) | CCAGGCTTTACCATAATGGC |
| DEN2-2420 (−) | CCAGCTGCACAACGCAACCAC |
| DEN4-685 (+) | AGAGCGGAGAACGGAGACGAG |
| DEN4-2454 (−) | TCCGCTTCCACACTTCAATTC |
Numbers indicate 5′ genomic nucleotide positions homologous or complementary to primer sequences.
Phylogenetic analyses.
The deduced E protein amino acid sequences and representative sequences from the GenBank library (Table 1) were aligned using the PILEUP program of the Genetics Computer Group with default gap penalties. Although sylvatic DEN-3 strains are not available, GenBank sequences of endemic DEN-3 strains were included in our analysis to provide information on the relative divergence levels in comparison to other DEN serotypes. Nucleotide sequences were aligned manually based on codon homology. Phylogenetic analyses of the aligned nucleotide and amino acid sequences were performed using the PAUP maximum-parsimony program (version 3.0; D. L. Swofford, Illinois Natural History Survey, Champaign) and neighbor-joining program implemented in the PHYLIP package (version 3.5p; J. Felsenstein, Department of Genetics, University of Washington, Seattle). Homologous sequences from Japanese encephalitis, Kunjin, and West Nile viruses (a sister group to DEN [20]) were used as the outgroup to root the DEN tree. Bootstrapping (8) with 200 to 1,000 replicates was used to place confidence values on groupings within the trees.
Estimation of DEN divergence times.
Divergence times for ancestral DEN lineages were estimated using synonymous or total nucleotide divergence rates estimated from maximum-parsimony trees using two methods: (i) a regression analysis where each sequence within a geographically restricted clade was compared to the predicted sequence of its hypothetical ancestor based on maximum-parsimony analysis, and the synonymous percent difference was plotted versus the year of isolation, and (ii) identification of sister sequences that were closely related and isolated at least 5 years apart in the same geographical region. The differences in synonymous changes depicted in branch lengths separating each sister sequence from the predicted common ancestor were divided by the number of years between isolations to yield rates expressed as synonymous changes per year, and several estimates were compared to provide an estimated mean and standard deviation.
Nucleotide sequence accession numbers.
Nucleotide sequences were submitted to GenBank under accession no. AF231715 to AF231726 (Table 1).
RESULTS
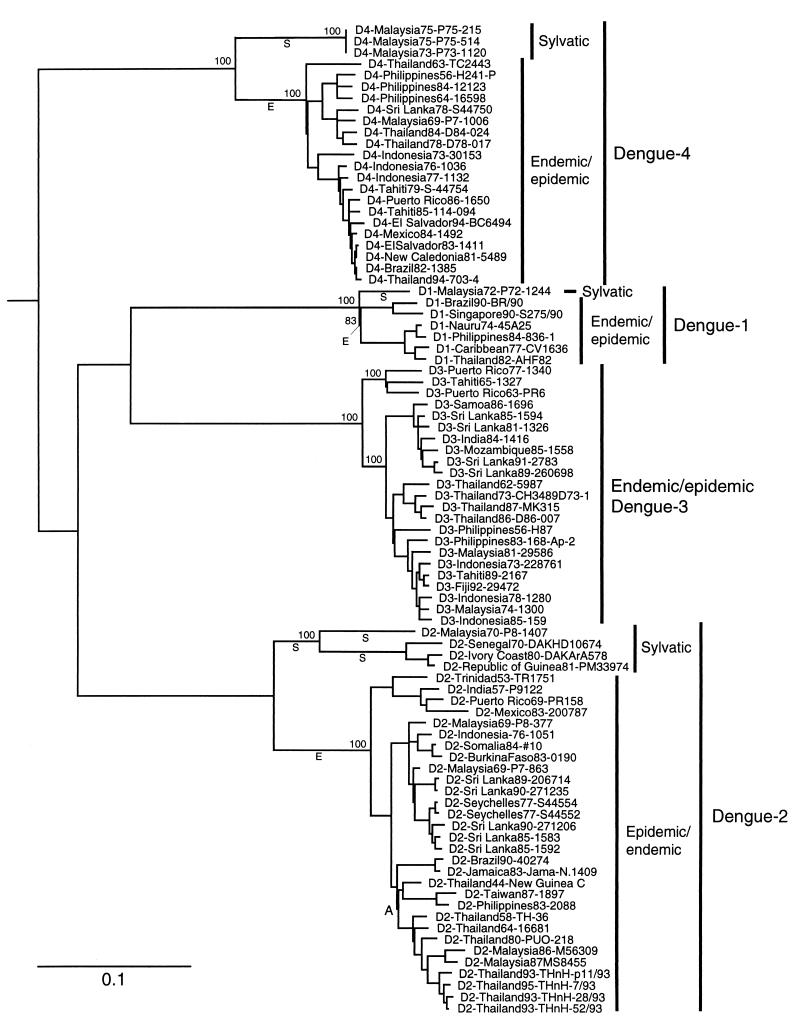
The E gene sequences we determined for DEN-1, -2, and -4 strains were 1,482 to 1,485, 1,485, and 1,485 nucleotides long, respectively. These lengths were identical to those of most other strains sequenced previously (6, 7, 11, 27, 42, 43). Phylogenetic trees produced with the maximum-parsimony and neighbor-joining methods produced very similar topologies, differing only within a few terminal groupings in the DEN-3 and DEN-4 groups. Maximum parsimony produced 36 trees of equal total length, also differing only in a few DEN-3 and DEN-4 terminal groupings. All trees indicated that the DEN are monophyletic, with 100% bootstrap support. DEN-1 and DEN-3 were sister groups, with 100% bootstrap support in amino acid trees, and DEN-4 was the basal group in all trees (Fig. 1). These overall relationships are generally consistent with previous findings (1, 24) but are inconsistent with the placement of DEN-2 at the base of the DEN clade by Kuno et al. (20). Our higher bootstrap values, presumably the result of longer sequences and more informative characters, suggest that DEN-4 is correctly placed in the basal position (Fig. 1).
FIG. 1.
Phylogenetic tree derived from E protein gene nucleotide sequences of sylvatic and representative endemic/epidemic DEN strains using maximum parsimony (PAUP, version 3.0) and drawn using branch lengths derived by neighbor joining using the Kimura two-parameter formula (PHYLIP, version 3.5p). The scale shows a genetic distance of 0.1, or 10% nucleotide sequence divergence. Homologous sequences from Japanese encephalitis, Kunjin, and West Nile viruses (a sister group to DEN [20]) were used as the outgroup to root the DEN tree. Numbers indicate bootstrap (8) values for monophyletic groups to the right. Branches labeled “E” indicate periods of evolution predicted to represent emergence of endemic/epidemic DEN and used to generate Table 3; branches labeled “S” represent continued sylvatic evolution during the same time period. Node A defines the clade used for the regression analysis of the DEN evolutionary rate.
All sylvatic DEN isolates from Malaysia and Africa were genetically distinct from the endemic/epidemic isolates; as in previous studies using other genome regions (31), the African sylvatic DEN-2 isolates were quite distinct from all endemic/epidemic isolates, differing by about 19% in nucleotide sequence and 5% in amino acid sequence (Fig. 1). The Malaysian sylvatic isolates of DEN-1, -2, and -4 were also distinct from both endemic/epidemic isolates and African sylvatic strains; the Malaysian DEN-2 isolates differ from endemic/epidemic strains by about 17% in nucleotide sequence and 4% in amino acid sequence, while the DEN-1 and -4 Malaysian sylvatic strains differed from their endemic/epidemic counterparts by about 7 and 14%, respectively, at the nucleotide level and 3 and 5%, respectively, at the amino acid level.
All phylogenetic trees placed the sylvatic isolates from Malaysia and Africa at the basal positions of their respective DEN serotype groups, with bootstrap values of 84 to 100% supporting these basal sylvatic positions. Trees constructed using deduced amino acid sequences yielded identical internal topology, although the resolution of terminal groups was reduced as expected due to the loss of informative synonymous differences. The geographic origin of the sylvatic DEN isolates is not a valid explanation for their phylogenetically distinct placement, because endemic/epidemic strains isolated from humans or mosquitoes in Malaysia grouped with the other endemic/epidemic isolates.
To determine when the sylvatic and endemic/epidemic forms diverged, we estimated the synonymous nucleotide sequence divergence rate using two methods. A regression analysis was conducted using sequences of all isolates within a geographically restricted clade of DEN-2 strains (isolated primarily from Asia and Malaysia from 1944 to 1995 [Fig. 1, clade A]). Each sequence within this clade was compared to the sequence of its predicted ancestor (the parsimony tree node with its predicted sequence), and the percent synonymous nucleotide sequence changes in the branches was plotted versus the year of isolation. This method yielded a slope of 0.05% divergence per year, or 5 × 10−4 substitutions per synonymous site per year (R2 − 0.72). The second method utilized sister sequences that were closely related and isolated at least 5 years apart in the same geographical region. The synonymous differences in branch lengths separating each sister isolate from the common ancestor (parsimony tree node) were divided by the number of years between isolations, and the mean and standard deviation were determined from these values. A divergence rate of 5 × 10−4 (standard deviation, ±3 × 10−4) substitutions per synonymous site per year was obtained. The nonsynonymous rate estimate was 6 × 10−5. This rate is similar to the nonsynonymous estimate of 7.5 × 10−5 obtained previously for mosquito-borne flaviviruses (44).
Using the synonymous nucleotide substitution rates and the formula of Li et al. (26) for correcting synonymous substitution distances for sequential substitutions of the same nucleotide, we estimated that the sylvatic DEN-2 genotypes diverged from the endemic/epidemic forms on the order of 1,000 ± 500 years ago (estimate ± standard deviation). The African and Malaysian sylvatic lineages of DEN-2 diverged on the order of 800 ± 400 years ago. Sylvatic and endemic/epidemic DEN-1 and -4 probably diverged on the order of 200 ± 100 and 600 ± 300 years ago, respectively. Estimates using nonsynonymous substitution rates applied directly to the branch lengths of parsimony trees, as well as Ka (nonsynonymous distance) values (26), yielded a similar time estimates (800 ± 400 years ago) for divergence of the sylvatic and endemic DEN-2 genotypes. The ancestor of all DEN probably occurred much earlier than the estimated times of divergence of the endemic/epidemic and sylvatic forms of each serotype. All of these estimates rely on the assumption of a constant rate of DEN sequence evolution, which may not be completely valid.
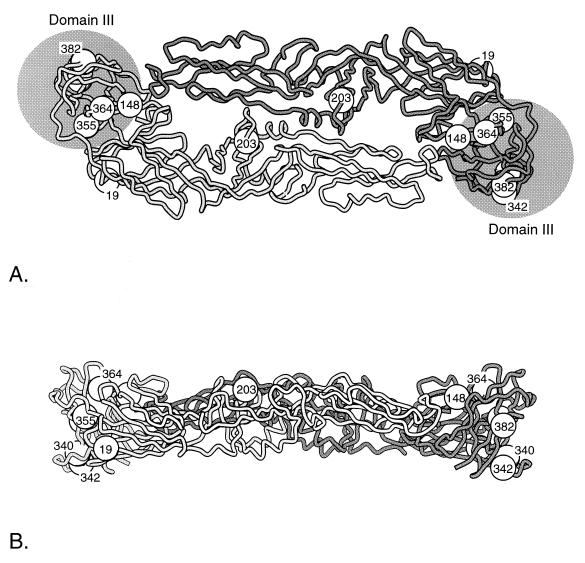
The three-dimensional structure of the soluble portion of the E protein of central European tick-borne encephalitis virus was determined by Rey et al. (30). Due to the conservation of cysteine residues and amino acid similarity among flavivirus E proteins, it is possible to model the E protein structure of DEN. To evaluate amino acid substitutions that may have accompanied the adaptation of ancestral, sylvatic DEN to human transmission involving peridomestic vectors, such as A. aegypti and A. albopictus, we analyzed amino acid changes on the branches of parsimony trees corresponding to endemic/epidemic emergence (Fig. 1 and Table 3). One (residue 352) of three DEN-1 amino acids, two (residues 352 and 388) of five DEN-2 amino acids, and five (residues 340, 342, 355, 364, and 382) of eight DEN-4 amino acids showing changes in the tree branches predicted to have accompanied endemic/epidemic emergence lie within domain III, which may be involved in cellular receptor binding. The three amino acids representing DEN-1 emergence are not predicted to be in spatial proximity, while two of the five amino acids for DEN-2 (residues 352 and 388) are predicted to be spatially close to one other within domain III. Of the eight amino acids for DEN-4, six (residues 148, 340, 342, 355, 364, and 382) are predicted to be close to one other within domain III (Fig. 2). Residue 148 is near a putative glycosylation site that is exposed on the surface of the virus.
TABLE 3.
Amino acid changes predicted to have accompanied E protein evolution of DENa
| Period of evolution | Amino acid position | Change |
|---|---|---|
| DEN-1 emergence | 6 | V→I |
| 120 | E→K | |
| 345 | I→V | |
| DEN-2 emergence | 122 | L→K |
| 181 | I→V | |
| 236 | M→T | |
| 345 | K→R | |
| 379 | V→I | |
| DEN-4 emergence | 19 | T→A |
| 148 | A→T | |
| 203 | G→K | |
| 340 | K→R | |
| 342 | M→V | |
| 355 | I→T | |
| 364 | I→V | |
| 382 | A→V | |
| Sylvatic DEN-1 evolution | 16 | S→L |
| 157 | E→G | |
| Sylvatic DEN-2 evolution | 59 | Y→F |
| 113 | I→V | |
| 203 | N→K | |
| 331 | S→A | |
| 367 | V→I | |
| 446 | A→V | |
| 481 | L→I | |
| Sylvatic DEN-4 evolution | 132 | I→V |
| 154 | D→S | |
| 162 | A→T | |
| 478 | T→S |
Unique synapomorphies (shared and derived amino acids) represented in the branches designated E and S in Fig. 1.
FIG. 2.
Three-dimensional model of the DEN E protein, based on the crystal structure of the soluble portion of the E protein of central European tick-borne encephalitis virus as determined by Rey et al. (30). Circles represent amino acids predicted to have changed during the period of evolution representing emergence of endemic/epidemic DEN-4 from a sylvatic ancestor (Fig. 1 and Table 3). (A) View of the E protein homodimer from above the virion; (B) side view of the E protein homodimer.
To determine whether the E protein changes predicted to have accompanied DEN emergence reflect only a general trend in the evolution of DEN toward domain III changes, we determined the amino acid changes predicted to have accompanied evolution of sylvatic strains during the same time period (Fig. 1); these changes are listed in Table 3. In contrast to the changes accompanying endemic/epidemic emergence, these showed no apparent clustering within a particular domain; i.e., they were distributed throughout the E protein amino acid sequence.
DISCUSSION
The relationships among sylvatic and endemic/epidemic DEN isolates that we have determined using nucleotide and amino acid-derived phylogenies are consistent with the ancestors of DEN-1, -2, and -4 being either sylvatic or endemic/epidemic strains. Although sylvatic DEN-3 were not isolated in Malaysia by Rudnick, the presence of DEN-3 antibodies in nonhuman canopy-dwelling primates indicated that a sylvatic DEN-3 cycle also existed there (35). The lack of large concentrated human populations prior to a few thousand years ago suggests that the sylvatic form of these viruses was ancestral; the endemic/epidemic forms probably evolved after urban human populations, sufficient to serve as reservoirs, arose (13). This suggests that all four epidemic/endemic DEN serotypes evolved independently from sylvatic progenitors. Estimates for the minimum human population size required to support endemic DEN transmission range from 10,000 to 1 million (19). Human populations in Asia are believed to have approached these sizes on the order of 4,000 years ago, when urban civilizations first arose (3). The DEN-2 divergence estimate of 1,000 ± 500 years ago is consistent with these estimates of human population sizes as well as the first historical record of dengue-like illness in China, dating over 1,000 years ago (13).
Although we did not attempt to estimate the divergence times of the different DEN serotypes, they probably diverged in the very distant past. Some rodent-borne RNA viruses with similar levels of sequence divergence have been estimated to have diverged over the course of millions of years during coevolution with their reservoir hosts (2, 29). The emergences of human DEN transmission are probably relatively recent events in the history of DEN, and most of the evolution of DEN probably occurred when they were sylvatic viruses utilizing only nonhuman primate reservoirs.
The sister group relationship of the African and Malaysian DEN-2 indicates that the sylvatic ancestor of endemic/epidemic DEN-2 could have occurred in either Africa or the Asian-Oceanic region. The greater diversity of sylvatic (ancestral) DEN in Malaysia (presence of all four serotypes) compared to Africa suggests that a sylvatic ancestor of all DEN arose in the Asian-Oceanic region and diverged into the four serotypes recognized today. The four independent evolutionary events leading to the emergence of endemic DEN-1 to -4 from sylvatic progenitors presumably involved switching from sylvatic, canopy-dwelling Aedes mosquito vectors to A. albopictus and later A. aegypti. This indicates that adaptation to new vectors and vertebrate hosts by DEN arboviruses occurred repeatedly. Because A. aegypti presumably did not occur in Asia and Oceania at that time, A. albopictus and/or other Aedes mosquitoes were probably the original human vectors (13). This hypothesis is also supported by the greater susceptibility of A. albopictus to infection by DEN, suggesting greater virus adaptation. Acquisition of A. aegypti as a vector may have occurred only during the past few centuries as commerce distributed this mosquito throughout the tropics from its origins in Africa (13).
The exact location where the four DEN serotypes evolved cannot be determined due to very limited sampling of sylvatic strains, especially in Asia and Oceania. However, considering the limited cross protection against heterologous challenge exhibited by current human DEN, it seems likely that more complete cross protection probably existed long ago when ancestral, sylvatic DEN strains had reached lesser levels of divergence than are reflected in contemporary strain sequences. Strong cross-reactivity of protective immunity is believed to result in direct competition among virus lineages and competitive exclusion if different strains occupy the same ecological niche (9). Therefore, it seems likely that the divergence of the four DEN serotypes occurred in different geographic regions or in transmission cycles relying on different hosts. The studies of Rudnick and colleagues in Malaysia (34–37) indicate that the four sylvatic DEN serotypes probably overlap in their host utilization. This suggests that DEN may have evolved allopatrically and later colonized common geographic regions after divergence had reached the level where strain variation could be maintained due to limited cross protection. Additional field studies to identify the natural reservoir host of sylvatic viruses, as well as cross protection studies in nonhuman primates, are needed to evaluate this hypothesis.
Overall, the structural model of the E protein (30) would predict that domain III of the E protein (amino acids 302 to 404) is implicated in the emergence of urban DEN. This domain is believed to interact with cellular receptors for virus entry. The data for DEN-4 are most compelling, as six of the eight amino acid changes are close to each other within domain III (Fig. 2); residue 382 lies in a four-amino-acid sequence that is specific to mosquito-borne flaviviruses, is predicted to be exposed on the surface of the virus, and may be involved in receptor binding. Similarly, residue 148 is near a putative glycosylation site that is exposed on the surface of the virus. It is not known if this glycosylation site is utilized by the sylvatic DEN strains. These modeling predictions are suggestive of different interactions between the viral E protein and host cells, either those of epidemic vectors such as A. aegypti and A. albopictus or those of human hosts. It is unlikely that the preponderance of amino acid changes in domain III reflects a general trend in the evolution of DEN; examination of the branches predicted to represent continuing evolution of sylvatic DEN during the same time period showed no similar clustering of E protein changes in domain III (Table 3). The potential adaptive value of some of the E protein changes associated with epidemic emergence should be assessed with reverse genetic approaches using cDNA clones to test the fitness of these mutations in epidemic versus sylvatic vectors.
ACKNOWLEDGMENTS
We thank Edward Holmes for helpful advice with the phylogenetic methods and divergence time estimates. Robert Tesh kindly provided some of the DEN strains that we studied.
This research was supported by National Institutes of Health grants AI39800, AI39508, and AI10984.
REFERENCES
- 1.Blok J, McWilliam S M, Butler H C, Gibbs A J, Weiller G, Herring B L, Hemsley A C, Aaskov J G, Yoksan S, Bhamarapravati N. Comparison of a dengue-2 virus and its candidate vaccine derivative: sequence relationships with the flaviviruses and other viruses. Virology. 1992;187:573–590. doi: 10.1016/0042-6822(92)90460-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bowen M D, Peters C J, Nichol S T. Phylogenetic analysis of the Arenaviridae: patterns of virus evolution and evidence for cospeciation between arenaviruses and their rodent hosts. Mol Phylogenet Evol. 1997;8:301–316. doi: 10.1006/mpev.1997.0436. [DOI] [PubMed] [Google Scholar]
- 3.Burns E M, Ralph P L, Lerner R E, Meacham S. World civilization. A. New York, N.Y: W. W. Norton & Co.; 1986. [Google Scholar]
- 4.Chu M C, O'Rourke E J, Trent D W. Genetic relatedness among structural protein genes of dengue 1 virus strains. J Gen Virol. 1989;70:1701–1712. doi: 10.1099/0022-1317-70-7-1701. [DOI] [PubMed] [Google Scholar]
- 5.Cordellier R, Bouchite B, Roche J C, Monteny N, Diaco B, Akoliba P. Circulation silvatique du virus Dengue 2 en 1980, dans les savannes sub-soudaniennes du Cote d'Ivoire. Cah O R S T O M Ser Entomol Med Parasitol. 1983;21:165. [Google Scholar]
- 6.Despres P, Frenkiel M P, Deubel V. Differences between cell membrane fusion activities of two dengue type-1 isolates reflect modifications of viral structure. Virology. 1993;196:209–219. doi: 10.1006/viro.1993.1469. [DOI] [PubMed] [Google Scholar]
- 7.Deubel V, Kinney R M, Trent D W. Nucleotide sequence and deduced amino acid sequence of the nonstructural proteins of dengue type 2 virus, Jamaica genotype: comparative analysis of the full-length genome. Virology. 1988;165:234–244. doi: 10.1016/0042-6822(88)90677-0. [DOI] [PubMed] [Google Scholar]
- 8.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 9.Ferguson N, Anderson R, Gupta S. The effect of antibody-dependent enhancement on the transmission dynamics and persistence of multiple-strain pathogens. Proc Natl Acad Sci USA. 1999;96:790–794. doi: 10.1073/pnas.96.2.790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fu J, Tan B H, Yap E H, Chan Y C, Tan Y H. Full-length cDNA sequence of dengue type 1 virus (Singapore strain S275/90) Virology. 1992;188:953–958. doi: 10.1016/0042-6822(92)90560-c. [DOI] [PubMed] [Google Scholar]
- 11.Gruenberg A, Woo W S, Biedrzycka A, Wright P J. Partial nucleotide sequence and deduced amino acid sequence of the structural proteins of dengue virus type 2, New Guinea C and PUO-218 strains. J Gen Virol. 1988;69:1391–1398. doi: 10.1099/0022-1317-69-6-1391. [DOI] [PubMed] [Google Scholar]
- 12.Gubler D J. Dengue. In: Monath T P, editor. The arboviruses: epidemiology and ecology. II. Boca Raton, Fla: CRC Press; 1988. pp. 223–260. [Google Scholar]
- 13.Gubler D J. Dengue and dengue hemorrhagic fever: its history and resurgence as a global public health problem. In: Gubler D J, Kuno G, editors. Dengue and dengue hemorrhagic fever. New York, N.Y: CAB International; 1997. pp. 1–22. [Google Scholar]
- 14.Gubler D J, Nalim S, Tan R, Saipan H, Sulianti Saroso J. Variation in susceptibility to oral infection with dengue viruses among geographic strains of Aedes aegypti. Am J Trop Med Hyg. 1979;28:1045–1052. doi: 10.4269/ajtmh.1979.28.1045. [DOI] [PubMed] [Google Scholar]
- 15.Gubler D J, Rosen L. Variation among geographic strains of Aedes albopictus in susceptibility to infection with dengue viruses. Am J Trop Med Hyg. 1976;25:318–325. doi: 10.4269/ajtmh.1976.25.318. [DOI] [PubMed] [Google Scholar]
- 16.Gubler D J, Trent D W. Emergence of epidemic dengue/dengue hemorrhagic fever as a public health problem in the Americas. Infect Agents Dis. 1994;2:383–393. [PubMed] [Google Scholar]
- 17.Kawano H, Rostapshov V, Rosen L, Lai C J. Genetic determinants of dengue type 4 virus neurovirulence for mice. J Virol. 1993;67:6567–6575. doi: 10.1128/jvi.67.11.6567-6575.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kinney R M, Butrapet S, Chang G J, Tsuchiya K R, Roehrig J T, Bhamarapravati N, Gubler D J. Construction of infectious cDNA clones for dengue 2 virus: strain 16681 and its attenuated vaccine derivative, strain PDK-53. Virology. 1997;230:300–308. doi: 10.1006/viro.1997.8500. [DOI] [PubMed] [Google Scholar]
- 19.Kuno G. Factors influencing the transmission of dengue viruses. In: Gubler D J, Kuno G, editors. Dengue and dengue hemorrhagic fever. New York, N.Y: CAB International; 1997. pp. 61–88. [Google Scholar]
- 20.Kuno G, Chang G-J J, Tsuchiya K R, Karabatsos N, Cropp C B. Phylogeny of the genus Flavivirus. J Virol. 1998;72:73–83. doi: 10.1128/jvi.72.1.73-83.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kurane I, Ennis F E. Immunity and immunopathology in dengue virus infections. Semin Immunol. 1992;4:121–127. [PubMed] [Google Scholar]
- 22.Lanciotti R S, Gubler D J, Trent D W. Molecular evolution and phylogeny of dengue-4 viruses. J Gen Virol. 1997;78:2279–2284. doi: 10.1099/0022-1317-78-9-2279. [DOI] [PubMed] [Google Scholar]
- 23.Lanciotti R S, Lewis J G, Gubler D J, Trent D W. Molecular evolution and epidemiology of dengue-3 viruses. J Gen Virol. 1994;75:65–75. doi: 10.1099/0022-1317-75-1-65. [DOI] [PubMed] [Google Scholar]
- 24.Lewis J A, Chang G J, Lanciotti R S, Kinney R M, Mayer L W, Trent D W. Phylogenetic relationships of dengue-2 viruses. Virology. 1993;197:216–224. doi: 10.1006/viro.1993.1582. [DOI] [PubMed] [Google Scholar]
- 25.Lewis J G, Chang G J, Lanciotti R S, Trent D W. Direct sequencing of large flavivirus PCR products for analysis of genome variation and molecular epidemiological investigations. J Virol Methods. 1992;38:11–23. doi: 10.1016/0166-0934(92)90165-a. [DOI] [PubMed] [Google Scholar]
- 26.Li W-H, Wu C-I, Luo C-C. A new method for estimating synonymous and nonsynonymous rates of nucleotide substitution considering the relative likelihood of nucleotide and codon changes. Mol Biol Evol. 1985;2:150–174. doi: 10.1093/oxfordjournals.molbev.a040343. [DOI] [PubMed] [Google Scholar]
- 27.McKee K T, Jr, Bancroft W H, Eckels K H, Redfield R R, Summers P L, Russell P K. Lack of attenuation of a candidate dengue 1 vaccine (45AZ5) in human volunteers. Am J Trop Med Hyg. 1987;36:435–442. doi: 10.4269/ajtmh.1987.36.435. [DOI] [PubMed] [Google Scholar]
- 28.Monath T P. Dengue: the risk to developed and developing countries. Proc Natl Acad Sci USA. 1994;91:2395–2400. doi: 10.1073/pnas.91.7.2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morzunov S P, Rowe J E, Ksiazek T G, Peters C J, St. Jeor S C, Nichol S T. Genetic analysis of the diversity and origin of hantaviruses in Peromyscus leucopus mice in North America. J Virol. 1998;72:57–64. doi: 10.1128/jvi.72.1.57-64.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rey F A, Heinz F X, Mandl C, Kunz C, Harrison S C. The envelope glycoprotein from tick-borne encephalitis virus at 2 Å resolution. Nature. 1995;375:291–298. doi: 10.1038/375291a0. [DOI] [PubMed] [Google Scholar]
- 31.Rico-Hesse R. Molecular evolution and distribution of dengue viruses type 1 and 2 in nature. Virology. 1990;174:479–493. doi: 10.1016/0042-6822(90)90102-w. [DOI] [PubMed] [Google Scholar]
- 32.Rico-Hesse R, Harrison L M, Salas R A, Tovar D, Nisalak A, Ramos C, Boshell J, de Mesa M T, Nogueira R M, da Rosa A T. Origins of dengue type 2 viruses associated with increased pathogenicity in the Americas. Virology. 1997;230:244–251. doi: 10.1006/viro.1997.8504. [DOI] [PubMed] [Google Scholar]
- 33.Roche J C, Cordellier R, Hervy J P, Digoutte J P, Monteny N. Isolement de 96 souches de virus dengue 2 a partir de moustiques captures en Cote d'Ivoire et Houte-Volta. Ann Virol. 1983;134E:233–234. [Google Scholar]
- 34.Rudnick A. Ecology of dengue virus. Asian J Infect Dis. 1978;2:156–160. [Google Scholar]
- 35.Rudnick A. The ecology of the dengue virus complex in peninsular Malaysia. In: Pang T, Pathmanathan R, editors. Proceedings of the International Conference on Dengue/DHF. Kuala Lumpur, Malaysia: University of Malaysia Press; 1984. p. 7. [Google Scholar]
- 36.Rudnick A. Studies of the ecology of dengue in Malaysia: a preliminary report. J Med Entomol. 1965;2:203–208. doi: 10.1093/jmedent/2.2.203. [DOI] [PubMed] [Google Scholar]
- 37.Rudnick A, Marchette N J, Garcia R. Possible jungle dengue—recent studies and hypotheses. Jpn J Med Sci Biol. 1967;20:69–74. [PubMed] [Google Scholar]
- 38.Sabin A B. Research on dengue during World War II. Am J Trop Med Hyg. 1952;1:30–50. doi: 10.4269/ajtmh.1952.1.30. [DOI] [PubMed] [Google Scholar]
- 39.Samuel S, Koh C L, Blok J, Pang T, Lam S K. Nucleotide sequence of the envelope protein gene of a Malaysian dengue-2 virus isolated from a patient with dengue haemorrhagic fever. Nucleic Acids Res. 1989;17:8875. doi: 10.1093/nar/17.21.8875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Samuel S, Koh C L, Blok J, Pang T, Lam S K. Nucleotide sequence of the envelope protein gene of a Malaysian dengue-2 virus isolated from a patient with dengue shock syndrome. Nucleic Acids Res. 1989;17:8888. doi: 10.1093/nar/17.21.8888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sanchez I J, Ruiz B H. A single nucleotide change in the E protein gene of dengue virus 2 Mexican strain affects neurovirulence in mice. J Gen Virol. 1996;77:2541–2545. doi: 10.1099/0022-1317-77-10-2541. [DOI] [PubMed] [Google Scholar]
- 42.Shiu S Y, Jiang W R, Porterfield J S, Gould E A. Envelope protein sequences of dengue virus isolates TH-36 and TH-Sman, and identification of a type-specific genetic marker for dengue and tick-borne flaviviruses. J Gen Virol. 1992;73:207–212. doi: 10.1099/0022-1317-73-1-207. [DOI] [PubMed] [Google Scholar]
- 43.Thant K Z, Morita K, Igarashi A. Detection of the disease severity-related molecular differences among new Thai dengue-2 isolates in 1993, based on their structural proteins and major non-structural protein NS1 sequences. Microbiol Immunol. 1996;40:205–216. doi: 10.1111/j.1348-0421.1996.tb03336.x. [DOI] [PubMed] [Google Scholar]
- 44.Zanotto P M, Gould E A, Gao G F, Harvey P H, Holmes E C. Population dynamics of flaviviruses revealed by molecular phylogenies. Proc Natl Acad Sci USA. 1996;93:548–553. doi: 10.1073/pnas.93.2.548. [DOI] [PMC free article] [PubMed] [Google Scholar]