ABCA1 is a plasma membrane protein that transfers membrane phospholipid (PL) and cholesterol to extracellular acceptors, such as apolipoprotein A-1 (APOA1) to form HDL particles. ABCA1 is also a key step in removal of excess cholesterol from cells. ABCA1 is unique among ABC transporters in that receptor is required for full transport of its lipid substrates. The concentration of ABCA1 on the surface is tightly regulated through synthesis, transport to cell surface, cellular internalization and degradation. Strong evidence indicates that ABCA1 dimerization is required for cholesterol transport, although monomeric ABCA1 and multimeric ABCA1 have been reported.
Key unresolved issues are: 1) How does ABCA1 facilitate the transport of cholesterol and PL to APOA1? 2) From what membrane pools are the cholesterol and PL derived? 3) How does APOA1 bind to ABCA1, and does it drive interconversion between ABCA1 monomer and dimer? 4) What cell structures are involved in the internalization and degradation of ABCA1 from the cell surface? Targeted structural analysis of the relationship of ABCA1 with the rest of the cell machinery would aid in teasing out answers to these questions.
To carry out structural investigations of cellular changes related to ABCA1 activity, we have undertaken scanning electron microscopy (SEM) and transmission electron microscopy (TEM) analyses of ABCA1 distribution and cellular interaction in BHK cells expressing mifepristone-inducible human ABCA1. Cells with upregulate ABCA1 expression or only basal ABCA1 expression were incubated with APOA1 protein, fixed with a solution of paraformaldehyde, and then immunolabeled with anti-APOA1 colloidal gold.
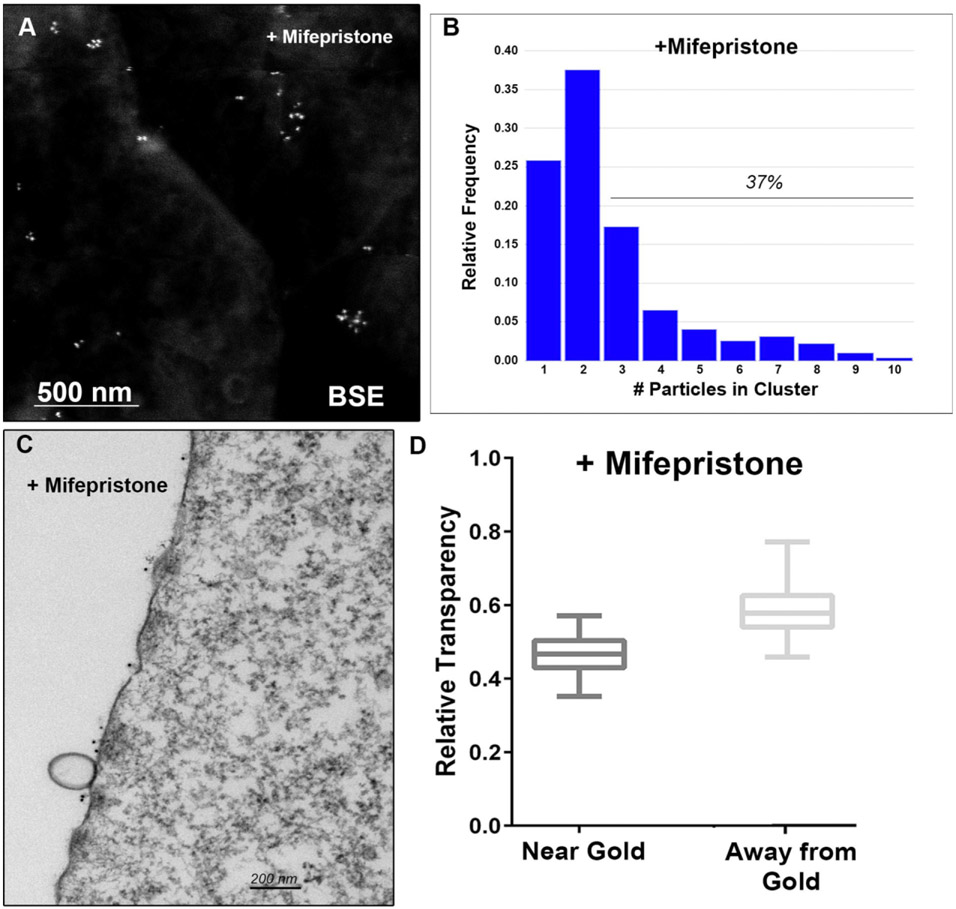
SEM showed that immunogold binding to bound APOA1 occurred in discrete clusters distributed around the plasma membrane (Figure 1A). Upregulation of ABCA1 with mifepristone increased APOA1 binding 27-fold, indicating its major role in observed binding. Calculation of the number of bound APOA1 in a cluster indicated 72% of gold within clusters and 37% in clusters of 3 or more particles (Figure 1B).
Fig. 1.
Analysis of ABCA1 clustering. A) SEM backscatter image of ABCA1 bound APOA1 distribution on cell surface. B) Quantitation of ABCA1 clustering on cell surface. C) TEM Image of cytoplasmic density below ABCA1 clusters. Note the membrane vesicles next to two clusters of ummunolabeled ABCA1. D) Quantitation of cytoplasmic density directly below ABCA1 clusters vs. >3 um away.
TEM showed that the cytoplasm directly below areas of APOA1 binding was denser and more compact than areas away from the APOA1 binding (Figure 1C and 1D). This was true both for cells with basal ABCA1 expression and when ABCA1 expression was upregulated. In both cases, cytoplasm transparency was statistically lower in regions of immunogold binding. Formation of exocytic microparticles from the plasma membrane was also enhanced (9X) in areas adjacent to ABCA1.
These studies reveal that ABCA1 is not homogenously distributed across the cell surface. Moreover, these surface microdomains of ABCA1 were associated with distinct alterations in the underlying cytoplasm and these areas were more likely to form exocytic vesicles than areas lacking ABCA1. Thus, the ABCA1-rich microdomains have specific membrane and cytoplasmic properties. Understanding these properties should help us answer the key questions remaining regarding ABCA1 structure and function [1].
Acknowledgments
1. Supported by NIH grant P01 HL116263 and Vanderbilt Cell Imaging Shared Resource, partially supported by NIH Grants CA68485, DK20593, DK58404, DK59637 and EY08126.