Abstract
Computer analysis of the Epstein-Barr virus (EBV) genome indicates there are ∼100 open reading frames (ORFs). Thus far about 30 EBV genes divided into the categories latent and lytic have been identified. The BamHI F region of EBV is abundantly transcribed during lytic replication. This region is highly conserved among herpesviruses, thus suggesting that some common function could be retained in the ORFs encompassed within this viral fragment. To identify putative novel proteins and possible new markers for viral replication, we focused our attention on the first rightward ORF in the BamHI F region (BFRF1). Histidine and glutathione S-transferase-tagged BFRF1 fusion proteins were synthesized to produce a mouse monoclonal antibody (MAb). Analysis of human sera revealed a high seroprevalence of antibodies to BFRF1 in patients affected by nasopharyngeal carcinoma or Burkitt's lymphoma, whereas no humoral response to BFRF1 could be detected among healthy donors. An anti-BFRF1 MAb recognizes a doublet migrating at 37 to 38 kDa in cells extracts from EBV-infected cell lines following lytic cycle activation and in an EBV-negative cell line (DG75) transfected with a plasmid expressing the BFRF1 gene. Northern blot analysis allowed the detection of a major transcript of 3.7 kb highly expressed in EBV-positive lytic cycle-induced cell lines. Treatment with inhibitors of viral DNA polymerase, such as phosphonoacetic acid and acyclovir, reduced but did not abolish the transcription of BFRF1, thus indicating that BFRF1 can be classified as an early gene. Cell fractionation experiments, as well as immunolocalization by immunofluorescence microscopy, immunohistochemistry, and immunoelectron microscopy, showed that BFRF1 is localized on the plasma membrane and nuclear compartments of the cells and is a structural component of the viral particle. Identification of BFRF1 provides a new marker with which to monitor EBV infection and might help us better understand the biology of the virus.
Epstein-Barr virus (EBV) is a member of the gammaherpesviruses that infects roughly 95% of adult individuals worldwide. EBV has been found to infect epithelial cells and B lymphocytes. Primary EBV infection is clinically inapparent in the vast majority of the population, resulting in a lifelong virus persistence. In a restricted group of individuals, primary infection causes a self-limiting lymphoproliferative disorder known as infectious mononucleosis (IM). Furthermore, the virus has been associated with human malignancies, such as Burkitt's lymphoma (BL) and nasopharyngeal carcinoma (NPC), and lymphoproliferative disorders that develop in immunodeficient subjects (11, 15, 25, 27, 41). In vitro EBV infects resting B lymphocytes, giving rise to lymphoblastoid cell lines (19, 21). In these cell lines, the virus establishes a latent infection in which only a subset of nine viral proteins, thus indicated as latent proteins, and two small nonpolyadenylated transcripts, known as EBER-1 and -2, are expressed. Six of the latent proteins belong to a family of nuclear antigens, designated as EBNA 1 to -6, while the three remaining are localized on plasma membrane and are indicated as LMP-1, -2A, and -2B. In a fraction of cells that ranges between 0.5 and 5%, spontaneous activation of the lytic cycle takes place. However the switch to the lytic cycle can be induced by different pleiotropic stimuli, such as phorbol esters, sodium butyrate, antiimmunoglobulin (anti-Ig), and calcium ionophores, as well as by the transfection of the EBV gene BZLF1 that drives the expression of the ZEBRA protein (6, 8, 16, 18, 32–33, 40) and of the BRLF1 gene encoding the Rta viral transactivator (26, 39). During the lytic phase, many genes required for virus production are induced. According to their sequential activation, they have been classified into three different groups: immediate early, early, and late.
The EBV genome has been completely sequenced, and computer-assisted analysis indicates the presence of ∼100 open reading frames (ORFs) (1). Thus far, about 20 lytic gene products have been identified and characterized. Among them, immediate-early proteins are transactivators of the lytic cycle, early proteins are mainly involved in the processes related to viral DNA replication, and late proteins are predominantly structural elements. However, the full cascade of events that leads to virus production is far from being fully understood. Previous studies have shown that the region encompassed within the BamHI F fragment is highly transcribed during the lytic phase of EBV (13). Computer analysis has indicated the hypothetical presence of two leftward ORFs (BFLF1 and BFLF2) and three rightward ORFs (BFRF1, BFRF2, and BFRF3). So far only the ORF designated as BFRF3 has been found to encode a protein. It is a late lytic gene product, whose molecular mass ranges around 21 kDa, belonging to the viral capsid antigen components (37). Antibodies to BFRF3 are detected in more than 95% of EBV-infected subjects, providing an additional marker with which to evaluate EBV infection (31, 36). The block encompassing the BamHI F region in EBV is highly conserved among human, murine, and equine herpesviruses. This seems to suggest that some function pivotal to herpesvirus biology could be retained in this group of ORFs.
In the present study, we focused our attention on the first rightward ORF of the BamHI F region (BFRF1) to possibly identify a novel protein that could help us to further understand the biology of EBV as well as to provide a new marker with which to study viral replication.
MATERIALS AND METHODS
Cell cultures.
DG75 is an EBV-negative human B-cell line derived from a BL (3). P3HR-1 is a human B-cell line derived from an EBV-positive BL which spontaneously produces EBV particles (12). Raji is a human B-cell line derived from a BL harboring a defective EBV genome that is unable to replicate viral DNA and express late viral genes (22). B95-8 is a marmoset B-cell line transformed with EBV which spontaneously undergoes production of viral particles (19). To induce EBV lytic-cycle gene expression, cells were treated with 20 ng of 12-tetradecanoylphorbol 13-acetate (TPA) per ml and 3 mM sodium butyrate for 48 h. Inhibition of viral DNA replication was obtained by addition to the cell cultures of either phosphonoacetic acid (PAA) to a final concentration of 0.8 mM or acyclovir (ACV) to a final concentration of 1 mM.
VLL is an EBV-positive human lymphoblastoid cell line spontaneously generated from peripheral blood mononuclear cells isolated from a healthy donor.
Akata is an EBV-positive cell line derived from a BL that can be induced to produce EBV by treatment with anti-Ig (32).
All cell lines were maintained in RPMI 1640 medium supplemented with 10% fetal calf serum.
RNA preparation and Northern blot analysis.
Total RNA was extracted with TRIzol (Life Technologies) according to the manufacturer's instructions. Ten micrograms of RNA was loaded for each sample and resolved by electrophoresis through a 1.2% agarose–6% formaldehyde gel in 20 mM morpholinepropanesulfonic acid (MOPS) (pH 7.0) (Sigma). After migration, RNA was then transferred to Nytran Plus (Schleicher & Schuell) membranes in 20× SSC (1× SSC is 0.15 M NaCl plus 15 mM sodium citrate) and UV cross-linked. For BFRF1, a probe was obtained from digestion of the PCR fragment of the BFRF1 gene with NarI (genomic coordinates 58875 to 59493), generating a 618-bp fragment. For BZLF1, the BamHI Z region was used as a probe (genomic coordinates 101947 to 103816). For BLRF2, a probe of 509 bp was generated by digesting the BamHI L region with DraI and XmaI (genomic coordinates 88865 to 89374). For BALF5, a 1,045-bp genomic probe was obtained from digestion with BamHI and EcoRV of a BamHI A fragment (genomic coordinates 154747 to 155792). Probes were [α-32P]dCTP labelled by Klenow fragment DNA polymerase according to standard procedures (29). Hybridizations were carried out in phosphate buffer (0.5 M NaH2PO4 [pH 6.8], 0.5 M Na2HPO4 [pH 6.8], 0.7% sodium dodecyl sulfate [SDS], 1% bovine serum albumin [BSA], 1 mM EDTA) at 60°C overnight. Filters were subsequently washed at 60°C twice in buffer A (0.5% BSA, 5% SDS, 40 mM NaH2PO4 [pH 6.8], 40 mM Na2HPO4 [pH 6.8], 1 mM EDTA) and twice in buffer B (1% SDS, 40 mM NaH2PO4 [pH 6.8], 40 mM Na2HPO4 [pH 6.8], 1 mM EDTA) and bands were traced by autoradiography. Strand specificity of the BFRF1 transcripts was assessed by reprobing the filters with an oligonucleotide designated NF13 (5′-CACTAATCATATCCATGACCCGAGAGGCCT-3′). As a control of RNA quality and equal loading, membranes were hybridized with a β-actin oligoprobe (5′-TGTTGGCGTACAGGTCTTTGCGGATGTCCA-3′).
Generation of BFRF1 expression plasmids and cell transfection.
The BFRF1 ORF (genomic coordinates 58891 to 59898) was amplified by PCR with Pfu polymerase (Stratagene) from the B95-8 genomic DNA by using the following primers: F1u (5′-CCTAGATCTCGAGAATCATG-3′); F1d (5′-CCTGGAGAATTCCCGCTCCC-3′). XhoI and EcoRI sites were inserted in the F1u primer and in the F1d primer, respectively (underlined). The amplified fragment was first digested with XhoI-EcoRI and then was cloned into pTrcHis B vector (Invitrogen) to obtain His-BFRF1 plasmid, as well as in the pGEX-1 vector (Pharmacia) to obtain a glutathione S-transferase (GST)-BFRF1 construct, carrying a six-histidine tag and a GST tag, respectively. The fidelity of the amplified product was confirmed by DNA sequencing of both plasmids. The cytomegalovirus (CMV) immediate-early gene promoter-driven eukaryotic expression vector pHD1013 has been used to generate plasmids for B-cell transfections (7). The BFRF1 ORF was subcloned from His-BFRF1 in the SmaI site of pHD1013 to generate the CMV-BFRF1 construct. The CMV-BZLF1 plasmid whose transfection in eukaryotic cells leads to the expression of the ZEBRA protein was a generous gift of G. Miller. Twenty micrograms of either the CMV-BFRF1 or CMV-BZLF1 construct was used to transfect 107 cells by electroporation with a Bio-Rad Gene Pulser. Twenty-four hours after transfection, cells were harvested, washed once with phosphate-buffered saline (PBS), and stained for indirect immunofluorescence assay (IFA) or Western blot analysis for protein expression.
Production of BFRF1 MAb.
Escherichia coli BL21(DE3)pLysS strain cells were transformed with the His-BFRF1 or GST-BFRF1 plasmid to produce BFRF1 fusion proteins that were then purified through column chromatography, according to the manufacturer's instructions. The predicted molecular mass of the histidine-tagged protein was 40.6 kDa, while the molecular mass of GST-BFRF1 was expected to be 63.6 kDa. The purity of the recombinant proteins was analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) and Coomassie blue staining. Four-week-old BALB/c mice were immunized twice by intraperitoneal injection with 25 μg of His-BFRF1-purified protein emulsified in RIBI adjuvant (RIBI Immunochemical Research). Mice were then given a booster immunization intravenously with 10 μg of the immunogen, and immune splenocytes were removed 3 days later. Somatic cell hybrids were prepared with the mouse nonsecreting myeloma cell line NS-1 as previously described (20). Hybridoma supernatants were screened for differential immunoreactivity between GST-BFRF1- and GST-purified proteins by enzyme-linked immunosorbent assay (ELISA). Positive hybridoma cell lines were cloned twice by limiting dilution. One monoclonal antibody (MAb), E7, which specifically recognizes the GST-BFRF1- and His-BFRF1-purified proteins by ELISA and Western blotting analyses, was selected. Tissue culture supernatant of MAb R4 recognizing the unrelated carcinoembryonic antigen was used as a negative control (2).
Immunoblotting.
Cells (106) were resuspended in 50 μl of SDS-sample buffer (5% SDS, 25 mM tris hydroxymethyl aminomethane [pH 6.8], 5% 2-mercaptoethanol) and lysed by sonication. Samples were then subjected to SDS-PAGE. Equal protein loading on SDS-PAGE was determined by spectrophotometric assay of the collected samples. Proteins were then transferred to nitrocellulose filters (0.45-μm pore size; Schleicher & Schuell) according to standard procedures (29). The membranes were incubated at least 1 h with blocking solution (5% nonfat dried milk–0.1% Tween 20 in PBS). Antigens were detected by incubation for 1 h at room temperature with either anti-BFRF1 MAb E7 (diluted 1:50), MAb R4 (1:50), or anti-ZEBRA MAb (Dako) (1:50). Filters were then washed twice in PT20 buffer (1× PBS, 0.1% Tween 20). Following a washing step, membranes were incubated with horseradish peroxidase-conjugated antirabbit or antimouse antibody (Sigma), and after two further washings in PT20 buffer, bands were visualized by enhanced chemiluminescence (ECL system) according to the manufacturer's specifications (Amersham).
IFA.
The IFA was performed as follows. Cells were harvested, washed in PBS, seeded onto multispot microscope slides (ICN), air dried, and fixed for 5 min in acetone-methanol (1:1). Fixed cells were incubated with tissue culture supernatant of the monoclonal anti-BFRF1 antibody E7 (diluted 1:20) for 1 h at 37°C. The slides were then washed in PBS and subsequently incubated for 45 min at 37°C with fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse antibody (diluted 1:80) (Cappel). Following a further wash in PBS, slides were observed with a fluorescence microscope. For double-staining immunofluorescence, cells were first incubated for 1 h with anti-BFRF1 monoclonal antibody and then were stained for 45 min with FITC-conjugated antimouse antibody, 1 h with anti-ZEBRA rabbit polyclonal antibody (kindly provided by G. Miller), followed by 45 min with goat anti-rabbit Texas red antibody (Cappel). MAb R4 and normal rabbit serum were used as negative controls.
Ultrathin cryosections.
TPA-induced B95-8 cells were fixed with a mixture of 2% paraformaldehyde and 0.2% glutaraldehyde in PBS for 1 h at 4°C, washed, and embedded in 12% gelatin (Merck) in 0.1 M phosphate buffer that was solidified on ice. Gelatin blocks were infused with 2.3 M sucrose for 3 h at 4°C, frozen in liquid nitrogen, and cryosectioned. Ultrathin cryosections were collected with sucrose and methyl cellulose and incubated with anti-BFRF1 monoclonal antibody diluted 1:20 in PBS–1% BSA. Following several washes in PBS–0.1% BSA, the sections were incubated with 18-nm-diameter colloidal gold particles (prepared by the citrate method) conjugated with protein A (Pharmacia) diluted 1:10 in PBS. Control experiments were performed by omission of the primary antibody from the labeling procedure. Finally, ultrathin cryosections were stained with a solution of 2% methyl cellulose and 0.4% uranyl acetate before electron microscopy examination.
Cell fractionation.
Cell fractionation was performed as described elsewhere (9). Briefly, B95-8 cells were harvested, washed with PBS, and resuspended in HEM buffer (20 mM HEPES [N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid], 1 mM EDTA, 1 mM 2-mercaptoethanol, protease inhibitors). Cells were Dounce homogenized, and nuclei were collected by centrifugation at 750 × g for 5 min. Cell extracts were kept at 4°C for 5 min, and the remaining intact nuclei were again collected by a further centrifugation at 750 × g for 5 min. The supernatant was recovered, and a crude membrane fraction was obtained by centrifugation at 43,000 × g for 20 min. The leftover supernatant represented the cytoplasmic fraction. The nuclear and membrane fractions were dispersed directly into SDS-sample buffer, whereas proteins in the soluble fraction were first precipitated by the addition of 6 volumes of acetone before being solubilized in SDS-sample buffer.
Screening of human sera.
Serum samples were collected with informed consent from 40 subjects affected by NPC, 15 patients affected by BL, 24 individuals affected by IM, and 58 healthy donors. Sera were kept at −20°C until use. Assayed sera were diluted 1:20 in PBS and screened for the presence of anti-BFRF1 antibodies by Western blot analysis with both GST-BFRF1- and His-BFRF1-purified proteins. As a negative control, we used GST-purified protein. The same sera were also tested by IFA in DG75 cells transfected with CMV-BFRF1 plasmid and in untreated DG75 cells as controls.
EBV whole-virion purification.
Virion purification was performed as described elsewhere (38). Two hundred milliliters of a B95-8 cell culture was starved for 16 days before collection of supernatant. Cells were fractured with 3 cycles of rapid freezing and thawing before collection of supernatant, which was then pelleted at 22,000 × g overnight and resuspended in 24 ml of TNE buffer (0.01 M Tris [pH 7.4], 0.1 M NaCl, 1 mM EDTA). The suspension was layered onto a Nycodenz (Nyegaard & Co) step gradient (20 and 40% [wt/vol]) made up in the TNE buffer and centrifuged at 54,000 × g for 2 h. The band visible at the interface was recovered, diluted in TNE buffer, and layered onto a continuous sucrose gradient (20 to 60% [wt/wt]) made up in TNE buffer and centrifuged at 56,000 × g for 2 h. The single visible band was collected, suspended in TNE buffer, layered onto a second sucrose gradient, and centrifuged as before. The single visible band was harvested and diluted in TNE buffer, and EBV virions were pelleted by ultracentrifugation at 155,000 × g for 1 h. Recovered virions were resuspended in 500 μl of lysis buffer and loaded onto SDS-PAGE gel.
IHC and ISH.
Immunohistochemistry (IHC) was performed with frozen and paraffin sections. Frozen sections were acetone fixed and then incubated with anti-BFRF1 MAb (diluted 1:100) or with anti-LMP-1 antibody CS1-4 (diluted 1:10) (Dako) for 10 min. After extensive washing, samples were treated with the Dako LSAB kit, followed by incubation with 0.03% H2O2 and 0.06% 3,3-diaminobenzidine (Sigma). The slides were counterstained with hematoxylin and mounted in Entellan resin solution (Merck). Paraffin sections were deparaffinized, rehydrated, and then incubated for 1 h with either anti-BFRF1 or anti-LMP-1 antibodies. Sections were then treated with the catalyzed signal amplification system (Dako), followed by incubation with 0.03% H2O2 and 0.06% 3,3-diaminobenzidine (Sigma). In situ hybridization (ISH) for EBERs was performed on paraffin sections. Following deparaffinization and rehydration, samples were predigested with proteinase K and then incubated at room temperature for 2 h with FITC-conjugated EBER-EBV probe (Dako), consisting of a cocktail of EBER-1 and -2 oligonucleotides, both 30 bp in length. Sections were then treated with alkaline phosphatase-conjugated rabbit F(ab′) anti-FITC antibody. The reaction product was revealed by the enzyme substrate 5-bromo-4-chloro-3-indolylphosphate–nitroblue tetrazolium (BCIP-NBT) in dimethylformamide solution. As a control for IHC on frozen and paraffin sections, cytosmears and paraffin cytoblocks were used. One hundred microliters of each of the B95-8 and DG75 cell lines at 106 cells/ml was cytocentrifuged to prepare cytosmears. Paraffin cytoblocks were prepared as follows. Two hundred milliliters of a pellet containing 50 × 106 cells was cytocentrifuged with Shandon cytoblock cassettes to obtain a paraffin-embedded cell suspension that was used to prepare paraffin sections.
RESULTS
Predicted features of the EBV ORF BFRF1.
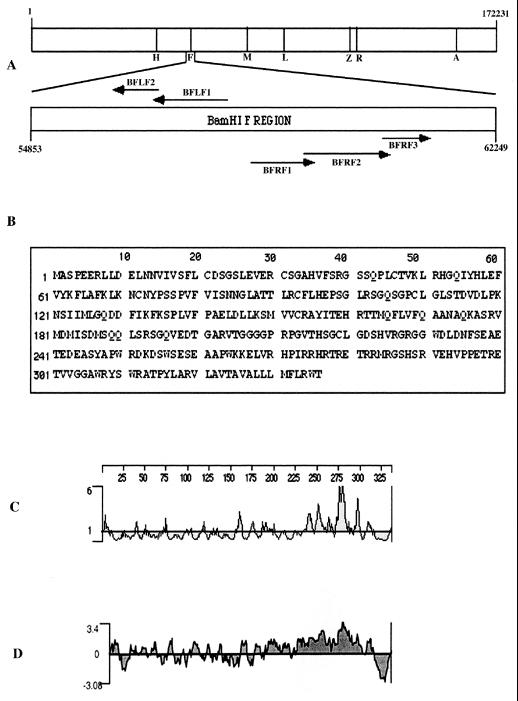
Figure 1 shows a diagramatic representation of the ORFs encompassed within the BamHI F region. A computer-assisted analysis of the EBV genome indicates that the BFRF1 ORF encodes a putative protein of 336 amino acids with an apparent molecular mass of 37.6 kDa. The protein is predicted to have an isoelectric point of 8.3; the primary amino acid sequence has one potential site for N glycosylation and one for O glycosylation. Furthermore, seven potential casein kinase II and four protein kinase C phosphorylation sites are present along the amino acid sequence. Kyte-Doolittle hydrophilicity analysis and Emini surface probability analysis revealed one potential transmembrane domain localized between residues 317 to 333, next to the C terminus of the protein (Fig. 1C to D). Therefore, the BFRF1 protein may retain the characteristics of a type II membrane protein, presenting a long cytoplasmic tail.
FIG. 1.
Computer analysis of the ORF BFRF1. (A) Schematic drawings of the genomic locations of the BamHI F region in the EBV genome and the ORFs encompassed within the region. (B) Putative 336-amino-acid sequence of the ORF BFRF1. (C) Emini analysis showing surface probability of the BFRF1 protein. (D) Kyte-Doolittle analysis showing hydrophilicity of the BFRF1 protein.
Generation of recombinant BFRF1 proteins and of a MAb to BFRF1.
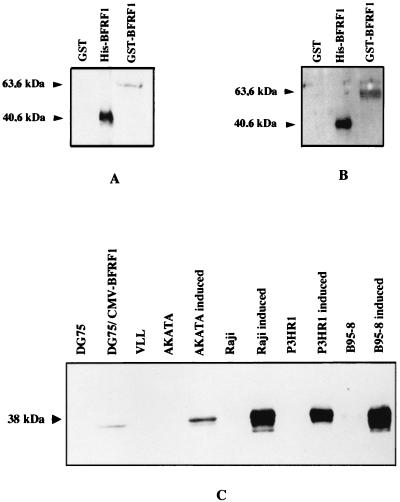
To produce a source of protein to use as immunogen and to analyze patient serum immunoreactivity for the BFRF1 protein, two recombinant fusion proteins (His-BFRF1 and GST-BFRF1) were generated in E. coli cells. His-BFRF1 and GST-BFRF1 were purified to homogeneity through column chromatography: the degree of purification was monitored by SDS-PAGE. Coomassie blue staining of the gel revealed single bands with apparent molecular masses of about 40 and 63 kDa for His-BFRF1 and GST-BFRF1, respectively. This finding was in agreement with the predicted molecular masses of the His-BFRF1 (40.6 kDa) and GST-BFRF1 (63.6 kDa) fusion proteins. To evaluate the structural and functional properties of the BFRF1 gene product, a BFRF1-specific MAb was developed by using the His-BFRF1-purified protein as an immunogen. MAb E7 was selected, by ELISA, based on its strong immunoreactivity with the GST-BFRF1 protein in the absence of GST protein immunoreactivity. MAb E7 was also able to detect by Western blot analysis the GST-BFRF1 and His-BFRF1 proteins, but not the GST protein used as a negative control (Fig. 2A). No immunoreactivity with the recombinant proteins was observed with MAb R4, recognizing the carcinoembryonic antigen (data not shown). Furthermore, MAb E7 immunoprecipitated both GST- and His-tagged BFRF1 from lysates (data not shown). Altogether, these results established the specificity of MAb E7 for the BFRF1 protein.
FIG. 2.
BFRF1 is recognized by MAb E7 as well as by human sera. (A) Specificity of the MAb E7 generated following immunization of mice with purified His-BFRF1 was confirmed by Western blot analysis. Samples representing purified His-BFRF1, GST-BFRF1, and GST were run, and the immunoblot was probed with anti-BFRF1 MAb E7. (B) Screening of the seroprevalence to BFRF1 by Western blot analysis. Fractions containing purified GST, His-BFRF1, and GST-BFRF1 were separated by SDS-PAGE, and immunoblots were performed with human sera (diluted 1:20) from NPC, BL, and IM patients as well as from healthy donors. A serum sample from a BFRF1-seropositive NPC patient is represented in panel B. (C) Expression of BFRF1 in different cell lines. Cell extracts from different cell lines were electrophoresed by SDS-PAGE (12% polyacrylamide). The following samples were analyzed: DG75 cells, DG75 cells transfected with CMV-BFRF1, VLL cells, Akata cells, Akata cells treated with antihuman Ig, Raji cells, P3HR-1 and B95-8 cells, and the same cell lines treated with TPA (20 ng/ml) and butyrate (3 mM). The immunoblot was probed with the anti-BFRF1 MAb E7.
Detection of antibodies to BFRF1 in sera of NPC and BL patients.
To test whether the BFRF1 gene product is expressed in vivo and to evaluate its ability to induce a permanent humoral response, immunoblots were performed with both histidine- and GST-tagged purified BFRF1 proteins produced in E. coli. Human sera from NPC patients, BL patients, individuals affected by IM, and healthy EBV-seropositive donors previously assayed by IFA for anti-EBV early antigens (EA) were screened.
Antibodies to BFRF1 were detected by immunoblotting (1:20 dilution) in sera of 31 of 40 (77.5%) NPC patients, 7 of 15 (47%) BL patients, and 1 of 24 (4%) individuals with IM, while no positivity could be detected in 58 healthy, EBV-seropositive controls (Fig. 2B). Among the NPC sera analyzed, a seroprevalence of 100% to BFRF1 could be detected in subjects showing an anti-EA titer higher than 1:40, whereas sera with titers equal to or lower than 1:40 showed a BFRF1 seroprevalence of 65%. The results of the screening were also confirmed by testing the sera by IFA on DG75 cells transfected with CMV-BFRF1. Thus, we concluded that BFRF1 protein is indeed produced in vivo and stimulates a humoral response.
BFRF1 encodes a lytic protein.
To determine whether BFRF1 protein is expressed in EBV-infected cells, cell extracts obtained from EBV-positive and EBV-negative cell lines were assayed by Western blot analysis. SDS lysates were produced from (i) three EBV-positive cell lines (Raji, P3HR-1, and B95-8) treated or not with EBV lytic cycle inducers TPA and sodium butyrate; (ii) the EBV-positive cell line Akata, either induced with monoclonal antihuman IgG or uninduced; (iii) the EBV-positive lymphoblastoid cell line VLL; or (iv) the EBV-negative cell line DG75, as well as the same cell line transiently transfected with CMV-BFRF1 plasmid. Following electrophoresis, the samples mentioned above were analyzed in immunoblots with the MAb E7. A doublet migrating at 37 to 38 kDa was detected in the four EBV-positive cell lines after induction of EBV replication. A weak band was also detected in CMV-BFRF1-transfected DG75 cells. A weaker doublet was observed in untreated B95-8 and P3HR-1 cells (at longer exposure), in keeping with the observation that, in these cell lines, about 0.5 to 5% of cells undergo spontaneous activation of the lytic cycle. Metabolic labeling with [3H]glucosamine, as well as experiments performed in the presence of endoglycosidases F and H, ruled out the possibility that BFRF1 is a glycoprotein. In addition, preliminary experiments with lambda phosphatase seem to suggest the presence of a phosphorylated form of the protein (data not shown). Experiments are in progress to further address this issue. No signal was present in untreated Raji and Akata cells or in VLL and in DG75 cells. Since in VLL cells and in uninduced Raji and Akata cells EBV expression is restricted to the latent proteins, the absence of BFRF1 gene product indicates that BFRF1 is a lytic protein (Fig. 2C).
BFRF1 is expressed as an early transcript.
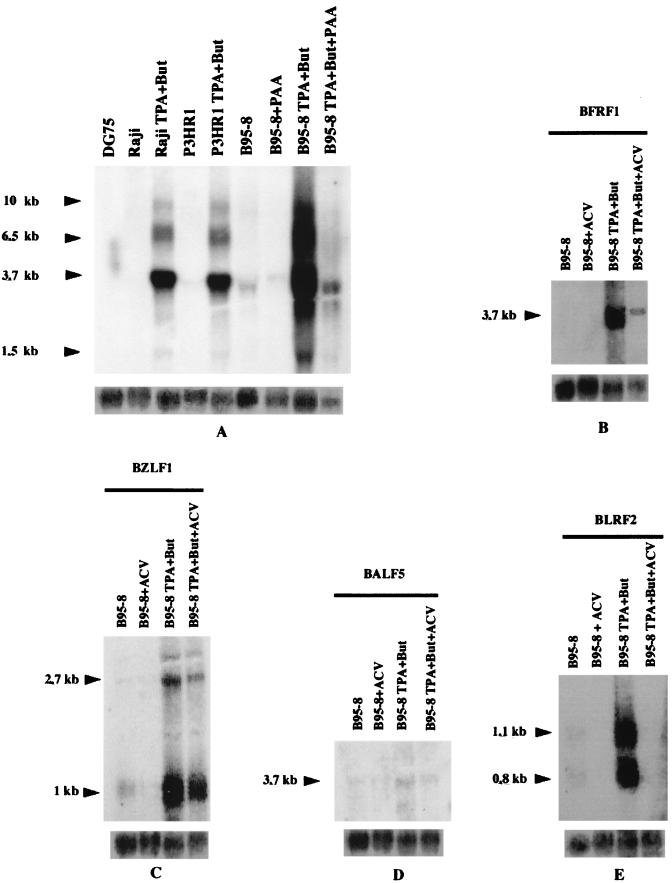
Northern blot analysis was carried out to study the regulation of BFRF1 expression. Following hybridization with a genomic probe to the BFRF1 ORF, a strong band migrating at 3.7 kb, as well as three additional bands of 1.5, 6.5, and 10.0 kb, were detected in Raji, B95-8, and P3HR-1 cells following treatment with TPA and sodium butyrate, whereas a faint band was present in uninduced B95-8 and P3HR-1 cells. No specific signal was revealed in uninduced Raji cells and in EBV-negative DG75 cells. These data are in keeping with a previous study showing the presence of multiple transcripts crossing the BamHI F region (13). The same transcripts were also detected in B95-8 cells induced by TPA and sodium butyrate in the presence of PAA, although a decrease of the signal was visible (Fig. 3A). To further investigate the behavior of BFRF1 in presence of a more specific viral DNA polymerase inhibitor, we analyzed the effect of ACV treatment of induced B95-8 cells. BFRF1 expression was only partially reduced by ACV treatment (Fig. 3B), in agreement with the results obtained with PAA. As a control of the effectiveness of ACV treatment, we analyzed expression of the immediate-early gene BZLF1 (Fig. 3C), of the EBV polymerase gene BALF5 (known as an early gene) (Fig. 3D), and of the late gene BLRF2 (30) (Fig. 3E). The strand specificity of the transcripts was confirmed with a 30-bp oligonucleotide indicated as NF13 (data not shown). From these experiments, we conclude that BFRF1 is expressed as an early gene, since it is only partially affected by treatment with PAA and ACV, which are known to inhibit completely the transcription of late lytic viral messengers, and is present in Raji cells, which harbor a defective viral strain that does not allow the expression of EBV genes belonging to the late phase of the lytic cycle. To rule out the possibility that BFRF1 might behave as an immediate-early gene, we carried out additional experiments with the protein synthesis inhibitor cycloheximide (CHX). CHX treatment (4 h) of TPA-induced B95-8 cells did not affect BFRF1 and BZLF1 expression, whereas, after 16 h of incubation with CHX, BFRF1 expression was dramatically reduced (data not shown).
FIG. 3.
Analysis of BFRF1 RNA expression. (A) Northern blot analysis in which 10 μg of total RNA extracted from different cell lines was separated on a 1.2% agarose–6% formaldehyde gel. The following cell lines were analyzed for BFRF1 expression: untreated Raji, P3HR-1, and B95-8 cells; the same cell lines chemically induced; or B95-8 cells treated with PAA and DG75 cells. The blot was hybridized with the BFRF1 probe. But, butyrate. (B to E) Northern blot analysis of untreated B95-8 cells and B95-8 cells chemically induced in absence or in presence of ACV. The blot in panel B was hybridized with anti-BFRF1. (C) Blot hybridized with anti-BZLF1. (D and E) Blots probed with anti-BALF5 and anti-BLRF2, respectively. Equal RNA loading was assessed by β-actin hybridization.
BFRF1 is mainly localized on cellular plasma membrane and is present in the virions.
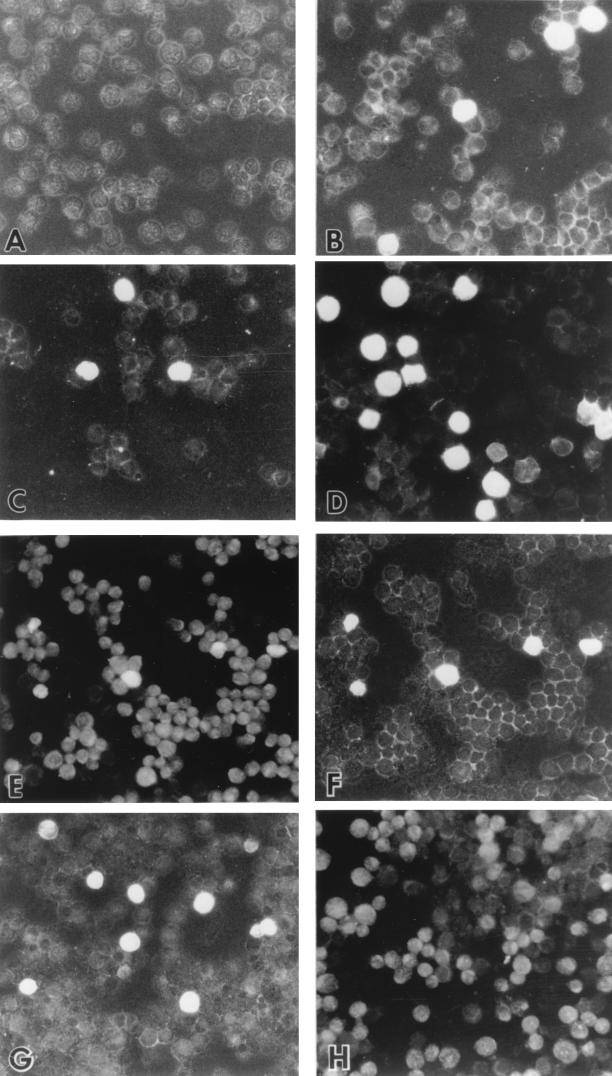
To visualize cellular localization of BFRF1, IFA was performed with Raji and B95-8 cells by using MAb E7. Following treatment with TPA and sodium butyrate, BFRF1 was detected predominantly on the plasma membrane, although a cytoplasmic staining was present (Fig. 4A to D). Moreover, in a good percentage of cells, staining of the nuclear membrane could be also detected. No reactivity with the unrelated MAb R4 was observed in these cell lines (data not shown). We also demonstrated that BFRF1 transactivation could be achieved following transfection of BZLF1, as shown in the panel reporting the double staining for ZEBRA and BFRF1 in Raji cells transfected with the CMV-BZLF1 plasmid (Fig. 4E to F). This is in agreement with the finding that activation of the lytic cycle is necessary for BFRF1 expression. On the other hand, the expression of BFRF1, but not that of ZEBRA, could be detected when Raji cells were transfected with the CMV-BFRF1 construct (Fig. 4G to H).
FIG. 4.
BFRF1 is detected by immunofluorescence on Raji and B95-8 cells. (A and B) show untreated Raji cells (A) or Raji cells treated with TPA (20 ng/ml) and butyrate (3 mM) (B). (C and D) Untreated B95-8 cells (C) or B95-8 cells treated with TPA (20 ng/ml) and butyrate (3 mM) (D). Cells in panels A to D were incubated with anti-BFRF1 MAb E7, followed by FITC-conjugated antimouse antibody. (E and F) Raji cells transfected with CMV-BZLF1 and double stained with MAb E7 followed by FITC-conjugated antimouse antibody (E) and anti-BZLF1 rabbit antibody followed by Texas red-conjugated antirabbit antibody (F; same field as in panel E). (G and H) Raji cells transfected with CMV-BFRF1 and double stained with MAb E7 followed by FITC-conjugated antimouse antibody (G) and rabbit anti-BZLF1 antibody followed by Texas red-conjugated antirabbit antibody (H; same field as in panel G).
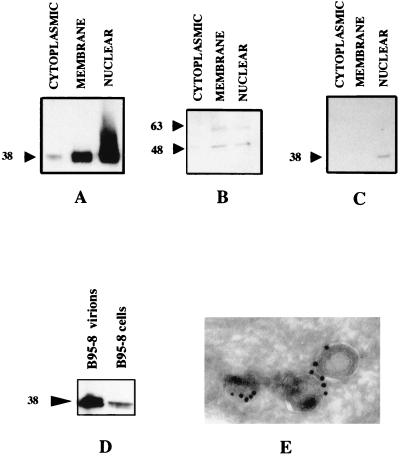
Data regarding cellular localization of BFRF1 were confirmed by experiments with cell fractionation. B95-8 cells treated with TPA and sodium butyrate were lysed and separated into membrane, cytoplasmic, and nuclear fractions before being immunoblotted. A strong band migrating at 38 kDa reacted with the anti-BFRF1 MAb E7 in the nuclear and membrane fractions, whereas trace amounts were detectable in the cytoplasmic fraction. The same fractions were analyzed with anti-LMP-1 and anti-ZEBRA antibodies. LMP-1 is present in the membrane and nuclear fractions, while ZEBRA is mainly detectable in the nuclear fraction (Fig. 5A to C).
FIG. 5.
BFRF1 is mainly localized in the membranes and nuclear fractions and is present in the virions. B95-8 cells were lysed and separated in membrane, cytoplasmic, and nuclear fractions. Samples were run on an SDS-PAGE (12% polyacrylamide) gel, and immunoblots were probed with anti-BFRF1 MAb E7 (A), anti-LMP-1 MAb (B), and rabbit monospecific anti-ZEBRA antibody (C). EBV virions were obtained from B95-8 cells, and after lysis, they were electrophoresed on an SDS-PAGE (12% polyacrylamide) gel. Immunoblotting was performed with MAb E7 (D). Molecular masses are shown to the left of panels A to D (kilodaltons). (E) Intracellular enveloped virions immunogold labeled with MAb E7 on an ultrathin cryosection of TPA-induced B95-8 cells (original magnification, ×120,000).
Furthermore, experiments were carried out to evaluate the presence of BFRF1 in the virions. B95-8 cells were starved to increase viral production, and subsequently virions were purified through centrifugations on gradients. Immunoblots performed with recovered and lysed virions with anti-BFRF1 MAb showed a strong positive signal, indicating that BFRF1 is abundantly present in EBV virions (Fig. 5D). In addition, immunoelectron microscopy revealed that intracellular enveloped virions are specifically labeled by anti-BFRF1 antibody (Fig. 5E).
BFRF1 protein is not detected in NPC or in HD.
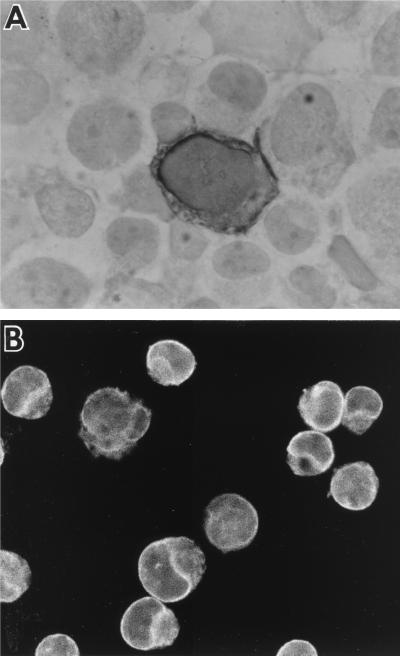
The possibility of using MAb E7 in IHC was tested with cytosmears of B95-8 and DG75 cells. BFRF1 was clearly observed in 10 to 15% of B95-8 cells treated with TPA and sodium butyrate and in 5% of untreated B95-8 cells, and it was consistently missing in DG75 cells. The staining was mainly localized in correspondence with the nuclear and cellular membranes, as observed in IFA (Fig. 6). Since NPC samples were available only in paraffin blocks, we tested the possibility of using MAb E7 on sections of paraffin-embedded pellets of centrifuged induced and uninduced B95-8 cells. By this technique, 14 to 20% of treated B95-8 cells and 6% of untreated B95-8 cells were immunostained for MAb E7. The staining pattern was similar to that observed on the cytosmears described above.
FIG. 6.
Immunolocalization of BFRF1. TPA-induced B95-8 cells immunolabeled with the anti-BFRF1 MAb E7 reveal preferential labeling over the nuclear and plasma membranes (A) IHC (original magnification, ×1,000). (B) Immunofluorescence (original magnification, ×600).
BFRF1 expression was then investigated with frozen and paraffin-embedded sections of tissues involved in Hodgkin's disease (HD) and NPC. Frozen sections obtained from 12 lymph nodes involved in HD of the nodular sclerosis (5 samples) and mixed cellularity (7 samples) subtypes were immunostained with MAb E7. Among them, 2 of 12 were LMP-1 positive, but none was immunostained for BFRF1, as expected. Since previous studies (35) demonstrated that EBV is present in almost all lymph nodes involved in HD in human immunodeficiency virus type 1 (HIV-1)-infected patients, we tested frozen sections of two HD-positive, HIV-1-positive lymph nodes for BFRF1 expression. Both were positive for LMP-1; however, they were consistently negative for BFRF1.
The expression of BFRF1 was then investigated with paraffin sections from 11 cases of well-differentiated (7 samples) and poorly differentiated (4 samples) NPC and from 9 further lymph nodes involved in HD of the nodular sclerosis (5 samples) and mixed cellularity (4 samples) subtypes. Five of 11 NPC samples were positive for EBER in in situ hybridization, and 2 of 11 were reactive with LMP-1. Three of nine HD samples were positive for EBER and LMP-1. However, all samples were consistently negative for BFRF1.
DISCUSSION
In the present study, we identified, cloned, and expressed a novel EBV-encoded protein. BFRF1 belongs to the lytic proteins, since its expression is achieved following activation of the EBV replication cycle. Furthermore, it can be classified as an early protein, given the fact that it is only partially inhibited by treatment with PAA and ACV (with the BFRF1 gene behaving like BALF5, a known early gene) and that it is present in Raji cells which harbor a defective EBV strain that does not allow expression of the late lytic genes.
To date, roughly a dozen EBV early lytic proteins are known: they are either transactivators of the EBV lytic cycle (ZEBRA or Rta) or proteins involved in the machinery of DNA replication. So far, no specific function can be ascribed to BFRF1.
Based on DNA sequence, BFRF1 homologs are present among other members of the herpesviruses. In particular, the best characterized homolog is the herpes simplex virus type 1 (HSV-1) gene UL34. Like BFRF1, the UL34 gene product is membrane anchored and is present in the mature virions (24). Furthermore, previous studies have shown that the UL34 product is a phosphoprotein and that it is the substrate of the HSV-encoded protein kinase US3 (23). Although it should be noticed that the consensus sequence (R)nX(S/T)YY, which has been described as being recognized by US3 and by other herpesvirus protein kinases, is not present in BFRF1, preliminary experiments seem to indicate that BFRF1, like UL34, is phosphorylated. A strong colinear homology of the region that encompasses the BFRF1 ORF is present among other members of the herpesvirus family besides HSV-1. Indeed, BFRF1 belongs to a family that groups together the following homologs: Human cytomegalovirus (UL50), Varicella-zoster virus (UL24), Human herpesvirus 6 (HHV-6) (UL34), HHV-7 (UL34), HHV-8 (ORF67), and Equine herpesvirus 1 EHV-1 (ORF26). All of them have been reported as putative ORFs, since no corresponding protein has been found thus far. Identification of BFRF1 protein adds a second member to this homolog family: BFRF1 and HSV-1 UL34 are actually expressed during viral infection, thus suggesting that the other related ORFs might be coding for proteins and that some function required for herpesvirus replication is retained in this group of proteins. Interestingly, HSV-1 UL34 has been shown to be required for viral replication, and recently it has been demonstrated to play a role in the viral envelopment process (28).
The localization of BFRF1 on cellular membranes is in keeping with the Kyte-Doolittle and Emini analyses predicting a potential transmembrane domain close to the carboxyl end of the protein. Furthermore, its presence on the virions seems to suggest that the protein could migrate to the intracellular membranes, such as the inner nuclear membrane, where the budding of EBV occurs. This event would then lead to the encapsidation of BFRF1 within the virions (34).
BFRF1 protein has not been detected in the NPC and HD specimens we analyzed. This observation is concordant with previous studies carried out with NPC in which IHC failed to show any viral lytic protein, even though transcripts to the corresponding EBV lytic proteins have been detected. As for HD, viral lytic proteins have been found in a very small percentage of cases (4). However, that BFRF1 protein is present, but under the threshold of detection, cannot be ruled out.
Moreover, BFRF1 protein is expressed in vivo, as demonstrated by the presence of antibodies in sera of patients affected by NPC and BL. The lack of antibodies to BFRF1 in sera of healthy individuals as well as in all but one of the IM patients should not be considered unexpected. The humoral response to BFRF1 may be a consequence of an active and prolonged viral replication going on in NPC and in BL that could be necessary to stimulate the production of BFRF1 antibodies to a detectable level. Moreover, previous studies have revealed antibodies to other EBV lytic proteins, such as ZEBRA, DNase, ribonucleotide reductase, and thymidine kinase, in NPC patients (5, 10, 14, 17). On the other hand, the same antibodies are less frequently detected in healthy individuals. However, we cannot exclude that individuals who scored seronegative for BFRF1 might be seropositive with a titer below the threshold of detection. It should also be noted that, at least among NPC patients, BFRF1 seropositivity strongly correlates with anti-EBV EA titer. Indeed, subjects with anti-EA titers equal to or higher than 1:40 show BFRF1 positivity invariably.
Identification of BFRF1 as a novel EBV protein could be of interest for at least two lines of research. First, even though virion proteins are of obvious importance for a detailed understanding of EBV biology, they have not been extensively investigated. The characterization of BFRF1 as a virionic protein represents a new element in this picture, and it could turn out to be helpful for a better understanding of EBV biology. Although at present we do not have any evidence to support a functional role of the BFRF1 gene product, our observation that the protein is localized over the cell nuclear membrane and the very recent demonstration that the BFRF1 homolog of HSV-1, UL34, is necessary for virus envelopment (28), may suggest a similar role for BFRF1. Experiments to study viral envelopment and BFRF1 localization by immunoelectron microscopy are currently ongoing in our laboratory. Second, even though BFRF1 seropositivity is not detectable in the totality of NPC and BL patients, it may still provide an additional marker with which to study individuals affected by these two EBV-associated neoplastic diseases.
ACKNOWLEDGMENTS
We thank George Miller for the generous gift of polyclonal anti-ZEBRA antibody and CMV-BZLF1 plasmid and Maria Rosaria Torrisi for helpful discussions. We thank L. Vestri, D. Galafate, and C. Talerico for skillful technical assistance.
This work was partially supported by grants from MURST; from Associazione Italiana per la Ricerca sul Cancro (AIRC); from Ministero della Sanità, Progetto AIDS; and from Istituto Pasteur Fondazione Cenci-Bolognetti, Università di Roma “La Sapienza.”
REFERENCES
- 1.Baer R, Bankier A T, Biggin M D, Deininger P L, Farrell P J, Gibson T J, Hatfull G, Hudson G S, Satchwell S C, Seguin C, Tuffnell P S, Barrell B G. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature (London) 1984;310:207–211. doi: 10.1038/310207a0. [DOI] [PubMed] [Google Scholar]
- 2.Bei R, Moretti A, Visco V, De Filippi R, Tsang K Y, Frati L, Muraro R. Cell mediated cytotoxicity of human colon carcinoma cells by a monoclonal antibody (R4) recognizing the carcinoembryonic antigen (CEA) and CEA-related molecules. Int J Oncol. 1996;8:1127–1135. doi: 10.3892/ijo.8.6.1127. [DOI] [PubMed] [Google Scholar]
- 3.Ben-Bassat H, Goldblum N, Mitrani S, Goldblum T, Yoffey J M, Cohen M M, Bentwich Z, Ramot B, Klein E, Klein G. Establishment in continuous culture of a new type of lymphocyte from a ‘Burkitt-Like’ malignant lymphoma (line DG75) Int J Cancer. 1977;19:27–33. doi: 10.1002/ijc.2910190105. [DOI] [PubMed] [Google Scholar]
- 4.Brousset P, Knecht H, Rubin B, Drouet E, Chittal S, Meggetto F, Saati T A, Bachmann E, Denoyel G, Sergeant A, Delsol G. Demonstration of Epstein-Barr virus replication in Reed-Sternberg cells of Hodgkin's disease. Blood. 1993;82:872–876. [PubMed] [Google Scholar]
- 5.Chen H F, Sauter M, Haiss P, Muller-Lantzsch N. Immunological characterization of the Epstein-Barr virus phosphoprotein PP58 and deoxyribonuclease expressed in the baculovirus expression system. Int J Cancer. 1991;48:879–888. doi: 10.1002/ijc.2910480615. [DOI] [PubMed] [Google Scholar]
- 6.Countryman J, Miller G. Activation of the expression of the latent Epstein-Barr herpesvirus after gene transfer with a small cloned subfragment of heterogenous viral DNA. Proc Natl Acad Sci USA. 1985;82:4085–4089. doi: 10.1073/pnas.82.12.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis M G, Huang E S. Transfer and expression of plasmids containing human cytomegalovirus immediate-early gene 1 promoter-enhancer sequences in eukaryotic and prokaryotic cells. Biotechnol Appl Biochem. 1988;10:6–12. [PubMed] [Google Scholar]
- 8.Faggioni A, Zompetta C, Grimaldi S, Barile G, Frati L, Lazdins J. Calcium modulation activates Epstein-Barr virus genome in latently infected cells. Science. 1986;232:1551–1556. doi: 10.1126/science.3012779. [DOI] [PubMed] [Google Scholar]
- 9.Fries K L, Sculley T B, Webster-Cyriaque J, Rajadurai P, Sadler R H, Raab-Traub N. Identification of a novel protein encoded by the BamHI A region of the Epstein-Barr virus. J Virol. 1997;71:2765–2771. doi: 10.1128/jvi.71.4.2765-2771.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ginsburg M. Antibodies against the large subunit of the EBV-encoded ribonucleotide reductase in patients with nasopharyngeal carcinoma. Int J Cancer. 1990;45:1048–1053. doi: 10.1002/ijc.2910450612. [DOI] [PubMed] [Google Scholar]
- 11.Hanto D W, Frizzera G F, Gajl-Peczalska K J, Sakamoto K, Purtilo D T, Balfour H H, Simmons R L, Najarian J S. Epstein-Barr virus-induced B-cell lymphoma after renal transplantation. N Engl J Med. 1982;306:913–918. doi: 10.1056/NEJM198204153061506. [DOI] [PubMed] [Google Scholar]
- 12.Hinuma Y, Konn M, Yamaguchi J, Wudarski D J, Blakeslee J R, Jr, Grace J T., Jr Immunofluorescence and herpes-type particles in the P3HR-1 Burkitt lymphoma cell line. J Virol. 1967;1:1045–1051. doi: 10.1128/jvi.1.5.1045-1051.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hudson G S, Gibson T J, Barrell B G. The BamHI F region of the B95-8 Epstein-Barr virus genome. Virology. 1985;147:99–109. doi: 10.1016/0042-6822(85)90230-2. [DOI] [PubMed] [Google Scholar]
- 14.Joab I, Nicolas J C, Schwaab G, de-The G, Clausse B, Perricaudet M, Zeng Y. Detection of anti-Epstein-Barr virus transactivator (ZEBRA) antibodies in sera from patients with nasopharyngeal carcinoma. Int J Cancer. 1991;48:647–649. doi: 10.1002/ijc.2910480503. [DOI] [PubMed] [Google Scholar]
- 15.Katz B Z, Raab-Traub N, Miller G. Latent and replicating forms of Epstein-Barr virus DNA in lymphomas and lymphoproliferative diseases. J Infect Dis. 1989;160:589–594. doi: 10.1093/infdis/160.4.589. [DOI] [PubMed] [Google Scholar]
- 16.Lin J C, Shaw J E, Smith M C, Pagano J S. Effect of 12-O-tetradecanoyl-phorbol-13-acetate on the replication of the Epstein-Barr virus. I. Characterization of viral DNA. Virology. 1979;99:183–187. doi: 10.1016/0042-6822(79)90052-7. [DOI] [PubMed] [Google Scholar]
- 17.Littler E, Newman W, Arrand J R. Immunological response of nasopharyngeal carcinoma patients to the Epstein-Barr virus coded thymidine kinase expressed in Escherichia coli. Int J Cancer. 1990;45:1028–1032. doi: 10.1002/ijc.2910450608. [DOI] [PubMed] [Google Scholar]
- 18.Luka J, Kallinn B, Klein G. Induction of the Epstein-Barr virus (EBV) cycle in latently infected cells by n-butyrate. Virology. 1979;94:228–231. doi: 10.1016/0042-6822(79)90455-0. [DOI] [PubMed] [Google Scholar]
- 19.Miller G, Lipman M. Release of infectious Epstein-Barr virus by transformed marmoset leukocytes. Proc Natl Acad Sci USA. 1973;70:190–194. doi: 10.1073/pnas.70.1.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muraro R, Wunderlich D, Thor A, Lundy J, Noguchi P, Cunningham R, Schlom J. Definition by monoclonal antibodies of a repertoire of epitopes on carcinoembryonic antigen differentially expressed in human colon carcinomas versus normal adult tissues. Cancer Res. 1985;45:5769–5780. [PubMed] [Google Scholar]
- 21.Pope J, Horne M, Scott W. Transformation of fetal human leukocytes in vitro by filtrates of a human leukemic cell line containing herpes-like virus. Int J Cancer. 1968;3:857–866. doi: 10.1002/ijc.2910030619. [DOI] [PubMed] [Google Scholar]
- 22.Pulvertaft R J V. Cytology of Burkitt's tumour (African lymphoma) Lancet. 1964;i:238–240. doi: 10.1016/s0140-6736(64)92345-1. [DOI] [PubMed] [Google Scholar]
- 23.Purves F C, Spector D, Roizman B. The herpes simplex virus 1 protein kinase encoded by the US3 gene mediates posttranslational modification of the phosphoprotein encoded by the UL34 gene. J Virol. 1991;65:5757–5764. doi: 10.1128/jvi.65.11.5757-5764.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Purves F C, Spector D, Roizman B. UL34, the target of the herpes simplex virus US3 protein kinase, is a membrane protein which in its unphosphorylated state associates with novel phosphoproteins. J Virol. 1992;66:4295–4303. doi: 10.1128/jvi.66.7.4295-4303.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raab-Traub N, Flynn K, Pearson G, Huang A, Levine P, Lenier A, Pagano J. The differentiated form of nasopharyngeal carcinoma contains Epstein-Barr virus DNA. Int J Cancer. 1987;39:25–29. doi: 10.1002/ijc.2910390106. [DOI] [PubMed] [Google Scholar]
- 26.Ragoczy T, Heston L, Miller G. The Epstein-Barr virus Rta protein activates lytic cycle genes and can disrupt latency in B lymphocytes. J Virol. 1998;72:7978–7984. doi: 10.1128/jvi.72.10.7978-7984.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rickinson A B, Kieff E. Epstein-Barr virus. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2397–2446. [Google Scholar]
- 28.Roller R J, Zhou Y, Schnetzer R, Ferguson J, DeSalvo D. Herpes simplex virus type 1 UL34 gene product is required for viral envelopment. J Virol. 2000;74:117–129. doi: 10.1128/jvi.74.1.117-129.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 30.Serio T R, Angeloni A, Kolman J L, Gradoville L, Sun R, Katz D A, Van Grunsven W, Middeldorp J, Miller G. Two 21-kilodalton components of the Epstein-Barr virus capsid antigen complex and their relationship to ZEBRA-associated protein p21 (ZAP21) J Virol. 1996;70:8047–8054. doi: 10.1128/jvi.70.11.8047-8054.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shedd D, Angeloni A, Niederman J, Miller G. Detection of human serum antibodies to the BFRF3 Epstein-Barr virus capsid component by means of a DNA-binding assay. J Infect Dis. 1995;172:1367–1370. doi: 10.1093/infdis/172.5.1367. [DOI] [PubMed] [Google Scholar]
- 32.Takada K. Cross-linking of cell surface immunoglobulin induces Epstein-Barr virus in Burkitt lymphoma lines. Int J Cancer. 1984;33:27–32. doi: 10.1002/ijc.2910330106. [DOI] [PubMed] [Google Scholar]
- 33.Takada K, Ono Y. Synchronous and sequential activation of latently infected Epstein-Barr virus genomes. J Virol. 1989;63:445–449. doi: 10.1128/jvi.63.1.445-449.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Torrisi M R, Cirone M, Pavan A, Zompetta C, Barile G, Frati L, Faggioni A. Localization of Epstein-Barr virus envelope glycoproteins on the inner nuclear membrane of virus-producing cells. J Virol. 1989;63:828–832. doi: 10.1128/jvi.63.2.828-832.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Uccini S, Monardo F, Stoppacciaro A, Gradilone A, Aglianò A M, Faggioni A, Manzari V, Vago L, Costanzi G, Ruco L P, Baroni C D. High frequency of Epstein-Barr virus genome detection in Hodgkin's disease of HIV-positive patients. Int J Cancer. 1990;46:581–585. doi: 10.1002/ijc.2910460405. [DOI] [PubMed] [Google Scholar]
- 36.van Grunsven W M, Spaan W J, Middeldorp J M. Localization and diagnostic application of immunodominant domains of the BFRF3-encoded Epstein-Barr virus capsid protein. J Infect Dis. 1994;170:13–19. doi: 10.1093/infdis/170.1.13. [DOI] [PubMed] [Google Scholar]
- 37.van Grunsven W M J, van Heerde E C, de Haard H J W, Spaan W J M, Middeldorp J M. Gene mapping and expression of two immunodominant Epstein-Barr virus capsid proteins. J Virol. 1993;67:3908–3916. doi: 10.1128/jvi.67.7.3908-3916.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamamoto M, Black J B, Stewart J A, Lopez C, Pellett P E. Identification of a nucleocapsid protein as a specific serological marker of human herpesvirus 6 infection. J Clin Microbiol. 1991;28:1957–1962. doi: 10.1128/jcm.28.9.1957-1962.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zalani S, Holley-Guthrie E, Kenney S. Epstein-Barr viral latency is disrupted by the immediate early BRLF1 protein through a cell-specific mechanism. Proc Natl Acad Sci USA. 1996;93:9194–9199. doi: 10.1073/pnas.93.17.9194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.zur Hausen H, O'Neil F J, Freese U K, Hecker E. Persisting oncogenic herpesvirus induced by the tumor promoter TPA. Nature (London) 1978;272:373–375. doi: 10.1038/272373a0. [DOI] [PubMed] [Google Scholar]
- 41.zur Hausen H, Schulte-Holthausen H, Klein G, Henle W, Henle G, Clifford P, Santesson L. EBV DNA in biopsies of Burkitt tumors and anaplastic carcinomas of the nasopharynx. Nature (London) 1970;228:1056–1058. doi: 10.1038/2281056a0. [DOI] [PubMed] [Google Scholar]