Abstract
The genes encoding the α (63-kDa) and β (95-kDa) subunits of Rous sarcoma virus (RSV) reverse transcriptase (RT) or the entire Pol polypeptide (99 kDa) were mutated in the conserved aspartic acid residue Asp 181 of the polymerase active site (YMDD) or in the conserved Asp 505 residue of the RNase H active site. We have analyzed heterodimeric recombinant RSV αβ and αPol RTs within which one subunit was selectively mutated. When αβ heterodimers contained the Asp 181→Asn mutation in their β subunits, about 42% of the wild-type polymerase activity was detected, whereas when the heterodimers contained the same mutation in their α subunits, only 7.5% of the wild-type polymerase activity was detected. Similar results were obtained when the conserved Asp 505 residue of the RNase H active site was mutated to Asn. RNase H activity was clearly detectable in αβ heterodimers mutated in the β subunit but was lost when the mutation was present in the α subunit. In summary, our data imply that the polymerase and RNase H active sites are located in the α subunit of the heterodimeric RSV RT αβ.
Reverse transcriptase (RT) of Rous sarcoma virus (RSV) is a component of the Gag-Pol precursor protein. Pol is composed of polymerase, RNase H, and integrase domains and an additional short 4.1-kDa protein located at the C terminus of the protein. Pol is processed into polypeptides of various lengths by the viral protease; its α polypeptide (63 kDa) contains the polymerase and RNase H domains, and its β polypeptide (95 kDa) consists of the polymerase, RNase H, and integrase domains but lacks the C-terminal 4.1-kDa protein (1, 7, 8, 17, 28, 30). In addition, the integrase domain (32 kDa) is also present and active as a separate enzyme (9, 30). Three forms of RT have been isolated from avian sarcoma and leukosis viruses (ASLV): α, β, and αβ, with the major form being the heterodimer (8, 11, 16). We have shown previously that the different forms of RSV RT can be expressed and purified from insect cells using the baculovirus expression system (37). In order to examine the subunit organization of RSV RT, the technique of subunit-selective mutagenesis was used (13, 21) to analyze the effect of RSV RTs carrying a mutation in only one of the two subunits constituting the αβ or αPol heterodimer.
It has been shown previously by biochemical and crystallographic data that heterodimeric human immunodeficiency virus type 1 (HIV-1) RT p66-p51 reveals an asymmetric subunit organization. The polymerase active site is present only in the larger p66 subunit of the heterodimer (13, 14, 19, 36), while the p51 subunit, which is identical in sequence to p66 but lacks the RNase H domain, is not directly involved in catalysis (21). However, p51 has to fulfill an important stabilizing function since the monomeric subunits are inactive (26, 27). Recent kinetic analyses we performed with homodimeric p51 RT from equine infectious anemia virus (EIAV) lacking the RNase H domain indicated an asymmetric subunit organization similar to that of heterodimeric p66-p51 RT, with the polymerase active site being present in only one of the subunits (32).
Taking these results into consideration, the question of whether RSV RTs possess similar subunit organizations arises, since the heterodimer is organized differently from lentivirus RTs. Heterodimeric RSV RTs αβ and αPol differ from heterodimeric lentivirus RTs by the presence of two RNase H domains instead of only one and by the presence of the integrase domain, which is located in the larger subunits of the heterodimers. In order to obtain more information about the subunit organization of heterodimeric RSV RT, we coinfected insect cells with two different types of baculoviruses harboring the gene coding for a wild-type or a mutant RSV RT subunit. This method allows purification of mutant heterodimeric RSV RT αβ or αPol possessing a mutation in only one subunit. Our results indicate that both the polymerase and RNase H active sites are located in the α subunit of heterodimeric RSV RT αβ or αPol.
MATERIALS AND METHODS
Mutagenesis of baculovirus transfer vectors.
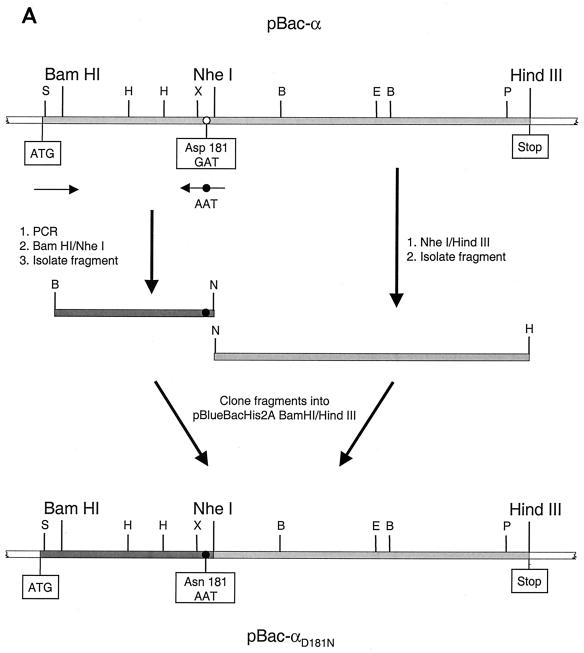
For mutagenesis of the RSV RT genes, we used the recombinant baculovirus transfer vectors described previously (37). The mutagenesis procedure of the transfer vector pBac-α is shown schematically in Fig. 1. The mutation in the active site of the polymerase of RSV RT α was created using an oligodeoxynucleotide created by PCR that contained the desired mutation. A fragment of about 580 bases harboring a mutated codon 181 in which Asp 181 was changed to Asn (GAT→AAT) was amplified by PCR. The resulting PCR fragment was treated with BamHI and NheI (Fig. 1A). An NheI/HindIII fragment containing the 3′-terminal region of the RT α gene was created by restriction of pBac-α. The two fragments were cloned into pBac-α restricted with BamHI and HindIII, yielding pBac-αD181N. The mutation was confirmed by sequencing. The resulting plasmid, pBac-αD181N, was used to obtain the corresponding mutation in the gene encoding β. pBac-αD181N was digested with XhoI and PmlI. The resulting XhoI/PmlI fragment containing the mutation was cloned into pBac-β that had been digested with XhoI and PmlI.
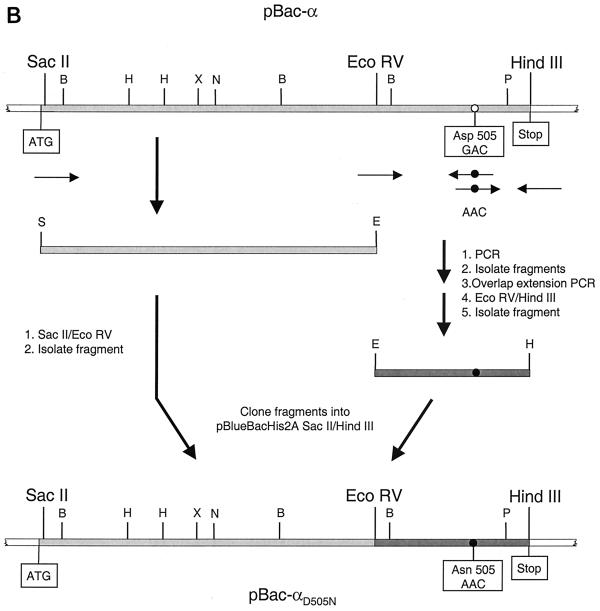
FIG. 1.
Mutagenesis of baculovirus transfer vectors. The open reading frame of the gene encoding RSV RT α is shaded in light gray. The start (ATG) and stop of the gene are indicated. The restriction enzymes relevant for the cloning procedure are shown. Abbreviations for other enzymes are as follows: B, BamHI; E, EcoRV; H, HindIII; P, PmlI; S, SacII; and X, XhoI. The locations of the PCR primers are indicated by black arrows. A white circle indicates the location of Asp 181 or Asp 505. A black circle indicates the mutagenized codon for Asn 181 or Asn 505. (A) Mutagenesis of the codon for Asp 181 in pBac-α. (B) Mutagenesis of the codon for Asp 505 in pBac-α.
To obtain the mutation Asn 505 in the RNase H active site of pBac-α, a combination of overlap extension PCR (12) and restriction fragment cloning was used. This is schematically shown in Fig. 1B. The PCR fragment was restricted with EcoRV and HindIII. The SalI/EcoRV fragment was obtained from pBac-α. Both fragments were purified on an agarose gel and cloned into pBac-α restricted with SacII and HindIII. The resulting plasmid, pBac-αD505N, was used to obtain pBac-βD505N. As described above, pBac-αD505N was digested with XhoI and PmlI and the fragment was cloned into pBac-β digested with XhoI and PmlI.
Deletion of the DNA sequence coding for the His6 tag at the N terminus of RSV RT.
In the transfer vector pBlueBacHis2A (Invitrogen), the DNA sequences for the N-terminal His6 tag and the enterokinase cleavage site are located between a SacII and a BamHI restriction site upstream of the corresponding RSV RT gene. The ATG start codon is located upstream of the SacII site (Fig. 1). The BamHI site represents the original start site for the RSV RT gene. The BamHI site was exchanged for a SacII site via PCR amplification of a fragment including the XhoI site at the 3′ end. The fragment was treated with SacII and XhoI and purified. pBac-α was also restricted with SacII and XhoI, thus deleting the DNA coding sequence for the His6 tag, the enterokinase site, and the 5′ end of the RSV RT. The purified plasmid backbone was ligated to the SacII/XhoI PCR fragment and used for transformation of Escherichia coli. Since the 5′ termini of the genes coding for Pol and β are identical to that of the gene coding for α, the same PCR fragment was cloned into the corresponding plasmids pBac-β and pBac-Pol, which contain the genes for Pol and β, respectively. Thus, enzymes lacking the His6 tag and the enterokinase cleavage site were obtained.
Isolation of recombinant virus.
The methods used to isolate recombinant virus and high-titer viral stocks and to quantify growth and infection of Sf21 insect cells have been described previously (37).
Purification of mutated RSV RT proteins.
Heterodimeric wild-type or mutant RSV RTs αβ and αPol harbored an N-terminal His6 extension on both subunits. The RTs were prepared by coinfection of Sf21 insect cells with two different types of baculoviruses, each harboring the gene for one of the subunits. Enzymes were purified by metal chelate chromatography as previously described (37). Subsequent purification over a heparin-Sepharose column via elution with an NaCl gradient allowed separation of the mutant heterodimer from the homodimeric α and β or Pol. Purification of the corresponding enzymes harboring the His6 tag only in the mutated subunit was performed in the same way, thus allowing separation of the heterodimer from the wild-type homodimer, which, due to the lack of a tag, does not bind to the Ni-chelate column. To identify DNase or RNase contaminations in our enzyme preparations, an aliquot was incubated with radioactively labeled DNA or RNA substrate for 10 min. The presence of degradation products was determined by analysis of the substrates on a denaturing sequencing gel. All enzymes were free of nuclease contamination.
HPLC gel filtration analysis.
High-performance liquid chromatography (HPLC) gel filtration analysis was performed as described previously (32, 37).
Evaluation of RT enzyme activities.
Quantitative RT activity assays and qualitative analysis of polymerization products and of RNase H activities were performed as described recently (37). For the quantitative RT activity assay whose results are shown in Table 1, poly(rA) · oligo(dT) was used as a substrate. Dimeric RSV RT enzyme (0.1 pmol) was added to a final volume of 30 μl of reaction mixture (37).
TABLE 1.
RT activities of selectively mutated RSV RTsa
| Enzyme | % of wild-type activityb |
|---|---|
| αD181NPol | 7.5 ± 2.5 |
| αβD181N | 42 ± 10 |
| αD181NβD181N | 1.5 ± 0.5 |
| αD505NPol | 34 ± 6 |
| αβD505N | 74 ± 15 |
| αD505NβD505N | 32 ± 7 |
RNA-dependent DNA polymerase activity was quantitated on a poly(rA) · oligo(dT)12–18 substrate as described previously (37). The activities of αPol and αβ were set to 100%.
Values are means ± standard deviations.
RESULTS
Strategy for construction and isolation of the selectively mutated heterodimeric RSV RTs αβ and αPol.
Sequence analyses of different polymerase genes show that the catalytic amino acid residues of the polymerase and RNase H active sites of RTs are highly conserved (2, 3, 15). In addition, comparison of the crystal structures of HIV-1 RT and the finger and palm domains of murine leukemia virus RT indicates that the geometry of the polymerase active sites of RTs is highly conserved (6, 14, 19, 29). We used this information for selecting the amino acids to be mutated in RSV RT to obtain enzymes impaired in their polymerase or RNase H activities.
We elected to mutate one aspartic acid residue (Asp 181) of the conserved YXDD motif of the polymerase active site into an asparagine. An exchange of the corresponding Asp 185 for His in HIV-1 RT has been shown to drastically reduce the polymerase activity of HIV-1 RT (20). Mutation of this residue to Asn in the p66 subunit of HIV-1 RT yields an enzyme that retains only about 2.1% of the wild-type polymerase activity (21). Kinetic analysis of the mutant HIV-1 RT enzyme revealed a 485-fold reduction in the catalytic constant for dTTP incorporation into a homopolymeric substrate (18). In the RNase H domain, catalytic Asp 505 of RSV RT was changed into an asparagine, since the corresponding Asp 498 of HIV-1 RT has been shown to be crucial for RNase H activity (4, 24).
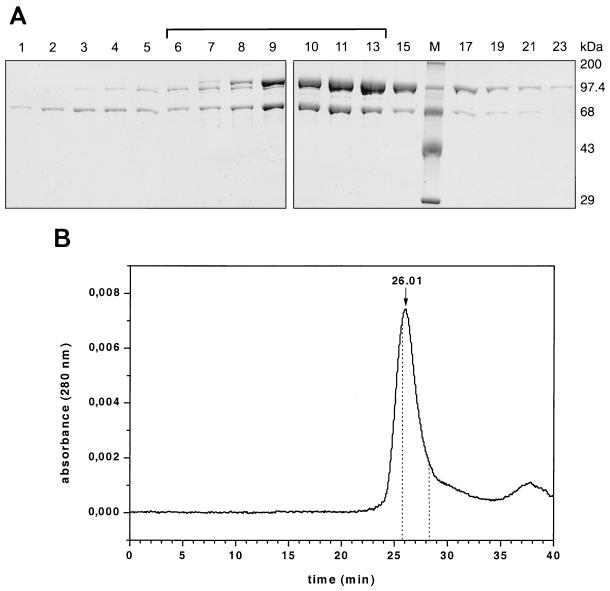
The mutations were introduced into the transfer vectors pBac-α and pBac-β (37), which harbor the genes coding for the α and β subunits of RSV RT, respectively (Fig. 1). These mutant transfer vectors were used to obtain recombinant baculoviruses that could be utilized to infect Sf21 insect cells. To achieve expression of heterodimeric RSV αβ and αPol RTs that were selectively mutated in only one subunit, insect cells were coinfected with wild-type and mutant virus. The proteins expressed contained a His6 tag on the N termini of both subunits of heterodimeric αβ or αPol. To eliminate homodimeric wild-type proteins, chromatography over a heparin-Sepharose column was performed after elution of the enzymes from the Ni-chelate column. By means of an NaCl gradient, we were able to separate the homodimers from the desired mutated heterodimeric protein. The analysis is demonstrated in Fig. 2A for enzyme αD181NPol and was carried out in a similar manner for all the other enzymes used. The fractions eluted from the heparin-Sepharose column were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie staining. Figure 2A shows the Coomassie stain of the eluted enzyme fractions of αD181NPol. The fractions containing the heterodimer were pooled as indicated by a bracket. With some of the enzymes the bands on the SDS gel imply that the subunit ratios are not 1:1 (Fig. 2A and 3). To verify that the enzyme is dimeric, analytical HPLC size exclusion chromatography of the pooled enzyme αD181NPol was performed. Our data confirm that the enzyme is a dimer and that the enzyme did not dissociate and reassociate to form homodimeric α or Pol. The retention time of the peak at 26.01 min corresponds to an apparent molecular mass of 175 kDa, which is in good agreement with the calculated molecular mass of 170 kDa for the heterodimer. The retention times of the homodimeric α and Pol are also indicated in Fig. 2B. Due to an overlap of the peaks representing αD181NPol and homodimeric Pol, we cannot completely rule out the possibility that there is a minor contamination with homodimeric Pol present in our preparation which is too small to be visible as a shoulder in the αD181NPol peak (Fig. 2B). Further experiments were performed to exclude this possibility (see below).
FIG. 2.
Analysis of RSV RT αD181NPol after elution from a heparin-Sepharose column. (A) SDS-PAGE of the eluted fractions. Proteins were detected by Coomassie staining. Lane M, protein molecular mass markers. The bracket indicates the pooled fractions. (B) HPLC size exclusion chromatography of the pooled fractions of αD181NPol (37). The peak with a retention time of 26.01 min corresponds to a molecular mass of ∼175 kDa. The retention times of homodimeric Pol (25.86 min) and α (28.26 min) were determined in separate runs with the corresponding enzymes and are indicated by dotted lines. The molecular mass of αD181NPol was determined using molecular mass standard proteins from U.S. Biochemical Corp. in the same buffer.
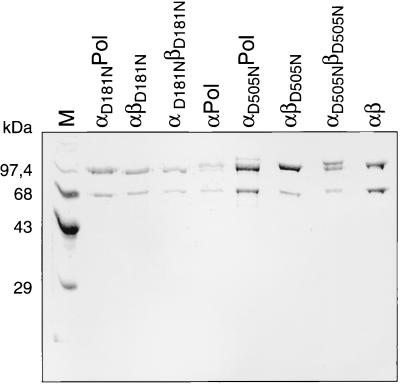
FIG. 3.
Analysis of the purified mutant RSV RT enzymes by SDS-PAGE. Proteins were detected by Coomassie staining. Lane M, protein molecular mass markers.
Figure 3 shows a Coomassie stain of all the RSV RT enzymes used in this study. They were all purified by the method described above. Mutant αD505NβD505N appears to contain a protein component larger than the normal β subunit. Western blot analysis of αD505NβD505N with a peptide antiserum directed against the C-terminal region of β (1) revealed that this band around 97 kDa is a contamination which does not bind the antibody (data not shown). However, as expected from the mutations introduced, the enzyme does not reveal any RNase H activity (see below) and appears to be free of nuclease contamination. We therefore decided to include it in our studies.
From the gel shown in Fig. 3 it also appears that there is more of the β subunit present in the αβD505N preparation. However, even if this was the case, it would not interfere with our investigations to determine the localization of the RNase H active site. Since homodimeric βD505NβD505N is inactive as an RNase H (our unpublished results), an excess of mutated βD505N will not influence the activity of the enzyme αβD505N.
The polymerase active site of the α subunit of the heterodimer is crucial for polymerase activity.
For unknown reasons, the expression of Pol is higher than that of β. However, we have shown previously that αβ and αPol do not show qualitative differences in their polymerization and RNase H activities (37). Therefore, we used mutant αPol instead of αβ for some of our experiments. To clarify the location of the polymerase active site, we produced mutant enzymes that contained the mutation D181N in only one subunit of the heterodimeric αβ or αPol. Table 1 shows the results of a quantitative polymerase activity assay using the homopolymeric substrate poly(rA) · oligo(dT)12–18. Our results indicate that enzymes containing Asn 181 in the α subunit or in both subunits display ≤7.5% of the corresponding wild-type polymerase activity. Enzyme containing the same mutation in the β subunit retained ∼42% of the wild-type activity. These results strongly suggest that the polymerase active site is located in the α subunit of the heterodimer. Reduction of activity within the β subunit was probably due to structural changes caused by the mutation. However, we cannot completely exclude the possibility that the catalytic residue of the β subunit contributed to the activity, although to a much lesser extent than α.
Table 1 shows that the mutants αD181NPol and αD181NβD181N still express polymerase activity, albeit at a very low level. This is not surprising, since we mutated only one of the amino acids forming the catalytic triad. However, in order to confirm that the residual activities of these enzymes were not due to trace amounts of homodimeric wild-type protein that was not detectable by HPLC gel filtration, control purifications were performed. Sf21 insect cells were coinfected with virus expressing the mutated subunit carrying the His6 tag together with virus expressing the corresponding wild-type subunit without the N-terminal His6 tag. Thus, only heterodimeric protein consisting of a His6-tagged mutated subunit and an untagged wild-type subunit was able to bind to the Ni-chelate column (e.g., His6-αD181N dimerized with Pol). In addition, homodimers consisting of two mutated subunits could bind but homodimers of the wild type could not bind due to the lack of a His6 tag. The homodimers were removed by further purification over heparin-Sepharose.
Similar to the corresponding enzyme carrying the His6 tag on both subunits, His6-αD181Nβ was found to express about 5% (±2.5) of the wild-type activity, indicating that the residual activity is not due to contaminating homodimeric wild-type protein. Rather, it is an intrinsic property of the mutated enzyme.
Therefore, for all subsequent experiments we used enzymes containing the His6 tag on both subunits, since we were not able to obtain sufficient amounts of protein when only one subunit was tagged. The low yield was due to the low expression of untagged proteins in comparison to that of the His6-tagged enzymes.
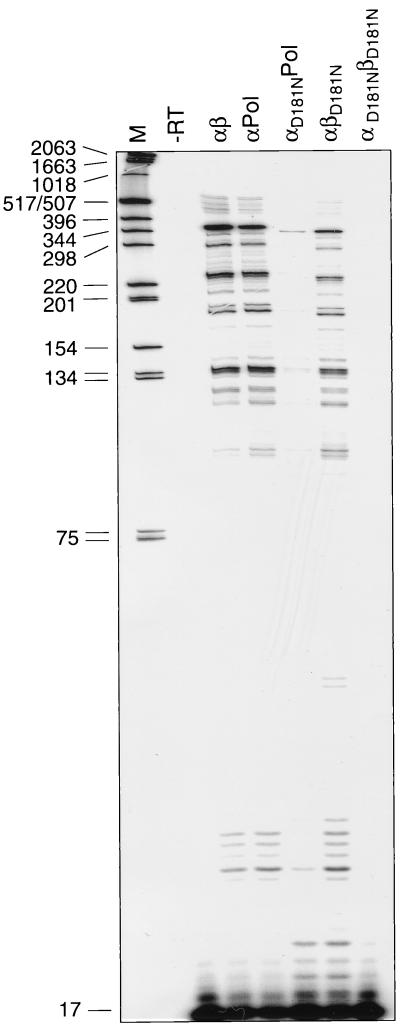
The DNA polymerase activities of the mutant enzymes were further investigated by a qualitative analysis. A radioactively labeled 17-mer DNA primer hybridized to single-stranded M13 DNA was used as a substrate. Figure 4 demonstrates that the activities of the two wild-type enzymes, αPol and αβ, were comparable. As expected from the results obtained with the RT assay (Table 1), somewhat less product was synthesized with αPolD181N than with the wild-type enzymes. However, the activity of αD181NPol was severely impaired and no extension products were detectable with the double mutant αD181NPolD181N in this experiment. This confirms our results shown in Table 1 that indicate the localization of the polymerase active site in the α subunit.
FIG. 4.
Polymerization activities on a DNA template catalyzed by RSV RTs selectively mutated in the polymerase active site. Reactions were performed for 10 min at 37°C in RT buffer with 10 nM M13 substrate and enzyme and a 250 μM concentration of each dNTP (37). Lane M shows DNA size markers (their sizes are indicated on the left), and lane -RT shows P-T without enzyme.
To analyze whether the mutations in the polymerase active site had an impact on the RNase H activities of the enzymes, we performed an RNase H assay with a 36-mer–127-mer DNA-RNA primer-template substrate (P-T) as described previously (37). The RNase H activities of the polymerase mutants are shown in Fig. 5C, together with the RNase H activities of the enzymes mutated in the RNase H active site (see below). Our results demonstrate that the mutation in the polymerase active site has only very little influence on RNase H activity.
FIG. 5.
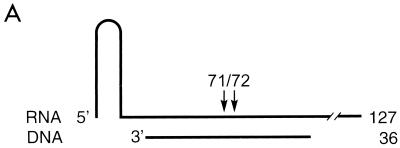
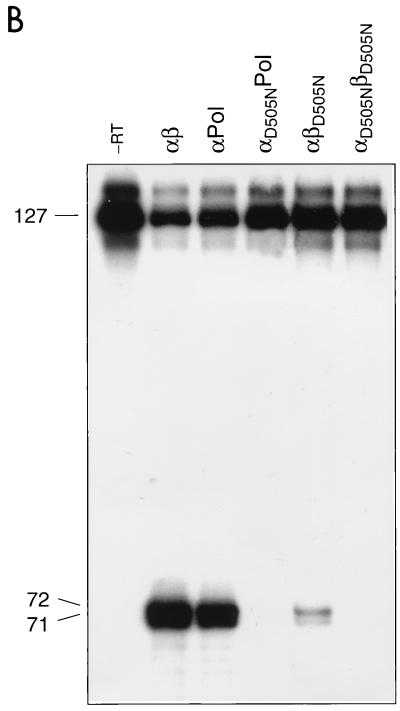
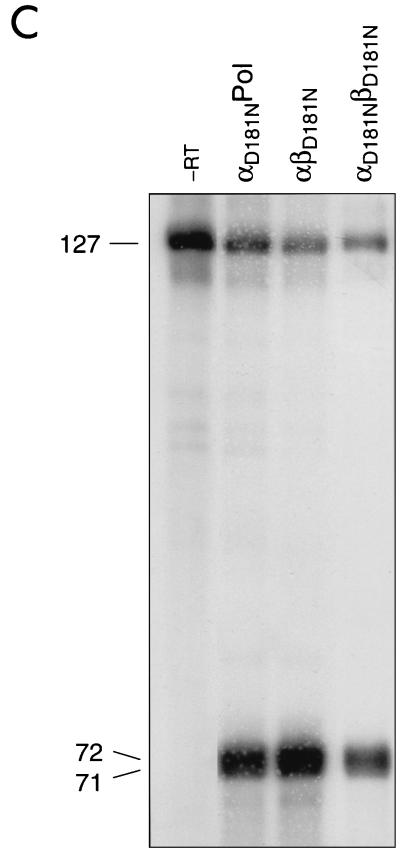
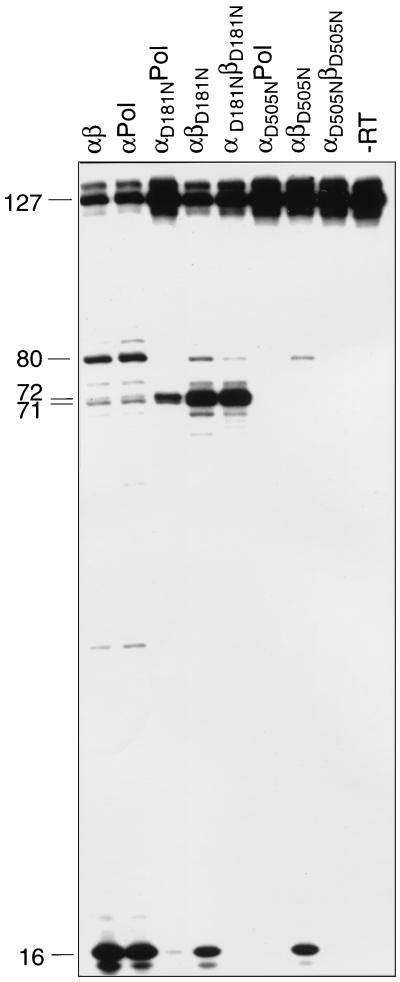
RNase H activities catalyzed by RSV RTs. (A) Schematic representation of the heteropolymeric DNA-RNA P-T substrate comprising a 5′-end labeled 127-mer RNA to which a 36-mer DNA primer was hybridized. The major cleavage sites at positions 71 and 72 are indicated by arrows. RNase H activities of RSV RTs mutated in the RNase H active site (B) or in the polymerase active site (C) are shown. Reactions were performed for 10 min at 37°C in RT buffer with 10 nM 36-mer–127-mer DNA-RNA P-T and 10 nM enzyme. The sizes of the 5′ RNA cleavage products are indicated on the left. Lane -RT contains P-T without enzyme.
The RNase H active site is located in the α subunit.
The location of the RNase H active site in the heterodimer was determined by changing Asp 505 to Asn. Selectively mutated heterodimeric proteins were purified and analyzed first in an RT activity assay to establish whether the mutation influences the polymerase activity (Table 1). The assay showed that the mutations in the RNase H active site have some impact on the polymerase activity, indicating possible structural changes caused by the mutations.
Qualitative analysis of the RNase H activities (Fig. 5B) demonstrated that when the mutation is present in the β subunit of the heterodimer, cleavage of the DNA-RNA hybrid is detectable. Compared to that in the wild-type enzyme, the amount of cleaved RNA was reduced. However, the observation that αD505NPol harboring the Asp 505 mutation in the α subunit exhibited no detectable RNase H activity suggests that the catalytic center of RNase H is also located in the α subunit of the heterodimer.
RNase H activity during polymerization.
During reverse transcription, RNase H hydrolysis and polymerization take place simultaneously. Therefore, we tested all the enzymes under conditions where the two enzyme functions could be observed. An RNase H assay was performed with the 36-mer–127-mer DNA-RNA substrate in the presence of deoxynucleoside triphosphates (dNTPs) to allow extension of the primer to the end of the template (Fig. 6). In this experiment the RNA of the DNA-RNA P-T was 5′-end labeled. Thus, complete extension of the primer could be detected by the presence of a short RNase H cleavage product which corresponded to a 5′-end labeled 16-mer RNA created by the RNase H when the RT reached the end of the RNA template (37).
FIG. 6.
Qualitative analysis of RNase H activities during DNA polymerization. The 127-mer RNA of the 36-mer–127-mer DNA-RNA was 5′-end labeled. Reactions were started by the addition of 10 nM enzyme to RT buffer containing a 10 nM concentration of the 36-mer–127-mer DNA-RNA P-T and a 50 μM concentration of each dNTP and stopped with formamide buffer after 10 min at 37°C. The sizes of the 5′ RNA cleavage products are indicated on the left. Complete primer extension in the presence of an active RNase H leads to a terminal RNA cleavage product of 16 nucleotides in length, as indicated at the bottom of the gel. Lane -RT, no enzyme added.
Interestingly, in the presence of dNTPs, some of the RT enzymes performed an additional cleavage around position 80 of the RNA template. This suggests a conformational rearrangement of the enzymes after dNTP binding that changed the position of the RNase H active site relative to that of the nucleic acid. Mutant αD181NPol performed the RNA cleavage at positions 71 and 72. However, only a few primer molecules were extended to the end of the RNA template, which was then cleaved by the RNase H activity of αD181NPol to create the 16-mer RNA.
No 16-mer RNA could be detected with the double mutant enzyme αD181NβD181N due to the lack of polymerization. However, in this case the RNase H cleavages at positions 71 and 72 of the template were visible. The cleavages at positions 71 and 72 reflect the RNase H cleavage positions obtained when no dNTPs are present (37). The wild-type enzymes αβ and αPol displayed cleavages at position 80 and positions 71 and 72. Due to their high polymerase activity, they were able to polymerize to the end of the template. Thus, the 16-mer RNA fragment at the end of the RNA template can be created by the RNase H. As implied from the results presented in Table 1, mutant αβD181N exhibited considerable polymerase activity, and due to its active RNase H, the 16-mer RNA was also present.
From the results shown in Fig. 5 and 6 and in Table 1, it can be concluded that the mutation in the RNase H catalytic site of the α subunit abrogates RNase H activity but not the concomitant polymerase activity. The mutant αβD505N showed a somewhat reduced RNase H activity, as indicated by the weak band at position 80. However, the 16-mer RNA fragment was produced after elongation of the template. As expected, no RNase H cleavage products were obtained with αD505NPol or αD505NβD505N.
DISCUSSION
Due to the difficulties in purifying recombinant RSV RTs, not much information has been available so far on mutated RSV RT enzymes. Here, we show for the first time the characterization of heterodimeric RSV RT selectively mutated in the active sites of the polymerase and RNase H domains of one subunit.
The structure of RSV RT is different from those of other retrovirus RTs since it harbors two RNase H domains and one integrase domain in the active heterodimer. Thus, it was interesting to analyze the structure-function relationships of this enzyme. Our results strongly suggest that in the heterodimeric RSV RT αβ or αPol the smaller α subunit contains the catalytic centers of the polymerase and RNase H domains. In contrast, the polymerase and RNase H domains of the larger subunit do not appear to play a catalytic role. Since we have mutated only one amino acid of the catalytic triads, it is not surprising that residual activities can be detected with the mutant enzymes.
The reduction of polymerase activity observed with αβD181N is probably due to structural changes since a similar reduction of polymerase activity is also found with the RNase H mutants αD505NPol and αD505NβD505N (Table 1). From the results obtained, we conclude that the β and Pol subunits fulfill mainly a structural function similar to that of the p51 subunits of HIV-1 RT and EIAV RT (13, 14, 19, 21, 29, 32). However, we cannot completely exclude the possibility that the large subunit fulfills some minor function in catalysis, since mutations in Asp 181 or Asp 505 of the β subunit appear to have some impact on the polymerase or RNase H activities. The α subunit harboring the active sites has a size of about 63 kDa and is comparable to the p66 subunit of HIV-1 RT p66-p51. Similar to HIV-1 p66, α harbors the polymerase and RNase H domains. The small p51 subunit of HIV-1 RT is an RNase H-deleted version of p66. The corresponding RNase H-deleted polypeptide of α is not found in ASLV virions.
The integrase domain of RSV RT is present only in the larger β subunit. Although β does not appear to contain the active sites of the polymerase and RNase H domains, previous results have shown that the integrase domain of heterodimeric ASLV RT expresses endonuclease activity (5, 22, 31), indicating a possible enzymatic function of the β subunit for integration. At present we are analyzing the integrase activities of our purified homodimeric and heterodimeric RSV RTs.
We have shown recently that RSV RT α is a homodimer (37). A dimeric organization has also been found for β and αβ, indicating that dimerization is a necessary feature for RSV RT enzyme activity (8, 11). The significance of dimerization has also been demonstrated for HIV-1 and EIAV RTs (26, 27, 32). Similar to what occurs with the RT from RSV, the catalytic sites of the polymerase and RNase H domains of these enzymes are formed by only one subunit. These results imply that an asymmetric subunit organization might be common for dimeric retroviral RTs, i.e., that the catalytic site is formed by only one of the two subunits. However, dimerization is apparently not important for all retrovirus RTs since previous experiments performed with the RTs from mouse mammary tumor virus and bovine leukemia virus indicate that these enzymes might be active as monomers (25, 34). The RT from murine leukemia virus is a monomeric ∼80-kDa protein that was suggested to dimerize upon binding to nucleic acid (10, 33, 35). However, recent data from a study using a chimeric HIV-1 RT with the RNase H domain of murine leukemia virus indicate that this chimeric protein is enzymatically active as a monomer (23). These results also demonstrate that there are significant differences in the mechanisms of polymerization and reverse transcription performed by RTs from different retroviruses.
ACKNOWLEDGMENTS
This work was supported by the Max-Planck-Gesellschaft and by a grant from the Deutsche Forschungsgemeinschaft to B.M.W.
We thank Martina Wischnewski and Karin Vogel-Bachmayr for skilled technical assistance with the protein purification and cloning procedures, Paul Rothwell for careful reading of the manuscript, and Roger Goody for support. We also thank Anna Marie Skalka (Fox Chase Cancer Center, Philadelphia, Pa.) for the kind gift of the peptide antiserum specific for the C terminus of β. We thank Erika Schlüter and Gesine Schulte for assistance in preparing the figures.
REFERENCES
- 1.Alexander F, Leis J, Soltis D A, Crowl R M, Danho W, Poonian M S, Pan Y-C E, Skalka A M. Proteolytic processing of avian sarcoma and leukosis viruses pol-endo recombinant proteins reveals another pol gene domain. J Virol. 1987;61:534–542. doi: 10.1128/jvi.61.2.534-542.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Argos P. A sequence motif in many polymerases. Nucleic Acids Res. 1988;16:9909–9916. doi: 10.1093/nar/16.21.9909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Delarue M, Poch O, Tordo N, Moras D, Argos P. An attempt to unify the structure of polymerases. Protein Eng. 1990;3:461–467. doi: 10.1093/protein/3.6.461. [DOI] [PubMed] [Google Scholar]
- 4.DeStefano J J, Wu W, Seehra J, McCoy J, Laston D, Albone E, Fay P J, Bambara R A. Characterization of an RNase H deficient mutant of human immunodeficiency virus-1 reverse transcriptase having an aspartate to asparagine change at position 498. Biochim Biophys Acta. 1994;1219:380–388. doi: 10.1016/0167-4781(94)90062-0. [DOI] [PubMed] [Google Scholar]
- 5.Duyk G, Leis J, Longiaru M, Skalka A M. Selective cleavage in the avian retroviral long terminal repeat sequence by the endonuclease associated with the αβ form of avian reverse transcriptase. Proc Natl Acad Sci USA. 1983;80:6745–6749. doi: 10.1073/pnas.80.22.6745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Georgiadis M M, Jessen S M, Ogata C M, Telesnitsky A, Goff S P, Hendrickson W A. Mechanistic implications from the structure of a catalytic fragment of Moloney murine leukemia virus reverse transcriptase. Structure. 1995;3:879–892. doi: 10.1016/S0969-2126(01)00223-4. [DOI] [PubMed] [Google Scholar]
- 7.Golomb M, Grandgenett D. Endonuclease activity of purified RNA-directed DNA polymerase from avian myeloblastosis virus. J Biol Chem. 1979;254:1606–1613. [PubMed] [Google Scholar]
- 8.Grandgenett D P, Gerard G F, Green M. A single subunit from avian myeloblastosis virus with both RNA-directed DNA polymerase and ribonuclease H activity. Proc Natl Acad Sci USA. 1973;70:230–234. doi: 10.1073/pnas.70.1.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grandgenett D P, Vora A C, Schiff R D. A 32,000-dalton nucleic acid-binding protein from avian retrovirus cores possesses DNA endonuclease activity. Virology. 1978;89:119–132. doi: 10.1016/0042-6822(78)90046-6. [DOI] [PubMed] [Google Scholar]
- 10.Guo J H, Wu W X, Yuan Z Y, Post K, Crouch R J, Levin J G. Defects in primer-template binding, processive DNA synthesis and RNase H activity associated with chimeric reverse transcriptases having the murine leukemia virus polymerase domain joined to Escherichia coli RNase H. Biochemistry. 1995;34:5018–5029. doi: 10.1021/bi00015a013. [DOI] [PubMed] [Google Scholar]
- 11.Hizi A, Joklik W K. RNA-dependent DNA polymerase of avian sarcoma virus B77. I. Isolation and partial characterization of the α, β2, and αβ forms of the enzyme. J Biol Chem. 1977;252:2281–2289. [PubMed] [Google Scholar]
- 12.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 13.Hostomsky Z, Hostomska Z, Fu T B, Taylor J. Reverse transcriptase of human immunodeficiency virus type 1: functionality of subunits of the heterodimer in DNA synthesis. J Virol. 1992;66:3179–3182. doi: 10.1128/jvi.66.5.3179-3182.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacobo-Molina A, Clark A D, Jr, Williams R L, Nanni R G, Clark P, Ferris A L, Hughes S H, Arnold E. Crystals of a ternary complex of human immunodeficiency virus type 1 reverse transcriptase with a monoclonal Fab fragment and double stranded DNA diffract to 3.5 Å resolution. Proc Natl Acad Sci USA. 1992;88:10895–10899. doi: 10.1073/pnas.88.23.10895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson M S, McClure M A, Feng D F, Gray J, Doolittle R F. Computer analysis of retroviral pol genes: assignment of enzymatic functions to specific sequences and homologies with nonviral enzymes. Proc Natl Acad Sci USA. 1986;83:7648–7652. doi: 10.1073/pnas.83.20.7648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kacian D L, Watson K F, Burny A, Spiegelman S. Purification of the DNA polymerase of avian myeloblastosis virus. Biochim Biophys Acta. 1971;246:365–383. doi: 10.1016/0005-2787(71)90773-8. [DOI] [PubMed] [Google Scholar]
- 17.Katz R A, Skalka A M. A C-terminal domain in the avian sarcoma-leukosis virus pol gene product is not essential for viral replication. J Virol. 1988;62:528–533. doi: 10.1128/jvi.62.2.528-533.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaushik N, Rege N, Yadav J S, Sarafianos S G, Modak M J, Pandey V N. Biochemical analysis of catalytically crucial aspartate mutants of human immunodeficiency virus type 1 reverse transcriptase. Biochemistry. 1996;35:11536–11546. doi: 10.1021/bi960364x. [DOI] [PubMed] [Google Scholar]
- 19.Kohlstaedt L A, Wang J, Friedman J M, Rice P A, Steitz T A. Crystal structure at 3.5 Å resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science. 1992;256:1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- 20.Larder B A, Purifoy D J, Powell K L, Darby G. Site-specific mutagenesis of AIDS virus reverse transcriptase. Nature. 1987;327:716–717. doi: 10.1038/327716a0. [DOI] [PubMed] [Google Scholar]
- 21.Le Grice S F, Naas T, Wohlgensinger B, Schatz O. Subunit-selective mutagenesis indicates minimal polymerase activity in heterodimer-associated p51 HIV-1 reverse transcriptase. EMBO J. 1991;10:3905–3911. doi: 10.1002/j.1460-2075.1991.tb04960.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leis J, Duyk G, Johnson S, Longiaru M, Skalka A. Mechanism of action of the endonuclease associated with the αβ and ββ forms of avian RNA tumor virus reverse transcriptase. J Virol. 1983;45:727–739. doi: 10.1128/jvi.45.2.727-739.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Misra H S, Pandey P K, Pandey V N. An enzymatically active chimeric HIV-1 reverse transcriptase (RT) with the RNase-H domain of murine leukemia virus RT exists as a monomer. J Biol Chem. 1998;273:9785–9789. doi: 10.1074/jbc.273.16.9785. [DOI] [PubMed] [Google Scholar]
- 24.Mizrahi V, Usdin M T, Harington A, Dudding L R. Site-directed mutagenesis of the conserved asp-443 and asp-498 carboxy-terminal residues of HIV-1 reverse transcriptase. Nucleic Acids Res. 1990;18:5359–5363. doi: 10.1093/nar/18.18.5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perach M, Hizi A. Catalytic features of the recombinant reverse transcriptase of bovine leukemia virus expressed in bacteria. Virology. 1999;259:176–189. doi: 10.1006/viro.1999.9761. [DOI] [PubMed] [Google Scholar]
- 26.Restle T, Müller B, Goody R S. Dimerization of human immunodeficiency virus type 1 reverse transcriptase. J Biol Chem. 1990;265:8986–8988. [PubMed] [Google Scholar]
- 27.Restle T, Müller B, Goody R S. RNase H activity of HIV reverse transcriptases is confined exclusively to the dimeric forms. FEBS Lett. 1992;300:97–100. doi: 10.1016/0014-5793(92)80172-d. [DOI] [PubMed] [Google Scholar]
- 28.Rho H M, Grandgenett D P, Green M. Sequence relatedness between the subunits of avian myeloblastosis virus reverse transcriptase. J Biol Chem. 1975;250:5278–5280. [PubMed] [Google Scholar]
- 29.Rodgers D W, Gamblin S J, Harris B A, Ray S, Culp J S, Hellmig B, Woolf D J, Debouck C, Harrison S C. The structure of unliganded reverse transcriptase from the human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1995;92:1222–1226. doi: 10.1073/pnas.92.4.1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schiff R D, Grandgenett D P. Virus-coded origin of a 32,000-dalton protein from avian retrovirus cores: structural relatedness of p32 and the beta polypeptide of the avian retrovirus DNA polymerase. J Virol. 1978;28:279–291. doi: 10.1128/jvi.28.1.279-291.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Skalka A M. Endonuclease activity associated with reverse transcriptase of avian sarcoma-leukosis viruses. In: Skalka A M, Goff S P, editors. Reverse transcriptase. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1993. pp. 193–204. [Google Scholar]
- 32.Souquet M, Restle T, Krebs R, Le Grice S F, Goody R S, Wöhrl B M. Analysis of the polymerization kinetics of homodimeric EIAV p51/51 reverse transcriptase implies the formation of a polymerase active site identical to heterodimeric EIAV p66/51 reverse transcriptase. Biochemistry. 1998;37:12144–12152. doi: 10.1021/bi9731596. [DOI] [PubMed] [Google Scholar]
- 33.Sun D, Jessen S, Liu C, Liu X, Najmudin S, Georgiadis M M. Cloning, expression, and purification of a catalytic fragment of Moloney murine leukemia virus reverse transcriptase: crystallization of nucleic acid complexes. Protein Sci. 1998;7:1575–1582. doi: 10.1002/pro.5560070711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taube R, Loya S, Avidan O, Perach M, Hizi A. Reverse transcriptase of mouse mammary tumour virus: expression in bacteria, purification and biochemical characterization. Biochem J. 1998;329:579–587. doi: 10.1042/bj3290579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Telesnitsky A, Goff S P. RNase H domain mutations affect the interaction between Moloney murine leukemia virus reverse transcriptase and its primer-template. Proc Natl Acad Sci USA. 1993;90:1276–1280. doi: 10.1073/pnas.90.4.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang J, Smerdon S J, Jäger J, Kohlstaedt L A, Rice P A, Friedman J M, Steitz T A. Structural basis of asymmetry in the human immunodeficiency virus type 1 reverse transcriptase heterodimer. Proc Natl Acad Sci USA. 1994;91:7242–7246. doi: 10.1073/pnas.91.15.7242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Werner S, Wöhrl B M. Soluble Rous sarcoma virus reverse transcriptases α, αβ and β purified from insect cells are processive DNA polymerases that lack an RNase H 3′→5′ directed processing activity. J Biol Chem. 1999;274:26329–26336. doi: 10.1074/jbc.274.37.26329. [DOI] [PubMed] [Google Scholar]